13.3
Impact Factor
Theranostics 2020; 10(11):5137-5153. doi:10.7150/thno.42325 This issue Cite
Research Paper
Selective targeting of the oncogenic KRAS G12S mutant allele by CRISPR/Cas9 induces efficient tumor regression
1. Guangdong Provincial Key Laboratory of Genome Read and Write, BGI-Shenzhen, Shenzhen 518083, China
2. China National GeneBank, BGI-Shenzhen, Jinsha Road, Shenzhen 518120, China
3. Guangdong Provincial Academician Workstation of BGI Synthetic Genomics, BGI-Shenzhen, Shenzhen, 518083, China
4. Department of Thoracic Surgery II, Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education/Beijing), Peking University Cancer Hospital and Institute, Beijing, China
5. BGI Education Center, University of Chinese Academy of Sciences, Shenzhen, 518083, China
6. James D. Watson Institute of Genome Sciences, Hangzhou 310058, China
7. Ab Vision, Inc, Milpitas, California, USA
*These authors contributed equally to this work.
Abstract
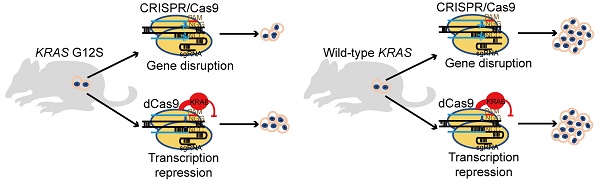
Rationale: KRAS is one of the most frequently mutated oncogenes in cancers. The protein's picomolar affinity for GTP/GDP and smooth protein structure resulting in the absence of known allosteric regulatory sites makes its genomic-level activating mutations a difficult but attractive target.
Methods: Two CRISPR systems, genome-editing CRISPR/SpCas9 and transcription-regulating dCas9-KRAB, were developed to deplete the KRAS G12S mutant allele or repress its transcription, respectively, with the goal of treating KRAS-driven cancers.
Results: SpCas9 and dCas9-KRAB systems with a sgRNA targeting the mutant allele blocked the expression of the mutant KRAS gene, leading to an inhibition of cancer cell proliferation. Local adenoviral injections using SpCas9 and dCas9-KRAB systems suppressed tumor growth in vivo. The gene-depletion system (SpCas9) performed more effectively than the transcription-suppressing system (dCas9-KRAB) on tumor inhibition. Application of both Cas9 systems to wild-type KRAS tumors did not affect cell proliferation. Furthermore, through bioinformatic analysis of 31555 SNP mutations of the top 20 cancer driver genes, the data showed that our mutant-specific editing strategy could be extended to a reference list of oncogenic mutations with high editing potentials. This pipeline could be applied to analyze the distribution of PAM sequences and survey the best alternative targets for gene editing.
Conclusion: We successfully developed both gene-depletion and transcription-suppressing systems to specifically target an oncogenic KRAS mutant allele that led to significant tumor regression. These findings show the potential of CRISPR-based strategies for the treatment of tumors with driver gene mutations.
Keywords: KRAS mutation, CRISPR/Cas9, dCas9-KRAB, mRNA-regulating, cancer therapy
 Global reach, higher impact
Global reach, higher impact