13.3
Impact Factor
Theranostics 2020; 10(10):4544-4556. doi:10.7150/thno.40532 This issue Cite
Review
Tumor circulome in the liquid biopsies for cancer diagnosis and prognosis
1. Cancer Institute (Key Laboratory of Cancer Prevention and Intervention, China National Ministry of Education), The Second Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou 310009, China
2. Institute of Translational Medicine, Zhejiang University, Hangzhou 310029, China
3. Department of Obstetrics, The Second Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou 310009, China
4. Department of Biochemistry, College of Biomedical Sciences, Zhejiang University School of Medicine, Hangzhou 310009, China
5. Department of Neurosurgery, The Second Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou 310009, China
* These authors contributed equally to this work.
Received 2019-9-22; Accepted 2020-1-7; Published 2020-3-15
Abstract
Liquid biopsy is a convenient, fast, non-invasive and reproducible sampling method that can dynamically reflect the changes in tumor gene expression profile, and provide a robust basis for individualized therapy and early diagnosis of cancer. Circulating tumor DNA (ctDNA) and circulating tumor cells (CTCs) are the currently approved diagnostic biomarkers for screening cancer patients. In addition, tumor-derived extracellular vesicles (tdEVs), circulating tumor-derived proteins, circulating tumor RNA (ctRNA) and tumor-bearing platelets (TEPs) are other components of liquid biopsies with diagnostic potential. In this review, we have discussed the clinical applications of these biomarkers, and the factors that limit their implementation in routine clinical practice. In addition, the most recent developments in the isolation and analysis of circulating tumor biomarkers have been summarized, and the potential of non-blood liquid biopsies in tumor diagnostics has also been discussed.
Keywords: liquid biopsy, tumor circulome, tumor screening
Introduction
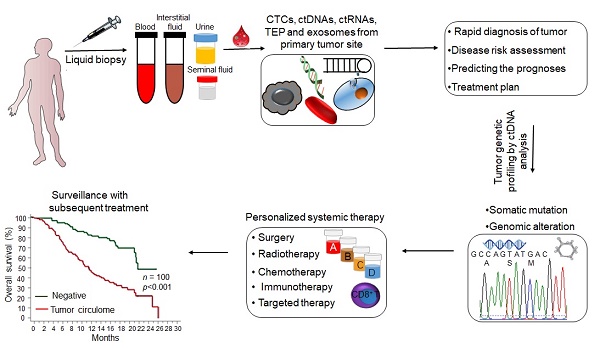
Early detection of cancer can significantly improve the therapeutic outcomes and patient prognosis. However, large-scale screening for early stage tumors is still not possible due to lack of suitable techniques. Although tissue biopsy is the gold standard of tumor detection and diagnosis, it is limited by the difficulties in obtaining tissue samples, poor sensitivity and accuracy, high procedural costs, inability to distinguish between heterogeneous tumors and invasiveness [1], and is therefore incompatible with longitudinal clinical monitoring. Liquid biopsies on the other hand can be collected in a non-invasive manner, and have gained considerable attention in recent years for early cancer screening, tumor progression monitoring, assessing therapeutic response and clinical prognosis, and detecting recurrent and refractory tumors [2, 3]. Common liquid biopsy markers (LBMs), including circulating tumor nucleic acids (ctDNA and ctRNA), circulating tumor cells (CTCs), tumor-derived extracellular vesicles (tdEVs) and tumor-educated platelets (TEPs), can be used as cancer biomarkers either directly or indirectly and have been summarized in Figure 1 [4].
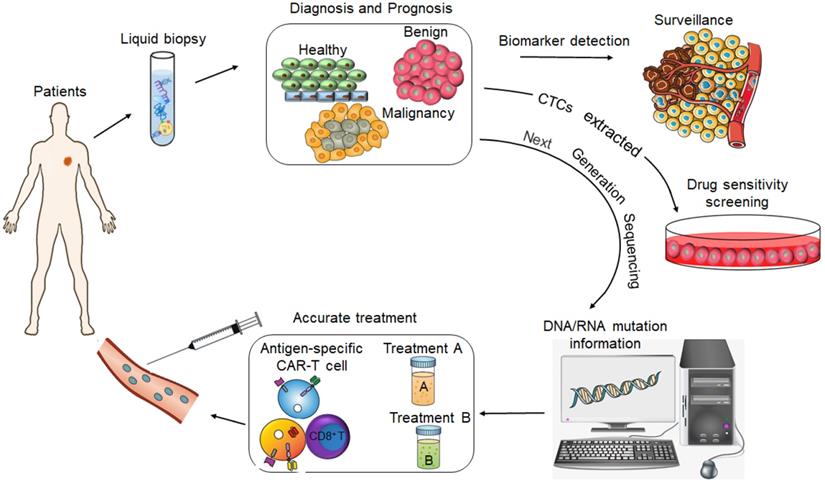
CtDNA was approved by FDA as a diagnostic marker for lung cancer and for screening colorectal cancer (CRC) in 2016 [5], and the CellSearch® CTCs capture system has also been approved by the FDA for diagnosing metastatic prostate, breast and colon cancers [6]. Although these are important milestones in the field of liquid biopsy and molecular diagnosis, such molecular-based tools have limited application in routine clinical practice since they are not approved worldwide and are not part of standard cancer diagnostics. The current focus is on developing more sensitive and high throughput early cancer diagnosis techniques, such as the mRNA Sentinel Principle Technologyi, new generation sequencing technology (NGS) [7], single-cell protein analysis and sequencing [8], high-resolution flow cytometers [9] and PCR- based assays [10], in order to expand the clinical applications of liquid biopsies. In fact, the identification of LBMs in liquid biopsy specimens has emerged as a potential means to improve prognostic prediction, guide risk-adaptive or precision therapy, and detect patients at the highest risk for disease relapse (Figure 2).
In this review, we have summarized the diagnostic and prognostic value of liquid biopsies in various cancers, the recent developments in the isolation of cancer biomarkers, potential clinical applications, and the limitations. In addition, the possibility of using body fluids other than blood, such as urine, salivary, cerebrospinal fluid (CSF), stool, sputum and lacuna liquid such as pleural effusion, as tumor-derived biomarkers has also been discussed.
Circulating protein markers
Circulating protein markers are the most established diagnostic tools for cancer, and include prostate specific antigen (PSA) for screening prostate cancer [11], cancer antigen (CA) 15-3 for postoperative follow-up of breast cancer, carbohydrate antigen 19-9 (CA 19-9), carcinoembryonic antigen (CEA) for intrahepatic cholangiocarcinoma (ICC), alpha-fetoprotein (AFP), and carbohydrate antigens for screening multiple malignancies [13, 14]. However, individual protein markers are significantly limited in terms of sensitivity and specificity, and associated with high false-positive rates. A panel of multiple protein biomarkers can improve the diagnostic and prognostic accuracy by reducing the number of both false positives and false negatives [15, 16]. In addition, diagnostic platforms using multiple circulating protein markers have been established in recent years for cancer screening. Matrix-assisted laser desorption /ionization time-of-flight mass spectrometry (MALDI-TOF MS) is a high-throughput, highly sensitive proteomics platform that can not only identify novel tumor-specific markers, but also enable early screening and diagnosis of tumors. Long et al. recently developed a nanoplatform for diagnosing multiple myeloma via detection of the urine Bence-Jones protein; macroporous ordered silica foams (MOSF) were firstly used to enrich the proteins and the resulting nanoparticle-protein composites were analyzed by MALDI-TOF MS. This panel diagnosed multiple myeloma with high sensitivity (95.24%, 20/21) and specificity (100%, 27/27), and is therefore a highly promising tool for the clinical diagnosis of Bence-Jones protein-related diseases [17]. Park et al. also developed a MALDI-TOF MS-based total serum protein fingerprinting tool for liver cancer diagnosis and confirmed its high sensitivity and specificity [18]. Taken together, diagnostic platforms based on circulating markers can be used for effective, high-throughput screening for various cancers and other diseases. These novel platforms have to be validated by testing on more clinical samples, and their performance can be further improved by extensive and reliable database matching, as well as intelligent and rapid information reporting systems.
The extracellular matrix (ECM) molecules, such as matrix metalloproteinases (MMPs), MMP- degraded collagens, collagen oligomeric matrix protein (COMP) and fibronectin, are released into the bloodstream from solid tumors, and have therefore emerged as promising surrogate markers of tumor development or clearance [19, 20]. In addition, these circulating ECM proteins reflect the tumor- microenvironment crosstalk, and can provide novel insights into tumor initiation and progression [21]. Each step in tumorigenesis is characterized by a distinct tumor microenvironment (TME) or tumor- associated inflammatory signature, which can affect tumor progression either favorably or adversely. Therefore, markers associated with tumor-associated inflammation or the TME can potentially increase the chances of monitoring the onset of tumor niches, metastatic growth, patient prognosis and anti-cancer drug efficacy. Lin et al. recently found that increased levels of CXCL8 and colony stimulating factor 2 (CSF-2), which facilitates macrophage-driven CXCL8 secretion, indicated poor clinical outcome and tumor progression in gastric cancer patients. Mechanistically, CXCL8 inhibits CD8+ T cells function by inducing the immunosuppresive PD-L1 on macrophages [22]. Lundgren et al. determined that high density of CD1a+ dendritic cells (DCs) was an independent prognostic factor for reduced OS in pancreatobiliary cancer, and increased prevalence of CD68+ and CD163+ tumor- associated macrophages (TAMs) were significantly associated with poor OS in periampullary adenocarcinoma patients [23]. The results indicate that markers associated with tumor inflammation and the microenvironment can be used for noninvasive diagnosis, screening, and postoperative follow-up of cancer.
The liquid biopsy markers (LBMs) commonly used in clinical or laboratory screening include circulating tumor proteins, circulating tumor ctDNA, circulating tumor cells (CTCs), tumor-derived extracellular vesicles (EV) and their components, circulating tumor ctRNA and tumor-cultured platelets (TEPs). All of them can be used directly or indirectly for cancer screening and diagnosis.
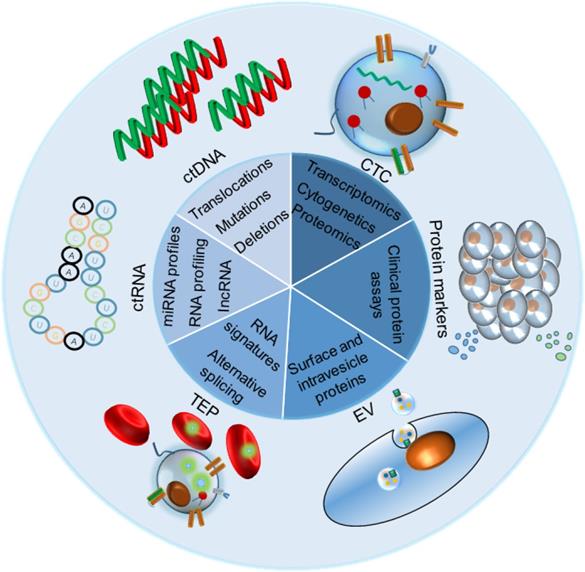
The value of liquid biopsy in tumor diagnosis and precision medicine.
Circulating tumor nucleic acids
Circulating or extracellular tumor nucleic acids are secreted from primary or metastatic cancer cells following apoptosis or necrosis [24], and differ significantly from the ctDNAs/RNAs in the sera of healthy individuals. Interestingly, the ctDNA isolated from the blood of patients with pancreatic cancer harbors the mutated KRAS gene [25], and N-ras gene mutations have been detected in the plasma of myelodysplastic syndrome and acute myelogenous leukemia (AML) patients [26], which further underscores the utility of ctDNA as a cancer biomarker. Several studies have subsequently reported the presence of extracellular-DNA (exDNA) in sera/plasma from patients with different cancers [27-30]. Similarly, the presence of exRNA, including messenger RNAs (mRNAs), microRNAs (miRNAs) and long non-coding RNAs (lncRNAs), have also gained attention in recent years as potential non-invasive cancer biomarkers [31]. The altered ctRNA levels in cancer patients often return to normal following surgery, and therefore are suitable indicators of therapeutic response [32, 33]. As a means of liquid biopsy, cfDNA/ctDNA technology can be further divided into two approaches: detection of gene mutations and methylation. The COBAS® EGFR Mutation Test v2 (Roche Diagnostics), the first ctDNA-based diagnostic test used in clinics, was approved by FDA in 2017 for monitoring the response to epidermal growth factor receptor (EGFR)-tyrosine kinase inhibitors (TKIs) in non-small cell lung cancer patients harboring EGFR-TKI sensitizing mutations [34]. In addition, EpiproColon®, a FDA-approved screening test for CRC, is based on the methylation pattern of the SEPT9 gene promoter [35]. However, Getz et al. recently suggested that detecting gene mutations in liquid biopsies may have limited use for early screening of cancer, since normal tissues also harbor mutations and somatic variations in multiple genes. They found that 95% of individuals have somatic mutations in at least one tissue, while 33% carry cancer-related mutations. Therefore, the methylation status of cfDNA/ctDNA is a better marker for early cancer screening, with greater sensitivity and specificity [36]. Besides, when using genetic mutations or molecular markers for tumor screening and diagnosis, genetic mutations or molecular markers expression in some special diseases, such as clonal erythropoiesis, need to be considered independently, as these diseases have mutations at specific genetic loci, or express a special molecular marker which may mislead the results when screening for molecular cancer-specific data onto liquid biopsy [37].
MicroRNAs are a class of small non-coding RNAs that regulate gene expression and are critical in several biological and pathological processes, including cancer initiation and progression. Several preclinical studies have reported circulating miRNA as tissue-specific cancer biomarkers that can not only monitor a prognosis but also determine the origin of tumor metastases [38, 39]. In addition, serum microRNA levels can also reflect other physiological conditions such as pregnancy, and can even be used to determine the pregnancy stage [40].
Currently, ctDNA analyses are based on PCR or next-generation sequencing (NGS). Allele-specific PCR was the earliest approach used in ctDNA detection [41], and a modified version is the technical basis of the COBAS® EGFR test [34]. More sensitive PCR technologies, such as digital PCR (dPCR), droplet digital PCR (ddPCR), and beads, emulsion, amplification and magnetics (BEAMing) have been developed in recent years and increase the accuracy of ctDNA detection [42]. Despite the high sensitivity, rapidity and relative low costs, these PCR techniques are limited by low multiplexing capacity since they can only analyze a restricted number of loci simultaneously rather than entire gene sequences. Compared to PCR-based technologies, NGS has lower sensitivity due to even fewer number of loci that it can analyze. Furthermore, since the mutant allele fraction (MAF) in a given locus is greater than 5%, whole- exome sequencing (WES) has the lowest sensitivity in analyzing ctDNA sequences compared to the other techniques [43]. The sensitivity of NGS can be increased by including patient or cancer-specific gene panels, e.g. the cancer personalized profiling deep sequencing (CAPP-Seq) technology [44]. In addition, the background signals can be minimized by tagging each template sequence with unique molecular identifiers (UMIs), and using selective nucleases on the non-mutated DNA [45]. In addition, artificial intelligence based on whole-genome sequencing (WGS) is a recent breakthrough in ctDNA analysis through liquid biopsies. In a more recent study, Velculescu et al. developed the artificial intelligence platform DELFI (DNA evaluation of fragments for early interception) for cancer specific screening, monitoring and diagnosis. They used known tumor-specific mutations to label tumor-derived ctDNA, compared them with the homologous healthy free DNA and determined the fragment length distribution, followed by introducing all information into the database. After establishing this intelligent platform, other variables such as GC abundance, position of chromosome arms, mutant alleles etc. were also introduced and each was assigned a specific score to improve detection. The information collated in the intelligent platform and the database can then be used to quickly and sensitively distinguish healthy patients from cancer patients on the basis of their ctDNA profile. This platform was used to test the plasma samples of 236 cancer patients and 245 healthy individuals, and showed respective accuracy rates of 91% and 98%. Thus, artificial intelligence platforms highlight the clinical importance of cell-free DNA and provide a proof-of-principle approach for the screening, early detection and monitoring of human cancer [46].
However, ctDNA detection has several limitations, such as low sensitivity at the early stages of cancer. The incipient tumors shed very low levels of ctDNA, which decrease the MAF and may escape detection [44]. To eliminate individual differences due to hereditary predispositions and improve the detection efficiency, the liquid biopsies should be collected as close to the tumor as possible, which unfortunately is technically challenging [47]. In addition, healthy individuals should also be screened for cancer-associated mutations to determine whether a particular mutation has a predictive value in early detection [42]. CancerSEEK, a recently developed high throughput screening test for liquid biopsies, can detect ctDNA in the blood of cancer patients with high sensitivity [48].
Circulating tumor cells
During malignant progression, the primary tumor masses shed a significant number of cells that invade adjacent tissues, and migrate to distant sites to establish metastatic clones and also regenerate blood vessel walls to support neo-angiogenesis [49]. These circulating tumor cells (CTCs) are therefore a reliable biomarker of cancer metastasis. A number of clinical trials, which mostly using the Cellsearch platform established prognostic and predictive value in patients, have confirmed that changes in CTC count between baseline and the second anti-cancer treatment coursed in advanced breast cancer [50] had been associated with an adverse prognostic and predictive value on the patient's outcome, with a reported good “negative predictive value” of CTCs. This finding has been reiterated in other metastatic cancer types, such as colorectal [51], prostate [52] and ovarian [53] cancers. Monitoring CTCs can not only improve the chances of early cancer detection and identify novel drug targets, but also predict patient prognoses and therapeutic responses. In addition, the CTC load can stratify patients into different risk groups for (neo) adjuvant therapies [54, 55]. CTCs are extremely rare in the peripheral blood of cancer patients, usually one per million blood cells. Therefore, it is technically challenging to detect them with high sensitivity and specificity. In addition, CTC isolation currently depends on the surface expression of epithelial markers; for instance, the CellSearch® system captures epithelial cells from the blood using EpCAM-coated magnetic beads, which are then identified using fluorescently-labeled antibodies against cytokeratins (CK 8, CK 18, CK 19). There are several hitherto non-approved systems as well that detect stem-like, mesenchymal-like and hybrid CTC subpopulations, which are clinically significant since CTCs show partial or complete epithelial- mesenchymal transition (EMT) and some even acquire stem cell-like characteristics [56, 57]. Therefore, the conventional epithelial marker-dependent CTC detection can neither distinguish between these subpopulations, nor can it identify the origin of micrometastatic and metastatic CTCs. Therefore, further research on these CTC subpopulations can provide new insights for anti- cancer therapeutics [58]. Ex vivo expansion of CTCs from individual patients can enable personalized drug screening, and assist in making more effective treatment decisions based on the unique tumor mutation profiles and drug sensitivity patterns [59]. Certainly, although in vitro culture of CTCs has important guiding value for clinical tumor treatment, this approach is fraught with several limitations. For instance, pre-sample processing, enrichment and sorting, and improper culture conditions can cause irreversible damage to the CTCs. Furthermore, long- term in vitro culture and multiple passages may alter the CTCs genetically and epigenetically, such that they no longer represent the phenotype of the original tumor. Therefore, it is essential to develop a more effective CTC sorting and enrichment system, as well as improve the in vitro culture conditions. In addition, establishing co- cultures of CTCs with immune cells and other blood cells can improve our understanding of the survival mechanism of CTCs in the peripheral blood. This in turn can help develop new intervention strategies and further promote the clinical usage of CTCs as an important tool for liquid biopsy. The isolated CTCs can be genetically analyzed using qPCR, dPCR-based mutational spectroscopy technology, target NGS and genome-wide sequencing technologies [60]. In addition, cytogenetic techniques such as fluorescence in situ hybridization (FISH) can be used to identify tumor-specific chromosomal rearrangements in the CTCs [61]. Since CTCs are involved in tumor progression and initiate metastases, mutiomics analyses at the single-cell level can help dissect the complex relationships between the tumor subpopulations and the surrounding normal tissue. Thus, single CTC genomics and transcriptomics are invaluable to the study of tumor heterogeneity and for comparative analysis with tissue biopsies [62-64].
CTC enrichment and detection methods are broadly classified as biological, physico-chemical and functional based on the specific CTC properties that are utilized. The biological methods of capturing CTCs depend on the expression of surface biomarkers; for example, CellSearch® is based on enriching the EpCAM+ cells [50] and can be combined with other cancer biomarkers or CD45+ depletion [65]. In addition, surface immuno-phenotyping can be synergized with microfluidics to further enhance CTC yields, such as the CTC-Chip [66] and NanoVelcro [67] platforms. CTCs can also be separated from normal blood cells based on their size, density, and dielectric properties through filtration [68], microfluidics [69], differential centrifugation [70], densitometry (MagDense) [71] and di-electrophoresis (DEPArray: a semiautomated system that allows the isolation of rare cells) [72]. Functional CTC captures assays include Vita-AssayTM [73], EPISPOT® assay [74] and TelomeScan® [75] that respectively exploit CAM digestion, protein release during culture and telomerase expression. All these methods have their own advantages but due to the rarity, vulnerability and heterogeneity of CTCs, any one method cannot capture sufficient number of CTCs for clinical and other applications. Therefore, a combination of two or more methods may improve CTC enrichment for liquid biopsies.
Tumor-derived extracellular vesicles (tdEVs)
Chargaff and West discovered back in 1946 that removal of the pelleted plasma fraction after high- speed centrifugation inhibited plasma clotting [76]. Years later, Peter Wolf determined that small (20-50 nm) lipid-bilayer-enclosed structures or vesicles that extruded from the platelets were the anti-coagulation factors [77]. Subsequent studies reported that the transferrin receptors on reticulocytes interacted with vesicles secreted from these cells [78-80]. Extracellular vesicles (EVs) are membranous granules released from all types of cells under physiological and pathological conditions, as well as in response to proteases, inflammatory cytokines, growth factors, biomechanical shear, stress-inducing factors and apoptotic signals. Based on their biogenesis, content and secretory pathways, EVs can be divided into exosomes and microvesicles [81]. Exosomes are exfoliated vesicles with ecto-enzyme activity and were first described by Trams et al. [82]. They originate during endocytic internalization from the inter 9nal budding of the plasma membrane. The early endosomes fuse with the Golgi complex to form late endosomes that then give rise to intraluminal vesicles contained within multi vesicular bodies (MVBs). The latter either fuse with the plasma membrane to release exosomes through exocytosis or are degraded upon fusing with the lysosomes [83].
EVs, especially the exosomes, mediate cell-cell communication by transporting cargo like proteins, DNA, mRNAs and miRNAs from the donor to target cells [84, 85]. For example, exosome-mediated glia-neuron communication maintains neuronal integrity [86]. Exosomes had increasingly gained attention as messengers of cancer cells wherein they can reprogram the transcriptome of target cells by transporting regulatory RNAs. For example, breast cancer cell-derived exosomes harbor components of the RISC (RNA-induced silencing complex)-loading complex (RLC) including pre-miRNAs, Dicer, Argonaute2 and TRBP (Trans-activation of transponder RNA binding proteins), and have a cell-independent capacity to process precursor miRNAs (pre-miRNAs) into mature miRNAs. The RISC-loaded exosomes are known to trigger the malignant transformation of the adjacent normal breast epithelial cells and drive tumor progression [87]. Studies show that cancer cells release significantly higher amounts of EVs compared to non-malignant cells, and are therefore suitable diagnostic markers as well as anti-cancer therapeutic targets [88-90]. Tumor-derived extracellular vesicles (tdEVs) not only influence the growth, progression and drug resistance of the parent tumors by altering the immediate microenvironment, but also create favorable conditions in distant organs to allow growth of disseminated tumor cells, a process known as pre-metastatic niche (PMN) formation [91]. For instance, metastatic melanomas release EVs that carry programmed death-ligand 1 (PD-L1) on their surface, which suppresses the function of CD8 T cells and facilitates tumor growth [92]. Furthermore, the distinct integrin expression patterns on tdEVs determine their adhesion to specific cell types and ECM proteins in specific organs [93].
The proteins and RNAs present in the lumen and membrane of EVs are potential cancer biomarkers that can allow early tumor diagnosis. Typical exosome- enriched proteins include members of the tetraspanin family (CD9, CD63 and CD81), members of the endosomal sorting complexes required for transport (TSG101 and Alix), and heat-shock proteins (Hsp60, Hsp70 and Hsp90) [94]. In addition, tumor-associated proteins such as CEA, EGFR VIII, HER2 and MelanA, along with a range of DNA, mRNAs and miRNAs have also been detected in exosomes. The exosomes harboring cancer-specific proteins like CD63, CD9 and CD81, and microRNA signatures have been isolated from the blood of breast cancer [95], ovarian cancer [96] and glioblastoma patients [97], and from the urine of prostate cancer patients [98, 99]. Exosomal miRNAs are protected from degradation of RNAse unlike the naked circulating miRNAs, and are therefore more reliable diagnostic biomarkers of tumor type and stage [100]. One study showed that compared to healthy controls, lung adenocarcinoma patients harbored twelve unique exosomal miRNAs (miR-17-3p, miR-21, miR-106a, miR-146, miR-155, miR-191, miR-192, miR-203, miR-205, miR-210, miR-212, and miR-214). In addition, four exosomal miRNAs (miR-378a, miR-379, miR-139-5p, and miR-200b-5p) were able to distinguish patients with lung carcinoma from healthy former smokers with 97.5% sensitivity and 72% specificity, and six miRNAs (miR-151a-5p, miR-30a-3p, miR-200b-5p, miR-629, miR-100 and miR-154-3p) could differentially diagnose between lung adenocarcinoma and granuloma [101]. Another study reported 80.65% and 83.33% sensitivity, and 91.67% and 90.32% specificity of exosomal miRNAs for diagnosing lung adenocarcinoma and squamous cell carcinoma (SCC) respectively. Taken together, these circulating miRNAs are potentially sensitive, non-invasive biomarkers for early NSCLC diagnosis [102]. Therefore, exosome-based assays have gained considerable attention for cancer diagnosis. For instance, peripheral blood-derived exosome screening is far less invasive compared to colonoscopy, and more specific compared to carcinoembryonic antigen (CEA) or carbohydrate antigenic determinant (CA 19-9) for diagnosing colon cancer.
EVs are released by several cell types, including B and T lymphocytes, dendritic cells, mast cells, intestinal epithelial cells, neurons, tumor cells and MSCs [103-108], and are present in physiological fluids such as urine, plasma, cerebrospinal fluid, milk and various exudates [98, 109-114]. However, although tumor-derived EVs (tdEVs) are promising blood biomarkers for cancer disease management, blood is a highly complex fluid that contains multiple particles of the same size range as tdEVs, which obscures their unimpeded analysis. Therefore, a highly sensitive and specific approach is required to capture and detect tdEVs. The isolation strategies of EVs are primarily based on their physical and biological properties. Almost all EVs, especially exosomes, can be extracted from body fluids by normal density-gradient centrifugation, ultracentrifugation, and the more advanced EV array and immuno-bead precipitation [90, 115]. Exosomes typically had diameters ranging from 30 to 100 nm and density 1.13-1.19 g/ml, and are usually saucer-shaped or hemispherical with a concave side [116, 117]. Although high exosome yield and purity can be achieved with filtration, the high pressure generated during the process can damage their structural integrity [118]. Therefore, immuno-affinity captured with antibodies targeting the surface proteins of EVs such as the tetraspanins are preferred for naive and intact EVs. This approach also allows high purity isolation of distinct immuno-phenotypic EV subsets. Beekman et al. [119] had recently developed a multi-modal analysis platform for the specific capture of tdEVs on antibody-functionalized stainless-steel substrates, followed by their analysis using SEM, Raman spectroscopy and AFM at the single EV level in terms of size and size distribution, and chemical fingerprint. A single surface marker however can miss significant subpopulations, which may be obviated by targeting multiple markers via microchip-based in situ immunoassay [120]. Microfluidics have emerged as a promising new method for rapidly capturing EVs based on their physical or biochemical features [121], and can boost affordable EV-based medical diagnostics. Size-exclusion chromatography (SEC), polymer precipitation, magnetic nanopore-sorting platform and alternating current electrokinetic (ACE) chip are some other techniques that have been tested for EV isolation, but are limited by low efficiency and purity. As already mentioned, the EVs can be profiled for somatic mutations, splice variants, gene fusions and aberrant gene or protein expression by RT-PCR, NGS, Western blotting, ELISA etc. However, these methods analyze the bulk EV population and therefore disregard the inherent heterogeneity. The micro-flow cytometry platform however can detect single circulating exosomes using beads [122], and nanoparticle tracking analysis (NTA) enables high resolution protein analysis of these individual EVs [123]. Several approaches have been examined to analyze the heterogeneity (DNA, RNA or proteins) of different exosomes and clarify their tissue of origin. We developed a robust microchip-based method for the selective and quantitative analysis of exosomes using digital detection integrated with nucleic acid amplification [124]. Kamali-Moghaddam et al. [125] employed a proximity-dependent barcoding assay to simultaneously profile multiple surface proteins on individual exosomes. This detection platform combines surface immuno-phenotyping with digital detection or NGS technology, and can further enhance the precision of heterogeneity analysis, which broadens its applications in basic research as well as disease diagnosis.
Compared to ctDNA and CTCs, the clinical translation of EVs is hampered due to the technically challenging isolation methods, low analytical sensitivity and poor stability. On the other hand, the amount of circulating EVs is significantly higher compared to that of CTCs [126], and can be indicative of the existence of tumors. For instance, circulating exosome levels increase in breast and pancreatic cancer patients [127], and the number of circulating microparticles (MPs) is significantly higher in multiple myeloma (MM) patients compared to healthy individuals. Moreover, the circulating MP level is also a potential marker for the diagnosis and prognosis of advanced NSCLC [128]. Due to the lack of unified standards for sample processing as well as EV separation and analysis, the clinical applications of EVs as a method of liquid biopsy are limited [129]. Another factor limiting EV-based diagnostics is the lack of standardized guidelines for defining EV characteristics, as well as the lack of appropriate controls for validation. The International Extracellular Vesicle Association has recently upgraded a comprehensive series of guidelines and recommendations to redefine EVs nomenclature [130], which will subsequently facilitate EV research and redefine the gold standard for EV extraction and analysis, and therefore expedite their clinical translation.
In addition to proteins, DNA, glycans, lipids and metabolites are also present on the surface of EVs, and are potential biomarkers for EV enrichment [131]. A database of EV-associated RNAs or proteins can greatly increase the chances of identifying novel circulating biomarkers specific for different cancer types. The exact role of EVs in various pathophysiological conditions also needs to be established in order to develop a standardized EV diagnostic system.
Tumor-educated platelets
Calverley et al. were the first to observe significant differences in the platelet genomic profiles of cancer patients and healthy individuals. They found that 197 platelet-associated genes were down- regulated in patients with metastatic lung cancer, and multiple genes were also spliced differentially between the control and patient groups [132]. Subsequently, Nilsson et al. reported that platelets from cancer patients can actively absorb tumor-derived EVs and take up RNA from tumor cells [133]. Therefore, these tumor-educated platelets (TEPs) are also potential biomarkers for cancer diagnosis and screening, and have been the focus of recent studies [134]. In 2015, Best et al. sequenced the transcriptomes of platelets derived from cancer patients in order to determine the diagnostic potential of TEPs by mRNA sequencing [135], and identified patients with six types of localized or metastatic tumors and healthy individuals with 96% accuracy, and the primary tumor location with 71% accuracy. The same group applied particle-swarm optimization (PSO)-enhanced algorithm to select possible biomarker panels from platelet RNA-sequencing libraries, and achieved accurate TEP-based detection of early- and late-stage NSCLC [136]. This algorithm may also enable optimization of diagnostics using other liquid biopsies. However, although many detection platforms for TEPs have been developed, patient and control samples ought to be collected under the same conditions to minimize any deviations. This is especially significant for platelet-related tests since they can be easily activated and alter their transcriptome profiles. In addition, exosome and cfDNA profiling can also be greatly affected by differences in sample collection and handling. Taken together, TEPs are potential biomarkers for the early diagnosis, screening and therapeutic monitoring of cancers, and their clinical translation would depend on developing appropriate isolation techniques.
Other biological specimens as liquid biopsies
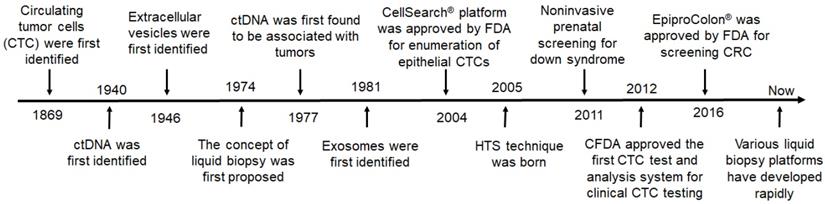
It has been 150 years since Ashworth first discovered CTCs, and now various liquid biopsy platforms and techniques have been developed for screening, detection, and diagnosis of tumors (Figure 3). During this period, various milestones of new discoveries or new technologies have greatly promoted the practical development of liquid biopsy.
The milestones in the development of liquid biopsies. HTS: high throughput sequencing; CRC: colorectal cancer; FDA: Food and Drug Administration; CFDA: China Food and Drug Administration.
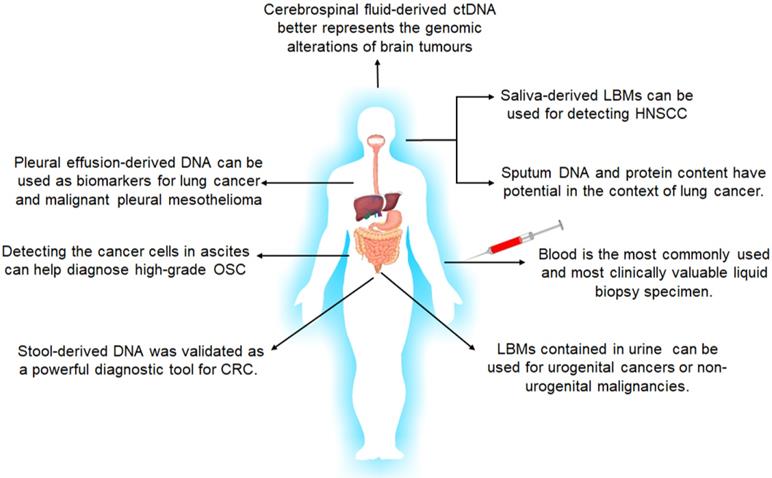
However, although liquid biopsy typically refers to utilization of blood samples, almost all body fluids, including but not limited to urine, saliva, sputum, feces, cerebrospinal fluid and lacuna liquid such as pleural effusion, are reliable cancer biomarkers suitable for liquid biopsies [137-139] (Figure 4). Since cancer biomarkers are typically expressed in the local tumors at the early stages of cancer, they usually do not appear in circulation, which further underscores the diagnostic significance of local liquid biopsies.
For genitourinary tumors such as prostate, the bladder and cervical cancer, urine is the ideal liquid biopsy for early diagnosis and treatment, and can be easily collected and analyzed for biomarker proteins, ctDNAs or EVs. Progensa® PCA3 Assay was approved by FDA in 2012ii for repetitive prostate biopsies in case of a negative first diagnosis [140]. ExoDx, SelectMDx and Michigan Prostate Score (MiPS) were recently developed as non-invasive screening tools for prostate cancer diagnosis and prognosis, and primarily detect androgen-related transmembrane protease serine 2 (TMPRSS2-ERG) or second chromosome locus associated with prostate-1 (SChLAP1) in the urine [141], along with PSA levels in the blood. In addition, the ExoDx® Prostate (IntelliScore)iii test can detect the expression levels of three exosome-associated RNAs in the urine with high sensitivity and specificity. Thus, urine analysis can obviate the need for invasive prostate tissue biopsies and digital rectal examination.
Salivary biomarkers include ctDNA, miRNAs and EV-associated miRNAs, and have significant clinical value in diagnosing head and neck squamous cell carcinoma and early oral cancers [142, 143]. Due to the presence of the blood-brain barrier, CSF is an important source of circulating biomarkers for CNS-restricted cancers. Ding et al. [144] reported that CSF-derived ctDNA can better reflect genetic alterations in brain tumors compared to ctDNA in the bloodstream. Akers et al. [144] also found that the altered miRNA profile of CSF is predictive of gliomas. The analysis of stool-derived DNA was recently validated as a powerful diagnostic tool for CRC [146], while sputum DNA and protein content have diagnostic potential in lung cancer [147]. Furthermore, presence of TP53-mutated cancer cells in ascites is indicative of high-grade serous ovarian carcinomas (HGSOCs) [148], and pleural effusion-derived DNA is a biomarker for lung cancer and malignant pleural mesothelioma [149]. Therefore, local liquid biopsies have greater diagnostic value compared to peripheral blood in some cancers, and further developments in the isolation and diagnostic techniques using these fluids will greatly improve early diagnosis of tumors.
The body fluids suitable for liquid biopsies and their applications in tumor diagnosis and screening. LBMs: liquid biopsy markers; HNSCC: head and neck squamous cell carcinoma; OSC: Ovarian Serous Carcinoma; CRC: colorectal cancer.
Summary and future directions
Liquid biopsies provide a cost-effective, fast, reproducible and non-invasive source for early cancer diagnosis and prognostic monitoring. In addition, analysis of circulating tumor-derived factors or the tumor circulome in the liquid biopsies can capture the clonal heterogeneity of these tumors unlike tissue biopsies. Various liquid biopsy samples can be combined to improve the chances of cancer diagnosis, and sequential real-time biopsies will further aid in the early identification of therapy-resistant tumors. Furthermore, detection and characterization of minimal residual disease after initial therapy can also be improved by analyzing liquid biopsies. Automated chip-based devices are particularly fitting for the high throughput analysis of biomarkers from whole blood and other body fluids, and obviates time consuming and costly purification steps. However, the lack of standardized pre-analytical and analytical variables is a significant limitation in this field, and has stymied large-scale clinical applications of liquid biopsies. In conclusion, liquid biopsies can be a powerful tool for cancer diagnosis, monitoring, prognosis and individualized treatment, and can completely change the current paradigms of cancer management. However, considerable research and development are still needed to improve the isolation, enrichment and downstream analysis of circulating biomarkers.
Abbreviations
FDA: food and drug administration; LBMs: liquid biopsy markers; CTCs: circulating tumor cells; ctDNA: circulating tumor DNA; tdEVs: tumor- derived extracellular vesicles; ctRNA: extracellular vesicles; ctRNA: circulating tumor RNA; TEPs: tumor- educated platelets; NGS: next generation sequence; PSA: prostate-specific antigen; CA: cancer antigen; CA19-9: carbohydrate antigen 19-9; CEA: carcinoembryonic antigen; MMPs: matrix metalloproteinases; COMP: collagen oligomeric matrix protein; lncRNAs: long non-coding RNAs; EGFR: epidermal growth factor receptor; dPCR: digital PCR; ddPCR: droplet digital PCR; WES: whole-exome sequence; UMIs: unique molecular identifiers; FISH: fluorescence in situ hybridization; PMN: pre-metastatic niche; PD-L1: programmed death-ligand 1; ASGPR1: asialoglycoprotein receptor-1; SEC: size-exclusion chromatography; ACE: alternating current electrokinetic; NTA: aanoparticle tracking analysis; TMPRSS2-ERG: androgen-related transmembrane protease serine 2; SChLAP1: second chromosome locus associated with prostate-1; MiPS: michigan prostate score.
Acknowledgements
This study was supported by the National S&T Major Project (NO.2018ZX10301201), the fundamental Research Funds for the Central Universities (2019FZJD009), the National Key R&D Program of China (2018YFA0108603), the Natural Science Foundation of China (81401541 and 81570168), the Distinguished Young Scientist Award of Natural Science Foundation of Zhejiang Province (LR16H180001), the Major Project in Science and Technology of Zhejiang Province (2014C03048-2), and Zhejiang Provincial Natural Science Foundation (No. LY19H040014).
Resources
iiwww.accessdata.fda.gov/scripts/cdrh/cfdocs/cfpma/pma.cfm?id=P100033
Competing Interests
The authors have declared that no competing interest exists.
References
1. Overman MJ, Modak J, Kopetz S, Murthy R, Yao JC, Hicks ME. et al. Use of research biopsies in clinical trials: are risks and benefits adequately discussed? J Clin Oncol. 2013;31:17-22
2. De Rubis G, Rajeev Krishnan S, Bebawy M. Liquid biopsies in cancer diagnosis, monitoring, and prognosis. Trends Pharmacol Sci. 2019;40:172-186
3. Zhang Y, Mi X, Tan X, Xiang R. Recent progress on liquid biopsy analysis using surface-enhanced Raman spectroscopy. Theranostics. 2019;9:491-525
4. Lopez A, Harada K, Mizrak Kaya D, Dong X, Song S, Ajani JA. Liquid biopsies in gastrointestinal malignancies: when is the big day? Expert Rev Anticancer Ther. 2018;18:19-38
5. Kwapisz D. The first liquid biopsy test approved. Is it a new era of mutation testing for non-small cell lung cancer? Ann Transl Med. 2017;5:46
6. Mathai RA, Vidya RVS, Reddy BS, Thomas L, Udupa K, Kolesar J. et al. Potential utility of liquid biopsy as a diagnostic and prognostic tool for the assessment of solid tumors: implications in the precision oncology. J Clin Med. 2019:8 pii: E373
7. Khotskaya YB, Mills GB, Mills Shaw KR. Next-generation sequencing and result interpretation in clinical oncology: challenges of personalized cancer therapy. Annu Rev Med. 2017;68:113-125
8. Dhar M, Lam JN, Walser T, Dubinett SM, Rettig MB, Di Carlo D. Functional profiling of circulating tumor cells with an integrated vortex capture and single-cell protease activity assay. Proc Natl Acad Sci USA. 2018;115:9986-9991
9. Kibria G, Ramos EK, Lee KE, Bedoyan S, Huang S, Samaeekia R. et al. A rapid, automated surface protein profiling of single circulating exosomes in human blood. Sci Rep. 2016;6:36502
10. Kim KY, Le QT, Yom SS, Pinsky BA, Bratman SV, Ng RH. et al. Current state of PCR-based epstein-barr virus DNA testing for nasopharyngeal cancer. J Natl Cancer Inst. 2017;109:djx007
11. Pinsky PF, Prorok PC, Kramer BS. Prostate cancer screening - a perspective on the current state of the evidence. N Engl J Med. 2017;376:1285-1289
12. Akbari Nakhjavani S, Khalilzadeh B, Samadi Pakchin P, Saber R, Ghahremani MH, Omidi Y. A highly sensitive and reliable detection of CA15-3 in patient plasma with electrochemical biosensor labeled with magnetic beads. Biosens Bioelectron. 2018;122:8-15
13. Qiu Y, He J, Chen X, Huang P, Hu K, Yan H. The diagnostic value of five serum tumor markers for patients with cholangiocarcinoma. Clin Chim Acta. 2018;480:186-192
14. Feng F, Tian Y, Xu G, Liu Z, Liu S, Zheng G. et al. Diagnostic and prognostic value of CEA, CA19-9, AFP and CA125 for early gastric cancer. BMC Cancer. 2017;17:737
15. Bhardwaj M, Gies A, Werner S, Schrotz-King P, Brenner H. Blood-based protein signatures for early detection of colorectal cancer: a systematic review. Clin Transl Gastroenterol. 2017;8:e128
16. Surinova S, Radová L, Choi M, Srovnal J, Brenner H, Vitek O. et al. Non-invasive prognostic protein biomarker signatures associated with colorectal cancer. EMBO Mol Med. 2015;7:1153-65
17. Long S, Qin Q, Wang Y, Yang Y, Wang Y, Deng A. et al. Nanoporous silica coupled MALDI-TOF MS detection of Bence-Jones proteins in human urine for diagnosis of multiple myeloma. Talanta. 2019;200:288-292
18. Park HG, Jang KS, Park HM, Song WS, Jeong YY, Ahn DH. et al. MALDI-TOF MS-based total serum protein fingerprinting for liver cancer diagnosis. Analyst. 2019;144:2231-2238
19. Willumsen N, Bager CL, Leeming DJ, Smith V, Karsdal MA, Dornan D. et al. Extracellular matrix specific protein fingerprints measured in serum can separate pancreatic cancer patients from healthy controls. BMC Cancer. 2013;13:554
20. Wang Z, Wang Y, Jia X, Han Q, Qian Y, Li Q. et al. MMP-2-controlled transforming micelles for heterogeneic targeting and programmable cancer therapy. Theranostics. 2019;9:1728-1740
21. Giussani M, Triulzi T, Sozzi G, Tagliabue E. Tumor extracellular matrix remodeling: new perspectives as a circulating tool in the diagnosis and prognosis of solid tumors. Cells. 2019:8 pii: E81
22. Lin C, He H, Liu H, Li R, Chen Y, Qi Y. et al. Tumour-associated macrophages-derived CXCL8 determines immune evasion through autonomous PD-L1 expression in gastric cancer. Gut. 2019;68:1764-1773
23. Lundgren S, Karnevi E, Elebro J, Nodin B, Karlsson MCI, Eberhard J. et al. The clinical importance of tumour-infiltrating macrophages and dendritic cells in periampullary adenocarcinoma differs by morphological subtype. J Transl Med. 2017;15:152
24. Stroun M, Maurice P, Vasioukhin V, Lyautey J, Lederrey C, Lefort F. et al. The origin and mechanism of circulating DNA. Ann N Y Acad Sci. 2000;906:161-168
25. Sorenson GD, Pribish DM, Valone FH, Memoli VA, Bzik DJ, Yao SL. Soluble normal and mutated DNA sequences from single-copy genes in human blood. Cancer Epidemiol Biomarkers Prev. 1994;3:67-71
26. Vasioukhin V, Anker P, Maurice P, Lyautey J, Lederrey C, Stroun M. Point mutations of the N-ras gene in the blood plasma DNA of patients with myelodysplastic syndrome or acute myelogenous leukaemia. Br J Haematol. 1994;86:774-779
27. Valpione S, Gremel G, Mundra P, Middlehurst P, Galvani E, Girotti MR. et al. Plasma total cell-free DNA (cfDNA) is a surrogate biomarker for tumour burden and a prognostic biomarker for survival in metastatic melanoma patients. Eur J Cancer. 2018;88:1-9
28. Passiglia F, Galvano A, Castiglia M, Incorvaia L, Calò V, Listì A. et al. Monitoring blood biomarkers to predict nivolumab effectiveness in NSCLC patients. Ther Adv Med Oncol. 2019;11:1-11
29. Molnár B, Galamb O, Kalmár A, Barták BK, Nagy ZB, Tóth K. et al. Circulating cell-free nucleic acids as biomarkers in colorectal cancer screening and diagnosis - an update. Expert Rev Mol Diagn. 2019;19:477-498
30. Leighl NB, Page RD, Raymond VM, Daniel DB, Divers SG, Reckamp KL. et al. Clinical utility of comprehensive cell-free DNA analysis to identify genomic biomarkers in patients with newly diagnosed metastatic non-small cell lung cancer. Clin Cancer Res. 2019;25:4691-4700
31. Mithraprabhu S, Morley R, Khong T, Kalff A, Bergin K, Hocking J. et al. Monitoring tumour burden and therapeutic response through analysis of circulating tumour DNA and extracellular RNA in multiple myeloma patients. Leukemia. 2019;33:2022-2033
32. Fernandez-Mercado M, Manterola L, Larrea E, Goicoechea I, Arestin M, Armesto M. et al. The circulating transcriptome as a source of non-invasive cancer biomarkers: concepts and controversies of non-coding and coding RNA in body fluids. J Cell Mol Med. 2015;19:2307-2323
33. Feng G, Li G, Gentil-Perret A, Tostain J, Genin C. Elevated serum-circulating RNA in patients with conventional renal cell cancer. Anticancer Res. 2008;28:321-326
34. Kwapisz D. The first liquid biopsy test approved. Is it a new era of mutation testing for non-small cell lung cancer? Ann Transl Med. 2017;5:46
35. Lamb YN, Dhillon S. Epi proColon® 2.0 CE: a blood-based screening test for colorectal cancer. Mol Diagn Ther. 2017:21 225-232
36. Yizhak K, Aguet F, Kim J, Hess JM, Kübler K, Grimsby J. et al. RNA sequence analysis reveals macroscopic somatic clonal expansion across normal tissues. Science. 2019:364 pii: eaaw0726
37. Kvasnicka HM. The differential diagnosis of classical myeloproliferative neoplasms (MPN): the updated WHO criteria. Rinsho Ketsueki. 2019;60:1166-1175
38. Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A. et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257-2261
39. Rosenfeld N, Aharonov R, Meiri E, Rosenwald S, Spector Y. et al. MicroRNAs accurately identify cancer tissue origin. Nat Biotechnol. 2008;26:462-469
40. Gilad S, Meiri E, Yogev Y, Benjamin S, Lebanony D, Yerushalmi N. et al. Serum microRNAs are promising novel biomarkers. PLoS One. 2008;3:e3148
41. Sorenson GD, Pribish DM, Valone FH, Memoli VA, Bzik DJ, Yao SL. Soluble normal and mutated DNA sequences from single-copy genes in human blood. Cancer Epidemiol. Biomarkers Prev. 1994;3:67-71
42. García-Foncillas J, Alba E, Aranda E, Díaz-Rubio E, López-López R, Tabernero J. et al. Incorporating BEAMing technology as a liquid biopsy into clinical practice for the management of colorectal cancer patients: an expert taskforce review. Ann Oncol. 2017;28:2943-2949
43. Wan JCM, Massie C, Garcia-Corbacho J, Mouliere F, Brenton JD, Caldas C. et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer. 2017;17:223-238
44. Newman AM, Bratman SV, To J, Wynne JF, Eclov NC, Modlin LA. et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med. 2014;20:548-554
45. Gale D, Lawson ARJ, Howarth K, Madi M, Durham B, Smalley S. et al. Development of a highly sensitive liquid biopsy platform to detect clinically-relevant cancer mutations at low allele fractions in cell-free DNA. PLoS One. 2018;13:e0194630
46. Cristiano S, Leal A, Phallen J, Fiksel J, Adleff V, Bruhm DC. et al. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature. 2019;570:385-389
47. Heitzer E, Perakis S, Geigl JB, Speicher MR. The potential of liquid biopsies for the early detection of cancer. NPJ Precis. 2017;1:36
48. Cohen JD, Li L, Wang Y, Thoburn C, Afsari B, Danilova L. et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. 2018;359:926-930
49. Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331:1559-1564
50. Giuliano M, Giordano A, Jackson S, De Giorgi U, Mego M, Cohen EN. et al. Circulating tumor cells as early predictors of metastatic spread in breast cancer patients with limited metastatic dissemination. Breast Cancer Res. 2014;16:440
51. Wang D, Yang Y, Jin L, Wang J, Zhao X, Wu G. et al. Prognostic models based on postoperative circulating tumor cells can predict poor tumor recurrence-free survival in patients with stage II-III colorectal cancer. J Cancer. 2019;10:4552-4563
52. Goodman OB Jr, Symanowski JT, Loudyi A, Fink LM, Ward DC, Vogelzang NJ. Circulating tumor cells as a predictive biomarker in patients with hormone-sensitive prostate cancer. Clin Genitourin Cancer. 2011;9:31-8
53. Poveda A, Kaye SB, McCormack R, Wang S, Parekh T, Ricci D. et al. Circulating tumor cells predict progression free survival and overall survival in patients with relapsed/recurrent advanced ovarian cancer. Gynecol Oncol. 2011;122:567-72
54. Lianidou ES, Mavroudis D, Sotiropoulou G, Agelaki S, Pantel K, Lianidou ES. et al. What's new on circulating tumor cells? A meeting report. Breast Cancer Res. 2010;12:307
55. Yu M, Bardia A, Aceto N, Bersani F, Madden MW, Donaldson MC. et al. Cancer therapy. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science. 2014;345:216-220
56. Satelli A, Batth I, Brownlee Z, Mitra A, Zhou S, Noh H. et al. EMT circulating tumor cells detected by cell-surface vimentin are associated with prostate cancer progression. Oncotarget. 2017;8:49329-37
57. Theodoropoulos PA, Polioudaki H, Agelaki S, Kallergi G, Saridaki Z, Mavroudis D. et al. Circulating tumor cells with a putative stem cell phenotype in peripheral blood of patients with breast cancer. Cancer Lett. 2010;288:99-106
58. de Wit S, Manicone M, Rossi E, Lampignano R, Yang L, Zill B. et al. EpCAMhigh and EpCAMlow circulating tumor cells in metastatic prostate and breast cancer patients. Oncotarget. 2018;9:35705-35716
59. Riethdorf S, O'Flaherty L, Hille C, Pantel K. Clinical applications of the CellSearch platform in cancer patients. Adv Drug Deliv Rev. 2018;125:102-121
60. Neumann MHD, Bender S, Krahn T, Schlange T. Detection of gene rearrangements in circulating tumor cells: examples of ALK-, ROS1-, RET-rear-rangements in non-small-cell lung cancer and ERG-rearrange-ments in prostate cancer. Adv Exp Med Biol. 2017;994:169-179
61. Zhu Z, Qiu S, Shao K, Hou Y. Progress and challenges of sequencing and analyzing circulating tumor cells. Cell Biol Toxicol. 2017;34:405-415
62. Gao W, Huang T, Yuan H, Yang J, Jin Q, Jia C. et al. Highly sensitive detection and mutational analysis of lung cancer circulating tumor cells using integrated combined immunomagnetic beads with a droplet digital PCR chip. Talanta. 2018;185:229-236
63. Lim SB, Lim CT, Lim WT. Single-Cell analysis of tirculating tumor cells: why heterogeneity matters. Cancers (Basel). 2019:11 pii: E1595
64. Keller L, Pantel K. Unravelling tumour heterogeneity by single-cell profiling of circulating tumour cells. Nat Rev Cancer. 2019;19:553-567
65. Liu Z, Fusi A, Klopocki E, Schmittel A, Tinhofer I, Nonnenmacher A. Negative enrichment by immunomagnetic nanobeads for unbiased characterization of circulating tumor cells from peripheral blood of cancer patients. J Transl Med. 2011;9:70
66. Sequist LV, Nagrath S, Toner M, Haber DA, Lynch TJ. The CTC-chip: an exciting new tool to detect circulating tumor cells in lung cancer patients. J Thorac Oncol. 2009;4:281-283
67. Varillas JI1, Zhang J, Chen K, Barnes II, Liu C, George TJ. et al. Microfluidic isolation of circulating tumor cells and cancer stem-like cells from patients with pancreatic ductal adenocarcinoma. Theranostics. 2019;9:1417-1425
68. Bobek V, Kacprzak G, Rzechonek A, Kolostova K. Detection and cultivation of circulating tumor cells in malignant pleural mesothelioma. Anticancer Res. 2014;34:2565-2569
69. Varillas JI, Zhang J, Chen K, Barnes II, Liu C, George TJ. et al. Microfluidic isolation of circulating tumor cells and cancer stem-like cells from patients with pancreatic ductal adenocarcinoma. Theranostics. 2019;9:1417-1425
70. Ramirez AB, U'Ren L, Campton DE, Stewart D, Nordberg JJ, Stilwell JL. et al. RareCyte® CTC analysis step 1: AccuCyte® sample preparation for the comprehensive recovery of nucleated cells from whole blood. Methods Mol Biol. 2017;1634:163-172
71. Durmus NG, Tekin HC, Guven S, Sridhar K, Arslan Yildiz A, Calibasi G. et al. Magnetic levitation of single cells. Proc Natl Acad Sci USA. 2015;112:E3661-8
72. Peeters DJ1, De Laere B, Van den Eynden GG, Van Laere SJ, Rothé F, Ignatiadis M. et al. Semiautomated isolation and molecular characterisation of single or highly purified tumour cells from CellSearch enriched blood samples using dielectrophoretic cell sorting. Br J Cancer. 2013;108:1358-67
73. Tulley S, Zhao Q, Dong H, Pearl ML, Chen WT. Vita-assay method of enrichment and identification of circulating cancer cells/circulating tumor cells (CTCs). Methods Mol Biol. 2016;1406:107-119
74. Alix-Panabieres C. EPISPOT assay: detection of viable DTCs/CTCs in solid tumor patients. Recent Results Cancer Res. 2012;195:69-76
75. Togo S, Katagiri N, Namba Y, Tulafu M, Nagahama K, Kadoya K. et al. Sensitive detection of viable circulating tumor cells using a novel conditionally telomerase-selective replicating adenovirus in non-small cell lung cancer patients. Oncotarget. 2017;8:34884-34895
76. Chargaff E, West R. The biological significance of the thromboplastic protein of blood. J Biol Chem. 1946;166:189-97
77. Wolf P. The nature and significance of platelet products in human plasma. Br J Haematol. 1967;13:269-288
78. Harding C, Heuser J, Stahl P. Receptor-mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes. J Cell Biol. 1983;97:329-339
79. Pan BT, Johnstone RM. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: selective externalization of the receptor. Cell. 1983;33:967-978
80. Pan BT, Teng K, Wu C, Adam M, Johnstone RM. Electron microscopic evidence for externalization of the transferrin receptor in vesicular form in sheep reticulocytes. J Cell Biol. 1985;101:942-948
81. Taylor J, Bebawy M. Front cover: proteins regulating microvesicle biogenesis and multidrug resistance in cancer. Proteomics. 2019;19:1-2
82. Trams EG, Lauter CJ, Salem N Jr, Heine U. Exfoliation of membrane ecto-enzymes in the form of micro-vesicles. Biochim Biophys Acta. 1981;645:63-70
83. He C, Zheng S, Luo Y3, Wang B. Exosome theranostics: biology and translational medicine. Theranostics. 2018;8:237-255
84. Siravegna G, Marsoni S, Siena S, Bardelli A. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol. 2017;14:531-548
85. Weidle UH, Birzele F, Kollmorgen G, Rüger R. The multiple roles of exosomes in metastasis. Cancer Genomics Proteomics. 2017;14:1-15
86. Frühbeis C, Fröhlich D, Kuo WP, Amphornrat J, Thilemann S, Saab AS. et al. Neurotransmitter-triggered transfer of exosomes mediates oligodendrocyte-neuron communication. PLoS Biol. 2013;11:e1001604
87. Melo SA, Sugimoto H1, O'Connell JT, Kato N, Villanueva A, Vidal A. et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell. 2014;26:707-721
88. van Niel G, D'Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol. 2018;19:213-228
89. Cocucci E, Meldolesi J. Ectosomes and exosomes: shedding the confusion between extracellular vesicles. Trends Cell Biol. 2015;25:364-372
90. Yáñez-Mó M. Siljander PR, Andreu Z, Zavec AB, Borràs FE, Buzas EI, et al. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles. 2015;4:27066
91. Fong MY, Zhou W, Liu L, Alontaga AY, Chandra M, Ashby J. Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat Cell Biol. 2015;17:183-194
92. Chen G, Huang AC, Zhang W, Zhang G, Wu M, Xu W. et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature. 2018;560:382-386
93. Hoshino A, Costa-Silva B, Shen TL, Rodrigues G, Hashimoto A, Tesic Mark M. et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527:329-335
94. Taylor DD, Gercel-Taylor C. Exosomes/microvesicles: mediators of cancer-associated immunosuppressive microenvironments. Semin Immunopathol. 2011;33:441-454
95. Toth B, Nieuwland R, Liebhardt S, Ditsch N, Steinig K, Stieber P. et al. Circulating microparticles in breast cancer patients: a comparative analysis with established biomarkers. Anticancer Res. 2008;28:1107-12
96. Tian XP, Wang CY, Jin XH, Li M, Wang FW, Huang WJ. et al. Acidic microenvironment up-regulates exosomal miR-21 and miR-10b in early-stage hepatocellular carcinoma to promote cancer cell proliferation and metastasis. Theranostics. 2019;9:1965-1979
97. Wang H, Jiang D, Li W, Xiang X, Zhao J, Yu B. et al. Evaluation of serum extracellular vesicles as noninvasive diagnostic markers of glioma. Theranostics. 2019;9:5347-5358
98. Nilsson J, Skog J, Nordstrand A, Baranov V, Mincheva-Nilsson L, Breakefield XO. et al. Prostate cancer-derived urine exosomes: a novel approach to biomarkers for prostate cancer. Br J Cancer. 2009;100:1603-1607
99. Mitchell PJ, Welton J, Staffurth J, Court J, Mason MD, Tabi Z. et al. Can urinary exosomes act as treatment response markers in prostate cancer? J Transl Med. 2009;7:4
100. Reclusa P, Taverna S, Pucci M, Durendez E, Calabuig S, Manca P. Exosomes as diagnostic and predictive biomarkers in lung cancer. J Thorac Dis. 2017;9:S1373-S1382
101. Cazzoli R, Buttitta F, Di Nicola M, Malatesta S, Marchetti A, Rom WN. et al. MicroRNAs derived from circulating exosomes as noninvasive biomarkers for screening and diagnosing lung cancer. J Thorac Oncol. 2013;8:1156-1162
102. Jin X, Chen Y, Chen H, Fei S, Chen D, Cai X. et al. Evaluation of tumor-derived exosomal miRNA as potential diagnostic biomarkers for early-stage non-small cell lung cancer using next-generation sequencing. Clin Cancer Res. 2017;23:5311-5319
103. Clayton A, Turkes A, Navabi H, Mason MD, Tabi Z. Induction of heat shock proteins in B-cell exosomes. J Cell Sci. 2005;118:3631-3638
104. Taylor DD, Gerçel-Taylor C. Tumour-derived exosomes and their role in cancer-associated T-cell signalling defects. Br J Cancer. 2005;92:305-311
105. Qiu X, Li Z1, Han X, Zhen L, Luo C, Liu M. et al. Tumor-derived nanovesicles promote lung distribution of the therapeutic nanovector through repression of kupffer cell-mediated phagocytosis. Theranostics. 2019;9:2618-2636
106. Fauré J, Lachenal G, Court M, Hirrlinger J, Chatellard-Causse C, Blot B. et al. Exosomes are released by cultured cortical neurones. Mol Cell Neurosci. 2006;31:642-648
107. Lai RC, Arslan F, Lee MM, Sze NS, Choo A, Chen TS. et al. Exosome secreted by MSC reduces myocardial ischemia/reperfusion injury. Stem Cell Res. 2010;4:214-222
108. Xian P, Hei Y, Wang R, Wang T, Yang J, Li J. et al. Mesenchymal stem cell-derived exosomes as a nanotherapeutic agent for amelioration of inflammation-induced astrocyte alterations in mice. Theranostics. 2019;9:5956-5975
109. Tetta C, Bruno S, Fonsato V, Deregibus MC, Camussi G. The role of microvesicles in tissue repair. Organogenesis. 2011;7:105-115
110. Michael A, Bajracharya SD, Yuen PS, Zhou H, Star RA, Illei GG. et al. Exosomes from human saliva as a source of microRNA biomarkers. Oral Dis. 2010;16:34-38
111. Lakkaraju A, Rodriguez-Boulan E. Itinerant exosomes: emerging roles in cell and tissue polarity. Trends Cell Biol. 2008;18:199-209
112. Kosaka N, Izumi H, Sekine K, Ochiya T. MicroRNA as a new immune-regulatory agent in breast milk. Silence. 2010;1:7
113. Keller S, Rupp C, Stoeck A, Runz S, Fogel M, Lugert S. et al. CD24 is a marker of exosomes secreted into urine and amniotic fluid. Kidney Int. 2007;72:1095-1102
114. Pisitkun T, Shen RF, Knepper MA. Identification and proteomic profiling of exosomes in human urine. Proc Natl Acad Sci USA. 2004;101:13368-13373
115. Rani S, O'Brien K, Kelleher FC, Corcoran C, Germano S, Radomski MW. et al. Isolation of exosomes for subsequent mRNA, microRNA, and protein profiling. Methods Mol Biol. 2011;784:181-195
116. Conde-Vancells J, Rodriguez-Suarez E, Embade N, Gil D, Matthiesen R, Valle M. et al. Characterization and comprehensive proteome profiling of exosomes secreted by hepatocytes. J Proteome Res. 2008;7:5157-5166
117. Liu C, Su C. Design strategies and application progress of therapeutic exosomes. Theranostics. 2019;9:1015-1028
118. Taylor DD, Shah S. Methods of isolating extracellular vesicles impact down-stream analyses of their cargoes. Methods. 2015;87:3-10
119. Beekman P, Enciso-Martinez A, Rho HS, Pujari SP, Lenferink A, Zuilhof H. et al. Immuno-capture of extracellular vesicles for individual multi-modal characterization using AFM, SEM and Raman spectroscopy. Lab Chip. 2019;19:2526-2536
120. Butvilovskaya VI, Tikhonov AA, Savvateeva EN, Ragimov AA, Salimov EL, Voloshin SA. et al. Hydrogel microchip as a tool for studying exosomes in human serum. Mol Biol (Mosk). 2017;51:817-823
121. Wang W, Luo J, Wang S. Recent progress in isolation and detection of extracellular vesicles for cancer diagnostics. Adv Healthc Mater. 2018;7:e1800484
122. Carnell-Morris P, Tannetta D, Siupa A, Hole P, Dragovic R. Analysis of extracellular vesicles using fluorescence nanoparticle tracking analysis. Methods Mol Biol. 2017;1660:153-173
123. Kahlert C, Melo SA, Protopopov A, Tang J, Seth S, Koch M. et al. Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J Biol Chem. 2014;289:3869-3875
124. Tian Q, He C, Liu G, Zhao Y, Hui L, Mu Y. et al. Nanoparticle ounting by microscopic digital detection: selective quantitative analysis of exosomes via surface-anchored nucleic acid amplification. Anal Chem. 2018;90:6556-6562
125. Wu D, Yan J, Shen X, Sun Y, Thulin M, Cai Y. et al. Profiling surface proteins on individual exosomes using a proximity barcoding assay. Nat Commun. 2019;10:3854
126. Garcia-Romero N, Esteban-Rubio S, Rackov G, Carrión-Navarro J, Belda-Iniesta C, Ayuso-Sacido A. Extracellular vesicles compartment in liquid biopsies: clinical application. Mol Aspects Med. 2018;60:27-37
127. Krishnan SR, Luk F, Brown RD, Suen H, Kwan Y, Bebawy M. Isolation of human CD138 (+) micro-particles from the plasma of patients with multiple myeloma. Neoplasia. 2016;18:25-32
128. Wang CC, Tseng CC, Chang HC, Huang KT, Fang WF, Chen YM. et al. Circulating microparticles are prognostic biomarkers in advanced non-small cell lung cancer patients. Oncotarget. 2017;8:75952-75967
129. Clayton A, Buschmann D, Byrd JB, Carter DRF, Cheng L, Compton C. et al. Summary of the ISEV workshop on extracellular vesicles as disease biomarkers, held in Birming-ham, UK, during December 2017. J Extracell Vesicles. 2018;7:1473707
130. Théry C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R. et al. Minimal information for studies of extra-cellular vesicles 2018 (MISEV2018): a position statement of the international society for extracellular vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7:1535750
131. Kosaka N, Kogure A, Yamamoto T, Urabe F, Usuba W, Prieto-Vila M. et al. Exploiting the message from cancer: the diagnostic value of extracellular vesicles for clinical applications. Exp Mol Med. 2019;51:31
132. Calverley DC, Phang TL, Choudhury QG, Gao B, Oton AB, Weyant MJ. et al. Significant downregulation of platelet gene expression in metastatic lung cancer. Clin Transl Sci. 2010;3:227-232
133. Nilsson RJ, Balaj L, Hulleman E, van Rijn S, Pegtel DM, Walraven M. et al. Blood platelets contain tumor-derived RNA biomarkers. Blood. 2011;118:3680-3683
134. Simon A Joosse, Klaus Pantel. Tumor-educated platelets as liquid biopsy in cancer patients. Cancer Cell. 2015;28:552-554
135. Best MG, Sol N, Kooi I, Tannous J, Westerman BA, Rustenburg F. et al. RNA-Seq of tumor-educated platelets enables blood-based pan-cancer, multiclass, and molecular pathway cancer diagnostics. Cancer Cell. 2015;28:666-676
136. Best MG, Sol N, In't Veld SGJG, Vancura A, Muller M, Niemeijer AN. et al. Swarm intelligence-enhanced detection of non-small-cell lung cancer using tumor-educated platelets. Cancer Cell. 2017;32:238-252
137. Ponti G, Manfredini M, Tomasi A. Non-blood sources of cell-free DNA for cancer molecular profiling in clinical pathology and oncology. Crit Rev Oncol Hematol. 2019;141:36-42
138. Lousada-Fernandez F, Rapado-Gonzalez O, Lopez-Cedrun JL, Lopez-Lopez R, Muinelo-Romay L, Suarez-Cunqueiro MM. Liquid biopsy in oral cancer. Int J Mol Sci. 2018:19 pii: E1704
139. Neoh KH, Hassan AA, Chen A, Sun Y, Liu P, Xu KF. et al. Rethinking liquid biopsy: Microfluidic assays for mobile tumor cells in human body fluids. Biomaterials. 2018;150:112-124
140. Durand X, Moutereau S, Xylinas E, de la Taille A. ProgensaTM PCA3 test for prostate cancer. Expert Rev Mol Diagn. 2011;11:137-144
141. Raja N, Russell CM, George AK. Urinary markers aiding in the detection and risk stratification of prostate cancer. Transl Androl Urol. 2018;7:S436-S442
142. Cao Y, Green K, Quattlebaum S, Milam B, Lu L, Gao D. et al. Methylated genomic loci encoding micro-RNA as a biomarker panel in tissue and saliva for head and neck squamous cell carcinoma. Clin Epigenetics. 2018;10:43
143. Gai C, Camussi F, Broccoletti R, Gambino A, Cabras M, Molinaro L. et al. Salivary extracellular vesicle-associated miRNAs as potential biomarkers in oral squamous cell carcinoma. BMC Cancer. 2018;18:439
144. De Mattos-Arruda L, Mayor R, Ng CKY, Weigelt B, Martínez-Ricarte F, Torrejon D. et al. Cerebrospinal fluid-derived circulating tumour DNA better represents the genomic alterations of brain tumours than plasma. Nat Commun. 2015;6:8839
145. Akers JC, Hua W, Li H, Ramakrishnan V, Yang Z, Quan K, Zhu W. et al. A cerebrospinal fluid microRNA signature as biomarker for glioblastoma. Oncotarget. 2017;8:68769-68779
146. Kisiel JB, Klepp P, Allawi HT, Taylor WR, Giakoumopoulos M, Sander T. et al. Analysis of DNA methylation at specific loci in stool samples detects colorectal cancer and high-grade dysplasia in patients with inflammatory bowel disease. Clin Gastroenterol Hepatol. 2019;17:914-921.e5
147. Rangel MP, Antonangelo L, Acencio MMP, Faria CS, de Sá VK, Leão PS. et al. Detection of sputum cofilin-1 as indicator of malignancy. Braz J Med Biol Res. 2018;51:e7138
148. Yeo CD, Kim JW, Kim KH, Ha JH, Rhee CK, Kim SJ. et al. Detection and comparison of EGFR mutations in matched tumor tissues, cell blocks, pleural effusions, and sera from patients with NSCLC with malignant pleural effusion, by PNA clamping and direct sequencing. Lung Cancer. 2018;81:207-212
149. Krimmel JD, Schmitt MW, Harrell MI, Agnew KJ, Kennedy SR, Emond MJ. et al. Ultra-deep sequencing detects ovarian cancer cells in peritoneal fluid and reveals somatic TP53 mutations in noncancerous tissues. Proc Natl Acad Sci USA. 2016;113:6005-6010
Author contact
![]() Corresponding authors: Ben Wang, PhD & Kefeng Ding, MD, Cancer Institute (Key Laboratory of Cancer Prevention and Intervention, China National Ministry of Education), The Second Affiliated Hospital & Institute of Translational Medicine, School of Medicine, Zhejiang University, Hangzhou, Zhejiang, China, 310009, China. Tel: 86-0571-86971790; Email: benwangedu.cn (B.W.), dingkefengedu.cn (K.D.); Liquan Wang, MD, Department of Obstetrics, The Second Affiliated Hospital, Zhejiang University, Hangzhou, Zhejiang, 310003, China. Email: wangliquanedu.cn (L.W.).
Corresponding authors: Ben Wang, PhD & Kefeng Ding, MD, Cancer Institute (Key Laboratory of Cancer Prevention and Intervention, China National Ministry of Education), The Second Affiliated Hospital & Institute of Translational Medicine, School of Medicine, Zhejiang University, Hangzhou, Zhejiang, China, 310009, China. Tel: 86-0571-86971790; Email: benwangedu.cn (B.W.), dingkefengedu.cn (K.D.); Liquan Wang, MD, Department of Obstetrics, The Second Affiliated Hospital, Zhejiang University, Hangzhou, Zhejiang, 310003, China. Email: wangliquanedu.cn (L.W.).
 Global reach, higher impact
Global reach, higher impact