13.3
Impact Factor
Theranostics 2020; 10(10):4507-4514. doi:10.7150/thno.41484 This issue Cite
Research Paper
Glucose oxidase-like activity of cerium oxide nanoparticles: use for personal glucose meter-based label-free target DNA detection
1. Department of Chemical and Biomolecular Engineering (BK21+ Program), KAIST, 291 Daehak-ro, Yuseong-gu, Daejeon 34141, Republic of Korea.
2. Department of Biological Engineering, College of Engineering, Konkuk University, Seoul 05029, Republic of Korea.
Received 2019-10-25; Accepted 2020-1-20; Published 2020-3-15
Abstract
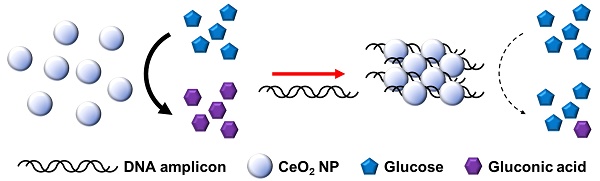
Recently, personal glucose meter (PGM) has been utilized for the detection of non-glucose targets for point-of-care (POC) testing. Aimed at this goal, we herein developed a new PGM-based label-free read-out method for polymerase chain reaction (PCR) based on our novel finding that cerium oxide nanoparticles (CeO2 NPs) exhibit glucose oxidase-like activity comparable to the natural glucose oxidase enzyme.
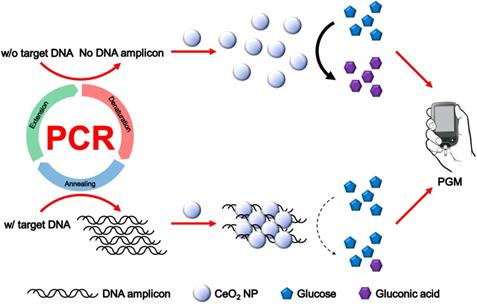
Methods: In principle, DNA amplicons produced by PCR in the presence of target DNA electrostatically bind to CeO2 NPs, leading to their aggregation and reducing the efficiency for CeO2 NP-catalyzed glucose oxidation reaction. Thus, glucose is hardly oxidized to gluconic acid, resulting in the maintenance of initial high glucose level. On the contrary, in the absence of target DNA or presence of non-target DNA, DNA amplicons are not produced and glucose is effectively oxidized by the glucose oxidase-like activity of CeO2 NPs, leading to the significant reduction of glucose level. Finally, the resulting glucose level is simply measured by using PGM.
Results: With this strategy, DNA amplicons were quantitatively examined within 5 min, realizing ultrafast analysis of PCR results without any cumbersome and labor-intensive procedures. In addition, the target genomic DNA derived from Escherichia coli (E. coli) was sensitively determined down to 10 copies with high selectivity.
Conclusion: Importantly, the use of PGM as a detection component enables its direct application in POC settings. Based on the meritorious features of PGM such as rapidity, simplicity, and cost-effectiveness, we expect that the devised system could serve as a core platform for the on-site read-out of PCR amplification.
Keywords: Polymerase chain reaction, Cerium oxide nanoparticle, Glucose oxidase-like activity, Personal glucose meter, Biosensor
Introduction
Personal glucose meter (PGM) that enables the self-monitoring of blood glucose level has been widely utilized as the home-kit for the self-diagnosis of diabetes. In recent years, many researchers have attempted to use PGM for the detection of non-glucose targets at point-of-care (POC) setting based on its advantageous features such as cost-effectiveness, simplicity, and portability [1-14]. The pioneering work was executed by Xiang et al. for the detection of various non-glucose analytes, which relies on the correlation between the concentration of non-glucose target and glucose level [15]. Specifically, the invertase-conjugated DNA probe was hybridized with DNA aptamer. Only in the presence of non-glucose target that forms the complex with DNA aptamer, the invertase-modified DNA probe was released and then used to catalyze the hydrolysis of sucrose to glucose after magnetic separation. The resulting glucose level that is proportional to the amount of non-glucose target was finally measured by using PGM. Based on this principle, diverse non-glucose analytes such as cocaine, adenosine, interferon-gamma, and uranium have been determined. However, this strategy still has the critical limitations that it requires the immobilization of DNA aptamer on the magnetic beads, the conjugation of DNA probe with invertase, and magnetic separation steps, which make the total assay quite complex and limit the practical application of PGM for biomolecular detection in facility-limited environments.
Since the discovery that magnetic nanoparticles (MNPs) have the peroxidase-mimicking activity, various metal nanoparticles including gold nanoparticles (AuNPs), platinum nanoparticles (PtNPs), cupric oxide nanoparticles (CuO NPs), iron oxide nanoparticles (Fe3O4 NPs), and cerium oxide nanoparticles (CeO2 NPs) have been found to possess the enzyme-mimicking activities such as peroxidase, catalase, and oxidase-like activities, which are even superior to those of natural enzymes [16-29]. Importantly, CeO2 NPs with oxidase-like activity exhibit the high stability and catalytic efficiency for substrate oxidation even without additional oxidizing agent. In addition, it was reported that the enzyme-mimicking activity of metal nanoparticles can be regulated by nucleic acids that bind to metal nanoparticles by the electrostatic interaction, inducing the aggregation of metal nanoparticles and thus decreasing their catalytic activity [24,26].
In this work, we developed a new PGM-utilized method for the on-site read-out of polymerase chain reaction (PCR) result based on our novel finding that CeO2 NPs exhibit the glucose oxidase-like activity enabling the correlation of DNA amplicon concentration with glucose level. In principle, the DNA amplicons produced by PCR in the presence of target DNA bind to CeO2 NPs through electrostatic interaction, inducing their aggregation and, accordingly, reducing the efficiency for CeO2 NP-catalyzed glucose oxidation reaction. Thus, the effective conversion of glucose to gluconic acid by CeO2 NPs is suppressed, keeping the initial high glucose level that can be simply measured by PGM. With this strategy, the target DNA was sensitively determined down to 10 copies with high selectivity. More importantly, the detection of DNA amplicons was completed in less than 5 min based on the high catalytic activity of CeO2 NPs, accomplishing ultrafast analysis of PCR results without any laborious and complex step by simply utilizing PGM as a detection component.
Methods
Materials
All DNA oligonucleotides used in this study were synthesized and purified with high performance liquid chromatography (HPLC) by Bioneer® (Daejeon, Korea). The sequence information of oligonucleotides employed in this study is in Table S1. The personal glucose meter (PGM) whose dynamic range for glucose detection is from 0.6 to 33 mM was purchased from Accu-Chek (Roche, Basel, Switzerland). Cerium (IV) oxide nanoparticle (CeO2 NP), sodium acetate, glucose, ethidium bromide (EtBr), hydroethidine, and 2,5-dihydroxybenzoic acid (DHB) were purchased from Sigma-Aldrich (St. Louis, MO, USA). The deoxynucleoside triphosphate (dNTP) and i-TaqTM DNA polymerase were purchased from Intron Biotechnology Inc. (Daejeon, Korea). Ultrapure DNase/RNase-free distilled water (DW) purchased from Bioneer® was used in all experiments. All chemicals used in this study were of analytical grade.
PGM-based label-free method for the read-out of PCR amplification using glucose oxidase-like activity of CeO2 NP
PCR for the amplification of 16S rRNA gene in Escherichia coli (E. coli) genomic DNA (gDNA) was first carried out on S1000TM thermal cycler (Bio-Rad, CA, USA) in a PCR solution (25 μL) containing 0.2 mM each dNTP, 0.4 μM each primer, 2.5 U i-TaqTM DNA polymerase, E. coli gDNA at different copy numbers, and 1X PCR buffer (10 mM Tris-HCl (pH 8.3), 50 mM KCl, and 2 mM MgCl2). The PCR solution was heated up to 95 °C for 5 min followed by 40 cycles of 95 °C for 15 s, 58 °C for 15 s, and 72 °C for 30 s and finalized at 72 °C for 5 min. The DNA amplicon (524 bp) was purified using NucleoSpin purification kit (Macherey-Nagel, Duren, Germany) according to the manufacturer's protocol. Next, the purified DNA amplicon (25 μL) was added to CeO2 NP solution containing 2.5 μL CeO2 NP (2 wt%) and 12.5 μL sodium acetate buffer (0.4 M, pH 4.2) and the mixture was incubated at room temperature for 2 min. After the addition of 10 μL glucose (300 mM), the reaction solution was further incubated at room temperature for 1.5 min. Finally, the resulting glucose level was measured by PGM. For the determination of E. coli gDNA in the human serum sample, PCR solution (25 μL) was first prepared by adding 0.2 mM each dNTP, 0.4 μM each primer, 2.5 U i-TaqTM DNA polymerase, E. coli gDNA at different copy numbers, and 1X PCR buffer (10 mM Tris-HCl (pH 8.3), 50 mM KCl, and 2 mM MgCl2) into 1% human serum sample, which was then analyzed according to the same procedure described above.
Investigation of glucose oxidase-like activity of CeO2 NP
The solution prepared by mixing 30 μL DW, 12.5 μL sodium acetate buffer (0.4 M, pH 4.2), 2.5 μL CeO2 NP (2 wt%), and 5 μL hydroethidine (10 μg/mL) was incubated at room temperature for 1.5 min. The fluorescent signal from hydroethidine was scanned in the range from 570 to 670 nm with 530 nm excitation wavelength by utilizing Tecan Infinite M200 pro-microplate reader (Mnnedorf, Switzerland). In addition, the solution for the matrix-assisted laser desorption/ionization time-of-flight (MALDI TOF) analysis prepared by mixing 25 μL DW, 12.5 μL sodium acetate buffer (0.4 M, pH 4.2), 2.5 μL CeO2 NP (2 wt%), and 10 μL glucose (300 mM) was incubated at room temperature for 1.5 min. Then, the solution was mixed with equal volume of matrix prepared by resolving 10 mg/mL DHB into 50% ethanol and the mixture was loaded on MTP 384 ground steel MALDI TOF MS plate (Hudson Surface Technology, NJ, USA). After drying of the loaded sample at room temperature for overnight, MALDI TOF analysis was conducted utilizing Autoflex III MALDI-TOF MS instrument equipped with ND-YAG laser operated at 355 nm wavelength (Bruker Daltonics, Germany). The voltage, delayed extraction time, grid voltage, laser rate, and laser shots were 20 kV, 10 ns, 70%, 1 - 200 Hz, and 1,500 per sample spot, respectively.
The kinetic parameters of CeO2 NP-catalyzed glucose oxidation reaction were obtained from the Michaelis-Menten model. The reaction velocity defined as L0 - L where L0 and L are glucose levels measured by PGM before and after CeO2 NP-catalyzed glucose oxidation reaction (30 min), respectively, was calculated from the solutions prepared by mixing 17.5 μL DW, 12.5 μL sodium acetate buffer (0.4 M, pH 4.2), 5 μL CeO2 NP (700 nM), 10 μL purified DNA amplicon (500 nM), and 5 μL glucose at varying concentrations and then incubated at room temperature for 30 min.
Confirmation of DNA-induced aggregation of CeO2 NP
We examined the crystalline nature of the employed CeO2 NP by using SmartLab X-ray diffractometer (Rigaku, Tokyo, Japan) with monochromatic Cu-Kα radiation. The scan range and rate were 20° - 65° and 5°/min, respectively. The surface state of CeO2 NP was also analyzed by using X-ray photoelectron spectrometer (Sigma Probe, Thermo Fisher Scientific, MA, USA) with microfocused monochromator X-ray source. In addition, the transmission electron microscopy (TEM) and energy-dispersive X-ray spectroscopy (EDS) data were obtained by using Cs-corrected STEM (JEM-ARM200F) in National NanoFab Center (NNFC, Daejeon, Korea). Samples for TEM and EDS analyses were prepared by casting 15 μL sample onto the copper grid (300 mesh) with a lacey carbon film (LC300-CU) purchased from Electron Microscopy Sciences (Hatfield, PA, USA). Then, the loaded sample was dried at room temperature overnight before the analysis. In addition, the zeta potential and dynamic light scattering (DLS) analysis were performed by using Zetasizer (Malvern, PA, USA). For zeta potential and DLS measurement, 1 mL sample prepared by adding 0.5 mg/mL CeO2 NP and 100 nM purified DNA amplicon into 0.1 M sodium acetate buffer (pH 4.2) was added in Zetasizer capillary cell and polystyrene cuvette, respectively, and the samples were scanned three times to obtain average zeta potential and diameter of CeO2 NP. The solution for the ultraviolet-visible spectroscopy (UV-Vis) analysis was prepared by mixing 25 μL DW, 12.5 μL sodium acetate buffer (0.4 M, pH 4.2), 2.5 μL CeO2 NP (2 wt%), and 10 μL purified DNA amplicon (500 nM). After the incubation at room temperature for 2 min, the solution was centrifuged at 10,000 g for 5 min. The precipitated CeO2 NP/DNA complex was dispersed in 0.1 M sodium acetate buffer (pH 4.2) and then the absorbance spectrum in the range from 220 to 440 nm was measured utilizing NanoDropTM One (Thermo Fisher Scientific, MA, USA).
Results & Discussion
PGM-based label-free method for the read-out of PCR amplification using glucose oxidase-like activity of CeO2 NP
The overall procedure for PGM-based label-free read-out of PCR amplification using glucose oxidase-like activity of CeO2 NPs is illustrated in Figure 1. The reaction system involves the PCR amplification with the target specific primer sets followed by the incubation with CeO2 NPs. In the presence of target genomic DNA (gDNA) derived from Escherichia coli (E. coli), the PCR process generates DNA amplicons that bind to CeO2 NPs through the electrostatic interaction with positively charged surface of CeO2 NPs, which induces the aggregation of CeO2 NPs. As a result, the glucose substrate has a limited access onto CeO2 NPs and initial high glucose level is maintained. In contrast, in the absence of target gDNA, DNA amplicons are not produced by PCR process. Thus, the CeO2 NPs are well dispersed in the solution and thus exert the effective glucose oxidase-like activity. Accordingly, glucose is effectively converted to gluconic acid, leading to significant reduction of glucose level. Finally, the resulting glucose level that is proportional to the amount of produced DNA amplicon is simply measured by using PGM.
Investigation of glucose oxidase-like activity of CeO2 NP
Since this is the first attempt to discover and utilize the glucose oxidase-like activity of CeO2 NPs, we examined the mechanism of CeO2 NP-catalyzed glucose oxidation reaction. As described in the following equation, the auto-catalytic redox switching of two oxidation states, Ce3+ and Ce4+, on the surface of CeO2 NP would be the key for the glucose oxidase-like activity of CeO2 NP [30-33].
2 CeO2 (Ce4+) + 2 H+ (aq) → Ce2O3 (Ce3+) + H2O (1)
Ce2O3 (Ce3+) + 1.5 O2 → 2 CeO2 (Ce4+) + O2•- (2)
O2•- + C6H12O6 + 2 H+ (aq) → C6H12O7 + H2O (3)
O2•- + Ce2O3 (Ce3+) + 2 H+ (aq) → 2 CeO2 (Ce4+) + H2O (4)
Under the acidic condition, the surface of CeO2 NP (Ce4+) is first reduced to Ce2O3 (Ce3+), generating oxygen vacancy (equation 1). Then, the oxygen molecule (O2) adjacent to the surface adsorbs onto the oxygen vacancy in Ce3+ state while being reduced to superoxide anion radical (O2•-) with the concomitant oxidation of Ce3+ to Ce4+ (equation 2). Next, O2•-, a powerful oxidizing agent, catalyzes the oxidation of glucose to gluconic acid (equation 3) and Ce3+ to Ce4+ (equation 4). We assume that this auto-catalytic redox reaction of CeO2 NP induces highly efficient glucose oxidation process.
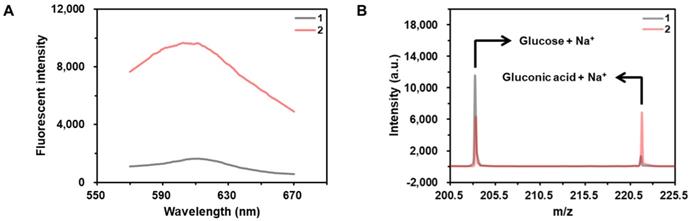
As shown in the results of Figure 2A, the fluorescent signal from hydroethidine, an O2•--specific fluorescent material, is highly enhanced in the presence of CeO2 NP under the acidic condition, demonstrating the production of O2•-. In addition, the results obtained by the matrix-assisted laser desorption/ionization time-of-flight (MALDI TOF) confirms that the glucose is converted to gluconic acid in the presence of CeO2 NP (Figure 2B).
We also investigated the kinetic parameters of CeO2 NPs by using the Michaelis-Menten model. As illustrated in Figure S1, Michaelis-Menten and Lineweaver-Burk plots were drawn by measuring the reaction velocities for the CeO2 NP-catalyzed glucose oxidation from the samples containing glucose at varying concentrations. The results show that the Michaelis-Menten constant (Km), maximum reaction velocity (Vmax), and catalytic constant (kcat) of CeO2 NP with 2 nm diameter were 5.93 mM, 0.59 μM/s, and 10.09 s-1, respectively, and these values are almost comparable to those of the glucose oxidase enzyme, demonstrating the high glucose oxidase-like activity of CeO2 NPs (Table 1) [34]. Considering that the CeO2 NPs are inorganic nanomaterials with higher stability than the natural enzymes, we believe that CeO2 NP could be better suited for the practical applications. Furthermore, we also examined the effect of particle size, pH, and temperature on the glucose oxidase-like activity of CeO2 NP by measuring the glucose level difference defined as L0 - L where L0 and L are glucose levels before and after CeO2 NP-catalyzed glucose oxidation reaction, respectively. As shown in Figure S2A, the glucose oxidase-like activity increased with decreasing size of CeO2 NP, which is consistent with the previous report that the smaller CeO2 NP shows the enhanced catalytic activity [35-37]. Moreover, the results in Figure S2B and S2C show that CeO2 NP exhibits the highest glucose oxidase-like activity under pH 4 and 20 °C.
Schematic illustration of PGM-based label-free read-out of PCR amplification using glucose oxidase-like activity of CeO2 NP.
Investigation of glucose oxidase-like activity of CeO2 NP. (A) Fluorescent signals from the samples containing (1) hydroethidine and (2) hydroethidine and CeO2 NP. The concentrations of hydroethidine and CeO2 NP were 0.1 wt% and 1 µg/mL, respectively. (B) MALDI TOF spectra of the samples containing (a) glucose and (b) glucose and CeO2 NP. Sodium adduct peaks of glucose and gluconic acid appeared at m/z equivalent to 203 and 221, respectively. The concentrations of glucose and CeO2 NP were 60 mM and 0.1 wt%, respectively.
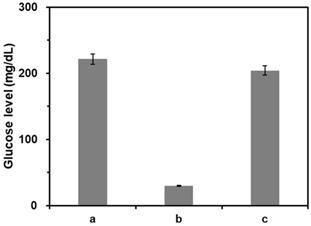
Feasibility of PGM-based label-free read-out of PCR amplification. The glucose levels were measured from reaction solutions containing (a) glucose only, (b) glucose and CeO2 NP, and (c) glucose, CeO2 NP, and purified DNA amplicon. The signal-to-noise ratios were calculated by dividing M by S where M and S are mean and standard deviation of the glucose levels based on triplicate measurements, which were (a) 28.5, (b) 41.7, and (c) 28.8. The concentrations of glucose, CeO2 NP, and purified DNA amplicon were 60 mM, 0.1 wt%, and 100 nM, respectively.
Comparison of Michaelis-Menten kinetic parameters of (a) CeO2 NP, (b) CeO2 NP/DNA amplicon complex, and (c) glucose oxidase. The concentrations of CeO2 NP, purified DNA amplicon, and glucose oxidase were 70, 100, and 70 nM
| Material | Km (mM) | Vmax (μM/s) | kcat (s-1) |
|---|---|---|---|
| a | 5.93 | 0.59 | 10.09 |
| b | 11.67 | 0.33 | 5.69 |
| c | 4.87 | 0.69 | 9.71 |
Detection feasibility
We first verified the feasibility of this strategy by measuring glucose levels from the samples at different reaction conditions. As shown in the results of Figure 3, the glucose level was significantly reduced by the presence of CeO2 NP due to its intrinsic glucose oxidase-like activity (b) compared to that in the absence of CeO2 NPs (a). On the other hand, when purified DNA amplicons was applied, the initial high glucose level was maintained (c). Notably, these results matched well with the Michaelis-Menten kinetic parameters. As shown in Table 1 and Figure S1, Vmax decreased while Km increased in the presence of purified DNA amplicon demonstrating the reduction of the efficiency for CeO2 NP-catalyzed glucose oxidation reaction, leading to the smaller catalytic constant (kcat) compared to that in the absence of DNA amplicon.
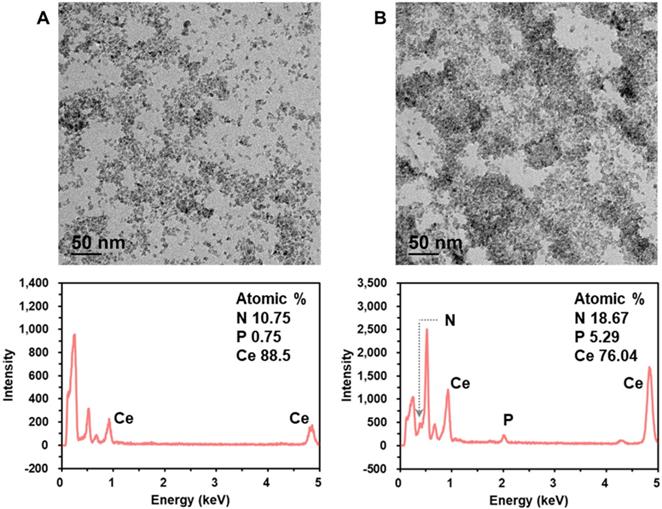
Next, we characterized the crystalline nature and surface state of the employed CeO2 NP by analyzing X-ray diffraction (XRD) and X-ray photoelectron spectroscopy (XPS) pattern. The XRD and XPS spectrum shown in Figure S3 are in good agreement with those in the previous reports [30,33]. In addition, we investigated the mechanism for DNA amplicon binding-induced aggregation of CeO2 NPs by using transmission electron microscopy (TEM), energy dispersive X-ray spectroscopy (EDS), zeta potential, dynamic light scattering (DLS), and ultraviolet-visible spectroscopy (UV-Vis) analyses. The TEM images and EDS spectra in Figure 4 clearly show that the CeO2 NPs were well dispersed in the absence of DNA amplicons, while the presence of purified DNA amplicons induced the aggregation of CeO2 NPs. In addition, the results of zeta potential (Figure S4A), DLS analysis (Figure S4B), EDS mapping (Figure S4C), and ultraviolet-visible spectroscopy (UV-Vis) analysis (Figure S5) supported our assumption that the surface of CeO2 NP shows positive charge which might results from the two oxidation states, Ce3+ and Ce4+, and thus the negatively charged DNA amplicons bind to the surface of CeO2 NPs, inducing the aggregation of CeO2 NPs [30-33].
Quantitative analysis of DNA amplicons
The reaction conditions were then optimized by examining the change of glucose level defined as L/L0 where L0 and L are glucose levels from the reaction solutions in the absence and presence of purified DNA amplicons, respectively. The results in Figure S6 show that the optimal concentrations of CeO2 NP and glucose were 0.1 wt% and 60 mM, respectively. In addition, the optimal times for DNA amplicon incubation and glucose oxidation were 2 and 1.5 min, respectively, which were employed for further experiments (Figure S7).
Under the optimized conditions, the capability of this strategy to quantitatively determine purified DNA amplicons was investigated by measuring glucose levels from the reaction solutions containing purified DNA amplicons at varying concentrations (Figure S8). The results show that the glucose level increased as the concentration of purified DNA amplicon increased in the range from 5 to 100 nM with excellent linear relationship (R2 = 0.9983). Importantly, it should be noted that the typical concentration range of DNA amplicon produced by PCR is from 10 to 100 nM, which is fully covered by the dynamic range of the proposed method [19,38]. In addition, it was confirmed that the glucose level increased as the concentration of the bound DNA amplicon increased, demonstrating the direct effect of the binding of DNA amplicon on the reduction of the efficiency for CeO2 NP-catalyzed glucose oxidation reaction.
Sensitivity and selectivity for target DNA detection
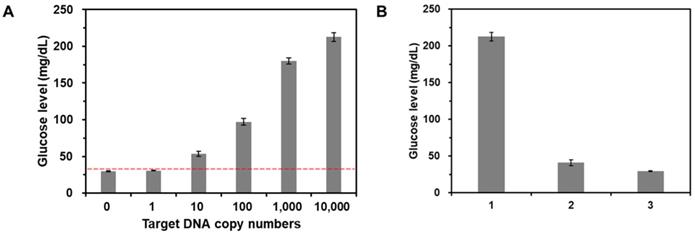
To examine the sensitivity of this method, target gDNA derived from E. coli with different copy numbers (0 - 104 copies) were amplified by PCR, which was then purified and incubated with CeO2 NPs. As shown in Figure 5A, the glucose level increased as the copy number of target gDNA increased. Importantly, the glucose level obtained from the reaction solution containing 10 copies of target DNA was clearly distinguished from the threshold line (red dotted line) defined as M + 3S where M and S are mean and standard deviation of glucose level obtained from the negative control without target gDNA, demonstrating high sensitivity of this strategy compared to those of previous methods (Table S2). Furthermore, in the same manner, we also successfully determined the target gDNA in the human serum down to 500 copies (Figure S9).
Furthermore, we also investigated the selectivity of this strategy by measuring the glucose levels from the reaction solutions containing non-target gDNA derived from Hepatitis B virus (HBV). The results in Figure 5B show that the glucose level in the presence of target gDNA (E. coli) (1) is distinctly higher than those in the presence of non-target gDNA (HBV) and from the negative control without target gDNA (c), verifying the high selectivity for target DNA detection.
Investigation of the mechanism for DNA amplicon binding-induced aggregation of CeO2 NP. TEM images and corresponding EDS spectra of CeO2 NP in the (A) absence and (B) presence of purified DNA amplicon. The concentrations of CeO2 NP and purified DNA amplicon were 0.1 wt% and 100 nM, respectively.
Sensitivity and selectivity for target DNA detection. (A) The glucose levels were measured from the reaction solutions containing target gDNA at varying amounts. The red dotted line is the threshold line defined as M + 3S where M and S are mean and standard deviation of glucose level obtained from the negative control without target DNA. The signal-to-noise ratios were calculated by dividing M by S where M and S are mean and standard deviation of the glucose levels based on triplicate measurements, which were 41.7, 53.1, 15.3, 21.2, 45, and 35.3 for the reaction solution with 0, 1, 10, 102, 103, and 104 copies target gDNA, respectively. The concentrations of glucose and CeO2 NP were 60 mM and 0.1 wt%, respectively. (B) The glucose levels were measured from the reaction solutions containing (1) target gDNA (E. coli), (2) non-target gDNA (HBV), and (3) no target DNA. The signal-to-noise ratios were calculated by dividing M by S where M and S are mean and standard deviation of the glucose levels based on triplicate measurements, which were (1) 35.3, (2) 10.1, and (3) 41.7. The concentrations of glucose, CeO2 NP, and target or non-target gDNA were 60 mM, 0.1 wt%, and 200 copies/µL, respectively.
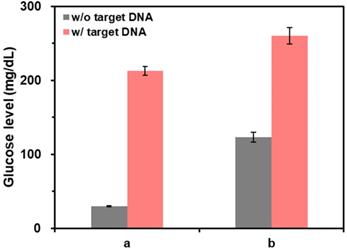
The effect of post-purification step on the detection of target DNA. The glucose levels were measured from samples containing (a) purified and (b) non-purified PCR solution. The signal-to-noise ratios were calculated by dividing M by S where M and S are mean and standard deviation of the glucose levels based on triplicate measurements, which were (a) 41.7 and 35.3 and (b) 18.1 and 23.5 for the reaction solution in the absence and presence of target DNA, respectively. The concentrations of glucose, CeO2 NP, and target DNA were 60 mM, 0.1 wt%, and 200 copies/µL, respectively.
Direct read-out of PCR amplification without post-purification steps
Finally, the capability of this method for the direct read-out of PCR amplification without post-purification of PCR solution was examined by measuring the glucose levels from the purified and non-purified PCR solutions. As shown in the results of Figure 6, when the PCR solutions were purified (a), the glucose level in the presence of target DNA was clearly distinguished from the one in the absence of target DNA. In contrast, when PCR solutions were not purified (b), the glucose level in the absence of target DNA was not decreased as effectively as the one in the case of purified reaction solutions. This result was attributed to the presence of DNA primers, dNTPs, and DNA polymerase in PCR solutions that can induce the aggregation of CeO2 NPs reducing their catalytic activity. However, the difference of glucose level in the absence and presence of target DNA was still high enough to determine the target DNA, demonstrating the capability of this method to directly examine PCR results without post-purification steps.
Conclusions
In this study, we developed a PGM-based label-free method for the on-site read-out of PCR amplification using glucose oxidase-like activity of CeO2 NPs that correlates the glucose level with the amount of DNA amplicon. With the proposed strategy, the target DNA was sensitively detected down to 10 copies with the high selectivity. In addition, after the completion of PCR, the result was rapidly examined in less than 5 min without any tedious and labor-intensive procedures. Importantly, this strategy employs the PGM as a detection component which would enable its facile application in the POC settings. It is noteworthy that this is the first report to discover the glucose oxidase-like activity of CeO2 NPs and utilize their activity for the label-free analysis of PCR amplification. Based on the advantageous features of rapidity, simplicity, cost-effectiveness, and portability, we expect that the devised system could serve as a core platform for the on-site read-out of PCR result.
Abbreviations
CeO2 NP: cerium oxide nanoparticle; DHB: 2,5-dihydroxybenzoic acid; DLS: dynamic light scattering; dNTP: deoxynucleoside triphosphate; EDS: energy-dispersive X-ray spectroscopy; gDNA: genomic DNA; MALDI TOF: matrix-assisted laser desorption/ionization time-of-flight; PCR: polymerase chain reaction; PGM: personal glucose meter; POC: point-of-care; TEM: transmission electron microscopy; UV-Vis: ultraviolet-visible spectroscopy; XRD: X-ray diffraction.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
This research was supported by BioNano Health-Guard Research Center funded by the Ministry of Science and ICT (MSIT) of Korea as Global Frontier Project (Grant number H-GUARD_2013M3A6B2078964). This research was also supported by the Mid-career Researcher Support Program of the National Research Foundation (NRF) funded by the MSIT of Korea (NRF-2018R1A2A1A05022355).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Carroll AE, Marrero DG, Downs SM. The HealthPia GlucoPack™ diabetes phone: a usability study. Diabetes Technol Ther. 2007;9:158-64
2. Xiang Y, Lu Y. Using commercially available personal glucose meters for portable quantification of DNA. Anal Chem. 2012;84:1975-80
3. Xiang Y, Lu Y. Portable and quantitative detection of protein biomarkers and small molecular toxins using antibodies and ubiquitous personal glucose meters. Anal Chem. 2012;84:4174-8
4. Ahn JK, Kim HY, Park KS, Park HG. A personal glucose meter for label-free and washing-free biomolecular detection. Anal Chem. 2018;90:11340-3
5. Wu T, Yang Y, Cao Y, Song Y, Xu LP, Zhang X. et al. Bioinspired DNA-inorganic hybrid nanoflowers combined with a personal glucose meter for onsite detection of miRNA. ACS Appl Mater Interfaces. 2018;10:42050-7
6. Fang J, Guo Y, Yang Y, Yu W, Tao Y, Dai T. et al. Portable and sensitive detection of DNA based on personal glucose meters and nanogold-functionalized PAMAM dendrimer. Sens Actuators B Chem. 2018;272:118-26
7. Yang X, Shi D, Zhu S, Wang B, Zhang X, Wang G. Portable aptasensor of aflatoxin B1 in bread based on a personal glucose meter and DNA walking machine. ACS Sens. 2018;3:1368-75
8. Taebi S, Keyhanfar M, Noorbakhsh A. A novel method for sensitive, low-cost and portable detection of hepatitis B surface antigen using a personal glucose meter. J Immunol Methods. 2018;458:26-32
9. Jia Y, Sun F, Na N, Ouyang J. Detection of p53 DNA using commercially available personal glucose meters based on rolling circle amplification coupled with nicking enzyme signal amplification. Anal Chim Acta. 2019;1060:64-70
10. Tang W, Yang J, Wang F, Wang J, Li Z. Thiocholine-triggered reaction in personal glucose meters for portable quantitative detection of organophosphorus pesticide. Anal Chim Acta. 2019;1060:97-102
11. Xu J, Qiao X, Zhang J, Cheng N, Sheng Q, Zheng J. et al. Point-of-care monitoring of intracellular glutathione and serum triglyceride levels using a versatile personal glucose meter. Anal Methods. 2019;11:1849-56
12. Shan Y, Zhang Y, Kang W, Wang B, Li J, Wu X. et al. Quantitative and selective DNA detection with portable personal glucose meter using loop-based DNA competitive hybridization strategy. Sens Actuators B Chem. 2019:197-203
13. Jang YJ, Lee KH, Yoo TH, Kim DM. Interfacing a personal glucose meter with cell-free protein synthesis for rapid analysis of amino acids. Anal Chem. 2019;91:2531-5
14. Zhu X, Sarwar M, Zhu JJ, Zhang C, Kaushik A, Li CZ. Using a glucose meter to quantitatively detect disease biomarkers through a universal nanozyme integrated lateral fluidic sensing platform. Biosens Bioelectron. 2019;126:690-6
15. Xiang Y, Lu Y. Using personal glucose meters and functional DNA sensors to quantify a variety of analytical targets. Nat Chem. 2011;3:697-703
16. Peng FF, Zhang Y, Gu N. Size-dependent peroxidase-like catalytic activity of Fe3O4 nanoparticles. Chin Chem Lett. 2008;19:730-3
17. Asati A, Santra S, Kaittanis C, Nath S, Perez JM. Oxidase-like activity of polymer-coated cerium oxide nanopartieles. Angew Chem Int Ed Engl. 2009;48:2308-12
18. Asati A, Kaittanis C, Santra S, Perez JM. pH-tunable oxidase-like activity of cerium oxide nanoparticles achieving sensitive fluorigenic detection of cancer biomarkers at neutral pH. Anal Chem. 2011;83:2547-53
19. Park KS, Kim MI, Cho DY, Park HG. Label-free colorimetric detection of nucleic acids based on target-induced shielding against the peroxidase-mimicking activity of magnetic nanoparticles. Small. 2011;7:1521-5
20. Yoshihisa Y, Zhao QL, Hassan MA, Wei ZL, Furuichi M, Miyamoto Y. et al. SOD/catalase mimetic platinum nanoparticles inhibit heat-induced apoptosis in human lymphoma U937 and HH cells. Free Radic Res. 2011;45:326-35
21. Chen W, Chen J, Feng YB, Hong L, Chen QY, Wu LF. et al. Peroxidase-like activity of water-soluble cupric oxide nanoparticles and its analytical application for detection of hydrogen peroxide and glucose. Analyst. 2012;137:1706-12
22. Wang S, Chen W, Liu AL, Hong L, Deng HH, Lin XH. Comparison of the peroxidase-like activity of unmodified, amino-modified, and citrate-capped gold nanoparticles. ChemPhysChem. 2012;13:1199-204
23. Wei H, Wang E. Nanomaterials with enzyme-like characteristics (nanozymes): next-generation artificial enzymes. Chem Soc Rev. 2013;42:6060-93
24. Kim MI, Park KS, Park HG. Ultrafast colorimetric detection of nucleic acids based on the inhibition of the oxidase activity of cerium oxide nanoparticles. Chem Commun (Camb). 2014;50:9577-80
25. He W, Wamer W, Xia Q, Yin JJ, Fu PP. Enzyme-like activity of nanomaterials. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev. 2014;32:186-211
26. Kim HY, Ahn JK, Kim MI, Park KS, Park HG. Rapid and label-free, electrochemical DNA detection utilizing the oxidase-mimicking activity of cerium oxide nanoparticles. Electrochem commun. 2019;99:5-10
27. Qin T, Ma R, Yin Y, Miao X, Chen S, Fan K. et al. Catalytic inactivation of influenza virus by iron oxide nanozyme. Theranostics. 2019;9:6920-35
28. Shi S, Wu S, Shen Y, Zhang S, Xiao Y, He X. et al. Iron oxide nanozyme suppresses intracellular Salmonella Enteritidis growth and alleviates infection in vivo. Theranostics. 2018;8:6149-62
29. Gao L, Fan K, Yan X. Iron oxide nanozyme: a multifunctional enzyme mimetic for biomedical applications. Theranostics. 2017;7:3207-27
30. Das M, Patil S, Bhargava N, Kang JF, Riedel LM, Seal S. et al. Auto-catalytic ceria nanoparticles offer neuroprotection to adult rat spinal cord neurons. Biomaterials. 2007;28:1918-25
31. Celardo I, Traversa E, Ghibelli L. Cerium oxide nanoparticles: a promise for applications in therapy. J Exp Ther Oncol. 2011;9:47-51
32. Cheng H, Lin S, Muhammad F, Lin YW, Wei H. Rationally modulate the oxidase-like activity of nanoceria for self-regulated bioassays. ACS Sens. 2016;1:1336-43
33. Vinothkumar G, Arunkumar P, Mahesh A, Dhayalan A, Suresh Babu K. Size- and defect-controlled anti-oxidant enzyme mimetic and radical scavenging properties of cerium oxide nanoparticles. New J Chem. 2018;42:18810-23
34. Luo W, Zhu C, Su S, Li D, He Y, Huang Q. et al. Self-catalyzed, self-limiting growth of glucose oxidase-mimicking gold nanoparticles. ACS Nano. 2010;4:7451-8
35. Conesa JC. Computer modeling of surfaces and defects on cerium dioxide. Surf Sci. 1995;339:337-52
36. Spanier JE, Robinson RD, Zhang F, Chan SW, Herman IP. Size-dependent properties of CeO2-y nanoparticles as studied by Raman scattering. Phys Rev B Condens Matter Mater Phys. 2001;64:2454071-8
37. Karakoti A, Singh S, Dowding JM, Seal S, Self WT. Redox-active radical scavenging nanomaterials. Chem Soc Rev. 2010;39:4422-32
38. Jung YK, Kim TW, Kim J, Kim JM, Park HG. Universal colorimetric detection of nucleic acids based on polydiacetylene (PDA) liposomes. Adv Funct Mater. 2008;18:701-8
Author contact
![]() Corresponding authors: E-mail: hgparkac.kr (H.G. Park); Phone: +82-42-350-3932; Fax: +82-42-350-3910. E-mail: kskonkukcom (K.S. Park); Phone: +82-2-350-3742; Fax: +82-2-350-3742.
Corresponding authors: E-mail: hgparkac.kr (H.G. Park); Phone: +82-42-350-3932; Fax: +82-42-350-3910. E-mail: kskonkukcom (K.S. Park); Phone: +82-2-350-3742; Fax: +82-2-350-3742.
 Global reach, higher impact
Global reach, higher impact