13.3
Impact Factor
Theranostics 2020; 10(7):3118-3137. doi:10.7150/thno.43298 This issue Cite
Review
Allele-specific genome targeting in the development of precision medicine
1. National Clinical Research Center for Geriatric Disorders, Department of Geriatrics, Xiangya Hospital, Central South University, Changsha, Hunan 410008, China.
2. Department of Rheumatology and Immunology, Xiangya Hospital, Central South University, Changsha, Hunan 410008, China
3. Department of Neurology, Xiangya Hospital, Central South University, Changsha, Hunan 410008, China
4. Key Laboratory of Hunan Province in Neurodegenerative Disorders, Central South University, Changsha, Hunan 410008, China
Received 2019-12-21; Accepted 2020-1-18; Published 2020-2-10
Abstract
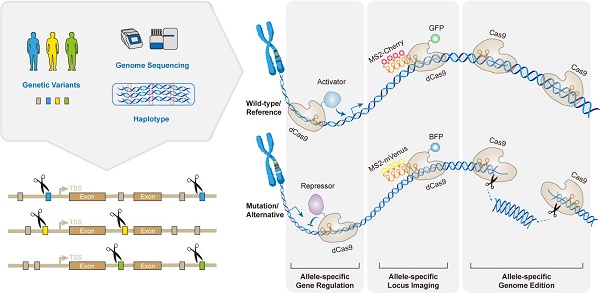
The CRISPR-based genome editing holds immense potential to fix disease-causing mutations, however, must also handle substantial natural genetic variations between individuals. Previous studies have shown that mismatches between the single guide RNA (sgRNA) and genomic DNA may negatively impact sgRNA efficiencies and lead to imprecise specificity prediction. Hence, the genetic variations bring about a great challenge for designing platinum sgRNAs in large human populations. However, they also provide a promising entry for designing allele-specific sgRNAs for the treatment of each individual. The CRISPR system is rather specific, with the potential ability to discriminate between similar alleles, even based on a single nucleotide difference. Genetic variants contribute to the discrimination capabilities, once they generate a novel protospacer adjacent motif (PAM) site or locate in the seed region near an available PAM. Therefore, it can be leveraged to establish allele-specific targeting in numerous dominant human disorders, by selectively ablating the deleterious alleles. So far, allele-specific CRISPR has been increasingly implemented not only in treating dominantly inherited diseases, but also in research areas such as genome imprinting, haploinsufficiency, spatiotemporal loci imaging and immunocompatible manipulations. In this review, we will describe the working principles of allele-specific genome manipulations by virtue of expanding engineering tools of CRISPR. And then we will review new advances in the versatile applications of allele-specific CRISPR targeting in treating human genetic diseases, as well as in a series of other interesting research areas. Lastly, we will discuss their potential therapeutic utilities and considerations in the era of precision medicine.
Keywords: genomic editing, CRISPR, genetic variants, SNP, allele-specific
Introduction
Inherited human diseases are caused by different types of gene mutations, insertions/deletions (indels), genomic structural variations, as well as pathogenic single nucleotide polymorphisms (SNPs) that are tightly associated with higher risks of diseases. Among those, dominant diseases present a great challenge for conducting gene therapies. Those patients inherited one pathogenic allele causing a disease phenotype, especially in a dominant-negative manner, and one normal allele as well. The treatment strategy thus typically involves the silence of the pathogenic alleles in an allele-specific manner, without affecting the wild-type ones.
During recent years, clustered regularly interspaced short palindromic repeats (CRISPR) has emerged to be a promising tool to treat human genetic diseases by disrupting deleterious gene sites. Basically, the CRISPR system is rather specific, with the potential to discriminate between similar alleles, even based on a single nucleotide difference. Therefore, it can be leveraged to establish allele-specific targeting of those heterogenous alleles.
So far, allele-specific CRISPR has been increasingly utilized not only in treating dominant negative diseases, but also in other multiple areas such as genome imprinting, haploinsufficiency, spatiotemporal loci imaging and immunocompatible manipulations. In this review, we will mainly focus on the versatile applications of allele-specific genome manipulations by virtue of the expanding toolbox of the CRISPR system, and further discuss their potential utilities and considerations in implementing precision medicine.
Genetic Diseases and Precision Medicine
A genetic disease is caused in whole or in part by an abnormality in the genetic makeup of an individual's genome. Those genetic abnormalities basically involve single-base mutations, indel mutations, duplication mutations, nucleotide repeat expansions and also chromosome structural abnormalities. In the human genome, the most frequent variations are SNPs and short indels [1-3]. The average human genome contains roughly 4-5 million SNPs that occur on average every 1000-2000 nucleotides [4], which presents at sufficient density for comprehensive haplotype analysis. Moreover, the recently released Exome Aggregation Consortium (ExAC) data set, with variants from 60,706 individuals, contains approximately one variant for every 8 nucleotides in the human exome [5].
Human genomes have now been sequenced at unprecedented rates with next-generation sequencing technologies, and the era of precision medicine is rapidly approaching. Over the last decade, the emerging next-generation sequencing has facilitated high-throughput genome-wide association studies (GAWS) that have identified numerous pathogenic SNPs [6, 7]. Notably, those pathogenic SNPs are by large kinds of gene mutations resided in both coding regions and regulatory regions, and also usually located at functional non-coding RNAs that participated in disease pathogenesis [8-11]. SNPs also underline differences in the susceptibility to a wide range of diseases and pharmacological treatment, that may help to guide the personalized drug usage [12-15]. Therefore, targeting variants like SNPs may provide a promising entry for silencing deleterious alleles in the era of precision medicine.
Genome Targeting and CRISPR
Although the concept of gene therapy emerged decades ago, it has been slow to achieve its full potential. This is partly due to concerns related to early clinical trials and the difficulty of safe and accurate targeting. However, with the advent of precise genome editing techniques such as CRISPR, we are on the verge of a gene therapy revolution. The CRISPR/Cas system was initially discovered as the adaptive immune system of the bacterial species by incorporating DNA of invaded phages or viruses into the host genome, which will be stored in an array in DNA [16-18]. And later this will be used to cleave the same invaded foreign DNA loci, based on complementarity to the encoded sequence. Over the past years, the CRISPR/Cas system has been extensively developed for targeting genes in mammalian cells [19, 20], and further transformed for curing genetic diseases and engineering desirable genetic traits.
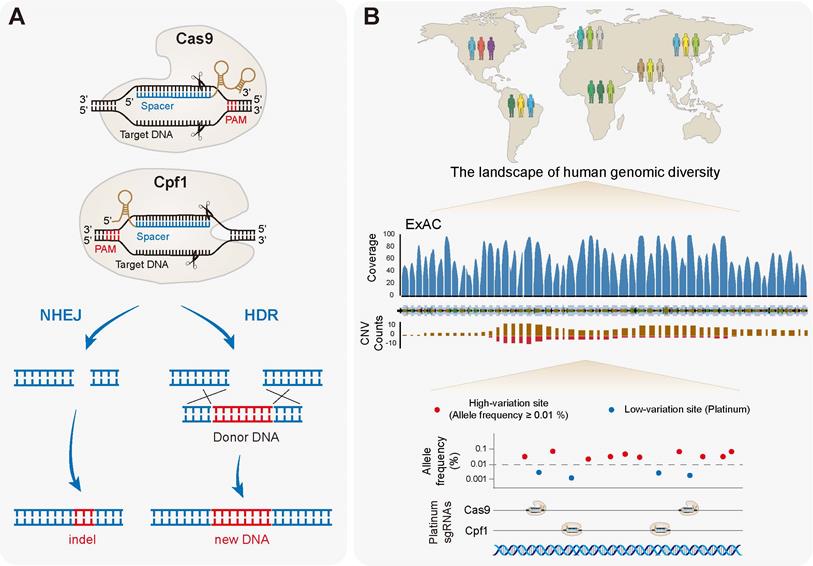
Basically, the CRISPR/Cas system consists of two components, Cas nucleases and single guide RNA (sgRNA), which form as a complex to bind and cleave double strand DNA (dsDNA) (Figure 1A). Cas binds to a DNA sequence complementary to the sgRNA spacer adjacent to a protospacer adjacent motif (PAM) site. Upon correct base pairing, Cas then activates its nuclease to cleave the dsDNA to form double-strand breaks (DSBs), which are later repaired either by non-homologous end joining (NHEJ) pathway or homology-directed repair (HDR) (Figure 1A). NHEJ results in random indels near the cleavage site that may cause frameshift mutations and premature stops. HDR typically occurs at lower and substantially more variable frequencies than NHEJ, but can be employed to introduce a specific DNA donor precisely at the target site.
Different types of Cas proteins have been discovered with distinct PAM recognition sites. So far, four major types of Cas nucleases including Cas9, Cpf1 (Cas12a), Cas12b (C2c1) and CasX (Cas12e) have been demonstrated to possess DNA targeting activities (Table 1), and available Cas nucleases from other bacterial species have been increasingly characterized [21-32]. Types of Cas nucleases possess different protein sizes, unique PAM constraints, cleavage patterns, as well as different lengths of seed regions that may determine targeting specificities (Table 1). Currently, the first- and best-characterized CRISPR system is that of Cas9 from Streptococcus pyogenes (SpCas9) [33, 34]. It requires at least one G in their PAMs, but is quite a large protein containing more than 1300 amino acids, which greatly hinders its usage in the package into adeno-associated virus (AAV) vectors for gene therapy (Table 1). The Cas9 from Staphylococcus aureus (SaCas9) [35] was then characterized with the advantage of its smaller size (with ~1000 amino acids), which is suitable for the AAV package. Similarly, other compact Cas9 orthologs, such as NmCas9 (1082 amino acids) [23] and CjCas9 (984 amino acids) [29], can be packaged in all-in-one AAV vectors for in vivo editings. Later discovered Cas nucleases such as Cpf1 [36-41], Cas12b [42, 43] and CasX [44] are with T-rich PAMs, and thus have broadened the range of genome editings, and are particularly useful in targeting AT-rich genomes or regions. Notably, the recently characterized Cas12b [42, 43] and CasX [44, 45] show to be with quite higher specificities, and also with smaller sizes. To further broaden the available targeting capabilities, Cas nucleases have been continuously engineered as multiple variants for either improved specificity (such as SpCas9-HF [46], eSpCas9 [47], HypaCas9 [48], SaCas9-HF [49] and enAsCpf1-HF1 [50]), or expanded available PAM sites [46, 50-58] (Table 1).
The CRISPR toolbox for genome DNA engineering
| Cas Nucleases | PAM Sequence | PAM Location | Size (aa) | TracrRNA Requirement | Cutting Manner | References | Notes |
|---|---|---|---|---|---|---|---|
| Type II System: Cas9 & Variants | |||||||
| SpCas9 | NGG | 3' end | 1368 | Yes | Blunt | [19, 20, 33, 34] | SpCas9 can also cleave sgRNA target sites followed by 'NAG', however with efficiency reduced to ∼20% |
| SpCas9-VRER | NGCG | 1368 | [51] | Has a stringent selectivity for an NGCG PAM sequence | |||
| SpCas9-EQR | NGAG | 1368 | [51] | Specific for an NGAG PAM | |||
| SpCas9-VQR | NGA | 1368 | [51] | Strongly recognizes sequences bearing the NGAN PAM | |||
| SpCas9-VRQR | NGA | 1368 | [46] | Has improved activities relative to the VQR variant on sites with NGAH (H = A, C, or T) PAMs | |||
| FnCas9 | NGG | 1629 | [52] | One of the largest Cas9 orthologs; exhibits slight activities toward those with the TGA and TAG PAMs | |||
| FnCas9-RHA | YG | 1629 | [52] | An engineered FnCas9 variant | |||
| NmCas9 | NNNNGHTT | 1082 | [21-25] | Preferred consensus PAM (5'-NNNNGATT-3') for NmeCas9 genome editing in human cells; Suitable for AAV package | |||
| Nme2Cas9 | NNNNCC | 1082 | [26] | Suitable for AAV package | |||
| GeoCas9 | NNNNCRAA NNNNGMAA | 1087 | [27] | A thermostable Cas9 | |||
| St1Cas9 | NNAGAAW | 1121 | [22, 28, 51] | ||||
| St3Cas9 | NGGNG | 1409 | [28] | ||||
| TdCas9 | NAAAAC | 1423 | [22] | ||||
| CjCas9 | NNNNACAC NNNNRYAC | 984 | [29-31] | Suitable for AAV package | |||
| ScCas9 | NNG | 1375 | [32] | ||||
| xCas9 | NG, GAA and GAT | 1368 | [53] | Used phage-assisted continuous evolution; an expanded PAM SpCas9 variant that can recognize a broad range of PAM sequences | |||
| SpCas9-NG | NG | 1368 | [54] | ||||
| SaCas9 | NNGRRT | 1053 | [35] | Suitable for AAV package | |||
| SaCas9-KKH | NNNRRT | 1053 | [55] | An engineered SaCas9 variant | |||
| cSaCas9 | Multiple PAMs | 1053 | [56] | Identified several chimeric SaCas9 variants with expanded recognition capability at NNVRRN, NNVACT, NNVATG, NNVATT, NNVGCT, NNVGTG, and NNVGTT PAM sequences | |||
| Type V-A System: Cpf1 (Cas12a) & Variants | |||||||
| AsCpf1 | TTTV | 5' end | 1307 | No | Staggered | [36-38] | Has a lower activity at a TTTT PAM. CTTA also led to high indel frequencies for both AsCpf1 and LbCpf1, which may be considered as a secondary PAM, especially for LbCpf1 [38] |
| LbCpf1 | TTTV | 1228 | [36, 38] | Has a lower activity at TTTT PAM | |||
| FnCpf1 | TTV | 1300 | [36, 39] | May manifest different activities depending on the organisms; KYTV reported in [39] | |||
| Mb3Cpf1 | TTV | 1261 | [40] | Exhibits comparable activity to AsCpf1 and LbCpf1 with TTTV PAMs; Can recognize a TTV PAM, but with lower efficiency | |||
| AsCpf1-RR | TYCV | 1307 | [57, 58] | Also cleaves ACCC and CCCC PAMs (and, to a lesser extent, VYCV) | |||
| AsCpf1-RVR | TATV | 1307 | [57, 58] | Also cleaves RATR PAMs | |||
| enAsCpf1 | TTYN VTTV TRTV | 1307 | [50] | ||||
| ArCpf1 BsCpf1 PrCpf1 | TTN | 1262 1206 1213 | [41] | ||||
| HkCpf1 | YTN TYYN | 1310 | [41] | ||||
| Type V-B System: Cas12b (C2c1) & Variants | |||||||
| AaCas12b | TTN | 5' end | 1129 | Yes | Staggered | [42] | |
| AkCas12b | TTTN | 1147 | [43] | ||||
| BhCas12b v4 | ATTN | 1108 | [43] | Also works at a subset of TTTN and GTTN PAMs, albeit with less robust activities | |||
| Type V-E System: CasX (Cas12e) & Variants | |||||||
| DpbCasX | TTCN | 5' end | 986 | Yes | Staggered | [44, 45] | |
| PlmCasX | TTCN | 978 | [44, 45] | ||||
The CRISPR/Cas system and identification of platinum targets. (A) The basic working principle of Cas9 (Class 2 type II) and Cpf1 (Class 2 type V). An sgRNA/crRNA (brown) encoding a spacer (blue) is bound to a target double strand DNA (black) proximal to a PAM site (red). Perfect base-pairing then activates the nuclease activity, cleaving both DNA strands (scissors) to form DSBs, which are later repaired either by the NHEJ pathway or the HDR pathway. (B) The CRISPR-based therapeutic genome editing must also handle with substantial natural genetic variations between individuals. Genetic variants may a great impact on sgRNA efficiencies, as well as both on- and off-target specificities. For the CRISPR-based therapy in large patient populations, genetic variations should be carefully taken into account in designing and evaluating sgRNAs. The solution would be identifying universal sgRNAs located in the low-variation regions. By searching the ExAC browser, for example, people can figure out exome-wide target sites, which lack variants occurring at allele frequencies of ≥ 0.01% (platinum targets). Selection of platinum sgRNAs should maximize the population efficacy of genome editing. The human LRRK2 gene was viewed as an example.
In the practice of genome engineering, the CRISPR system has been versatile in multiple areas. Initially it was used to disrupt gene expression by introducing DSBs, no matter with single sgRNAs at coding regions, or dual flanking sgRNAs at coding or non-coding regions. Later on, it was used to delete a large DNA fragment [59, 60], or even one deleterious chromosome by multiple sgRNAs that may be useful for treating diseases like Turner syndrome and Down syndrome [61].
As part of expanded applications, the Cas nuclease was also engineered to be catalytically inactive (dead Cas; dCas) that silenced its cleavage activity without compromising DNA binding activity [36, 62]. Due to this unique property, dCas could then be serving as programmable nucleic acid binding scaffolds for the recruitment of a variety of effector proteins/domains that may facilitate chromatin imaging [63, 64], purification of specific genomic loci [65, 66] and proximity labeling [67, 68], epigenetically gene activation [69-72] and gene repression [73, 74], or more excitingly, the base editing [75, 76]. Since the expanded engineered Cas variants or expanded tool of the CRISPR system, as well as their respective applications have been recently comprehensively reviewed elsewhere (please refer to [77-84]), we are not going to describe them in much more details in this review.
Genetic Variations and CRISPR-based Therapeutics
The CRISPR-based genome editing holds immense potential to fix disease-causing mutations, however, must also handle with substantial natural genetic variations between individuals. The designed sgRNA based on the reference genome may introduce mismatches between the guide RNA and the genomic DNA from each individual, which will negatively impact sgRNA efficiencies and lead to imprecise specificity prediction [85]. Further studies have conducted a more comprehensive analysis of the datasets of ExAC and 1000 Genomes Project (1000GP), and showed that genetic variations may significantly affect the sgRNA efficiencies, as well as both on- and off-target specificities at therapeutic sites [86, 87]. Therefore, genetic variants should be carefully assessed in the design and evaluation of sgRNAs for CRISPR-based treatments with large patient populations, otherwise, it may confound clinical trials and also lead to adverse consequences. The key would be identifying universal sgRNAs located in the low-variation regions [86]. For instance, by searching the ExAC browser [88], or the recent Genome Aggregation Database (gnomAD) browser [89], people can figure out exome-wide target sites, which lack variants occurring at allele frequencies of ≥ 0.01% (called "platinum" targets) (Figure 1B). Selection of platinum targets should maximize the population efficacy, and even facilitate the therapeutics for super populations of patients with specific variants, based on the demographic information provided by 1000GP [86].
Although the genetic variations would be a challenge for designing platinum sgRNAs in large populations, they may provide a promising entry for designing allele-specific sgRNAs for the treatment of each individual, taking advantage of heterozygous variants. Therefore, the exploitation of genetic variations brings about a concept of “seeking common ground while reserving differences” that not only helps to select the universal platinum targets for large populations, but also facilitates the design of unique targets in individuals for precision medicine.
Allele-specific Genome Targeting
The notion of “allele-specific intervention” has been originally raised for treating dominant diseases [90]. Those patients inherited a pathogenic allele and a normal allele on the chromosome pairs. As caused by harmful gain-of-function mutations, gene augmentation is unlikely to effectively alleviate the disease phenotype. The treatment strategy thus typically includes an allele-specific intervention by silencing or ablation of pathogenic alleles without any abnormal effect on wild-type alleles, especially in cases where wild-type allele expression is pivotal for cellular survival. Compared to the conventional editing of both alleles, the sustained expression of healthy alleles will minimize the potential negative effects on human physiology, prevent disease progression, and may even allow for the recovery of normal phenotypes.
It has to be noted that the allele-specific intervention has been widely achieved in the last decade by using either short-interfering RNAs (siRNAs) [91-101] or antisense oligonucleotides (ASOs) [102-106]. This potential of allele-specific siRNAs depends on its highly sequence-specific knockdown manner. The imperfect complementarity of siRNAs with mRNAs gives rise to the discrimination ability of suppressing either allele [107, 108]. Numerous studies have been successfully conducted to assess the potency and specificity of allele-specific gene suppression for various disorders including skin disorders [97, 98], multiple neurodegenerative diseases [99-105] and retinitis pigmentosa [106], among others, and produced immense therapeutic benefits.
In recent years, allele-specific CRISPR genome editing has emerged to be a potential approach to treat those dominant diseases, although the application of allele-specific Transcription Activator-Like Effector Nucleases (TALEN) has also been reported [109]. Similarly, the CRISPR system provides a highly specific genome editing capable of distinguishing pathogenic alleles from wild-type ones, partly based on the imperfect complementarity of sgRNAs with wild-type sequences. However, the differences, or particularly the advantages, of allele-specific CRISPR over siRNAs would be (i) the possibilities of novel PAMs derived from genetic variants, and (ii) the capabilities of targeting any gene locus of interest rather than merely transcribed mRNAs.
Based on the properties of discriminating sgRNAs, allele-specific CRISPR is intended to be used for targeting multiple types of genetic variants, including (i) Single-base mutations and SNPs; (ii) Short indels and nucleotides gaps; (iii) Reversed fragments; (iv) Virus integration (or other element integration); and (v) Chromosome-specific genomic indels.
Working Models of Allele-specific CRISPR
Allele-specific CRISPR works largely depending on the structures of discriminating sgRNAs, which contain several critical elements such as PAM sites and spacer sequences that provide the entry points for Cas binding. Genetic variants may present on both PAM and spacers, manipulation of which can be exploited for allele-specific genome targeting. Accordingly, there are basically two types of working models:
(1) The “In PAM” model. Gene mutations or SNPs that can create novel PAM sites at only one allele (Figure 2A). And vice versa, they may also eliminate PAM sites at either allele that renders discrimination ability. This model confers the most stringent allele-specific cleavages, since the binding of Cas with target DNA may not even happen without matching PAMs. Exploiting the various PAM sites recognized by different Cas variants produces alterative choices for the allele-specific strategy. For example, SpCas9 recognize a classical 5'-NGG-3' PAM, in which GG would allow for allele specificity, whereas SpCas9-VRER, one of its variants, has a stringent selectivity for a 5'-NGCG-3' PAM site. Interestingly, Scott et al. catalogued variants present among all possible targets in human reference exome using ExAC datasets, and observed that the total ratios of targets containing “In PAM” variants was similar (roughly 21-35%) for SpCas9, SpCas9-VQR, SaCas9 and AsCpf1, whereas 80% of targets were affected by “In PAM” variants for SpCas9-VRER [86]. This is due to the fact that the PAM for SpCas9-VRER contains a CpG motif that has been shown to be highly mutable in human exomes [5]. Thus, Cas enzymes using PAMs containing CG motifs might be considerably more flexible for allele-specific targeting.
The proposed model for allele-specific genome targeting by CRISPR. The “In PAM” model (A) and “Near PAM” model (B) for allele-specific genomic targeting. The “In PAM” model confers the most stringent allele-specific cleavages, since the binding of Cas with its target DNA is diminished. The “Near PAM” model exploits the discrimination ability of sgRNAs, with mutations or SNPs that locate within the spacer region, particularly the seed region. The variant is denoted by a red asterisk.
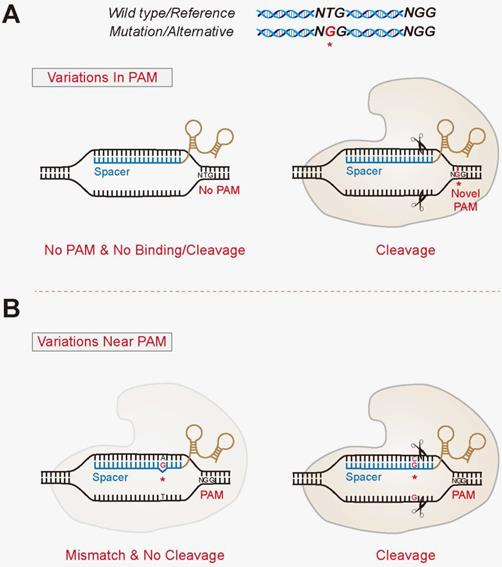
(2) The “Near PAM” model. Many pathogenic alleles, however, do not contain mutations that generate novel PAM sites, making them refractory to the “In PAM” model. One potential approach of expanding the applicability is to develop strategies in which the gene mutations or SNPs locate within the spacer region, particularly the seed region, of the discriminating sgRNAs (Figure 2B). Importantly, the seed sequence confers a rather stringent specificity of CRISPR. Studies have demonstrated that base pair mismatches between sgRNAs and their target sequences could significantly reduce or even abolish the efficacy of CRISPR-mediated editing [33, 34]. Cong et al. have previously reported that even single-base mismatches in the PAM-proximal seed region can eliminate genome cleavage of SpCas9 [19]. Also as previously demonstrated by Smith et al. in human induced pluripotent stem cells (iPSCs) and Yoshimi et al. in animal models, merely changing one single nucleotide within sgRNA spacers renders it unable to bind and cleave the target DNA [110, 111]. Similarly, in plants, Zhou et al. examined a total of 717 sequences harboring one SNP in each allele near/within the PAM, both of which completely abolished the editing events by the discriminating sgRNAs [112]. This feature could thus be exploited to specifically target disease- causing single mutations or SNPs to disable the mutant alleles, whilst insulating against genome modification in the wild-type counterparts.
Different types of Cas nucleases are basically endowed with different lengths of seed regions, which must be taken into account when practicing allele-specific targeting. For instance, the targeting specificity of SpCas9 is largely determined by the 10-12 nucleotides of proximal seeds of 3' PAMs [33]; whereas Cpf1 nucleases possess much shorter seeds with 5-6 nucleotides downstream of 5' PAMs [36, 38]. Interestingly, another two nucleases, Cas12b and CasX, appear to possess with much longer seed sequences and thus render higher targeting specificities [42-44].
Notably, a comprehensive analysis was recently performed on the genome-wide variants from more than 2500 individuals from the 1000GP and exome variants from more than 60,000 individuals in the ExAC [113]. The results showed that most variants reside in or near a PAM site that could affect sgRNA sites recognized by at least one of 11 chosen Cas types [113], which pretty well underpins the practicability of allele-specific targeting.
Major applications of allele-specific genome editing by CRISPR. (A) Apart from allele-specific gene knockout by wild-type Cas through its DNA cleavage activity, catalytically inactive dCas9 fused with various effector proteins/domains has been repurposed to achieve allele-specific loci imaging, gene activation, and gene repression. (B) Common SNPs representing a given haplotype can all be used for allele-specific targeting in the era of precision medicine. This strategy uses pairs of custom-designed haplotype-specific sgRNAs, which may permanently ablate any gain-of-function mutations in the human genome. (C) Scheme of the concept of correcting imprinting diseases by allele-specific epigenome editing. Normally, paternal and maternal alleles show different DNA methylation patterns (indicated by open and filled circles) at imprinted loci, however, they are lost in imprinting disorders. This epimutation could be corrected in an allele-specific manner, discriminating by the polymorphisms (denoted by red asterisks). The paternal allele showed here is selectively bound by the dCas9 fused with DNA methyltransferase DNMT that may restore the DNA methylation of this allele. (D) Allele-specific CRISPR manipulates the immunocompatibility by selectively targeting HLA haplotype. This strategy could greatly increase donor compatibility for regenerative medicine, and is anticipated to serve more pseudo-homozygous cells donors including iPSCs.
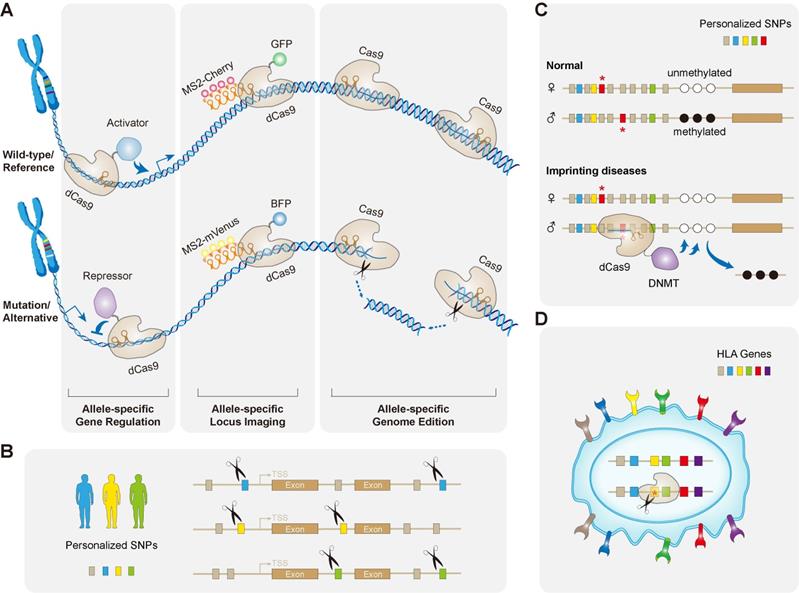
Applications of Allele-specific CRISPR
As it was mentioned earlier, allele-specific CRISPR works not only on gene mutations, but also on common or rare SNPs, exploiting of which contributes to its versatile applications (Figure 3). For example, by applying an allele-specific strategy, disease loci with mutations could be successfully modified to create isogenic iPSC lines derived from patients with frontotemporal dementia (FTD), for better disease modeling and cell therapy [114]. Similarly, allele-specific targeting was used to create animal models in several studies [115, 116]. In the zebrafish model, the polymorphisms were utilized to create deletions on specific chromosomes, and enhance the efficiency of a chromosome-specific loxP site in cis [116]. This allele-specific strategy may hence improve the utility of the zebrafish model for genetic studies.
Interestingly, a recent study developed an allele-specific CRISPR live-cell DNA imaging technique (termed SNP-CLING) that is able to resolve allelic positioning relative to nuclear sub-compartments and allele-specific interactions between non-homologous chromosomes [117]. To visualize each allele for a given locus simultaneously in living cells, they leveraged dCas9 with two to three sgRNAs by targeting possible SNPs genome-wide in PAM motifs. Briefly, they appended sgRNAs with RNA-aptamer motifs (such as MS2 and PP7) and co-transfected with their corresponding RNA-binding proteins fused to a fluorescent protein (such as mVenus and mCherry), for allele-specific labeling [117]. They particularly applied SNP-CLING to two long noncoding RNA (lncRNA) loci, Firre and CISTR-ACT, which are associated with heterozygous structural aberrations on higher-order nuclear architecture that eventually cause Mendelian diseases [117]. The technique of SNP-CLING thus overcomes the limitations of previous imaging and chromatin capture techniques in resolving allele-specific spatiotemporal properties of genomic loci in living cells. Importantly, using heterozygous SNPs in haplotypes of human pedigrees may render this imaging technique to be widely applicable to the study of gene locations at the allele-specific resolution (Figure 3A).
In the following part, we will mainly focus on the major applications of allele-specific CRISPR in the field of gene therapies (Table 2).
Summary of studies on disease treatment by allele-specific CRISPR
| Targeted Genes | Variants Types | Variants Locations | Cas Nucleases | PAM | Disease Types | Targeting Specificity | Functional Outcomes | References |
|---|---|---|---|---|---|---|---|---|
| RHO | P23H | -3 nt PAM | SaCas9 | NNGRRT | Retinitis pigmentosa (RP) | Indel formation was detected in the mutant His allele only | Delivered to both patient iPSCs in vitro and pig retina in vivo, and created a frameshift or premature stop that would prevent P23H transcription | [122] |
| RHO | P23H | -4 nt PAM | SaCas9-KKH | NNNRRT | Retinitis pigmentosa (RP) | No detectable cleavage was found either at WT or P23H allele | [123] | |
| RHO | P23H | -4 nt PAM | SpCas9-VQR | NGA | Retinitis pigmentosa (RP) | Presented a high rate of cleavage in the P23H but not WT allele | Slowed photoreceptor degeneration and improved retinal functions | [123] |
| RHO | P23H | -12 nt PAM | SaCas9-KKH | NNNRRT | Retinitis pigmentosa (RP) | (i) Unable to distinguish the mutant P23H allele from the wild-type one; (ii) Robust cutting efficiencies of 37.8% were observed in the injected WT mice, even though SaCas9-KHH preferentially targeted the mutant allele | [124] | |
| RHO | P23H | -4 nt PAM | SpCas9-VQR | NGA | Retinitis pigmentosa (RP) | (i) Unable to distinguish the mutant P23H allele from the wild-type one; (ii) Robust cutting efficiencies of 40% were observed in the injected WT mice, even though SpCas9-VQR preferentially targeted the mutant allele; (iii) Truncated sgRNA (17 nt) improved allele discrimination with a cleavage efficiency of 28%, and no detectable cleavage in the WT controls | [124] | |
| RHO | P23H | -4 nt PAM | SpCas9- VRQR | NGA | Retinitis pigmentosa (RP) | Truncated sgRNA (17 nt) paired with SpCas9-VRQR cleaved the P23H allele with greater efficiency (~ 2 fold) compared that with SpCas9-VQR, but also brought about an increase in targeting of the WT allele from 0% to 1.3 ± 0.3% | (i) Significantly delayed progression of photoreceptor cell degeneration in the outer nuclear layer; (ii) The low-level disruption of the WT allele did not abrogate the observed therapeutic benefit | [124] |
| RHO | S334ter | Novel PAM | SpCas9 | NGG | Retinitis pigmentosa (RP) | No cleavage was detected at the RHO WT allele | Prevented retinal degeneration and improved visual function in rat model | [125] |
| KRAS | G13A | Novel PAM | SpCas9 | NGG | Colorectal cancer | Completely silenced the mutant allele; No aberrant effects on the WT allele | Reversal of drug resistance to the MEK inhibitor | [126] |
| EGFR | L858R | Novel PAM | SpCas9 | NGG | Non-small cell lung cancer (NSCLC) | Small indels were detected in the EGFR mutant allele with a frequency of 3.6% (± 0.1%) at 2 days post-transfection; no mutations were detectably induced in the WT allele | Enhanced cancer cell killing and inhibition of tumor growth | [127] |
| BRAF | V600E | +1 nt PAM | As/LbCpf1 | TTTN | Melanoma | The efficiency of AsCpf1 was very weak, and no activity was detected using LbCpf1 | [128] | |
| BRAF | V600E | +13 nt PAM | As/LbCpf1 | TTTN | Melanoma | Both AsCpf1 and LbCpf1 show robust cleavage activities only in mutant sequence | [128] | |
| BRAF | V600E | -11 nt PAM | SpCas9 | NGG | Melanoma | SpCas9 cut the mutant allele ~ 4-fold more efficiently than the WT allele | [128] | |
| BRAF | V600E | Novel PAM | SpCas9-EQR | NGAG | Melanoma | No cleavage events were unexpectedly observed by Cas9-EQR for both WT and mutant allele | [128] | |
| TMC1 | M412K | -6 nt PAM | SpCas9 | NGG | Hearing loss | Modified the mutant TMC1 allele 23-fold more efficiently than the WT allele. Edited the WT TMC1 locus much less efficiently (0.066-1.6% indels). A truncated sgRNA decreased indel % on the mutant allele and further dampened its discrimination ability | Reduced progressive hearing loss and improved acoustic startle response | [130] |
| TMC1 | M412K | Novel PAM | SaCas9-KKH | NNNRRT | Hearing loss | Indel formation only in the Tmc1 allele; very little (0.0075%) in the WT one | Injected mice exhibit normal or near-normal thresholds of auditory brainstem responses | [131] |
| APP | KM670/671NL (APPswe) | -1/-2 nt PAM | SpCas9 | NGG | Early-Onset Alzheimer's Disease (EOAD) | (i) CRISPR-induced indels were only detected in APPSW alleles but not in APPWT alleles after deep sequencing detection; (ii) A truncated sgRNA (17 nt) was inefficient in disrupting the APPSW allele | Decreased the secretion of Aβ40 and Aβ42 | [132] |
| TGFBI | L527R | Novel PAM | SpCas9 | NGG | Corneal dystrophy | (i) Only resulted in cleavage of the mutant reporter; the WT reporter remained intact; (ii) sgRNA truncation did not improve specificity | [133] | |
| TGFBI | R555W | Novel PAM | SaCas9 | NNGRRT | Corneal dystrophy | Unable to distinguish between WT (NNGRRC) and mutant TGFBI (NNGRRT) sequence, due to the comparable efficiencies of recognizing NNGRRT/V | [133] | |
| TGFBI | R124L | Novel PAM | AsCpf1 | VYCV | Corneal dystrophy | Can distinguish between WT and mutant TGFBI sequence, but with a low efficiency | [133] | |
| TGFBI | R124C, R124H, R124L, R555Q, R555W | Differ by a single base pair in the spacer | SpCas9 | NGG | Corneal dystrophy | (i) Cut WT alleles with varying efficiencies; (ii) Truncated sgRNAs did not provide marked improvements of specificity, for most cases, maximal discrimination occurred with 20 or 19 nt guides; (iii) The additional G at the 5' end of the guide sequence did not provide an improved specificity in any case | [133] | |
| CRYGC | Leu160Stop | Novel PAM | SpCas9 | NGG | Nuclear cataracts | No gene editing events in the WT allele | The targeted mutant allele repaired by HDR | [134] |
| COL7A1 | c.8068_8084delinsGA | Short Indels: Different base pairs since -5 nt PAM | SpCas9 | NGG | Dystrophic epidermolysis bullosa (DDEB) | Specifically targeted only the mutant sequence of COL7A1 | Edited COL7 degraded at the protein level and could not undergo collagen triple helix formation | [135] |
| CALM2 | N98S | -1 nt PAM | SpCas9n | NGG | Early-onset long-QT syndrome (LQTS) | Obtained 7 of 18 clones with mutant allele-specific genome modification, and another 7 clones with both WT and mutant allele targeted. Hard to tell the specificity, due to the Cas9 double nickase system containing both upstream WT sgRNA and downstream mutant-specific sgRNA | Rescued the abnormal electrophysiological properties of iPSC derived cardiomyocytes | [136] |
| KRT12 | L132P | Novel PAM | SpCas9 | NGG | Meesmann's epithelial corneal dystrophy (MECD) | No effects on the WT allele | Achieved both in vitro and in vivo KRT12 mutation-specific targeting | [137] |
| DNM2 | R465W | -1 nt PAM | SpCas9 | NGG | Centronuclear myopathies (CNMs) | Used 18 bp truncated gRNAs. Analysis of single-cell clones showed 60% NHEJ in diseased fibroblasts. All of the NHEJ events detected occurred exclusively on the mutated allele | Targeting the mutated allele ameliorated disease-related phenotypes including the alterations in endocytosis and transferrin uptake, as well as autophagy defects | [138] |
| HTT | mHTT (CAG180, CAG69) | Novel PAM (derived from the promoter and intron 3) | SpCas9 | NGG | Huntington's disease (HD) | The combined usage of two allele-specific gRNAs selectively excised ~44 kb DNA spanning promoter region, transcription start site, and CAG expansion mutation of HTT, resulting in complete inactivation of the mutant allele without affecting the normal one | Completely prevented the generation of mHTT mRNAs and proteins | [59] |
| HTT | mHTT (CAG57, CAG97) | Novel PAM (derived from the promoter) | SpCas9 | NGG | Huntington's disease (HD) | Targeted only on the PAM-containing mutant allele | Reduced mHTT expressions in both primary fibroblast and BacHD transgenic mice | [60] |
Allele-specific CRISPR and Dominant Disorders
For dominant genetic diseases, only one allele harboring a mutation can cause disease phenotypes. Thus, the specific silencing of the disease-causing allele would be therapeutically desirable. Basically there are three scenarios that may provoke autosomal-dominant phenotypes: (i) Haploinsufficiency: a situation in which individuals whose mutations are heterozygous at a particular locus, are clinically affected because a single copy of the normal allele does not provide sufficient protein production to maintain normal functions [5, 118]; (ii) Dominant negative effect: a nonfunctional mutant whose expression adversely affects the function of the normal gene [119]; (iii) Gain-of-function mutation: a type of mutation that alters the gene product to acquire new abnormal functions that eventually lead to the phenotype [120]. As you may also read with details described below, allele-specific CRISPR can actually work well for each scenario to ameliorate the disease phenotypes: (i) by treating haploinsufficiency with selectively activation of wild-type allele expression; or (ii) by effectively disrupting the deleterious alleles with dominant negative effects or with gain-of-abnormal-functions.
So far, retinitis pigmentosa, a group of related eye disorders that cause progressive vision loss, was most treated by allele-specific targeting (Table 2). Mutations in the Rhodopsin (RHO) gene are the most common cause of autosomal dominant retinitis pigmentosa (adRP), accounting for 30 to 40 percent of all cases [121]. Several pieces of studies have converged to focus on the RHO mutations both in vitro and in vivo [122-125]. Burnight et al. first utilized SaCas9 and delivered discriminating sgRNAs (“Near PAM”) to iPSCs derived from a patient with the RHO P23H genotype, and observed that modifications only occurred in the mutant allele among the 86 clones sequenced, causing a frameshift and premature stop that would prevent transcription [122]. Later, they also packaged the sgRNA-SaCas9 cassette into AAV5, which was injected sub-retinally into a transgenic pig model that carries the human P23H mutation. Similarly, no indels were found in the wild-type counterpart [122]. Other studies led by Giannelli et al. and Li et al. alternatively chose the engineered variant SaCas9-KKH, SpCas9-VQR and SpCas9-VRQR for treating retinitis pigmentosa in murine RHO+/P23H mutant retinae, by the intravitreal AAV9-based delivery [123]. They tested several discriminating sgRNAs, although some of which did not really discriminate, photoreceptor cell degenerations were successfully slowed down by certain working sgRNAs [124]. Those divergent therapeutic results also remind us of the importance of selecting optimal Cas proteins and their matched sgRNAs.
Allele-specific CRISPR was also applied in the treatment of cancers [126-128] (Table 2). One of gain-of-function mutations in oncogene KRAS leads to the drug resistance to the MEK inhibitor AZD6244, which plays a critical role in anti-proliferation and pro-apoptosis in various tumor cell lines [129]. Allele-specific targeting by SpCas9 was then employed taking advantage of a novel PAM site (“In PAM”) created by the heterozygous G13A mutation (GGC > GCC). It come out that the G13A mutation was selectively and completely silenced in colorectal cancer cells, resulting in the reversal of drug resistance [126]. Around 15% of non-small cell lung cancer (NSCLC) cases are linked to mutations in the oncogene EGFR, which contributes to tumor progression [127]. Similarly, Koo et al., targeted EGFR harboring a single-nucleotide missense mutation L858R (CTG > CGG) that creates a novel PAM (“In PAM”) recognized by SpCas9. The combination of adenovirus delivery of SpCas9 and mutation-specific sgRNA resulted in precise destruction of the oncogenic allele with high specificity, and further promoted the killing of cancer cells and the reduction of tumor size in a xenograft mouse model [127]. Those results suggest that selective targeting of oncogenic mutations using CRISPR provides a robust surgical strategy to treat cancers.
Recently, another two excellent work have focused on the therapy of dominant progressive hearing loss. Gao et al. designed and validated that allele-specific editing preferentially disrupted the dominant deafness-related allele in the Tmc1 Beethoven mouse model, although the mutant Tmc1 allele differs from the wild-type allele at only one base pair [130].
Injection of SpCas9-sgRNA-lipid complexes targeting the Tmc1 allele into the cochlea of neonatal Tmc1 Beethoven mice significantly reduced progressive hearing loss [130]. The other work led by György et al. further screened 14 Cas9/gRNA combinations for specific and efficient disruption in fibroblasts from Tmc1Bth/WT mice, to improve allele selectivity [131]. They failed to use SpCas9 working with a “Near PAM” model, but instead turned to another Cas9 variant, SaCas9-KKH, that recognizes a novel PAM created by the Tmc1 mutation [131]. They then packaged AAVs containing SaCas9-KKH and the discriminating sgRNA and delivered them to sensory hair cells of the cochlea. This allele-specific strategy successfully targeted the mutant Tmc1 allele, and restored the hearing sensitivity in Beethoven mice, up to even one-year post injection [131].
Not surprisingly, a variety of other diseases were also studied using this allele-specific strategy (Table 2). For example, György et al. selectively disrupted the APPsw mutation allele both ex vivo and in vivo, and thereby decreased pathogenic Aβ secretion for treating familial Alzheimer's disease (AD) [132]. Christie et al. have tried to use different Cas proteins including SpCas9, SaCas9 and AsCpf1 respectively, based on the diverse mutations of TGFBI in treating autosomal dominant corneal dystrophy [133]. Wu et al. successfully used the allele-specific CRISPR to help correct a dominant mutation of CGYGC gene that causes cataracts, since the stage of mice zygotes [134]. The work by Shinkuma et al. successfully applied this strategy to dominant dystrophic epidermolysis bullosa (DDEB), caused by a dominant negative mutation in the COL7A1 gene encoding type VII collagen in iPSCs [135]. And Yamamoto et al. performed an allele-specific ablation that could rescue electrophysiological abnormalities of cardiomyocytes derived from a human iPSC model of long-QT syndrome (LQTS) with a CALM2 mutation [136]. Moreover, Courtney et al. established allele-specific cleavages of the dominant-negative KRT12 L132P mutated allele, which causes Meesmann's epithelial corneal dystrophy (MECD) [137]. Recently, Rabai et al. also examined the allele-specific inactivation or correction of a heterozygous mutation in the DNM2 gene, which causes the autosomal dominant centronuclear myopathies (CNMs) [138]. All in all, the allele-specific strategy has been increasingly implemented, and thus shows tremendous therapeutic potentials for dominantly inherited diseases.
Allele-specific CRISPR and SNPs Exploitation
Common SNPs representing a given haplotype can all be used for allele-specific targeting of mutant alleles caused by different mutations, which can make allele-specific therapy strategies more versatile and cost-effective (Figure 3B). Recently, two proof-of-principle studies have documented this strategy for treating Huntington's disease (HD) [59, 60]. HD is a dominantly inherited neurodegenerative disorder caused by CAG repeat expansions in the huntingtin (HTT) gene that results in an elongated polyglutamine (polyQ) tract in the huntingtin protein [139-141]. The pathogenic HTT basically consists of more than 35 CAG repeats and produces intracellular polyQ aggregates, preferentially causing the death of medium spiny neurons in the striatum, via a gain of toxic function [142, 143]. Deletion of the dominant mutant allele would leave one normal allele intact, which is sufficient for normal function and prevent the onset of the disease. Therefore, they deigned the allele-selective CRISPR/Cas9 strategy hopefully for selectively inactivating mutant HTT allele. They first screened for possible “In PAM” SNPs that are present merely on the mutant but not on normal chromosome haplotype. They then designed two allele-specific sgRNAs (“In PAM”) flanking the region of mutant allele [59, 60]. A large DNA stretch spanning the promoter region, transcription start site and the CAG repeat mutation, was eventually selectively excised. This excision completely silenced the generation of mutant HTT mRNA and protein in HD primary fibroblasts [59, 60] and later confirmed in vivo by AAV injection of discriminating Cas9/sgRNAs into a BacHD mouse model [60]. Those results of perfectly allele-specific genome targeting would definitely inspire us to treat dominant disorders like HD, by using SNP based allele-specific personalized therapy.
Allele-specific CRISPR and Haploinsufficiency
Apart from working on gain-of-function mutations, CRISPR has also been applied to activate specific genes that may be promising for loss-of-function treatments. Loss-of-function mutations in one allele can lead to haploinsufficiency, a condition that reduced protein doses may not be able to assure normal functions and consequently cause human diseases. It is now estimated that there are more than 660 genes associated with human diseases due to haploinsufficiency [144]. Although delivering additional copies of the gene via recombinant AAV (rAAV) is an effective and safe gene therapy, it still has some limitations, including limited size of DNA packaging (~5.0 kb, including inverted terminal repeats) and ectopic expression [145]. However, increasing the endogenous expression levels of the normal allele could overcome those limitations and potentially correct haploinsufficiency (Figure 3A).
Matharu et al. recently deployed a strategy of CRISPR-mediated activation (CRISPRa) for treating obesity, whereby dCas9 is fused with a transcriptional activator, VP64, so as to target one gene's regulatory element [146]. They focused on two genes, SIM1 and MC4R, which are both expressed in the paraventricular nuclei (PVN) of the hypothalamus and are necessary for the regulation of body weight. The haploinsufficiency of SIM1 and MC4R due to loss-of-function mutations would lead to hyperphagic obesity [147]. They thus designed discriminating sgRNAs that specifically recognize the SIM1 promoter (Prm-CRISPRa) or its distant hypothalamic enhancer (Enh-CRISPRa) [146]. Using the SIM1+/- mice as the obesity model, they observed that both Prm-CRISPRa and Enh-CRISPRa mice showed obviously reduced body fat and food intake [146]. Interestingly, the upregulated levels of SIM1 by CRISPRa targeting turned to be in a tissue-specific manner. They only observed significantly elevated mRNA levels in the hypothalamus or kidney, two tissues where SIM1 is expressed, but not in the lung and liver, where the regulatory elements are not active and SMI1 is not expressed [146]. This tissue-specificity endows an extra safety feature to the CRISPRa therapeutics.
They further employed rAAV to deliver CRISPRa (with both SpCas9 and SaCas9) into the hypothalamus of SIM1+/- mice or MC4R+/- mice respectively. It gave rise to several fold increase in SIM1 mRNA expression, and similarly rescues the weight gain phenotypes of mice model [146]. Those findings demonstrated that the CRISPRa system can rescue a haploinsufficient phenotype in vivo. Importantly, this CRISPRa strategy only targets endogenous regulatory elements to enhance the expression of the existing functional genes, without DNA breaks. Therefore, it can overcome the problem of ectopic gene expression, and can also be applied for genes that are not amenable to conventional gene therapy, due to that they have longer coding sequences beyond the rAAV packaging limitations.
Notably, one of the possible side effects after allele-specific targeting in dominant diseases, might be that the remaining normal alleles are causing haploinsufficiency. This would be diseases- and mutations-dependent. For targeting HTT alleles as mentioned above [59], loss of one copy of HTT did not actually generate typical HD symptoms, and one functional copy of HTT is sufficient to maintain normal cellular physiology. And also in the case of KRT12 [137], the expression of the dominant-negative mutant L132P is abolished by NHEJ, which could be considered a therapeutic success, as KRT12 has been shown not to demonstrate haploinsufficiency [148]. However, for those situations that may cause haploinsufficiency, it might be practical to combine CRISPRa to activate the normal copy simultaneously when selectively targeting of the mutant allele. Additionally, the CRISPR-mediated interference (CRISPRi) could also be employed to target the mutants and thereby suppressing their expression levels (Figure 3A). This CRISPRa/i strategy might contribute a fine-tuned control of expression levels, which may be appended to the current “all-or-none” paradigm of allele-specific targeting.
Allele-specific CRISPR and Epigenome Editing
In most cases of genome editing, both the paternal and maternal alleles of a given locus are targeted. For cases such as X-chromosome inactivetion and genomic imprinting diseases, however, one of the alleles is silenced due to the CpG methylation. Specifically, genomic imprinting is a unique epigenetic process in mammals, characterized by differential DNA methylation in the parental alleles at imprinting control regions (ICRs) [149, 150]. This differential methylation results in a mono-allelic expression of genes (paternal or maternal) without altering the DNA sequence. Genomic imprinting controls the expression of around 100 human genes, which directly or indirectly modulate fetal growth, frequently manifesting as severe developmental and neurological diseases, such as Silver-Russel syndrome, Beckwith-Wiedemann syndrome and Angelman syndrome [151].
Instead of binding directly on methylated DNA strands, epigenetic editing is a powerful way to alter DNA methylation at implicated loci. Allele-specific targeting is thus particularly applicable for treating those disorders. The easy and flexible targeting of the CRISPR-dCas9 system may allow to recruit particular DNA binding proteins/domains, which specifically bind to either allele of an imprinted locus in the patients [83]. The polymorphisms between maternal and paternal alleles would confer their discriminations. For example, the dCas9 fused with chromatin-modifying proteins such as DNA methyltransferases DNMT3A or demethylase domains, may facilitate an allele-specific binding of discriminating sgRNA at imprinted loci, and further reverses their methylated status [152, 153] (Figure 3C).
Allele-specific CRISPR and Immunocompatibility
When implementing cell transplantations, one major concern that must be overcome before clinical application is the potential human leukocyte antigen (HLA) mismatch, which can lead to immune rejection [154]. The cell therapies using iPSCs show to be promising in treating a variety of diseases, due to their capabilities of differentiating into a wide range of specialized cell types and tissues. However, it is time-consuming and costly to produce autologous clinical grade iPSCs for individual patients [155]. One alternative to personalized iPSC therapy is to establish a bank of clinically grade iPSC lines that can be differentiated for use in a broader number of patients [156]. HLA homozygous donors are being considered for covering most HLA haplotypes, however, recruiting HLA homozygous donors for the entire population is challenging. In a recent study, Xu et al. developed tailored HLA-editing strategies to facilitate the immunocompatibility of iPSCs [157]. They first generated HLA pseudo-homozygous iPSCs, after the allele-specific editing in HLA heterozygous iPSCs. HLA genes are highly polymorphic, so that the HLA codes of target alleles must be considered in designing the genome editing strategy. Moreover, all HLA genes have sequence similarity, which increases the risk of targeting non-specific HLA alleles. They thus generated a customized gRNA database for the SpCas9 system to involve all of the available HLA haplotype sequences from the IPD-IMGT/HLA database [158]. Among the 7,955 gRNAs with an ''NGG'' PAM sequence, they figured out 7,118 gRNAs that can target individual HLA genes, of which only 2,388 are able to target single alleles. They succeeded in generating class I HLA haploid iPSCs by allele-specific HLA targeting of heterozygous cells, thus producing so-called pseudo-homozygous iPSCs, which should have similar donor potential as HLA homozygous iPSCs [157]. This strategy could significantly improve donor compatibility in regenerative medicine, and is expected to serve pseudo-homozygous iPSCs for the majority of the world's population (Figure 3D).
Specificity and Off-Target
Truncation of sgRNAs
It is believed that there should be a balance between the on-target efficiency and specificity for a given sgRNA, because increasing specificity may also lead to a reduction in on-target cleavage. In terms of genome targeting for therapy, even with the allele-specific method, specificity is a very critical element that must be taken into account. So far, enormous efforts have been made to minimize potential off-target activities.
One of the strategies is the usage of a truncated sgRNA (17-18 nt rather than 20 nt in length), which was demonstrated to improve specificity [159, 160]. However, this conclusion appears to be quite controversial in the practice of genome editing by allele-specific CRISPR. Several studies have conducted allele-specific targeting with truncated sgRNAs. Christie et al. used both the mutant 18 and 20 nt sgRNAs for the cleavage of the mutant allele [133]. While the truncated 18 nt sgRNAs did not provide obvious improvements of specificity, maximal discrimination occurred with 20 or 19 nt sgRNAs for most cases [133]. Budde et al. made genome-editing efforts on correcting patient iPSCs harboring heterozygous dominant mutations that cause FTD [114]. They compared the effects of different lengths of allele-specific sgRNAs. A truncated sgRNA (17 nt) produced 6 fold lower NHEJ frequency than the full-length sgRNA (20 nt) and also fewer corrected iPSC clones. Moreover, whole genome sequencing showed that the mutational burden in both corrected iPSCs was similar, demonstrating that truncated sgRNAs at this site do not really enhance allele-specific targeting [114]. Similarity was also observed in a recent report by Gao et al. [130]. The full-length sgRNA (Tmc1-mut3) modified the mutant Tmc1 allele 23-fold more efficiently than the wild-type one, whereas the corresponding truncated 17 nt sgRNA (Tmc1-mut4) almost diminished its discrimination ability [130]. To the contrary, there were also reports echoing this strategy of using truncated sgRNAs. For instance, Li et al. successfully documented a truncated sgRNA (17 nt) that can improve allele discrimination with a cleavage efficiency of 28% of RHO P23H, without detectable cleavages in the wild-type alleles [124].
Those controversial results might be due to the status of the binding energy between RNA and DNA. The truncated gRNA works because the binding energy is reduced enough to bind a perfect target rather than to include mismatched targets [159]. Therefore, it reminds us that the usage of truncated gRNAs might be locus-dependent and empirically evaluated.
Beside the truncation of sgRNAs, there are other various approaches with the potential to improve targeting specificity, by using (1) low level or short-term exposure of Cas9: direct delivery of Cas mRNA/proteins [34, 161-163]; (2) Cas9 nickase (Cas9n) [164], or the engineered Cas nucleases with increased fidelity (such as SpCas9-HF [46], eSpCas9 [47], HypaCas9 [48], SaCas9-HF [49] and enAsCpf1-HF1 [50]); (3) chemically modified sgRNAs [165, 166]; (4) controllable Cas9 [167, 168]; (5) fusions of dCas9 with FokI nuclease domain [169, 170], and so forth. As this topic is not within the scope of this review and has already been well-described elsewhere [81, 171, 172], hence we will not discuss it with much more details. In the preclinical practice, we should try all the best to lower off-target activities of Cas nucleases, probably by the combinatorial approaches to optimize both Cas9 and sgRNA design [173].
Single-nucleotide Skipping
Another important phenomenon termed single-nucleotide skipping may also intervene with the allele-specific CRISPR. Although it is believed that even single nucleotide difference can confer the discrimination for allele-specific targeting, studies have shown that nucleotide mismatches at certain positions can be tolerated by CRISPR via single-nucleotide skipping [174, 175]. When DNA sequence contain single base insertion ("DNA bulge") or deletion ("RNA bulge"), the genomic locus can also be cleaved by Cas9 [174].
In another study, frequent off-target were observed in genome sites with a single-base bulge or up to 13 mismatches between the sgRNA and its genomic target [175], suggesting that single-nucleotide skipping from either the sgRNA or its genomic target can be tolerated.
Furthermore, a recent study conducted allele-specific genome editing, using the single G insertion in the p16ink4a gene model, and tested whether a single-nucleotide gap could also be exploited for allele-specific editing by CRISPR [176]. Interestingly, they found that the cleavage of target sites caused by single-nucleotide skipping can be observed if the gap occurs at the 1st or 2nd base upstream of the PAM [176]. Whereas skipping between the 3rd and 7th bases is unlikely to occur, or to be less effective in tolerating mismatches [176]. The general Cas9 tolerated DNA bulges in target sites lied in three regions: 7 bases from PAM, the 5'-end (PAM-distal) and the 3'-end (PAM-proximal) [176]. Although more studies are required to determine whether these positions are common for different target sequences, single-base DNA-bulges may be considered during off-target analysis. And also this brings a caveat for manipulating single-base variants in allele-specific targeting, if without an extra positional consideration of the variants.
Designing Allele-specific CRISPR
Designing Guidance
To edit the genomes in an allele-specific manner, basically an analysis of genetic variants in a given haplotype needs to be performed to examine if (i) they generate novel PAM sites; or (ii) they have PAM sites nearby, placing the variants within the seed region (Figure 4). This echoes the two working models of allele-specific CRSIPR. To allow the best discrimination, the “In PAM” model is the priority recommendation that contributes to the most stringent allele-specific cleavages. While taking a closer look at the discriminating sgRNAs, their features per se such as high/low GC contents, consecutive T bases or self-complementarity, may greatly dampen their on-target cleavage efficiencies [177, 178]. Therefore, the combined “In PAM” possibilities and sgRNA properties has to be taken into account. With the increasing choices of engineered Cas and PAM sites, it is worth designing multiple discriminating sgRNAs to be validated, before they are applied to gene therapies (Figure 4).
Bioinformatics Tools
Figuring out appropriate sgRNAs that may discriminate between two alleles is labor intensive and time consuming [60, 133], whereas not even approaching complete options. To aid the rational design of discriminating sgRNAs, therefore, a robust bioinformatics tool is highly desired (Figure 4). Although numerous tools are now available to assist with sgRNA selection for a reference genome [178], few have been implemented for allele-specific purposes. So far, tools like AlleleAnalyzer [113] and AsCRISPR [179] were recently developed for allele-specific and personalized sgRNA design. AlleleAnalyzer is a software that involves single-nucleotide variants and short indels to design dual sgRNAs for editing one or multiple haplotypes [113]. It also leverages patterns of shared genetic variations annotated by the ExAC and 1000GP to optimize sgRNA design for different human populations [64, 113].
It is worth noting that an increasing amount of non-coding RNAs or regulatory elements are identified, dual-sgRNA deletion of a large DNA fragment or gene locus might bring unpredictable risks for gene therapy. Genome targeting with a single allele-specific sgRNA should be an effective alternative strategy. The other tool, AsCRISPR, is a web server recently implemented in our lab that can process with either heterozygous allele sequences or Reference SNP cluster IDs, and would like to provide a complete list of potential single allele-specific sgRNAs working with more than 20 Cas proteins [179]. It is thus more like a query-driven tool focusing on the research studies and clinical therapeutics for the CRISPR community.
Therapeutic Perspectives
NHEJ vs. HDR
Conventional gene therapy shows great promise in the treatment of recessive diseases. However, their use in dominant diseases is limited because of the need for silencing or ablation of gain-of-function or dominant-negative mutant alleles, as well as introducing wild-type copies as replenishments that may even beyond the maximal packaging capacity of AAVs. In situ DNA repair by precise knock-in is an alternative avenue, involving the HDR pathway and a stretch of DNA donor. The low efficiency of HDR by CRISPR (generally less than 10%), however, still represents a practical challenge that hinders its applications [180, 181]. One potential advantage of allele-specific CRISPR would be that it merely employed the NHEJ pathway, instead of HDR, for efficiently targeting deleterious alleles. Without the need of providing DNA donors or viral vectors, it may enormously augment the therapeutic efficacy and reduce the complexity and cost of the therapy.
Moreover, HDR only occurs during late S and G2 phases when DNA replications are completed and sister chromatids can serve as repair templates, whereas NHEJ dominates DNA repair during G1, S and G2 phases [182]. Therefore, the knock-in method requiring HDR is less suited for post-mitotic cells such as neurons, whereas the allele-specific CRISPR using the NHEJ machinery could circumvent this issue and thus be applied to specifically target neurons in treating neurological diseases.
Personal Genomes and PAM Constraints
As a quite novel personalized strategy, allele-specific genomic targeting requires a thorough analysis of patients' haplotype patterns. This recapitulates the importance of personal genome sequencing (or genetic diagnosis) in implementing personalized therapy. Allele-specific CRISPR will then be applied to treating individual patients, starting with the identification of suitable polymorphisms that meet the designing criteria (Figure 4).
However, one limitation of the current technique is that it will require a unique variant-derived PAM or a PAM site close enough to the variant of interest. In some cases, there may be no appropriate choices of discriminating sgRNAs, and the cleavage efficiency of candidate sgRNAs might be even compromised. It urgently needs the growing expansion of the CRISPR-Cas toolbox with broader choices of PAM sites. The goal is to re-engineer Cas nucleases for desirable characteristics, including altered PAM sequences, better packaging into virus, better binding and cutting efficacy and higher specificity. And hopefully it can achieve highly efficient and allele-specific knockout of most, if not all, human dominant alleles in the practice of gene therapies.
Dosage Considerations
As mentioned earlier, allele-specific silencing of one mutant allele in dominant diseases gives rise to a 50% reduction in the expression level of the normal copy, if no compensation happens, and might probably cause haploinsufficiency, depending on the properties of given mutations (Figure 4). This reminds us of the potential dosage considerations during allele-specific targeting. It brings up several critical concerns, before the implementation of genomic engineering, that (i) whether the half amount of proteins from the remaining normal allele suffices to ameliorate or restore a disease phenotype; or vice versa (ii) to which extent would the augmentation/ correction of normal gene expression levels can produce therapeutic benefits in treating haploinsufficiency?
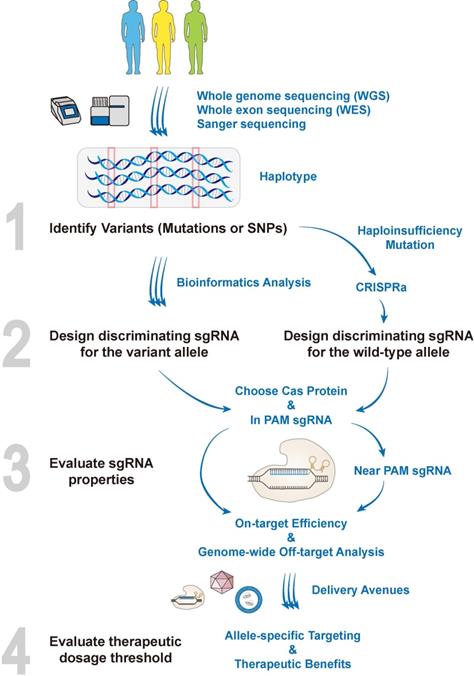
The pipeline of designing allele-specific CRISPR. In the era of precision medicine, haplotypes of patients are revealed by kinds of sequencing techniques. The personalized SNPs that represent a given haplotype, as well as genetic mutations, can all be exploited for allele-specific targeting. Bioinformatics tools are then employed for designing discriminating allele-specific sgRNAs combined with the optimal Cas protein. Notably, for genetic variants that may cause haploinsufficiency, it might be practical to combine CRISPRa to activate the wild-type allele simultaneously when selectively targeting of the variant allele. Basically, designing the discriminating sgRNAs needs to examine if (i) genetic variants generate novel PAM sites (In PAM sgRNA); or (ii) genetic variants have PAM sites nearby, placing the variants within the seed region (Near PAM sgRNA). To allow the best discrimination, the “In PAM sgRNA” is the priority recommendation that contributes to the most stringent allele-specific cleavages. The properties of discriminating sgRNAs such as GC contents and base constitutions, would then be taken into the evaluation of their on-target cleavage efficiencies. Delivery avenues such as AAVs, ribonucleoprotein (RNPs) or lipid nanoparticles (LNPs) will be employed to carry the Cas protein and discriminating sgRNAs into targeting sites. Importantly, the safety should be assessed by unbiased whole-genome off-target analysis at the preclinical stage. Finally, the dosage threshold for achieving therapeutic benefits after allele-specific targeting is disease-dependent, which needs to be empirically evaluated.
Certain diseases like Hemophilia B (HB), an X-linked genetic bleeding disorder caused by deficiency of coagulator factor IX (FIX), can be significantly restored by as low as 0.56% genetic correction [183]. Likewise, another study employed CRISPR to correct the dystrophin gene mutation in the germ line of mdx mice, the animal model of Duchenne muscular dystrophy (DMD) [184]. They generated several genetically mosaic animals containing 2% to 100% correction of the dystrophin gene. Strikingly, correcting just 17% of the mutant alleles was sufficient to allow dystrophin expression in most myofibers, and the muscles exhibited less histopathological features than mdx muscle [184]. The percentage of muscle phenotypic rescue (47% to 60%) in mosaic mice even exceeded the efficiency of gene correction (17%), demonstrating the advantage of corrected cells and their contribution to regenerating muscles [184]. Currently, one clinical trial is using SaCas9 and delivery by AAV into the retina to treat Type 10 Leber congenital amaurosis (LCA10), a severe retinal dystrophy caused by mutations in the CEP290 gene, by removing the aberrant splice donor created by the IVS26 mutation and thereby restoring normal CEP290 expression [185]. It has shown that it would be necessary to rescue 10% of foveal cones to achieve clinical benefits that are sufficient for near-normal visual acuity [185].
Importantly, the proliferative advantage of target cells after genome editing may also allow that correcting a low number of cells would be sufficient to reverse the disease phenotypes. For example, in treating patients with Fanconi anemia (FA), NHEJ-mediated gene editing was employed to efficiently edit multiple FANCA mutations in long-term hematopoietic stem cells (HSCs) and lymphoblastic cell lines (LCLs) [186]. Those corrected FA-HSCs showed significant proliferation advantage and phenotypic correction both in vitro and after transplantation in vivo, due to that NHEJ was positively employed to generate compensatory therapeutic mutations that may restore FANCA functions [186]. Specifically, they showed that therapeutic indels were initially at a low ratio (0.20%) and were greatly enriched during proliferation (47.36% at day 60), which successfully restored nuclear FANCD2 foci formation, increased mitomycin C resistance, reduced chromosomal fragility upon diepoxibutane challenge, and reduced reactive oxygen species levels in edited cells [186]. This study also echoes that, instead of HDR, NHEJ-based gene targeting could provide a simple therapeutic avenue for treating a specific group of patients with monogenic lympho-hematopoietic diseases.
Altogether, those delightful results may somehow relieve the researchers' anxiety for searching applicable sgRNAs with the highest efficiencies, but meanwhile remind us of a necessary pre-evaluation with the dosage “threshold” of therapeutic effects after a genome editing-based therapy [187]. It is also worth noting that the required dosage is quite dependent on given disease or mutation types, therefore, the therapeutic dosage of allele-specific targeting needs to be empirically evaluated.
An extra specific concern is that wild-type allele expressions may also be slightly dampened under less stringent allele-specific cleavages directed by certain discriminating sgRNAs [124, 130]. We are trying our best to avoid this, however in some cases, we may have no better choices. Theoretically, the reduction of 5%-10% of normal proteins leading to minor hypofunction might be tolerated in biological systems. Hence, it might be an acceptable trade-off if the dominant pathogenic alleles are potently disrupted.
Germline Editing and Ethical Dilemma
So far, almost all endeavors on allele-specific genome targeting were achieved either ex vivo or in vivo in affected somatic tissues. Ex vivo approaches are generally restricted to certain cell types that can be edited in the lab and subsequently transplanted, such as HSCs [186]. In vivo approaches, to the contrary, could be employed to a wider range of tissues, but the potential side effects induced by off-targets still represent a major safety concern. Delightfully, great efforts have been devoting to achieve tissue-specific genome targeting, to lower the risk of possible deleterious off-target indels in non-pathogenic tissues [188, 189].
The somatic gene therapy, however, cannot prevent transmission of mutated genes from parents to offspring, which may persist the entire family's medical burden. As such, it looks like that germline editing in reproductive cells (sperm and eggs) or preimplantation embryos holds greater potential for correcting disease-causing mutations at an early stage that may prevent the inheritance of genetic disease to all future generations. However, genome editing of human embryos is far from mature and will also raise numerous grave safety, social and ethical concerns. One recent study ever tried to edit β-Globin gene in nonviable human embryos, and it turned out that the targeting efficiency was low and, most critically, it brought about substantial off-target effects and mosaicism [190], which is a common issue occurred in germline editing that leads to the mixed untargeted and targeted cells within a multicellular embryo [191]. Better technologies for improving targeting specificities, thoroughly detecting off-targets and eliminating mosaicism have to be developed to secure the human applicability of germline editing. However, even with great technical advancements in future, human germline editing may raise other social concerns such as new forms of social inequality and discrimination that still needs to be extensively debated.
Summary
Overall, incorporating genetic variations into sgRNA designs successfully enabled the allele-specific genome engineering. The strategy of allele-specific CRISPR is now increasingly believed to be promising for treating genetic diseases for individual patient. Allele-specific genomic targeting has been versatile in increasing research areas, including the treatment of dominant negative diseases, genome imprinting, haploinsufficiency, spatiotemporal loci imaging and immunocompatible manipulations. Combined with other emerging tools such as CRISPR base editors, more advanced applications are likely to be exploited. However, more endeavors on improving sgRNA specificities, expanding recognized PAM sites, better packaging into AAVs, better binding and cutting efficacy and safer delivery of Cas nucleases, have to be addressed before it can be applied to humans.
Abbreviations
1000GP: 1000 Genomes Project; AAV: adeno-associated virus; AD: Alzheimer's disease; adRP: autosomal dominant retinitis pigmentosa; ASOs: antisense oligonucleotides; Cas9n: cas9 nickase; CNMs: centronuclear myopathies; CRISPR: clustered regularly interspaced short palindromic repeats; CRISPRa: CRISPR-mediated activation; CRISPRi: CRISPR-mediated interference; DDEB: dominant dystrophic epidermolysis bullosa; DMD: Duchenne muscular dystrophy; DSBs: double-strand breaks; dsDNA: double strand DNA; ExAC: Exome Aggregation Consortium; FA: Fanconi anemia; FIX: factor IX; FTD: frontotemporal dementia; gnomAD: Genome Aggregation Database; GWAS: genome-wide association studies; HB: hemophilia B; HD: Huntington's disease; HDR: homology-directed repair; HLA: human leukocyte antigen; HSCs: hematopoietic stem cells; HTT: huntingtin; ICRs: imprinting control regions; iPSCs: induced pluripotent stem cells; LCA10: Type 10 Leber congenital amaurosis; LCLs: lymphoblastic cell lines; LncRNA: long noncoding RNA; LQTS: long-QT syndrome; MECD: Meesmann's epithelial corneal dystrophy; NHEJ: non-homology end joining; NSCLC: non-small cell lung cancer; PAM: protospacer adjacent motif; polyQ: polyglutamine; PVN: paraventricular nuclei; RHO: rhodopsin; sgRNA: single guide RNA; siRNAs: short-interfering RNAs; SNPs: single nucleotide polymorphisms; TALEN: transcription activator-like effector nucleases.
Acknowledgements
This study was supported by grants from the National Natural Sciences Foundation of China [No. 81801200], Hunan Provincial Natural Science Foundation of China [No. 2019JJ40476], and Talents Startup Funds [No. 2209090550] of Xiangya Hospital, Central South University, Changsha, China. Although we have tried our best to include the related primary studies, we apologize that we may have unintentionally omitted other excellent findings in the area.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Mills RE, Luttig CT, Larkins CE, Beauchamp A, Tsui C, Pittard WS. et al. An initial map of insertion and deletion (INDEL) variation in the human genome. Genome Res. 2006;16:1182-90
2. Mullaney JM, Mills RE, Pittard WS, Devine SE. Small insertions and deletions (INDELs) in human genomes. Hum Mol Genet. 2010;19:R131-6
3. Genomes Project C, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM. et al. A global reference for human genetic variation. Nature. 2015;526:68-74
4. Sachidanandam R, Weissman D, Schmidt SC, Kakol JM, Stein LD, Marth G. et al. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 2001;409:928-33
5. Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T. et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285-91
6. Visscher PM, Brown MA, McCarthy MI, Yang J. Five years of GWAS discovery. Am J Hum Genet. 2012;90:7-24
7. Sud A, Kinnersley B, Houlston RS. Genome-wide association studies of cancer: current insights and future perspectives. Nat Rev Cancer. 2017;17:692-704
8. Zheng J, Huang X, Tan W, Yu D, Du Z, Chang J. et al. Pancreatic cancer risk variant in LINC00673 creates a miR-1231 binding site and interferes with PTPN11 degradation. Nat Genet. 2016;48:747-57
9. Hua JT, Ahmed M, Guo H, Zhang Y, Chen S, Soares F. et al. Risk SNP-Mediated Promoter-Enhancer Switching Drives Prostate Cancer through lncRNA PCAT19. Cell. 2018;174:564-75 e18
10. Miao YR, Liu W, Zhang Q, Guo AY. lncRNASNP2: an updated database of functional SNPs and mutations in human and mouse lncRNAs. Nucleic Acids Res. 2018;46:D276-D80
11. Wang Y, Guo Z, Zhao Y, Jin Y, An L, Wu B. et al. Genetic polymorphisms of lncRNA-p53 regulatory network genes are associated with concurrent chemoradiotherapy toxicities and efficacy in nasopharyngeal carcinoma patients. Sci Rep. 2017;7:8320
12. Hu W, Jiang C, Guan D, Dierickx P, Zhang R, Moscati A. et al. Patient Adipose Stem Cell-Derived Adipocytes Reveal Genetic Variation that Predicts Antidiabetic Drug Response. Cell Stem Cell. 2019;24:299-308 e6
13. Guo C, Lin X, Yin J, Xie X, Li J, Meng X. et al. Pharmacogenomics signature: A novel strategy on the individual differences in drug response. Cancer Lett. 2018;420:190-4
14. Xu X, Han L, Yang H, Duan L, Zhou B, Zhao Y. et al. The A/G allele of eIF3a rs3740556 predicts platinum-based chemotherapy resistance in lung cancer patients. Lung Cancer. 2013;79:65-72
15. Wang Y, Yin JY, Li XP, Chen J, Qian CY, Zheng Y. et al. The association of transporter genes polymorphisms and lung cancer chemotherapy response. PLoS One. 2014;9:e91967
16. Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60:174-82
17. Pourcel C, Salvignol G, Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology. 2005;151:653-63
18. Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S. et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709-12
19. Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N. et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819-23
20. Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE. et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823-6
21. Hou Z, Zhang Y, Propson NE, Howden SE, Chu LF, Sontheimer EJ. et al. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc Natl Acad Sci U S A. 2013;110:15644-9
22. Esvelt KM, Mali P, Braff JL, Moosburner M, Yaung SJ, Church GM. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Methods. 2013;10:1116-21
23. Ibraheim R, Song CQ, Mir A, Amrani N, Xue W, Sontheimer EJ. All-in-one adeno-associated virus delivery and genome editing by Neisseria meningitidis Cas9 in vivo. Genome Biol. 2018;19:137
24. Lee CM, Cradick TJ, Bao G. The Neisseria meningitidis CRISPR-Cas9 System Enables Specific Genome Editing in Mammalian Cells. Mol Ther. 2016;24:645-54
25. Amrani N, Gao XD, Liu P, Edraki A, Mir A, Ibraheim R. et al. NmeCas9 is an intrinsically high-fidelity genome-editing platform. Genome Biol. 2018;19:214
26. Edraki A, Mir A, Ibraheim R, Gainetdinov I, Yoon Y, Song CQ. et al. A Compact, High-Accuracy Cas9 with a Dinucleotide PAM for In Vivo Genome Editing. Mol Cell. 2019;73:714-26 e4
27. Harrington LB, Paez-Espino D, Staahl BT, Chen JS, Ma E, Kyrpides NC. et al. A thermostable Cas9 with increased lifetime in human plasma. Nat Commun. 2017;8:1424
28. Muller M, Lee CM, Gasiunas G, Davis TH, Cradick TJ, Siksnys V. et al. Streptococcus thermophilus CRISPR-Cas9 Systems Enable Specific Editing of the Human Genome. Mol Ther. 2016;24:636-44
29. Kim E, Koo T, Park SW, Kim D, Kim K, Cho HY. et al. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat Commun. 2017;8:14500
30. Yamada M, Watanabe Y, Gootenberg JS, Hirano H, Ran FA, Nakane T. et al. Crystal Structure of the Minimal Cas9 from Campylobacter jejuni Reveals the Molecular Diversity in the CRISPR-Cas9 Systems. Mol Cell. 2017;65:1109-21 e3
31. Koo T, Lu-Nguyen NB, Malerba A, Kim E, Kim D, Cappellari O. et al. Functional Rescue of Dystrophin Deficiency in Mice Caused by Frameshift Mutations Using Campylobacter jejuni Cas9. Mol Ther. 2018;26:1529-38
32. Chatterjee P, Jakimo N, Jacobson JM. Minimal PAM specificity of a highly similar SpCas9 ortholog. Sci Adv. 2018;4:eaau0766
33. Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816-21
34. Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V. et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827-32
35. Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ. et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186-91
36. Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P. et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759-71
37. Yamano T, Nishimasu H, Zetsche B, Hirano H, Slaymaker IM, Li Y. et al. Crystal Structure of Cpf1 in Complex with Guide RNA and Target DNA. Cell. 2016;165:949-62
38. Kim HK, Song M, Lee J, Menon AV, Jung S, Kang YM. et al. In vivo high-throughput profiling of CRISPR-Cpf1 activity. Nat Methods. 2017;14:153-9
39. Tu M, Lin L, Cheng Y, He X, Sun H, Xie H. et al. A 'new lease of life': FnCpf1 possesses DNA cleavage activity for genome editing in human cells. Nucleic Acids Res. 2017;45:11295-304
40. Zetsche B, Strecker J, Abudayyeh OO, Gootenberg JS, Scott DA, Zhang F. A Survey of Genome Editing Activity for 16 Cpf1 orthologs. bioRxiv. 2017 doi: 10.1101/134015
41. Teng F, Li J, Cui T, Xu K, Guo L, Gao Q. et al. Enhanced mammalian genome editing by new Cas12a orthologs with optimized crRNA scaffolds. Genome Biol. 2019;20:15
42. Teng F, Cui T, Feng G, Guo L, Xu K, Gao Q. et al. Repurposing CRISPR-Cas12b for mammalian genome engineering. Cell Discov. 2018;4:63
43. Strecker J, Jones S, Koopal B, Schmid-Burgk J, Zetsche B, Gao L. et al. Engineering of CRISPR-Cas12b for human genome editing. Nat Commun. 2019;10:212
44. Liu JJ, Orlova N, Oakes BL, Ma E, Spinner HB, Baney KLM. et al. CasX enzymes comprise a distinct family of RNA-guided genome editors. Nature. 2019;566:218-23
45. Burstein D, Harrington LB, Strutt SC, Probst AJ, Anantharaman K, Thomas BC. et al. New CRISPR-Cas systems from uncultivated microbes. Nature. 2017;542:237-41
46. Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z. et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490-5
47. Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84-8
48. Chen JS, Dagdas YS, Kleinstiver BP, Welch MM, Sousa AA, Harrington LB. et al. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature. 2017;550:407-10
49. Tan Y, Chu AHY, Bao S, Hoang DA, Kebede FT, Xiong W. et al. Rationally engineered Staphylococcus aureus Cas9 nucleases with high genome-wide specificity. Proc Natl Acad Sci U S A. 2019;116:20969-76
50. Kleinstiver BP, Sousa AA, Walton RT, Tak YE, Hsu JY, Clement K. et al. Engineered CRISPR-Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing. Nat Biotechnol. 2019;37:276-82
51. Kleinstiver BP, Prew MS, Tsai SQ, Topkar VV, Nguyen NT, Zheng Z. et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523:481-5
52. Hirano H, Gootenberg JS, Horii T, Abudayyeh OO, Kimura M, Hsu PD. et al. Structure and Engineering of Francisella novicida Cas9. Cell. 2016;164:950-61
53. Hu JH, Miller SM, Geurts MH, Tang W, Chen L, Sun N. et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature. 2018;556:57-63
54. Nishimasu H, Shi X, Ishiguro S, Gao L, Hirano S, Okazaki S. et al. Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science. 2018;361:1259-62
55. Kleinstiver BP, Prew MS, Tsai SQ, Nguyen NT, Topkar VV, Zheng Z. et al. Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition. Nat Biotechnol. 2015;33:1293-8
56. Ma D, Xu Z, Zhang Z, Chen X, Zeng X, Zhang Y. et al. Engineer chimeric Cas9 to expand PAM recognition based on evolutionary information. Nat Commun. 2019;10:560
57. Gao L, Cox DBT, Yan WX, Manteiga JC, Schneider MW, Yamano T. et al. Engineered Cpf1 variants with altered PAM specificities. Nat Biotechnol. 2017;35:789-92
58. Nishimasu H, Yamano T, Gao L, Zhang F, Ishitani R, Nureki O. Structural Basis for the Altered PAM Recognition by Engineered CRISPR-Cpf1. Mol Cell. 2017;67:139-47 e2
59. Shin JW, Kim KH, Chao MJ, Atwal RS, Gillis T, MacDonald ME. et al. Permanent inactivation of Huntington's disease mutation by personalized allele-specific CRISPR/Cas9. Hum Mol Genet. 2016;25:4566-76
60. Monteys AM, Ebanks SA, Keiser MS, Davidson BL. CRISPR/Cas9 Editing of the Mutant Huntingtin Allele In Vitro and In Vivo. Mol Ther. 2017;25:12-23
61. Zuo E, Huo X, Yao X, Hu X, Sun Y, Yin J. et al. CRISPR/Cas9-mediated targeted chromosome elimination. Genome Biol. 2017;18:224
62. Sapranauskas R, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 2011;39:9275-82
63. Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li GW. et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155:1479-91
64. Qin P, Parlak M, Kuscu C, Bandaria J, Mir M, Szlachta K. et al. Live cell imaging of low- and non-repetitive chromosome loci using CRISPR-Cas9. Nat Commun. 2017;8:14725
65. Fujita T, Fujii H. Efficient isolation of specific genomic regions and identification of associated proteins by engineered DNA-binding molecule-mediated chromatin immunoprecipitation (enChIP) using CRISPR. Biochem Biophys Res Commun. 2013;439:132-6
66. Fujita T, Asano Y, Ohtsuka J, Takada Y, Saito K, Ohki R. et al. Identification of telomere-associated molecules by engineered DNA-binding molecule-mediated chromatin immunoprecipitation (enChIP). Sci Rep. 2013;3:3171
67. Myers SA, Wright J, Peckner R, Kalish BT, Zhang F, Carr SA. Discovery of proteins associated with a predefined genomic locus via dCas9-APEX-mediated proximity labeling. Nat Methods. 2018;15:437-9
68. Qiu W, Xu Z, Zhang M, Zhang D, Fan H, Li T. et al. Determination of local chromatin interactions using a combined CRISPR and peroxidase APEX2 system. Nucleic Acids Res. 2019;47:e52
69. Perez-Pinera P, Kocak DD, Vockley CM, Adler AF, Kabadi AM, Polstein LR. et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat Methods. 2013;10:973-6
70. Maeder ML, Linder SJ, Cascio VM, Fu Y, Ho QH, Joung JK. CRISPR RNA-guided activation of endogenous human genes. Nat Methods. 2013;10:977-9
71. Hilton IB, D'Ippolito AM, Vockley CM, Thakore PI, Crawford GE, Reddy TE. et al. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol. 2015;33:510-7
72. Hsu MN, Liao HT, Truong VA, Huang KL, Yu FJ, Chen HH. et al. CRISPR-based Activation of Endogenous Neurotrophic Genes in Adipose Stem Cell Sheets to Stimulate Peripheral Nerve Regeneration. Theranostics. 2019;9:6099-111
73. Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE. et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442-51
74. Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP. et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173-83
75. Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420-4
76. Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI. et al. Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature. 2017;551:464-71
77. Adli M. The CRISPR tool kit for genome editing and beyond. Nat Commun. 2018;9:1911
78. Knott GJ, Doudna JA. CRISPR-Cas guides the future of genetic engineering. Science. 2018;361:866-9
79. Pyzocha NK, Chen S. Diverse Class 2 CRISPR-Cas Effector Proteins for Genome Engineering Applications. ACS Chem Biol. 2018;13:347-56
80. Wang F, Qi LS. Applications of CRISPR Genome Engineering in Cell Biology. Trends Cell Biol. 2016;26:875-88
81. Zhang F. Development of CRISPR-Cas systems for genome editing and beyond. Q Rev Biophys. 2019:52
82. Xu X, Qi LS. A CRISPR-dCas Toolbox for Genetic Engineering and Synthetic Biology. J Mol Biol. 2019;431:34-47
83. Pulecio J, Verma N, Mejia-Ramirez E, Huangfu D, Raya A. CRISPR/Cas9-Based Engineering of the Epigenome. Cell Stem Cell. 2017;21:431-47
84. Chaterji S, Ahn EH, Kim DH. CRISPR Genome Engineering for Human Pluripotent Stem Cell Research. Theranostics. 2017;7:4445-69
85. Yang L, Grishin D, Wang G, Aach J, Zhang CZ, Chari R. et al. Targeted and genome-wide sequencing reveal single nucleotide variations impacting specificity of Cas9 in human stem cells. Nat Commun. 2014;5:5507
86. Scott DA, Zhang F. Implications of human genetic variation in CRISPR-based therapeutic genome editing. Nat Med. 2017;23:1095-101
87. Lessard S, Francioli L, Alfoldi J, Tardif JC, Ellinor PT, MacArthur DG. et al. Human genetic variation alters CRISPR-Cas9 on- and off-targeting specificity at therapeutically implicated loci. Proc Natl Acad Sci U S A. 2017;114:E11257-E66
88. Karczewski KJ, Weisburd B, Thomas B, Solomonson M, Ruderfer DM, Kavanagh D. et al. The ExAC browser: displaying reference data information from over 60 000 exomes. Nucleic Acids Res. 2017;45:D840-D5
89. Karczewski KJ, Francioli LC, Tiao G, Cummings BB. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. bioRxiv. 2019 doi: 10.1101/531210
90. Miller VM, Xia H, Marrs GL, Gouvion CM, Lee G, Davidson BL. et al. Allele-specific silencing of dominant disease genes. Proc Natl Acad Sci U S A. 2003;100:7195-200
91. Trochet D, Prudhon B, Beuvin M, Peccate C, Lorain S, Julien L. et al. Allele-specific silencing therapy for Dynamin 2-related dominant centronuclear myopathy. EMBO Mol Med. 2018;10:239-53
92. Bongianino R, Denegri M, Mazzanti A, Lodola F, Vollero A, Boncompagni S. et al. Allele-Specific Silencing of Mutant mRNA Rescues Ultrastructural and Arrhythmic Phenotype in Mice Carriers of the R4496C Mutation in the Ryanodine Receptor Gene (RYR2). Circ Res. 2017;121:525-36
93. Novelli F, Lena AM, Panatta E, Nasser W, Shalom-Feuerstein R, Candi E. et al. Allele-specific silencing of EEC p63 mutant R304W restores p63 transcriptional activity. Cell Death Dis. 2016;7:e2227
94. Monga I, Qureshi A, Thakur N, Gupta AK, Kumar M. ASPsiRNA: A Resource of ASP-siRNAs Having Therapeutic Potential for Human Genetic Disorders and Algorithm for Prediction of Their Inhibitory Efficacy. G3 (Bethesda). 2017;7:2931-43
95. Zaleta-Rivera K, Dainis A, Ribeiro AJS, Sanchez Cordero P, Rubio G, Shang C. et al. Allele-Specific Silencing Ameliorates Restrictive Cardiomyopathy Due to a Human Myosin Regulatory Light Chain Mutation. Circulation. 2019
96. Giorgio E, Lorenzati M, Rivetti di Val Cervo P, Brussino A, Cernigoj M, Della Sala E. et al. Allele-specific silencing as treatment for gene duplication disorders: proof-of-principle in autosomal dominant leukodystrophy. Brain. 2019
97. Atkinson SD, McGilligan VE, Liao H, Szeverenyi I, Smith FJ, Moore CB. et al. Development of allele-specific therapeutic siRNA for keratin 5 mutations in epidermolysis bullosa simplex. J Invest Dermatol. 2011;131:2079-86
98. Lyu YS, Shi PL, Chen XL, Tang YX, Wang YF, Liu RR. et al. A Small Indel Mutant Mouse Model of Epidermolytic Palmoplantar Keratoderma and Its Application to Mutant-specific shRNA Therapy. Mol Ther Nucleic Acids. 2016;5:e299
99. Schwarz DS, Ding H, Kennington L, Moore JT, Schelter J, Burchard J. et al. Designing siRNA that distinguish between genes that differ by a single nucleotide. PLoS Genet. 2006;2:e140
100. Nishimura AL, Shum C, Scotter EL, Abdelgany A, Sardone V, Wright J. et al. Allele-specific knockdown of ALS-associated mutant TDP-43 in neural stem cells derived from induced pluripotent stem cells. PLoS One. 2014;9:e91269
101. Gonzalez-Alegre P, Bode N, Davidson BL, Paulson HL. Silencing primary dystonia: lentiviral-mediated RNA interference therapy for DYT1 dystonia. J Neurosci. 2005;25:10502-9
102. Southwell AL, Kordasiewicz HB, Langbehn D, Skotte NH, Parsons MP, Villanueva EB. et al. Huntingtin suppression restores cognitive function in a mouse model of Huntington's disease. Sci Transl Med. 2018:10
103. Southwell AL, Skotte NH, Kordasiewicz HB, Ostergaard ME, Watt AT, Carroll JB. et al. In vivo evaluation of candidate allele-specific mutant huntingtin gene silencing antisense oligonucleotides. Mol Ther. 2014;22:2093-106
104. Ostergaard ME, Southwell AL, Kordasiewicz H, Watt AT, Skotte NH, Doty CN. et al. Rational design of antisense oligonucleotides targeting single nucleotide polymorphisms for potent and allele selective suppression of mutant Huntingtin in the CNS. Nucleic Acids Res. 2013;41:9634-50
105. Carroll JB, Warby SC, Southwell AL, Doty CN, Greenlee S, Skotte N. et al. Potent and selective antisense oligonucleotides targeting single-nucleotide polymorphisms in the Huntington disease gene / allele-specific silencing of mutant huntingtin. Mol Ther. 2011;19:2178-85
106. Murray SF, Jazayeri A, Matthes MT, Yasumura D, Yang H, Peralta R. et al. Allele-Specific Inhibition of Rhodopsin With an Antisense Oligonucleotide Slows Photoreceptor Cell Degeneration. Invest Ophthalmol Vis Sci. 2015;56:6362-75
107. Hohjoh H. Disease-causing allele-specific silencing by RNA interference. Pharmaceuticals (Basel). 2013;6:522-35
108. Huang H, Qiao R, Zhao D, Zhang T, Li Y, Yi F. et al. Profiling of mismatch discrimination in RNAi enabled rational design of allele-specific siRNAs. Nucleic Acids Res. 2009;37:7560-9
109. Fink KD, Deng P, Gutierrez J, Anderson JS, Torrest A, Komarla A. et al. Allele-Specific Reduction of the Mutant Huntingtin Allele Using Transcription Activator-Like Effectors in Human Huntington's Disease Fibroblasts. Cell Transplant. 2016;25:677-86
110. Smith C, Abalde-Atristain L, He C, Brodsky BR, Braunstein EM, Chaudhari P. et al. Efficient and allele-specific genome editing of disease loci in human iPSCs. Mol Ther. 2015;23:570-7
111. Yoshimi K, Kaneko T, Voigt B, Mashimo T. Allele-specific genome editing and correction of disease-associated phenotypes in rats using the CRISPR-Cas platform. Nat Commun. 2014;5:4240
112. Zhou X, Jacobs TB, Xue LJ, Harding SA, Tsai CJ. Exploiting SNPs for biallelic CRISPR mutations in the outcrossing woody perennial Populus reveals 4-coumarate:CoA ligase specificity and redundancy. New Phytol. 2015;208:298-301
113. Keough KC, Lyalina S, Olvera MP, Whalen S, Conklin BR, Pollard KS. AlleleAnalyzer: a tool for personalized and allele-specific sgRNA design. Genome Biol. 2019;20:167
114. Budde JP, Martinez R, Hsu S, Wen N, Chen JA, Coppola G. et al. Precision genome-editing with CRISPR/Cas9 in human induced pluripotent stem cells. bioRxiv. 2017 doi: 10.1101/187377
115. Chao T, Liu Z, Zhang Y, Zhang L, Huang R, He L. et al. Precise and Rapid Validation of Candidate Gene by Allele Specific Knockout With CRISPR/Cas9 in Wild Mice. Front Genet. 2019;10:124
116. Capon SJ, Baillie GJ, Bower NI, da Silva JA, Paterson S, Hogan BM. et al. Utilising polymorphisms to achieve allele-specific genome editing in zebrafish. Biol Open. 2017;6:125-31
117. Maass PG, Barutcu AR, Shechner DM, Weiner CL, Mele M, Rinn JL. Spatiotemporal allele organization by allele-specific CRISPR live-cell imaging (SNP-CLING). Nat Struct Mol Biol. 2018;25:176-84
118. Huang N, Lee I, Marcotte EM, Hurles ME. Characterising and predicting haploinsufficiency in the human genome. PLoS Genet. 2010;6:e1001154
119. Veitia RA. Exploring the molecular etiology of dominant-negative mutations. Plant Cell. 2007;19:3843-51
120. Li Y, Zhang Y, Li X, Yi S, Xu J. Gain-of-Function Mutations: An Emerging Advantage for Cancer Biology. Trends Biochem Sci. 2019;44:659-74
121. Ferrari S, Di Iorio E, Barbaro V, Ponzin D, Sorrentino FS, Parmeggiani F. Retinitis pigmentosa: genes and disease mechanisms. Curr Genomics. 2011;12:238-49
122. Burnight ER, Gupta M, Wiley LA, Anfinson KR, Tran A, Triboulet R. et al. Using CRISPR-Cas9 to Generate Gene-Corrected Autologous iPSCs for the Treatment of Inherited Retinal Degeneration. Mol Ther. 2017;25:1999-2013
123. Giannelli SG, Luoni M, Castoldi V, Massimino L, Cabassi T, Angeloni D. et al. Cas9/sgRNA selective targeting of the P23H Rhodopsin mutant allele for treating retinitis pigmentosa by intravitreal AAV9.PHP.B-based delivery. Hum Mol Genet. 2018;27:761-79
124. Li P, Kleinstiver BP, Leon MY, Prew MS, Navarro-Gomez D, Greenwald SH. et al. Allele-Specific CRISPR-Cas9 Genome Editing of the Single-Base P23H Mutation for Rhodopsin-Associated Dominant Retinitis Pigmentosa. CRISPR J. 2018;1:55-64
125. Bakondi B, Lv W, Lu B, Jones MK, Tsai Y, Kim KJ. et al. In Vivo CRISPR/Cas9 Gene Editing Corrects Retinal Dystrophy in the S334ter-3 Rat Model of Autosomal Dominant Retinitis Pigmentosa. Mol Ther. 2016;24:556-63
126. Li Y, Mendiratta S, Ehrhardt K, Kashyap N, White MA, Bleris L. Exploiting the CRISPR/Cas9 PAM Constraint for Single-Nucleotide Resolution Interventions. PLoS One. 2016;11:e0144970
127. Koo T, Yoon AR, Cho HY, Bae S, Yun CO, Kim JS. Selective disruption of an oncogenic mutant allele by CRISPR/Cas9 induces efficient tumor regression. Nucleic Acids Res. 2017;45:7897-908
128. Yang M, Wei H, Wang Y, Deng J, Tang Y, Zhou L. et al. Targeted Disruption of V600E-Mutant BRAF Gene by CRISPR-Cpf1. Mol Ther Nucleic Acids. 2017;8:450-8
129. Little AS, Balmanno K, Sale MJ, Newman S, Dry JR, Hampson M. et al. Amplification of the driving oncogene, KRAS or BRAF, underpins acquired resistance to MEK1/2 inhibitors in colorectal cancer cells. Sci Signal. 2011;4:ra17
130. Gao X, Tao Y, Lamas V, Huang M, Yeh WH, Pan B. et al. Treatment of autosomal dominant hearing loss by in vivo delivery of genome editing agents. Nature. 2018;553:217-21
131. Gyorgy B, Nist-Lund C, Pan B, Asai Y, Karavitaki KD, Kleinstiver BP. et al. Allele-specific gene editing prevents deafness in a model of dominant progressive hearing loss. Nat Med. 2019;25:1123-30
132. Gyorgy B, Loov C, Zaborowski MP, Takeda S, Kleinstiver BP, Commins C. et al. CRISPR/Cas9 Mediated Disruption of the Swedish APP Allele as a Therapeutic Approach for Early-Onset Alzheimer's Disease. Mol Ther Nucleic Acids. 2018;11:429-40
133. Christie KA, Courtney DG, DeDionisio LA, Shern CC, De Majumdar S, Mairs LC. et al. Towards personalised allele-specific CRISPR gene editing to treat autosomal dominant disorders. Sci Rep. 2017;7:16174
134. Wu Y, Liang D, Wang Y, Bai M, Tang W, Bao S. et al. Correction of a genetic disease in mouse via use of CRISPR-Cas9. Cell Stem Cell. 2013;13:659-62
135. Shinkuma S, Guo Z, Christiano AM. Site-specific genome editing for correction of induced pluripotent stem cells derived from dominant dystrophic epidermolysis bullosa. Proc Natl Acad Sci U S A. 2016;113:5676-81
136. Yamamoto Y, Makiyama T, Harita T, Sasaki K, Wuriyanghai Y, Hayano M. et al. Allele-specific ablation rescues electrophysiological abnormalities in a human iPS cell model of long-QT syndrome with a CALM2 mutation. Hum Mol Genet. 2017;26:1670-7
137. Courtney DG, Moore JE, Atkinson SD, Maurizi E, Allen EH, Pedrioli DM. et al. CRISPR/Cas9 DNA cleavage at SNP-derived PAM enables both in vitro and in vivo KRT12 mutation-specific targeting. Gene Ther. 2016;23:108-12
138. Rabai A, Reisser L, Reina-San-Martin B, Mamchaoui K, Cowling BS, Nicot AS. et al. Allele-Specific CRISPR/Cas9 Correction of a Heterozygous DNM2 Mutation Rescues Centronuclear Myopathy Cell Phenotypes. Mol Ther Nucleic Acids. 2019;16:246-56
139. Zuccato C, Valenza M, Cattaneo E. Molecular mechanisms and potential therapeutical targets in Huntington's disease. Physiol Rev. 2010;90:905-81
140. Yang HM, Yang S, Huang SS, Tang BS, Guo JF. Microglial Activation in the Pathogenesis of Huntington's Disease. Front Aging Neurosci. 2017;9:193
141. Wu J, Tang Y, Zhang CL. Targeting N-Terminal Huntingtin with a Dual-sgRNA Strategy by CRISPR/Cas9. Biomed Res Int. 2019;2019:1039623
142. Saudou F, Humbert S. The Biology of Huntingtin. Neuron. 2016;89:910-26
143. Jiang H, Sun YM, Hao Y, Yan YP, Chen K, Xin SH. et al. Huntingtin gene CAG repeat numbers in Chinese patients with Huntington's disease and controls. Eur J Neurol. 2014;21:637-42
144. Landrum MJ, Lee JM, Benson M, Brown G, Chao C, Chitipiralla S. et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44:D862-8
145. Wang D, Tai PWL, Gao G. Adeno-associated virus vector as a platform for gene therapy delivery. Nat Rev Drug Discov. 2019;18:358-78
146. Matharu N, Rattanasopha S, Tamura S, Maliskova L, Wang Y, Bernard A. et al. CRISPR-mediated activation of a promoter or enhancer rescues obesity caused by haploinsufficiency. Science. 2019:363
147. Holder JL Jr, Butte NF, Zinn AR. Profound obesity associated with a balanced translocation that disrupts the SIM1 gene. Hum Mol Genet. 2000;9:101-8
148. Kao WW, Liu CY, Converse RL, Shiraishi A, Kao CW, Ishizaki M. et al. Keratin 12-deficient mice have fragile corneal epithelia. Invest Ophthalmol Vis Sci. 1996;37:2572-84
149. Tucci V, Isles AR, Kelsey G, Ferguson-Smith AC, Erice Imprinting G. Genomic Imprinting and Physiological Processes in Mammals. Cell. 2019;176:952-65
150. Monk D, Mackay DJG, Eggermann T, Maher ER, Riccio A. Genomic imprinting disorders: lessons on how genome, epigenome and environment interact. Nat Rev Genet. 2019;20:235-48
151. Peters J. The role of genomic imprinting in biology and disease: an expanding view. Nat Rev Genet. 2014;15:517-30
152. Bashtrykov P, Kungulovski G, Jeltsch A. Correction of aberrant imprinting by allele-specific epigenome editing. Clin Pharmacol Ther. 2016;99:482-4
153. Bashtrykov P, Jeltsch A. Allele-Specific Epigenome Editing. Methods Mol Biol. 2018;1767:137-46
154. Bradley JA, Bolton EM, Pedersen RA. Stem cell medicine encounters the immune system. Nat Rev Immunol. 2002;2:859-71
155. Neofytou E, O'Brien CG, Couture LA, Wu JC. Hurdles to clinical translation of human induced pluripotent stem cells. J Clin Invest. 2015;125:2551-7
156. Taylor CJ, Peacock S, Chaudhry AN, Bradley JA, Bolton EM. Generating an iPSC bank for HLA-matched tissue transplantation based on known donor and recipient HLA types. Cell Stem Cell. 2012;11:147-52
157. Xu H, Wang B, Ono M, Kagita A, Fujii K, Sasakawa N. et al. Targeted Disruption of HLA Genes via CRISPR-Cas9 Generates iPSCs with Enhanced Immune Compatibility. Cell Stem Cell. 2019;24:566-78 e7
158. Robinson J, Halliwell JA, Hayhurst JD, Flicek P, Parham P, Marsh SG. The IPD and IMGT/HLA database: allele variant databases. Nucleic Acids Res. 2015;43:D423-31
159. Fu Y, Reyon D, Joung JK. Targeted genome editing in human cells using CRISPR/Cas nucleases and truncated guide RNAs. Methods Enzymol. 2014;546:21-45
160. Fu YF, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32:279-84
161. Kuscu C, Arslan S, Singh R, Thorpe J, Adli M. Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat Biotechnol. 2014;32:677-83
162. Wang M, Zuris JA, Meng F, Rees H, Sun S, Deng P. et al. Efficient delivery of genome-editing proteins using bioreducible lipid nanoparticles. Proc Natl Acad Sci U S A. 2016;113:2868-73
163. Zuris JA, Thompson DB, Shu Y, Guilinger JP, Bessen JL, Hu JH. et al. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat Biotechnol. 2015;33:73-80
164. Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE. et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380-9
165. Hendel A, Bak RO, Clark JT, Kennedy AB, Ryan DE, Roy S. et al. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat Biotechnol. 2015;33:985-9
166. Ryan DE, Taussig D, Steinfeld I, Phadnis SM, Lunstad BD, Singh M. et al. Improving CRISPR-Cas specificity with chemical modifications in single-guide RNAs. Nucleic Acids Res. 2018;46:792-803
167. Konermann S, Brigham MD, Trevino A, Hsu PD, Heidenreich M, Cong L. et al. Optical control of mammalian endogenous transcription and epigenetic states. Nature. 2013;500:472-6
168. Davis KM, Pattanayak V, Thompson DB, Zuris JA, Liu DR. Small molecule-triggered Cas9 protein with improved genome-editing specificity. Nat Chem Biol. 2015;11:316-8
169. Guilinger JP, Thompson DB, Liu DR. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat Biotechnol. 2014;32:577-82
170. Tsai SQ, Wyvekens N, Khayter C, Foden JA, Thapar V, Reyon D. et al. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol. 2014;32:569-76
171. Zhang XH, Tee LY, Wang XG, Huang QS, Yang SH. Off-target Effects in CRISPR/Cas9-mediated Genome Engineering. Mol Ther Nucleic Acids. 2015;4:e264
172. Lee M, Kim H. Therapeutic application of the CRISPR system: current issues and new prospects. Hum Genet. 2019;138:563-90
173. Wyvekens N, Topkar VV, Khayter C, Joung JK, Tsai SQ. Dimeric CRISPR RNA-Guided FokI-dCas9 Nucleases Directed by Truncated gRNAs for Highly Specific Genome Editing. Hum Gene Ther. 2015;26:425-31
174. Lin YN, Cradick TJ, Brown MT, Deshmukh H, Ranjan P, Sarode N. et al. CRISPR/Cas9 systems have off-target activity with insertions or deletions between target DNA and guide RNA sequences. Nucleic Acids Res. 2014;42:7473-85
175. Wang XL, Wang YB, Wu XW, Wang JH, Wang YJ, Qiu ZJ. et al. Unbiased detection of off-target cleavage by CRISPR-Cas9 and TALENs using integrase-defective lentiviral vectors. Nat Biotechnol. 2015;33:175-8
176. Fujita T, Yuno M, Fujii H. Allele-specific locus binding and genome editing by CRISPR at the p16INK4a locus. Sci Rep. 2016;6:30485
177. Thyme SB, Akhmetova L, Montague TG, Valen E, Schier AF. Internal guide RNA interactions interfere with Cas9-mediated cleavage. Nat Commun. 2016;7:11750
178. Graham DB, Root DE. Resources for the design of CRISPR gene editing experiments. Genome Biol. 2015;16:260
179. Zhao G, Li J, Tang B, Tang Y. AsCRISPR: a web server for allele-specific sgRNA design in precision medicine. bioRxiv. 2019 doi: 10.1101/672634
180. Zhang JP, Li XL, Li GH, Chen W, Arakaki C, Botimer GD. et al. Efficient precise knockin with a double cut HDR donor after CRISPR/Cas9-mediated double-stranded DNA cleavage. Genome Biol. 2017;18:35
181. Liu M, Rehman S, Tang X, Gu K, Fan Q, Chen D. et al. Methodologies for Improving HDR Efficiency. Front Genet. 2018;9:691
182. Heyer WD, Ehmsen KT, Liu J. Regulation of homologous recombination in eukaryotes. Annu Rev Genet. 2010;44:113-39
183. Guan Y, Ma Y, Li Q, Sun Z, Ma L, Wu L. et al. CRISPR/Cas9-mediated somatic correction of a novel coagulator factor IX gene mutation ameliorates hemophilia in mouse. EMBO Mol Med. 2016;8:477-88
184. Long C, McAnally JR, Shelton JM, Mireault AA, Bassel-Duby R, Olson EN. Prevention of muscular dystrophy in mice by CRISPR/Cas9-mediated editing of germline DNA. Science. 2014;345:1184-8
185. Maeder ML, Stefanidakis M, Wilson CJ, Baral R, Barrera LA, Bounoutas GS. et al. Development of a gene-editing approach to restore vision loss in Leber congenital amaurosis type 10. Nat Med. 2019;25:229-33
186. Roman-Rodriguez FJ, Ugalde L, Alvarez L, Diez B, Ramirez MJ, Risueno C. et al. NHEJ-Mediated Repair of CRISPR-Cas9-Induced DNA Breaks Efficiently Corrects Mutations in HSPCs from Patients with Fanconi Anemia. Cell Stem Cell. 2019;25:607-21 e7
187. Cox DB, Platt RJ, Zhang F. Therapeutic genome editing: prospects and challenges. Nat Med. 2015;21:121-31
188. Finn JD, Smith AR, Patel MC, Shaw L, Youniss MR, van Heteren J. et al. A Single Administration of CRISPR/Cas9 Lipid Nanoparticles Achieves Robust and Persistent In Vivo Genome Editing. Cell Rep. 2018;22:2227-35
189. Lee J, Mou H, Ibraheim R, Liang SQ, Liu P, Xue W. et al. Tissue-restricted genome editing in vivo specified by microRNA-repressible anti-CRISPR proteins. RNA. 2019;25:1421-31
190. Liang P, Xu Y, Zhang X, Ding C, Huang R, Zhang Z. et al. CRISPR/Cas9- mediated gene editing in human tripronuclear zygotes. Protein Cell. 2015;6:363-72
191. Mehravar M, Shirazi A, Nazari M, Banan M. Mosaicism in CRISPR/Cas9- mediated genome editing. Dev Biol. 2019;445:156-62
Author contact
![]() Corresponding author: Dr. Yu Tang, tangyuskyedu.cn or tangyu-skycom
Corresponding author: Dr. Yu Tang, tangyuskyedu.cn or tangyu-skycom
 Global reach, higher impact
Global reach, higher impact