13.3
Impact Factor
Theranostics 2020; 10(7):3035-3048. doi:10.7150/thno.42559 This issue Cite
Research Paper
Inter- and intratumor DNA methylation heterogeneity associated with lymph node metastasis and prognosis of esophageal squamous cell carcinoma
1. Institute of Genomic Medicine, Wenzhou Medical University, Wenzhou, China
2. Beijing Institutes of Life Science, Chinese Academy of Sciences, Beijing, China
3. Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education/Beijing), Department of Radiation Oncology, Peking University Cancer Hospital & Institute, Beijing, China
4. University of Chinese Academy of Sciences, Beijing, China
5. Institute of Digestive Diseases, Tongji University School of Medicine, Shanghai, China
6. National Engineering Center for Biochip at Shanghai, Shanghai, China
#Authors contributed equally to this work.
Received 2019-11-28; Accepted 2020-1-22; Published 2020-2-10
Abstract
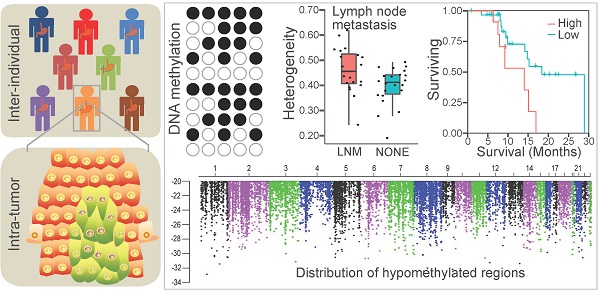
Background: Esophageal squamous cell carcinoma (ESCC), one of the leading causes of cancer mortality worldwide, is a heterogeneous cancer with diverse clinical manifestations. However, little is known about the epigenetic heterogeneity and its clinical relevance for this prevalent cancer.
Methods: We generated 7.56 Tb single-base resolution whole-genome bisulfite sequencing data for 84 ESCC and paired paraneoplastic tissues. The analysis identified inter- and intratumor DNA methylation (DNAm) heterogeneity, epigenome-wide DNAm alterations together with the functional regulators involved in the hyper- or hypomethylated regions, and their association with clinical features. We then validated the correlation between the methylation level of specific regions and clinical outcomes of 96 ESCC patients in an independent cohort.
Results: ESCC manifested substantial inter- and intratumor DNAm heterogeneity. The high intratumor DNAm heterogeneity was associated with lymph node metastasis and worse overall survival. Interestingly, hypermethylated regions in ESCC were enriched in promoters of numerous transcription factors, and demethylated noncoding regions related to RXR transcription factor binding appeared to contribute to the development of ESCC. Furthermore, we identified numerous DNAm alterations associated with carcinogenesis and lymph node metastasis of ESCC. We also validated three novel prognostic markers for ESCC, including one each in the promoter of CLK1, the 3' untranslated region of ZEB2, and the intergenic locus surrounded by several lncRNAs.
Conclusions: This study presents the first population-level resource for dissecting base-resolution DNAm variation in ESCC and provides novel insights into the ESCC pathogenesis and progression, which might facilitate diagnosis and prognosis for this prevalent malignancy.
Keywords: epigenetic heterogeneity, cancer mortality, whole-genome bisulfite sequencing, prognostic markers, personalized medicine
Introduction
Esophageal cancer is the ninth most prevalent malignancy and the sixth leading cause of cancer mortality worldwide [1]. Esophageal squamous cell carcinoma (ESCC), which constitutes greater than 80% of esophageal cancers, represents the most frequent histological type of esophageal cancer [2], and the incidence and mortality of ESCC exhibit considerable geographic variation [3, 4]. The majority of ESCC patients experience lymph node metastasis (LNM) or distal metastases at diagnosis, leading to poor outcomes for these patients (five-year survival rate < 20%) [2, 5]. Large-scale exome and genome sequencing have identified numerous genomic alterations in ESCC, including somatic mutations in TP53, PIK3CA, NOTCH1, and copy number alterations (CNAs) in pivotal RTK-MAPK-PI3K pathway genes [4, 6-10]. This information has yielded profound insights for precision diagnosis and treatment of this common cancer. However, genetic alterations alone are inadequate in explaining the complexity, prevalence, and pathogenesis of ESCC.
ESCC has been reported to harbor abundant inactivating mutations in histone-modifying and chromatin-remodeling regulators, KMT2D (MLL2), KDM6A (UTX), and KMT2C (MLL3) [11]. Genetic mutations in these epigenetic regulators might disrupt the entire epigenetic regulatory network, highlighting the significance of adjusting our focus beyond genetic alterations in ESCC pathogenesis. DNA methylation (DNAm), one of the well-characterized epigenetic modifications, coordinates various biological processes by regulating gene expression and posttranscriptional activity [12-14]. Some specific DNAm alterations have been established as hallmarks in specific cancers and can be used in the diagnosis of specific cancer types [15-20]. In particular, abnormal DNAm in oncogenes and tumor-suppressor genes are generally involved in all steps of tumorigenesis [21-23]. Compared to traditional irreversible genetic changes, DNAm can be reversed with drugs [24, 25], indicating their potential role as molecular targets for therapeutic intervention. Previous studies have investigated DNAm changes in ESCC based on methylation array data from The Cancer Genome Atlas (TCGA) project, identifying several specific DNAm alterations associated with clinical outcomes in ESCC patients [26, 27]. However, array-based technologies limit exhaustive screening of epigenome-wide DNAm alterations due to low genome coverage and low sensitivity of probe cross-hybridization. Thus, it is imperative to decipher the epigenome-wide high-resolution DNAm signatures of ESCC for clinical applications.
Inter- and intratumor genetic heterogeneity is a fundamental property of human cancers that confers a formidable barrier to cancer treatment [28]. Spatial genetic heterogeneity and clonal evolution in esophageal cancer have been demonstrated [29-31]; however, inter- and intratumoral epigenetic heterogeneity and their clinical relevance for ESCC is less well defined. Recent data in other cancer types have shown the power of DNAm sequencing for analyzing epigenetic heterogeneity. DNAm heterogeneity, quantified using the proportion of discordantly methylated reads (PDR), entropy, or epipolymorphism, has been linked to clinical variables in acute myeloid leukemia [32], chronic lymphocytic leukemia [33], glioblastoma [34], and Ewing sarcoma [35], suggesting the extensive involvement of DNAm heterogeneity in tumorigenesis and progression.
In this study, we performed whole-genome bisulfite sequencing (WGBS) on 84 ESCCs and paired paraneoplastic tissues to elucidate epigenetic heterogeneity at both inter- and intratumor levels as well as their association with clinical outcomes. Furthermore, we identified epigenome-wide DNAm alterations associated with carcinogenesis and LNM of ESCC. Our study provides novel insights into the ESCC pathogenesis and progression, which might facilitate its diagnosis and prognosis.
Materials and Methods
Clinical patients and samples
Eighty-four ESCC and paired paraneoplastic samples were provided by the National Engineering Center for Biochip at Shanghai (Shanghai, China), whose pathological and clinical features are shown in Supplementary Table S1. All tumor samples were examined by two experienced pathologists and ensured the carcinoma content greater than 70%. Samples were snap-frozen in liquid nitrogen and stored at -80°C for WGBS and targeted DNA sequencing. The study was approved by the Ethical Review Board of National Engineering Center for Biochip at Shanghai (ID: YB M-05-02), and clinical data were collected after patients provided informed consent.
Clinical data of 96 ESCC patients and their matched methylation array data were downloaded from the TCGA-ESCA cohort (https://portal.gdc.cancer.gov).
DNA extraction, targeted DNA sequencing, and identification of somatic mutations
Genomic DNA was extracted from frozen tissues using the DNeasy Blood & Tissue Kit (Qiagen, Shanghai, China) and was then used for targeted DNA sequencing and WGBS library construction. A mutation hotspot panel was designed to target 32 ESCC-related genes (Supplementary Table S2), which were reported to be frequently mutated in two cohorts [9, 11] or Cosmic database. Approximately 1 μg of genomic DNA was sheared using a Covaris S220 focused-ultrasonicator, resulting in fragments of 150~250 bp. End repair, dA-tailing, and adapter ligation were performed using KAPA Hyper Prep Kits. The ligation product was cleaned up and size-selected using Beckman Ampure XP Beads (Beckman Coulter, Brea, CA), followed by PCR to generate the whole-genome library, which was then hybridized with biotin-labeled probes of target regions and captured using Dynabeads™ MyOne™ Streptavidin T1, and amplified to generate targeted DNA library. Subsequently, the library was sequenced for Illumina 150-bp paired-end reads, and clean reads were aligned to a reference genome (hg38, GRCh38) using the Burrows-Wheeler Aligner (BWA) (http://bio-bwa.sourceforge.net/). After local realignment, somatic single nucleotide variants were identified using MuTect (https://software.broadinstitute.org/cancer/cga/mutect), and annotated using VarCards [36].
WGBS and methylation analysis
MethylC-seq protocol was used to prepare WGBS sequencing libraries. First, approximately 1 μg genomic DNA was fragmented into 400 bp fragments using a Covaris S220 focused-ultrasonicator. Fragmented DNA was end-repaired, and dA tailed and then ligated to Illumina TruSeq adapter (all Cs methylated) using the KAPA Hyper Prep Kit (KAPA Biosystems, Wilmington, MA, USA). The adapter-ligated DNA was purified with beads and eluted with elution buffer. The EpiTect Fast DNA Bisulfite Kit (Qiagen, Shanghai, China) was used to convert the adapter-ligated DNA. Finally, bisulfite-treated DNA was amplified using 2× KAPA HiFi Uracil+ Readymix (KAPA Biosystems, Wilmington, MA, USA) to produce the WGBS library. Each library was sequenced using the Illumina HiSeq X Ten platform and generated 150 bp paired-end reads. Raw sequencing datasets were deposited in the Sequence Read Archive of NCBI (http://www.ncbi.nlm.nih.gov/sra) under accession number PRJNA523898. Sequencing quality was assessed using the FastQC software (Babraham Bioinformatics, Cambridge, UK). All sequencing reads were aligned to the human reference genome GRCh38/hg38 using BsMap (http://code.google.com/p/bsmap/). Methylation site calling and methylation levels of each CpG site were determined using BisSNPV.0.82.2 software (https://sourceforge.net/projects/bissnp). Whole-genome differentially methylated regions (DMRs) were detected using metilene software [37]. The dimension reduction analysis of tumor and normal tissues based on the methylation levels of 53,995 reference genes (Ensembl v78) was performed using multidimensional scaling (MDS) in Euclidean distance with the R package RnBeads [38].
Chromatin immunoprecipitation sequencing data analysis
ChIP-seq data of two ESCC cell lines (TE7, KYSE510) from a previous study [39] were mapped to the human reference genome (GRCh38/hg38) using Bowtie (http://bowtie-bio.sourceforge.net/index.shtml). ChIP-Seq peaks were determined with read pileups for every 50 bp bins using MACS (http://liulab.dfci.harvard.edu/MACS/). The generated wiggle files were normalized in terms of reads per million (rpm) and then transformed into bigwig format files using the wigToBigWig tool (http://hgdownload.cse.ucsc.edu/admin/exe/). H3K27ac bigwig tracks were visualized in the UCSC Genome Browser (http://genome.ucsc.edu/cgi-bin/hgTracks).
Bioinformatic analysis of heterogeneity among tumor samples
Inter-individual DNAm heterogeneity quantified by the coefficient of variation (CV) of the tumor and paraneoplastic tissues was measured according to the previously described method [35]. The methylation level of each 5-kb tiling region was averaged only when the region was covered by more than ten sequencing reads, and then the CV for each sample was calculated according to the methylation levels of these genome-wide tiling regions.
Bioinformatic analysis of heterogeneity within individual tumors
Intratumor DNAm heterogeneity was quantified by PDR, entropy, or epipolymorphism. The PDR score was calculated as the proportion of discordant reads, containing both methylated and unmethylated CpGs, among all WGBS reads within a local region that covered at least four CpGs [33]. PDR was determined only when each CpG was covered by more than ten reads. The epi-allele entropy and epipolymorphism were calculated using a slightly modified version of methclone (https://code.google.com/p/methclone). Input files to methclone were created by aligning the WGBS reads to the human reference genome (hg38) using Bismark (https://www.bioinformatics.babraham.ac.uk/projects/bismark/).
Region set and functional enrichment analysis
Region set enrichment analysis among differentially methylated regions was determined using Locus Overlap Analysis (LOLA) [40]. P-values were corrected for multiple testing using the Benjamini and Yekutieli method, and all enrichments with an adjusted p-value below 0.05 were considered significant. Functional enrichment analysis for host genes of promoter-associated DMRs was performed using BiNGO (https://www.psb.ugent.be/cbd/papers/BiNGO/Home.html) with an adjusted p-value below 0.05.
Patient survival analysis
Kaplan-Meier survival was performed using the R package 'survival'. Significance in overall or disease-free survival was calculated using the log-rank test. Cox proportional hazards regression was performed using the function coxph () from the R package 'survival'.
Somatic copy-number alteration analysis
Somatic CNAs of each paired tumor and paraneoplastic samples was performed using R package SaasCNV (https://zhangz05.u.hpc.mssm.edu/saasCNV/). Recurrent focal somatic CNAs were identified by the GISTIC 2.0 algorithm (http://software.broadinstitute.org/cancer/software/genepattern/modules/docs/GISTIC_2.0).
Results
Epigenome-wide alterations of DNAm in ESCC
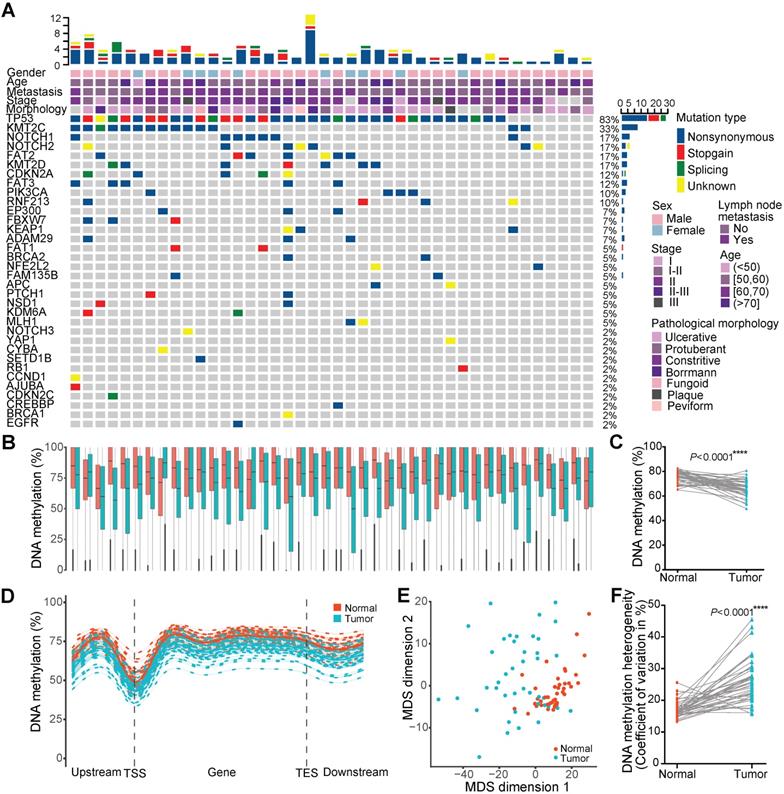
Single-base resolution DNAm profiles can provide an unbiased global view of the DNAm landscape. To gain a comprehensive insight into variations in the DNA methylome in ESCC, we performed WGBS on 84 ESCCs and paired paraneoplastic tissues from 42 patients with a median age of 58 and median overall survival of 9.18 months (Figure 1A). Seventy-one percent of these patients were TNM stage II, and 48% had LNM. Targeted DNA amplicon sequencing showed 83% of them harbored TP53 mutations (Figure 1A). Over 7.56 Tb sequencing data produced by WGBS from these patients were mapped to a reference genome (Hg38) using Bsmap, providing a median coverage of 24.92× per sample (range 17.04-39.77×; Supplementary Table 1). Epigenome-wide alterations of DNAm were observed in ESCC tissues, and the average CpG methylation levels of normal paraneoplastic tissues and cancer tissues were 76.31% and 66.36%, respectively (Figure 1B, C). In general, the methylation levels of transcription start sites were the lowest, but with the marked elevation in the gene body region in both tissues. The global methylation levels in all regions were systematically reduced in cancer tissues compared to paraneoplastic tissues (Figure 1D). Multidimensional scaling analysis based on the methylation level of all reference genes was performed to assess the similarity of individuals within the dataset. The multidimensional scaling plot discriminated the majority of malignant and adjacent benign tissues from ESCC patients, with normal tissues clustering closer together than cancer tissues (Figure 1E), indicating highly variable methylation patterns across cancer tissues. These findings were confirmed by interindividual heterogeneity quantified using the coefficient of variation for DNAm level throughout the genome (Figure 1F). The average DNAm heterogeneity level in ESCC patients was 0.26, with a range from 0.16 to 0.46, which is higher than that of several recognized heterogeneous cancer types, including prostate cancer and chronic lymphocytic leukemia (CLL) [35]. These results revealed substantial DNAm heterogeneity among ESCC patients, highlighting the importance of considering nongenetic aspects of tumor heterogeneity in pathogenesis and therapy of ESCC.
DNAm alterations associated with ESCC carcinogenesis
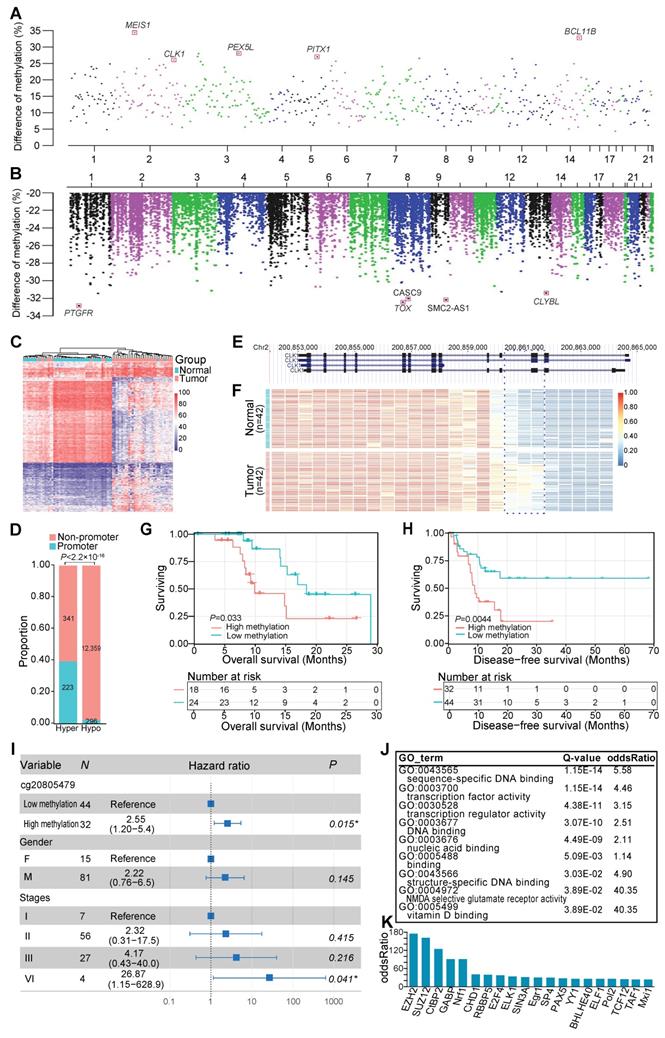
To dissect the DNAm changes associated with ESCC carcinogenesis, we performed differential DNAm analysis on all matched tumor and normal tissues. Genome-wide DNAm changes between cancer and normal tissues revealed 13,219 differentially methylated regions (DMRs) (Figure 2A, B), 95.73% of which were hypomethylated in cancer tissues (Figure 2B). In light of the importance of promoter regions in regulating the gene expression, we performed hierarchical cluster analysis based on methylation values of 519 aberrantly methylated promoter regions and observed that it could discriminate the majority of malignant and adjacent tissues (Figure 2C). Interestingly, our data showed that 39.54% of 564 hypermethylated regions resided in promoter regions, whereas only 3% of 12,655 hypomethylated regions were identified in the promoter regions (Figure 2D). These data indicated that hyper- and hypomethylated changes had distinct genomic distribution, and increased DNAm alteration tended to occur within promoters. For example, one of the strongest tumor-specific hypermethylated signals on chromosome 2 was located in the promoter or the intron of CDC-like kinase 1 (CLK1) transcripts (Figure 2 A, E, F). The methylation level of the CLK1 locus was predicted to be negatively correlated with overall survival of the ESCC patients (Figure 2G) and disease-free survival of ESCC patients in the TCGA-ESCA cohort (Figure 2H), even after adjusting for multiple clinical factors, including gender and TNM staging (Figure 2I). To understand the overall functional relevance of genes containing hypermethylated promoters, we performed gene ontology enrichment analysis. Genes related to sequence-specific DNA binding and transcription factor activity were found to be significantly enriched (Figure 2J), and some of them were well-known tumor suppressors, such as BCL11B [41] and PITX1 [42]. Furthermore, transcription factor binding site enrichment analysis for all identified hypermethylated regions revealed 20 significantly enriched transcription factors including several well-established ESCC-associated genes (Figure 2K), such as EZH2 [43], SUZ12, and CtBP2 [44]. These findings highlighted the widespread involvement of transcriptional regulators in the pathogenesis of ESCC.
Epigenome-wide changes of DNA methylation in ESCC. (A) Landscape of somatic genetic mutations in 42 ESCC patients. Each row denotes a gene, and each column represents an individual tumor. The uppermost scale indicates the number of identified mutations (y-axis) for each patient (x-axis). The top five rows of the x-axis indicate key clinical parameters for each patient. The right side of the y-axis shows the percentage of samples with mutations for each gene. Mutation type and clinical characteristics are represented by different colors. DNA methylation (B) and global DNA methylation (C) levels between 42 tumors and paired normal tissues. (D) Metaplot of CpG methylation levels across gene bodies. TSS, transcription start site; TES, transcription end site. (E) Multidimensional scaling plot of the tumor and normal tissues based on the methylation levels of all reference genes. (F) Interindividual DNA methylation heterogeneity quantified by the coefficient of variation between 42 tumors and paired normal tissues.
DNA methylation alteration within promoters in ESCC. Genome-wide distribution of significantly hypermethylated (A) and hypomethylated (B) regions in ESCC. (C) Hierarchical clustering for methylation values of aberrantly methylated promoters. (D) Proportion of promoter-associated DMRs to aberrantly hypermethylated (hyper) or hypomethylated (hypo) regions. Example of tumor-specific hypermethylation at the promoter or the intron of CLK1 transcripts (E, F), one of the strongest genome-wide signals on chromosome 2 (A). Kaplan-Meier plot showing overall survival (G) and disease-free survival (H) stratified by ESCC patients of our sequenced (G) and TCGA-ESCA cohort (H) according to methylation levels of the CLK1 promoter locus, respectively. (I) Multivariate Cox regression analysis of methylation levels based on a probe (cg20805479) within the CLK1 promoter locus after controlling for gender and TNM stage. (J) Functional enrichment of genes with differentially hypermethylated promoters in ESCC. (K) Transcription factors binding site enrichment of aberrantly hypermethylated promoter regions.
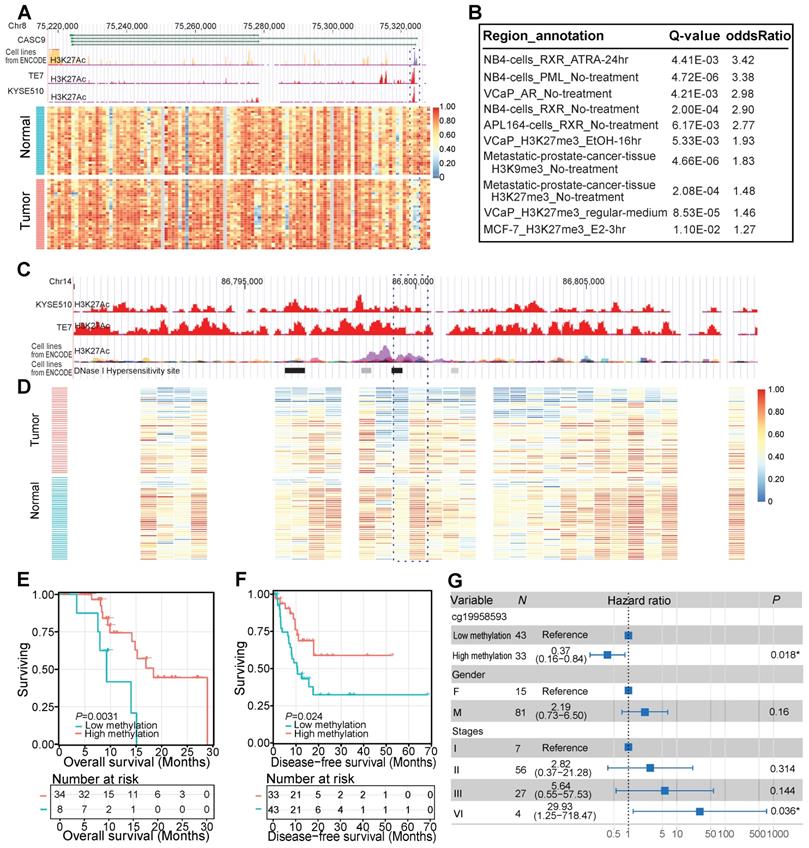
As mentioned above, the majority of all identified DMRs were hypomethylated in cancer tissues. One example of such a tumor-specific hypomethylated region was located in the promoter of a well-known ESCC-implicated long noncoding RNA (lncRNA), CASC9 (Figure 3A). This tumor-associated lncRNA was reported to function as an oncogene by downregulating the expression of PDCD4 through recruiting EZH2 and altering trimethylation levels of H3K27 in ESCC cells [45]. From the perspective of DNAm, our study highlighted the critical role of CASC9 as a valuable marker for ESCC diagnosis and prognosis [45, 46]. Furthermore, we observed that 55.93% of identified hypomethylated regions resided in distal intergenic regions. We performed region set enrichment analysis of these DMRs using LOLA [40] to detect enriched regulators based on collected regulatory region sets of chromatin immunoprecipitation sequencing peaks [47, 48] and DNaseI hypersensitive elements [49]. As expected, DMRs located within these intergenic regions were enriched for H3K27me3 modification sites (Figure 3B), a mark associated with chromosome inactivation [50]. Interestingly, DMRs within these noncoding regions were also heavily enriched for binding sites of retinoid X receptor (RXR) (Figure 3B), which co-occupies the active enhancers defined by H3K27ac [51], suggesting the modulating role of RXR during carcinogenesis and development of ESCC. One example of such DMRs was located on Chr14: 86,799,804-86,800,434 (Figure 3C, D) surrounded by several lncRNAs, including LINC01148 and LINC02309. Decreased DNAm of this region was observed in ESCC tissues of our sequenced samples (Figure 3D). Another observation in ESCC cell lines in this region was of the high level of histone H3K27 acetylation (Figure 3C), a chromatin marker associated with gene activation and active enhancers [52]. Also, a peak of the DNase I hypersensitivity site, a reflection of chromatin accessibility, was observed in this region in cell lines from the ENCODE database (Figure 3C). The methylation levels of this intergenic locus were predicted to be positively correlated with the overall survival of the sequenced ESCC patients (Figure 3E). This contention was further validated in ESCC patients from another independent TCGA-ESCA cohort (Figure 3F) even after adjusting for multiple clinical factors, including gender and TNM staging (Figure 3G). These data, together with other reports [39, 53, 54], suggest a regulatory role for these noncoding regions in ESCC pathogenesis, highlighting the importance of further analysis of these regions.
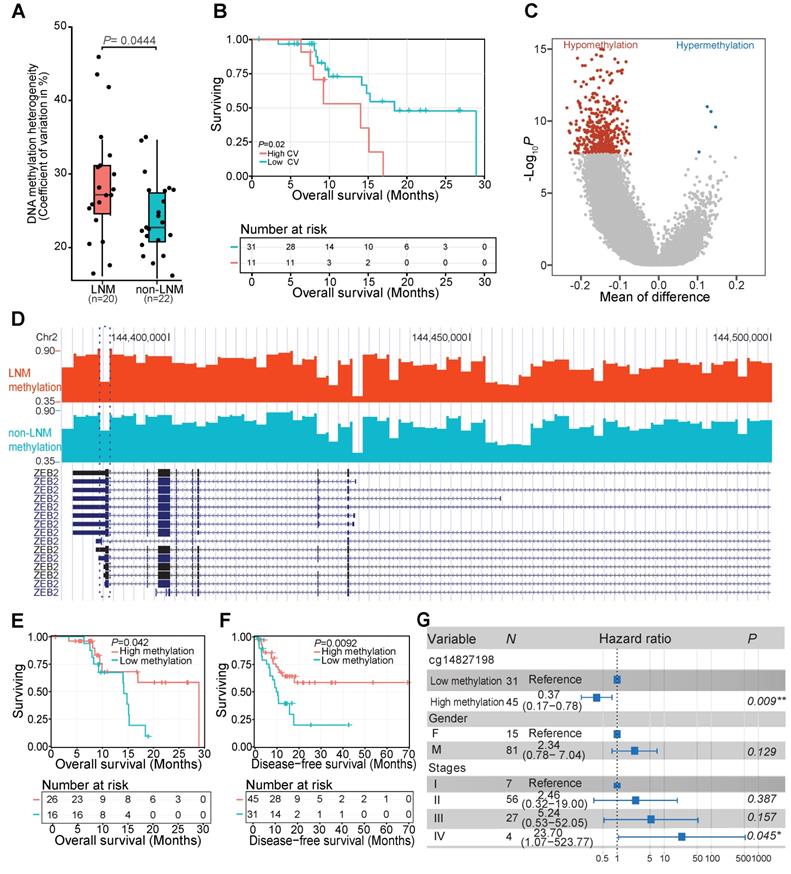
DNAm changes associated with LNM of ESCC patients
To further identify the relationship between DNAm variation and clinical variables, we explored the association between interindividual DNAm heterogeneity and clinical data. We found that patients with LNM, an early event associated with poor prognosis in ESCC patients [2, 5], exhibited higher interindividual DNAm heterogeneity than patients without LNM (Figure 4A). Kaplan-Meier plot showed that ESCC patients with high inter-individual DNAm heterogeneity experienced a worse overall survival (Figure 4B). We then compared the methylomes of patients with or without LNM, and identified 490 DMRs associated with LNM features of ESCC (Figure 4C) (Supplementary Table S3). For example, decreased DNAm in patients with LNM was observed at 3' untranslated region of ZEB2 (Figure 4D). The low methylation levels of this locus were predicted to correlate with worse overall survival of ESCC patients in our sequenced (Figure 4E) and TCGA-ESCA cohorts (Figure 4F) even after adjusting for multiple clinical factors (Figure 4G). It was previously demonstrated that ZEB2 could promote metastasis of gastric cancer and colorectal cancer, a modulated epithelial-mesenchymal transition of gastric cancer cells, and was associated with poor prognosis of colorectal cancer [55, 56]. Identification of these DMRs provides valuable markers associated with LNM status that might help predict ESCC prognosis in patients with LNM.
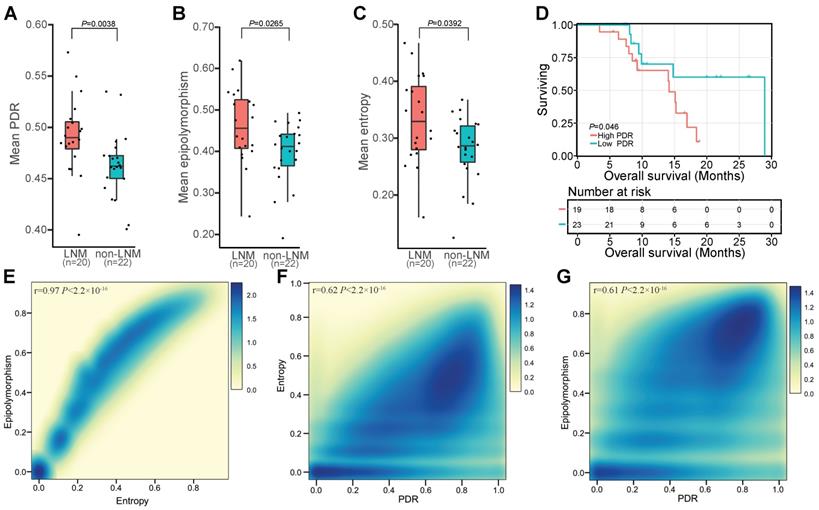
Widespread intratumor DNAm heterogeneity in patients with LNM
In view of the widespread inter-individual DNAm heterogeneity in ESCC, we investigated whether DNAm differences existed between cells within the same tumor. Three methods, PDR, entropy, and epipolymorphism, were used to evaluate intratumor heterogeneity in ESCC. The PDR score is considered to be an indicator of epigenetic instability that might contribute to the clonal selection of individual cells [33, 35]. We observed higher locally disordered DNAm heterogeneity in patients with LNM than without LNM (Figure 5A). Epipolymorphism is a measurement of the observed consistency of a given methylation pattern within a region versus the expected random pattern and can be used to measure the level of overall epigenetic dysregulation of a specific sample [57-59]. Elevated overall epipolymorphism values, reflecting higher heterogeneity, were observed in patients with LNM (Figure 5B). Increased sub-clonal variety measured by the epiallele entropy was also observed in patients with LNM (Figure 5C). Regions with high epiallele entropy and high epipolymorphism also exhibited higher PDR values, indicating agreement between these methods (Figure 5E, F, G) and supporting the conclusion that intratumor DNAm heterogeneity was higher in patients with LNM than in patients without LNM (Figure 5A, B, C). To further elucidate the impact of intratumor DNAm heterogeneity on clinical outcome, we performed Kaplan-Meier survival analysis of our sequenced patients stratified according to the PDR score. The PDR level was predicted to correlate with the overall survival of ESCC patients, and patients with a high PDR level exhibited worse overall survival (Figure 5D). This finding suggested the prognostic value of intratumor DNAm heterogeneity for ESCC patients.
Hypomethylated changes within noncoding regions in ESCC. (A) Tumor-specific hypomethylation at the CASC9 promoter locus with decreased DNA methylation in the sequenced ESCC tumors and high histone H3K27 acetylation in ESCC cell lines. H3K27ac profiles include a cross-tissue consensus track from the ENCODE database and ChIP-seq data from two ESCC cell lines (TE7, KYSE510) from a previous study [39]. (B) Significant overlap of aberrantly hypomethylated intergenic regions with public annotation data of cancer cell lines, based on LOLA Core enrichment analysis [40]. Tumor-specific hypomethylation at the intergenic locus (Chr14: 86,799,804-86,800,434) with decreased DNA methylation in the sequenced ESCC tumors (D) and high histone H3K27 acetylation in ESCC and ENCODE cell lines (C). Kaplan-Meier plots showing overall survival (E) and disease-free survival (F) stratified for the sequenced ESCC patients (E) and the TCGA-ESCA cohort (F) according to methylation levels of the intergenic locus (Chr14: 86,799,804-86,800,434). (G) Multivariate Cox regression analysis based on methylation levels of a probe (cg19958593) within the intergenic locus (Chr14: 86,799,804-86,800,434) after controlling for gender and TNM stage.
DNA methylation changes associated with LNM in ESCC patients. (A) Interindividual DNA methylation heterogeneity quantified by the coefficient of variation (CV) between patients with or without LNM. The coefficient of variation across each genome was calculated as a measure of heterogeneity between samples. (B) Kaplan-Meier plot showing overall survival stratified by subjects from the sequenced ESCC patients according to the interindividual DNA methylation heterogeneity level. (C) Distribution of differentially methylated regions between tissues of patients with or without LNM. Red and blue dots correspond to aberrantly hypomethylated and hypermethylated regions, respectively. (D) LNM-specific hypomethylation at the 3' untranslated region of ZEB2 in patients with LNM. Kaplan-Meier plots showing overall survival (E) and disease-free survival (F) stratified according to the methylation level of 3' untranslated region of ZEB2 in the sequenced ESCC patients (E) and TCGA-ESCA cohort (F). (G) Multivariate Cox regression analysis based on the methylation level of a probe (cg14827198) within the 3' untranslated region of ZEB2 after controlling for gender and TNM stage.
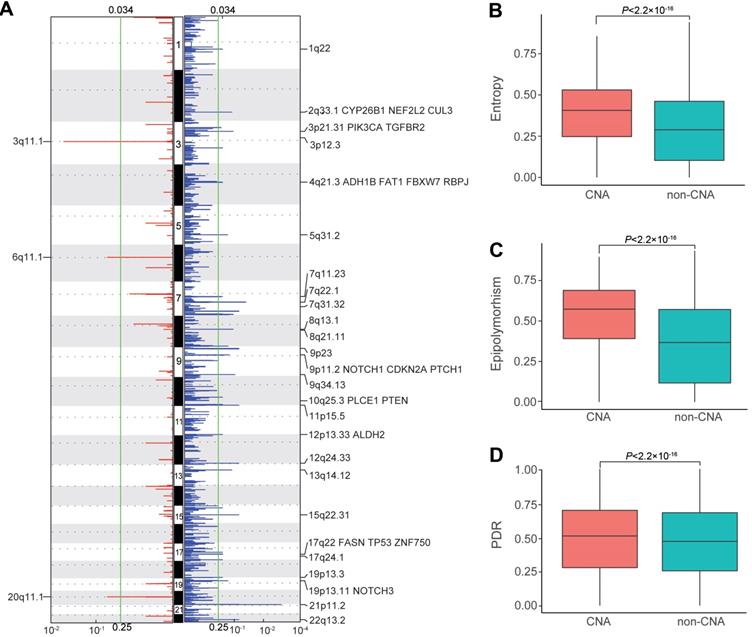
Intratumor DNAm heterogeneity within copy number alteration (CNA) regions of ESCC patients
Considering the pivotal role of copy number alterations in the pathogenesis of ESCC [4, 6-10], we compared intratumor DNAm heterogeneity within CNA and non-CNA regions of ESCC patients. We first identified somatic CNAs of each patient from the WGBS data and then analyzed somatic CNA data with Gistic2 to define recurrently amplified and deleted regions. Consistent with previous studies, we detected several well-defined ESCC-associated CNAs (Figure 6A), including CYP26B1 [60], CUL3 [8], ADH1B, and ALDH2 [61]. Tumor tissues had an average entropy level of 0.3812 within CNA regions but displayed a much-decreased level of 0.2924 in the non-CNA regions. Also, CNA regions with higher entropy exhibited higher epipolymorphism and PDR values (Figure 6B, C, D). These results revealed that intratumor DNAm heterogeneity varied across genome segments of ESCC patients, and was higher in CNA regions compared to that in non-CNA regions, indicating a close link between epigenetic variations and structural variability.
DNA methylation patterns identify widespread intratumor heterogeneity in patients with LNM. Distribution of sample-wise discordantly methylated read (PDR) scores (A), epipolymorphism (B), and entropy (C) between patients with or without LNM. (D) Kaplan-Meier plots showing overall survival stratified according to PDR score in subjects from the sequenced patients. Density scatterplot showing the relationship between epipolymorphism and entropy (E), entropy and PDR (F), and epipolymorphism and PDR (G) for 5-kb tiling regions.
Discussion
Inter- and intratumor heterogeneity fuels resistance to therapy in multiple cancers [28]. Thus, detailed knowledge of tumor heterogeneity is beneficial for the clinical management of cancer patients. ESCC, one of the leading causes of cancer mortality worldwide and a heterogeneous cancer with diverse clinical manifestations [62], represents an ideal model to investigate tumor heterogeneity and progression. Recently, it has been reported that ESCC displays higher intratumor mutational heterogeneity than several other cancer types, including esophageal adenocarcinoma (EAC), which is another predominant histopathological subtype of esophageal cancer [63]. Also, analysis of ESCC by multiple region whole-exome sequencing to analyze intratumor heterogeneity identified several heterogeneous somatic mutations in tumor-suppressor genes such as TP53, ZNF750, and KMT2D [29, 64]. A higher intratumor heterogeneity in EAC has been reported to be associated with a poor response to neoadjuvant chemotherapy [30]. However, when mutant-allele tumor heterogeneity (MATH) [65] based on whole-exome sequencing data of ESCC patients in the TCGA database was analyzed, no significant association was observed between intratumor heterogeneity and clinical outcomes, including overall survival, progression-free survival, and disease-specific survival (Supplementary Figure S1). Thus, comprehensive analysis beyond the level of genetic heterogeneity is required to explain the poor overall 5-year survival rates of ESCC. Herein, using single-base resolution WGBS, we identified population-level DNAm variations in ESCC, which manifested greater inter-individual DNAm heterogeneity than prostate cancer or chronic lymphocytic leukemia, two well-recognized heterogeneous cancer types [35]. We also observed increased inter-individual and intra-tumor DNAm heterogeneity in patients with LNM, which is a clinical feature tightly correlated with ESCC patient prognosis [2, 5]. This was consistent with the observation that patients with more aggressive disease tend to exhibit high tumor heterogeneity [66]. Furthermore, our data demonstrated that ESCC patients with high inter- or intratumor DNAm heterogeneity experienced worse overall survival. Similarly, a high level of DNAm heterogeneity was also observed to be associated with adverse clinical outcomes of chronic lymphocytic leukemia patients [58]. Together, these data suggest the prognostic value of inter- and intratumor epi-heterogeneity for ESCC patients, although a larger cohort is still needed to validate these findings.
Intratumor DNA methylation heterogeneity within copy number alteration (CNAs) regions of ESCC patients. (A) Regions of recurrent focal amplifications (left) and focal deletions (right) are plotted by the false discovery rate (x-axis) for each chromosome (y-axis). Distribution of entropy (B), epipolymorphism (C), and discordantly methylated read (PDR) scores (D) across the genome of ESCC patients between CNA and non-CNA regions
Consistent with the systematically reduced DNAm level in cancers, we observed that the majority of identified DMRs were hypomethylated in ESCC tissues. Compared with the hypomethylated regions in ESCC, the identified hypermethylated regions were enriched in promoters of transcription factors, including BCL11B and PITX1, two well-known tumor suppressors [41, 42]. Furthermore, transcription factor binding site enrichment analysis for all identified hypermethylated regions revealed several well-known ESCC-associated genes, including EZH2 [43], SUZ12, and CtBP2 [44]. These data suggested the essential role of transcription factors during the pathogenesis of ESCC.
Interestingly, we observed that more than half of the identified hypomethylated regions in ESCC were located in the distal intergenic regions, which were greatly enriched for binding sites of retinoid X receptor (RXR) that co-occupies active enhancers defined by H3K27ac [51]. Regional enrichment of these demethylated regions suggests a correlation between DNA hypomethylation and activation of histone markers in ESCC. This notion is consistent with a previous study showing that DNA hypermethylation in super-enhancers of ESCC reduced active histone markers [39]. The methylation levels of several intergenic loci were predicted to be correlated with the overall survival of our sequenced ESCC patients, which was further validated in ESCC patients from another independent cohort, the TCGA-ESCA, even after adjusting for multiple clinical factors, including gender and TNM staging. Additionally, increased lncRNA expression in noncoding regions is reported to promote ESCC cell proliferation, migration, invasion, and growth of xenograft tumors [53]. Our data, together with previous reports [39, 53, 54], suggest a regulatory role for these noncoding regions in ESCC pathogenesis, highlighting the importance of further analysis of these regions.
Conclusions
In summary, we presented high-resolution, population-level DNAm variations in ESCC, provided evidence for widespread intratumor DNAm heterogeneity among ESCC patients, and identified numerous DNAm alterations associated with ESCC carcinogenesis and progression. Our study might facilitate a better prognosis of this prevalent disease and provide a translational basis for designing personalized medicine strategies.
Abbreviations
ESCC: Esophageal squamous cell carcinoma; DNAm: DNA methylation; LNM: lymph node metastasis; CNAs: copy number alterations; TCGA: The Cancer Genome Atlas; PDR: discordantly methylated reads; WGBS: whole-genome bisulfite sequencing; DMRs: differentially methylated regions; MDS: multidimensional scaling; CV: coefficient of variation; CLL: chronic lymphocytic leukemia; CLK1: CDC like kinase 1.
Supplementary Material
Supplementary figure and tables.
Acknowledgements
This work was supported by grants from the National Key R&D Program of China (No.2016YFC0900400) and the National Natural Science Foundation of China (No.31872237).
Availability of data and materials
Raw sequencing datasets were deposited in the Sequence Read Archive of NCBI (http://www.ncbi.nlm.nih.gov/sra) under accession number PRJNA523898.
Study approval
This study was approved by the Ethical Review Board of National Engineering Center for Biochip at Shanghai (ID: YB M-05-02), and clinical data were collected after patients provided informed consent.
Author Contributions
HT, MY, LW, WW, CL, XR, XS, and WW performed data analysis. HG provided samples. JL, XW, ZZ, QL, and WC performed experiments. HT, HG, and ZS designed experiments and drafted the initial manuscript. All authors read and approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424
2. Enzinger PC, Mayer RJ. Esophageal cancer. N Engl J Med. 2003;349:2241-52
3. Rustgi AK, El-Serag HB. Esophageal carcinoma. N Engl J Med. 2014;371:2499-509
4. Zhang L, Zhou Y, Cheng C, Cui H, Cheng L, Kong P. et al. Genomic analyses reveal mutational signatures and frequently altered genes in esophageal squamous cell carcinoma. Am J Hum Genet. 2015;96:597-611
5. Arnold M, Soerjomataram I, Ferlay J, Forman D. Global incidence of oesophageal cancer by histological subtype in 2012. Gut. 2015;64:381-7
6. Agrawal N, Jiao Y, Bettegowda C, Hutfless SM, Wang Y, David S. et al. Comparative genomic analysis of esophageal adenocarcinoma and squamous cell carcinoma. Cancer Discov. 2012;2:899-905
7. Wu C, Wang Z, Song X, Feng XS, Abnet CC, He J. et al. Joint analysis of three genome-wide association studies of esophageal squamous cell carcinoma in Chinese populations. Nat Genet. 2014;46:1001-6
8. Chang J, Tan W, Ling Z, Xi R, Shao M, Chen M. et al. Genomic analysis of oesophageal squamous-cell carcinoma identifies alcohol drinking-related mutation signature and genomic alterations. Nat Commun. 2017;8:15290
9. Lin DC, Hao JJ, Nagata Y, Xu L, Shang L, Meng X. et al. Genomic and molecular characterization of esophageal squamous cell carcinoma. Nat Genet. 2014;46:467-73
10. Sawada G, Niida A, Uchi R, Hirata H, Shimamura T, Suzuki Y. et al. Genomic Landscape of Esophageal Squamous Cell Carcinoma in a Japanese Population. Gastroenterology. 2016;150:1171-82
11. Song Y, Li L, Ou Y, Gao Z, Li E, Li X. et al. Identification of genomic alterations in oesophageal squamous cell cancer. Nature. 2014;509:91-5
12. Pan XY, You HM, Wang L, Bi YH, Yang Y, Meng HW. et al. Methylation of RCAN1.4 mediated by DNMT1 and DNMT3b enhances hepatic stellate cell activation and liver fibrogenesis through Calcineurin/NFAT3 signaling. Theranostics. 2019;9:4308-23
13. Zhao T, Bao Y, Gan X, Wang J, Chen Q, Dai Z. et al. DNA methylation-regulated QPCT promotes sunitinib resistance by increasing HRAS stability in renal cell carcinoma. Theranostics. 2019;9:6175-90
14. Chen K, Liu MX, Mak CS, Yung MM, Leung TH, Xu D. et al. Methylation-associated silencing of miR-193a-3p promotes ovarian cancer aggressiveness by targeting GRB7 and MAPK/ERK pathways. Theranostics. 2018;8:423-36
15. Yu M, Maden SK, Stachler M, Kaz AM, Ayers J, Guo Y. et al. Subtypes of Barrett's oesophagus and oesophageal adenocarcinoma based on genome-wide methylation analysis. Gut. 2018
16. Wong CC, Kang W, Xu J, Qian Y, Luk STY, Chen H. et al. Prostaglandin E2 induces DNA hypermethylation in gastric cancer in vitro and in vivo. Theranostics. 2019;9:6256-68
17. Long J, Chen P, Lin J, Bai Y, Yang X, Bian J. et al. DNA methylation-driven genes for constructing diagnostic, prognostic, and recurrence models for hepatocellular carcinoma. Theranostics. 2019;9:7251-67
18. Hlady RA, Zhao X, Pan X, Yang JD, Ahmed F, Antwi SO. et al. Genome-wide discovery and validation of diagnostic DNA methylation-based biomarkers for hepatocellular cancer detection in circulating cell free DNA. Theranostics. 2019;9:7239-50
19. Liang W, Zhao Y, Huang W, Gao Y, Xu W, Tao J. et al. Non-invasive diagnosis of early-stage lung cancer using high-throughput targeted DNA methylation sequencing of circulating tumor DNA (ctDNA). Theranostics. 2019;9:2056-70
20. Gampenrieder SP, Rinnerthaler G, Hackl H, Pulverer W, Weinhaeusel A, Ilic S. et al. DNA Methylation Signatures Predicting Bevacizumab Efficacy in Metastatic Breast Cancer. Theranostics. 2018;8:2278-88
21. Heyn H, Vidal E, Ferreira HJ, Vizoso M, Sayols S, Gomez A. et al. Epigenomic analysis detects aberrant super-enhancer DNA methylation in human cancer. Genome Biol. 2016;17:11
22. Li L, Xu J, Qiu G, Ying J, Du Z, Xiang T. et al. Epigenomic characterization of a p53-regulated 3p22.2 tumor suppressor that inhibits STAT3 phosphorylation via protein docking and is frequently methylated in esophageal and other carcinomas. Theranostics. 2018;8:61-77
23. Wang T, Liu Q, Li X, Wang X, Li J, Zhu X. et al. RRBS-analyser: a comprehensive web server for reduced representation bisulfite sequencing data analysis. Hum Mutat. 2013;34:1606-10
24. Yang X, Lay F, Han H, Jones PA. Targeting DNA methylation for epigenetic therapy. Trends Pharmacol Sci. 2010;31:536-46
25. Yeon A, You S, Kim M, Gupta A, Park MH, Weisenberger DJ. et al. Rewiring of cisplatin-resistant bladder cancer cells through epigenetic regulation of genes involved in amino acid metabolism. Theranostics. 2018;8:4520-34
26. Hoshimoto S, Takeuchi H, Ono S, Sim MS, Huynh JL, Huang SK. et al. Genome-wide hypomethylation and specific tumor-related gene hypermethylation are associated with esophageal squamous cell carcinoma outcome. J Thorac Oncol. 2015;10:509-17
27. Cancer Genome Atlas Research N, Analysis Working Group. Asan U, Agency BCC, Brigham, Women's H, Broad I, et al. Integrated genomic characterization of oesophageal carcinoma. Nature. 2017;541:169-75
28. Dagogo-Jack I, Shaw AT. Tumour heterogeneity and resistance to cancer therapies. Nat Rev Clin Oncol. 2018;15:81-94
29. Hao JJ, Lin DC, Dinh HQ, Mayakonda A, Jiang YY, Chang C. et al. Spatial intratumoral heterogeneity and temporal clonal evolution in esophageal squamous cell carcinoma. Nat Genet. 2016;48:1500-7
30. Murugaesu N, Wilson GA, Birkbak NJ, Watkins T, McGranahan N, Kumar S. et al. Tracking the genomic evolution of esophageal adenocarcinoma through neoadjuvant chemotherapy. Cancer Discov. 2015;5:821-31
31. Findlay JM, Castro-Giner F, Makino S, Rayner E, Kartsonaki C, Cross W. et al. Differential clonal evolution in oesophageal cancers in response to neo-adjuvant chemotherapy. Nat Commun. 2016;7:11111
32. Li S, Garrett-Bakelman FE, Chung SS, Sanders MA, Hricik T, Rapaport F. et al. Distinct evolution and dynamics of epigenetic and genetic heterogeneity in acute myeloid leukemia. Nat Med. 2016;22:792-9
33. Landau DA, Clement K, Ziller MJ, Boyle P, Fan J, Gu H. et al. Locally disordered methylation forms the basis of intratumor methylome variation in chronic lymphocytic leukemia. Cancer Cell. 2014;26:813-25
34. Klughammer J, Kiesel B, Roetzer T, Fortelny N, Nemc A, Nenning KH. et al. The DNA methylation landscape of glioblastoma disease progression shows extensive heterogeneity in time and space. Nat Med. 2018;24:1611-24
35. Sheffield NC, Pierron G, Klughammer J, Datlinger P, Schonegger A, Schuster M. et al. DNA methylation heterogeneity defines a disease spectrum in Ewing sarcoma. Nat Med. 2017;23:386-95
36. Li J, Shi L, Zhang K, Zhang Y, Hu S, Zhao T. et al. VarCards: an integrated genetic and clinical database for coding variants in the human genome. Nucleic Acids Res. 2018;46:D1039-D48
37. Juhling F, Kretzmer H, Bernhart SH, Otto C, Stadler PF, Hoffmann S. metilene: fast and sensitive calling of differentially methylated regions from bisulfite sequencing data. Genome Res. 2016;26:256-62
38. Assenov Y, Muller F, Lutsik P, Walter J, Lengauer T, Bock C. Comprehensive analysis of DNA methylation data with RnBeads. Nature Methods. 2014;11:1138-40
39. Lin DC, Dinh HQ, Xie JJ, Mayakonda A, Silva TC, Jiang YY. et al. Identification of distinct mutational patterns and new driver genes in oesophageal squamous cell carcinomas and adenocarcinomas. Gut. 2018;67:1769-79
40. Sheffield NC, Bock C. LOLA: enrichment analysis for genomic region sets and regulatory elements in R and Bioconductor. Bioinformatics. 2016;32:587-9
41. Gutierrez A, Kentsis A, Sanda T, Holmfeldt L, Chen SC, Zhang J. et al. The BCL11B tumor suppressor is mutated across the major molecular subtypes of T-cell acute lymphoblastic leukemia. Blood. 2011;118:4169-73
42. Kolfschoten IG, van Leeuwen B, Berns K, Mullenders J, Beijersbergen RL, Bernards R. et al. A genetic screen identifies PITX1 as a suppressor of RAS activity and tumorigenicity. Cell. 2005;121:849-58
43. He LR, Liu MZ, Li BK, Jia WH, Zhang Y, Liao YJ. et al. High expression of EZH2 is associated with tumor aggressiveness and poor prognosis in patients with esophageal squamous cell carcinoma treated with definitive chemoradiotherapy. Int J Cancer. 2010;127:138-47
44. Guan C, Shi H, Wang H, Zhang J, Ni W, Chen B. et al. CtBP2 contributes to malignant development of human esophageal squamous cell carcinoma by regulation of p16INK4A. J Cell Biochem. 2013;114:1343-54
45. Wu Y, Hu L, Liang Y, Li J, Wang K, Chen X. et al. Up-regulation of lncRNA CASC9 promotes esophageal squamous cell carcinoma growth by negatively regulating PDCD4 expression through EZH2. Mol Cancer. 2017;16:150
46. Liang Y, Chen X, Wu Y, Li J, Zhang S, Wang K. et al. LncRNA CASC9 promotes esophageal squamous cell carcinoma metastasis through upregulating LAMC2 expression by interacting with the CREB-binding protein. Cell Death Differ. 2018;25:1980-95
47. Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis C, Doyle F. et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57-74
48. Roadmap Epigenomics C, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A. et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317-30
49. Sheffield NC, Thurman RE, Song L, Safi A, Stamatoyannopoulos JA, Lenhard B. et al. Patterns of regulatory activity across diverse human cell types predict tissue identity, transcription factor binding, and long-range interactions. Genome Res. 2013;23:777-88
50. Rougeulle C, Chaumeil J, Sarma K, Allis CD, Reinberg D, Avner P. et al. Differential histone H3 Lys-9 and Lys-27 methylation profiles on the X chromosome. Mol Cell Biol. 2004;24:5475-84
51. Daniel B, Nagy G, Hah N, Horvath A, Czimmerer Z, Poliska S. et al. The active enhancer network operated by liganded RXR supports angiogenic activity in macrophages. Genes Dev. 2014;28:1562-77
52. Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ. et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107:21931-6
53. Jiang YY, Lin DC, Mayakonda A, Hazawa M, Ding LW, Chien WW. et al. Targeting super-enhancer-associated oncogenes in oesophageal squamous cell carcinoma. Gut. 2017;66:1358-68
54. Xie JJ, Jiang YY, Jiang Y, Li CQ, Lim MC, An O. et al. Super-Enhancer-Driven Long Non-Coding RNA LINC01503, Regulated by TP63, Is Over-Expressed and Oncogenic in Squamous Cell Carcinoma. Gastroenterology. 2018;154:2137-51
55. Dai YH, Tang YP, Zhu HY, Lv L, Chu Y, Zhou YQ. et al. ZEB2 promotes the metastasis of gastric cancer and modulates epithelial mesenchymal transition of gastric cancer cells. Dig Dis Sci. 2012;57:1253-60
56. Li MZ, Wang JJ, Yang SB, Li WF, Xiao LB, He YL. et al. ZEB2 promotes tumor metastasis and correlates with poor prognosis of human colorectal cancer. Am J Transl Res. 2017;9:2838-51
57. Li S, Garrett-Bakelman F, Perl AE, Luger SM, Zhang C, To BL. et al. Dynamic evolution of clonal epialleles revealed by methclone. Genome Biol. 2014;15:472
58. Oakes CC, Claus R, Gu L, Assenov Y, Hullein J, Zucknick M. et al. Evolution of DNA methylation is linked to genetic aberrations in chronic lymphocytic leukemia. Cancer Discov. 2014;4:348-61
59. Landan G, Cohen NM, Mukamel Z, Bar A, Molchadsky A, Brosh R. et al. Epigenetic polymorphism and the stochastic formation of differentially methylated regions in normal and cancerous tissues. Nat Genet. 2012;44:1207-14
60. Chang J, Zhong R, Tian J, Li J, Zhai K, Ke J. et al. Exome-wide analyses identify low-frequency variant in CYP26B1 and additional coding variants associated with esophageal squamous cell carcinoma. Nat Genet. 2018;50:338-43
61. Cui R, Kamatani Y, Takahashi A, Usami M, Hosono N, Kawaguchi T. et al. Functional variants in ADH1B and ALDH2 coupled with alcohol and smoking synergistically enhance esophageal cancer risk. Gastroenterology. 2009;137:1768-75
62. Pennathur A, Gibson MK, Jobe BA, Luketich JD. Oesophageal carcinoma. Lancet. 2013;381:400-12
63. Cao W, Wu W, Yan M, Tian F, Ma C, Zhang Q. et al. Multiple region whole-exome sequencing reveals dramatically evolving intratumor genomic heterogeneity in esophageal squamous cell carcinoma. Oncogenesis. 2015;4:e175
64. Chen XX, Zhong Q, Liu Y, Yan SM, Chen ZH, Jin SZ. et al. Genomic comparison of esophageal squamous cell carcinoma and its precursor lesions by multi-region whole-exome sequencing. Nat Commun. 2017;8:524
65. Mroz EA, Rocco JW. MATH, a novel measure of intratumor genetic heterogeneity, is high in poor-outcome classes of head and neck squamous cell carcinoma. Oral Oncol. 2013;49:211-5
66. Tabassum DP, Polyak K. Tumorigenesis: it takes a village. Nat Rev Cancer. 2015;15:473-83
Author contact
![]() Corresponding authors: Zhongsheng Sun, Beijing Institutes of Life Science, Chinese Academy of Sciences, Beichen West Road, Chao Yang District, Beijing 100101, China. Tel.: +86 10 64864959; Fax: +86 10 84504120; E-mail: sunzsac.cn or Hengjun Gao, Institute of Digestive Diseases, Tongji University School of Medicine, Shanghai 200065, China. E-mail: hengjun_gaoedu.cn
Corresponding authors: Zhongsheng Sun, Beijing Institutes of Life Science, Chinese Academy of Sciences, Beichen West Road, Chao Yang District, Beijing 100101, China. Tel.: +86 10 64864959; Fax: +86 10 84504120; E-mail: sunzsac.cn or Hengjun Gao, Institute of Digestive Diseases, Tongji University School of Medicine, Shanghai 200065, China. E-mail: hengjun_gaoedu.cn
 Global reach, higher impact
Global reach, higher impact