13.3
Impact Factor
Theranostics 2020; 10(5):2029-2046. doi:10.7150/thno.41106 This issue Cite
Research Paper
A multi-omics investigation of the molecular characteristics and classification of six metabolic syndrome relevant diseases
1. CAS Key Laboratory of Separation Science for Analytical Chemistry, Dalian Institute of Chemical Physics, Chinese Academy of Sciences, Dalian 116023, China.
2. Laboratory of High-Resolution Mass Spectrometry Technologies, Dalian Institute of Chemical Physics, Chinese Academy of Sciences (CAS), Dalian 116023, China
3. The University of Chinese Academy of Sciences, Beijing 100049, China
4. Key Laboratory of Systems Biology, CAS Center for Excellence in Molecular Cell Science, Institute of Biochemistry and Cell Biology, University of Chinese Academy of Sciences, Chinese Academy of Sciences, 320 Yue-Yang Road, Shanghai 200031, China
5. Shanghai Diabetes Institute, Shanghai Key Laboratory of Diabetes Mellitus, Shanghai Key Clinical Center for Metabolic Diseases, Shanghai Jiao Tong University Affiliated Sixth People's Hospital, 600 Yishan Road, Shanghai, 200233, People's Republic of China
# Equal contribution
Abstract
Metabolic syndrome (MTS) is a cluster of concurrent metabolic abnormal conditions. MTS and its component metabolic diseases are heterogeneous and closely related, making their relationships complicated, thus hindering precision treatment.
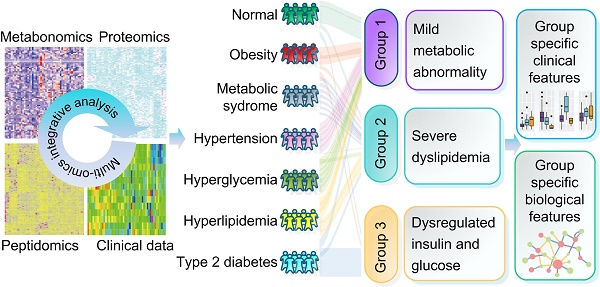
Methods: We collected seven groups of samples (group a: healthy individuals; group b: obesity; group c: MTS; group d: hyperglycemia, group e: hypertension, group f: hyperlipidemia; group g: type II diabetes, n=7 for each group). We examined the molecular characteristics of each sample by metabolomic, proteomic and peptidomic profiling analysis. The differential molecules (including metabolites, proteins and peptides) between each disease group and the healthy group were recognized by statistical analyses. Furthermore, a two-step clustering workflow which combines multi-omics and clinical information was used to redefine molecularly and clinically differential groups. Meanwhile, molecular, clinical, network and pathway based analyses were used to identify the group-specific biological features.
Results: Both shared and disease-specific molecular profiles among the six types of diseases were identified. Meanwhile, the patients were stratified into three distinct groups which were different from original disease definitions but presented significant differences in glucose and lipid metabolism (Group 1: relatively favorable metabolic conditions; Group 2: severe dyslipidemia; Group 3: dysregulated insulin and glucose). Group specific biological signatures were also systematically described. The dyslipidemia group showed higher levels in multiple lipid metabolites like phosphatidylserine and phosphatidylcholine, and showed significant up-regulations in lipid and amino acid metabolism pathways. The glucose dysregulated group showed higher levels in many polypeptides from proteins contributing to immune response. The another group, with better glucose/lipid metabolism ability, showed higher levels in lipid regulating enzymes like the lecithin cholesterol acyltransferase and proteins involved in complement and coagulation cascades.
Conclusions: This multi-omics based study provides a general view of the complex relationships and an alternative classification for various metabolic diseases where the cross-talk or compensatory mechanism between the immune and metabolism systems plays a critical role.
Keywords: metabolic syndrome, metabolic diseases, multi-omics data, lipid and glucose metabolism, disease subtype identification
 Global reach, higher impact
Global reach, higher impact