13.3
Impact Factor
Theranostics 2020; 10(26):12060-12071. doi:10.7150/thno.48918 This issue Cite
Review
Circulating tumor cells in hepatocellular carcinoma: single-cell based analysis, preclinical models, and clinical applications
Department of Laboratory Medicine, Zhongnan Hospital of Wuhan University, Wuhan 430071, P. R. China.
#Equal contributions to this work.
Received 2020-5-31; Accepted 2020-9-23; Published 2020-10-26
Abstract
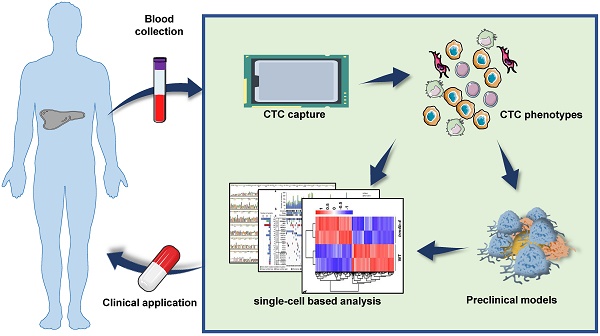
Circulating tumor cells (CTCs) are shed into the bloodstream from primary tumors and metastatic lesions and provide significant information about tumor progression and metastasis. CTCs contribute to tumor metastasis through the epithelial-to-mesenchymal transition (EMT). CTC clusters and stem-like phenotypes lead to a more aggressive and metastatic potential. CTCs retain the heterogeneity and imitate the nature of corresponding primary tumors. Therefore, it is important to use single-cell based analysis to obtain information on tumor heterogeneity and biology. CTCs are also good candidates for building preclinical models (especially 3D organoid cultures) for drug screening, disease modeling, genome editing, tumor immunity research, and organ-like biobank establishment. In this article, we summarize the current CTC capture technology, dissect the phenotypes associated with CTC metastasis, and review the progress in single-cell based analysis and preclinical modeling of the pattern and kinetics of CTCs. In particular, we discuss the use of CTCs to assess the progression of hepatocellular carcinoma (HCC).
Keywords: circulating tumor cells, single-cell based analysis, preclinical models, hepatocellular carcinoma, clinical application
Introduction
Circulating tumor cells (CTCs), which carry valuable tumor information, arise from the hematogenous diffusion of metastatic tumors [1] and have always been the focus of clinical research. In the past decades, various CTC capture technologies have been developed based on the biological features of CTCs, such as immune affinity and physical characteristics. In breast, prostate, and colorectal cancers, using the FDA-approved CellSearch platform, the CTC numbers have been correlated with progression-free survival (PFS) and overall survival (OS) [2, 3].
With emerging technological developments, CTC research is not limited to enumeration. Different CTC phenotypes have been analyzed, including the epithelial, mesenchymal, and stem cell types and CTC clusters, which were associated with distinct kinetics and functions [4]. In particular, advances in sequencing technology have facilitated the analysis of the genomes, transcriptomes, and proteins of individual CTCs, deepening our understanding of tumor heterogeneity for companion diagnostics [5, 6]. Besides, in vitro and in vivo CTC culture approaches have provided significant insights into tumor development and metastasis [7, 8].
Hepatocellular carcinoma (HCC) is the seventh most common cancer and the second leading cause of cancer-related deaths worldwide [9]. Current therapies for HCC include radical surgical resection, local ablation, or liver transplantation, which only apply to early-stage HCC, and their efficacy is not satisfactory due to the high recurrence rate [10]. Therefore, the approaches for diagnosing and monitoring HCC are important. Currently, diagnosing and monitoring HCC mainly depend on serum biomarker detection, biopsy, and imaging analysis. Each of these approaches suffers from drawbacks. Almost one-third of HCC patients show no significant changes in alpha-fetoprotein (AFP), an important serum biomarker [11]. A biopsy is invasive and may cause injury to patients and does not apply to dynamic monitoring. The sensitivity and specificity of medical imaging are low when a lesion is less than 2 cm.
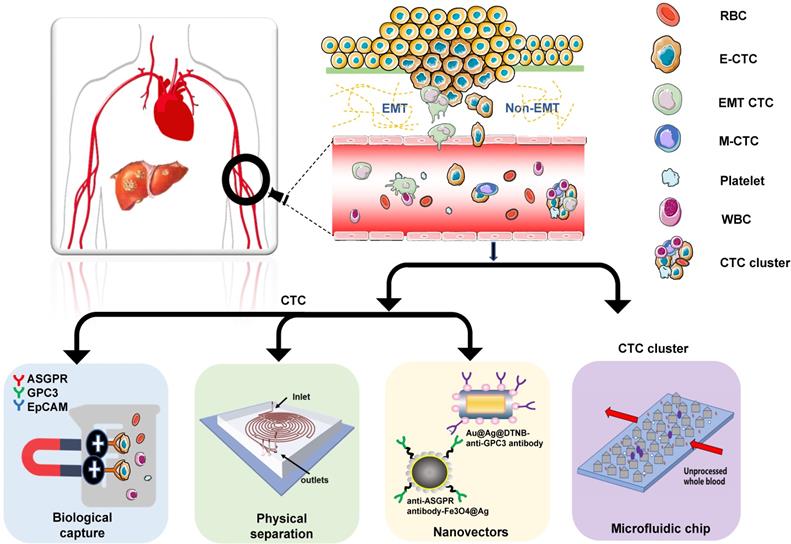
Overview of CTC capture techniques: CTCs are obtained from patients' blood samples in a non-invasive way. Many techniques distinguish CTCs from normal blood components (mainly red blood cells and white blood cells). HCC CTCs are primarily isolated based on their unique biological or physical properties. Currently, nanomaterials and microfluidic chips are widely used for CTCs capture.
In recent years, a series of “liquid biopsy” techniques have been developed to evaluate HCC biomarkers [12]. Liquid biopsy uses a non-invasive approach to obtain tumor information for tumor diagnosis and dynamic monitoring by assessing CTCs, circulating tumor DNA (ctDNA), microRNA (miRNA), and extracellular vesicles (EVs). CTCs obtained from HCC patients represent live tumor cells, which carry more comprehensive tumor information. Therefore, it is necessary to retrospectively analyze the clinical application of CTCs in the diagnosis and treatment of HCC.
Strategies for CTCs enrichment
CTCs, first discovered by Thomas Ashworth 150 years ago, can be obtained multiple times from tumor patients without an invasive approach. The recent development of new and powerful technologies has remarkably facilitated the precise capture of CTCs [13]. Current CTC capture techniques are summarized in Figure 1.
CTCs from HCC patients are primarily isolated based on their unique biological or physical properties. The biological methods include immune magnetic bead capture and nucleic acid aptamer capture. Court et al. [14] optimized HCC CTC capture using the NanoVelcro method that recognizes several cell-surface markers, such as asialoglycoprotein receptor (ASGPR), glypican-3 (GPC3), and epithelial cell adhesion molecule (EpCAM). In this study, HCC-CTCs were identified in 97% of HCC, and accurately discriminated HCC and non-HCC patients. Wang et al. used sLex-AP (aptamer for carbohydrate sialyl Lewis X) to coat the HA/CTS nanofilm to identify HCC CTCs in 59.5% of HCC patients [15]. These studies confirmed that both the CTC positive rate and number significantly correlated with tumor size, portal vein tumor thrombus, and the tumor-node-metastasis (TNM) stage. The physical properties applicable to CTC detection include size [16, 17], density [18], deformability, migratory capacity [19], and electric charge [20]. Wan et al. developed a novel Labyrinth microfluidic device, offering unique features over other inertial devices to efficiently isolate CTCs from the peripheral blood of HCC patients [21]. It incorporates 56 sharp corners to allow focusing on smaller cells that were previously difficult to target. Using this device, CTCs were detected in 75% of patients with early-stage HCC (TNM 0/I) and 96.2% of patients with advanced-stage HCC (TNM II-IV).
Currently, nanomaterials and microfluidic chips are widely used for CTC capture. According to the study by Ozkumur et al., “CTC-iChip”, an inertial focusing-enhanced microfluidic CTC capture platform, is capable of isolating CTCs with or without tumour membrane epitopes [22]. This technique applies to virtually all tumors. Zhang et al. proposed an on‐chip strategy combining multiplex SERS nano-vectors and multivariate analysis for in situ profiling of circulating tumor cell phenotypes on microfluidic chips [23]. Besides, Pang et al. reported a magnetically assisted surface‐enhanced Raman scattering (SERS) biosensor for HCC CTC detection [24]. This biosensor consists of anti‐ASGPR antibody‐Fe3O4@Ag magnetic nanoparticles and anti‐GPC3 antibody‐Au@Ag@DTNB nanorods. Because of the dual‐selectivity of the two antibodies, and the dual‐enhancement SERS signal of the MNP silver shell and the Au@Ag NRs SERS tags, a limit of detection of 1 cell/mL for HCC CTC in human peripheral blood with a linear relationship from 1 to 100 cells/mL can be achieved. The increasingly extensive research also proved the importance of CTC cluster analysis, and developed efficient CTC cluster technology. For example, a unique microfluidic chip, termed Cluster-Chip, which exploits the unique geometries of cellular aggregates to distinguish CTC clusters from single cells in the blood, was introduced by Sarioglu et al. [25]. This technique enables specific and label-free isolation of CTC clusters from patients with different cancers. Although there are many emerging capture technologies. Non-label capture and in-situ inspection methods are still urgently needed to facilitate subsequent culture and single-cell analysis of CTCs.
Metastasis-associated CTC phenotypes
Epithelial-mesenchymal transition (EMT)
EMT is a process that initiates metastasis [26-28]. Liu et al. showed that tumor cells undergo gradual or complete EMT (E, E/m, M/e, and M) that is associated with slow proliferation, loss of epithelial cell adhesion molecule EpCAM expression, and increased migration [29]. In the metastatic cascade, tumor cells undergoing EMT have an increased ability to intravasate into the lympho-vascular system. Once tumor cells migrate into blood vessels, they become CTCs. E-type and E/m-type CTCs have enhanced capacity to adhere and extravasate to distant sites. Importantly, E-type and E/m-type cells are associated with higher proliferation and metastatic outgrowth, while M/e- and M-type cells are related to long-term tumor recurrence. In HCC, Liu et al. showed that mixed CTCs might be vital for intrahepatic metastasis, and mesenchymal CTCs could predict extrahepatic metastasis [30]. Qi et al. revealed that total CTC count and the proportion of M-CTCs are negative factors for postoperative HCC recurrence [31]. However, the spatial heterogeneity of phenotypic and molecular characteristics of CTCs within the circulatory system remains unclear. Therefore, Sun et al. mapped the distribution and characterized the biological features of HCC CTCs along the transportation route [32]. Single-cell characterization demonstrated that the EMT status of CTCs was heterogeneous across different vascular compartments. CTCs were predominantly epithelial at release but switched to EMT-activated phenotype during hematogenous transit via the Smad2 and β-catenin pathways. Besides, EMT activation in the primary tumor correlated with total CTC number, rather than epithelial or EMT-activated subsets in hepatic veins (HV) [32].
CTC clusters
Besides EMT, CTC clusters have been proposed to indicate the initiation of tumor metastasis [33]. Although CTC clusters are rare in circulation relative to single CTCs, they have 23- to 50-fold increased metastatic potential [34, 35]. It was first demonstrated by Aceto et al. that CTC clusters originate from oligoclonal tumor cell groupings rather than cell aggregation in the blood vessels [34]. They also provided evidence that plakoglobin-dependent intercellular adhesion is crucial for the formation of CTC clusters. Sun et al. prospectively measured CTCs at five key vascular sites in patients with localized HCC [32]. Circulating tumor microemboli (CTM) were detected in tumor efferent vessels in approximately half of the patients, indicating that CTM originate from oligoclonal tumor cell groupings. However, it is worth noting that CTM were identified in hepatic veins (HV) but not peripheral artery (PA) in 15 patients, whereas 5 of them exhibited reemergence of CTM in peripheral vein (PV). This suggests that singular CTCs might be able to reaggregate in the bloodstream, which is contrary to the conclusion drawn by Aceto et al. [36]. They compared methylated DNA regions and found that CTC clusters and CTCs have distinctive DNA methylation status at differentially methylated regions (DMRs). The DMRs of CTC clusters feature hypomethylation at the binding sites of stemness- and proliferation-associated transcription factors, such as Oct4, Nanog, Sox2, and Sin3A [36].
Stem-like phenotypes
It has been more than a century since Cohnheim proposed the “embryonic theory” of cancer. The cancer stem cell (CSC) hypothesis argues that cancers arise from a subset of malignant cells that possess stem cell characteristics [37]. Although both CSCs and mature cancer cells can enter the bloodstream, CSCs are more prone to survive in the circulation and reside in distant organs or recirculate back to the liver [37]. Currently, CD133 and CD90 have been used to distinguish and isolate CTCs or CSCs in HCC. However, so far, no good circulating tumor stem cell (CTSC) marker has been identified in HCC. Liu et al. showed that ICAM-1 is a marker of HCC stem cells. They quantified circulating CD45-ICAM-1+ tumor cells from 60 HCC patients using flow cytometry and found that higher frequencies of circulating CD45-ICAM-1+ cells in HCC patients correlated with more aggressive tumor behavior and worse clinical outcomes [38]. Sun et al. found that EpCAM+ CTCs show stem cell characteristics and indicate poor prognosis of HCC after curative resection [39].
In animal models, CD90+CXCR4+ HCC cells could be CTSCs, as reported by Zhu et al. [40]. Selective elimination of these cells may substantially improve the efficacy of current HCC therapy by reducing metastasis. The oncogene Sox12 may be a novel marker for CSCs in HCC as proposed by Zou et al. [41]. Compared to Sox12- HCC cells, Sox12+ HCC cells were more frequently detected in circulation, had a higher efficiency of generating tumor spheres in culture or forming distal metastasis, and displayed greater chemo-resistance to cisplatin [41]. Although there is no current consensus on the CSC phenotype in HCC, these studies provide a guideline for building a molecular panel to assess CTC stemness in clinical applications. Such a CTC panel, including four putative stem cell biomarkers (EpCAM, CD133, CD90, and CK19) was constructed by Guo et al. [42]. The panel performed well in detecting early-stage and AFP-negative HCC, as well as differentiating HCC in hepatitis B infection (CHB), liver cirrhosis (LC), and benign hepatic lesions (BHL) [42].
Although the tumor metastasis mechanism remains unclear [34], CTC phenotypes can be classified to evaluate their malignant potential before and during tumor therapies. Wang et al. systematically investigated the clinical significance of diverse subtypes of CTCs and showed that the presence of EpCAM+ multiploid CTSCs (cut-off: ≥1 cell in 6 ml of blood), EpCAM- small triploid CTCs (≥5 cells), CTM (≥1), and increased triploid CTCs, were highly relevant for poor prognosis [4].
Single-cell based analysis
Single-cell sequencing technology has been a major breakthrough in sequencing history. CTCs can be processed and analyzed as single cells and then subjected to single-cell sequencing. Given the considerable heterogeneity of CTCs, it is essential to analyze the molecular and genetic characteristics of single CTCs [5, 6]. The schematic of single-cell analysis of CTCs is shown in Figure 2.
Single-cell genomic and transcriptomic analysis
With advances in the next-generation sequencing (NGS) and single-cell sequencing (SCS) technology, it is possible to obtain the complete genomes of CTCs and compare them with the corresponding primary and metastatic tumors. Several clinically relevant genomic alterations have been discovered, such as single nucleotide variations (SNV), microsatellite instability, and copy-number variations, providing valuable information for the companion diagnostics [43]. At the transcriptomic level, gene expression profiles of individual CTCs can reveal complex expression patterns within and across patients. Importantly, chemo-resistant tumor cell subsets and relevant signaling pathways could also be identified [44, 45]. D'Avola et al. developed a new technology combining image flow cytometry and high-density single-cell mRNA sequencing to identify CTCs in HCC patients [46]. Genome-wide expression profiling of CTCs using this approach demonstrated CTC heterogeneity and detected known oncogenic drivers in HCC such as Oct4. In another study, Sun et al. collected blood from the peripheral vein, peripheral artery, hepatic vein, inferior hepatic vena cava (IHIVC) and portal vein (PoV) before tumor resection. They analyzed the EMT phenotypes of CTCs using the 4-channel immunofluorescence CellSearch assay and microflow quantitative RT-PCR. The study demonstrated the heterogeneity of EMT status in CTCs across different vascular compartments [32]. CTCs were predominantly epithelial at release but switched to EMT-activated phenotype during hematogenous transit via Smad2 and β-catenin signaling. Thus, single-cell analysis provides a novel tool for biomarker identification in HCC and would ultimately help customize therapeutic interventions.
Epigenetic analysis
Epigenetic events, including histone modification and DNA base modifications (such as methylation and hydroxy-methylation), also contribute to tumor cell heterogeneity. ChIP-seq (Illumina) is used for studying histone modifications at the single-cell level [47], and the whole-genome bisulfite sequencing (WGBS) offers the most comprehensive profile of DNA methylation [48]. Pixberg et al. established a CTC line using the peripheral blood from a mouse HCC model [49]. To explore the mechanisms regulating the expression of HGF and c-MET, DNA methylation at the promoters of both genes was evaluated using the bisulfite-conversion and high-resolution melting analysis. The overexpression of HGF and c-MET in CTCs was associated with decreased DNA methylation at their promoters. The study concluded that during hematogenous dissemination, HCC CTCs undergo EMT under the influence of up-regulated HGF. This process also involves the up-regulation of c-MET induced by demethylation at 6 CpG sites. So far, the epigenetic characteristics of CTCs remain largely unexplored. However, existing data strongly encourage epigenetic analysis of CTCs to understand the molecular mechanisms of HCC metastasis.
Schematic representation of single-cell analysis of CTCs in HCC: Genomic, transcriptomic, proteomic, and epigenetic analyses are included. In the genomic analysis, tumor-related gene mutations are analyzed for companion diagnostics. In the transcriptome analysis, RNA levels are analyzed to assess tumor heterogeneity, identify tumor cell subsets, and dissect signaling pathways related to chemo-resistance. The epigenetic and proteomic analyses offer additional information about tumor heterogeneity.
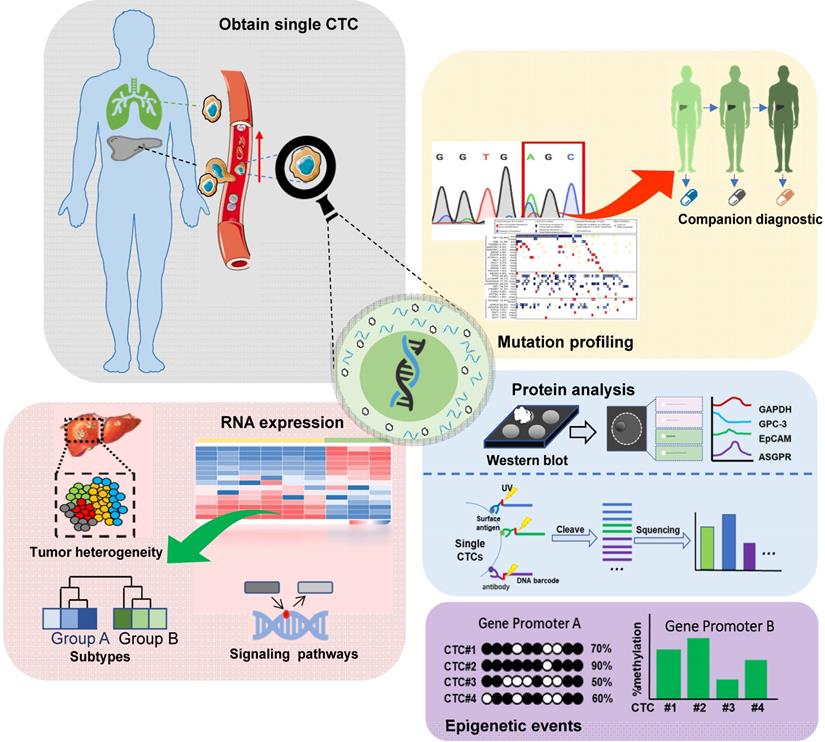
Protein analysis
Single-cell protein analysis identifies the heterogeneity of seemingly similar tumor cells, providing critical insights into the mechanisms underlying tumor heterogeneity [50, 51]. For this, Western blotting is the most popular protein analysis approach. Recently, multiple single-cell protein quantification methods have been developed. Flow cytometry has a high flux [52-54] but limited multiplexing capability due to the spectral overlap of fluorescent-labeled antibodies. By using the transition metal mass marker, mass spectrometry significantly improves (> 30) the detection of multiple proteins [55]. However, these methods are ineffective in finding rare target cells, especially in the presence of overwhelming background cells [56, 57]. Based on DNA barcoding and high-throughput sequencing, Wang et al. developed a microchip-assisted single-cell proteomic method for profiling CTC surface antigens facilitating the phenotypic and functional analysis of single CTCs [58].
Preclinical models
Although the techniques mentioned above have provided abundant information about CTCs, their functions still need to be validated in preclinical models. CTC-based preclinical models include 2D cultures, CTC spheroids, CTC-derived organoids, and xenografts [8]. Many studies have successfully established 2D cultures and CTC spheroids from cancer patients [59-62]. For example, Zhang et al. isolated CTCs from 31 patients and cultured them into spheroids [63]. Furthermore, the sensitivity of CTCs to chemotherapeutic agents (sorafenib or oxaliplatin) can be effectively tested utilizing the spheroids. 2D cultures and spheroid formation are easily conducted at low costs. However, cells generated by these two methods lack genomic and spatial heterogeneity. To address this issue, CTC-derived xenografts and organoids have emerged recently.
CTC- derived xenograft (CDX)
CTC-derived xenograft (CDX) is an in vivo model in which CTCs enriched from patient blood are injected into immunocompromised mice to generate tumors and expand the number of original tumor cells [64]. CDX is a good method for imitating tumor evolution and studying the metastatic process [65, 66]. Baccelli et al. had developed a xenograft assay and shown that primary human luminal breast cancer CTCs contain metastasis-initiating cells (MICs) that cause metastasis in the bone, lung, and liver. These MIC-containing CTCs expressed EpCAM, CD44, CD47, and MET [67]. In a small cohort of patients with metastasis, the number of EpCAM+CD44+CD47+MET+ CTCs correlated with increased metastatic sites and a poor survival rate. Recently, Vishnoi et al. established a novel triple- negative breast cancer (TNBC) liver metastasis‐specific CDX model that selectively recapitulates CTC biology for four sequential generations of mice [68]. Using multi-parametric flow cytometry analysis, immune phenotyping, and genomic sequencing, 597 genes specific to TNBC liver metastasis were identified in isolated CDX‐derived cells. This study provided a remarkable insight into the mechanism of TNBC disease progression in the liver. CDX can not only study the metastasis process, but also mirror the response of small cell lung cancer patients to platinum and etoposide chemotherapy, providing tractable systems for therapy testing and understanding drug resistance mechanisms [69]. Although CDX represents classical preclinical mouse models that are relatively easy to handle, CDX development still presents an enormous challenge due to low CTC prevalence in several cancers. The generation efficiency of CDX is much lower than that of patient-derived xenograft (PDX) [70]. Currently, the CDX model has not been successfully established in HCC so continuous endeavor is still required.
CTC-derived organoid (CDO)
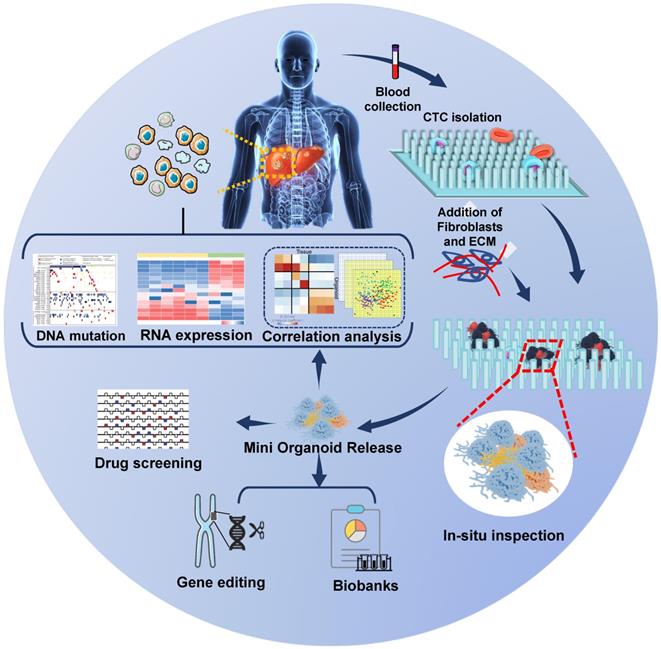
The establishment of CDX models is time-consuming and inapplicable to large-scale drug screening. CDO is the alternative that allows quick molecular analysis and high-throughput drug screening. An organoid is a special 3D culture model harboring a semisolid extracellular matrix supplemented with growth factors for tissues [71, 72]. Organoids have been successfully developed from primary tumors and metastatic lesions from various organs and their practicality has been demonstrated [73, 74]. They can be genetically modified using retroviruses, inhibitors, and/or the CRISPR/Cas9 system and can be used to build cancer models and identify key oncogenic “driver mutations” [75-77]. More significantly, organoids meet the demand for high-throughput drug screening needed to develop tumor therapeutics [78]. As CTCs are rare in the blood, the first successful establishment of CDO models by Gao et al. was a significant advance [79]. They established seven organoid lines from prostate cancer biopsy specimens and CTCs and reported frequent dysfunction of RB and TP53 pathways in castration-resistant prostate cancer (CRPC) organoid lines, suggesting that drugs targeting these pathways could be promising for therapy. In lung cancer, Zhang et al. [80] developed a novel in situ capture method for ex vivo CTC expansion in a 3D co-culture model simulating tumor microenvironment. They successfully expanded CTCs from 14 out of 19 early-stage lung cancer patients and revealed several mutations, including TP53 in both cultured CTCs and primary lung cancer. This strategy sets the stage to further characterize CTC biology and metastatic factors in patients with early-stage cancers. Although CDOs have not been established from HCC patients yet, we propose a strategy for analyzing HCC CDOs (Figure 3).
Clinical application in HCC
Early detection and neoplasm staging
There is considerable evidence for a crucial role of CTCs as initiators of metastasis, suggesting CTCs as a biomarker for the early detection of HCC. In previous CTC studies, early detection of HCC was mainly based on the assessment of CTC numbers, showing a significant positive correlation between the CTC number and the standard Barcelona Clinic Liver Cancer (BCLC) stage as well as the serum AFP level [81]. Qi et al. used an advanced CanPatrol CTC-enrichment technique and in situ hybridization to sort and classify CTCs from blood samples and found 90.18% of HCC patients to be CTC positive, even at the early stage of HCC [31]. CTCs were also detected in 2 of 12 patients with hepatitis B virus (HBV) infection, and both patients developed small HCC tumors within 5 months. Another study by Wang et al. implicated CTCs in tumor staging [15]. The presence of CTCs correlated highly with TNM staging from 66% in stage I to 100% in stage IV with CTC number being 1±1 in stage I to 4±2 in stage IV. Therefore, CTC testing can serve as a supplement to classical TNM staging for improving diagnostic efficiency. However, other studies point out that CTC testing is not an ideal independent diagnostic tool for HCC [82, 83] and simultaneous detection of CTCs and AFP might improve the detection accuracy of HCC patients [83].
Speculative strategy for analyzing CTC-derived organoids in HCC: Patients' blood samples are collected, and CTCs are captured and cultured in situ on a specific chip to form mini organoids. The mini CTC-derived organoids provide in situ observation of cell phenotypes, and the released cells are used for genetic analysis, drug screening, gene editing, and biobanks construction.
Prognostic evaluation and recurrence monitoring
In patients diagnosed with HCC, CTCs not only contribute to neoplasm staging but are also useful for prognosis [81, 84]. Ye et al. [85] studied the relationship between the CTC count and the clinical outcome of HCC patients after radical resection and found that both disease-free survival and overall survival were significantly better in patients with lower CTC counts (≤2/5 mL) compared with those with higher counts (>2/5 mL), implicating a high CTC count in the poor prognosis of HCC patients. Yu et al. also reported the association of increased postoperative CTC numbers with worse prognosis of HCC patients [86]. In a study by Ou et al., CTCs were classified using EMT markers and the presence of mesenchymal CTCs, together with mixed phenotypic and epithelial CTCs predicted the shortest relapse-free survival [87]. These observations indicated that the CTC phenotypes might serve as a prognostic indicator for HCC patients, and some specific CTC phenotypes may be more significant for the prognosis than others.
Since CTC numbers may change after anti-tumor treatment, they could be used for predicting or evaluating therapeutic efficacy before or after treatments. Ye et al. used CanPatrol™ system to count CTCs 1 day prior to and 30 days after surgical excision of HCC [88]. The study showed that postoperative CTC counts (> 2 and > 5) and pre/postoperative change in CTC counts were significantly associated with PFS and were a better predictor of performance than absolute counts. PCR and FACS were used by Zhou et al. to detect the preoperative levels of EpCAMmRNA+ CTCs and CD4+CD25+Foxp3+ Treg cells in 49 HCC patients. The data showed that elevated CTC/Treg levels implied a higher risk of postoperative recurrence [89].
Companion diagnostic
Mutation profiles and drug-resistant molecular expression profiles have been widely identified in many solid tumors. In this context, CTC analysis allows the determination of therapeutic targets and resistance mechanisms to cancer therapies at the DNA, RNA, and protein levels and has great potential to identify the patient population most likely to respond to specific treatments. Li et al. presented a novel system to provide quantitative information of sorafenib-related targets through simultaneously detecting phosphorylated ERK (pERK) and Akt (pAkt) in HCC CTCs [90]. This study demonstrated that CTCs could substitute tumor tissues in the characterization of pERK/pAkt. pERK+/pAkt- CTCs, the most sorafenib-sensitive cells that could serve as an independent PFS indicator in sorafenib-treated HCC patients.
Immune checkpoint inhibitors have launched a new era in immunotherapy, with exceptional long-term remissions in some patients across diverse tumor entities. However, only a few patients respond to this modality, with many experiencing severe side effects [91]. Therefore, it is necessary to monitor the expression of molecular targets (such as PD-L1) of immune checkpoint inhibitor therapy. It was first reported by Winograd et al. that evaluation of PD-L1+ CTCs discriminated between HCC patients with early-stage and advanced/metastatic disease [92]. Of 6 patients receiving anti-PD1 therapy, 3 demonstrated a good response had PD-L1+ CTCs. In contrast, only 1 of 3 non-responders had PD-L1+ CTCs, suggesting that PD-L1+ CTCs might be predictive of immunotherapy response. A more comprehensive clinical application of CTCs in HCC is summarized in Table 1 and Figure 4.
Future perspectives
CTCs offer valuable diagnostic and therapeutic information on HCC, although some challenges still exist in their identification, quantification, and/or characterization. Currently, new CTC capture technologies are emerging, while the CellSearch system is still the only FDA-approved CTC detection and quantification method. So, we urgently need novel and certified tools for quick isolation and characterization of CTCs. There are various sources of CTCs including primary tumors and metastatic lesions. Identifying of the sources of captured CTCs is beneficial for a more in-depth analysis. For a comprehensive understanding of CTC biology, multiple omic disciplines should be combined for single-cell analysis. With the development of CDO approaches, we can co-culture CTCs with immune cells to simulate the tumor microenvironment to monitor tumor progression and associated molecular events. Furthermore, as part of liquid biopsy, CTC testing should be combined with other liquid biopsies such as analysis of ctDNA and exosomes to promote the efficiency of clinical CTC tests. Ongoing and future research on CTC capture technology, molecular profiling, sing-cell analysis, and preclinical models are expected to considerably improve CTC testing for early diagnosis, efficacious treatment, and effective prognostic management of HCC. The overview of CTC research is showed in the Supplementary Figure.
Clinical application of CTCs in HCC: Peripheral blood is obtained from HCC patients using a non-invasive approach, followed by CTC capture on various platforms. Early detection, prognostic evaluation, recurrence monitoring, and companion diagnostic of HCC are achieved by enumerating CTCs, analyzing their molecular phenotypes, and testing targeted genes.
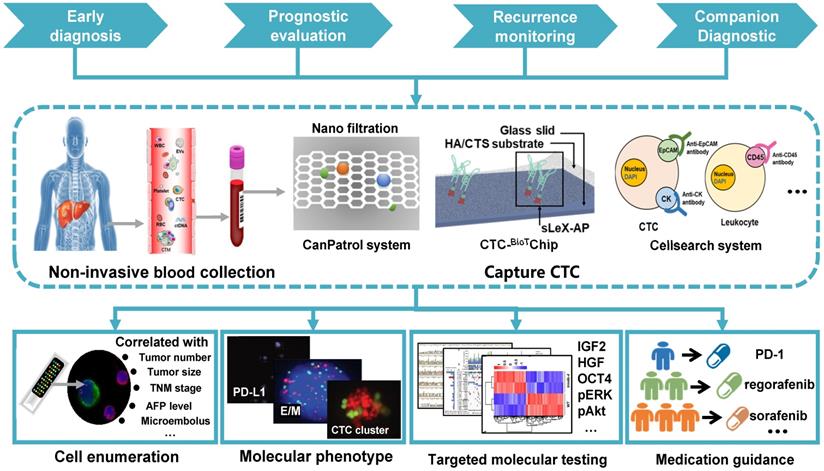
Summary of clinical application of CTCs
| Study | Parents | Method | CTC Marker | Main finding | |
|---|---|---|---|---|---|
| Early diagnosis and neoplasm staging | |||||
| Wang S et al [15]. 2016 | 42 HCC | CTC-BioTChip | EpCAM | The platform identified CTCs (2±2 per 2 mL) in 59.5% HCC patients; CTCs were significantly correlated with tumor size, portal vein tumor thrombus, and the TNM (tumor-node-metastasis) stage. | |
| Qi LN et al [31]. 2018 | 112 HCC treated with R0 resection | CanPatrolTM RNA-ISH | EpCAM, E-cadherin, CK8/18/19, vimentin, Twist | 90.18% patients with HCC were CTC positive, even with early-stage disease; CTCs were also detected in 2 of 12 patients with hepatitis B virus (HBV), both of whom had small HCC tumors detected within 5 months. | |
| Prognostic evaluation and recurrence monitoring | |||||
| Sun YF et al [93]. 2013 | 123 HCC | CellSearch | EpCAM | CTCs were present in 66.67% of patients; A preoperative CTC 7.5 of ≥2 was an independent prognostic factor for tumor recurrence. | |
| Yu JJ et al [86]. 2018 | 139 HCC; 23BHT | CellSearch | EpCAM | Patients with increased postoperative CTC counts (from preoperative CTC < 2 to postoperative CTC ≥ 2) had significantly shorter DFS and OS. | |
| Chen J et al [81]. 2017 | 195 HCC | CanPatrolTM | CK, EpCAM, Twist, Cadherin, Snail, Vimentin, AKT2 | CTCs were detected in 95% HCC patients. The AUC of the ROC curve was 0.861 in discriminating metastatic and non-metastatic patients. The proportion of hybrid and mesenchymal CTCs was associated with age, BCLC stages, metastasis and AFP levels and recurrence. | |
| Yin LC et al [84]. 2018 | 80 HCC; 10HV | CanPatrolTM | Twist | Twist+ CTCs were detected in 67.5% HCC patients. The positive ratios of Twist+ CTCs correlated with portal vein tumor thrombi, TNM staging, AFP, cirrhosis, tumor number, tumor size, microvascular invasion, as well as postop recurrence and the mortality. | |
| Ye XP et al [88]. 2018 | 42 HCC | CanPatrolTM | TP53 | Postoperative CTC counts (> 2 and > 5) and changes in CTC counts may be independent prognostic indicators for PFS in patients with HBV-related HCC | |
| Shi J et al [94]. 2016 | 47 HCC undergoing cryoablation | MACS, RT-qPCR, MACS, RT-qPCR | MAGE-3, Survivin, CEA | Average CTCs decreased significantly following cryosurgery. | |
| Zhou Y et al [89]. 2016 | 49 HCC undergoing curative resection; 50 HV | RosetteSep, qRT-PCR | EpCAM, CD4+ CD25+, Foxp3 | Patients with high CTC/Treg levels showed a significantly higher risk of developing postoperative HCC and higher 1-year recurrence rates. | |
| Wang Z et al [95]. 2018 | 62 HCC undergoing radical resection | CanPatrolTM | CK, EpCAM, Vimentin, Twist | The total number of CTCs, mesenchymal CTCs, and mixed CTCs in the recurrence group was significantly higher than in the non-recurrence group; Mesenchymal CTCs associated with shortened postoperative DFS. | |
| Shen J et al [96]. 2018 | 89 HCC undergoing TACE | Cell Search | EpCAM | CTC count is an independent predictor of OS and progression-free survival in patients treated with chemoembolization; A higher number of CTCs associated with mortality and progression. | |
| von Felden J et al [97]. 2017 | 58 HCC undergoing resection | CellSearch | EpCAM | CTC-positive patients had a significantly higher risk of recurrence with a HR of 2.3, and a shorter RFS. | |
| Qi LN et al [98]. 2018 | 112 HCC treated with R0 resection; 12 HBV; 20 HV | CanPatrolTM | EpCAM, CK, Vimentin, Twist | CTC count ≥16 and mesenchymal-CTC (M-CTC) percentage ≥2% prior to resection was significantly associated with early recurrence, multi-intrahepatic recurrence, and lung metastasis. | |
| Ou H et al [87]. 2018 | 165 HCC undergoing radical resection | CanPatrolTM | EpCAM, CK, Vimentin, Twist | Mesenchymal CTCs were significantly correlated with high AFP levels, multiple tumors, advanced TNM and BCLC stage, presence of embolus or microembolus, and earlier recurrence. | |
| Sun YF et al [32]. 2018 | 73 HCC undergoing curative resection | CellSearch, qRT-PCR | EpCAM, E-cadherin, N-cadherin, Vimentin, Snail, Slug | CTC and circulating tumor microemboli burden in hepatic veins and peripheral circulation prognosticated postoperative lung metastasis and intrahepatic recurrence, respectively. | |
| Wang L et al [4]. 2018 | 14 HCC; 16 CCA; 4 GBC undergoing resection | SE-iFISH | aneuploid chromosome 8 | Postsurgical quantity of small triploid CTCs (≥5 cells/6 ml blood), multiploid (≥pentasomy 8) CTSCs or CTM (either one ≥ 1) significantly correlated to HCC patients' poor prognosis, | |
| Companion Diagnostics | |||||
| Li J et al [90]. 2016 | 109 HCC on sorafenib | Ficoll-Paque | pERK | CTCs could be used in place of tumor tissue for the characterization of pERK/pAkt expression; pERK+/pAkt- CTCs were most sensitive to sorafenib and an independent predictive factor of PFS in HCC patients treated with sorafenib. | |
| Winograd P et al [92]. 2018 | 73 HCC: 8 HV;11 BLD | CTC-iChip | PD-L1 | PD-L1 CTC phenotype may help guide selection of patients likely to benefit from immune checkpoint inhibitors. | |
BHT, benign hepatic tumor; BLD, benign liver disease; CCA, cholangiocarcinoma; CTC, circulating tumor cell; CTM, circulating tumor microemboli; DFS, disease-free survival; EMT, epithelial to mesenchymal transition; GBC, gallbladder cancer; HBV, hepatitis B virus; HCC, hepatocellular carcinoma; HV, healthy volunteers; IVC, inferior vena cava; LC, liver cirrhosis; OS, overall survival; PFS, progression-free survival; RFS, recurrence-free survival; HR, hazard ratio.
Abbreviations
AFP: alpha-fetoprotein; ASGPR: asialoglycoprotein receptor; CD44: cluster of differentiation 44; CD45: cluster of differentiation 45; CD47: cluster of differentiation 47; CD90: cluster of differentiation 90; CD133: cluster of differentiation 133; CTC: circulating tumor cell; CTM: circulating tumor microemboli; CDO: CTC-derived organoid; CDX: CTC-derived xenograft; CK19: cytokeratin 19; CTSC: circulating tumor stem cell; CSC: cancer stem cell; CXCR4: chemokine (C-X-C motif) receptor 4; EpCAM: epithelial cell adhesion molecule; EMT: epithelial-mesenchymal transition; GPC3: glypican-3; HBV: hepatitis B virus; HGF: hepatocyte growth factor; HCC: hepatocellular carcinoma; IGF-2: insulin like growth factor 2; Oct4: octamer-binding transcription factor 4; PD‐L1: programmed cell death ligand 1; Sox2: SRY-related high-mobility-group (HMG)-box protein-2; TNM: tumor-node-metastasis; TNBC: triple- negative breast cancer.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
This work was supported by the Improvement Project for Theranostic ability on Difficulty Miscellaneous disease (Tumor) and the Science & Technology Innovation Fostering Foundation of Zhongnan Hospital of Wuhan University (No. znpy2018027, ZLYNXM202008), and National Natural Science Foundation of China (No. 81672114, 81702627). This work was also funded by Medical Talented Youth Development Project in Health Commission of Hubei Province (No. WJ2019Q049).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Pantel K, Speicher MR. The biology of circulating tumor cells. Oncogene. 2016;35:1216-24
2. Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC. et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351:781-91
3. Bauernhofer T, Zenahlik S, Hofmann G, Balic M, Resel M, Pirchmoser R. et al. Association of disease progression and poor overall survival with detection of circulating tumor cells in peripheral blood of patients with metastatic breast cancer. Oncol Rep. 2005;13:179-84
4. Wang L, Li Y, Xu J, Zhang A, Wang X, Tang R. et al. Quantified postsurgical small cell size CTCs and EpCAM(+) circulating tumor stem cells with cytogenetic abnormalities in hepatocellular carcinoma patients determine cancer relapse. Cancer Lett. 2018;412:99-107
5. Lim SB, Di Lee W, Vasudevan J, Lim WT, Lim CT. Liquid biopsy: one cell at a time. NPJ Precis Oncol. 2019;3:23
6. Keller L, Pantel K. Unravelling tumour heterogeneity by single-cell profiling of circulating tumour cells. Nat Rev Cancer. 2019;19:553-67
7. Praharaj PP, Bhutia SK, Nagrath S, Bitting RL, Deep G. Circulating tumor cell-derived organoids: Current challenges and promises in medical research and precision medicine. Biochim Biophys Acta Rev Cancer. 2018;1869:117-27
8. Tellez-Gabriel M, Cochonneau D, Cade M, Jubellin C, Heymann MF, Heymann D. Circulating Tumor Cell-Derived Pre-Clinical Models for Personalized Medicine. Cancers (Basel). 2018 11
9. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424
10. Llovet JM, Zucman-Rossi J, Pikarsky E, Sangro B, Schwartz M, Sherman M. et al. Hepatocellular carcinoma. Nat Rev Dis Primers. 2016;2:16018
11. Seo SI, Kim HS, Kim WJ, Shin WG, Kim DJ, Kim KH. et al. Diagnostic value of PIVKA-II and alpha-fetoprotein in hepatitis B virus-associated hepatocellular carcinoma. World J Gastroenterol. 2015;21:3928-35
12. Mann J, Reeves HL, Feldstein AE. Liquid biopsy for liver diseases. Gut. 2018;67:2204-12
13. Low WS, Wan Abas WA. Benchtop technologies for circulating tumor cells separation based on biophysical properties. Biomed Res Int. 2015;2015:239362
14. Court CM, Hou S, Winograd P, Segel NH, Li QW, Zhu Y. et al. A novel multimarker assay for the phenotypic profiling of circulating tumor cells in hepatocellular carcinoma. Liver Transpl. 2018;24:946-60
15. Wang S, Zhang C, Wang G, Cheng B, Wang Y, Chen F. et al. Aptamer-Mediated Transparent-Biocompatible Nanostructured Surfaces for Hepatocellular Circulating Tumor Cells Enrichment. Theranostics. 2016;6:1877-86
16. Lin HK, Zheng S, Williams AJ, Balic M, Groshen S, Scher HI. et al. Portable filter-based microdevice for detection and characterization of circulating tumor cells. Clin Cancer Res. 2010;16:5011-8
17. Hosokawa M, Hayata T, Fukuda Y, Arakaki A, Yoshino T, Tanaka T. et al. Size-selective microcavity array for rapid and efficient detection of circulating tumor cells. Anal Chem. 2010;82:6629-35
18. Gertler R, Rosenberg R, Fuehrer K, Dahm M, Nekarda H, Siewert JR. Detection of circulating tumor cells in blood using an optimized density gradient centrifugation. Recent Res Cancer. 2003;162:149-55
19. Liu Z, Zhang W, Huang F, Feng H, Shu W, Xu X. et al. High throughput capture of circulating tumor cells using an integrated microfluidic system. Biosens Bioelectron. 2013;47:113-9
20. Gascoyne PR, Noshari J, Anderson TJ, Becker FF. Isolation of rare cells from cell mixtures by dielectrophoresis. Electrophoresis. 2009;30:1388-98
21. Wan S, Kim TH, Smith KJ, Delaney R, Park GS, Guo H. et al. New Labyrinth Microfluidic Device Detects Circulating Tumor Cells Expressing Cancer Stem Cell Marker and Circulating Tumor Microemboli in Hepatocellular Carcinoma. Sci Rep. 2019;9:18575
22. Ozkumur E, Shah AM, Ciciliano JC, Emmink BL, Miyamoto DT, Brachtel E. et al. Inertial focusing for tumor antigen-dependent and -independent sorting of rare circulating tumor cells. Sci Transl Med. 2013;5:179ra47
23. Zhang Y, Wang Z, Wu L, Zong S, Yun B, Cui Y. Combining Multiplex SERS Nanovectors and Multivariate Analysis for In situ Profiling of Circulating Tumor Cell Phenotype Using a Microfluidic Chip. Small. 2018;14:e1704433
24. Pang Y, Wang C, Xiao R, Sun Z. Dual-Selective and Dual-Enhanced SERS Nanoprobes Strategy for Circulating Hepatocellular Carcinoma Cells Detection. Chemistry. 2018;24:7060-7
25. Sarioglu AF, Aceto N, Kojic N, Donaldson MC, Zeinali M, Hamza B. et al. A microfluidic device for label-free, physical capture of circulating tumor cell clusters. Nat Methods. 2015;12:685-91
26. Ledford H. Cancer theory faces doubts. Nature. 2011;472:273
27. Lim J, Thiery JP. Epithelial-mesenchymal transitions: insights from development. Development. 2012;139:3471-86
28. Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442-54
29. Liu X, Li J, Cadilha BL, Markota A, Voigt C, Huang Z. et al. Epithelial-type systemic breast carcinoma cells with a restricted mesenchymal transition are a major source of metastasis. Sci Adv. 2019;5:eaav4275
30. Liu YK, Hu BS, Li ZL, He X, Li Y, Lu LG. An improved strategy to detect the epithelial-mesenchymal transition process in circulating tumor cells in hepatocellular carcinoma patients. Hepatol Int. 2016;10:640-6
31. Qi LN, Xiang BD, Wu FX, Ye JZ, Zhong JH, Wang YY. et al. Circulating Tumor Cells Undergoing EMT Provide a Metric for Diagnosis and Prognosis of Patients with Hepatocellular Carcinoma. Cancer Res. 2018;78:4731-44
32. Sun YF, Guo W, Xu Y, Shi YH, Gong ZJ, Ji Y. et al. Circulating Tumor Cells from Different Vascular Sites Exhibit Spatial Heterogeneity in Epithelial and Mesenchymal Composition and Distinct Clinical Significance in Hepatocellular Carcinoma. Clin Cancer Res. 2018;24:547-59
33. Liotta LA, Saidel MG, Kleinerman J. The significance of hematogenous tumor cell clumps in the metastatic process. Cancer Res. 1976;36:889-94
34. Aceto N, Bardia A, Miyamoto DT, Donaldson MC, Wittner BS, Spencer JA. et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell. 2014;158:1110-22
35. Cheung KJ, Ewald AJ. A collective route to metastasis: Seeding by tumor cell clusters. Science. 2016;352:167-9
36. Gkountela S, Castro-Giner F, Szczerba BM, Vetter M, Landin J, Scherrer R. et al. Circulating Tumor Cell Clustering Shapes DNA Methylation to Enable Metastasis Seeding. Cell. 2019;176:98 -+
37. Li F, Tiede B, Massague J, Kang Y. Beyond tumorigenesis: cancer stem cells in metastasis. Cell Res. 2007;17:3-14
38. Liu S, Li N, Yu X, Xiao X, Cheng K, Hu J. et al. Expression of intercellular adhesion molecule 1 by hepatocellular carcinoma stem cells and circulating tumor cells. Gastroenterology. 2013;144:1031-41 e10
39. Sun YF, Xu Y, Yang XR, Guo W, Zhang X, Qiu SJ. et al. Circulating stem cell-like epithelial cell adhesion molecule-positive tumor cells indicate poor prognosis of hepatocellular carcinoma after curative resection. Hepatology. 2013;57:1458-68
40. Zhu L, Zhang W, Wang J, Liu R. Evidence of CD90+CXCR4+ cells as circulating tumor stem cells in hepatocellular carcinoma. Tumour Biol. 2015;36:5353-60
41. Zou S, Wang C, Liu J, Wang Q, Zhang D, Zhu S. et al. Sox12 Is a Cancer Stem-Like Cell Marker in Hepatocellular Carcinoma. Mol Cells. 2017;40:847-54
42. Guo W, Sun YF, Shen MN, Ma XL, Wu J, Zhang CY. et al. Circulating Tumor Cells with Stem-Like Phenotypes for Diagnosis, Prognosis, and Therapeutic Response Evaluation in Hepatocellular Carcinoma. Clin Cancer Res. 2018;24:2203-13
43. Pailler E, Faugeroux V, Oulhen M, Mezquita L, Laporte M, Honore A. et al. Acquired Resistance Mutations to ALK Inhibitors Identified by Single Circulating Tumor Cell Sequencing in ALK-Rearranged Non-Small-Cell Lung Cancer. Clin Cancer Res. 2019;25:6671-82
44. Lohr JG, Kim S, Gould J, Knoechel B, Drier Y, Cotton MJ. et al. Genetic interrogation of circulating multiple myeloma cells at single-cell resolution. 2016; 8: 363ra147-363ra147.
45. Miyamoto DT, Zheng Y, Wittner BS, Lee RJ, Zhu H, Broderick KT. et al. RNA-Seq of single prostate CTCs implicates noncanonical Wnt signaling in antiandrogen resistance. Science. 2015;349:1351-6
46. D'Avola D, Villacorta-Martin C, Martins-Filho SN, Craig A, Labgaa I, von Felden J. et al. High-density single cell mRNA sequencing to characterize circulating tumor cells in hepatocellular carcinoma. Sci Rep. 2018;8:11570
47. Lang JE, Scott JH, Wolf DM, Novak P, Punj V, Magbanua MJ. et al. Expression profiling of circulating tumor cells in metastatic breast cancer. Breast Cancer Res Treat. 2015;149:121-31
48. Farlik M, Sheffield NC, Nuzzo A, Datlinger P, Schonegger A, Klughammer J. et al. Single-cell DNA methylome sequencing and bioinformatic inference of epigenomic cell-state dynamics. Cell Rep. 2015;10:1386-97
49. Ogunwobi OO, Puszyk W, Dong HJ, Liu C. Epigenetic upregulation of HGF and c-Met drives metastasis in hepatocellular carcinoma. PLoS One. 2013;8:e63765
50. Heath JR, Ribas A, Mischel PS. Single-cell analysis tools for drug discovery and development. Nat Rev Drug Discov. 2016;15:204-16
51. Heath JR, Davis ME. Nanotechnology and cancer. Annu Rev Med. 2008;59:251-65
52. Perez OD, Nolan GP. Simultaneous measurement of multiple active kinase states using polychromatic flow cytometry. Nat Biotechnol. 2002;20:155-62
53. De Rosa SC, Herzenberg LA, Herzenberg LA, Roederer M. 11-color, 13-parameter flow cytometry: identification of human naive T cells by phenotype, function, and T-cell receptor diversity. Nat Med. 2001;7:245-8
54. Perfetto SP, Chattopadhyay PK, Roederer M. Seventeen-colour flow cytometry: unravelling the immune system. Nat Rev Immunol. 2004;4:648-55
55. Bendall SC, Simonds EF, Qiu P, Amir el AD, Krutzik PO, Finck R. et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332:687-96
56. Lu Y, Yang L, Wei W, Shi Q. Microchip-based single-cell functional proteomics for biomedical applications. Lab Chip. 2017;17:1250-63
57. Su Y, Shi Q, Wei W. Single cell proteomics in biomedicine: High-dimensional data acquisition, visualization, and analysis. Proteomics. 2017 17
58. Wang C, Yang L, Wang Z, He J, Shi Q. Highly multiplexed profiling of cell surface proteins on single circulating tumor cells based on antibody and cellular barcoding. Anal Bioanal Chem. 2019;411:5373-82
59. Hamilton G, Burghuber O, Zeillinger R. Circulating tumor cells in small cell lung cancer: ex vivo expansion. Lung. 2015;193:451-2
60. Cegan M, Kolostova K, Matkowski R, Broul M, Schraml J, Fiutowski M. et al. In vitro culturing of viable circulating tumor cells of urinary bladder cancer. Int J Clin Exp Pathol. 2014;7:7164-71
61. Yu M, Bardia A, Aceto N, Bersani F, Madden MW, Donaldson MC. et al. Cancer therapy. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science. 2014;345:216-20
62. Klameth L, Rath B, Hochmaier M, Moser D, Redl M, Mungenast F. et al. Small cell lung cancer: model of circulating tumor cell tumorospheres in chemoresistance. Sci Rep. 2017;7:5337
63. Zhang Y, Zhang X, Zhang J, Sun B, Zheng L, Li J. et al. Microfluidic chip for isolation of viable circulating tumor cells of hepatocellular carcinoma for their culture and drug sensitivity assay. Cancer Biol Ther. 2016;17:1177-87
64. Hodgkinson CL, Morrow CJ, Li Y, Metcalf RL, Rothwell DG, Trapani F. et al. Tumorigenicity and genetic profiling of circulating tumor cells in small-cell lung cancer. Nat Med. 2014;20:897-903
65. Tran NH, Cavalcante LL, Lubner SJ, Mulkerin DL, LoConte NK, Clipson L. et al. Precision medicine in colorectal cancer: the molecular profile alters treatment strategies. Ther Adv Med Oncol. 2015;7:252-62
66. Jin K, Teng L, Shen Y, He K, Xu Z, Li G. Patient-derived human tumour tissue xenografts in immunodeficient mice: a systematic review. Clin Transl Oncol. 2010;12:473-80
67. Baccelli I, Schneeweiss A, Riethdorf S, Stenzinger A, Schillert A, Vogel V. et al. Identification of a population of blood circulating tumor cells from breast cancer patients that initiates metastasis in a xenograft assay. Nat Biotechnol. 2013;31:539-44
68. Vishnoi M, Liu NH, Yin W, Boral D, Scamardo A, Hong D. et al. The identification of a TNBC liver metastasis gene signature by sequential CTC-xenograft modeling. Mol Oncol. 2019;13:1913-26
69. Hodgkinson CL, Morrow CJ, Li Y, Metcalf RL, Rothwell DG, Trapani F. et al. Tumorigenicity and genetic profiling of circulating tumor cells in small-cell lung cancer. Nature Medicine. 2014;20:897-903
70. Drapkin BJ, George J, Christensen CL, Mino-Kenudson M, Dries R, Sundaresan T. et al. Genomic and Functional Fidelity of Small Cell Lung Cancer Patient-Derived Xenografts. Cancer Discovery. 2018;8:600-15
71. Sato T, Stange DE, Ferrante M, Vries RG, Van Es JH, Van den Brink S. et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology. 2011;141:1762-72
72. Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE. et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262-5
73. Sachs N, Clevers H. Organoid cultures for the analysis of cancer phenotypes. Current Opinion in Genetics & Development. 2014;24:68-73
74. Weeber F, van de Wetering M, Hoogstraat M, Dijkstra KK, Krijgsman O, Kuilman T. et al. Preserved genetic diversity in organoids cultured from biopsies of human colorectal cancer metastases. P Natl Acad Sci USA. 2015;112:13308-11
75. Drost J, van Jaarsveld RH, Ponsioen B, Zimberlin C, van Boxtel R, Buijs A. et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature. 2015;521:43-7
76. Fumagalli A, Drost J, Suijkerbuijk SJ, van Boxtel R, de Ligt J, Offerhaus GJ. et al. Genetic dissection of colorectal cancer progression by orthotopic transplantation of engineered cancer organoids. Proc Natl Acad Sci U S A. 2017;114:E2357-E64
77. Boj SF, Hwang CI, Baker LA. et al. Organoid models of human and mouse ductal pancreatic cancer. Cell. 2015;160:324-38
78. Zhang Z, Shiratsuchi H, Palanisamy N, Nagrath S, Ramnath N. Expanded Circulating Tumor Cells from a Patient with ALK-Positive Lung Cancer Present with EML4-ALK Rearrangement Along with Resistance Mutation and Enable Drug Sensitivity Testing: A Case Study. J Thorac Oncol. 2017;12:397-402
79. Gao D, Vela I, Sboner A, Iaquinta PJ, Karthaus WR, Gopalan A. et al. Organoid cultures derived from patients with advanced prostate cancer. Cell. 2014;159:176-87
80. Zhang Z, Shiratsuchi H, Lin J, Chen GA, Reddy RM, Azizi E. et al. Expansion of CTCs from early stage lung cancer patients using a microfluidic co-culture model. Cancer Research. 2015 75
81. Chen J, Cao SW, Cai Z, Zheng L, Wang Q. Epithelial-mesenchymal transition phenotypes of circulating tumor cells correlate with the clinical stages and cancer metastasis in hepatocellular carcinoma patients. Cancer Biomarkers. 2017;20:487-98
82. Sun C, Liao WJ, Deng ZF, Li EL, Feng Q, Lei J. et al. The diagnostic value of assays for circulating tumor cells in hepatocellular carcinoma A meta-analysis. Medicine. 2017 96
83. Chen VL, Xu D, Wicha MS, Lok AS, Parikh ND. Utility of Liquid Biopsy Analysis in Detection of Hepatocellular Carcinoma, Determination of Prognosis, and Disease Monitoring: A Systematic Review. Clin Gastroenterol Hepatol. 2020
84. Yin LC, Luo ZC, Gao YX, Li Y, Peng Q, Gao Y. Twist Expression in Circulating Hepatocellular Carcinoma Cells Predicts Metastasis and Prognoses. Biomed Res Int. 2018;2018:3789613
85. Ye X, Li G, Han C, Han Q, Shang L, Su H. et al. Circulating tumor cells as a potential biomarker for postoperative clinical outcome in HBV-related hepatocellular carcinoma. Cancer Manag Res. 2018;10:5639-47
86. Yu JJ, Xiao W, Dong SL, Liang HF, Zhang ZW, Zhang BX. et al. Effect of surgical liver resection on circulating tumor cells in patients with hepatocellular carcinoma. Bmc Cancer. 2018 18
87. Ou H, Huang Y, Xiang L, Chen Z, Fang Y, Lin Y. et al. Circulating Tumor Cell Phenotype Indicates Poor Survival and Recurrence After Surgery for Hepatocellular Carcinoma. Dig Dis Sci. 2018;63:2373-80
88. Ye XP, Li GH, Han CY, Han QF, Shang LM, Su H. et al. Circulating tumor cells as a potential biomarker for postoperative clinical outcome in HBV-related hepatocellular carcinoma. Cancer Management and Research. 2018;10:5639-47
89. Zhou Y, Wang B, Wu J, Zhang C, Zhou Y, Yang X. et al. Association of preoperative EpCAM Circulating Tumor Cells and peripheral Treg cell levels with early recurrence of hepatocellular carcinoma following radical hepatic resection. BMC Cancer. 2016;16:506
90. Li J, Shi LH, Zhang XF, Sun B, Yang YF, Ge NJ. et al. pERK/pAkt phenotyping in circulating tumor cells as a biomarker for sorafenib efficacy in patients with advanced hepatocellular carcinoma. Oncotarget. 2016;7:2646-59
91. Baxi S, Yang A, Gennarelli RL, Khan N, Wang Z, Boyce L. et al. Immune-related adverse events for anti-PD-1 and anti-PD-L1 drugs: systematic review and meta-analysis. BMJ. 2018;360:k793
92. Winograd P, Hou S, Court CM, Sadeghi S, Finn RS, DiPardo B. et al. Evaluation of hepatocellular carcinoma circulating tumor cells expressing programmed death-ligand 1. Hpb. 2018;20:S2-S3
93. Sun YF, Xu Y, Yang XR, Guo W, Zhang X, Qiu SJ. et al. Circulating stem cell-like epithelial cell adhesion molecule-positive tumor cells indicate poor prognosis of hepatocellular carcinoma after curative resection. Hepatology. 2013;57:1458-68
94. Shi J, Li Y, Liang SZ, Zeng JY, Liu GF, Mu F. et al. Circulating tumour cells as biomarkers for evaluating cryosurgery on unresectable hepatocellular carcinoma. Oncology Reports. 2016;36:1845-51
95. Wang Z, Luo L, Cheng Y, He GL, Peng BJ, Gao Y. et al. Correlation Between Postoperative Early Recurrence of Hepatocellular Carcinoma and Mesenchymal Circulating Tumor Cells in Peripheral Blood. Journal of Gastrointestinal Surgery. 2018;22:633-9
96. Shen J, Wang WS, Zhu XL, Ni CF. High Epithelial Cell Adhesion Molecule-Positive Circulating Tumor Cell Count Predicts Poor Survival of Patients with Unresectable Hepatocellular Carcinoma Treated with Transcatheter Arterial Chemoembolization. J Vasc Interv Radiol. 2018;29:1678-84
97. von Felden J, Schulze K, Krech T, Ewald F, Nashan B, Pantel K. et al. Circulating tumor cells as liquid biomarker for high HCC recurrence risk after curative liver resection. Oncotarget. 2017;8:89978-87
98. Qi LN, Xiang BD, Wu FX, Ye JZ, Zhong JH, Wang YY. et al. Circulating Tumor Cells Undergoing EMT Provide a Metric for Diagnosis and Prognosis of Patients with Hepatocellular Carcinoma. Cancer Research. 2018;78:4731-44
Author contact
![]() Corresponding author: Fu-Bing Wang, Department of Laboratory Medicine, Zhongnan Hospital of Wuhan University, No.169 Donghu Road, Wuchang District, Wuhan 430071, P.R. China. Phone: +86-27-67813517; E-mail: wfb20042002com.
Corresponding author: Fu-Bing Wang, Department of Laboratory Medicine, Zhongnan Hospital of Wuhan University, No.169 Donghu Road, Wuchang District, Wuhan 430071, P.R. China. Phone: +86-27-67813517; E-mail: wfb20042002com.
 Global reach, higher impact
Global reach, higher impact