13.3
Impact Factor
Theranostics 2020; 10(24):11080-11091. doi:10.7150/thno.49864 This issue Cite
Research Paper
Development and interpretation of a pathomics-based model for the prediction of microsatellite instability in Colorectal Cancer
1. Information Management and Big Data Center, Nanfang Hospital, Southern Medical University, Guangzhou, China.
2. AI Lab, Tencent, Shenzhen, China.
3. Department of Radiation Oncology, Nanfang Hospital, Southern Medical University, Guangzhou, China.
4. Department of Pathology, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, Hubei, China.
5. Indiana University Bloomington, Bloomington, USA.
6. Tongshu Biotechnology Co., Ltd. Shanghai, China.
7. Tencent Healthcare, Shenzhen, China.
8. Department of Medicine, Indiana University School of Medicine, Indianapolis, IN, USA.
9. Regenstrief Institute, Indianapolis, IN, USA.
10. Department of Computer Science, Technical University of Munich, Munich, Germany.
#These authors contributed equally to this study.
Abstract
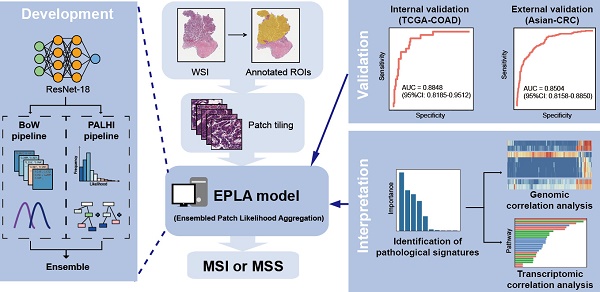
Microsatellite instability (MSI) has been approved as a pan-cancer biomarker for immune checkpoint blockade (ICB) therapy. However, current MSI identification methods are not available for all patients. We proposed an ensemble multiple instance deep learning model to predict microsatellite status based on histopathology images, and interpreted the pathomics-based model with multi-omics correlation.
Methods: Two cohorts of patients were collected, including 429 from The Cancer Genome Atlas (TCGA-COAD) and 785 from an Asian colorectal cancer (CRC) cohort (Asian-CRC). We established the pathomics model, named Ensembled Patch Likelihood Aggregation (EPLA), based on two consecutive stages: patch-level prediction and WSI-level prediction. The initial model was developed and validated in TCGA-COAD, and then generalized in Asian-CRC through transfer learning. The pathological signatures extracted from the model were analyzed with genomic and transcriptomic profiles for model interpretation.
Results: The EPLA model achieved an area-under-the-curve (AUC) of 0.8848 (95% CI: 0.8185-0.9512) in the TCGA-COAD test set and an AUC of 0.8504 (95% CI: 0.7591-0.9323) in the external validation set Asian-CRC after transfer learning. Notably, EPLA captured the relationship between pathological phenotype of poor differentiation and MSI (P < 0.001). Furthermore, the five pathological imaging signatures identified from the EPLA model were associated with mutation burden and DNA damage repair related genotype in the genomic profiles, and antitumor immunity activated pathway in the transcriptomic profiles.
Conclusions: Our pathomics-based deep learning model can effectively predict MSI from histopathology images and is transferable to a new patient cohort. The interpretability of our model by association with pathological, genomic and transcriptomic phenotypes lays the foundation for prospective clinical trials of the application of this artificial intelligence (AI) platform in ICB therapy.
Keywords: microsatellite instability, colorectal cancer, pathomics, multi-omics, ensembled patch likelihood aggregation (EPLA)
 Global reach, higher impact
Global reach, higher impact