13.3
Impact Factor
Theranostics 2020; 10(24):11049-11062. doi:10.7150/thno.49168 This issue Cite
Review
The emerging role of super enhancer-derived noncoding RNAs in human cancer
1. Department of Pathology, Xiangya Hospital, Central South University, Changsha, Hunan, 410008, China.
2. Department of Endocrinology, Xiangya Hospital, Central South University, Changsha, Hunan 410008, China.
3. Department of Oncology, Xiangya Hospital, Central South University, Changsha 410008, Hunan, China.
4. National Clinical Research Center for Geriatric Disorders, Xiangya Hospital, Central South University, Changsha 410008, Hunan, China.
#Co-first authors with equal contributions to this work.
Received 2020-6-6; Accepted 2020-8-23; Published 2020-9-2
Abstract
Super enhancers (SEs) are large clusters of adjacent enhancers that drive the expression of genes which regulate cellular identity; SE regions can be enriched with a high density of transcription factors, co-factors, and enhancer-associated epigenetic modifications. Through enhanced activation of their target genes, SEs play an important role in various diseases and conditions, including cancer. Recent studies have shown that SEs not only activate the transcriptional expression of coding genes to directly regulate biological functions, but also drive the transcriptional expression of non-coding RNAs (ncRNAs) to indirectly regulate biological functions. SE-derived ncRNAs play critical roles in tumorigenesis, including malignant proliferation, metastasis, drug resistance, and inflammatory response. Moreover, the abnormal expression of SE-derived ncRNAs is closely related to the clinical and pathological characterization of tumors. In this review, we summarize the functions and roles of SE-derived ncRNAs in tumorigenesis and discuss their prospective applications in tumor therapy. A deeper understanding of the potential mechanism underlying the action of SE-derived ncRNAs in tumorigenesis may provide new strategies for the early diagnosis of tumors and targeted therapy.
Keywords: Super enhancers, Noncoding RNAs, Tumorigenesis, Inflammatory response, Therapy
Introduction
A tumor is a malignant mass of cancerous cells that undergoes uncontrolled growth and replication. Developing tumors invade surrounding tissues and cause organ failure. Although significant advances have been made in the understanding and treatment of tumors in recent years, the morbidity and mortality rates remain high due to tumor recurrence and metastasis [1]. Therefore, to improve the overall survival of patients with cancer, early screening and tumor marker detection are important clinical strategies. Tumor biomarkers are molecules expressed in cancer cells and tissues that can reflect the progression and prognosis of malignant tumors [2]. They can be divided into five groups according to their biological functions: (1) oncofetal proteins, (2) tumor antigens, (3) enzymes, (4) hormones, and (5) special plasma proteins. In recent years, studies have found that non-coding RNAs (ncRNAs) have potential tumor marker characteristics. It is possible that analyzing ncRNAs may help diagnose many malignant tumors, such as gastric cancer, bladder cancer, prostate cancer, pancreatic cancer and cholangiocarcinoma [3-6].
With the advent of the post-genomic era, humans have been gaining a greater understanding of the genome and enhancers. In 2013, Young [7] proposed the concept of super enhancers (SEs). SEs are large clusters of adjacent enhancers that drive the expression of genes which regulate cellular identity; SE regions can be enriched with a high density of transcription factors (TFs), co-factors, and enhancer-associated epigenetic modifications. SEs can activate the expression of genes that determine cellular identity, thus affecting the occurrence of tumors and other diseases. It has been revealed in recent years that SEs not only directly activate exon-encoded genes to regulate biological functions, but they also activate the expression of ncRNAs, which regulate biological processes indirectly. In 2017, Suzuki et al. [8] reported that SEs are the core of tissue-specific miRNA networks and influence the progress of multiple tumors by regulating miRNA production. During the process of cell transformation, activated SEs usually regulate the generation of pro-carcinogenic miRNAs, and the inhibited SEs are often associated with tumor suppressor miRNAs. Therefore, the combination of SEs with multiple downstream miRNAs has the potential to serve as a cancer-related biomarker. The "SE-TF-ncRNA-target gene" regulatory networks have been widely studied. This review aims to describe and summarize several kinds of SE-derived ncRNAs and their functions in tumorigenesis, and primarily discuss their prospective clinical applications, aiming at providing new strategies for tumor-targeted therapies.
Schematic diagram of super enhancers (SEs). SEs are large clusters of adjacent enhancers that drive the expression of genes that regulate cellular identity, and SE regions can be enriched with a high density of transcription factors, co-factors, and enhancer-associated epigenetic modifications.
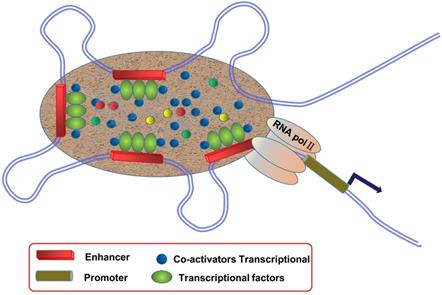
Biogenesis and characteristics of SEs
SEs are large clusters of adjacent enhancers, and SE regions can be enriched with a high density of TFs, co-factors, and enhancer-associated epigenetic modifications [9, 10] (Figure 1). Compared to typical enhancers (TEs), SEs have the following functional characteristics: (1) modified with a high density of H3K27ac and H3K4me1, as well as binding mediator complex and Bromodomain-containing protein 4 (BRD4); (2) binding with TFs and transcriptional activity-associated chromosome markers; (3) regulation of gene expression; and (4) sensitivity to the effect of transcriptional inhibitors [11-13]. SEs exist in many cell types and are closely related to biological growth and cancer. They are usually enriched in genetic susceptibility regions of the genome, which are closely associated with a variety of disease pedigrees, suggesting that they may play an important role in disease diagnosis and treatment.
At present, SEs are identified based on enhancers, involving three steps: (1) identifying the active enhancer sites, (2) stitching the enhancers, and (3) determining the threshold between the SEs and normal enhancers. First, chromatin immunoprecipitation (ChIP-seq) is used to detect factors or histone modifications associated with active enhancers, such as TFs, transcriptional coactivators (e.g. Mediator, p300, etc.), and histone modifications H3K27ac and H3K4me1. Next, the obtained enhancers are stitched. Researchers concatenate the enhancers within 12.5 kb of each other to define a single entity called the “stitched enhancer.” Lastly, to determine the threshold, they rank the stitched enhancer entities and the remaining individual enhancers (those without a neighboring enhancer within 12.5 kb) based on the total background-normalized level of the Med1 signal within the genomic region to obtain a resultant curve. The signal value obtained at the tangential point of the line with slope 1 on the curve is used as the threshold to distinguish SEs from normal enhancers. SEs are defined as regions to the right of the threshold of the resulting curve, while normal enhancers are defined, as those to the left [14]. With the development of next-generation sequencing technologies, more methods of SE identification have been used (Table 1). After identifying SEs, the expression of protein-coding genes and ncRNAs regulated by SEs can be predicted according to the gene location. Through RNA-seq, it is possible to establish a network between SEs and abnormally expressed mRNAs and ncRNAs. In addition, we can speculate on the key SEs and SE-derived ncRNAs in tumors. Information on SE-derived ncRNAs may provide a theoretical basis for future studies on the mechanisms underlying tumors and their treatment.
Classification of SE-derived ncRNAs
SEs not only directly activate exon-encoded genes to regulate biological functions, but also activate the expression of ncRNAs, which regulate biological processes indirectly. These ncRNAs mainly include microRNAs (miRNAs), long non-coding RNAs (lncRNAs), circular RNAs (circRNAs), and enhancer RNAs (eRNAs) (Table 2). The activation of SEs can induce ncRNAs to regulate target genes in various ways. On one hand, they can promote the transcription and maturation of miRNAs, transcription and generation of lncRNAs, and transcription and ring formation of circRNAs. On the other hand, the transcription products of SEs themselves, that is eRNAs, also play a synergistic role in regulating gene expression. miRNAs were the first ncRNAs to be identified. After transcription, the initial product of a miRNA undergoes two shears, one in the nucleus and one in the cytoplasm, to form a mature miRNA [23, 24]. Suzuki et al. [8] reported that SEs promote the maturity of pri-miRNAs by recruiting Drosha/DGCR 8 to regulate the transcription and synthesis of tissue-specific miRNAs (miR-290-295 and miR-106a-363). SE-derived lncRNAs usually have two forms, one of which is eRNA. An eRNA is an ncRNA formed after the self-transcription of the enhancer itself, with a sequence length of 0.5-5 kb; therefore, it is also classified as a lncRNA. There are many mechanisms by which eRNAs affect gene expression [25, 26]. The other form is lncRNA, which are transcribed from the promoter region through the regulation of SEs; we normally call these SE-derived lncRNAs [27]. Recent studies suggested that SE-derived lncRNAs can interact with exon-sensitive lncRNAs, and this interaction activates promoters and enhancers, thereby promoting chromatin loop integration and nuclear topological domain formation of the target genes, contributing to their expression [28]. At present, more researchers are examining the mechanism underlying the action of SE-derived lncRNAs in tumors and other diseases. circRNAs are a class of recently discovered ncRNAs. Studies on the mechanism of the formation of SE-derived circRNAs and the role of SEs remain scarce. However, by using polymerase chain reaction (PCR) and in situ hybridization (ISH), scientists have detected the expression levels of circRNAs in various human tissues and found that SE-derived circRNAs have higher expression levels and stronger tissue specificity than TE-derived circRNAs [29].
Methods of super enhancer identification
| Methods | Description | Advantages | Disadvantages | Ref. |
|---|---|---|---|---|
| ChIP-seq (Chromatin Immunoprecipitation sequencing) | A method for detecting genome-wide DNA segments interacting with histones and transcription factors. | 1) Provides a high-resolution map of genomic expression regulation with less samples; 2) No signal noise deviations as direct sequencing method is used. | 1) Unstable data accuracy as it is greatly influenced by the quality of antibodies; 2) High cost of building ChIP-seq workflow. | [14, 15] |
| 3C-seq (Chromosome conformation capture) | A method for detecting the DNA-DNA interactions between enhancer regions and one other transcriptional regulatory elements. | 1) Combine with quantitative PCR (qPCR) to reveal the results quantitatively; 2) No sequencing is required so the cost is low. | 1) Low-throughput because interactions must be tested one at a time; 2) Not unbiased because genomic positions must be chosen to test for interactions. | [16, 17] |
| 4C-seq (Circularized chromosome conformation capture) | A method for detecting genome-wide DNA-DNA interactions with a single chosen genomic location of interest. | 1) Provides a high-resolution map of chromatin interactions with a chosen 'viewpoint'; 2) Fewer samples are needed for sufficient sequencing depth. | 1) Inefficient because primers must be redesigned specifically before each 'viewpoint' tested; 2) Improvement is needed for data normalization and unbiased estimate. | [17, 18] |
| Hi-C (High-throughput chromosome conformation capture) | A method for detecting pairwise contacts between virtually any pair of genomic loci. | 1) A matrix-balancing normalization method associated with high-resolution sequencing is developed; 2) Combines with visualization platforms to construct the 3D structure of chromatin interaction. | 1) High signal noise because of polymerization state and dynamic chromatin interactions; 2) Insufficient genome-wide resolution to bridge 3D information to gene function perfectly. | [19, 20] |
| STARR-seq (self-transcribing active regulatory region sequencing) | A method to identify transcriptional enhancers and to assess their activity quantitatively by cloning DNA fragments downstream of a core promoter. | 1) Provides genome-wide cell type-specific quantitative enhancer activity maps of any cell type; 2) Not affected by the location of the sequences. | Repeated identification may exist because of lack of accurate context markers. | [21, 22] |
Biological characteristics of SE-ncRNAs
| SE-ncRNAs | Definition | Biogenesis mechanism | Functions | Ref. |
|---|---|---|---|---|
| miRNAs | MicroRNAs are small endogenous RNAs which are 19 to 25 nucleotides in size that regulate post-transcriptional gene expression. | MicroRNAs are transcribed from endogenous gene sequences to form hairpin pri-miRNAs, which are processed by Drosha/DGCR8 and further cleaved by Dicer to form mature miRNAs. SEs enhance the transcription and promote the maturity of pri-miRNAs by recruiting Drosha/DGCR8. | 1) MicroRNAs can bind to the 3'-UTR region of the target mRNA and inhibit the target genes' expression at the translation level; 2) miRNAs can bind to the coding region or ORF region of the target mRNA to affect its stability; 3) miRNAs enter the nucleus and regulate the target genes' expression at the transcriptional level. | [8, 30] |
| lncRNAs | Long non-coding RNAs have a transcribing length of 200-100000 nt, lack a completely functional open reading frame (ORF), rarely encode a functional short peptide, and are located in nucleus or cytoplasm. | Five main mechanisms of lncRNA biogenesis:1) Transformation from a protein-coding gene that acquires frame disruptions; 2) Chromosome rearrangement; 3, 4) Neighboring repeats originating from two tandem duplications; 5) Insertion by a transposable element to become a functional ncRNA. | The molecular functions of lncRNAs at the epigenetic, transcriptional, and post-transcriptional levels are subdivided as follows: 1) recruiting and interacting with proteins; 2) acting as a co-regulator or a co-repressor; 3) acting as a decoy; 4) acting as host genes for miRNA; 5) interacting with miRNA. | [31, 32] |
| circRNAs | Circular RNAs are composed of >200 nucleotides and have a covalent closed loop structure without a 5' cap and/or a 3' poly (A), which can encode a small amount of polypeptide. | They are mainly produced by cyclization of exons and/or introns. They can be divided into different types, according to the method of cyclization: 1) Formation by spliceosome-dependent cable tail patching; 2) Cis-acting elements promoting formation; 3) RNA binding proteins regulating circRNA formation. | 1) Circular RNAs can act as miRNA sponges. They can indirectly regulate miRNA downstream target genes' expression by preventing miRNAs from binding to the 3' untranslated regions of the mRNA; 2) They combine with RNA binding proteins (RBP), playing an important role in changing RNA splicing modes and mRNA stability; 3) They can also act as “miRNA reservoirs,” which can release large amounts of miRNAs under certain circumstances to inhibit the expression of target genes. | [33-35] |
| eRNAs | Enhancer RNA was identified as a self-transcription of the enhancer itself, with a sequence length of 0.5-5 kb. | Enhancer RNAs are transcribed from putative enhancer regions marked by histone modifications, such as H3K4m1/2 and H3K27Ac, and enriched with many transcription factors, such as LDTFs, P300, CBP, BRD4, and MED1. Recently eRNAs transcribed from super enhancers were named super-enhancer RNAs (seRNAs). | 1) They synchronously combine with enhancers and promoters and enhance their interaction to stabilize the chromatin loop; 2) eRNAs initiate the transcription of targets by binding to promoters directly or indirectly via recruitment of RNA polymerase II; 3) eRNAs promote target transcription by enhancing the binding of RNA polymerase II; 4) eRNAs act as a decoy for the negative elongation factor (NELF) complex and prompt the elongation of the paused RNA polymerase II. | [36, 37] |
Biological functions of SE-derived ncRNAs
SE-derived ncRNAs are widely expressed in a variety of cells and are involved in various diseases, such as diabetes, rheumatoid arthritis, and tumors. SE-derived ncRNAs also play an important role in regulating the following biological activities via a series of action modes (Table 2).
Determination of tissue specificity
Song et al. [38] pointed out that the methylation of enhancer DNA is closely related to the heterogeneity of cells. DNA methylation of Sox2 and Mir290 SEs can guide the differentiation of embryonic stem cells. Suzuki et al. [8] reported that SEs are highly correlated with miRNAs in tissue-specific determination. Several tissue-specific SE-derived miRNAs have been detected: miR-290-295 in mouse embryonic cells (mESCs), miR-142 in mouse precursor B cells, miR-1/133a2 in myotube tissues, and miR-142/210 in Th cells. It has been shown that these tissue-specific SE-derived miRNAs are part of a gene regulatory network that controls cell-type specificity, and tissue specificity may be attributed to target avoidance phenomena between miRNAs and target genes [39]. Ounzain et al. [40] found a cardiomyocyte-specific SE-derived lncRNA, CARMEN. Research has shown that CARMEN can affect the fate of cardiomyocytes and interact with TFs SUZ12 and EZH2.
Regulating growth and development
SE-derived ncRNAs play an important role in the growth and development of neonates. Ounzain et al. [41] reported that Novlnc6, an SE-derived lncRNA, is associated with cardiac development. Novlnc6 is regulated by many important major TFs and regulatory proteins in the heart, including TBX20 and MEF2A. Anderson et al. [42] reported that SE-derived lncRNAs upregulate cardiac development by interacting with the TF HAND2. Other studies found that the SE-derived lncRNA Whisper can induce myocardial remodeling [43], whereas SE-circNfix inhibits the repair and regeneration of mouse cardiomyocytes [29]. SE-miR140 may cause skeletal dysplasia [44], and Degirmenci et al. [45] reported that the eRNA lncASIR is associated with insulin-induced adipocyte metabolism. The eRNA Bloodlnc can promote erythroblast proliferation and enucleation of red blood cells and participate in erythrocyte proliferation and differentiation [46].
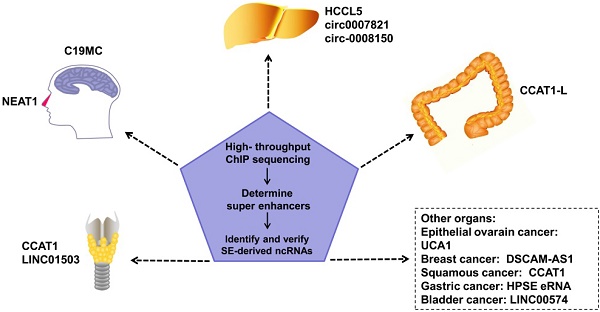
An increasing number of SE-derived ncRNAs have been reported to be aberrantly expressed in human cancers, including esophageal squamous cell carcinoma (ESCC), hepatocellular Carcinoma (HCC), epithelial ovarian cancer (EOC), embryonal tumor with multilayered rosettes (ETMRs), colorectal cancer (CRC), breast cancer, squamous cancer, lymphocytic leukemia, nasopharynx cancer.
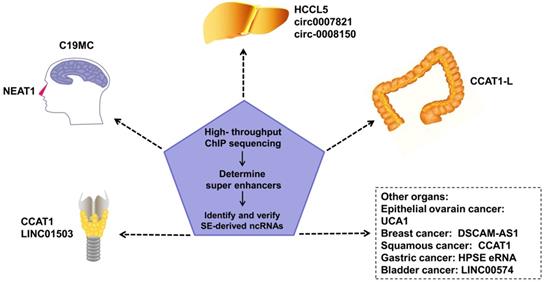
Regulating tumor progression
SEs in tumor cells can drive oncogene expression by mutating to produce new SEs, chromosomal translocations of proto-SEs, local amplification, overexpression of oncogenic TFs, and other genetic mechanisms. SEs regulate the malignant biological phenotype of tumors via these mechanisms [47-49]. SE-derived ncRNAs have been reported to be driven by SEs in many tumors (Figure 2). Estrogen receptor α in the absence of hormones (apoERα) can induce the expression of the SE-derived lncRNA DSCAM-AS1 and affect breast cancer tumorigenesis [50]. There are many TNF-β-regulating SE-derived lncRNAs. Considering that hepatic stellate cells are involved in hepatic fibrosis and cirrhosis, this finding suggests that these lncRNAs are involved in liver cancer [51]. In addition, regions of active enhancers or SEs can be transcribed to form eRNAs to synergize with the SEs of proto-oncogenes to further accelerate tumor occurrence and development [52]. However, not all tumor-related SEs play a carcinogenic role. Some SE-derived miRNAs inhibit tumor progression [8].
SE-derived ncRNAs, temperature regulation, and circadian rhythm
Raymond et al. [53] found that under cold stimulation, SE-miR-32 activates the p38/MAPK signaling pathway by inhibiting the expression of target gene Tob1 and promoting the expression of FGF21, to drive brown adipose tissue (BAT) thermogenesis and white fat browning in mice. Fan et al. [54] found that the SE-derived lncRNA lnc-Crot is a long-range circadian lncRNA, and its role is influenced by the TFs BMAL1 and REV-ERBα.
SE-derived ncRNAs and immune cells
SE-derived ncRNAs are closely associated with inflammation-related diseases and immune cells. Vahedi et al. [55] indicated that one-third of the ncRNAs (501/1524) in T cells are formed from the self-transcription of SEs; they are known as eRNAs, and most of them have a Bach2-binding site. It is noteworthy that genetic variation at this site is associated with many immune diseases, including rheumatoid arthritis, Crohn's disease, multiple sclerosis, asthma, and type I diabetes mellitus (T1DM). Agirre et al. [56] reported that eRNAs from SE transcription are involved in B cell receptor activation and the humoral immune response. There is an interaction between activation-induced deaminase (AID) and SE-derived ncRNAs, which affects the production and classification of antibodies in B cells [57, 58]. SE is also involved in the pathogenesis of autoimmune diseases. Many novel SE-derived lncRNAs have been found in leukocytes from patients with autoimmune diseases, such as Crohn disease and irritable bowel syndrome [59]. In peripheral mononuclear erythrocytes from patients with multiple sclerosis (MS), SE enrichment and circRNA (has_circ_0043813) expression increase simultaneously [60], but the specific mechanism underlying this phenomenon has not yet been elucidated.
Role of SE-derived ncRNAs in cancer
The article “Hallmarks of cancer: the next generation” published by Hanahan and Weinberg in 2011 in Cell summarizes the ten main characteristics of tumors. Recent studies have shown that SE-derived ncRNAs are closely related to the occurrence and development of tumors. SE-derived ncRNAs are involved in malignant biological behaviors such as uncontrolled proliferation, invasion and metastasis, chemoresistance, and tumor inflammatory response.
SE-derived ncRNAs regulate malignant tumor proliferation. A.The transcription factor SOX2 and p63 can bind to the SE region upstream of lncRNA CCAT1 to promote its expression. The obtained CCAT1 can form transcription complexes with SOX2 and p63 to regulate the expression of EGFR, leading to the malignant proliferation of SCC cells through the MEK/ERK1/2 and PI3K/AKT pathways. B. That KLF6 super enhancer was deleted will induce the over-expression of miR-1301, thereby inhibiting hepatoma cell proliferation by inducing p21and p53 in a p53-dependent manner. C. SE-derived lncRNA UCA1 interacts with AMOT to promote YAP activation and nuclear translocation and induces the expression of YAP's target genes, thereby driving the development of ovarian cancer. D. The transcription factor TP63 binds to the SE region upstream of LINC01503 to activate its expression; activated SE-derived LINC01503, in combination with EBP-1 and ERK2, stimulates the malignant proliferation of esophageal squamous cell carcinoma (ESCC) cells by activating the PI3K/AKT1 and ERK/MAPK signaling pathways.
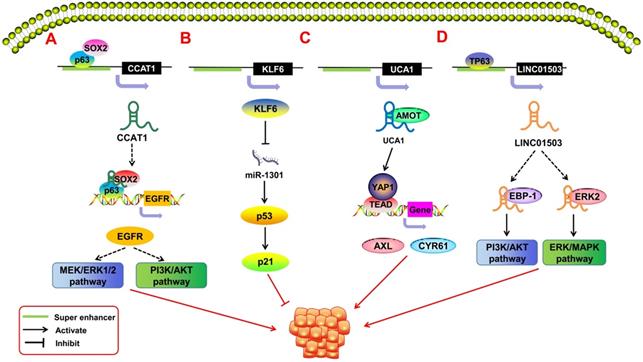
SE-derived ncRNAs and malignant proliferation
Sustaining proliferative signaling is a fundamental feature of tumors. Normal cells tightly regulate growth-promoting and death-inducing signals to guide cell cycle and cell growth and maintain the homeostatic cell number and tissue architecture. However, these signals are deregulated in cancerous cells, thus leading to unlimited proliferation [61]. The malignant proliferation of tumors involves the regulation of multiple signaling pathways (e.g. the PI3K pathway) and a variety of key proteins (e.g. cyclin).
In recent years, SE-derived ncRNAs have been found to play an important role in promoting the malignant proliferation of tumors (Figure 3). Some SE-derived ncRNAs affect proliferation-related signaling pathways, thus leading to the unlimited proliferation of tumor cells [62]. Xie et al. [63] reported a squamous cell carcinoma (SCC)-specific SE-derived lncRNA LINC01503. The TF TP63 binds to the SE region upstream of LINC01503 in esophageal squamous cell carcinoma (ESCC) and activates it. Activated SE-LINC01503, in combination with EBP-1 and ERK2, stimulates the malignant proliferation of ESCC cells by activating the PI3K/AKT and ERK/MAPK signaling pathways. Similarly, Jiang et al. [64] found that in SCC, the tissue-specific lncRNA CCAT1 is regulated by SEs. CCAT1 has been shown to promote malignant proliferation in many types of tumors [6, 65, 66]. SOX2 and p63 can bind to the SE region upstream of CCAT1 to regulate the transcriptional expression of CCAT1. The obtained CCAT1 can form transcription complexes with SOX2 and p63 to regulate the expression of EGFR and then lead to the malignant proliferation of SCC cells through the MEK/ERK1/2 and PI3K/AKT pathways. Xiang et al. [67] pointed out that SE-derived CCAT1-L can also promote tumor proliferation by enhancing the transcription of the oncogene MYC. The Hippo/YAP signaling pathway has also been associated with tumor proliferation [68, 69]. Lin et al. [70] showed that in epithelial ovarian cancer (EOC), the SE-derived lncRNA UCA1 interacts with the Hippo/YAP1 signaling pathway and reduces the phosphorylation level of YAP1 in the cytoplasm of tumor cells. This encourages the entry of YAP1 into the nucleus and promotes the expression of the target genes AXL and CYR61, which promote cell proliferation. SE-derived ncRNAs also play an important role in virus-associated tumors. Liang et al. [71] showed that Epstein-Barr virus (EBV) SEs can affect the expression of the proto-oncogene MYC in the regulation of the growth and survival of lymphoblastoid cell lines, and it is an important risk factor for EBV-associated malignant proliferation.
SE-derived ncRNAs regulate tumor metastasis. A. The transcription factor ZEB1 binds to the SE region upstream of lncRNA HCCL5 to activate its expression; activated SE-derived HCCL5 accelerates the EMT phenotype of hepatocellular carcinoma (HCC) by upregulating the expression of Snail, Slug, ZEB1, and Twist1. B. C19MC is overexpressed when transcription factor TTYH1 binds to the SE region and initiates the C19MC-LIN28A-MYCN circuit by inhibiting LIN28A and upregulating MYCN to regulate the progression of embryonal tumors with multilayered rosettes (ETMRs). C. The transcription factors FOS, ZEB1, and MAX bind to the SE region potentially upstream of the LINC00152 promoter; knocking out LINC00152 inhibits the invasion and metastasis of breast cancer cells. D. Transcription factor YY1 combines p65/p300 to form a transcription complex to promote quaking (QKI) expression by binding to the SE regions of QKI; meanwhile, QKI can cause the formation of circ-0008150 and circ-0007821 to promote the expression of EMT-related markers vimentin and zeb1 by adsorbing miR-615-5p and miR-381-3p.
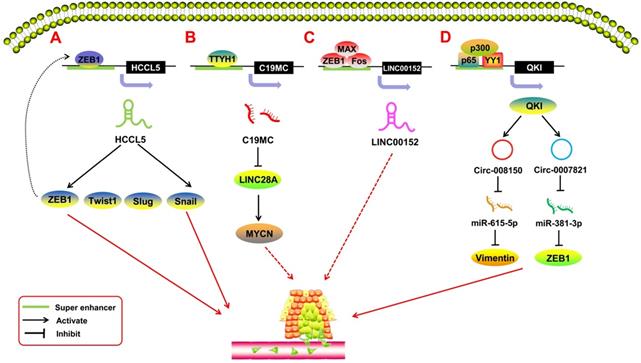
SE-derived ncRNAs and metastasis
Tumor metastasis is a dynamic process in which malignant cells spread to different tissues and organs from the primary tumor site and continue to proliferate and form new secondary tumors. The secondary and primary tumor tissues always have the same histological type [61, 72, 73]. Tumor metastasis usually involves the following mechanisms: (1) epithelial-mesenchymal transition (EMT), (2) tumor heterogeneity and cancer stem cells, and (3) angiogenesis and hypoxia induction [74, 75].
SE-derived ncRNAs have been reported to play a crucial role in the process of tumor metastasis (Figure 4). EMT is the most common mechanism of tumor metastasis. Peng et al. [76] reported a SE-derived lncRNA, HCCL5, which is hepatocellular carcinoma (HCC)-specific. The TF ZEB1 can bind to the SE region upstream of HCCL5 to regulate its transcription. By upregulating the expression of the TFs Snail, Slug, ZEB1, and Twist1, SE-HCCL5 promotes EMT in HCC cells. Xu et al. [77] assessed the regulatory network of LINC00152 in pan-cancer through bioinformatics analysis and found that the TFs FOS, ZEB1, and MAX bind to the SE region potentially upstream of the LINC00152 promoter, and knocking out LINC00152 inhibits the invasion and metastasis of the breast cancer cell line MDA-MB-231. Xie et al. [63] indicated that SE-LINC01503 not only promotes proliferation, but also plays a positive role in SCC metastasis. Upon abolishing the expression of SE-LINC01503, the migration and invasion of SCC cells was markedly inhibited. In terms of SE-derived miRNAs, Chan et al. [78] reported on C19MC, a large SE-derived miRNA cluster. The SE region upstream of C19MC can be regulated by TTYH1, and overexpressed C19MC drives the C19MC-LIN28A-MYCN circuit in embryonal tumors with multilayered rosettes (ETMRs). This oncogenic circuit promotes tumor metastasis in ETMRs [79]. With regard to SE-derived circRNAs, Han et al. [80] showed that the TF Yin-Yang 1 (YY1), combined with p65/p300, forms a transcription complex that promotes quaking (QKI) expression by binding to the SE regions of QKI. Meanwhile, QKI can cause the formation of circ-0008150 and circ-0007821 to promote the expression of EMT-related markers vimentin and zeb1 by adsorbing miR-615-5p and miR-381-3p. This process is highly activated during EMT in HCC, and the oncogenic SE-derived circRNAs would, in turn, promote the metastasis of HCC cells.
SE-derived ncRNAs and drug resistance
Chemoresistance is the ability of tumor cells to evade or cope with the presence of chemotherapeutics and is a key challenge in cancer treatment. Chemoresistance mechanisms are complex and usually depend on tumor stem cells, apoptosis, EMT, the tumor microenvironment, and many other factors [81]. The increased consumption of rapidly growing tumor cells leads to hypoxia. A hypoxic microenvironment enhances malignancy and chemoresistance of tumor cells. Hypoxia affects intracellular signaling through the TF hypoxia-inducible factor-1a (HIF-1α) which binds to the hypoxia response elements of many genes, including hexokinase and glucose transporter 1 (GLUT1). This forces the tumor cell to produce ATP through glycolysis and create a lower pH to prevent drug entry [82]. Moreau et al. [83] reported that hypoxia can induce the activation of 358 SE regions in tumor cells, and database analysis showed that for 20% of SEs, the closest RefSeq gene was a lncRNA, as exemplified by MALAT1 and LUCAT1. MALAT1 can not only achieve chemoresistance by competitively inhibiting miR-23b-3p or miR-203, but can also bind with EZH2 to regulate CDH1 transcription expression and promote E-cadherin expression and oxaliplatin-induced EMT [84-86]. Xu et al. [77] investigated the chemoresistance of SE-derived LINC00152 across all cancers. Through microarray technology, they found that LINC00152 may be related to methotrexate resistance in pancreatic, colon, and osteosarcoma cell lines. Yue's study [87] showed that LINC00152 sponged miR-193a-3p to block its inhibitory effect on erbB-4 and mediated the activation of the AKT signaling pathway, which is involved in tumor chemoresistance.
In addition, chemoresistance is usually associated with the inhibition of apoptosis in tumor cells. Inducing apoptosis is one of the anti-tumor mechanisms of chemotherapy drugs, while anti-apoptosis has become an effective pathway for chemoresistance [88, 89]. Anti-apoptosis activity in tumors can occur in the following ways: (1) shearing pro-apoptotic genes and inhibiting TF recruitment to inactivate pro-apoptotic genes (e.g. p53) and (2) overexpression of anti-apoptotic genes (e.g. Bcl-2 and survivin) through transcriptional regulation networks [90]. Studies have shown that SE-derived ncRNAs are closely related to the anti-apoptosis characteristics of tumors. Lin et al. [70] found that after SE-UCA1 activates the Hippo/YAP1 pathway, proliferation and cell survival are simultaneously promoted in ovarian cancer cells, showing some anti-apoptosis characteristics. In addition, Moreau et al. [83] found that hypoxia can regulate SE activity, which in turn, mediates tumor apoptosis, and can also activate the expression of some lncRNAs. However, whether SE regulates apoptosis by regulating the expression of lncRNAs has not been further analyzed. SE-miR-200a is a proven epithelial-specific SE-derived ncRNA [8], and miR-200a can inhibit cell apoptosis in many tumors [91-93]. Therefore, we speculate that SE-miR-200a is involved in the chemoresistance mechanism whereby apoptosis is inhibited in tumor cells. To date, the mechanism underlying the action of SE-derived ncRNAs in tumor chemoresistance has not been fully elucidated, and findings obtained from high-throughput tests and bioinformatics analysis require further verification.
SE-derived ncRNAs and inflammatory response
Tumor inflammatory response is an important feature of tumors. Approximately 25% of tumors are caused by inflammation, and inflammatory cell infiltration occurs in almost all tumor microenvironments, and inflammatory cells and factors affect every step of tumor development [94]. Recent studies reported that SE-derived ncRNAs play a crucial role in regulating tumor inflammatory response (Figure 5).
Immune cells such as T regulatory cells (Tregs) are important components of the tumor inflammatory microenvironment; they are involved in immune escape and immune tolerance of the tumors. If these Tregs are too active, they suppress the immune response and accelerate tumor invasion in the body. Anandagoda et al. [95] found that in Tregs, the TF FoxP3 can bind to the SE regions of pri-miR-142 to promote its transcription, thereby inhibiting the expression of its downstream target gene PDE3b. SE-derived miR-142 leads to immune tolerance. In addition, Duan et al. [96], through ChIP-seq and relevant data analysis, found that TFs and the co-factors NF-κB, BRD4, and RNA Pol II can bind to the SE region upstream of pri-miR-146a and pri-miR-155 and promote their transcription. Both miR-146a and miR-155 are inflammation-associated miRNAs. miR-146a can mediate tumor angiogenesis by participating in the recruitment and activation of tumor-associated macrophages [97]. It can also promote the secretion of epidermal growth factor (EGF) and colony stimulating factor-1 (CSF-1) [98]. In contrast, miR-155 can be released into the tumor microenvironment by malignant cells and transferred into normal cells via exosomes. Exosomal miR-155 regulates the expression of inflammatory factors such as IL-6 and IL-8 [99]. Angiotensin II (Ang II) is also an inflammatory factor in the tumor microenvironment, and it can mediate angiogenesis and tumor metastasis [100]. Das et al. [101] reported that Ang II can induce transcription of SE-derived lnc-Ang383 in vascular smooth muscle cells (VSMCs), and it promotes VSMC proliferation and angiogenesis. However, whether Ang II/SE-derived lncRNAs can participate in tumor growth requires further exploration. Above all, it is clear that SE-derived ncRNAs play an important role in the tumor inflammatory response.
SE-derived ncRNAs regulate tumor-associated inflammatory response. A. The transcription factor FOXP3 can bind to the SE regions of pri-miR-142 to promote its transcription, thereby inhibiting the expression of its downstream target gene PDE3b. SE-derived miR-142 leads to immune tolerance. B. Transcription factors and the co-factors NF-κB, BRD4, and RNA Pol II can bind to the SE region upstream of pri-miR-146a and pri-miR-155 and promote their transcription. Tumor angiogenesis can be mediated by miR-146a, which participates in the recruitment and activation of tumor-associated macrophages to induce the secretion of epidermal growth factor (EGF) and colony stimulating factor-1 (CSF-1); miR-155 can be released into the tumor microenvironment by malignant cells and transferred into normal cells via exosomes. C. SE-derived lnc-Ang383 is induced to transcript in vascular smooth muscle cells (VSMCs) and promotes VSMC proliferation and angiogenesis.
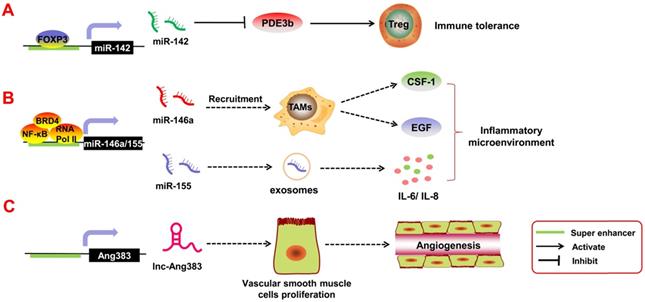
Clinical applications of SE-derived ncRNAs in cancers
With the development of high-throughput sequencing technologies and bioinformatics prediction software, an increasing number of SE-derived ncRNAs have been identified. SE-derived ncRNAs have shown increasing significance in the early diagnosis of tumors, evaluation of tumor prognosis, targeting of therapies for tumors, and many other tumor-related clinical applications (Table 3).
In recent years, many studies have reported that the abnormal expression of SE-derived ncRNAs in a variety of tumor tissues is closely related to the stage, malignancy, infiltration, and other clinicopathological characteristics of patients with cancer. SE-derived ncRNAs are potential tumor markers, especially SE-derived lncRNAs. Xie et al. [63] reported that levels of the SE-derived lncRNA LINC01503 were substantially higher in cancerous tissues than in the adjacent non-malignant esophageal epithelium; high LINC01503 expression was significantly correlated with shorter overall survival (OS) and disease-free survival (DFS) in patients with cancer. Meanwhile, based on the Cox proportional hazard model, LINC01503 serves as an independent prognostic factor for poor survival by multivariate regression analysis. Similarly, Peng et al. [76] demonstrated that the expression of the SE-derived lncRNA HCCL5 was significantly higher in HCC tissues than in adjacent non-malignant tissues by ISH analysis. The expression of HCCL5 was closely related to the gender, pathological diagnosis, and tumor grade of patients with HCC; and cancer patients with high HCCL5 expression had significantly shorter OS and DFS, according to the Cancer Genome Atlas (TCGA) database analysis. Furthermore, CCAT1-L is reported to be an SE-derived lncRNA in colorectal cancer (CRC), and the expression of CCAT1-L was higher in CRC tissues than in paired normal mucosa tissues [67]. Similarly, the SE-derived lncRNA NEAT1 was found to be higher in nasopharynx cancer (NPC) tissues than in the normal mucosa [104]. Lin et al. [70] reported that the SE-derived lncRNA UCA1 was highly expressed in patients with EOC, and the expression of UCA1 was positively correlated with high-grade cancer, as determined by TCGA analysis. Therefore, UCA1 could be used as a diagnostic marker. Although some SE-derived lncRNAs have not been reported to be involved in the clinicopathological characteristics of tumors, lncRNAs have been shown to be associated with the clinicopathological characteristics of a variety of tumors in other studies. Elevated expression of SE-MALAT1 is related to TNM stage, lymph node metastasis, and poor prognosis in tumors such as lung cancer, gastric cancer, and thyroid carcinoma [105-109]. SE-LINC00152 was discovered using a high-throughput pan-cancer method [77]. In multiple studies and meta-analyses, the expression of LINC00152 was shown to be closely related to the clinical outcome, OS, and DFS in a variety of tumors, such as lung, liver, and stomach cancer [110-113].
As pivotal tumor markers, miRNAs and circRNAs have clinical significance in the diagnosis and prognosis of tumors such as lung cancers and colon cancers [114-116] However, there are few relevant reports on SE-derived miRNAs and SE-derived circRNAs in the clinicopathological characteristics of tumors. Patrick et al. [78] reported that the high expression of SE-C19MC in ETMRs was related to high malignancy and poor prognosis. Although C19MC was demonstrated to be involved in the T stage and vascular invasion in hepatocellular carcinoma [117], the clinicopathological characteristics of SE-C19MC in hepatocellular carcinoma remain unclear. MiR-155, a SE-derived miRNA [96], is overexpressed in breast cancer, cervical cancer, malignant B-cell lymphoma, and many other tumors, and the expression of miR-155 has been reported to be associated with some malignant phenotypes, such as tumor location, tumor grade, TNM staging, and distant metastasis [118-124]. Derived from epithelium-specific SEs, miR-200a has also been reported in several studies to be related to the clinical characteristics and prognosis of head and neck squamous cell carcinoma and ovarian carcinoma [125-129]. However, none of these studies have directly linked SE with the clinical applications of miRNAs. Thus, the clinical applications of these SE-ncRNAs deserve further studies.
Conclusions and perspectives
SEs and ncRNAs are both hotspots in the field of tumor research, and studies have shown how SE-derived ncRNAs play a role in tumor development. SE-derived ncRNAs could become a new entry point for tumor therapeutics. Currently, there are two main types of targeted inhibitors for SE-derived ncRNAs. (1) RNA-targeting inhibitors: For example, an anti-miR-155 inhibitor and modified miR-155-DOPC liposome nanoparticles have significant inhibitory effects on tumor cells [130, 131]. Furthermore, in vivo tumorigenesis experiments in mice showed that antisense oligonucleotides of MALAT1 could significantly inhibit tumor invasion and metastasis [132, 133]. (2) SE-targeting inhibitors: The SEs inhibitors ThZ1 and JQ1 can effectively resist the expression of MYC and inhibit the proliferation, migration, and invasion of osteosarcoma cells [134]. Interestingly, ThZ1 and JQ1 are broad-spectrum inhibitors; hence, whether they will remain effective towards SE-derived ncRNAs is unclear. Once demonstrated experimentally, SE inhibitors will be a new option for cancer treatment. In addition, immunotherapy has emerged as a new attractive treatment for tumors [135]. An increasing number of immunosuppressors have been found; Xu et al. [77] demonstrated that SE-derived ncRNAs may become the next breakthrough for the immunotherapy of solid tumors.
The correlation between SE-ncRNAs and Clinicopathological features of tumors
| Tumor types | SE-ncRNA | Expression | Clinicopathology features | Ref. |
|---|---|---|---|---|
| Esophageal squamous cell carcinoma | LINC01503 | Upregulated | Shorter overall survival and disease-free survival | [63] |
| CCAT1 | Upregulated | Lymph node metastasis and TNM staging | [65] | |
| Hepatocellular carcinoma | HCCL5 | Upregulated | Gender, pathological diagnosis and tumor grade, and shorter overall survival and disease-free survival | [76] |
| circ-0008150; circ-0007821 | — | [80] | ||
| Epithelial ovarian cancer | UCA1 | Upregulated | Grade, and poorer survival | [70] |
| Embryonal tumor with multilayered rosettes | C19MC (miRNA family) | Upregulated | Frequent copy-number aberrations, and diagnosis indicator | [78] |
| Gastric cancer | HPSE eRNA | Upregulated | Local invasion, lymph node metastasis, advanced TNM stage, and shorter overall survival | [102] |
| Bladder cancer | LINC00574 | Upregulated | Shorter overall survival | [103] |
| Colorectal cancer | CCAT1-L | Upregulated | — | [67] |
| Breast cancer | DSCAM- AS1 | Upregulated | — | [50] |
| Squamous cancer | CCAT1 | Upregulated | — | [64] |
| Lymphocytic leukemia | ESE RNA | Upregulated | — | [71] |
| Nasopharynx cancer | NEAT1 | Upregulated | — | [104] |
The SE-ncRNAs associated databases
| Database | Functions of database | Website | Ref. |
|---|---|---|---|
| dbSUPER | The first integrated interactive database of SEs in transcriptional regulation of cellular identities and diseases | http://bioinfo.au.tsinghua.edu.cn/dbsuper/ | [136] |
| SEA | Includes SEs in multiple species and their roles in cellular identities | http://sea.edbc.org | [137] |
| SEdb | A wide range of human genome SEs and their potential roles in gene regulation | http://www.licpathway.net/sedb | [138] |
| SEanalysis | Provides a comprehensive analysis of SE-associated regulatory networks, the relationship between SE-associated genes and TFs | http://licpathway.net/SEanalysis | [139] |
| SEA version 3.0 | A comprehensive extension and update of the Super-Enhancer archive. | http://sea.edbc.org | [140] |
| DEEPSEN | A convolutional neural network based method for super-enhancer prediction. | https://github.com/1991Troy/DEEPSEN | [141] |
| dbCoRC | A database of core transcriptional regulatory circuitries modeled by H3K27ac ChIP-seq signals. | http://dbcorc.cam-su.org | [142] |
| Cistrome Cancer | A comprehensive resource for predicted transcription factor (TF) targets and enhancer profiles and "super-enhancer" target genes | http://cistrome.org/CistromeCancer/ | [143] |
| SELER | Transcriptional regulation of SE-lncRNAs in human tumors | http://www.seler.cn | [144] |
| TRCirc | Mainly includes transcription of circRNAs and partial SE-circRNAs | http://www.licpathway.net/TRCirc | [145] |
However, there are also some limitations in the present study. First, specific SE-derived ncRNAs have different expression levels in different tumors and regulate different tumor phenotypes. This may be related to tissue specificity, but the concrete mechanism underlying this phenomenon has not been clarified. Second, databases of SE-derived ncRNAs need further improvement. There are some databases of SEs and their disease-related phenotypes [136-143] (Table 4), but when this article was written, there were only two databases summarizing SE-derived ncRNAs [144, 145]. The database SELER established by Guo et al. [144] mainly includes the transcriptional regulation pathways of SE-associated lncRNAs in human tumors, and TRCirc, a circRNA database established by Tang et al. [145], provides resources for the efficient retrieval, browsing, and visualization of transcriptional regulation information for related sequences. The shortcomings of relevant databases will become an obstacle for future research. There is limited knowledge of SE-derived ncRNAs in clinical research. Although correlation analysis has been carried out on some SE-derived ncRNAs and tumor clinicopathological features and prognostic indicators, the guiding significance of most SE-derived ncRNAs as prognostic factors was obtained through meta-analysis and other data mining approaches, needing further clinical verification. Evidence on SE-derived ncRNAs as a therapeutic target for cancer is insufficient. Although some studies have used inhibitors to target the corresponding SE-derived ncRNAs in tumor progression experiments, most of them used animal models. The inhibitory effects of SE-derived ncRNAs in humans lack supporting data. Therefore, whether these basic research results can be translated into clinical applications remains to be elucidated.
Abbreviations
SE: super enhancer; TE: typical enhancer; ncRNA: non-coding RNA; miRNA: microRNA; lncRNA: long non-coding RNA; circRNA: circular RNA; TF: transcription factor; BRD4: Bromodomain containing 4; ChIP-seq: Chromatin immunoprecipitation; eRNA: enhancer RNA; PCR: Polymerase Chain Reaction; ISH: in situ hybridization; mESC: mouse embryonic stem cell; apoERα: Estrogen Receptor α in absence of hormones; BAT: Brown adipose tissue; AID: Activation-induced deaminase; ESCC: esophageal squamous cell carcinoma; ESE: EBV super enhancer; LCL: lymphoblastoid cell lines; EMT: epithelial-mesenchymal; ETMRs: Embryonal tumors with multilayered rosettes; QKI: Quaking; HIF-1a: hypoxia inducible factor-1a; HRE: hypoxia responsible element; GLUT1: Glucose transporter 1; Treg: T regulatory cells; TAMs: tumor-associated macrophages; EGF: epidermal growth factor; CSF-1: colony stimulating factor-1; AngⅡ: Angiotensin Ⅱ; VSMCs: Vascular Smooth Muscle Cells; OS: overall survival; DFS: disease-free survival; ISH: in situ hybridization; CRC: Colorectal Cancer; ASO: antisense oligonucleotide.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (81903032), the National Key Research and Development Program of China (2016YFC1201800), the China Postdoctoral Science Foundation (2020M672520), the student innovation project of Central south university (1053320191093), and the Youth Fund of Xiangya Hospital (2018Q011).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7-30
2. Cooner WH. Definition of the ideal tumor marker. Urol Clin North Am. 1993;20:575-9
3. He X, Li S, Yu B, Kuang G, Wu Y, Zhang M. et al. Up-regulation of LINC00467 promotes the tumourigenesis in colorectal cancer. J Cancer. 2019;10:6405-13
4. Kanwal R, Plaga AR, Liu X, Shukla GC, Gupta S. MicroRNAs in prostate cancer: Functional role as biomarkers. Cancer Lett. 2017;407:9-20
5. Galardi A, Colletti M, Businaro P, Quintarelli C, Locatelli F, Di Giannatale A. MicroRNAs in Neuroblastoma: Biomarkers with Therapeutic Potential. Curr Med Chem. 2018;25:584-600
6. Kojima M, Sudo H, Kawauchi J, Takizawa S, Kondou S, Nobumasa H. et al. MicroRNA markers for the diagnosis of pancreatic and biliary-tract cancers. PLoS One. 2015;10:e0118220
7. Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH. et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307-19
8. Suzuki HI, Young RA, Sharp PA. Super-Enhancer-Mediated RNA Processing Revealed by Integrative MicroRNA Network Analysis. Cell. 2017;168:1000-14 e15
9. Cai J, Chen S, Yi M, Tan Y, Peng Q, Ban Y. et al. DeltaNp63alpha is a super enhancer-enriched master factor controlling the basal-to-luminal differentiation transcriptional program and gene regulatory networks in nasopharyngeal carcinoma. Carcinogenesis. 2019 bgz203
10. He Y, Long W, Liu Q. Targeting Super-Enhancers as a Therapeutic Strategy for Cancer Treatment. Front Pharmacol. 2019;10:361
11. Fukaya T, Lim B, Levine M. Enhancer Control of Transcriptional Bursting. Cell. 2016;166:358-68
12. Hnisz D, Shrinivas K, Young RA, Chakraborty AK, Sharp PA. A Phase Separation Model for Transcriptional Control. Cell. 2017;169:13-23
13. Sabari BR, Dall'Agnese A, Boija A, Klein IA, Coffey EL, Shrinivas K. et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science. 2018 361
14. Pott S, Lieb JD. What are super-enhancers? Nat Genet. 2015;47:8-12
15. Chen X, Bhadauria V, Ma B. ChIP-Seq: A Powerful Tool for Studying Protein-DNA Interactions in Plants. Curr Issues Mol Biol. 2018;27:171-80
16. Cao F, Fang Y, Tan HK, Goh Y, Choy JYH, Koh BTH. et al. Super-Enhancers and Broad H3K4me3 Domains Form Complex Gene Regulatory Circuits Involving Chromatin Interactions. Sci Rep. 2017;7:2186
17. Davies JO, Oudelaar AM, Higgs DR, Hughes JR. How best to identify chromosomal interactions: a comparison of approaches. Nat Methods. 2017;14:125-34
18. Jiang YY, Jiang Y, Li CQ, Zhang Y, Dakle P, Kaur H. et al. TP63, SOX2, and KLF5 Establish a Core Regulatory Circuitry That Controls Epigenetic and Transcription Patterns in Esophageal Squamous Cell Carcinoma Cell Lines. Gastroenterology. 2020 S0016-5085(20)34854-X.19
19. Kong S, Zhang Y. Deciphering Hi-C: from 3D genome to function. Cell Biol Toxicol. 2019;35:15-32
20. Huang J, Li K, Cai W, Liu X, Zhang Y, Orkin SH. et al. Dissecting super-enhancer hierarchy based on chromatin interactions. Nat Commun. 2018;9:943
21. Barakat TS, Halbritter F, Zhang M, Rendeiro AF, Perenthaler E, Bock C. et al. Functional Dissection of the Enhancer Repertoire in Human Embryonic Stem Cells. Cell Stem Cell. 2018;23:276-88 e8
22. Muerdter F, Boryn LM, Arnold CD. STARR-seq - principles and applications. Genomics. 2015;106:145-50
23. Ou C, Sun Z, Li X, Li X, Ren W, Qin Z. et al. MiR-590-5p, a density-sensitive microRNA, inhibits tumorigenesis by targeting YAP1 in colorectal cancer. Cancer Lett. 2017;399:53-63
24. Zheng X, Wang J, Wei L, Peng Q, Gao Y, Fu Y. et al. Epstein-Barr Virus MicroRNA miR-BART5-3p Inhibits p53 Expression. J Virol. 2018 92
25. Chen H, Du G, Song X, Li L. Non-coding Transcripts from Enhancers: New Insights into Enhancer Activity and Gene Expression Regulation. Genomics Proteomics Bioinformatics. 2017;15:201-7
26. Mousavi K, Zare H, Dell'orso S, Grontved L, Gutierrez-Cruz G, Derfoul A. et al. eRNAs promote transcription by establishing chromatin accessibility at defined genomic loci. Mol Cell. 2013;51:606-17
27. Hsieh CL, Fei T, Chen Y, Li T, Gao Y, Wang X. et al. Enhancer RNAs participate in androgen receptor-driven looping that selectively enhances gene activation. Proc Natl Acad Sci U S A. 2014;111:7319-24
28. Pefanis E, Wang J, Rothschild G, Lim J, Kazadi D, Sun J. et al. RNA exosome-regulated long non-coding RNA transcription controls super-enhancer activity. Cell. 2015;161:774-89
29. Huang S, Li X, Zheng H, Si X, Li B, Wei G. et al. Loss of Super-Enhancer-Regulated circRNA Nfix Induces Cardiac Regeneration After Myocardial Infarction in Adult Mice. Circulation. 2019;139:2857-76
30. Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597-610
31. Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629-41
32. Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904-14
33. Zhang Z, Yang T, Xiao J. Circular RNAs: Promising Biomarkers for Human Diseases. EBioMedicine. 2018;34:267-74
34. Ebbesen KK, Kjems J, Hansen TB. Circular RNAs: Identification, biogenesis and function. Biochim Biophys Acta. 2016;1859:163-8
35. Nie H, Wang Y, Liao Z, Zhou J, Ou C. The function and mechanism of circular RNAs in gastrointestinal tumours. Cell Prolif. 2020;53(7):e12815
36. Mao R, Wu Y, Ming Y, Xu Y, Wang S, Chen X. et al. Enhancer RNAs: a missing regulatory layer in gene transcription. Sci China Life Sci. 2019;62:905-12
37. Lee JH, Xiong F, Li W. Enhancer RNAs in cancer: regulation, mechanisms and therapeutic potential. RNA Biol. 2020 p: 1-10
38. Song Y, van den Berg PR, Markoulaki S, Soldner F, Dall'Agnese A, Henninger JE. et al. Dynamic Enhancer DNA Methylation as Basis for Transcriptional and Cellular Heterogeneity of ESCs. Mol Cell. 2019;75:905-20 e6
39. Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP. et al. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310:1817-21
40. Ounzain S, Micheletti R, Arnan C, Plaisance I, Cecchi D, Schroen B. et al. CARMEN, a human super enhancer-associated long noncoding RNA controlling cardiac specification, differentiation and homeostasis. J Mol Cell Cardiol. 2015;89:98-112
41. Ounzain S, Micheletti R, Beckmann T, Schroen B, Alexanian M, Pezzuto I. et al. Genome-wide profiling of the cardiac transcriptome after myocardial infarction identifies novel heart-specific long non-coding RNAs. Eur Heart J. 2015;36:353-68a
42. Anderson KM, Anderson DM, McAnally JR, Shelton JM, Bassel-Duby R, Olson EN. Transcription of the non-coding RNA upperhand controls Hand2 expression and heart development. Nature. 2016;539:433-6
43. Micheletti R, Plaisance I, Abraham BJ, Sarre A, Ting CC, Alexanian M. et al. The long noncoding RNA Wisper controls cardiac fibrosis and remodeling. Sci Transl Med. 2017;9(395):eaai9118
44. Grigelioniene G, Suzuki HI, Taylan F, Mirzamohammadi F, Borochowitz ZU, Ayturk UM. et al. Gain-of-function mutation of microRNA-140 in human skeletal dysplasia. Nat Med. 2019;25:583-90
45. Degirmenci U, Li J, Lim YC, Siang DTC, Lin S, Liang H. et al. Silencing an insulin-induced lncRNA, LncASIR, impairs the transcriptional response to insulin signalling in adipocytes. Sci Rep. 2019;9:5608
46. Alvarez-Dominguez JR, Knoll M, Gromatzky AA, Lodish HF. The Super-Enhancer-Derived alncRNA-EC7/Bloodlinc Potentiates Red Blood Cell Development in trans. Cell Rep. 2017;19:2503-14
47. Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA. et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934-47
48. Mansour MR, Abraham BJ, Anders L, Berezovskaya A, Gutierrez A, Durbin AD. et al. Oncogene regulation. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science. 2014;346:1373-7
49. Sengupta S, George RE. Super-Enhancer-Driven Transcriptional Dependencies in Cancer. Trends Cancer. 2017;3:269-81
50. Miano V, Ferrero G, Rosti V, Manitta E, Elhasnaoui J, Basile G. et al. Luminal lncRNAs Regulation by ERalpha-Controlled Enhancers in a Ligand-Independent Manner in Breast Cancer Cells. Int J Mol Sci. 2018 p:19
51. Zhou C, York SR, Chen JY, Pondick JV, Motola DL, Chung RT. et al. Long noncoding RNAs expressed in human hepatic stellate cells form networks with extracellular matrix proteins. Genome Med. 2016;8:31
52. Rothschild G, Basu U. Lingering Questions about Enhancer RNA and Enhancer Transcription-Coupled Genomic Instability. Trends Genet. 2017;33:143-54
53. Ng R, Hussain NA, Zhang Q, Chang C, Li H, Fu Y. et al. miRNA-32 Drives Brown Fat Thermogenesis and Trans-activates Subcutaneous White Fat Browning in Mice. Cell Rep. 2017;19:1229-46
54. Fan Z, Zhao M, Joshi PD, Li P, Zhang Y, Guo W. et al. A class of circadian long non-coding RNAs mark enhancers modulating long-range circadian gene regulation. Nucleic Acids Res. 2017;45:5720-38
55. Vahedi G, Kanno Y, Furumoto Y, Jiang K, Parker SC, Erdos MR. et al. Super-enhancers delineate disease-associated regulatory nodes in T cells. Nature. 2015;520:558-62
56. Agirre X, Meydan C, Jiang Y, Garate L, Doane AS, Li Z. et al. Long non-coding RNAs discriminate the stages and gene regulatory states of human humoral immune response. Nat Commun. 2019;10:821
57. Meng FL, Du Z, Federation A, Hu J, Wang Q, Kieffer-Kwon KR. et al. Convergent transcription at intragenic super-enhancers targets AID-initiated genomic instability. Cell. 2014;159:1538-48
58. Pefanis E, Wang J, Rothschild G, Lim J, Chao J, Rabadan R. et al. Noncoding RNA transcription targets AID to divergently transcribed loci in B cells. Nature. 2014;514:389-93
59. Aune TM, Crooke PS 3rd, Patrick AE, Tossberg JT, Olsen NJ, Spurlock CF 3rd. Expression of long non-coding RNAs in autoimmunity and linkage to enhancer function and autoimmune disease risk genetic variants. J Autoimmun. 2017;81:99-109
60. Paraboschi EM, Cardamone G, Solda G, Duga S, Asselta R. Interpreting Non-coding Genetic Variation in Multiple Sclerosis Genome-Wide Associated Regions. Front Genet. 2018;9:647
61. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646-74
62. Ri K, Kim C, Pak C, Ri P, Om H. The KLF6 Super Enhancer Modulates Cell Proliferation via MiR-1301 in Human Hepatoma Cells. Microrna. 2020;9:64-9
63. Xie JJ, Jiang YY, Jiang Y, Li CQ, Lim MC, An O. et al. Super-Enhancer-Driven Long Non-Coding RNA LINC01503, Regulated by TP63, Is Over-Expressed and Oncogenic in Squamous Cell Carcinoma. Gastroenterology. 2018;154:2137-51 e1
64. Jiang Y, Jiang YY, Xie JJ, Mayakonda A, Hazawa M, Chen L. et al. Co-activation of super-enhancer-driven CCAT1 by TP63 and SOX2 promotes squamous cancer progression. Nat Commun. 2018;9:3619
65. Zhang E, Han L, Yin D, He X, Hong L, Si X. et al. H3K27 acetylation activated-long non-coding RNA CCAT1 affects cell proliferation and migration by regulating SPRY4 and HOXB13 expression in esophageal squamous cell carcinoma. Nucleic Acids Res. 2017;45:3086-101
66. Zhu H, Zhou X, Chang H, Li H, Liu F, Ma C. et al. CCAT1 promotes hepatocellular carcinoma cell proliferation and invasion. Int J Clin Exp Pathol. 2015;8:5427-34
67. Xiang JF, Yin QF, Chen T, Zhang Y, Zhang XO, Wu Z. et al. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res. 2014;24:513-31
68. Ou C, Sun Z, Li S, Li G, Li X, Ma J. Dual roles of yes-associated protein (YAP) in colorectal cancer. Oncotarget. 2017;8:75727-41
69. Ou C, Sun Z, He X, Li X, Fan S, Zheng X. et al. Targeting YAP1/LINC00152/FSCN1 Signaling Axis Prevents the Progression of Colorectal Cancer. Adv Sci (Weinh). 2020;7:1901380
70. Lin X, Spindler TJ, de Souza Fonseca MA, Corona RI, Seo JH, Dezem FS. et al. Super-Enhancer-Associated LncRNA UCA1 Interacts Directly with AMOT to Activate YAP Target Genes in Epithelial Ovarian Cancer. iScience. 2019;17:242-55
71. Liang J, Zhou H, Gerdt C, Tan M, Colson T, Kaye KM. et al. Epstein-Barr virus super-enhancer eRNAs are essential for MYC oncogene expression and lymphoblast proliferation. Proceedings of the National Academy of Sciences. 2016;113:14121-6
72. Li J, Wang W, Chen S, Cai J, Ban Y, Peng Q. et al. FOXA1 reprograms the TGF-beta-stimulated transcriptional program from a metastasis promoter to a tumor suppressor in nasopharyngeal carcinoma. Cancer Lett. 2019;442:1-14
73. Jiang L, Wang R, Fang L, Ge X, Chen L, Zhou M. et al. HCP5 is a SMAD3-responsive long non-coding RNA that promotes lung adenocarcinoma metastasis via miR-203/SNAI axis. Theranostics. 2019;9:2460-74
74. Sun Z, Ou C, Liu J, Chen C, Zhou Q, Yang S. et al. YAP1-induced MALAT1 promotes epithelial-mesenchymal transition and angiogenesis by sponging miR-126-5p in colorectal cancer. Oncogene. 2019;38:2627-44
75. Ge X, Li GY, Jiang L, Jia L, Zhang Z, Li X. et al. Long noncoding RNA CAR10 promotes lung adenocarcinoma metastasis via miR-203/30/SNAI axis. Oncogene. 2019;38:3061-76
76. Peng L, Jiang B, Yuan X, Qiu Y, Peng J, Huang Y. et al. Super-Enhancer-Associated Long Noncoding RNA HCCL5 Is Activated by ZEB1 and Promotes the Malignancy of Hepatocellular Carcinoma. Cancer Res. 2019;79:572-84
77. Xu S, Wan L, Yin H, Xu H, Zheng W, Shen M. et al. Long Noncoding RNA Linc00152 Functions as a Tumor Propellant in Pan-Cancer. Cell Physiol Biochem. 2017;44:2476-90
78. Sin-Chan P, Mumal I, Suwal T, Ho B, Fan X, Singh I. et al. A C19MC-LIN28A-MYCN Oncogenic Circuit Driven by Hijacked Super-enhancers Is a Distinct Therapeutic Vulnerability in ETMRs: A Lethal Brain Tumor. Cancer Cell. 2019;36:51-67 e7
79. Li M, Lee KF, Lu Y, Clarke I, Shih D, Eberhart C. et al. Frequent amplification of a chr19q13.41 microRNA polycistron in aggressive primitive neuroectodermal brain tumors. Cancer Cell. 2009;16:533-46
80. Han J, Meng J, Chen S, Wang X, Yin S, Zhang Q. et al. YY1 Complex Promotes Quaking Expression via Super-Enhancer Binding during EMT of Hepatocellular Carcinoma. Cancer Res. 2019;79:1451-64
81. Yan Y, Xu Z, Chen X, Wang X, Zeng S, Zhao Z. et al. Novel Function of lncRNA ADAMTS9-AS2 in Promoting Temozolomide Resistance in Glioblastoma via Upregulating the FUS/MDM2 Ubiquitination Axis. Front Cell Dev Biol. 2019;7:217
82. Yeldag G, Rice A, Del Rio Hernandez A. Chemoresistance and the Self-Maintaining Tumor Microenvironment. Cancers (Basel). 2018 p:10
83. Moreau PR, Ord T, Downes NL, Niskanen H, Bouvy-Liivrand M, Aavik E. et al. Transcriptional Profiling of Hypoxia-Regulated Non-coding RNAs in Human Primary Endothelial Cells. Front Cardiovasc Med. 2018;5:159
84. YiRen H, YingCong Y, Sunwu Y, Keqin L, Xiaochun T, Senrui C. et al. Long noncoding RNA MALAT1 regulates autophagy associated chemoresistance via miR-23b-3p sequestration in gastric cancer. Mol Cancer. 2017;16:174
85. Li P, Zhang X, Wang H, Wang L, Liu T, Du L. et al. MALAT1 Is Associated with Poor Response to Oxaliplatin-Based Chemotherapy in Colorectal Cancer Patients and Promotes Chemoresistance through EZH2. Mol Cancer Ther. 2017;16:739-51
86. Chen W, Xu XK, Li JL, Kong KK, Li H, Chen C. et al. MALAT1 is a prognostic factor in glioblastoma multiforme and induces chemoresistance to temozolomide through suppressing miR-203 and promoting thymidylate synthase expression. Oncotarget. 2017;8:22783-99
87. Yue B, Cai D, Liu C, Fang C, Yan D. Linc00152 Functions as a Competing Endogenous RNA to Confer Oxaliplatin Resistance and Holds Prognostic Values in Colon Cancer. Mol Ther. 2016;24:2064-77
88. Pistritto G, Trisciuoglio D, Ceci C, Garufi A, D'Orazi G. Apoptosis as anticancer mechanism: function and dysfunction of its modulators and targeted therapeutic strategies. Aging (Albany NY). 2016;8:603-19
89. Xu H, Lin F, Wang Z, Yang L, Meng J, Ou Z. et al. CXCR2 promotes breast cancer metastasis and chemoresistance via suppression of AKT1 and activation of COX2. Cancer Lett. 2018;412:69-80
90. Renganathan A, Felley-Bosco E. Long Noncoding RNAs in Cancer and Therapeutic Potential. Adv Exp Med Biol. 2017;1008:199-222
91. Hu B, Qiu-Lan H, Lei RE, Shi C, Jiang HX, Qin SY. Interleukin-9 Promotes Pancreatic Cancer Cells Proliferation and Migration via the miR-200a/Beta-Catenin Axis. Biomed Res Int. 2017;2017:2831056
92. Li R, He JL, Chen XM, Long CL, Yang DH, Ding YB. et al. MiR-200a is involved in proliferation and apoptosis in the human endometrial adenocarcinoma cell line HEC-1B by targeting the tumor suppressor PTEN. Mol Biol Rep. 2014;41:1977-84
93. Huang C, Zeng X, Jiang G, Liao X, Liu C, Li J. et al. XIAP BIR domain suppresses miR-200a expression and subsequently promotes EGFR protein translation and anchorage-independent growth of bladder cancer cell. J Hematol Oncol. 2017;10:6
94. Murata M. Inflammation and cancer. Environ Health Prev Med. 2018;23:50
95. Anandagoda N, Willis JC, Hertweck A, Roberts LB, Jackson I, Gokmen MR. et al. microRNA-142-mediated repression of phosphodiesterase 3B critically regulates peripheral immune tolerance. J Clin Invest. 2019;129:1257-71
96. Duan Q, Mao X, Xiao Y, Liu Z, Wang Y, Zhou H. et al. Super enhancers at the miR-146a and miR-155 genes contribute to self-regulation of inflammation. Biochim Biophys Acta. 2016;1859:564-71
97. Simanovich E, Brod V, Rahat MM, Rahat MA. Function of miR-146a-5p in Tumor Cells As a Regulatory Switch between Cell Death and Angiogenesis: Macrophage Therapy Revisited. Front Immunol. 2017;8:1931
98. Szebeni GJ, Vizler C, Kitajka K, Puskas LG. Inflammation and Cancer: Extra- and Intracellular Determinants of Tumor-Associated Macrophages as Tumor Promoters. Mediators Inflamm. 2017;2017:9294018
99. Chen C, Luo F, Liu X, Lu L, Xu H, Yang Q. et al. NF-kB-regulated exosomal miR-155 promotes the inflammation associated with arsenite carcinogenesis. Cancer Lett. 2017;388:21-33
100. Lu Q, Ma Z, Ding Y, Bedarida T, Chen L, Xie Z. et al. Circulating miR-103a-3p contributes to angiotensin II-induced renal inflammation and fibrosis via a SNRK/NF-kappaB/p65 regulatory axis. Nat Commun. 2019;10:2145
101. Das S, Senapati P, Chen Z, Reddy MA, Ganguly R, Lanting L. et al. Regulation of angiotensin II actions by enhancers and super-enhancers in vascular smooth muscle cells. Nat Commun. 2017;8:1467
102. Jiao W, Chen Y, Song H, Li D, Mei H, Yang F. et al. HPSE enhancer RNA promotes cancer progression through driving chromatin looping and regulating hnRNPU/p300/EGR1/HPSE axis. Oncogene. 2018;37:2728-45
103. Zhang S, Cao H, Ye L, Wen X, Wang S, Zheng W. et al. Cancer-associated methylated lncRNAs in patients with bladder cancer. Am J Transl Res. 2019;11:3790-800
104. Yuan J, Jiang YY, Mayakonda A, Huang M, Ding LW, Lin H. et al. Super-Enhancers Promote Transcriptional Dysregulation in Nasopharyngeal Carcinoma. Cancer Res. 2017;77:6614-26
105. Chen Y, Xiao Z, Hu M, Luo X, Cui Z. Diagnostic efficacy of long non-coding RNA MALAT-1 in human cancers: a meta-analysis study. Oncotarget. 2017;8:102291-300
106. Tian X, Xu G. Clinical value of lncRNA MALAT1 as a prognostic marker in human cancer: systematic review and meta-analysis. BMJ Open. 2015;5:e008653
107. Chen W, Zhao W, Zhang L, Wang L, Wang J, Wan Z. et al. MALAT1-miR-101-SOX9 feedback loop modulates the chemo-resistance of lung cancer cell to DDP via Wnt signaling pathway. Oncotarget. 2017;8:94317-29
108. Li Y, Wu Z, Yuan J, Sun L, Lin L, Huang N. et al. Long non-coding RNA MALAT1 promotes gastric cancer tumorigenicity and metastasis by regulating vasculogenic mimicry and angiogenesis. Cancer Lett. 2017;395:31-44
109. Li C, Cui Y, Liu LF, Ren WB, Li QQ, Zhou X. et al. High Expression of Long Noncoding RNA MALAT1 Indicates a Poor Prognosis and Promotes Clinical Progression and Metastasis in Bladder Cancer. Clin Genitourin Cancer. 2017;15:570-6
110. Quan FY, Jiang J, Zhai YF, Li B, Wu XH, Nie W. The prognostic effect of LINC00152 for cancer: a meta-analysis. Oncotarget. 2017;8:75427-33
111. Li N, Feng XB, Tan Q, Luo P, Jing W, Zhu M. et al. Identification of Circulating Long Noncoding RNA Linc00152 as a Novel Biomarker for Diagnosis and Monitoring of Non-Small-Cell Lung Cancer. Dis Markers. 2017;2017:7439698
112. Zhao L, Chi WW, Cao H, Meng WX, Cui WN, Wang BS. [Expression of long-chain non-coding RNA LINC00152 in laryngeal squamous cell carcinoma and its clinical significance]. Lin Chung Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 2019;33:721-5
113. Li J, Wang X, Tang J, Jiang R, Zhang W, Ji J. et al. HULC and Linc00152 Act as Novel Biomarkers in Predicting Diagnosis of Hepatocellular Carcinoma. Cell Physiol Biochem. 2015;37:687-96
114. Zhang HD, Jiang LH, Sun DW, Hou JC, Ji ZL. CircRNA: a novel type of biomarker for cancer. Breast Cancer. 2018;25:1-7
115. Chen S, Li T, Zhao Q, Xiao B, Guo J. Using circular RNA hsa_circ_0000190 as a new biomarker in the diagnosis of gastric cancer. Clin Chim Acta. 2017;466:167-71
116. Yao JT, Zhao SH, Liu QP, Lv MQ, Zhou DX, Liao ZJ. et al. Over-expression of CircRNA_100876 in non-small cell lung cancer and its prognostic value. Pathol Res Pract. 2017;213:453-6
117. Augello C, Vaira V, Caruso L, Destro A, Maggioni M, Park YN. et al. MicroRNA profiling of hepatocarcinogenesis identifies C19MC cluster as a novel prognostic biomarker in hepatocellular carcinoma. Liver Int. 2012;32:772-82
118. Moyal L, Barzilai A, Gorovitz B, Hirshberg A, Amariglio N, Jacob-Hirsch J. et al. miR-155 is involved in tumor progression of mycosis fungoides. Exp Dermatol. 2013;22:431-3
119. Khalighfard S, Alizadeh AM, Irani S, Omranipour R. Plasma miR-21, miR-155, miR-10b, and Let-7a as the potential biomarkers for the monitoring of breast cancer patients. Sci Rep. 2018;8:17981
120. Park S, Eom K, Kim J, Bang H, Wang HY, Ahn S. et al. MiR-9, miR-21, and miR-155 as potential biomarkers for HPV positive and negative cervical cancer. BMC Cancer. 2017;17:658
121. Due H, Svendsen P, Bodker JS, Schmitz A, Bogsted M, Johnsen HE. et al. miR-155 as a Biomarker in B-Cell Malignancies. Biomed Res Int. 2016;2016:9513037
122. Dusilkova N, Basova P, Polivka J, Kodet O, Kulvait V, Pesta M. et al. Plasma miR-155, miR-203, and miR-205 are Biomarkers for Monitoring of Primary Cutaneous T-Cell Lymphomas. Int J Mol Sci. 2017 p:18
123. Liu J, Mao Q, Liu Y, Hao X, Zhang S, Zhang J. Analysis of miR-205 and miR-155 expression in the blood of breast cancer patients. Chin J Cancer Res. 2013;25:46-54
124. Qu YL, Wang HF, Sun ZQ, Tang Y, Han XN, Yu XB. et al. Up-regulated miR-155-5p promotes cell proliferation, invasion and metastasis in colorectal carcinoma. Int J Clin Exp Pathol. 2015;8:6988-94
125. Greither T, Vorwerk F, Kappler M, Bache M, Taubert H, Kuhnt T. et al. Salivary miR-93 and miR-200a as post-radiotherapy biomarkers in head and neck squamous cell carcinoma. Oncol Rep. 2017;38:1268-75
126. Suo HB, Zhang KC, Zhao J. MiR-200a promotes cell invasion and migration of ovarian carcinoma by targeting PTEN. Eur Rev Med Pharmacol Sci. 2018;22:4080-9
127. Jia C, Zhang Y, Xie Y, Ren Y, Zhang H, Zhou Y. et al. miR-200a-3p plays tumor suppressor roles in gastric cancer cells by targeting KLF12. Artif Cells Nanomed Biotechnol. 2019;47:3697-703
128. Meng X, Muller V, Milde-Langosch K, Trillsch F, Pantel K, Schwarzenbach H. Diagnostic and prognostic relevance of circulating exosomal miR-373, miR-200a, miR-200b and miR-200c in patients with epithelial ovarian cancer. Oncotarget. 2016;7:16923-35
129. Feng J, Wang J, Chen M, Chen G, Wu Z, Ying L. et al. miR-200a suppresses cell growth and migration by targeting MACC1 and predicts prognosis in hepatocellular carcinoma. Oncol Rep. 2015;33:713-20
130. Feng M, Luo X, Gu C, Fei J. Seed targeting with tiny anti-miR-155 inhibits malignant progression of multiple myeloma cells. J Drug Target. 2015;23:59-66
131. Bayraktar R, Van Roosbroeck K. miR-155 in cancer drug resistance and as target for miRNA-based therapeutics. Cancer Metastasis Rev. 2018;37:33-44
132. Gutschner T, Hammerle M, Eissmann M, Hsu J, Kim Y, Hung G. et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73:1180-9
133. Gong N, Teng X, Li J, Liang XJ. Antisense Oligonucleotide-Conjugated Nanostructure-Targeting lncRNA MALAT1 Inhibits Cancer Metastasis. ACS Appl Mater Interfaces. 2019;11:37-42
134. Chen D, Zhao Z, Huang Z, Chen DC, Zhu XX, Wang YZ. et al. Super enhancer inhibitors suppress MYC driven transcriptional amplification and tumor progression in osteosarcoma. Bone Res. 2018;6:11
135. Desrichard A, Snyder A, Chan TA. Cancer Neoantigens and Applications for Immunotherapy. Clin Cancer Res. 2016;22:807-12
136. Khan A, Zhang X. dbSUPER: a database of super-enhancers in mouse and human genome. Nucleic Acids Res. 2016;44:D164-71
137. Wei Y, Zhang S, Shang S, Zhang B, Li S, Wang X. et al. SEA: a super-enhancer archive. Nucleic Acids Res. 2016;44:D172-9
138. Jiang Y, Qian F, Bai X, Liu Y, Wang Q, Ai B. et al. SEdb: a comprehensive human super-enhancer database. Nucleic Acids Res. 2019;47:D235-D43
139. Qian FC, Li XC, Guo JC, Zhao JM, Li YY, Tang ZD. et al. SEanalysis: a web tool for super-enhancer associated regulatory analysis. Nucleic Acids Res. 2019;47:W248-W55
140. Chen C, Zhou D, Gu Y, Wang C, Zhang M, Lin X. et al. SEA version 3.0: a comprehensive extension and update of the Super-Enhancer archive. Nucleic Acids Res. 2020;48:D198-D203
141. Bu H, Hao J, Gan Y, Zhou S, Guan J. DEEPSEN: a convolutional neural network based method for super-enhancer prediction. BMC Bioinformatics. 2019;20:598
142. Huang M, Chen Y, Yang M, Guo A, Xu Y, Xu L. et al. dbCoRC: a database of core transcriptional regulatory circuitries modeled by H3K27ac ChIP-seq signals. Nucleic Acids Res. 2018;46:D71-D7
143. Mei S, Meyer CA, Zheng R, Qin Q, Wu Q, Jiang P. et al. Cistrome Cancer: A Web Resource for Integrative Gene Regulation Modeling in Cancer. Cancer Res. 2017;77:e19-e22
144. Guo ZW, Xie C, Li K, Zhai XM, Cai GX, Yang XX. et al. SELER: a database of super-enhancer-associated lncRNA- directed transcriptional regulation in human cancers. Database (Oxford). 2019. 2019
145. Tang Z, Li X, Zhao J, Qian F, Feng C, Li Y. et al. TRCirc: a resource for transcriptional regulation information of circRNAs. Brief Bioinform. 2018
Author contact
![]() Corresponding authors: E-mail: ouchunlinedu.cn; zhoujh15com. Joint-supervision by J.Z & C.O.
Corresponding authors: E-mail: ouchunlinedu.cn; zhoujh15com. Joint-supervision by J.Z & C.O.
 Global reach, higher impact
Global reach, higher impact