13.3
Impact Factor
Theranostics 2020; 10(24):10993-11012. doi:10.7150/thno.43094 This issue Cite
Research Paper
CRISPR-Sunspot: Imaging of endogenous low-abundance RNA at the single-molecule level in live cells
1. Department of Physiology, School of Basic Medical Sciences, Nanjing Medical University, Nanjing, 211166, China.
2. Key Laboratory of Cardiovascular & Cerebrovascular Medicine, Drug Target and Drug Discovery Center, School of Pharmacy, Nanjing Medical University, Nanjing, 211166, China.
3. Institute of Pharmacology and Toxicology, College of Pharmaceutical Sciences, Zhejiang University, Hangzhou, 310058, China.
4. Department of Genetics, Yale University School of Medicine, New Haven, CT, USA.
5. Department of Cell Biology, School of Medicine, Zhejiang University, Hangzhou, 310058, China.
6. Center for Global Health of Nanjing Medical University, Nanjing, 211166, China.
*These authors contributed equally to this work.
Abstract
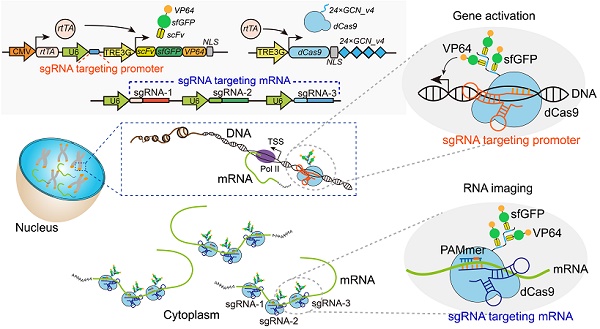
CRISPR/Cas-based mRNA imaging has been developed to labeling of high-abundance mRNAs. A lack of non-genetically encoded mRNA-tagged imaging tools has limited our ability to explore the functional distributions of endogenous low-abundance mRNAs in cells. Here, we developed a CRISPR-Sunspot method based on the SunTag signal amplification system that allows efficient imaging of low-abundance mRNAs with CRISPR/Cas9.
Methods: We created a stable TRE3G-dCas9-EGFP cell line and generated an Inducible dCas9-EGFP imaging system for assessment of two factors, sgRNA and dCas9, which influence imaging quality. Based on SunTag system, we established a CRISPR-Sunspot imaging system for amplifying signals from single-molecule mRNA in live cells. CRISPR-Sunspot was used to track co-localization of Camk2a mRNA with regulatory protein Xlr3b in neurons. CRISPR-Sunspot combined with CRISPRa was used to determine elevated mRNA molecules.
Results: Our results showed that manipulating the expression of fluorescent proteins and sgRNA increased the efficiency of RNA imaging in cells. CRISPR-Sunspot could target endogenous mRNAs in the cytoplasm and amplified signals from single-molecule mRNA. Furthermore, CRISPR-Sunspot was also applied to visualize mRNA distributions with its regulating proteins in neurons. CRISPR-Sunspot detected the co-localization of Camk2a mRNA with overexpressed Xlr3b proteins in the neuronal dendrites. Moreover, we also manipulated CRISPR-Sunspot to detect transcriptional activation of target gene such as HBG1 in live cells.
Conclusion: Our findings suggest that CRISPR-Sunspot is a novel applicable imaging tool for visualizing the distributions of low-abundance mRNAs in cells. This study provides a novel strategy to unravel the molecular mechanisms of diseases caused by aberrant mRNA molecules.
Keywords: CRISPR/Cas9, mRNA, imaging, neuron, transcriptional activation
 Global reach, higher impact
Global reach, higher impact