13.3
Impact Factor
Theranostics 2020; 10(24):10908-10924. doi:10.7150/thno.48264 This issue Cite
Research Paper
Circular RNA circPPM1F modulates M1 macrophage activation and pancreatic islet inflammation in type 1 diabetes mellitus
1. Institute of Pediatrics, Children's Hospital of Fudan University, and the Shanghai Key Laboratory of Medical Epigenetics, International Co-laboratory of Medical Epigenetics and Metabolism, Ministry of Science and Technology, Institutes of Biomedical Sciences, Fudan University, Shanghai 200032, China.
2. National Health Commission (NHC) Key Laboratory of Neonatal Diseases, Fudan University, Shanghai, China.
3. Department of Pediatric Endocrinology and Inherited Metabolic Diseases, Children's Hospital of Fudan University, Shanghai, 201102, China.
#These authors contributed equally to this work.
Received 2020-5-16; Accepted 2020-8-21; Published 2020-8-29
Abstract
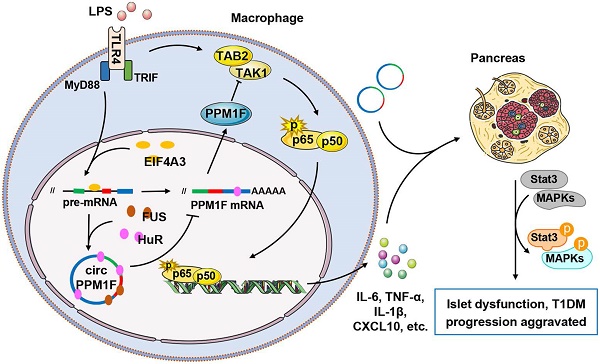
Rationale: Macrophages play critical roles in the pathogenesis of type 1 diabetes mellitus (T1DM). Circular RNAs (circRNAs) are a novel class of endogenous RNAs with covalently closed loop structures, implicated in various disease processes. However, their impact on macrophage activation and T1DM pathogenesis remains elusive.
Methods: circRNA expression profiles of peripheral blood mononuclear cells (PBMCs) from T1DM children were determined by whole transcriptome microarray. Bioinformatics, quantitative real-time PCR, Western blot, RNA immunoprecipitation (RIP), cell co-culture, cell proliferation, and cell apoptosis assays were performed to investigate the expression, function, and regulatory mechanisms of circPPM1F in vitro. The regulatory role of circPPM1F in vivo was evaluated in the streptozocin-induced diabetic mouse model.
Results: We identified 27 upregulated and 31 downregulated differentially expressed circRNAs in T1DM patients. circPPM1F, a circRNA with unknown function, was dominantly expressed in monocytes and significantly upregulated in T1DM patients. Functionally, circPPM1F promoted lipopolysaccharide (LPS)-induced M1 macrophage activation via enhancement of the NF-κB signaling pathway. Mechanistically, circPPM1F competitively interacted with HuR to impair the translation of protein phosphatase, Mg2+/Mn2+ dependent 1F (PPM1F), thus alleviating the inhibitory effect of PPM1F on the NF-κB pathway. Moreover, eukaryotic initiation factor 4A-III (EIF4A3) and fused in sarcoma (FUS) coordinately regulated circPPM1F expression during M1 macrophage activation. In addition, circPPM1F could exacerbate pancreas injury in the streptozocin-induced diabetic mice by activation of M1 macrophages in vivo.
Conclusions: circPPM1F is a novel positive regulator of M1 macrophage activation through the circPPM1F-HuR-PPM1F-NF-κB axis. Overexpression of circPPM1F could promote pancreatic islet injury by enhancing M1 macrophage activation and circPPM1F may serve as a novel potential therapeutic target for T1DM in children.
Keywords: Circular RNA, Type 1 diabetes mellitus, Macrophage activation, RNA-binding protein, Islet injury
Introduction
Type 1 diabetes mellitus (T1DM, also known as insulin-dependent diabetes) is a chronic autoimmune disease, driven by the interplay between individual genetics and environmental triggers [1, 2]. T1DM is diagnosed at all ages [3, 4], and is characterized by aberrant autoimmune destruction of pancreatic islet β cells, resulting in a complete lack of insulin synthesis and necessitating lifelong hormone replacement therapy [5]. Recent studies have revealed that a spike in the incidence of T1DM was found at ages 10-14 years, and worldwide estimates of numbers of children and adolescents with T1DM continue to increase [6].
Emerging evidence has suggested that, throughout the immune system, macrophages play a critical role in insulitis, and support autoimmune T cells to aggravate the infiltration of inflammatory cells during T1DM [7]. Macrophages have been classified as two extreme examples on the activation spectrum: classically activated macrophages (M1) with a pro-inflammatory phenotype and alternatively activated macrophages (M2) with an anti-inflammatory phenotype. Generally, lipopolysaccharide (LPS) and interferon (IFN)-γ are the main stimulating factors for M1 macrophage activation, while interleukin (IL)-4 and IL-13 can activate macrophages to M2 [8, 9]. Notably, M1 macrophage activation drives pathogenesis and progression of diabetes through exacerbation of inflammatory responses via secretion of inflammatory cytokines [10, 11]. For example, Arnush et al. found M1 macrophages promoted destruction of β cells in T1DM mice through excessive production of IL-1β [12]. In addition, Tim-3 exacerbated podocyte injury via M1 macrophage activation in streptozocin (STZ)-induced diabetic nephropathy [13]. Importantly, blocking macrophage infiltration into the pancreas or restraining macrophage activation in diabetic mice maintained pancreas function and prevented T1DM initiation [14, 15]. Therefore, elucidating the underlying mechanism of M1 macrophage activation is likely to lead to a better understanding of the pathogenesis and therapy of T1DM.
Circular RNA (circRNA) is a covalently closed loop molecule ligated by a 3′-5′ phosphodiester bond at the junction site, which is generated through back-splicing and often considered to be a byproduct of aberrant splicing [16, 17]. Recently, high-throughput sequencing has been used to identify thousands of endogenous circRNA species in mammalian cells, which are stable, conserved, and abundant [18, 19]. By modulation of gene expression, interaction with RNA binding proteins (RBPs), circRNAs directly or indirectly participate in autoimmune diseases and cancers. For instance, circ-RasGEF1B promotes the antigen presentation process by positively regulating the stability of intercellular adhesion molecule 1 (ICAM-1) mRNA in macrophages exposed to LPS [20]. Additionally, circZKSCAN1 negatively regulates cancer stem cells by physically binding fragile X mental retardation protein (FMRP) against cell cycle and apoptosis regulator 1 (CCAR1) complex in hepatocellular carcinoma [21]. However, the roles of circRNAs in regulating M1 macrophage activation and T1DM are currently undefined.
In this study, we conducted a genome-wide analysis of circRNA expression profiles in peripheral blood mononuclear cells (PBMCs) from T1DM patients and healthy controls and identified a novel circRNA, circPPM1F, which was significantly upregulated in the PBMCs of T1DM patients. Functionally, circPPM1F could promote LPS-induced M1 macrophage activation via enhancement of NF-κB signaling. Mechanistically, circPPM1F competitively interacted with HuR to impair the translation of PPM1F, thus alleviating the inhibitory effect of PPM1F on the NF-κB pathway. Moreover, we found that EIF4A3 and FUS participated in the maintenance of high levels of circPPM1F expression. In addition, circPPM1F could exacerbate pancreas injury in STZ-induced diabetic mice by activating M1 macrophages. Overall, our study indicates that circPPM1F plays an important role in the development of T1DM and suggests a potential therapeutic target for T1DM.
Materials and Methods
Human study subjects
T1DM patients were diagnosed according to the criteria of American Diabetes Association (ADA) [22]. Human peripheral blood samples were collected from T1DM patients (8.5 ± 0.6 y) and age-matched healthy controls (8.0 ± 1.2 y), following informed consent from all patients, at the Children′s Hospital of Fudan University, Shanghai, China. Detailed characteristics of the study subjects are presented in Table S1. The study was approved by the Research Ethics Board of the Children′s Hospital of Fudan University [No. (2016) 96].
PBMCs were isolated using Ficoll-Hypaque (GE Healthcare, USA). Briefly, peripheral blood was mixed with phosphate-buffered saline (PBS) and overlaid on the Ficoll-Hypaque solution (density: 1.077 g/mL). Following centrifugation at 400 ×g for 30 min, PBMCs were aspirated from the interface. The cell pellet was washed twice with PBS and resuspended in TRIzol for subsequent RNA extraction and quantitative real-time PCR (qRT-PCR). In addition, CD14+, CD3+, and CD19+ cells were sorted from PBMCs with a magnetic cell sorting system (Miltenyi Biotec, Germany). Sorted cells were subjected to RNA extraction and qRT-PCR.
RNA extraction and quantitative real-time PCR
The nuclear and cytoplasmic fractions or total RNA were extracted using TRIzol (Invitrogen, USA) followed by reverse transcription of mRNAs and circRNAs using PrimeScript II 1st Strand cDNA Synthesis Kit (Takara, Japan) according to the standard manufacturer′s instructions. A qRT-PCR assay was performed to measure mRNAs and circRNAs expression with SYBR® Premix Ex Taq™ II (Takara, Japan) using the Roche 480 Real Time PCR System. β-actin served as internal control for mRNAs and circRNAs. Relative quantification (2-ΔΔCT) was used for results analysis. The primers sequences in qRT-PCR are listed in Table S2.
Microarray
The circRNA microarray analysis was performed at the Shanghai Biotechnology Corporation. In brief, Total RNA of PBMCs from T1DM patients (n = 4) and age-matched healthy controls (n = 4) were extracted and purified followed by amplification and labeling with a Low Input Quick Amp WT Labeling Kit. Labeled cRNAs were purified using the RNeasy mini kit. Each slide was hybridized with 1.65 μg Cy3-labeled cRNA for 17 h using a gene expression hybridization kit, and then scanned with an Agilent microarray scanner using default settings. Data were extracted with the Feature Extraction software 10.7 (Agilent Technologies, Santa Clara, CA, US). Raw data were normalized by the Quantile algorithm and limma package in R. Overall, 88750 circRNAs were tested.
Cell culture and transfection
The human THP1 cell line was obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA), and MIN6 and Raw264.7 cells were obtained from the Fudan IBS cell resource center (FDCC, Shanghai, China). THP1 and MIN6 cells were maintained in RPMI-1640 medium (Gibco, Gaithersburg, MD, USA) with 10% fetal bovine serum (FBS, Gibco) and 1% penicillin/streptomycin (Gibco), and the MIN6 cell culture medium was supplemented with 50 μM 2-mercaptoethanol. Raw264.7 cells were maintained in Dulbecco′s modified Eagle′s medium with high glucose (DMEM, Gibco) supplemented with 10% FBS. All cells were kept in a humidified cell incubator with 5% CO2 at 37 °C.
Cells were transiently transfected with Lipofectamine RNAiMAX reagent (Invitrogen, USA) and chemically synthesized si-circPPM1F, siPPM1F, siHuR, siEIF4A3, or siFUS (GenePharma, Shanghai, China) according to standard protocols. Cells were transiently transfected with 500 ng/mL ectopic expression vector of circPPM1F with Lipofectamine 2000 reagent (Invitrogen, USA), according to the manufacturer's instructions. qRT-PCR assay was used to assess RNA expression levels 48 h after transfection. Relative sequences of siRNA are listed in Table S3.
For the activation of THP1-derived macrophage, THP1 cells were treated as previously described [23]. Briefly, THP1 were treated with 500 ng/mL phorbol 12-myristate 13-acetate (PMA) for 48 h to induce the differentiation of THP1 into macrophages (THP1 macrophages), and then 200 ng/mL LPS was used to induce M1 macrophage activation.
Western blot
Total protein was extracted using Radio Immunoprecipitation Assay (RIPA) lysis buffer (Thermo Scientific, USA). Lysates were resolved by electrophoresis, transferred to polyvinylidene fluoride (PVDF) membranes, and probed with antibodies directed against PPM1F (Abcam); HuR, phosphorylated NF-κB p65 (Ser536) (p-p65), p65, phosphorylated p38 (T180/Y182) (p-p38), p38, phosphorylated JNK (T183/Y185) (p-JNK), JNK, phosphorylated ERK1/2 (T202/Y204) (p-ERK1/2), ERK1/2, phosphorylated m-TOR (Ser2448) (p-mTOR), mTOR, phosphorylated Stat3 (Y705) (p-Stat3), Stat3, Bcl2 (Cell Signaling Technology, USA); Bax (EMD Millipore, USA), and β-tubulin (Abcam, USA). The bands were detected by developing with chemiluminescent HRP substrate (Thermo Scientific, USA), and intensity of bands was determined by imaging with a Molecular Imager® (Bio-RAD, ChemiDocTM XRS+ Imaging System, USA). All results were normalized to those of β-tubulin, which was used as a loading control. Detailed characteristics of the antibodies are presented in Table S4.
RNase R digestion
Four micrograms total RNA from THP1 cells was either untreated (control) or treated with 20 units of RNase R (Epicenter; USA, RNR07250) in the presence of 1 × Reaction Buffer, and incubated for 30 min at 37 °C. The digested RNA was isolated using acid phenol-chloroform (5:1). Then reverse transcription and qRT-PCR were performed, as described in the RNA extraction and qRT-PCR section.
RNA stability
THP1 cells (1 × 105) were placed in 24-well plates and treated with 250 ng/mL actinomycin D (Act D, Sigma) added to the cell culture medium. The levels of circPPM1F and PPM1F were detected at 0, 3, 6, 12, and 24 h.
Subcellular fractionation and localization
Nuclear and cytosolic fractions were separated using the Nuclear and Cytoplasmic Extraction Kit (Cwbio, China). A total of 1 × 107 cells were harvested, re-suspended in 1 mL of Nc-buffer A and 55 µL Nc-Buffer B, and incubated for 20 min on ice. Cells were then centrifuged for 15 min at 12, 000 ×g; the resulting supernatants (containing the cytoplasmic component) and nuclear pellets were used for RNA extraction.
RNA immunoprecipitation
The RNA immunoprecipitation (RIP) assay was performed using the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (EMD Millipore Corp., Billerica MA, USA). Two 10-cm culture dishes of THP1 macrophages (1 × 107/dish) were harvested, centrifuged and re-suspended using 100 μL RIP lysis buffer supplemented with protease and RNase inhibitors, and 5 μg IgG or HuR antibody-coated beads were incubated with the cell lysates under rotary agitation at 4 °C overnight. Following proteinase K treatment, the immunoprecipitated RNAs were extracted and reversely transcribed as described in the RNA extraction and qRT-PCR section. Levels of circPPM1F and PPM1F were detected by qRT-PCR assay.
circPPM1F vector construction
3D5 is a modified plasmid based on the pZW1 vector. Two reverse complementary sequences helped to form a circular structure derived from the POLR2A gene and were inserted upstream of the restriction site Xho I and downstream of Pac I. Primer 1 or primer 2 (Table S2) was used to amplify the sequence of circPPM1F upstream (~340 bp) or downstream (~3800 bp) of cDNA or gDNA. Two PCR segments were inserted into the 3D5 vector following digestion with restriction enzymes Xho I and Pac I using a seamless cloning assay (Figure S1), and the product was transformed into STBL3 competent cells. The recombinant vector sequences were validated by Sanger sequencing.
Conditional media collection and cell treatment
Raw264.7 cells were seeded in six-well plates, and transfected with 3D5-circPPM1F, or corresponding negative control plasmids according to the cell transfection method used. Next, 48 h post transfection, the transfected cells were stimulated with 200 ng/mL LPS for 20 h. Culture media were gathered and centrifuged at 3000 rpm at 4 °C for 30 min, after which the supernatants (conditional media) were collected. The conditional media were used for treatment of MIN6 cells, at a ratio of 3:1 with complete medium.
Enzyme-linked immunosorbent assay (ELISA)
The protein levels of IL-6 and TNF-α in the conditional media were measured with the mouse IL-6 and TNF-α DuoSet ELISA kit (eBioscience) according to the manufacturer's instructions. A microplate reader (Synergy4; BioTek, Winooski, VT, USA) was used to read the absorbance at 450/570 nm.
Cell proliferation assay
The CCK8 assay was performed to assess the proliferative ability of MIN6 cells according to the manufacturer's instructions (DOJINDO Molecular Technologies, Inc., Kumamoto, Japan). Briefly, MIN6 cells (1.8 × 103) were placed in 96-well plates and cultured in conditional medium. Each sample was assayed in triplicate. Cell viability was determined at 0, 24, 48, and 72 h using 10 μL CCK8 solution treatment for 2 h. The optical density of each well was assessed using a Microplate reader (Synergy4; BioTek, Winooski, VT, USA) at 450 nm.
Cell apoptosis assay
MIN6 cells were cultured in six-well plates (1 × 105 cells/well) for 48 h using conditional media, followed by H2O2 (500 μΜ in FBS-free medium) treatment for 15 h, and then pooled together after trypsin without EDTA digestion. Cell apoptosis was analyzed using the Annexin V-FITC/Propidium Iodide (PI) Apoptosis Detection Kit (BD Pharmingen, New York, USA, #556547) according to the manufacturer's instructions. MIN6 cells were stained with FITC and PI and then analyzed by fluorescence-activated cell sorting using FACS Canto II (BD Biosciences, San Jose, CA, USA). The cell apoptosis data were analyzed by FlowJo V10 software (Tree Star, San Francisco, CA, USA). Cells of quadrant 4 that were considered viable were FITC Annexin V and PI negative; cells of quadrant 3 that were in early apoptosis were FITC Annexin V positive and PI negative; cells of quadrant 2 that were in late apoptosis were both FITC Annexin V and PI positive; and cells of quadrant 1 that were necrotic were FITC Annexin V negative and PI positive. The percentages of cell apoptosis were the sum of that from early apoptosis and late apoptosis.
Animal studies in the STZ-induced diabetic mouse model
C57BL/6 mice (8-week-old, male) were purchased from Shanghai Slac Laboratory Animal Co. Ltd. All mice were housed at the Experiment Animal Center of Children′s Hospital of Fudan University at room temperature 22 °C under a 12:12 h light/dark cycle, and were provided with rodent chow and tap water. The mice were randomly divided into four experimental groups: Control, STZ, STZ+pZW1, and STZ+circPPM1F group. On day 0, STZ+circPPM1F and STZ+pZW1 group mice were intraperitoneally (i.p.) injected with circPPM1F plasmid (8 μg/mouse, 150 μL) and an equal amount of pZW1 using the Entranster™-in vivo Transfection Reagent (Engreen, China), respectively; control and STZ group mice were injected with PBS. From day one, STZ, STZ+pZW1, and STZ+circPPM1F group mice were i.p. injected with repeated low doses of STZ (50 mg/kg body weight/day, 300 μL/mouse for five consecutive days); control group mice were injected with sodium citrate buffer (Fankew, FK4006). On day seven, mice were injected circPPM1F, control plasmids, or PBS again, and then received two injections per week. The animal experiments were repeated independently 3 times.
Body weight was measured weekly from day one. Blood glucose was also detected weekly from the tail vein blood using a Roche glucose reader (Roche Diagnostics GmbH, Germany). All mice were fasted for 12 h before glucose detection. On day 29, mice were sacrificed and pancreas tissues dissected and analyzed for pathology.
This project complied with the institutional guidelines and laws for the care and use of laboratory animals. The study was approved and overseen by the Animal Studies Committee of the Children′s Hospital of Fudan University [No. (2016) 96].
Immunohistochemistry and Immunofluorescence studies
Pancreas tissues were fixed with 4% paraformaldehyde and then embedded in paraffin and cut into slices. For immunohistochemical analysis, a portion of paraffin sections were routinely stained with hematoxylin and eosin (H&E), while others were incubated with antibodies against Ki-67 (Signalway Antibody, USA), Insulin (Proteintech, USA), and F4/80 (Cell Signaling Technology, USA). Islets from consecutive tissue cross-sections were photographed at identical exposure conditions and magnification (400×) using a microscope (Leica). For immunofluorescence analysis, paraffin slides of 3-4 μm were prepared, and then incubated with rabbit anti-mouse F4/80 and rabbit anti-mouse iNOS (Servicebio, China). FITC (green) and Cy3 (red)-conjugated goat anti-rabbit IgG (Servicebio) were used to visualize F4/80 and iNOS, respectively. DAPI (4′, 6-diamidino-2-phenylindole) was used to stain the cell nuclei (blue). Images were captured with a fluorescence microscope (Leica). Detailed characteristics of the antibodies used are presented in Table S4.
Preparation of pancreas single cell suspensions and flow cytometry
Pancreas tissues were collected into the gentleMACS C Tubes (#130-093-237; Miltenyi Biotec, Germany) containing the enzyme mix of the Multi Tissue Dissociation Kit 1 (#130-110-201, Miltenyi Biotec,Germany) in serum-free RPM-1640 and cut into 2 × 4-mm pieces. Tissues were subjected to the run program Multi_37C_m (30 min) on the gentleMACS Octo Dissociator with Heaters (#130-096-427, Miltenyi Biotec, Germany). The cell suspensions were passed through a 70-μm MACS SmartStrainer (#130-098-462, Miltenyi Biotec, Germany), followed by centrifuging at 300 ×g for 7 min at 4 °C. All isolated cells were suspended in 100 μL iced PBS supplemented with 2% FBS. Cells were then counted with trypan blue and processed for flow cytometry in as indicated below.
For surface marker analysis, cells were stained with anti-mouse F4/80 (eBioscience, USA) for 30 min at 4 °C. For intracellular cytokine staining, cells were fixed and permeabilized and labeled with anti-mouse iNOS (eBioscience, USA) after anti-F4/80 staining. The concentration of each antibody was used according to the recommended product protocol. Cells were examined by flow cytometry using a BD FACSCanto II instrument (BD Biosciences, San Jose, CA, USA) and analyzed with FlowJo V10 software (Tree Star, San Francisco, CA, USA). Detailed characteristics of the antibodies used are presented in Table S4.
Statistical analysis
Results from three independent experiments are expressed as mean ± standard error of the mean (SEM). The two-tailed Student's t-tests were used for comparisons between two groups, and one-way analysis of variance (ANOVA) was used for multifactorial comparisons. A p-value of < 0.05 was considered statistically significant. The relationship between circPPM1F and IL-6, IL-1β, TNF-α, EIF4A3 or FUS was tested using Pearson's correlation and linear regression. Statistical analyses were performed with SPSS v.19.0 software or GraphPad Prism 7.0 software.
Results
circPPM1F was upregulated in PBMCs from children with type 1 diabetes mellitus
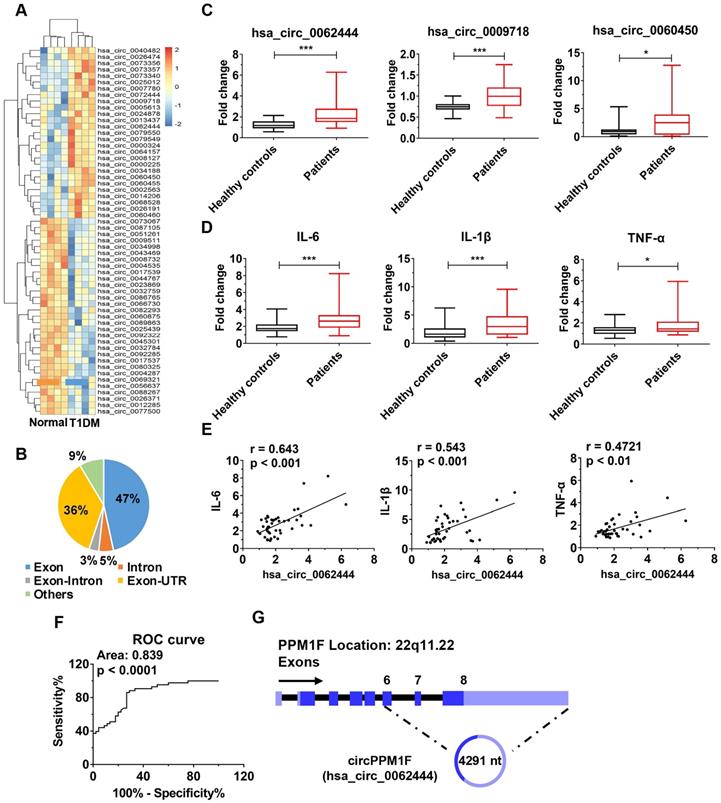
To identify differentially expressed circRNAs in T1DM, we first analyzed circRNA transcript profiles of PBMCs from T1DM children (n = 4) and age-matched healthy controls (n = 4) by circRNA microarray. Using a two-fold change and p < 0.05 as the threshold to define up- or down-regulated circRNAs, we identified 27 upregulated and 31 downregulated differentially expressed circRNAs in T1DM patients compared with healthy controls (Figure 1A). We analyzed the composition of the differentially expressed circRNAs in light of the positions of circRNAs in the transcripts; the profile consisted of 27 exonic circRNAs (47%), three intronic circRNAs (5%), two exonic-intronic circRNAs (3%), 21 exonic-UTR circRNA (36%), and others (9%) (Figure 1B). Next, we evaluated the expression of 27 upregulated circRNAs in PBMCs from children with T1DM and healthy control subjects in an expanded cohort. The results showed that only hsa_circ_0062444, hsa_circ_0009718 and hsa_circ_0060450 were detectable and significantly upregulated in PBMCs from T1DM patients compared with healthy controls (Figure 1C). Furthermore, higher expression levels of IL-6, IL-1β, and TNF-α were also observed in T1DM patients (Figure 1D). Importantly, the positive correlation between hsa_circ_0062444 and IL-6, IL-1β or TNF-α was validated in human patients with T1DM, while no correlations between the hsa_circ_0009718 or hsa_circ_0060450 and IL-6, IL-1β or TNF-α were identified (Figure 1E and Figure S2A). Furthermore, the ability of hsa_circ_0062444 to differentiate T1DM patients from healthy subjects was assessed by receiver operating curve (ROC) analysis, which yielded an area under the curve of 0.839 (Figure 1F). Together, these findings implied that hsa_circ_0062444 might have important role in T1DM pathogenesis.
Next, bioinformatics analysis showed that hsa_circ_0062444 was 4291 nt long and was the back-spliced circular product of the last three exons of the PPM1F transcript; thus, as an exonic circRNA, it was named as circPPM1F (Figure 1G). To assess the role of circPPM1F in the regulation of subsets of PBMCs, we separated human PBMCs into T cells, B cells, and monocytes to detect the expression levels of circPPM1F. Surprisingly, circPPM1F was mainly expressed in monocytes rather than T and B cells (Figure S2B). Considering monocytes can transform into macrophages in inflammatory tissues and circPPM1F was closely associated with inflammatory cytokines in T1DM patients, we subsequently focused on the regulatory role of circPPM1F in LPS-induced M1 macrophage activation.
circPPM1F promoted M1 macrophage activation through enhancement of NF-κB signaling
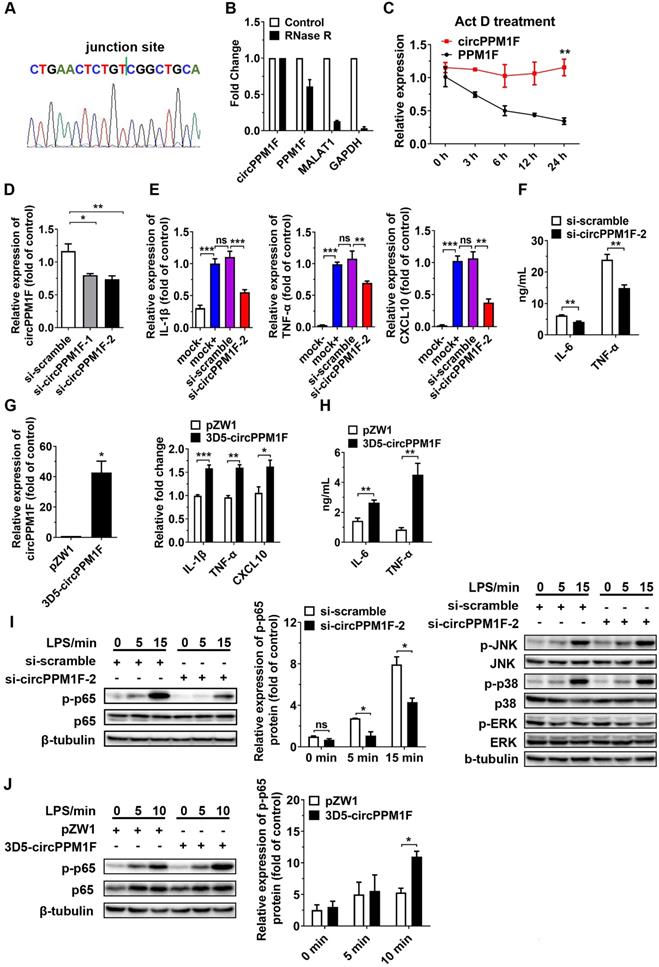
To verify whether circPPM1F was a truly circRNA and not a linear RNA, we performed RT-PCR and Sanger sequencing assays. A strict concordance between the sequencing results and the public circPPM1F sequence in circBase (http://www.circbase.org/cgi-bin/simplesearch.cgi) was observed (Figure 2A). Furthermore, we found the endogenous expression of circPPM1F was resistant to excessive ribonuclease R (RNase R) digestion while linear mRNAs were severely degraded (Figure 2B). Additionally, the stability of circPPM1F after Act D treatment in THP1 macrophages was examined. We found that circPPM1F was highly stable, with a half-life > 24 h, whereas PPM1F mRNA was readily degraded and had a half-life < 6 h (Figure 2C). Collectively, this evidence suggested that endogenous circPPM1F was truly a circular RNA.
To determine the functional influence of circPPM1F on M1 macrophage activation, we first knocked down the expression of circPPM1F in THP1-derived macrophages with two small interfering RNAs (siRNAs) targeting the circPPM1F junction site; the knockdown efficiency was confirmed by qRT-PCR and si-circPPM1F-2 was selected for the later study (Figure 2D). We found the expression of M1 macrophage associated genes (IL-1β, TNF-α and CXCL10) was downregulated in the cells with circPPM1F knockdown after LPS stimulation (Figure 2E). Meanwhile, the IL-6 and TNF-α protein expression were suppressed in the supernatant (Figure 2F). Further, in the “gain-of-function” studies with ectopically expressing circPPM1F, we found the expression of IL-1β, TNF-α, and CXCL10 were upregulated following LPS stimulation (Figure 2G), concomitant with significantly enhanced IL-6 and TNF-α protein levels in the supernatant (Figure 2H). Taken together, these findings suggested that circPPM1F can promote M1 macrophage activation.
Upon activation, LPS-TLR4 augments macrophage activity through the production of inflammatory cytokines, and activation of NF-κB and MAPK pathways [24]. To explore signaling pathways involved in circPPM1F regulation of M1 macrophage activation, we investigated the effects of circPPM1F on NF-κB and MAPK pathways. Intriguingly, western blotting showed that circPPM1F knockdown did indeed result in markedly reduced levels of phosphorylated p65 in THP1 macrophages, whereas ectopic expression of circPPM1F significantly increased the levels of phosphorylated p65. However, THP1 macrophages with circPPM1F knockdown exhibited no differences in phosphorylation levels of JNK, p38, and ERK (Figure 2I-J). Overall, these findings implied that circPPM1F can promote M1 macrophage activation by enhancing NF-κB signaling.
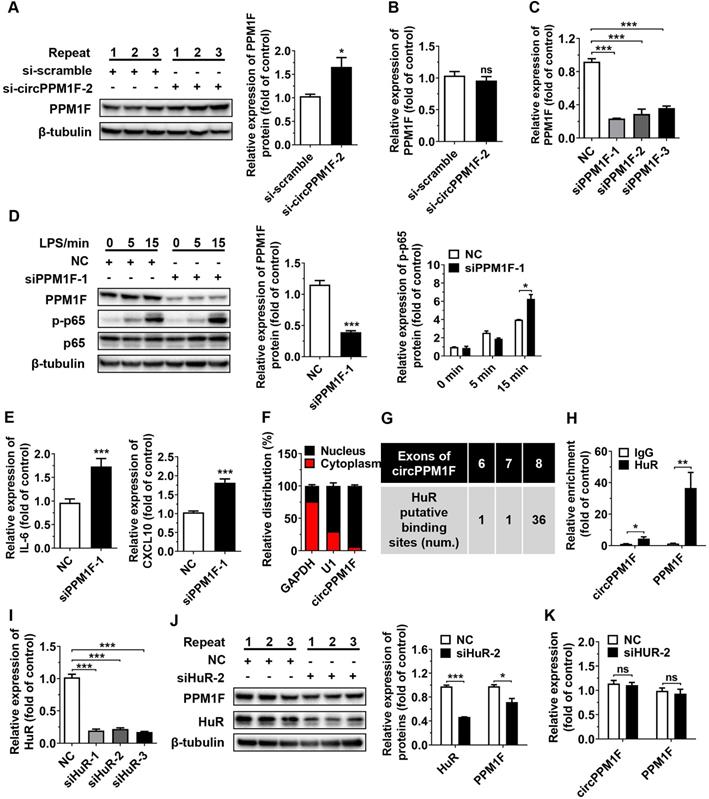
circPPM1F inhibited PPM1F translation through competitively binding to HuR
PPM1F, a member of the PP2C family of Ser/Thr protein phosphatases, is a negative regulator of the IKK-NF-κB pathway through its effects on the dephosphorylation of TAK1 [25]. As circPPM1F is the back-spliced circular product of the coding gene, PPM1F, and circRNAs have been reported to exert regulatory effects on host genes at both transcriptional and post-transcriptional levels [26, 27], we questioned whether the effects of circPPM1F on NF-κB signaling activation were due to its influence on PPM1F expression. To verify our hypothesis, the levels of PPM1F protein and mRNA expression were determined in THP1 macrophages following circPPM1F knockdown. Surprisingly, knockdown of circPPM1F resulted in a significant increase of PPM1F protein expression, with no changes in its mRNA expression (Figure 3A-B), indicating that circPPM1F negatively regulated the expression of PPM1F at the translational stage. Furthermore, we knocked down the expression of PPM1F in macrophages and found phosphorylation of p65 and the expression of IL-6, and CXCL10 were increased following LPS stimulation (Figure 3C-E). Taken together, these data implied that circPPM1F suppressed PPM1F translation, thereby facilitating NF-κB pathway and M1 macrophage activation.
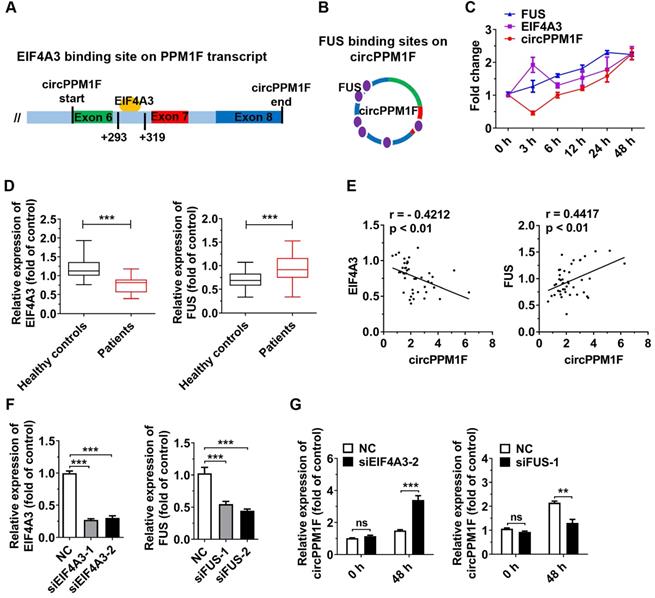
circPPM1F is upregulated and is associated with inflammatory cytokines in peripheral blood mononuclear cells from type 1 diabetes mellitus patients. A. Heatmap showing 27 upregulated and 31 downregulated differentially expressed circRNAs in peripheral blood mononuclear cell (PBMC)s of type 1 diabetes mellitus (T1DM) patients (n = 4) and age-matched healthy controls (n = 4) (fold change > 2.0, p < 0.05). B. Composition of the circRNAs according to the position of the gene in the transcript. C, D. Analyses of the expression levels of hsa_circ_0062444, hsa_circ_0009718, hsa_circ_0060450, IL-6, IL-1β, and TNF-α (normalized to β-actin) in PBMCs from 43 T1DM patients and 45 healthy controls, as determined by qRT-PCR. E. Correlation analysis of the expression of hsa_circ_0062444 and IL-6, IL-1β or TNF-α in 43 T1DM patients (Pearson's correlation). F. Receiver operating curve (ROC) analysis of circPPM1F levels in the study population. G. Schematic illustration showing the location of hsa_circ_0062444 in host gene PPM1F. Data are presented as mean ± SEM. *p ≤ 0.05, ***p ≤ 0.001.
circPPM1F promotes M1 macrophage activation by enhancing NF-κB signaling. A. The location of circPPM1F in the PPM1F transcript was validated by Sanger sequencing. B. Total RNA was digested with or without RNase R, followed by quantitative real-time PCR (qRT-PCR) measurements of circPPM1F, PPM1F, MALAT1, and GAPDH. C. The stability of circPPM1F was detected by qRT-PCR in THP1 macrophages after actinomycin D (Act D) treatment. D. qRT-PCR analysis of circPPM1F expression levels in THP1 macrophages following circPPM1F knockdown by two distinct siRNAs. E. qRT-PCR analyses of IL‐1β, TNF‐α, and CXCL10 in THP1 macrophages with conditional treatment. Mock-, untransfected, and unstimulated cells; mock+, LPS stimulated alone cells; si-scramble, LPS-stimulated cells following transfection with si-scramble; si-circPPM1F-2, LPS-stimulated cells following transfection with si-circPPM1F-2. F. ELISA analyses of secreted cytokine levels in THP1 macrophages with circPPM1F knockdown, followed by LPS treatment. G. qRT-PCR analyses of circPPM1F in THP1 macrophages with 3D5-circPPM1F or pZW1 transfection (left); M1-associated gene expressions in LPS stimulated THP1 macrophages overexpressing circPPM1F were quantified by qRT-PCR analysis (right). H. ELISA analyses of secreted cytokine levels in circPPM1F-overexpressed THP1 macrophages, followed by LPS treatment. I. Western blot showing total p65, p38, ERK1/2, JNK and their phosphorylation levels in THP1 macrophages with or without circPPM1F knockdown after LPS treatment. J. Western blotting analysis to evaluate levels of total p65, phosphorylated p65 in circPPM1F-overexpressed THP1 macrophages. The levels of p-p65 were normalized to that of β-tubulin and quantified using Image J software. Data are presented as mean ± SEM from three independent experiments. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ns indicates no significance.
circPPM1F competitively binds to HuR to impair PPM1F translation. A. Western blotting analysis to evaluate levels of PPM1F in circPPM1F knocked down-THP1 macrophages. B. qRT-PCR analyses of PPM1F expression in THP1 macrophages with circPPM1F knockdown. C. Analysis of PPM1F expression in THP1 macrophages transfected with PPM1F siRNA (200 nM) or control siRNA by qRT-PCR. D. Western blot analysis of PPM1F, p65, and p-p65 in THP1 macrophages with PPM1F knockdown, followed by LPS stimulation. E. Quantitative real-time PCR (qRT-PCR) analysis of M1-associated gene expression in THP1 macrophages with PPM1F knockdown, followed by LPS stimulation. F. qRT-PCR results showing the distribution of circPPM1F in the cytoplasmic and nuclear fractions of THP1 macrophages. GAPDH as cytoplasm control transcript, and U1 as nuclear control transcript. G. Putative HuR binding sites within circPPM1F full-length sequence. H. The enrichment levels of circPPM1F and PPM1F in the products of the RNA immunoprecipitation (RIP) assay (HuR IP compared with IgG IP) as detected by qRT-PCR. I. qRT-PCR analysis of HuR in THP1 macrophages transfected with HuR siRNA (200 nM) or control siRNA. J. Protein levels of PPM1F and HuR were detected by western blotting in THP1 macrophages with HuR knockdown. K. qRT-PCR analysis of circPPM1F and PPM1F in THP1 macrophages with HuR knockdown. The levels of PPM1F, p-p65 and HuR were normalized to those of β-tubulin and quantified using Image J software. Data are presented as mean ± SEM from three independent experiments. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ns indicates no significance.
Subcellular localization contributes to the modulatory mechanism of circRNAs on their targets. To further explore the mechanism underlying circPPM1F-mediated effects on PPM1F, we performed subcellular fractionation and localization assays. Surprisingly, in contrast to the cytoplasmic localization observed for a large number of verified exonic circRNAs, circPPM1F was primarily localized to the nucleus of THP1 macrophages (Figure 3F). circRNAs in the nucleus are reported to regulate gene translation via interaction with RNA-binding proteins (RBPs). We therefore searched for all putative RBPs binding to circPPM1F using the circRNA interactome database (https://circinteractome.nia.nih.gov/RNA_Binding_Protein/rna_binding_protein.html). Notably, the analysis revealed that the RBP containing the most potential binding sites with the circPPM1F was HuR (Figure 3G). HuR, a member of the ELAVL family of RBPs and selectively bind to AU-rich elements (AREs) within the 3′-untranslated regions (UTRs) of mRNA, stabilizing ARE-containing mRNA to regulate gene translation [28]. To determine whether the interaction between circPPM1F and HuR was responsible for circPPM1F-mediated translational inhibition of PPM1F, we conducted RIP using an antibody specific for HuR and found a 5-fold enrichment of circPPM1F and 30-fold enrichment of PPM1F when the anti-HuR antibody was used, relative to use the IgG control (Figure 3H). These results thus implied a direct interaction of HuR with circPPM1F or PPM1F.
EIF4A3 and FUS cooperatively regulate circPPM1F expression. A, B. Bioinformatic prediction of binding sites of EIF4A3 (A) and FUS (B) on circPPM1F. C. Expression levels of circPPM1F, EIF4A3, and FUS were detected by quantitative real-time PCR (qRT-PCR) in THP1 macrophages treated with LPS. D. qRT-PCR analyses of the expression levels of EIF4A3 and FUS in PBMCs from 43 T1DM patients and 45 healthy controls. E. Correlation analysis of the expression of circPPM1F and EIF4A3, or FUS levels in patients with type 1 diabetes mellitus (T1DM) (Pearson's correlation). F. qRT-PCR was used to measure the expression levels of EIF4A3 and FUS in THP1 macrophages with EIF4A3 or FUS knockdown. G. qRT-PCR analyses of the expression levels of circPPM1F in EIF4A3 or FUS knocked down THP1 macrophages with or without lipopolysaccharide (LPS) treatment. Data are presented as mean ± SEM from three independent experiments. **p ≤ 0.01, ***p ≤ 0.001, ns indicates no significance.
Next, we explored whether HuR is involved in the effects of circPPM1F on PPM1F translation; thus, siRNA directed at HuR was transfected to knock down HuR expression in THP1 macrophages. The knockdown of HuR resulted in a significant decrease in protein expression of PPM1F in THP1 macrophages (Figure 3I-J), whereas no changes in circPPM1F and PPM1F mRNA expression were detected (Figure 3K). Collectively, these studies strongly suggested that circPPM1F could suppress PPM1F translation through competitive interaction with HuR, which subsequently promotes activation of M1 macrophages.
EIF4A3 and FUS coordinately regulated circPPM1F expression
Next, we explored the biogenesis of circPPM1F. Previous studies have shown that the biogenesis of circRNA depends on the spliceosomal machinery containing with cis- and trans-regulatory elements. Reverse intronic complementary sequences (ICS) induced “head-to-tail” splicing by bringing the 5′- and 3′-termini of an exon or of consecutive exons into spatial proximity [29, 30]. However, there were no ICS in either of the intronic flanking regions of the splice junction sites of circPPM1F. Therefore, RBPs might be responsible for expression of circPPM1F. Using a bioinformatics method (https://circinteractome.nia.nih.gov/index.html), we queried all RBPs that were reported to be involved in circRNA generation. The result revealed that a binding site for EIF4A3 is present in the downstream region of the circPPM1F transcript, and FUS-binding sites were identified in the mature circPPM1F (Figure 4A-B). We then measured the levels of circPPM1F, EIF4A3, and FUS expression over time in THP1 macrophages stimulated by LPS. Interestingly, FUS exhibited a time-dependent increased expression following LPS treatment, while EIF4A3 was upregulated at 3 h after stimulation, dropped to a basic level at 6 h, and thereafter was gradually upregulated. circPPM1F was downregulated at 3 h, but afterward its expression appeared to be gradually upregulated (Figure 4C). Furthermore, we analyzed the levels of EIF4A3 and FUS expression in PBMCs from T1DM patients and healthy controls. The results indicated that the expression level of EIF4A3 was significantly decreased, whereas that of FUS was increased in T1DM patients compared with healthy controls (Figure 4D). Importantly, there was a negative correlation between EIF4A3 and circPPM1F, while a positive correlation was observed between FUS and circPPM1F (Figure 4E). The above results implied that EIF4A3 and FUS may play important roles in circPPM1F expression.
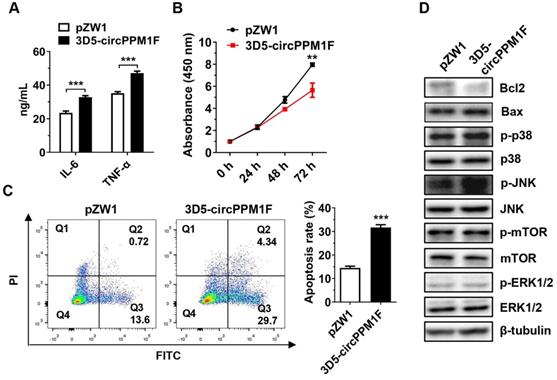
circPPM1F induces pancreatic β cell apoptosis through M1 macrophage activation. A. ELISA analyses of secreted cytokine levels in conditional media from circPPM1F-overexpressed Raw264.7 cells, followed by LPS treatment. B. Following incubation with conditional media, cell proliferation of MIN6 cells was assessed by the CCK-8 assay. C. After the incubation with conditional media, MIN6 cells were stained with Annexin V-FITC and propidium iodide (PI) before fluorescence analysis by flow cytometry. The percentage of cells in the four different quadrants was calculated and the results presented in different histograms indicating the fraction of apoptotic cells were Annexin V+/PI- and Annexin V+/PI+. D. Western blotting analysis to detect expression levels of Bcl2, Bax, p38, p-p38, JNK, p-JNK, ERK1/2, p-ERK1/2, mTOR, and p-mTOR in MIN6 cells cultured with conditional media. Data are presented as means ± SEM from three independent experiments. **p ≤ 0.01, ***p ≤ 0.001.
To further investigate the roles of EIF4A3 and FUS in circPPM1F expression, we knocked down EIF4A3 and FUS expression in THP1 macrophages by transfection with their individual siRNAs (Figure 4F). Surprisingly, knockdown of EIF4A3 or FUS resulted in no differences in the expression level of circPPM1F in THP1 macrophages without LPS treatment. However, knockdown of EIF4A3 significantly elevated the expression level of circPPM1F, while knockdown of FUS decreased its level in THP1 macrophages treated with LPS (Figure 4G). Overall, these results implied that EIF4A3 and FUS coordinately regulate circPPM1F expression during M1 macrophage activation.
circPPM1F induced pancreatic β-cell apoptosis by promoting M1 macrophage activation in vitro
To assess the pathological effects of circPPM1F-induced M1 macrophage activation on pancreatic β-cells, we performed a cell co-culture assay with Raw264.7 cells overexpressing circPPM1F and murine pancreatic β-cells MIN6. Consistent with the mRNA levels, the IL-6 and TNF-α protein levels were enhanced in conditional media from Raw264.7 cells overexpressing circPPM1F (Figure 5A). Notably, we found that the proliferation rate of MIN6 cells was significantly inhibited by the conditional media (Figure 5B). In addition, the media markedly enhanced the rate of apoptosis in MIN6 cells (Figure 5C). Previous studies have indicated that mTOR and MAPK pathways are crucial for pancreatic β-cell apoptosis [31, 32]. To investigate the potential molecular mechanisms inducing MIN6 cell apoptosis, we confirmed the effects of the conditional media on mTOR and MAPK pathways in MIN6 cells via western blotting. Decreased expression levels of Bcl2 and elevated Bax were observed in MIN6 cells cultured with media from circPPM1F overexpressing Raw264.7, and this was accompanied by increased levels of p38 and JNK phosphorylation (Figure 5D). However, there were no apparent changes in the phosphorylation levels of mTOR and ERK (Figure 5D). Collectively, these data suggested that circPPM1F-mediated M1 macrophage activation could induce pancreatic β cell apoptosis through the MAPK pathway in vitro.
circPPM1F exacerbated pancreas injury in STZ-induced diabetic mice through M1 macrophage activation
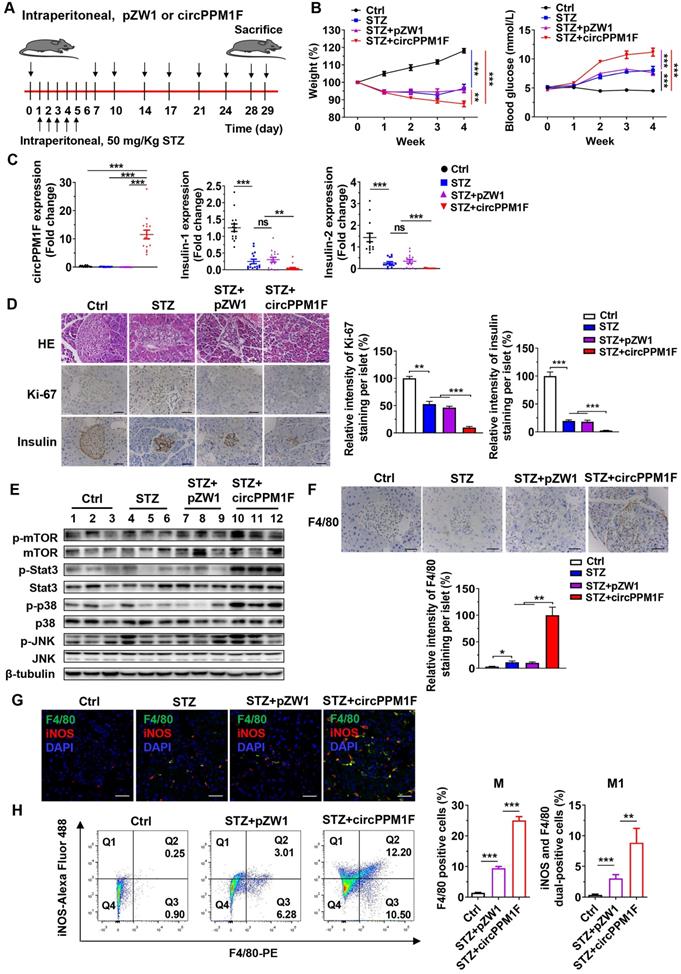
To assess the role of circPPM1F in pancreas injury of diabetic mice, we generated a diabetic mouse model using intraperitoneal (i.p.) injection of STZ (Figure 6A). We first investigated the dynamic expression profiles of circPPM1F in the pancreas, liver, spleen, and PBMCs of mice injected with circPPM1F or pZW1 plasmid for 3, 5, and 7 days. The results showed that the level of circPPM1F expression was highest in pancreas compared with that in the liver, spleen and PBMCs, and peaked on day 5 (Figure S3A). Next, successful induction of the diabetic mouse model was evidenced by significant weight loss one week after initial injection of STZ, and hyperglycemia two weeks after the injection. Importantly, the STZ+circPPM1F group mice achieved more severe weight reduction and hyperglycemia than STZ or STZ+pZW1 treated mice groups (Figure 6B). Further, the levels of circPPM1F and insulin expression in pancreas tissues were measured. Mice in the STZ+circPPM1F group displayed significantly enhanced circPPM1F and decreased insulin expression in the pancreas compared with the other three groups (Figure 6C). Furthermore, we assessed the effects of circPPM1F on pancreatic injury. The expression levels of inflammatory cytokines, oxidative stress indicators, indicators related to insulin secretion and cell apoptosis were detected. Specifically, STZ+circPPM1F group mice exhibited significantly higher levels of IL-6, IL-1β, iNOS, and Bax expression, accompanied by lower levels of Sod, Cat, GSH-Px, Glut2, Gck, and Bcl2 expression, relative to control, STZ, and STZ+pZW1 groups (Figure S3B). Similar to the results in Bcl2 and Bax at the mRNA level, decreased levels of Bcl2 and enhanced Bax protein were detected in mice of the STZ+circPPM1F group compared with those of the STZ+pZW1 group (Figure S3C).
Pancreatic dysfunction usually represents pathological changes in the diabetic pancreas. Notably, Histological analysis showed that STZ and STZ+pZW1 groups displayed distinctly abnormal islets structure as compared with the control group, appearing as small islets, inhomogeneous islet cells, and cytoplasmic vacuolation. Specifically, STZ+circPPM1F group mice exerted more severe damage of islets. Furthermore, Ki-67 staining indicated decreased proliferation of pancreas cells in the STZ and STZ+pZW1 groups, and a greater decrease in the STZ+circPPM1F group, compared with the control group. In addition, similar to the mRNA expression patterns observed for insulin in the experimental mice, protein levels of insulin were significantly lower in mice overexpressing circPPM1F (Figure 6D). Altogether, these findings indicated that circPPM1F could aggravate the pancreas injury of STZ-induced diabetic mice.
Subsequently, we wanted to examine the molecular mechanism of pancreas injury. Considering that the Stat3, MAPK, and Akt-mTOR pathways play a prominent role in oxidative stress, inflammatory response, and cell apoptosis [33, 34], we measured the levels of Stat3, p38/JNK, mTOR and their corresponding phosphorylated protein expression. Importantly, the results showed that the mice of the STZ+circPPM1F group exhibited significantly enhanced levels of phosphorylated Stat3, p38, and JNK expression compared with control, STZ and STZ+pZW1 groups. However, there were no differences in the phosphorylation levels of mTOR (Figure 6E). Therefore, these data suggested that circPPM1F could increase pancreas injury by activating MAPK and Stat3 pathways in vivo.
In order to further evaluate effects of circPPM1F on macrophage activation in pancreas islets of diabetic mice, we assessed the infiltration of macrophages into islets cells. Compared with control mice, increased levels of F4/80+ cells were observed in pancreas islets in STZ and STZ+pZW1 group mice, and the highest levels of F4/80+ cells were detected in the STZ+circPPM1F group (Figure 6F). Moreover, to demonstrate whether circPPM1F promoted infiltration of macrophages into the islet cells due to M1 macrophage activation, we detected levels of F4/80+/iNOS+ cells by immunofluorescence staining. Notably, elevated levels of F4/80+/iNOS+ cells were observed in pancreas islets of mice from the STZ+circPPM1F group compared with that from mice with or without STZ treatment (Figure 6G). In addition, the levels of M1 macrophage activation in the pancreas were validated by flow cytometry. Similar to the positive patterns of F4/80 and iNOS observed in immunohistochemistry (IHC) and immunofluorescence assays, the STZ+circPPM1F group mice displayed higher frequencies of macrophages (F4/80+) and M1 macrophages (F4/80+/iNOS+) in comparison to the other groups (Figure 6H). Taken together, these findings implied that circPPM1F could facilitate injury of pancreatic islets in diabetic mice by promoting M1 macrophage activation.
circPPM1F facilitates pancreatic islet injury in diabetic mice through M1 macrophage activation. A. Treatment of circPPM1F in the STZ-induced diabetic mouse model (15 mice per group). B. Mean weekly body weight (left) and fasting blood glucose (right) change in four-group mice. C. Levels of circPPM1F and insulin in pancreas tissues were detected by quantitative real-time PCR (qRT-PCR). D. Representative hematoxylin and eosin (H&E)-stained pancreas tissues, immunohistochemistry (IHC) images of Ki-67 and insulin expression in pancreatic islets from experimental mice. Scale bar indicates 50 µm. Semi-quantification of Ki-67 and insulin staining of per islet were done by using Image J software. E. The levels of total p38, JNK, Stat3, mTOR, and their corresponding phosphorylated forms in pancreas tissues from experimental models were quantified by western blot. F. Representative IHC images of F4/80 expression in pancreatic islets from experimental mice. Scale bar indicates 50 µm. Semi-quantification of F4/80 staining of per islet were done by using Image J software. G. Immunofluorescence staining of infiltrated F4/80+/iNOS+ M1 macrophages in mice pancreatic islets. Green represents anti-F4/80 Ab; red represents anti-iNOS Ab; yellow represents F4/80 and iNOS merged; blue represents DAPI. Scale bar indicates 50 µm. H. The percentages of M1 macrophages in pancreas tissue cells from STZ-treated mice with or without circPPM1F overexpression and control mice were determined by flow cytometry (left). Quantification analyses of macrophages (F4/80+) and M1 macrophages (F4/80+/iNOS+) in pancreas tissue cells (right). *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001.
Discussion
In this study, we showed for the first time that circPPM1F was overexpressed in PBMCs from T1DM patients, and it increased M1 macrophage activation through the circPPM1F-HuR-PPM1F-NF-κB axis. Moreover, EIF4A3 and FUS were critical for coordinately controlling circPPM1F expression. In addition, circPPM1F could accelerate pancreatic islet cell apoptosis in diabetic mice by promoting the activation of M1 macrophages. Thus, we speculate that circPPM1F might potential represent a new therapeutic target for T1DM, adding a new dimension to the functional importance of circRNA regulation in diabetes mellitus.
Exonic circRNAs are usually abundant in the cytoplasm and function mainly through the microRNA “sponge” mechanism [35]. In contrast, intron-containing circRNAs (intronic circRNAs and exon-intron circRNAs), in general, are enriched in the nucleus and are involved in regulation of host gene expression [36]. However, Errichelli et al. found that three completely spliced exonic circRNAs were almost exclusively located to the nucleus [37]. Consistent with this finding, in our study, as an exonic circRNA, circPPM1F was also constitutively expressed in the nucleus rather than in the cytoplasm of THP1 macrophages. Among the predicted RBPs of circPPM1F, we found 14 binding sites for fragile X mental retardation 1 (FMR1, also known as FMRP) were present in circPPM1F. Interestingly, FMRP participated in RNA trafficking from the nucleus to the cytoplasm, displaying different subcellular distribution due to alternative splicing [38, 39]. We speculated that FMRP might be responsible for the nuclear and cytoplasmic localization of circPPM1F. It would be helpful to investigate the interaction of FMRP with circPPM1F to better understand the nuclear location of circPPM1F.
Recently, circRNAs have attracted increasing attention for their potential roles in regulating parental gene expression [26, 27]. Meanwhile, parental genes may also be involved in circRNA biosynthesis [40]. PPM1F has been reported to regulate cancer cell growth and metastasis [41], whereas its roles in the pathogenesis of T1DM and M1 macrophage activation remain unclear. Notably, our findings provide evidence supporting the hypothesis that circPPM1F-mediated M1 macrophage activation may be attributable to reduced protein levels of the PPM1F gene. Recently, HuR is a well-studied RBP that positively augments stability of a number of linear mRNAs and ncRNAs, but also binds to introns of pre-mRNAs to modulate splicing [28]. It has been revealed that the interaction of HuR and circPABPN1 impaired the normal interaction of HuR's with linear mRNAs, especially parental pre-mRNA of circPABPN1, which consequently suppressed the production of PABPN1 protein [42]. Importantly, our data provide further evidence that HuR is a key contributor to circPPM1F and PPM1F interactions. Instead of impacting on circPPM1F transcription, the interaction with HuR and circPPM1F prevented HuR binding to PPM1F mRNA, resulting in a reduction of PPM1F translation. Such a relationship between circRNA and host mRNA is conceptually intriguing, and it might be generalizable to other RBPs. Additionally, our study was the first to reveal the role of PPM1F in M1 macrophage activation, implying that the circPPM1F-HuR-PPM1F axis may represent a novel potential therapeutic target in T1DM.
RBPs are required for regulation of the biogenesis specific circRNAs in a positive or negative way. In this study, our data showed that EIF4A3 and FUS oppositely regulated circPPM1F expression in response to external stimuli. EIF4A3, a member of the DEAD box protein family, has been implicated in nuclear and mitochondrial splicing, ribosome and spliceosome assembly, and translation initiation. Recent studies have revealed that EIF4A3 promoted circMMP9 and circSEPT9 expression via binding to parental pre-mRNAs [43, 44]. However, in our study we found that EIF4A3 could inhibit circPPM1F expression. Unexpectedly, we identified a binding site for EIF4A3 in the downstream region (i.e., at intron 6 of PPM1F transcript) of circPPM1F. Therefore, we assume that this unusual binding site might be responsible for the suppression of EIF4A3 on circPPM1F biogenesis. In addition, Errichelli et al. found FUS either increased or repressed circRNA biogenesis by binding to the introns flanking the back-splicing junctions, and the interaction machinery could control a complex interplay between linear and back-splicing [37]. In contrast to back-splicing regulation, our data indicated that FUS might also bind to mature circPPM1F at the post-transcriptional level and positively mediate its expression upon stimulation. Overall, these findings demonstrated that diverse RBPs participate in circRNA biogenesis, and that some may contribute to diverse regulatory mechanisms, even antagonistically. Hopefully, our study will prompt future work exploring RBP-based diverse regulation of circRNA biogenesis.
STZ is a broad-spectrum antibiotic possessing antitumor, oncogenic, and diabetogenic properties, and multiple small dose injections of STZ in mice produce pancreatic insulitis, with progression to nearly complete β-cell destruction and diabetes mellitus [45]. Although the timing and appearance of the inflammatory islet lesions do not demonstrate that STZ stimulation acts by initiating a cell-mediated immune reaction, multiple low doses of STZ have been shown to selectively destruct β-cells, which in turn induces immune reactions against pancreatic islets, leading to β-cell apoptosis and subsequently diabetes mellitus. This model resembles the key features of T1DM patients with a loss of β-cell function and the development of hyperglycemia [46-48]. At present, the STZ-induced mouse model is one of the most widely used animal models of human autoimmune diabetes in T1DM studies [46, 49-51]. In our study, we focused the effect of circPPM1F on development of STZ-induced diabetes mellitus. Consistent with in vitro studies of increased pancreatic β-cell apoptosis, we found that circPPM1F-mediated M1 macrophage activation could also facilitate pancreas injury in diabetic mice. However, it remains to be clarified whether macrophage-specific expression of circPPM1F facilitates the development of T1DM. Treatment of circPPM1F with a macrophage-specific promoter in the STZ-treated NOD mouse model, or generating a chimeric, macrophage-specific circPPM1F knocked-in T1DM mouse model would be helpful to address the issue.
In summary, our studies demonstrate the positive role of circPPM1F in LPS-induced M1 macrophage activation through the circPPM1F-HuR-PPM1F-NF-κB axis. In vivo, circPPM1F facilitated pancreas injury in STZ-induced diabetic mice by promoting M1 macrophage activation. Additionally, EIF4A3 and FUS might be required for the maintenance of circPPM1F expression during the progression of T1DM. Taken together, our work provides new insights into the pathogenesis of T1DM and suggests a potential novel biomarker or therapeutic target for T1DM.
Abbreviations
Act D: actinomycin D; circRNAs: circular RNAs; EIF4A3: eukaryotic initiation factor 4A-III; FMR1: fragile X mental retardation 1 (also known as FMRP); FUS: fused in sarcoma; GEO: Gene Expression Omnibus; HuR: human antigen R; MAPK: mitogen-activated protein kinase; NF-κB: nuclear factor kappa B; mTOR: mammalian target of rapamycin; PPM1F: protein phosphatase, Mg2+/Mn2+ dependent 1F; PBMCs: peripheral blood mononuclear cells; qRT-PCR: quantitative real-time PCR; RBP: RNA-binding protein; RIP: RNA immunoprecipitation; ROC: receiver operating curve; siRNA: small interfering RNA; Stat3: signal transducers and activators of transcription 3; STZ: streptozocin; T1DM: type 1 diabetes mellitus.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
Availability of data and material
The datasets and computer code produced in this study are available in the following databases: circBase (http://www.circbase.org/cgi-bin/simplesearch.cgi), and circRNA interactome database (https://circinteractome.nia.nih.gov/RNA_Binding_Protein/rna_binding_protein.html).
Accession numbers
Microarray data have been deposited in the Gene Expression Omnibus (GEO) under accession numbers GSE133225.
Ethics approval and consent to participate
T1DM patients and age-matched healthy controls were recruited from the Children's Hospital of Fudan University following informed consent from their parents was obtained. The study was approved by the Research Ethics Board of the Children′s Hospital of Fudan University [No. (2016) 96].
Funding
This work was supported by grants from the National Key R&D Program of China (2016YFC1305102 to YZ), National Natural Science Foundation of China (81671561, 81974248 to YZ, 81900751 to HX), the International Joint Laboratory Program of National Children's Medical Center (EK1125180109 to YZ), Program for Outstanding Medical Academic Leader (2019LJ19 to YZ) and Shanghai Municipal Planning Commission of Science and Research Fund (201740065 to YZ and 20174Y0079 to HX). Shanghai Pujiang Program (16PJ1401600 to FJ). Shanghai Committee of Science and Technology (19ZR1406400 to FJ).
Authors' contributions
CZ, XH, LY, JR, SH, WX, YG, QL and XW designed and carried out experiments, and analyzed data. CS, FL and WL recruited and characterized the human participants. CZ and YZ wrote the manuscript. YZ planned, designed, supervised, and coordinated the overall research efforts.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Patterson CC, Dahlquist GG, Gyurus E, Green A, Soltesz G, Group ES. Incidence trends for childhood type 1 diabetes in Europe during 1989-2003 and predicted new cases 2005-20: a multicentre prospective registration study. Lancet. 2009;373:2027-33
2. Rewers M, Ludvigsson J. Environmental risk factors for type 1 diabetes. Lancet. 2016;387:2340-8
3. Diaz-Valencia PA, Bougneres P, Valleron AJ. Global epidemiology of type 1 diabetes in young adults and adults: a systematic review. BMC Public Health. 2015;15:255
4. Thomas NJ, Jones SE, Weedon MN, Shields BM, Oram RA, Hattersley AT. Frequency and phenotype of type 1 diabetes in the first six decades of life: a cross-sectional, genetically stratified survival analysis from UK Biobank. Lancet Diabetes Endocrinol. 2018;6:122-9
5. Hughes JW, Bao YK, Salam M, Joshi P, Kilpatrick CR, Juneja K. et al. Late-onset T1DM and older age predict risk of additional autoimmune disease. Diabetes Care. 2019;42:32-8
6. Patterson CC, Karuranga S, Salpea P, Saeedi P, Dahlquist G, Soltesz G. et al. Worldwide estimates of incidence, prevalence and mortality of type 1 diabetes in children and adolescents: results from the International Diabetes Federation Diabetes Atlas, 9th edition. Diabetes Res Clin Pract. 2019;157:107842
7. Cnop M, Welsh N, Jonas JC, Jorns A, Lenzen S, Eizirik DL. Mechanisms of pancreatic beta-cell death in type 1 and type 2 diabetes: many differences, few similarities. Diabetes. 2005;54(Suppl 2):S97-107
8. Martinez FO, Sica A, Mantovani A, Locati M. Macrophage activation and polarization. Front Biosci. 2008;13:453-61
9. Murray PJ, Allen JE, Biswas SK, Fisher EA, Gilroy DW, Goerdt S. et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 2014;41:14-20
10. Calderon B, Suri A, Unanue ER. In CD4+ T-cell-induced diabetes, macrophages are the final effector cells that mediate islet beta-cell killing: studies from an acute model. Am J Pathol. 2006;169:2137-47
11. Wang F, Sun F, Luo J, Yue T, Chen L, Zhou H. et al. Loss of ubiquitin-conjugating enzyme E2 (Ubc9) in macrophages exacerbates multiple low-dose streptozotocin-induced diabetes by attenuating M2 macrophage polarization. Cell Death Dis. 2019;10:892
12. Arnush M, Scarim AL, Heitmeier MR, Kelly CB, Corbett JA. Potential role of resident islet macrophage activation in the initiation of autoimmune diabetes. J Immunol. 1998;160:2684-91
13. Yang H, Xie T, Li D, Du X, Wang T, Li C. et al. Tim-3 aggravates podocyte injury in diabetic nephropathy by promoting macrophage activation via the NF-kappaB/TNF-alpha pathway. Mol Metab. 2019;23:24-36
14. Jun HS, Yoon CS, Zbytnuik L, van Rooijen N, Yoon JW. The role of macrophages in T cell-mediated autoimmune diabetes in nonobese diabetic mice. J Exp Med. 1999;189:347-58
15. Thayer TC, Delano M, Liu C, Chen J, Padgett LE, Tse HM. et al. Superoxide production by macrophages and T cells is critical for the induction of autoreactivity and type 1 diabetes. Diabetes. 2011;60:2144-51
16. Pasman Z, Been MD, Garcia-Blanco MA. Exon circularization in mammalian nuclear extracts. RNA. 1996;2:603-10
17. Wilusz JE. A 360 degrees view of circular RNAs: From biogenesis to functions. Wiley Interdiscip Rev RNA. 2018;9:e1478
18. Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J. et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141-57
19. Rybak-Wolf A, Stottmeister C, Glazar P, Jens M, Pino N, Giusti S. et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58:870-85
20. Ng WL, Marinov GK, Liau ES, Lam YL, Lim YY, Ea CK. Inducible RasGEF1B circular RNA is a positive regulator of ICAM-1 in the TLR4/LPS pathway. RNA Biol. 2016;13:861-71
21. Zhu YJ, Zheng B, Luo GJ, Ma XK, Lu XY, Lin XM. et al. Circular RNAs negatively regulate cancer stem cells by physically binding FMRP against CCAR1 complex in hepatocellular carcinoma. Theranostics. 2019;9:3526-40
22. American Diabetes A. (2) Classification and diagnosis of diabetes. Diabetes Care. 2015;38(Suppl):S8-S16
23. Han X, Huang S, Xue P, Fu J, Liu L, Zhang C. et al. LncRNA PTPRE-AS1 modulates M2 macrophage activation and inflammatory diseases by epigenetic promotion of PTPRE. Sci Adv. 2019;5:eaax9230
24. Nyati KK, Masuda K, Zaman MM, Dubey PK, Millrine D, Chalise JP. et al. TLR4-induced NF-kappaB and MAPK signaling regulate the IL-6 mRNA stabilizing protein Arid5a. Nucleic Acids Res. 2017;45:2687-703
25. Weng T, Koh CG. POPX2 phosphatase regulates apoptosis through the TAK1-IKK-NF-kappaB pathway. Cell Death Dis. 2017;8:e3051
26. Yang F, Hu A, Li D, Wang J, Guo Y, Liu Y. et al. Circ-HuR suppresses HuR expression and gastric cancer progression by inhibiting CNBP transactivation. Mol Cancer. 2019;18:158
27. Zhou J, Zhang S, Chen Z, He Z, Xu Y, Li Z. CircRNA-ENO1 promoted glycolysis and tumor progression in lung adenocarcinoma through upregulating its host gene ENO1. Cell Death Dis. 2019;10:885
28. Lebedeva S, Jens M, Theil K, Schwanhausser B, Selbach M, Landthaler M. et al. Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol Cell. 2011;43:340-52
29. Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12:381-8
30. Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159:134-47
31. Liu Y, Han J, Zhou Z, Li D. Paeoniflorin protects pancreatic beta cells from STZ-induced damage through inhibition of the p38 MAPK and JNK signaling pathways. Eur J Pharmacol. 2019;853:18-24
32. Yang Z, Liu F, Qu H, Wang H, Xiao X, Deng H. 1, 25(OH)2D3 protects beta cell against high glucose-induced apoptosis through mTOR suppressing. Mol Cell Endocrinol. 2015;414:111-9
33. Li J, He J, Zhang X, Li J, Zhao P, Fei P. TSP1 ameliorates age-related macular degeneration by regulating the STAT3-iNOS signaling pathway. Exp Cell Res. 2020;388:111811
34. Wu J, Yang C, Liu J, Chen J, Huang C, Wang J. et al. Betulinic acid attenuates T-2-Toxin-induced testis oxidative damage through regulation of the JAK2/STAT3 signaling pathway in mice. Biomolecules. 2019 9
35. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK. et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384-8
36. Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH. et al. Circular intronic long noncoding RNAs. Mol Cell. 2013;51:792-806
37. Errichelli L, Dini Modigliani S, Laneve P, Colantoni A, Legnini I, Capauto D. et al. FUS affects circular RNA expression in murine embryonic stem cell-derived motor neurons. Nat Commun. 2017;8:14741
38. Bagni C, Greenough WT. From mRNP trafficking to spine dysmorphogenesis: the roots of fragile X syndrome. Nat Rev Neurosci. 2005;6:376-87
39. Fu XG, Yan AZ, Xu YJ, Liao J, Guo XY, Zhang D. et al. Splicing of exon 9a in FMR1 transcripts results in a truncated FMRP with altered subcellular distribution. Gene. 2020;731:144359
40. Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M. et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55-66
41. Wang H, Chen W, Jin M, Hou L, Chen X, Zhang R. et al. CircSLC3A2 functions as an oncogenic factor in hepatocellular carcinoma by sponging miR-490-3p and regulating PPM1F expression. Mol Cancer. 2018;17:165
42. Abdelmohsen K, Panda AC, Munk R, Grammatikakis I, Dudekula DB, De S. et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 2017;14:361-9
43. Wang R, Zhang S, Chen X, Li N, Li J, Jia R. et al. EIF4A3-induced circular RNA MMP9 (circMMP9) acts as a sponge of miR-124 and promotes glioblastoma multiforme cell tumorigenesis. Mol Cancer. 2018;17:166
44. Zheng X, Huang M, Xing L, Yang R, Wang X, Jiang R. et al. The circRNA circSEPT9 mediated by E2F1 and EIF4A3 facilitates the carcinogenesis and development of triple-negative breast cancer. Mol Cancer. 2020;19:73
45. Like AA, Rossini AA. Streptozotocin-induced pancreatic insulitis: new model of diabetes mellitus. Science. 1976;193:415-7
46. Fidler TP, Marti A, Gerth K, Middleton EA, Campbell RA, Rondina MT. et al. Glucose metabolism is required for platelet hyperactivation in a murine model of type 1 diabetes. Diabetes. 2019;68:932-8
47. Lin Y, Sun Z. Antiaging gene klotho attenuates pancreatic beta-cell apoptosis in type 1 diabetes. Diabetes. 2015;64:4298-311
48. O'Brien BA, Harmon BV, Cameron DP, Allan DJ. Beta-cell apoptosis is responsible for the development of IDDM in the multiple low-dose streptozotocin model. J Pathol. 1996;178:176-81
49. Tekula S, Khurana A, Anchi P, Godugu C. Withaferin-A attenuates multiple low doses of Streptozotocin (MLD-STZ) induced type 1 diabetes. Biomed Pharmacother. 2018;106:1428-40
50. Wu M, Chen W, Zhang S, Huang S, Zhang A, Zhang Y. et al. Rotenone protects against beta-cell apoptosis and attenuates type 1 diabetes mellitus. Apoptosis. 2019;24:879-91
51. Cheng RX, Feng Y, Liu D, Wang ZH, Zhang JT, Chen LH. et al. The role of Nav1.7 and methylglyoxal-mediated activation of TRPA1 in itch and hypoalgesia in a murine model of type 1 diabetes. Theranostics. 2019;9:4287-307
Author contact
![]() Corresponding author: Yufeng Zhou, M.D., Ph.D., 399 Wanyuan Rd, Minhang, Shanghai 201102, China. Telephone: 86-021-64932907; E-mail: yfzhou1edu.cn.
Corresponding author: Yufeng Zhou, M.D., Ph.D., 399 Wanyuan Rd, Minhang, Shanghai 201102, China. Telephone: 86-021-64932907; E-mail: yfzhou1edu.cn.
 Global reach, higher impact
Global reach, higher impact