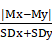13.3
Impact Factor
Theranostics 2020; 10(1):50-61. doi:10.7150/thno.36274 This issue Cite
Research Paper
Theranostic application of miR-429 in HER2+ breast cancer
1. Institute of Molecular Bioimaging and Physiology, National Research Council (IBFM-CNR), Via F.Cervi 93, 20090 Segrate-Milan, Milan, Italy.
2. Department of Pathophysiology and Transplantation, University of Milan, Milan, Italy.
3. Laboratory of Nanomedicine and Molecular Imaging, Istituti Clinici Scientifici Maugeri IRCCS, via Maugeri 4, 27100, Pavia, Italy.
4. Department of Biomedical and Clinical Sciences “L. Sacco”, Università degli studi di Milano, via G. B. Grassi 74, 20157 Milano, Italy.
5. Breast Unit, Surgery Department, Istituti Clinici Scientifici Maugeri IRCCS, via Maugeri 4, 27100, Pavia, Italy.
* These authors contributed equally to this work
Received 2019-5-3; Accepted 2019-9-3; Published 2020-1-1
Abstract
Human epidermal growth factor receptor 2 (HER2) is overexpressed/amplified in one third of breast cancers (BCs), and is associated with the poorer prognosis and the higher metastatic potential in BC. Emerging evidences highlight the role of microRNAs (miRNAs) in the regulation of several cellular processes, including BC.
Methods: Here we identified, by in silico approach, a group of three miRNAs with central biological role (high degree centrality) in HER2+ BC. We validated their dysregulation in HER2+ BC and we analysed their functional role by in vitro approaches on selected cell lines and by in vivo experiments in an animal model.
Results: We found that their expression is dysregulated in both HER2+ BC cell lines and human samples. Focusing our study on the only upregulated miRNA, miR-429, we discovered that it acts as an oncogene and its upregulation is required for HER2+ cell proliferation. It controls the metastatic potential of HER2+ BC subtype by regulating migration and invasion of the cell.
Conclusions: In HER2+ BC oncogenic miR-429 is able to regulate HIF1α pathway by directly targeting VHL mRNA, a molecule important for the degradation of HIF1α. The overexpression of miR-429, observed in HER2+ BC, causes increased proliferation and migration of the BC cells. More important, silencing miR-429 succeeds in delaying tumor growth, thus miR-429 could be proposed as a therapeutic probe in HER2+ BC tumors.
Keywords: Theranostics, HER2+ breast cancer, microRNAs, diagnosis, therapeutic tool
Introduction
The St.Gallen International Breast Cancer Conference classifies breast cancer (BC) into five intrinsic molecular subtypes: luminal A, luminal B, HER2-enriched, basal-like, and normal-like [1]. BC patients with overexpression of Human epidermal growth factor receptor 2 (HER2) have shown a poorer and shorter overall survival (OS) [2, 3] being HER2 expression a predictor of metastatic BC [4]. Indeed, patients with HER2+ BC frequently develop metastases in the brain, liver and lung [5].
HER2 was discovered as a human proto-oncogene in 1985 through its homology with v-erbB and with the human epidermal growth factor receptor (EGFR) gene. HER2 gene encodes for a tyrosine kinase receptor, whose amplification and activation lead to uncontrolled cell proliferation. In 1987, D. Slamon described amplification of HER2 gene in ~30% of all clinical samples of BC [6]. As a result, HER2 receptor is overexpressed in HER2+ BC patients up to 20 times more than in normal tissues [6]. This overexpression is involved in the cell proliferation maintenance, avoiding apoptosis and in favoring cell spreading and metastasis formation [7].
The actual methods for HER2+ BC diagnosis are based on the analysis of tissue samples by immunohistochemistry (IHC) of HER2 protein or in situ hybridization of HER2 specific transcript. This method is widely used as it is cost-effective, easy to be used for rapid evaluation in primary diagnostics and for rapid screens [8]. Although the optimization guides help the pathologists in the diagnosis [9, 10], the IHC approach is limited by technical features, such as the correct identification of the tumor areas within a complex tissue section, or the interpretation of the visual expression pattern and intensities in the tumor cells and in surrounding tissue [11]. Poor reproducibility among different laboratories, or among observers and pathologists in the same laboratory has emerged (i.e. [12]). Moreover, some studies highlight the existence of discordance in 25% of HER2+ BC cases between two different pathologists [13-15]. These observations pushed the research towards the discovery of new reliable diagnostic biomarkers to be used in precision medicine for HER2+ diagnosis.
The main methods to define new robust biomarker are based on the analysis of high-throughput database, in order to identify differentially expressed genes, and on the integration of this information with functional pathways and with the biological relationships among cellular components (network) [16-19]. These integrative approaches consider also epigenetic features of the tumors, with a particular attention to microRNA (miRNA) molecules. miRNAs are small non coding RNA with a main role in BC development, being able to regulate several mRNA targets. A small change in miRNA expression levels could have a high impact on the phenotype of the cell [20].
In our laboratory we developed a new in silico integrative approach to put all the information on differentially expressed genes, network and functional pathways in relation with miRNA molecules, in order to identify a specific group of miRNAs with a main role in the development of HER2+ BC. In particular, we identified three miRNAs, namely Hsa-miR-190, Hsa-miR-429 and Hsa-miR-584 (indicated from here on as miR-190, miR-429 and miR-584, respectively), having a higher degree centrality in HER2+ BC, being able to control the highest number of genes in coupled functional pathways.
In this paper, we described the bioinformatics approach and validated the ability of these three miRNAs to perform differential diagnosis of HER2+ BC. Moreover, we focused on miR-429, the only upregulated miRNA of the three with a highest number of connections with the functional pathways, and we analyzed its role in controlling specific function altered in HER2+ BC. miR-429 demonstrated to be a diagnostic molecule involved in the control of HIF1α pathway, having as a direct target VHL gene. Indeed, in vitro experiments revealed that miR-429 is involved in the regulation of proliferation and metastatic potential of highly aggressive BC. In addition to its diagnostic properties, in vivo studies with xenografted mouse, showed that the treatment with a silencer oligonucleotide of miR-429 is able to cause a delay of tumor proliferation, thus making miR-429 a possible candidate molecule for the development of new therapeutic tools in HER2+ BC.
Material and Methods
Datasets for computational analysis
We applied the computational approach on HER2+ BC dataset of IlluminaHiSeq RNASeqV2 derived from The Cancer Genome Atlas (TCGA). We selected 43 HER2+ BC samples and 113 normal samples (NS). The expression levels of 1046 miRNAs and 15243 genes (excluding genes with a small variance) were considered. We used BC matched samples of mRNA and miRNA.
Computational approach
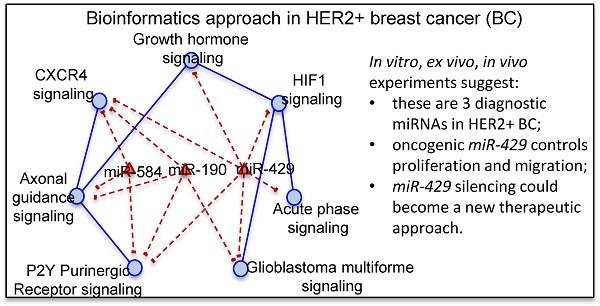
The computational analysis consists of 4 steps. In the first step we applied a differential expression analysis identifying pathways enriched with differentially expressed genes between HER2+ BC samples and NS. In the second step we calculated a measure of pathway cross-talk among pairwise pathways. In the third step we selected pairwise pathways with a machine learning approach to distinguish HER2+ BC samples vs NS with the best performance, generating a pathway cross-talk network. In the fourth step we applied a mutual information analysis in order to identify differentially expressed miRNAs regulating pathway cross-talk.
The computational approach was based on a Monte Carlo Cross-Validation, dividing the original dataset into training (60%) and testing (40%) dataset. The first three steps were repeated 50 times on different generated training data set. The fourth step was applied on the whole dataset. To avoid problems of unbalanced classes, we generated randomly an equal number of samples for each class (HER2+ BC and NS). Figure 1 shows the workflow of the proposed computational approach.
Workflow of the proposed computational approach.
Pathway enrichment analysis
Differentially expressed genes between HER2+ BC and NS were identified by a statistical analysis using TCGAbiolinks package [21] (|logFC| >1 and p-value<0.01). The p-values were adjusted by the Benjamin-Hochberg method for multiple testing correction [22].
Considering 589 pathways from the Ingenuity Pathway Analysis (IPA) database, we carried out a pathway enrichment analysis. The enrichment was performed using the Fisher's Exact Test; we considered a pathway to be enriched with differentially expressed genes if the associated p-value was <0.01 [23].
Pathway cross-talk quantification
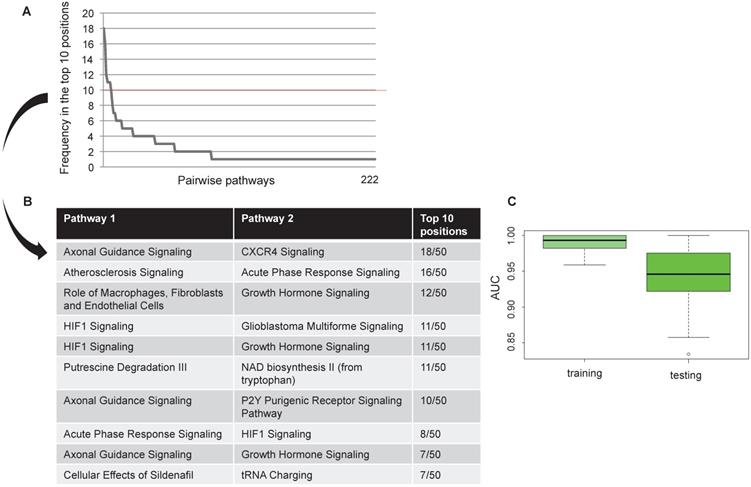
Interactions between enriched pathways were evaluated through a discriminating score (DS) [24]. This score was calculated considering the mean and standard deviation of the gene expression levels for each couple of pathways. Mx and My represent the mean of gene expression levels for the pathway x and y; SDx and SDy are the standard deviation for the pathway x and y.
DS =
Machine learning approach: pathway cross-talk network
We implemented a machine learning model using a Random Forest classification of the R-package [25](k-cross validation, k=10) in order to select the pairwise pathways that achieved the best performance in the classification of HER2+ BC and NS. The performances were valuated using the Area Under Curve (AUC). The classification was repeated 50 times for each different training dataset, selecting the top 10 pairs of pathways for each bootstrap. Finally, we selected the pathways that achieved the top 10 positions in a major number of boots. The AUC performances were validated also using the testing dataset.
miRNAs regulating pathway cross-talk network
Differential expression analysis of miRNAs between HER2+ BC and NS was performed using TCGAbiolinks package (|logFC| >1 and adjusted p-value <0.01) [21].
Mutual information was applied between differentially expressed miRNAs in HER2+ BC vs NS and 15243 genes of the original mRNA dataset, using the R-package parmigene [26].
Since we obtained a list of genes potentially regulated by miRNAs, we evaluated the pathways enriched with target genes for each miRNA. In particular, using Fisher's exact test, we selected miRNAs that are able to regulate the network of pathways (p-values<0.01). Furthermore, we selected the miRNAs with the highest value of degree centrality in the network, namely those able to regulate the major number of genes in the pathways [27].
We generated a pathway cross talk network, considering only the pathways in the top 10 pairs of pathways that present at least 2 or more interactions with the other pathways and miRNAs.
BC human tissue samples for ex vivo studies
We used ten samples of HER2+ BC human tissue and corresponding non-tumoral clear margins from surgical resections performed from 2011 to 2013 at the Breast Unit of Istituti Clinici Scientifici Maugeri IRCCS, Pavia, Italy. Samples referred to the biological collection of the “Bruno Boerci” Oncological Biobank for research applications (Istituti Clinici Scientifici Maugeri IRCCS, Pavia), a biobank certified to ISO 9001:2015 and member of BBMRI.it (the italian node of the BBMRI-ERIC, Biobanking and BioMolecular resources Research Infrastructure - European Research Infrastructure Consortium). Upon informed consent from patients, samples were collected, processed and stored at -80°C as snap-frozen aliquotes immediately after surgery, according to the best practices in biobanking (certification ISO 9001:2015). At the time of collection, HER2 enrichment was assessed through IHC molecular characterization by the Pathology Service (Istituti Clinici Scientifici Maugeri IRCCS, Pavia), according to the clinical guidelines on BC (ASCO - American Society of Clinical Oncology). Molecular characterization of the tumor samples is shown in Table S1. The samples were used for the isolation of total RNA and miRNA analysis in real-time PCR (as described in the methods below).
BC cell culture for in vitro studies
We used SKBR3 cells (ICLC, Genova, Italy), that are the reference HER2+, human, BC epithelial cell line [28] and MCF10A cells, as human, normal-like epithelial breast cells [29]. We maintained the cell line within a humidified atmosphere containing 5% CO2 at 37 °C in DMEM high glucose cell culture medium (Gibco, Life Technologies), with 10% fetal bovine serum (FBS), 1% penicillin/streptomycin, 2mM L-Glutamine (Euroclone) following the manufacturer's recommendation. Dulbecco Phosphate-Buffered Saline (D-PBS), and trypsin were obtained by Lonza (Euroclone).
RNA extraction and real time-PCR analysis
Total RNA of both cell lines and human samples was isolated using TRIzol reagent (Life Technologies) following the manufacturer's recommendations and as already described in [29]. For miRNA quantification, total RNA was reverse transcribed using MystiCq microRNA cDNA synthesis kit (Sigma Aldrich), following manufacturer's recommendations. miRNAs were amplified in Eco real time-PCR (RT-PCR) (Illumina, Euroclone) using Power Up Sybr green mix (Applied Biosystem) in combination with homemade designed primers (Sequences in the Table S2). For each RT-PCR analysis the results are presented as 2^-ΔΔCt method [30] normalizing the results on the expression of a miR-103-3p (Table S2) and the positive control of the MystiCq microRNA cDNA synthesis kit.
miRNA modulation
miRNA increase or decrease were obtained in SKBR3 cell line by transfection of sense or antisense oligonucleotide (Sigma Aldrich) with metafectene reagents (Biontex, Germany), following manufacturer's instruction. The sequences for miRNA modulation are in Table S2. All miRNA oligonucleotides were designed on the sequences from miRbase database [31].
MTT assay
SKBR3 cells were seeded in 96 wells at 5000 cells/well. 48h after seeding the cells were treated with Scramble S or As miR-429 oligonucleotide for miRNA modulation. At 24, 48 and 72h after treatment, the cells were stained with 50ug/ul of thiazolyl blue tetrazolium bromide (MTT) (Sigma Aldrich) for 4 h at 37°C and then lysed in dimethyl sulfoxide (DMSO; Euroclone, Italy). The staining was quantified at 540nm in FLUOstar Omega microplate reader (Euroclone). The presented results are the average±standard deviation (sd) of three independent experiments.
Wound-healing assay
The SKBR3 cells were seeded at a confluence of 100000/well in 24-well plate (Euroclone). After 48 h, necessary to reach 80% confluency, a scratch was performed in the middle of each well, as described in [29]. After washing the cells, new fresh medium was added, and miRNAs were modulated by transfection, as described before. Pictures of each well were taken every 24 h. The effect on cell migration was quantified by ImageJ software, following the protocol described in [32]. Experiments were performed in triplicate (n = 9) and a t test was calculated.
Cell invasion in Boyden's chamber
To study the invasion ability of the cells, we performed the Boyden's chamber test on SKBR3 in the presence of miR-429 silencing. Fifty thousand SKBR3 cells were seeded on the top of the 70ul coating Matrigel (BD) and covered with 350ul of complete medium except for 2% FBS. The lower chamber was filled with complete medium. One hour after seeding, the cells were treated with 150nM scramble or As miR-429 oligonucleotides. Twenty-four hours later, cells migrated through the Boyden's chamber, were fixed in 4% PFA and stained with cresyl violet for 5 minutes (min.). Ten pictures for each condition were taken and the stained cells were count. The experiment was performed in triplicate and the average of the migrated cells is calculated.
HER2+ BC animal model for in vivo treatment studies
Animal experiments were carried out in compliance with the institutional guidelines for the care and use of experimental animals (European Directive 2010/63/UE and the Italian law 26/2014), authorized by the Italian Ministry of Health and approved by the Animal Use and Care Committee of the University of Milan. Ten female nude mice (athymic nude-Foxn1 Nu/Nu, Envigo), 7-8 weeks old, were all xenografted to model HER2+ BC with 5 million SKBR3 cells in Matrigel injected in mammary gland. During in vivo study, mice were maintained on a 12-h light-dark cycle in cages of five animals with water and food ad libitum (environment of 23 ± 1°C and 50 ± 5% humidity). After detection of the tumors (about 20 days), the animals were divided in two groups: the first group was treated with 5 pM scramble S oligonucleotide in the presence of Atelogene as vehicle; the second group was treated with 5 pM As miR-429 in the presence on Atelogene as vehicle. One animal was treated with vehicle alone. Tumor growth was followed over time by measurements of the three dimensions of the palpable masses (height x length x depth) and the tumor volume was calculated by multiplying the three dimensions of the tumor measured by caliper.
For ex vivo studies, at the end of the experiments, mice were sacrificed by cervical dislocation after sedation, and tumours were explanted for immune-histochemical and molecular analyses.
Luciferase assay
To assess if VHL is a direct target of miR-429, pGL3-control vector or pGL3-VHL-WT was transfected with lipofectamine 3000 (Invitrogen) in SKBR3 cells, according to the manufacturer's suggestion, in combination with 100 nM mimic miR-429 or miR-142 (a known regulator of VHL) or scramble miR sequence. After 24 h, Gaussia Luciferase (GLuc) activity was measured in the collected medium and normalized on the Secreted Alkaline Phosphatase (SEAP) content. Results were detected by Glomax (GloMax-Multi Detection System; Promega, Madison, WI, USA).
Western blot analysis
Western blot on total protein lysates was performed as described in [29]. Anti VHL or anti actin antibodies (both Santa Cruz Biotechnologies) was used 1:500 overnight in 3% skimmed milk and the secondary anti-mouse HRP (Santa Cruz Biotechnologies) was used 1:1000 1 h at room temperature in 3% skimmed milk. Membranes were developed using the SuperSignal West Pico Chemiluminescent Substrate (ThermoScientific, France) on Amersham Hyperfilm MP (Ge Helthcare, Italy). The blots of both proteins were quantified by ImageJ software (http://imagej.nih.gov/ij/). Experiments were performed two times in triplicate.
Hypoxia induction
In order to study the effects of hypoxia on miR-429 expression, we used a hypoxic chamber [33]. During hypoxia experiments, SKBR3 cells were seeded at 150000 cells/w in a 6 wells' plate and incubated in the hypoxic chamber containing 1% O2 gas mixture for 24 h (h). After this treatment, the cells were used for RNA extraction and RT-PCR analysis of miR-429 expression.
Results
Computational approach predicted that miR-190, miR-429 and miR-584 have a central role in the diagnosis of HER2+ BC
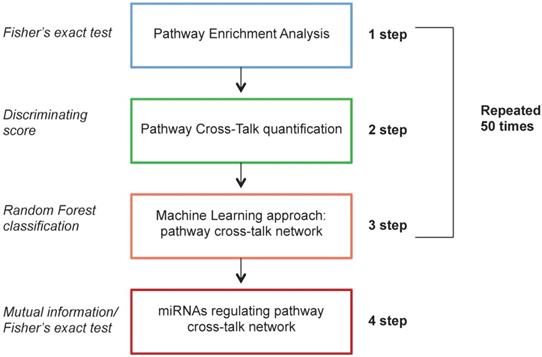
We found 222 pairwise pathways that obtained an AUC value in the top 10 positions in at least one bootstrap in discriminating HER2+ BC versus NC. Figure 2A shows the distribution of the 222 pairwise pathways in the top 10 positions for each boot. Then, we selected the 10 pairwise pathways that achieved the best performances in all bootstraps (Figure 2B). Overall, the selected pairwise pathways obtained an AUC value of 0.98 in the training and 0.94 in the testing data set (Figure 2C).
We identified miRNAs regulating the selected pairwise pathways generating a diagnostic pathway-miRNA network.
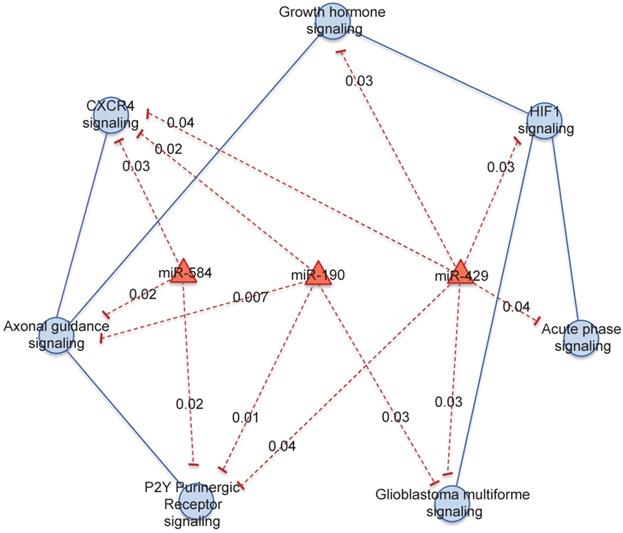
We focused on 3 miRNAs altered in HER2+ BC indicating the number of genes potentially regulated by each miRNA obtained by mutual information approach, on the total number of genes in the coupled pathway. In our study we considered those miRNAs with the highest degree centrality, miR-190, miR-429 and miR-584. From our analysis miR-429 control 33 genes over 484 in 3 coupled pathways, miR-190 and miR-584 controls 30 and 49 genes over 693 in 3 and 2 coupled pathways, respectively (Table S3). We selected the pathways and miRNAs with the highest degree centrality (HDC) obtaining a network of 7 pathways and 3 miRNAs (Figure 3).
miR-429 regulates the major number of pathways: "HIF1 signaling", "Acute Phase Response signaling", "Glioblastoma Multiforme signaling", "P2Y Purigenic Receptor signaling", "CXCR4 signaling " and “Growth hormone signaling”. miR-584 regulates "P2Y Purigenic Receptor signaling", "CXCR4 signaling" and "Axonal Guidance signaling". miR-190 regulates "P2Y Purigenic Receptor signaling", "Axonal Guidance signaling", "CXCR4 signaling" and "Glioblastoma Multiforme signaling".
In vitro studies confirmed that miR-190, miR-429 and miR-584 are diagnostic molecules for HER2+ BC
To evaluate if the expression levels of miR-190, -429 and -584 are different in HER2+ versus normal-like epithelial cells, as predicted by in silico analysis, we performed RT-PCR assay of these three miRNAs in both cell line and human BC samples. The SKBR3 HER2+ cells compared to MCF10A cells have significantly higher levels of miR-429 compared to normal-like cells (Figure S1A and Figure S2) and lower levels of both miR-190, and miR-584 (Figure S1B and C, respectively).
ex vivo studies confirmed that miR-190, miR-429 and miR-584 are diagnostic molecules for HER2+ BC
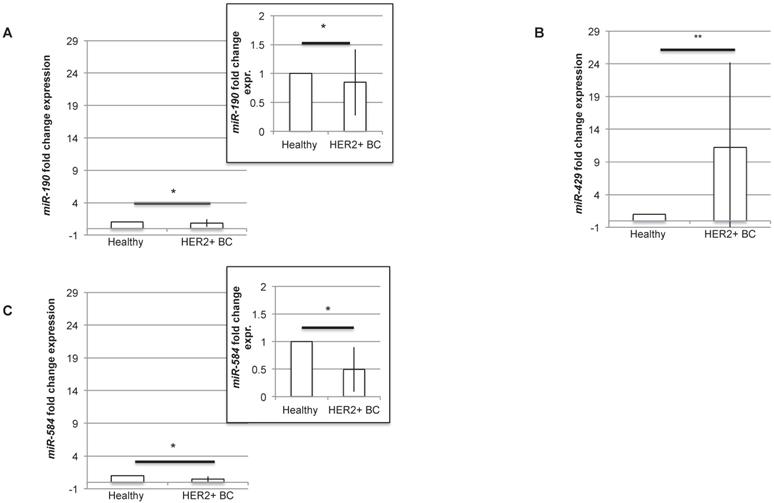
In order to understand if the three miRNAs could be proposed as diagnostic molecules for HER2+ BC, we analysed the level of expression of miR-190, miR-429 and miR-584 in human BC HER2+ samples compared to normal surrounding epithelial mammary tissue. The RT-PCR experiment revealed that all the miRNAs are differentially expressed, being miR-429 significantly upregulated and miR-190 and miR-584 significantly downregulated in human HER2+ BC samples (Figure 4A-C). In particular, considering the expression level of each miRNA in healthy tissue, miR-190 and miR-584 are downregulated to 0.84±0.57 and 0.49±0.40 respectively, while miR-429 is upregulated to 11.23±12.95.
In vitro downregulation of miR-429 blocked the proliferation and decreased migration and invasion of HER2+ BC cells
As miR-429 role in HER2+ BC is unknown and as it is the only miRNA upregulated in HER2+ BC, we analysed the effect of miR-429 silencing in HER2+ BC cell line. No correlation was observed between the different stages of BC and the miR-429 expression level (Figure S3). Several of the target genes of miR-429 are involved in the control of proliferation (i.e. 'HIF1 signaling'). We thus analyzed the effect of miRNA reduction on proliferation, by MTT assay. The significant downregulation of miR-429, obtained by 200nM As oligonucleotide treatment in SKBR3 cells for 72 h (Figure S4A), caused a delay in the proliferation rate (Figure 5A), that was statistically significant compared to scramble-treated cells (t test p value <0.05 *, <0.01 **). Similar results were obtained on another HER2+ BC cell line (MDA-MB-453, data in Figure S4C).
Computational approach results Frequency of pairwise pathways in the top 10 for all 50 bootstraps (A). Pathways achieving the best performance in all 50 bootstraps (B) Boxplot representation of the AUC values in the training and testing dataset (C).
Pathway cross-talk networks in HER2 BC and their regulatory miRNAs. In red are indicated miRNAs and their connections, in blue are the functional pathways and their connections with the corresponding p-values.
miR-429 is overexpressed, while miR-190 and miR-584 are downregulated in HER2+ human BC. RT-PCR analysis of the fold change expression levels of miR-190 (A), miR-429 (B) and miR-584 (C) in HER2+ BC human samples compared to their corresponding healthy mammary tissue. The smaller panels in plot 4A and 4C represent an enlargement of the starting plot. The average value is indicated in red (n=10 samples, T test, p value<0.05, *).
miR-429 controls HER2+ BC cell proliferation , migration and invasion. MTT assay was performed on SKBR-3 cell at 0, 24, 48 h after 200nM Scramble or As miR-429 oligonucleotide treatment. The results are the average of three independent experiments in triplicate (t test compared to scramble treated cells, p value <0.05, *; <0.01, **)(A). 200nM 72h treatment with As miR-429 reduced the expression of CDK4 significantly (t test n=2 experiments in triplicate, p value <0.01**)(B). Wound healing test was performed on SKBR-3 treated with 200nM scramble oligonucleotide or As miR-429 in culture for 9 days. In each sample, the wound area was quantified by Image J following [31](t test compared to scramble-treated cells; n=3 experiments in triplicate, p value <0.01, **) (C). Boyden's chamber test was performed on SKBR-3 in the presence of 150nM scramble or As miR-429 in culture for 24h. The count of the cells was performed on 10 images from each sample (t test compared to scramble-treated cells; n=3 experiments, p value<0.01, **) (D).
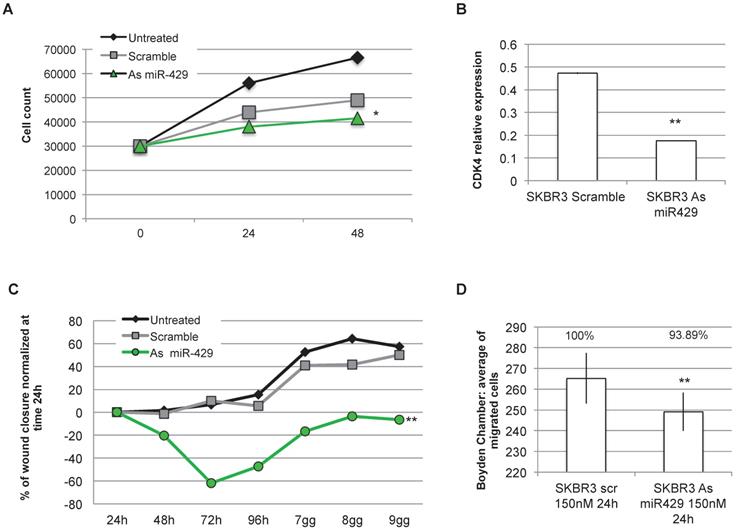
The observed reduction of proliferation rate observed in SKBR3 cells treated with As miR-429 could be due to a reduction of CDK4 expression, thus possibly causing a block in G1 to S phase transition (Figure 5B).
Considering that miR-429 is predicted by in silico analysis to regulate also pathways involved in cell migration and invasion (i.e CXCR4 pathway), we focused on the effects of As miR-429 modulation in wound healing and Boyden's chamber tests. In the first assay, we quantify the area of the wound at each time point in SKBR3 cells treated with Scramble S or As miR-429 oligonucleotides and normalized on that of the 24h (as suggested in [32]) (Figure S4B), calculating the percentage of wound closure at each time point. The delay of the As miR-429 treated SKBR3 cells in wound closure was significantly different from the scramble-treated cells (Figure 5C).
In the second assay, we observed that the treatment of 150nM As miR-429 reduced significantly the migration ability of HER2+ SKBR3 cells compared to the scramble-treated cells (Figure 5D).
In vivo downregulation of miR-429 blocked tumor proliferation
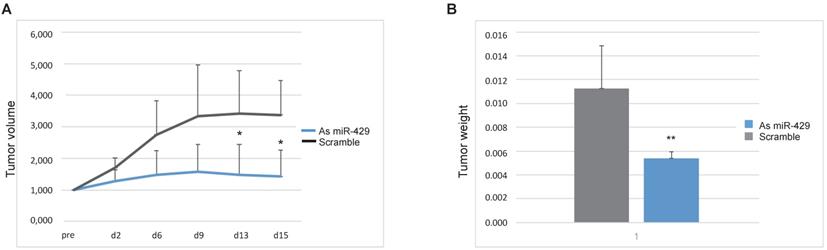
To propose miR-429 as a target for therapeutic tool development for HER2+ BC, we tested the efficacy of As miR-429 treatment on the in vivo growth of HER2+ BC tumors. In particular, we treated the tumors formed by 5 million SKBR3 xenografted in ten NOD/SCID mouse mammary glands with different condition (Figure S5A). The animals treated with atelogene vehicle (n=1), or scramble oligonucleotide and atelogene (n=4) developed rapidly growing tumors. In those treated with As miR-429 and atelogene (n=5) the growth of the tumor was delayed, with a significant difference at day 13 and 15 after therapeutic treatment, measured by tumor volume (Figure 6A), and by tumor weight at day 15 (Figure 6B), when the tumors were explanted and the mice were sacrificed. Representative pictures of treated tumors are presented in Figure S5B. In these tumors, we observed that the reduction of miR-429, obtained by As treatment, induced VHL increase and a parallel decrease of HIF1α targets, such as SLUG, VEGF and SNAIL (Figure S7).
As miR-429 counteracts the growth of HER2+ BC tumors xenografted in mouse. Tumors formed by SKBR3 in NOD/SCID mouse were treated with As miR-429 or scramble S oligonucleotide in the presence of atelogene reagent (vehicle). The volume of each tumor (height x length x depth) was measured at different time points and the curves represent the average volume in 15 days of measurements (A). The weights of the five Scramble S- or As miR-429-treated tumors are represented (t test p value<0.01, **, n=5) (B).
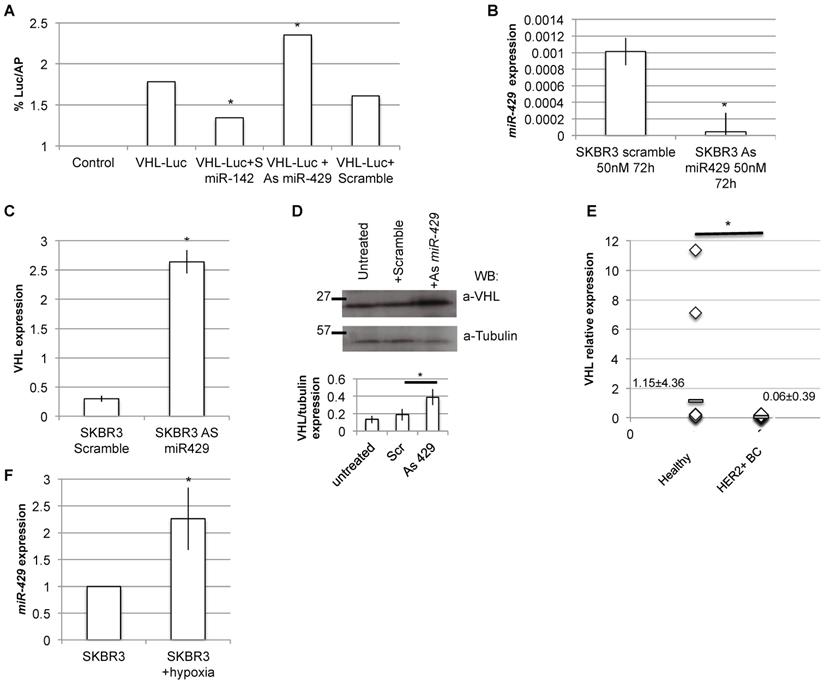
VHL is the direct target of miR-429. Luciferase assay: HER2+ cells were transfected with the Luciferase+ 3'UTR VHL (pGL3-VHL) construct alone, with 100nM 48h S Scramble or S miR-142 or As miR-429. The luciferase expression (LUC) was normalized on alkaline phosphatase content (AP). The chart represents the percentage of LUC/AP ratio (T test, p value<0.05, *; n=2 independent experiment)(A). RT-PCR analysis revealed that miR-429 modulation, obtained with 50nM As miR-429 for 72h (B), caused a significant increase of VHL expression, both at mRNA (C) and protein levels (D) (T test, p value<0.05, *; n=3 independent experiment). RT-PCR analysis on human HER2+ BC samples revealed that miR-429 is downregulated, compared to healthy tissue (E).
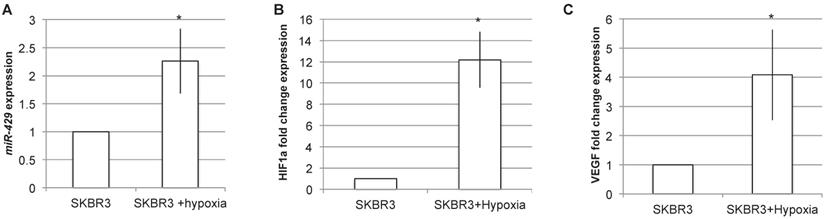
miR-429 is induced by hypoxia. RT-PCR analysis on SKBR3 revealed that 24h of hypoxia is able to induce miR-429 increased expression (A). The hypoxia induction has been verified by the RT-PCR analysis of HIF1α and its target VEGF mRNA induction (B).
VHL signaling pathway is target of miR-429
As VHL is predicted to be a putative miR-429 direct target (TargetScan and miRbase bioinformatics tools), we analyzed in detail the 3' untranslated regions (3'UTR) of human VHL target mRNA, and we found a possible seed for miR-429 (Figure S6). Thus, the dual luciferase reporter assay was performed to test if miR-429 is able to directly interact with 3'UTR of VHL. When we overexpressed the luciferase-3'UTR VHL construct in the presence of As miR-429, we observed a significant increase in luciferase activity compared to that observed in the cells co-trasfected with the same construct and the scramble oligonucleotide (Figure 7A). As a control, we performed the same transfection using the S miR-142, a known regulator of 3'UTR of human VHL [34].
Moreover, the in vitro downregulation of miR-429 in HER2+ SKBR3 (Figure 7B) cells also caused a parallel upregulation of VHL mRNA and protein expression (Figure 7C and D, respectively).
To confirm the possible relation among miR-429 and VHL target, we performed RT-PCR analysis of VHL expression on the human BC samples: as expected, VHL is downregulated in human HER2+ BC samples, compared to healthy surrounding tissue (Figure 7E).
miR-429 is induced by hypoxia
Being VHL a key component of hypoxia pathway, we analyzed the effect of hypoxia induction on miR-429 expression levels (Figure 8). The RT-PCR analysis of miR-429 expression in SKR3 after 24h of hypoxia treatment revealed that this condition is able to statistically increase miR-429 expression (Figure 8A). The efficacy of hypoxia induction was evaluated by analyzing HIF1α and its target VEGF mRNA expression, which are upregulated after hypoxia treatment (Figure 8B and C).
Discussion
Different miRNA signatures have been already proposed to have diagnostic or prognostic properties (i.e.[35, 36]). In this paper, we evaluated the diagnostic potential of three miRNAs, miR-190, miR-429 and miR-584, and the theranostic role of miR-429 in HER2+ BC. Although these miRNAs could be de-regulated in other BC subtypes, only in HER2+ they are altered being regulators of a specific pathway cross-talk network. Indeed, these miRNAs have been selected with an integrative bioinformatics approach relying on those miRNAs with a higher degree centrality in controlling a group of functional pathways containing differentially expressed genes, altered in HER2+ BC versus normal mammary tissues. RT-PCR analysis confirmed that miR-429, as well as miR-190 and miR-584, are differentially expressed in both HER2+ BC cell lines, and in human HER2+ BC tissues (miR-190 and miR-584 downregulated and miR-429 upregulated). Although miR-584 and miR-190 are already known tumor suppressor (i.e. [37, 38]), our results suggested a possible use of all these 3 miRNAs for diagnostic purpose.
We selected miR-429 as a possible therapeutic molecule, being the only upregulated miRNA among the three, and having a high degree centrality in controlling differentially expressed genes involved in "HIF1 signaling", "Acute Phase Response signaling", "Glioblastoma Multiforme signaling", "P2Y Purigenic Receptor signaling", "CXCR4 signaling " and “Growth hormone signaling”. In other tumors, as colorectal cancer, non-small cell lung cancer and triple negative BC, miR-429 has been proposed as a regulator of metastatic properties of the cells [39, 40]. Our in vitro experiments on HER+ cells suggested that miR-429 has a role in controlling cell proliferation (MTT assay), possibly via Cyclin-dependent kinase 4 (CDK4) as in colorectal cancer [41], and confirmed also that miR-429 is involved in migration and invasion (wound healing and Boyden's chamber tests, respectively). These results, obtained by As miR-429, could be due to its direct targeting of VHL gene (dual luciferase reporter assay). VHL codifies for a tumor suppressor protein involved in the degradation of hypoxia-inducible factor alpha (HIF1α) and the stromal-derived factor-1 receptor (CXCR4) proteins, which leads to the block of angiogenesis and tumor cell migration [42]. It is already known that VHL is underexpressed in highly aggressive and grade 2-grade 3 BC tumors, with a specific significant reduction of VHL expression in those tumors that form recurrence [43]. As VHL is a component of the HIF1α pathway, as hypoxia is one of the main functions affected in solid tumors (i.e. [44]), and having VHL increase an impact on triple negative BC cell growth [45], we speculated that the As miR-429 treatment, increasing mRNA of VHL, impacts on HIF1α protein degradation, leading to cell proliferation inhibition and to the block of metastatic potential of the HER2+ cells. Our in vitro experiments proved that As miR-429 caused a delay of HER2+ cell growth and block invasion and migration ability of the cells. Our in vivo experiments on HER2+ BC mouse models confirmed that the silencing of miR-429 impact on tumor cell growth and on tumor volume. These effects on cell proliferation and cell migration/invasion are possibly due to the decrease of HIF1α target mRNAs, such as SNAIL, VEGF and SLUG, (as shown in Figure S5) suggesting the use of As miR-429 as a new therapeutic option for HER2+ BC patients. Moreover, we showed that the hypoxia treatment, which is a common state for several HER2+ BC tumors, induces miR-429 expression; indeed, it has been demonstrated that overexpression of HER2 is able to induce HIF1 pathway components [46]. Further research is needed to understand the in vivo effects of As miR-429 on metastatic potential of HER2+ BC tumors. However, the use of As miR-429 could be considered as a novel tool to be combined with classical chemotherapeutic drugs, such as trastuzumab, for HER2+ BC treatment. Some limitations of the use of miRNA-based therapy need to be overcome, in particular those regarding the systemic toxicity and the tissue targeting of the miRNA [47]. Nevertheless, the combination of two drugs, one miRNA-based and one chemotherapeutic agent, could increase benefits to the patient by reducing the dose of chemotherapy and thus lowering its systemic toxicity.
In order to reduce the miRNA-mediated off-target effects and to better address the miRNA-based therapy to HER2+ BC tissues, we could take adantage of new emerging approach, i.e. those that use lipo-complexes conjugated with tumor targeting molecules, cationic polymers, conjugation of miRNA with lipids or receptor-binding molecules to increase cellular uptake, limiting the spreading of the drug in other organs [48].
In conclusion, from our results we proposed miR-190, miR-429 and miR-584 as diagnostic molecules for HER2+ BC, being miR-190 and miR-584 downregulated and miR-429 upregulated in HER2+ BC. The miR-429 increase could be induced by hypoxia, a common phenomenon for HER2+ BC. The silencing of miR-429, by regulating cell proliferation and invasion by targeting VHL mRNA, could be proposed a new anti-proliferative and anti-metastatic therapeutic strategy, alone or in combination with other drugs, for HER2+ BC patients' treatment.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
INTEROMICS flagship project (Interomics PB05), National Research Council CUP Grant B91JI2000190001, and the Italian Ministry of University and Research (MIUR) SYSBIONET-Italian ROADMAP ESFRI Infrastructures.
Funding
This work was supported by Italian Ministry of University and Research (MIUR) SYSBIONET-Italian ROADMAP ESFRI Infrastructures; Departments of Excellence 2018 -2022 Grant awarded by the Italian Ministry of Education, University and Research (MIUR) [L.232/2016]. INTEROMICS flagship project (Interomics PB05), National Research Council CUP Grant B91JI2000190001.
Author Contributions
C.C. and B.G. made substantial contributions to the conception and design of the work and to data analysis. N.C. and B.G. designed the experimental plan and performed the experiment. M.C., L.A. O.L. designed and performed the experiment in animal models. P.F. and C.F. characterized and selected the human samples for the experimental validation. C.C., N.C., M.C., L.A. O.L. P.F., T.M., C.F., B.G., C.I. revised the work critically and drafted the manuscript, B.G. and C.I. answering the reviewers' comments, and all giving final approval of the version to be published. All authors have read and approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61-70
2. Tandon AK, Clark GM, Chamness GC, Ullrich A, McGuire WL. HER-2/neu oncogene protein and prognosis in breast cancer. J Clin Oncol. 1989;7:1120-8
3. Goldhirsch A, Wood WC, Coates AS, Gelber RD, Thurlimann B, Senn HJ. et al. Strategies for subtypes-dealing with the diversity of breast cancer: highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann Oncol. 2011;22:1736-47
4. Ahmed AR. HER2 expression is a strong independent predictor of nodal metastasis in breast cancer. J Egypt Natl Canc Inst. 2016;28:219-27
5. Aversa C, Rossi V, Geuna E, Martinello R, Milani A, Redana S. et al. Metastatic breast cancer subtypes and central nervous system metastases. Breast. 2014;23:623-8
6. Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177-82
7. Wahler J, Suh N. Targeting HER2 Positive Breast Cancer with Chemopreventive Agents. Curr Pharmacol Rep. 2015;1:324-35
8. Mendes D, Alves C, Afonso N, Cardoso F, Passos-Coelho JL, Costa L. et al. The benefit of HER2-targeted therapies on overall survival of patients with metastatic HER2-positive breast cancer-a systematic review. Breast Cancer Res. 2015;17:140-54
9. Maxwell P, Salto-Tellez M. Validation of immunocytochemistry as a morphomolecular technique. Cancer Cytopathol. 2016;124:540-5
10. Elliott K, McQuaid S, Salto-Tellez M, Maxwell P. Immunohistochemistry should undergo robust validation equivalent to that of molecular diagnostics. J Clin Pathol. 2015;68:766-70
11. Varga Z, Diebold J, Dommann-Scherrer C, Frick H, Kaup D, Noske A. et al. How reliable is Ki-67 immunohistochemistry in grade 2 breast carcinomas? A QA study of the Swiss Working Group of Breast- and Gynecopathologists. PLoS One. 2012;7:e37379-91
12. Polley MY, Leung SC, McShane LM, Gao D, Hugh JC, Mastropasqua MG. et al. An international Ki67 reproducibility study. J Natl Cancer Inst. 2013;105:1897-906
13. Gancberg D, Di Leo A, Cardoso F, Rouas G, Pedrocchi M, Paesmans M. et al. Comparison of HER-2 status between primary breast cancer and corresponding distant metastatic sites. Ann Oncol. 2002;13:1036-43
14. Gong Y, Booser DJ, Sneige N. Comparison of HER-2 status determined by fluorescence in situ hybridization in primary and metastatic breast carcinoma. Cancer. 2005;103:1763-9
15. Zidan J, Dashkovsky I, Stayerman C, Basher W, Cozacov C, Hadary A. Comparison of HER-2 overexpression in primary breast cancer and metastatic sites and its effect on biological targeting therapy of metastatic disease. Br J Cancer. 2005;93:552-6
16. Cava C, Bertoli G, Colaprico A, Olsen C, Bontempi G, Castiglioni I. Integration of multiple networks and pathways identifies cancer driver genes in pan-cancer analysis. BMC Genomics. 2018;19:25-41
17. Cava C, Colaprico A, Bertoli G, Bontempi G, Mauri G, Castiglioni I. How interacting pathways are regulated by miRNAs in breast cancer subtypes. BMC Bioinformatics. 2016;17:183-206
18. Hadi NI, Jamal Q. "OMIC" tumor markers for breast cancer: A review. Pak J Med Sci. 2015;31:1256-62
19. Koo J, Zhang J, Chaterji S. Tiresias: Context-sensitive Approach to Decipher the Presence and Strength of MicroRNA Regulatory Interactions. Theranostics. 2018;8:277-91
20. Cava C, Colaprico A, Bertoli G, Graudenzi A, Silva TC, Olsen C. et al. SpidermiR: An R/Bioconductor Package for Integrative Analysis with miRNA Data. Int J Mol Sci. 2017;18:274-88
21. Colaprico A, Silva TC, Olsen C, Garofano L, Cava C, Garolini D. et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016;44:e71-82
22. Benjamini Ya, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. 1995;57:289-300
23. Colaprico A, Cava C, Bertoli G, Bontempi G, Castiglioni I. Integrative Analysis with Monte Carlo Cross-Validation Reveals miRNAs Regulating Pathways Cross-Talk in Aggressive Breast Cancer. Biomed Res Int. 2015;2015:831314-31
24. Cava C, Colaprico A, Bertoli G, Bontempi G, Mauri G, Castiglioni I. How interacting pathways are regulated by miRNAs in breast cancer subtypes. BMC Bioinformatics. 2016;17:183-206
25. Liaw A, Wiener M. Classification and Regression by randomForest. R News. 2002:2 /3: 18-22
26. Sales G, Romualdi C. parmigene-a parallel R package for mutual information estimation and gene network reconstruction. Bioinformatics. 2011;27:1876-7
27. Cava C, Bertoli G, Colaprico A, Bontempi G, Mauri G, Castiglioni I. In-Silico Integration Approach to Identify a Key miRNA Regulating a Gene Network in Aggressive Prostate Cancer. Int J Mol Sci. 2018;19:910-32
28. Holliday DL, Speirs V. Choosing the right cell line for breast cancer research. Breast Cancer Res. 2011;13:215-22
29. Bertoli G, Cava C, Diceglie C, Martelli C, Rizzo G, Piccotti F. et al. MicroRNA-567 dysregulation contributes to carcinogenesis of breast cancer, targeting tumor cell proliferation, and migration. Breast Cancer Res Treat. 2017;161:605-16
30. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402-8
31. Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68-73
32. Grada A, Otero-Vinas M, Prieto-Castrillo F, Obagi Z, Falanga V. Research Techniques Made Simple: Analysis of Collective Cell Migration Using the Wound Healing Assay. J Invest Dermatol. 2017;137:e11-e16
33. Wu D, Yotnda P. Induction and testing of hypoxia in cell culture. J Vis Exp. 2011 e2899: 1-4
34. Lu Y, Ji N, Wei W, Sun W, Gong X, Wang X. MiR-142 modulates human pancreatic cancer proliferation and invasion by targeting hypoxia-inducible factor 1 (HIF-1alpha) in the tumor microenvironments. Biol Open. 2017;6:252-9
35. Wang G, Gormley M, Qiao J, Zhao Q, Wang M, Di Sante G. et al. Cyclin D1-mediated microRNA expression signature predicts breast cancer outcome. Theranostics. 2018;8:2251-63
36. Bertoli G, Cava C, Castiglioni I. MicroRNAs: New Biomarkers for Diagnosis, Prognosis, Therapy Prediction and Therapeutic Tools for Breast Cancer. Theranostics. 2015;5:1122-43
37. Zhang Y, Wang Y, Wang J. MicroRNA-584 inhibits cell proliferation and invasion in non-small cell lung cancer by directly targeting MTDH. Exp Ther Med. 2018;15:2203-11
38. Yu Y, Luo W, Yang ZJ, Chi JR, Li YR, Ding Y. et al. miR-190 suppresses breast cancer metastasis by regulation of TGF-beta-induced epithelial-mesenchymal transition. Mol Cancer. 2018;17:70-82
39. Xiao P, Liu W, Zhou H. miR-429 promotes the proliferation of non-small cell lung cancer cells via targeting DLC-1. Oncol Lett. 2016;12:2163-8
40. Ye ZB, Ma G, Zhao YH, Xiao Y, Zhan Y, Jing C. et al. miR-429 inhibits migration and invasion of breast cancer cells in vitro. Int J Oncol. 2015;46:531-8
41. Han Y, Zhao Q, Zhou J, Shi R. miR-429 mediates tumor growth and metastasis in colorectal cancer. Am J Cancer Res. 2017;7:218-33
42. Haase VH. The VHL tumor suppressor: master regulator of HIF. Curr Pharm Des. 2009;15:3895-903
43. Zia MK, Rmali KA, Watkins G, Mansel RE, Jiang WG. The expression of the von Hippel-Lindau gene product and its impact on invasiveness of human breast cancer cells. Int J Mol Med. 2007;20:605-11
44. Mack FA, Rathmell WK, Arsham AM, Gnarra J, Keith B, Simon MC. Loss of pVHL is sufficient to cause HIF dysregulation in primary cells but does not promote tumor growth. Cancer Cell. 2003;3:75-88
45. Liu N, Xia WY, Liu SS, Chen HY, Sun L, Liu MY. et al. MicroRNA-101 targets von Hippel-Lindau tumor suppressor (VHL) to induce HIF1alpha mediated apoptosis and cell cycle arrest in normoxia condition. Sci Rep. 2016;6:20489-502
46. Jarman EJ, Ward C, Turnbull AK, Martinez-Perez C, Meehan J, Xintaropoulou C. et al. HER2 regulates HIF-2alpha and drives an increased hypoxic response in breast cancer. Breast Cancer Res. 2019;21:10-28
47. Rupaimoole R, Han HD, Lopez-Berestein G, Sood AK. MicroRNA therapeutics: principles, expectations, and challenges. Chin J Cancer. 2011;30:368-70
48. Baumann V, Winkler J. miRNA-based therapies: strategies and delivery platforms for oligonucleotide and non-oligonucleotide agents. Future Med Chem. 2014;6:1967-84
Author contact
![]() Corresponding author: Tel: +390221717542. Email: gloria.bertolicnr.it
Corresponding author: Tel: +390221717542. Email: gloria.bertolicnr.it
 Global reach, higher impact
Global reach, higher impact