13.3
Impact Factor
Theranostics 2019; 9(25):7556-7565. doi:10.7150/thno.38065 This issue Cite
Review
Current status and future trends of clinical diagnoses via image-based deep learning
1. Beijing Institute of Ophthalmology, Beijing Tongren Eye Center, Beijing Tongren Hospital, Capital Medical University, Beijing Ophthalmology and Visual Science Key Lab, Beijing, China
2. Royal Victorian Eye and Ear Hospital, Melbourne, Victoria 3002, Australia
3. Faculty of Medicine, Macau University of Science and Technology, Taipa, Macau
Received 2019-7-1; Accepted 2019-7-28; Published 2019-10-12
Abstract

With the recent developments in deep learning technologies, artificial intelligence (AI) has gradually been transformed from cutting-edge technology into practical applications. AI plays an important role in disease diagnosis and treatment, health management, drug research and development, and precision medicine. Interdisciplinary collaborations will be crucial to develop new AI algorithms for medical applications. In this paper, we review the basic workflow for building an AI model, identify publicly available databases of ocular fundus images, and summarize over 60 papers contributing to the field of AI development.
Keywords: artificial intelligence, deep learning, machine learning, ophthalmology
Introduction
Artificial intelligence (AI) has recently experienced an era of explosive growth across many industries, and healthcare is no exception [1]. AI will have particular utility in healthcare and will dramatically change the diagnostic and treatment pathways for many, if not most, diseases. Regardless of the specific technique, the general aim of these technologies in medicine is to use computer algorithms to uncover relevant information from data and to assist clinical decision making [2]. In many developed countries and China, the application of AI technology in healthcare has developed quickly, at least in part because it enhances human resources and abilities and improves the accuracy of medical treatment. As many countries that support the development of advanced technologies welcome the incoming era of AI, they will begin to develop the necessary specifications of governance by law, regulation, technology, and standards to fully optimize this developing field of technology.
Ophthalmology is a discipline that is highly dependent on technological development. Modern ophthalmology currently makes full use of mechanical, electrical, magnetic, acoustic, optical, and other imaging technologies, and it will lead in fully implementing and adapting new technological developments such as AI. Ophthalmologists should enthusiastically embrace the development of AI technology and use it to promote advances in ocular medicine as much as possible.
Workflow of deep learning
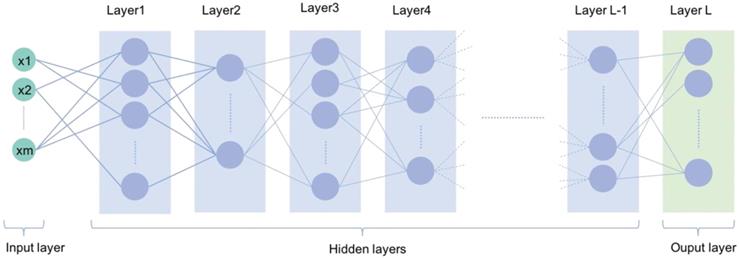
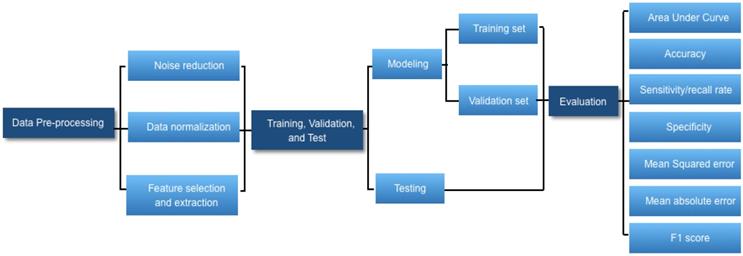
AI is broadly used in both the technical and popular lexicons to encompass a spectrum of learning, including but not limited to machine learning, representation learning, deep learning, and natural language processing [1]. Deep learning is making major advances in solving problems that have resisted the best attempts of the AI community for many years. It is very good at discovering intricate structures in high-dimensional data and is therefore applicable to multiple medical domains [3]. Deep learning discovers intricate structure in large data sets by using multiple intermediate layers positioned between the input and output layers, allowing each level to learn to transform its input signal into the following layer (Fig 1). The application of deep learning, particularly in images of the retina include classification, e.g., detection of diabetic retinopathy (DR) and diabetic macular edema (DME) in fundus photographs [4]; segmentation, e.g., segmentation of the lungs [5], brain [6], cell mitosis [7]; and prediction, e.g., prediction of myopia development and progression [8]. The workflow of deep learning can be defined in three stages: (1) pre-processing of the image data; (2) training of the model, validation, and model testing; and (3) evaluation (Fig. 2). Data pre-processing is a critical step that is necessary to build accurate machine learning models. The pre-processed work includes noise reduction, data normalization, feature selection, and extraction [9]. For training a model, we initially split the model into three sections: data training, validation, and testing. The training set enables the model to learn to fit the data parameters of the classifier. The validation set is used to prevent overfitting, and the test set is used to evaluate the performance of the trained model. Evaluation is an integral part of the development process. It helps to determine if the model will do a good job of predicting the target on new and future data.
Common open database of retina images
Many public databases have been published, and most include instructions for use by researchers in analysis and testing. For diseases of the retina, the databases include basic pathological features that usually provide information about the seven layers of the retina and about the choroid and sclera. This type of information is compiled by a process referred to as “segmentation”, which historically has been achieved by manual image processing, but increasingly it is done by computer algorithms. This information allows comparison of the performance of different algorithms analyzing the same fundus image, with reference to the reliable implementation of a gold-standard procedure [10]. Commonly used fundus databases includes DRIVE [11], STARE [12], Image-Ret [13,14], e-ophtha [15], HEI-MED [16], Retinopathy Online Challenge [17], Messidor [18], RIM-ONE [19], and DRION-DB [20]. Among them, DRIVE, STARE, Image-Ret, and Messidor are used mostly to diagnose DR, while DRION-DB and RIM-ONE are used mostly for segmentation of the optic nerve head in the diagnosis of glaucoma (Table 1).
A typical deep learning neural network with multiple deep layers between input and output layers
Workflow diagram of developing a deep learning-based medical diagnostic algorithm.
Summary of publicly available databases of ocular retinal images
| Database | Number of images | Camera Model | Image Resolution (pixels) | Field of View | Application |
|---|---|---|---|---|---|
| DRIVE [11] | 40 | Canon CR5 | 768×584 | 45° | Blood vessel segmentation |
| STARE [12] | 400 | Topcon trv-50 | 605×700 | 35° | Blood vessel segmentation; Optic disk detection |
| Image-Ret [13,14] | |||||
| DIARETDB0 | 130 | unknown | 1500×1152 | 50° | Diabetic retinopathy detection |
| DIARETDB1 | 89 | unknown | 1500×1152 | 50° | Diabetic retinopathy detection |
| e-ophtha [15] | |||||
| e-ophtha_EX | 82 | OPHDIAT Tele-medical network | 2048×1360; 1440×960 | - | Diabetic retinopathy detection |
| e-ophtha_MA | 381 | 1440×960; 2544×1696 | Diabetic retinopathy detection | ||
| HEI-MED [16] | 169 | Zeiss Visucam PRO fundus camera | 2196×1958 | 45° | Hard exudate detection; Diabetic macular edema assessment |
| Retinopathy Online Challenge [17] | 100 | Canon CR5-45-NM | 768×576; 1058×061; 1389×1383 | 45° | Microaneurysms detection |
| Messidor [18] | 1200 | TopCon TRC NW6 | 1440×960; 2240×1488; 2304×1536 | 45° | Diabetic retinopathy detection |
| RIM-ONE [19] | 169 | Nidek AFC-210 with a body of a Canon EOS 5D | 2144 × 1424 | - | Optic nerve head segmentation |
| DRIONS-DB [20] | 110 | Color analogue fundus camera digitized by HP-PhotoSmart-S20 scanner | 600×400 | - | Optic nerve head segmentation |
Important research studies applying artificial intelligence to ophthalmic conditions
Image classification is a long-term research topic in the field of computer vision and pattern recognition. Recent advances of deep learning techniques have greatly facilitated the research of image classification. Many deep learning models have demonstrated performances comparable with well-trained human experts in the classification of natural images, and some have outperformed the experts. The flourish of deep learning-based image classification started with the work of AlexNet [21], where an eight-layered convolutional neural network (CNN) was designed for the classification task in ImageNet Large Scale Visual Recognition Challenge (ILSVRC). Subsequently, a series of deeper neural networks continuously refreshed the record of ILSVRC, including GoogLeNet (22 layers) [22], VGGNet (16 or 19 layers) [23], and Deep Residual Net (18, 34, 50, 101, or 152 layers) [24]. These neural networks are the most widely used architectures that can achieve accurate classification for natural images by training deep models with millions of annotated images.
Diabetic retinopathy
Diabetic retinopathy is the most common organ complication and can manifest as the earliest sign of complication of diabetes mellitus. Early detection and continuous monitoring of DR is essential to control the disease in the early stage to prevent blindness. The automatic detection of DR has attracted a lot of attention. Most automated methods use fundus images as an input. These photographs are examined for the presence of lesions indicative of DR, including microaneurysms, hemorrhages, exudates, and cotton wool spots. The application of automated image analysis to fundus images may reduce the workload and costs by minimizing the number of photographs that need to be manually graded [25]. Gulshan et al [4] were the first to present a deep learning algorithm for the detection of DR in retinal fundus photographs. In 2 validation sets of 9963 images and 1748 images, at the operating point selected for high specificity, the algorithm had 90.3%and 87.0%sensitivity and 98.1%and 98.5%specificity for detecting referable diabetic retinopathy, defined as moderate or worse diabetic retinopathy or referable macular edema by the majority decision of a panel of at least 7 US board-certified ophthalmologists. Subsequently, Ting et al[26] developed a deep learning system to detect multiple related eye diseases, including DR, possible glaucoma, and age-related macular degeneration(AMD), the Area under the receiver operating characteristic curve (AUC) of 0.936 for referable DR ,sensitivity and specificity were 90.5% and 91.6%, For vision-threatening diabetic retinopathy, AUC was 0.958,sensitivity and specificity were 100% and 91.1%. More recently, deep learning was applied to automated segmentation of optical coherence tomography (OCT) images. Kermany et al [27] developed an OCT imaging diagnostic tool based on a deep learning framework for screening patients with AMD, DME, and drusen. The classifier distinguishing DME images from normal images achieved an accuracy of 98.2%, with a sensitivity of 96.8% and specificity of 99.6%. In April 2018, the first AI diagnostic system to receive US Food and Drug Administration (FDA) approval for marketing was IDx-DR, the case of IDx-DR highlights one of the earliest successes of an AI-based technology completing the regulatory process in the United States.
Glaucoma
Glaucoma is a group of eye diseases that damage the optic nerve and can result in irreversible vision loss and blindness and is the second leading cause of blindness worldwide. It is estimated that the disease affected 60.5 million people in 2010, and this figure is expected to reach 79.6 million by 2020 [28]. Currently, there is no cure for glaucoma, and vision loss, once it has occurred, is permanent. However, early detection and treatment are helpful to slow or stop the disease progression and can protect against serious vision loss. Many researchers have studied how to diagnose glaucoma automatically based on retinal images. These studies can be separated into two types. The first type outputs the glaucoma diagnosis results directly through deep learning models. Li et al [29] trained a CNN on LabelMe datasets for glaucoma diagnosis, In the validation dataset, this DL system achieved an AUC of 0.986 with sensitivity of 95.6% and specificity of 92.0%. The most common reasons for false-negative grading were glaucomatous optic neuropathy with coexisting eye conditions, including pathologic or high myopia, DR, and AMD. The leading reason for false-positive results was having other eye conditions, mainly including physiologic cupping. The second type of studies uses deep learning models to segment the glaucoma related tissues such as optic disc and optic cup, and then calculates medical measures (e.g., cup-to-disc ratio) for diagnosis. Previous studies have used various special forms of perimetry to discriminate preperimetric glaucoma from healthy eyes[30]. Asaoka et al [31] applied a DL method to differentiate the visual fields of preperimetric open-angle glaucoma patients from the healthy eyes, the AUC was 0.926.
Age-Related Macular Degeneration
AMD is a leading cause of irreversible visual loss in the aging population; the meta-analysis conducted by Wong et al [32] suggested that AMD, was responsible for 8.7% of all global blindness. Fortunately, the anti-vascular endothelial growth factor (anti-VEGF) medications have revolutionized the treatment of exudative retinal diseases, OCT is critical to guiding the administration of anti-VEGF therapy by providing a clear cross-sectional representation of the retinal pathology in these conditions. Kermany et al [27] developed an OCT imaging diagnostic tool based on a deep learning framework for screening patients with AMD, DME, and drusen. This AI system categorized images with choroidal neovascularization and images with diabetic macular edema as “urgent referrals”, drusen as “routine referrals”, normal images were labeled for “observation”. They achieved an accuracy of 96.6%, with a sensitivity of 97.8%, a specificity of 97.4%, and a weighted error of 6.6%. The classifier distinguishing choroidal neovascularization images from normal images achieved an accuracy of 100.0%, with a sensitivity of 100.0% and specificity of 100.0%. Recently, DeepMind and the Moorfields Eye Hospital [33] developed an AI system was trained on 14 884 OCT scans to detect 9 different OCT pathologies (choroidal neovascularization, macular edema, drusen, geographic atrophy, epiretinal membrane, vitreomacular traction, full-thickness macular hole, partial thickness macular hole, and central serous retinopathy). The system was then able to recommend a referral decision based on the most urgent conditions detected, the correct referral decision with 94% accuracy, matching world-leading eye experts.
In addition to detecting and monitoring common blinding eye diseases, deep learning is also being expanded to the field of rare diseases, such as congenital cataracts and retinopathy of prematurity (ROP) in newborns. Long et al [34] constructed a CNN-based computer-aided diagnosis framework (CC-Cruiser) to classify and grade congenital cataract. In the clinical trial, CC-Cruiser achieved 98.25% accuracy with the identification networks; 100%, 92.86% and 100% accuracy for opacity areas, densities and locations, respectively, with the evaluation networks; and 92.86% accuracy with the strategist networks. Brown et al [35] reported the results of a fully automated DL system that could diagnose plus disease, the most important feature of severe ROP, for diagnosis of plus disease, the algorithm achieved a sensitivity of 93% with 94% specificity. For detection of pre-plus disease or worse, the sensitivity and specificity were 100% and 94%, respectively. In addition, retinal microvascular changes and retinopathy provide important clinical indicators for predicting the occurrence, development, therapeutic effect and prognosis of cardiovascular and cerebrovascular diseases. Poplin et al[36] using deep-learning models trained on data from 284,335 patients and validated on two independent datasets of 12,026 and 999 patients, they predicted cardiovascular risk factors not previously thought to be present or quantifiable in retinal images, such as age (mean absolute error within 3.26 years), gender(AUC) = 0.97, smoking status (AUC = 0.71), systolic blood pressure (mean absolute error within 11.23 mmHg) and major adverse cardiac events (AUC = 0.70).Current AI studies using deep learning techniques for DR, AMD, glaucoma, cataract, and anterior ocular segment diseases are summarized in Table 2 [4,26,27,29,31,34,37-63].
Summary of influential papers on ophthalmic image analysis
| Disease | Procedures / examinations | Data sets | Deep learning techniques | Performance | Reference | |
|---|---|---|---|---|---|---|
| Keratoconus | Pentacam | 194 normal, 454 keratoconus, 67 forme fruste, 28 astigmatic, 117 after refractive surgery | SVM | Acc: 98.9%; Sen: 99.1%; Spe: 98.5%; AUC:0.99 | Hidalgo et al. [37] | |
| Dual Scheimpflug Analyzer | 177 normal, 148 keratoconus | Decision Tree | Sen: 100%; Spe: 99.5% | Smadja et al. [38] | ||
| Pentacam HR | 30 normal, 15 unilateral keratoconus, 30 bilateral keratoconus | FNN | Bilateral keratoconus versus normal; AUC: 0.99; Sen: 100%; Spe: 95% | Kovacs et al. [39] | ||
| Pterygium | Anterior segment photographed images | 2,692 non-pterygium, 325 pterygium | Shape features + SVM/ANN | Acc: 91.27%; AUC: 0.956; Sen: 88.7%; Spe: 88.3% | Zaki et al. [40] | |
| Cataract | Slit-lamp image | 5,378 images with decimal grading scores ranging from 0.1 to 5.0. | CRNN | 70.7% exact integral agreement ratio (R0); 88.4% decimal grading error ≤ 0.5 (Re0.5); 99.0% decimal grading error ≤ 1.0 (Re1.0 ). | Gao et al. [41] | |
| Slit-lamp image | 476 normal, 410 cataract | DCNN | Cataract vs Normal: Acc:98.87%;Sen:98.78% Spe:98.95% | Long et al. [34] | ||
| Fundus image | 767 normal, 472 cataract (246 mild cataract,128 moderate cataract, and 98 severe cataract) | SVM, BPNN | Acc: 93.2% for detection,84.5% for grading; Sen:94.2% for detection,74.6-89.3% for grading; Spe:91.5% for detection,90.4-98.9% for grading | Yang et al. [42] | ||
| Slit-lamp image | 476 normal, 410 pediatric cataract | CNN, SVM | Acc, Sen, and Spe: classification (97.07%, 97.28%, and 96.83%,) three-degree grading area (89.02%, 86.63%, and 90.75%) density (92.68%, 91.05%, and 93.94%) location (89.28%, 82.70%, and 93.08%) | Liu et al. [43] | ||
| POAG | Fundus image | Training set:125,189; Validation set: 71,896 | DLS | AUC: 0.942; Sen: 96.4%; Spe: 87.2% | Ting et al. [26] | |
| Fundus image | Training set: 31,745; Validation set: 8,000 | DCNN | AUC: 0.98; Acc: 92.9%; Sen: 95.6%; Spe: 92.0%; AUC: 0.986 | Li et al. [29] | ||
| Fundus image | 589 normal, 837 glaucoma | CNN | Acc: 98.13%; Sen: 98%; Spe: 98.3% | Raghavendra et al. [44] | ||
| Fundus image | 30 normal, 30 open-angle glaucoma | SVW | Acc:91.67; Sen:90%;Spe:93.33% | Krishnan et al. [45] | ||
| Visual field | Training set:257; Test set: 129 | ANN | AUC: 0.890; Sen: 78.3%; Spe: 89.5% | Oh et al. [46] | ||
| Fundus image | 266 normal, 72 mild, 86 moderate, 86 severe glaucoma | SVM | Acc: 93.1%; Sen: 89.75%; Spe: 96.2% | Acharya et al. [47] | ||
| Fundus image and SLO image | Normal/glaucoma Fundus images:85/39; Normal/glaucoma SLO images: 46/19 | RIFM | Acc for Fundus images: 94.4%,SLO images: 93.9%;Sen for Fundus images: 92.3%,SLO images: 89.5%;Spe for Fundus images: 95.3%,SLO images: 93.5% | Haleem et al. [48] | ||
| Visual fields | 53 glaucoma eyes, 108 normal eyes | FNN | AUC: 92.6%, The sensitivity was 77.8%,54.6%, and 50.0%, respectively, at the specificity of 90%, 95%,and 99% | Asaoka et al. [31] | ||
| DR | Fundus image | Training set:76,370; Validation set: 112,648 | DLS | For referable DR: AUC: 0.936; Sen: 90.5; Spe: 91.6%; For vision-threatening DR: AUC: 0.958; Sen: 100%; Spe: 91.1% | Ting et al. [26] | |
| Fundus image | Development Data Set (EyePACS in the United States and 3 eye hospitals in India): 128,175 Validation Data Set (EyePACS-1: 9,963; Messidor-2: 1,748) | DCNN | AUC: 0.991 for EyePACS,0.990 for Messidor;Sen: 90.3% for EyePACS,87% for Messidor;Spe: 98.1% for EyePACS,98.5% for Messidor | Gulshan et al. [4] | ||
| Fundus image | 170 DR, 170 normal | PNN-GA, SVM quadratic kernels | PNN-GA: Acc:99.41%,Sen:99.41% Spe:99.41%; SVM: Acc:99.12 % Sen:98.82%,Spe:99.41% | Ganesan et al. [49] | ||
| Fundus image | EyePACS: 75,137 DR images; External validation: MESSIDOR 2 and E-Ophtha | DCNN | AUC 0.94 for Messidor 2, 0.95 for E-Ophtha;Sen 93% for Messidor 2,87% for E-Ophtha;Spe 90% for Messidor 2,94% for E-Ophtha | Gargeya et al. [50] | ||
| Fundus image | Training set: 327 diabetic patients; Validation set: 725 diabetic patients | LASSO | Acc: 89.2%; AUC: 0.90; Sen: 75%; Spe: 89.6% | Oh et al. [51] | ||
| Fundus image | Training set: 400; Testing set: 9,954 | Ensemble of classifiers with hidden Markov chain for context information, trained by genetic algorithm | Sen: 92.2%; Spe: 90.4% | Tang et al. [52] | ||
| Fundus image | Messidor-2 dataset: 1,748 | CNN | Referable DR: AUC: 0.980; Sen: 96.8%; Spe: 87%; Vision threatening DR: AUC: 0.989; Sen: 100%; Spe: 90.8% | Abramoff et al. [53] | ||
| Fundus image | 4,445 DR; 5,494 normal | DCNN | Acc: 0.81 | Takahashi et al. [54] | ||
| Fundus image | DIARETDB1, FAZ, MESSIDOR, Private dataset: 750 (Normal: 150, mild NPDR: 150, moderate NPDR: 150, severe NPDR: 150, PDR: 150) | DNN | AUC: 0.924; Sen: 92.18%; Spe: 94.50% | Abbas et al. [55] | ||
| DME | SD-OCT | Training set: 11,349; DEM; 51,140 normal; Validation set: 250 DME, 250 normal | CNN | Acc: 98.2%; Sen: 96.8%; Spe: 99.6% | Kermany et al. [27] | |
| DME | Fundus image | 283 DR; 1,086 normal | Ensemble of Gaussian mixture model and SVM with RBF kernel | Acc: 96.8%; Sen: 97.3%; Spe: 95.9% | Akram et al. [56] | |
| AMD | Fundus image | Training set 72,610; Validation set: 35,948 | DLS | AUC: 0.931; Sen: 93.2%; Spe: 88.7% | Ting et al. [26] | |
| Fundus image | AREDS dataset: >130,000 | DCNN | AUC: 0.94∼0.96 Acc: 88.4%∼91.6% Sen: 71%∼88.4% Spe: 91.4%∼94.1% | Burlina et al. [57] | ||
| Fundus image | AREDS dataset: 5,664 | DCNN | Acc 79.4% (4-class) 81.5% (3-class); 93.4% (2-class) | Burlina et al. [58] | ||
| SD-OCT | Training and validation sets: 1,012 (AMD: 701; normal: 311); Test:100 (AMD: 50, normal: 50) | DCNN | Acc: 96%; Sen: 100%; Spe: 92% | Treder et al. [59] | ||
| Fundus image | 135 AMD subjects, 135 normal subjects | Feature extracted by Discrete wavelet transform and others for feature selection and classification | Average Acc: 93.7%; Sen: 91.11%; Spe: 96.3% | Mookiah et al. [60] | ||
| OCT | 48,312 AMD; 52,690 normal | DCNN | AUC: 0.975; Sen: 92.6%; Spe: 93.7% | Lee et al. [61] | ||
| SD-OCT | 1,289 | CNN | The mean Dice coefficient for human interrater reliability and deep learning were 0.750 and 0.729, respectively. | Lee et al. [62] | ||
| CNV | SD-OCT | Training set: 37,206 CNV, 51,140 normal; Validation set: 250 CNV, 250 normal | CNN | Acc: 100%; Sen:100%; Spe:100% | Kermany et al. [27] | |
| CNV | Fluorescein angiography | 33 | AdaBoost | Accuracy: 83.26% | Tsai et al. [63] |
RBFNN, radial basis function neural network; SVM, support vector machine; MLP, multi-layer perceptron; CRNN, convolutional-recursive neural networks; DCNN, Deep-learning convolutional neural network; BPNN, Back propagation neural network; DLS, deep learning system; CNN-FE, convolutional neural networks feature-exaggerated; MLP-BP, Multilayer Perceptron with Back Propagation; RIFM, Regional Image Features Model; FNN, feed-forward neural network; PNN-GA, probabilistic neural network-genetic algorithms; LASSO, least absolute shrinkage and selection operator; NB, naive Bayes; PNN, probabilistic neural network; RBF, Radial basis function; SD-OCT, spectral domain optical coherence tomography; SLO, Scanning Laser Ophthalmoscopy. Acc, accuracy; Sen, sensitivity; Spe, specificity; Vs, versus; AUC, area under the curve; POAG, primary open-angle glaucoma; AMD, age-related macular degeneration; OCT, optical coherence tomography; DR, diabetic retinopathy.
Status of AI applications in clinical diagnoses
On April 2, 2019, the FDA issued a discussion paper that proposed a regulatory framework for modifications to artificial intelligence/machine learning (AI/ML)-based software as medical devices (SaMD) [64]. This document explains the principles for developing artificial intelligence software, the new framework for equipment, the principles of the total product lifecycle (TPLC) certification methodology, and examples of potential real-world AI software modifications that may or may not be allowed under the proposed framework. The idea of the proposal was that with appropriately tailored regulatory oversight, AI/ML-based SaMD will deliver safe and effective software functionality that improves the quality of care that patients receive.
To date, two AI algorithms have been fully approved by the FDA and used clinically. One is IDx-DR for detecting DR, and the other is Viz.AI for analyzing images for indicators associated with a stroke. These two devices are described as “locked” algorithms, meaning that they can only be modified by the manufacturer at intervals for the purpose of “training” with new data, followed by manual verification and validation of the updated algorithm. However, there is much promise beyond locked algorithms that is ripe for health care applications. These opportunities require careful oversight to ensure that the benefits of these advanced technologies outweigh the risks to patients. These machine learning algorithms can continually evolve and are often referred to as “adaptive” or “continuously learning” algorithms. Adaptive algorithms can learn from new user data presented to the algorithm through real-world use. The FDA is exploring a framework that would allow for modifications to algorithms to be made from real-world learning and adaptation, while ensuring that the safety and effectiveness of the software as a medical device is maintained [65].
Main challenges in the application of AI
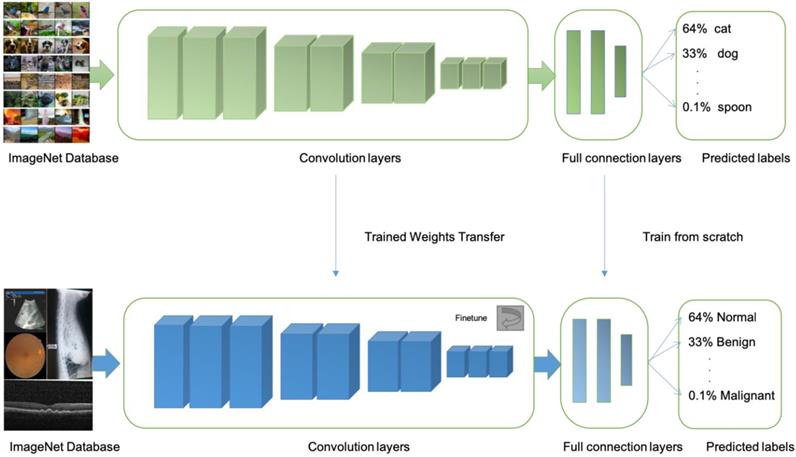
At present, there are at least four limitations in AI technology that is based on machine learning, First, most machine learning methods have too few training sets and verification sets. More image data training is needed to further improve accuracy, sensitivity, and specificity. Transfer learning is an approach is more suitable when limited training data is available for the problem under consideration. In transfer learning, one can learn a complex model using data from a source domain where large-scale annotated images are available (e.g. natural images). Then, the model is further fine-tuned with data of the target domain where only a small number of annotated images are available (e.g. medical images) [66] (Fig 3).
The second limitation is that the examination/detection equipment used in different countries, regions, and medical institutions is not uniform; therefore, the acquired images have differences in quality and resolution, which will inevitably affect the accuracy of image analysis and thus affect the accuracy of the diagnosis. These differences will present certain obstacles in the wide-scale applications of AI technology. One solution to this problem is to unify and standardize the examination equipment. This will be difficult to achieve. Another method is to further improve AI machine learning methods at the framework and algorithm level so that they can be flexibly applied to images of different qualities while simultaneously ensuring the accuracy of intelligent diagnosis. This will increase the applicability of AI in different regions and medical institutions. However, a lot of research support is still needed in this area.
Illustrations of transfer learning: a neural network is pretrained on ImageNet and subsequently trained on retinal, OCT, X-ray images, B-scans for different disease classifications
The third limitation is that the current machine learning methods for disease diagnosis lack “explanation capacity”. They do not have the ability to provide the clinician or other users with the reason for the diagnosis. The output result is based only on training and intensive learning. Thus, it is only a simple statement based upon the differences in the patient and normal images. There is no explanation for why the differences exist or the pathological basis of the differences. This, along with other issues, will affect to some extent the acceptance of these devices by doctors in clinical applications and could even provoke confusion among clinicians.
Finally, the fourth limitation in machine learning-based AI technology is that the diagnosis of some rare diseases is still unreliable. Because of the scarcity of these diseases, there are not enough cases to meet the requirements of the training and verification sets. It is difficult or impossible to ensure the accuracy of the learning model in diagnosing rare diseases. To improve the diagnosis of rare diseases, it will be necessary to optimize machine learning algorithms. This can be done by transitioning from reliance on the number of learning samples for accuracy training to utilizing combinations of various training modes and types.
Suggestions for the application and development of AI in medicine
1. Consolidate the data foundation of AI applications
AI must first collect a certain amount of data in the medical industry. The structure of current medical data is very complex, in part, because there is no uniformity in the standards for compilation, and this leads to widespread information islands. It is necessary to establish a mechanism for circulating and sharing medical data. Development of data desensitization methods will promote data standardization and normalization. These methods can be used to establish standard test data sets to consolidate the data foundations of AI applications.
2. Define the positioning of AI development
In medicine, AI aims to help doctors (rather than substitute for doctors) to reduce the morbidity and mortality rates of patients waiting for professionals. Because doctors will not be replaced by AI, the diagnostic result of AI is only a reference for a clinical diagnosis, and doctors will always be responsible for the result. Currently, AI products in medicine play only an auxiliary role in the clinic, such as the diagnosis of DR, cancer screening, medical image recognition, disease rehabilitation, and in other fields.
3. Formulate regulations and laws for the application of AI in medicine
To achieve the uniform standards necessary for effective medical AI applications, laws, regulations, and other levels of governance must be established at the national level. The implementation of the national standards in industry and in routine and research clinical settings will ensure that the technology can be made widely and quickly available in the safest and most rational way. This will prudently promote the application of AI in the medical field.
4. Strengthen data security of AI data applications
As with the collection of any personal and medical data, the risk of inadvertent or pirated disclosure is a major concern. To reduce these risks, it is necessary to strengthen the construction of privacy protection, desensitize the data, and collect the data according to different levels and different granularities to reduce the risk of privacy disclosure. The United States has extensive experience in privacy protection of medical data. While enhancing privacy protection, it encourages the rational access and meaningful use of data and makes a selected portion of it available for research in real time, open access databases.
5. Promote the cultivation of interdisciplinary talents
Future development of AI in medicine can be enhanced by focusing on the integration of disciplines such as medicine, information science, and engineering. This goal can be achieved by encouraging universities, research institutes, and enterprises to cooperate with each other, set up appropriate scholarships, and establish training bases and local pilot programs. Finally, an innovative talent introduction system and mechanism should be developed to attract highly talented students to carry out innovation and entrepreneurship in the field of medical AI, thus driving the further development of the field.
Future of AI application in clinic
Machine learning has shown its great potential in ophthalmology. Most of the current studies regarding intelligent diagnosis of eye diseases focus on dual classification problems, whereas many patients suffer from multiple categorical retinal diseases in the clinical setting. It is therefore necessary to have a model for detecting and distinguishing DR, AMD, glaucoma, and other retinal disorders simultaneously [67].
With a new generation of AI developed as a broad strategy, the applications of AI in the medical field will increase and improve. AI plays an important role in disease diagnosis and treatment, health management, drug research and development, precision medicine, etc. It can contribute significantly to solving problems of the uneven distribution of medical resources, reducing costs, and improving treatment efficiency. Applying AI helps to make up for the shortcomings of insufficient medical resources, enhance the fairness of medical services, and improve the construction of hierarchical diagnosis and treatment. In the future, AI will also offer important support for establishing an integrated medical service system. A qualified and efficient integrated medical service system can be built with the help of information-based systems.
Laws and regulations to define the legal status, responsibility sharing mechanisms, and supervision of automated systems are not yet enacted in China and most other countries. Given the complex ethical boundaries of medical AI application, the excessive control of medical AI will hinder innovation and development. On the other hand, the lack of management brings the risk of unclear subject responsibility in AI applications. Therefore, it is necessary to reasonably define AI in the medical field. The current laws on medical AI regulation are either non-existent or are in only the most primitive stages of development. There are no sound laws that regulate medical big data, the basis for medical AI. Further, there are no clear legal instructions regarding the ownership of AI data, the right to use it, privacy standards, data security, accountability norms, and whether laws can protect researchers, clinicians, and engineers from errors of innovation.
Summary
Deep learning has the ability to probe more deeply into and discern more discriminative features from extremely large datasets. It has been applied in many research and clinical fields that rely on medical image analysis, making breakthrough progress in those disciplines. Due to unique features in ophthalmology, the diagnosis of eye diseases in clinical practice requires interpretation of many imaging studies for auxiliary diagnosis. However, detection resolution of the human eye is limited and so is human attention span. Proficiency levels of ophthalmologists also differ and it's inevitable for human errors to occur. As documented in the existing literature review, most of the current deep learning methods representing the leading level are the use of supervised learning, especially the CNN-based framework. Preliminary researches mainly focused on pre-training CNN and taking CNN as feature extractor. These pre-training networks could be downloaded directly, and conveniently applied to the analysis of any medical images. In recent two years, end-to-end training CNN has become a prioritized approach for the analysis of medical images. However, obtaining well-annotated data used for supervised learning is another major challenge for the application of deep learning to the analysis of medical images. As annotated data were usually limited at present, how to utilize unannotated images to achieve a high diagnostic accuracy using a combination of unsupervised and supervised learning will be another important development direction. In addition, electronic medical records (EMRs) contain a wealth of clinical diagnostic and treatment information that can be extracted and used to form diagnoses using natural language processing and deep learning. This information can be used to supplement the image data to formulate a complete diagnosis mimicking a human physician [68]. Thus, in the near foreseeable future, AI relying on deep learning will combine image analysis with EMRs, further advancing the diagnostic power and ability to monitor disease progression and response to treatment in ways never before anticipated.
Acknowledgements
This study was funded in part by the: Beijing Hospitals Authority Youth Programme, code: QML20170206; The priming scientific research foundation for the junior researcher in Beijing Tongren Hospital, Capital Medical University (No. 2018-YJJ-ZZL-052).
Competing Interests
The authors have declared that no competing interest exists.
References
1. He J, Baxter SL, Xu J, Xu J, Zhou X, Zhang K. The practical implementation of artificial intelligence technologies in medicine. Nat Med. 2019;25:30-36
2. Murdoch TB, Detsky AS. The inevitable application of big data to health care. JAMA. 2013;309:1351-1352
3. LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436-444
4. Gulshan V, Peng L, Coram M, Stumpe MC, Wu D, Narayanaswamy A. et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA. 2016;316:2402-2410
5. Middleton I, Damper RI. Segmentation of magnetic resonance images using a combination of neural networks and active contour models. Med Eng Phys. 2004;26:71-86
6. Pereira S, Pinto A, Alves V, Silva CA. Brain tumor segmentation using convolutional neural networks in MRI images. IEEE Trans Med Imaging. 2016;35:1240-1251
7. Ciresan DC, Giusti A, Gambardella LM, Schmidhuber J. Mitosis detection in breast cancer histology images with deep neural networks. Med Image Comput Comput Assist Interv. 2013;16:411-418
8. Lin H, Long E, Ding X, Diao H, Chen Z, Liu R. et al. Prediction of myopia development among Chinese school-aged children using refraction data from electronic medical records: A retrospective, multicentre machine learning study. PLoS Med. 2018;15:e1002674
9. Caixinha M, Nunes S. Machine learning techniques in clinical vision sciences. Curr Eye Res. 2017;42:1-15
10. Jordan KC, Menolotto M, Bolster NM, Livingstone IAT, Giardini ME. A review of feature-based retinal image analysis. Exp Rev Ophthalmol. 2017;12:207-220
11. Niemeijer M, Staal JJ, Ginneken BV, Loog M. DRIVE: digital retinal images for vessel extraction. 2004. http://www.isi.uu.nl/Research/Databases/DRIVE
12. McCormick B, Goldbaum M. The STARE Project. 1975. http://cecas.clemson.edu/~ahoover/stare/
13. Kauppi T, Kalesnykiene V, Kamarainen J-K, Lensu L,Sorri I,Pietileta J,et al. DIARETDB0: Evaluation database and methodology for diabetic retinopathy algorithms. 2006. http://www.it.lut.fi/project/imageret/diaretdb0/
14. Kauppi T, Kalesnykiene V, Kamarainen J-K, Lensu L,Sorri I,Raninen A,et al. DIARETDB1 Standard Diabetic Retinopathy Database. 2007. http://www2.it.lut.fi/project/imageret/diaretdb1/
15. Decencière E, Cazuguel G, Zhang X, G Thibault, J-C.Klein, F Meyer. et al. TeleOphta: machine learning and image processing methods for teleophthalmology. IRBM. 2013;34:196-203
16. Niemeijer M, van Ginneken B, Cree MJ, Mizutani A, Quellec G, Sanchez C. et al. Retinopathy online challenge: automatic detection of microaneurysms in digital color fundus photographs. IEEE Trans Med Imaging. 2010;29:185-195
17. Giancardo L, Meriaudeau F, Karnowski T P, Li Y, Garg S, Tobin Jr K.W. et al. Exudate-based diabetic macular edema detection in fundus images using publicly available datasets. Medical Image Analysis. 2012;16:216-226
18. Messidor T-V. MESSIDOR: methods to evaluate segmentation and indexing techniques in the field of retinal ophthalmology. 2014. http://messidor.crihan.fr/index-en.php
19. Fumero F, Alayon S, Sanchez JL, Sigut J, Gonzalez-Hernandez M. RIM-ONE: An open retinal image database for optic nerve evaluation. Comp Med Sy. 2011:24 th International Symposium. 2011;1-6
20. Carmona EJ, Rincón M, García-Feijoó J, Martínez-de-la-Casa JM. Identification of the optic nerve head with genetic algorithms. Artif Intell Med. 2008;43:243-259
21. Krizhevsky A, Sutskever I, Hinton GE. Imagenet classification with deep convolutional neural networks.In Proceedings of the 25th International Conference on Neural Information Processing Systems - Volume 1 (NIPS'12), F. Pereira, C. J. C. Burges, L. Bottou, and K. Q. Weinberger (Eds.), Curran Associates Inc, USA,2012:1097-1105.
22. Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, Erhan D, Vanhoucke V, Rabinovich A. et al. Going deeper with convolutions. CVPR. 2015:1-9
23. Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556.
24. He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. CVPR. 2016:770-778
25. Scotland GS, McNamee P, Fleming AD, Goatman KA, Philip S, Prescott GJ. et al. Costs and consequences of automated algorithms versus manual grading for the detection of referable diabetic retinopathy. Br J Ophthalmol. 2010;94:712-719
26. Ting DSW, Cheung CY, Lim G, Tan GSW, Quang ND, Gan A. et al. Development and validation of a deep learning system for diabetic retinopathy and related eye diseases using retinal images from multiethnic populations with diabetes. JAMA. 2017;318:2211-2223
27. Kermany DS, Goldbaum M, Cai W, Valentim CCS, Liang H, Baxter SL. et al. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell. 2018;172:1122-1131
28. H.A. Quigley, A.T. Broman. The number of people with glaucoma worldwide in 2010 and 2020. Br J Ophthalmol. 2006;90:262-267
29. Li Z, He Y, Keel S, Meng W, Chang RT, He M. Efficacy of a deep learning system for detecting glaucomatous optic neuropathy based on color fundus photographs. Ophthalmology. 2018;125:1199-1206
30. Choi JA, Lee NY, Park CK. Interpretation of the Humphrey Matrix 24-2 test in the diagnosis of preperimetric glaucoma. Jpn J Ophthalmol. 2009;53:24-30
31. Asaoka R, Murata H, Iwase A, Araie M. Detecting preperimetric glaucoma with standard automated perimetry using a deep learning classifier. Ophthalmology. 2016;123:1974-1980
32. Wong WL, Su X, Li X, Cheung CM, Klein R, Cheng CY. et al. Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: a systematic review and meta-analysis. Lancet Glob Health. 2014;2:e106-116
33. De Fauw J, Ledsam JR, Romera-Paredes B, Nikolov S, Tomasev N, Blackwell S. et al. Clinically applicable deep learning for diagnosis and referral in retinal disease. Nat Med. 2018;24:1342-1350
34. Long E, Lin H, Liu Z, Wu X, Wang L, Jiang J. et al. An artificial intelligence platform for the multihospital collaborative management of congenital cataracts. Nat Biomed Eng. 2017:1 0024
35. Brown JM, Campbell JP, Beers A, Chang K, Ostmo S, Chan RVP. et al. Automated Diagnosis of Plus Disease in Retinopathy of Prematurity Using Deep Convolutional Neural Networks. JAMA Ophthalmol. 2018;136:803-810
36. Poplin R, Varadarajan AV, Blumer K, Liu Y, McConnell MV, Corrado GS. et al. Prediction of cardiovascular risk factors from retinal fundus photographs via deep learning. Nat Biomed Eng. 2018;2:158-164
37. Hidalgo R I, Rodriguez P, Rozema JJ, Dhubhghaill S, Zakaria N. et al. Evaluation of a machine-learning classifier for keratoconus detection based on Scheimpflug tomography. Cornea. 2016;35:827-832
38. Smadja D, Touboul D, Cohen A, Doveh E, Santhiago MR, Mello GR. et al. Detection of subclinical keratoconus using an automated decision tree classification. Am J Ophthalmol. 2013;156:237-246
39. Kovács I, Miháltz K, Kránitz K, Juhász E, Takács A, Dienes L. et al. Accuracy of machine learning classifiers using bilateral data from Scheimpflug camera for identifying eyes with preclinical signs of keratoconus. J Cataract Refract Surg. 2016;42:275-283
40. Zaki WMDW, Daud MM, Abdani SR, Hussain A, Mutalib HA. Automated pterygium detection method of anterior segment photographed images. Comp Methods Programs Biomed. 2018;154:71-78
41. Gao X, Lin S, Wong TY. Automatic Feature Learning to Grade Nuclear Cataracts Based on Deep Learning. IEEE Trans Biomed Eng. 2015;62:2693-2701
42. Yang JJ, Li J, Shen R, Zeng Y, He J, Bi J. et al. Exploiting ensemble learning for automatic cataract detection and grading. Comput Methods Programs Biomed. 2016;124:45-57
43. Liu X, Jiang J, Zhang K, Long E, Cui J, Zhu M. et al. Localization and diagnosis framework for pediatric cataracts based on slit-lamp images using deep features of a convolutional neural network. PLoS One. 2017;12:e0168606
44. Raghavendra U, Fujita H, Bhandary SV, Gudigar A, Tan JH, Acharya UR. Deep convolution neural network for accurate diagnosis of glaucoma using digital fundus images. Inf Sci. 2018;441:41-49
45. Krishnan MMR, Faust O. Automated glaucoma detection using hybrid feature extraction in retinal fundus images. J Mech Med Biol. 2013;13:1350011
46. Oh E, Yoo TK, Hong S. Artificial neural network approach for differentiating open-angle glaucoma from glaucoma suspect without a visual field test. Invest Ophthalmol Vis Sci. 2015;56:3957-3966
47. Acharya UR, Ng EYK, Eugene LWJ, Noronha KP, Min LC, Nayak KP. et al. Decision support system for the glaucoma using Gabor transformation. Biomed Signal Process Control. 2015;15:18-26
48. Haleem MS, Han L, Hemert Jv, Fleming A, Pasquale LR, Silva PS. et al. Regional image features model for automatic classification between normal and glaucoma in fundus and scanning laser ophthalmoscopy (SLO) images. J Med Syst. 2016;40:132
49. Ganesan K, Martis RJ, Acharya UR, Chua CK, Min LC, Ng EYK. et al. Computer-aided diabetic retinopathy detection using trace transforms on digital fundus images. Med Biol Eng Comput. 2014;52:663-672
50. Gargeya R, Leng T. Automated identification of diabetic retinopathy using deep learning. Ophthalmology. 2017;124:962-969
51. Oh E, Yoo TK, Park EC. Diabetic retinopathy risk prediction for fundus examination using sparse learning: a cross-sectional study. BMC Med Inf Decis Mak. 2013;13:106
52. Tang HL, Goh J, Peto T, Ling BW, Al Turk LI, Hu Y. et al. The reading of components of diabetic retinopathy: an evolutionary approach for filtering normal digital fundus imaging in screening and population based studies. PLoS One. 2013;8:e66730
53. Abramoff MD, Lou Y, Erginay A, Clarida W, Amelon R, Folk JC. et al. Improved automated detection of diabetic retinopathy on a publicly available dataset through integration of deep learning. Invest Opthalmol Vis Sci. 2016;57:5200-5206
54. Takahashi H, Tampo H, Arai Y, Inoue Y, Kawashima H. Applying artificial intelligence to disease staging: deep learning for improved staging of diabetic retinopathy. PLoS One. 2017;12:e179790
55. Abbas Q, Fondon I, Sarmiento A, Jimenez S, Alemany P. Automatic recognition of severity level for diagnosis of diabetic retinopathy using deep visual features. Med Biol Eng Comput. 2017;55:1959-1974
56. Akram MU, Tariq A, Khan SA, Javed MY. Automated detection of exudates and macula for grading of diabetic macular edema. Comput Methods Programs Biomed. 2014;114:141-152
57. Burlina PM, Joshi N, Pekala M, Pacheco KD, Freund DE, Bressler NM. Automated grading of age-related macular degeneration from color fundus images using deep convolutional neural networks. JAMA Ophthalmol. 2017;135:1170-1176
58. Burlina P, Pacheco KD, Joshi N, Freund DE, Bressler NM. Comparing humans and deep learning performance for grading AMD: a study in using universal deep features and transfer learning for automated AMD analysis. Comput Biol Med. 2017;82:80-86
59. Treder M, Lauermann JL, Eter N. Automated detection of exudative age-related macular degeneration in spectral domain optical coherence tomography using deep learning. Graefes Arch Clin Exp Ophthalmol. 2018;256:259-265
60. Mookiah MR, Acharya UR, Koh JE, Chua CK, Tan JH, Chandran V. et al. Decision support system for age-related macular degeneration using discrete wavelet transform. Med Biol Eng Comput. 2014;52:781-796
61. Lee CS, Baughman DM, Lee AY. Deep learning is effective for the classification of OCT images of normal versus age-related macular degeneration. Ophthalmol Retina. 2017;1:322-327
62. Lee CS, Tyring AJ, Deruyter NP, Wu Y, Rokem A, Lee AY. Deep-learning based, automated segmentation of macular edema in optical coherence tomography. Biomed Opt Express. 2017;8:3440-3448
63. Tsai CL, Yang YL, Chen SJ, Lin KS, Chan CH, Lin WY. Automatic characterization of classic choroidal neovascularization by using AdaBoost for supervised learning. Invest Ophthalmol Vis Sci. 2011;52:2767-2774
64. Statement from FDA Commissioner Scott Gottlieb, M.D. on steps toward a new, tailored review framework for artificial intelligence-based medical devices. 2019. https://www.fda.gov/news-events/press-announcements/statement-fda-commissioner-scott-gottlieb-md-steps-toward-new-tailored-review-framework-artificial?utm_campaign=2019-04-03+CDRH+New
65. U.S. Food & Drug Administration. Proposed regulatory framework for modifications to artificial intelligence/machine learning (AI/ML) based software as medical device (SaMD). 2019. https://www.fda.gov/medical-devices/software-medical-device-samd/artificial-intelligence-and-machine-learning-software-medical-device
66. Going Deep in Medical Image Analysis: Concepts, Methods, Challenges and Future Directions. 2019. https://arxiv.org/abs/1902.05655
67. Balyen L, Peto T. Promising Artificial Intelligence-Machine Learning-Deep Learning Algorithms in Ophthalmology. Asia Pac J Ophthalmol (Phila). 2019;8:264-272
68. Liang H, Tsui BY, Ni H, Valentim CCS, Baxter SL, Liu G. et al. Evaluation and accurate diagnoses of pediatric diseases using artificial intelligence. Nat Med. 2019;25:433-438
Author contact
![]() Corresponding author: Kang Zhang; e-mail: kang.zhangcom
Corresponding author: Kang Zhang; e-mail: kang.zhangcom
 Global reach, higher impact
Global reach, higher impact