13.3
Impact Factor
Theranostics 2019; 9(15):4342-4353. doi:10.7150/thno.34090 This issue Cite
Research Paper
Up-regulation of PCOLCE by TWIST1 promotes metastasis in Osteosarcoma
1. State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Department of Experimental Research, Sun Yat-sen University Cancer Center, Guangzhou, China
2. Endoscopic and Laser Department, Sun Yat-sen University Cancer Center, Guangzhou, China
3. Department of Pathology, Xiangya Hospital, School of Basic Medicine, Central South University, Changsha, Hunan 410078 China.
4. Department of Musculoskeletal Oncology, The First Affiliated Hospital of Sun Yat-Sen University, Guangzhou, China.
5. Cancer center, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan 430022, China.
#These authors contributed equally to this work.
Received 2019-2-13; Accepted 2019-5-26; Published 2019-6-9
Abstract
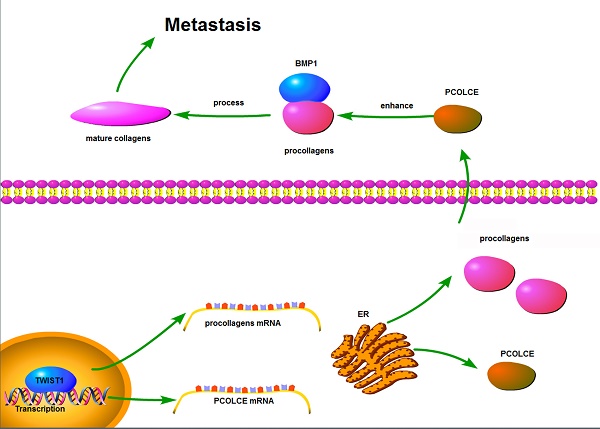
Procollagen C-proteinase enhancer protein (PCOLCE) was originally identified as an enhancer to facilitate the catalysis of procollagens by BMP1. PCOLCE participates in the reconstitution of extracellular and corneal repair. The elevation of PCOLCE in blood indicates that breast cancer has metastasized into the bones. However, direct research on PCOLCE has not been reported.
Methods: ECM candidates were identified by RNA-seq analysis from 4 normal and 16 osteosarcoma tissues. The in vitro migration and invasion abilities of osteosarcoma cells were determined by a Transwell assay. A spontaneous metastatic osteosarcoma model was established to assess osteosarcoma metastasis in vivo. The N-linked glycosylated amino acids were identified by PNGase F treatment combined with Western blotting. The mechanism of TWIST1 regulating PCOLCE transcription was elucidated by luciferase, qPCR and ChIP assays.
Results: PCOLCE was markedly up-regulated in human osteosarcoma tissues compared to its expression in noncancerous adjacent tissues; high PCOLCE expression in tissues correlated with a poor patient prognosis, and the knockdown of PCOLCE by shRNAs impaired the migration, invasion and lung metastasis of osteosarcoma cells. The overexpression of wild-type PCOLCE, but not its N29Q mutant, promoted migration, invasion and metastasis, indicating that the glycosylation of PCOLCE at Asn29 is necessary for its functions in osteosarcoma. TWIST1, a key transcription factor in metastasis, was also overexpressed in osteosarcoma tissues and positively correlated with either PCOLCE or its potential procollagen substrates, such as COL1A1, COL1A2, COL5A1, COL8A2 and COL10A1.
Conclusion: Our findings are the first to provide evidence that PCOLCE plays a critical role in promoting the lung metastasis of osteosarcoma, and this up-regulation of PCOLCE by TWIST1 may lead to a new therapeutic strategy to treat patients with metastatic osteosarcoma.
Keywords: PCOLCE, TWIST1, Osteosarcoma, Metastasis, N-glycosylation
Introduction
Osteosarcoma, the most common primary malignant bone tumor, is composed of malignant osteoblasts that produce immature bone or osteoid tissues [1]. It arises primarily in adolescence and childhood and has a second peak incidence in those over the age of 50 [2]. Despite the improvement of treatment strategies, such as surgery or chemotherapy, the overall 5-year survival is approximately 75% for nonmetastatic osteosarcoma but has remained at 20% for metastatic cases over the past 30 years [3, 4]. Thus, it is urgent to understand the molecular mechanism of osteosarcoma metastasis.
The extracellular matrix (ECM) is composed of numerous biochemically distinct proteins that participate in various biological processes, such as embryonic development, tissue homeostasis and cell differentiation [5, 6]. Abnormal ECM dynamics also play a critical role in cancer progression and metastasis. For instance, matrix metalloproteinases (MMPs), the main enzymes involved in ECM degradation, are highly expressed in cancer and promote cancer metastasis [7, 8]. Lysyl oxidase (LOX) crosslinks collagens and elastins in the extracellular matrix and promotes bone metastasis in hypoxic cancers by inducing premetastatic bone lesions [9]. Collagens, the most abundant components of the ECM, are excessively deposited in the extracellular matrix to form the tumor microenvironment and to modulate cancer cell polarity and migration [9]. The down-regulation of BMP-1 (bone morphogenetic protein-1), which processes fibrillar collagen types I-III, leads to the suppression of TGFβ in non-small cell lung cancer cells [10].
Procollagen C-proteinase enhancer protein (PCOLCE) is a secreted glycoprotein that enhances the activity of procollagen C-proteinases to participate in ECM reconstruction [11-13]. PCOLCE binds to the C-propeptide of procollagen III and heparin sulfate through its CUB and NTR domains, respectively, leading to the enhancement of BMP-1 activity and collagen precursor maturation [14, 15]. It has been reported that the dysregulation of PCOLCE is involved in numerous diseases. For instance, the expression of PCOLCE positively correlates with muscle and liver fibrosis [16]. A PCOLCE deficiency contributes to deficient corneal repair [17]. Mutated PABPN1 binds to PCOLCE and entraps it within the nuclear compartment, leading to oculopharyngeal muscular dystrophy [18]. However, the role of PCOLCE in cancer remains to be explored. In the present study, we found that PCOLCE was highly expressed in osteosarcoma and may play an important role in promoting the lung metastasis of osteosarcoma.
Materials and methods
RNA preparation, RNA-seq and analysis
The total RNA from osteosarcoma tissues was extracted using the TRIzol Reagent (Invitrogen). The cDNA was synthesized with mRNA enriched by Oligo(dT) beads. Libraries for next-generation sequencing were constructed by blunting the ends of the cDNA; this was followed by A-tailing, ligation to sequencing adapters, and PCR with indexed sequencing primers. RNA-seq was carried out using Illumina HiSeqTM 2500 by Gene Denovo Biotechnology Co. (Guangzhou, China), and 10 Gb of the RNA-seq data of every sample are available from the GEO database (GEO: GSE00000). The low-quality reads were trimmed by Trimmomatic (Version 0.32)[19], and the clean reads were aligned to the human genome (GRCh38) using Tophat2 (Version 2.0.13)[20]. The expression levels of the genes were quantified by Cuffquant in the Cufflinks (Version 2.2.1)[21] suite. All the processes were processed by Gene Denovo Co. (Guangzhou, China).
Clinical samples
Paracancerous tissues and tumor samples were obtained from recruited osteosarcoma patients. The patients were diagnosed according to their clinicopathologic characteristics at the Department of Musculoskeletal Oncology at The First Affiliated Hospital of Sun Yat-Sen University (Guangzhou, China). No patient had received radiotherapy and/or chemotherapy prior to surgery. The samples were immediately frozen in liquid nitrogen for RNA extraction and qRT-PCR. The clinical information of 20 patients, whose samples were collected for RNA-seq, are listed as Supplementary Table S1 (sheet 3), whereas the clinical information of 68 patients, whose samples were collected for qRT-PCR, are listed as Supplementary Table S2.
Cell culture and transfection
U2OS and HOS were obtained from American Type Culture Collection. U2OS/MTX300 is methotrexate-resistant and is derived from U2OS. All cell lines were cultured in 10% fetal bovine serum containing DMEM (Dulbecco's Modified Eagle Medium) at 37°C and 5% CO2. Stable cell lines were cultured in complete medium supplemented with 0.5 μg/mL puromycin. The transfection experiments were performed according to the manufacturer's instructions included with Lipofectamine 2000TM (Invitrogen).
His-tagged protein purification
Stable cell lines expressing PCOLCE-His or N29Q-His were lysed by cell lysis buffer (phosphate 20 mM, NaCl 0.5 M, imidazole 20 mM, Triton X-100 2%, pH 7.4) at 4°C for 1 h, and the lysate was centrifuged to remove the cell debris before it was transferred to HiTrap (GE, 17040501). After washing with 5 ml binding buffer (phosphate 20 mM, NaCl 0.5 M, and imidazole 20 mM, pH 7.4), the protein was eluted with 1 ml elution buffer (phosphate 20 mM, NaCl 0.5 M, and imidazole 500 mM, pH 7.4). The elution product was diluted by adding 14 ml of PBS before it was concentrated to a 0.5 ml product by 10 KD Amicon Ultra-4 (Millipore). All buffer used in this process was supplemented with a protease inhibitor cocktail (Sigma).
Construct and plasmids
PCOLCE and N29Q CDSs were amplified in a thermal cycler using PrimeSTAR (TaKaRa, Japan) and were then infused with psin-EF1, which is used for producing lentiviruses, using the Clone Express II One Step Cloning Kit (Vazyme Biotech). The primers used for cloning PCOLCE and the N29Q mutant are as follows:
psin-PCOLCE-F: 5'-ATT GGG ATC CCC GGA CGG CCA CCA TGC TGC CTG CAG CCA CA-3';
psin-PCOLCE-R: 5'-GCA TGC GGA TCA CTA GTG TCA GTC CTG GGA CGC AGC A-3';
N29QF: 5'-AGA CCC CCC AAT ACA CCA GAC CCG TGT T-3';
N29QR: 5'-TGT ATT GGG GGG TCT GGC CCT GGG CAA-3'.
The PCOLCE CDS was amplified from cDNA derived from U2OS, and the N29Q mutant was generated by an overlap-extension strategy.
pLKO.1-puro was inserted with double-stranded oligonucleotides, which generates shRNA in the cell line, and the sequences are as follows:
PCOLCE-sh#1-s: 5'-CCG GTG AAG AAA GGA GTC AGT TAT CCT CGA GGA TAA CTG ACT CCT TTC TTC ATT TTT-3';
PCOLCE-sh#1-as: 5'-AAT TAA AAA TGA AGA AAG GAG TCA GTT ATC CTC GAG GAT AAC TGA CTC CTT TCT TCA-3';
PCOLCE-sh#2-s: 5'-CCG GCG CTG ACC TTC GAG AAG TTT GCT CGA GCA AAC TTC TCG AAG GTC AGC GTT TTT-3';
PCOLCE-sh#2-as: 5'-AAT TAA AAA CGC TGA CCT TCG AGA AGT TTG CTC GAG CAA ACT TCT CGA AGG TCA GCG-3'.
RNA extraction and qRT-PCR
The total RNA was extracted using the RNAprep Pure Cell/Bacteria Kit (Tiangen). The total RNA (500 ng) was reverse-transcribed using a HiScript II 1st Strand cDNA Synthesis Kit (Vazyme). qRT-PCR was performed using the ChamQ Universal SYBR qPCR Master Mix (Vazyme) on LightCycler 480 (Roche). All qRT-PCR samples were repeated at least 3 times. The primer sequences are as follows:
qGAPDH-F: 5'-GGA GCG AGA TCC CTC CAA AAT-3';
qGAPDH-R: 5'-GGC TGT TGT CAT ACT TCT CAT GG-3';
qPCOLCE-F: 5'-GTG CGG AGG GGA TGT GAA G-3';
qPCOLCE-R: 5'-CGA AGA CTC GGA ATG AGA GGG-3';
qTWIST1-F: 5'-GTC CGC AGT CTT ACG AGG AG-3';
qTWIST1-R: 5'-GCT TGA GGG TCT GAA TCT TGC T-3';
qCOL1A1-F: 5'-GAG GGC CAA GAC GAA GAC ATC-3';
qCOL1A1-R: 5'-CAG ATC ACG TCA TCG CAC AAC-3';
qCOL1A2-F: 5'-GGC CCT CAA GGT TTC CAA GG-3';
qCOL1A2-R: 5'-CAC CCT GTG GTC CAA CAA CTC-3';
qRT-col5A1-F: 5'-TAC AAC GAG CAG GGT ATC CAG-3';
qRT-col5A1-R: 5'-ACT TGC CAT CTG ACA GGT TGA-3';
qRT-col8A2-F: 5'-GCT GGC TTA GGC AAA CCT G-3';
qRT-col8A2-R: 5'-GGA CTC CCA CAC CGT CTA CT-3';
qRT-col10A1-F: 5'-ATG CTG CCA CAA ATA CCC TTT-3';
qRT-col10A1-R: 5'-GGT AGT GGG CCT TTT ATG CCT-3'.
Dual-Luciferase assay
293T cells were plated in 24-well plates and were transiently transfected with 200 ng of PCOLCE promoter-containing pGL-3 plasmids together with pTK-cLuc as the normalization control. 48 h later, the luciferase activity was measured for 3 independent experiments using the Dual-Luciferase Assay Kit (Promega).
Assessment of cell migration and invasion
Cells were seeded into Boyden chambers containing 24-well Transwell plates with a pore size of 8 μm (BD Bioscience). The upper chamber was either left uncoated for the migration assay or precoated with 50 μl 1:8 diluted Matrigel (BD Bioscience) for the invasion assay. U2OS (5×104), U2OS/MTX300 (5×104) and HOS (3×104) cells were seeded into the upper chamber. The upper chamber was filled with 200 μL serum-free specified medium, whereas the lower chamber was filled with complete medium containing 10% FBS. Following incubation for 12 h (U2OS, HOS) and 24 h (U2OS/MTX300), the cells that had invaded into the lower chamber were fixed with 4% paraformaldehyde and were stained with crystal violet for 1 h at room temperature. The cells in the upper chamber were removed by a cotton swab, and the remaining cells were counted in 5 randomly selected microscopic fields. All experiments were performed in triplicate.
Chromatin immunoprecipitation assay
The chromatin immunoprecipitation assay was performed using a ChIP Kit according to the manufacturer's instructions [22]. Briefly, each cell line was seeded into a 15 cm plate. The cells were fixed by adding 10% of the volume of the growth medium. The fixation was stopped by adding 5% of the volume of the stop solution to the existing culture medium. The cells were scratched, collected and lysed to obtain the nuclear pellet. The nuclear pellet was resuspended and sonicated. Five micrograms of antibodies together with protein A/G agarose beads was incubated overnight at 4°C. The bound DNA-protein complexes were eluted after a series of washes. Purified DNA was resuspended in TE buffer for qRT-PCR.
The PCR primer sequences are as follows:
CHIP-PCOLCE-EBX1-F1: 5'-AAC AAG GAC ACT CCC TTC AT-3';
CHIP-PCOLCE-EBX2-R1: 5'-TAG AGG CAG GGT TTC AGC AT-3'.
Western Blotting
Western blotting was performed as previously described [22]. The cells were collected and lysed in RIPA buffer (150 mM NaCl, 0.5% EDTA, 50 mM Tris, 0.5% NP40) and centrifuged for 20 min at 14,000 x g and 4°C. Before being loaded onto the polyacrylamide gel, the protein lysate or concentrated. cell culture supernatant was denatured at 105°C for 10 min in loading buffer (Tris-HCl pH 6.8 60 mM; SDS 2%; bromophenol blue 0.1%; glycerol 25%) supplemented with or without 14.4 mM beta-mercaptoethanol for a reduced or nonreduced reaction, respectively. A total of 20 μg of harvested protein was loaded and separated on a 10% SDS-polyacrylamide gradient gel. The proteins were then transferred onto polyvinylidene difluoride membranes and were blocked with 5% fat-free milk for 2 h at room temperature. The membranes were incubated with the primary antibody and the horseradish peroxidase-conjugated secondary antibody; then, the proteins were detected using an ECL chemiluminescence system (Pierce). The primary antibodies used were as follows: the antibody against GAPDH (13937-1-AP) was from Proteintech, and the antibody against HA (3724) was obtained from Cell Signaling Technology.
PNGase F Treatment
In this study, PNGase F was purchased from NEB corporation and was used to remove almost all N-linked oligosaccharides from the glycoproteins. The cell lysates were prepared as described in the Western blotting process. According to the PNGase F denaturing protocol, 5 μg of cell lysate was mixed with 1 μl of glycoprotein denaturing buffer (10X) and H2O to make a 10 μl total reaction volume. The mixture was then incubated at 100°C for 10 min and was then chilled on ice for 10 sec. Then, 2 μl GlycoBuffer 2 (10X), 2 μl 10% NP-40, 6 μl H2O, and 1 μl PNGase F was added into the mixture and incubated at 37°C for 1 h.
Supernatant concentration
The cells were cultured in 15 cm dishes with complete media. The media were removed and replaced by 25 ml of DMEM without antibiotics or FBS. After 48 h, the cell supernatant was loaded onto an Ultracel-10 centrifugal Filter Unit for centrifugation at 4000 rpm at 4°C for 1 h. The concentrated supernatants were lysed in reduced and nonreduced reactions for subsequent Western blotting.
Animal experiments
All animal experiments were performed according to the protocols approved by the Research Animal Resource Center of Sun Yat-Sen University and complied with the guidelines. The animal ethics number of this project is GZR2015-137. All animals were obtained from Beijing Vital River Laboratory Animal Technology Co., Ltd. For the spontaneous metastasis osteosarcoma model in vivo, U2OS/MTX300 cells stably expressing luciferase, including the vector, PCOLCE, N29Q, shNC, PCOLCE-sh#1, and PCOLCE-sh#2, were used as described. Six mice were injected separately with 1x106 cells in each group. When the tumor reached 1.5 cm in diameter, the mice were injected with a luciferase substrate, and the IVIS Lumina Imaging System (Xenogen) was used to capture the luciferase signal by taking photographs. The mice were then sacrificed, and the lungs were fixed in formalin (4 μmol/L) and stained with hematoxylin and eosin (H&E).
Statistical analysis
The SPSS software was used for statistical analysis. The statistical significance was tested by Student's t-test or a chi-square test. The curve of correlation between PCOLCE expression level and the overall survival was plotted by the Kaplan-Meier method. Differences were considered statistically significant when p values were <0.05.
Results
PCOLCE is up-regulated in osteosarcoma tissues
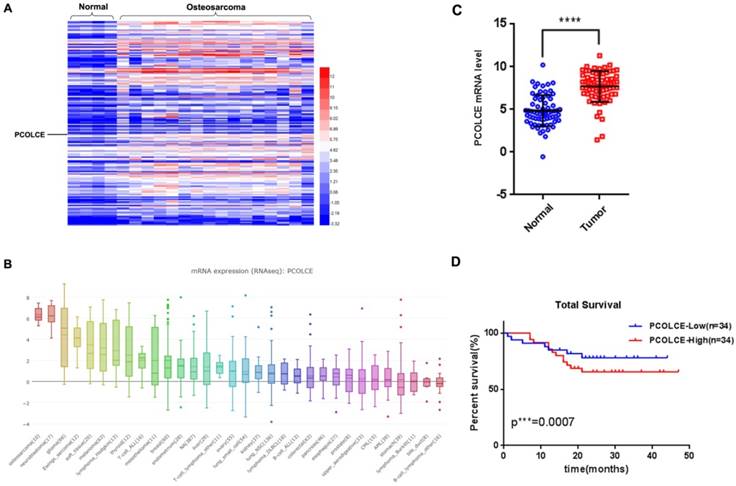
By analyzing our RNA-seq data from 4 noncancerous adjacent tissues and 16 osteosarcoma tissues (Fig. S1), we found that 169 genes encoding ECM proteins were overexpressed in osteosarcoma tissues compared to the expression in noncancerous adjacent tissues (sheets 1-2 of Table S1). Notably, PCOLCE, a procollagen C-proteinase enhancer protein for stimulating procollagen maturation, was one of the most up-regulated genes in osteosarcoma (Fig. 1A). Interestingly, data from CCLE (https://portals.broadinstitute.org/ccle/page?gene=PCOLCE) also indicated that the PCOLCE mRNA was dramatically elevated in osteosarcoma cell lines, such as U2OS and HOS cells (Fig. 1B). The up-regulation of PCOLCE was further confirmed by qRT-PCR using 68 pairs of human osteosarcoma tissues and their adjacent nontumor tissues (Fig. 1C), whose clinical information are listed as Supplementary Table S2. More importantly, the osteosarcoma patients with high levels of PCOLCE had poorer total survivals compare those with low levels of PCOLCE (Fig. 1D). The results indicate that PCOLCE is up-regulated in osteosarcoma tissues and may be involved in osteosarcoma progression.
Knockdown of PCOLCE by shRNA inhibits the lung metastasis of osteosarcoma
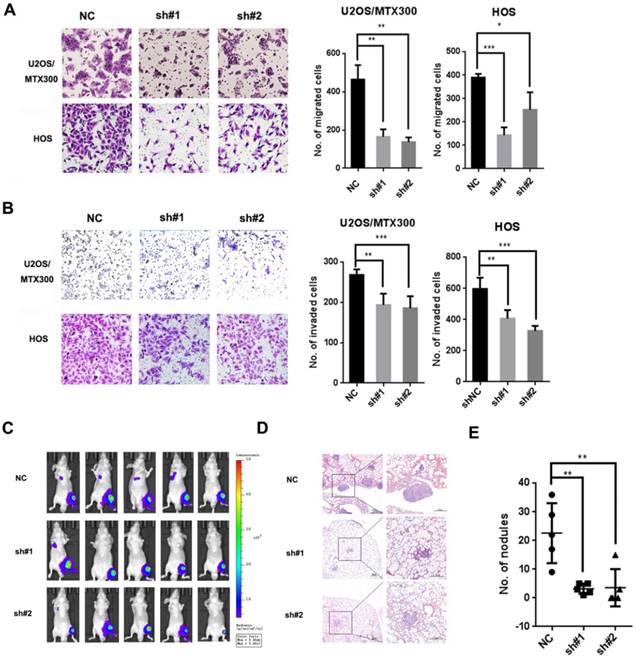
To determine the function of PCOLCE in osteosarcoma, we generated two independent short hairpin RNAs against PCOLCE, which could efficiently reduce the endogenous expression of PCOLCE in U2OS, U2OS/MTX300 and HOS cells (Fig. S2A-C). The knockdown of PCOLCE significantly inhibited cell migration and invasion in vitro (Fig. 2A and 2B, S3A and S3B), but did not affect cell viability (Fig. S2D and S2E). More importantly, in the orthotopic osteosarcoma metastasis mouse model using U2OS/MTX300-Luc cells with the stable knockdown of PCOLCE, only one or two out of five mice in the PCOLCE-shRNAs groups, while four out of five mice in the control group had luminescent signals within the lungs (Fig. 2C). Consistently, the mice bearing PCOLCE-sh#1 or PCOLCE-sh#2 cells had significant decreases in lung metastatic burden compared to those bearing control cells (Fig. 2D and 2E).
PCOLCE is up-regulated in osteosarcoma tissues. (A) The expression profiles of extracellular matrix proteins in 4 nontumor adjacent tissues and 16 osteosarcoma tissues are presented by a heatmap. (B) The PCOLCE mRNA levels in different cancer type cell lines from the CCLE database. (C) The mRNA levels of PCOLCE in the indicated tissues were analyzed by qRT-PCR (mean ± SD. ****p < 0.0001). (D). The total survival curves were generated based on the PCOLCE mRNA levels from 68 osteosarcoma tissues. The actuarial probabilities were calculated using the Kaplan-Meier method. The patients with high levels of PCOLCE had poorer total survivals.
PCOLCE promotes osteosarcoma cell migration and invasion
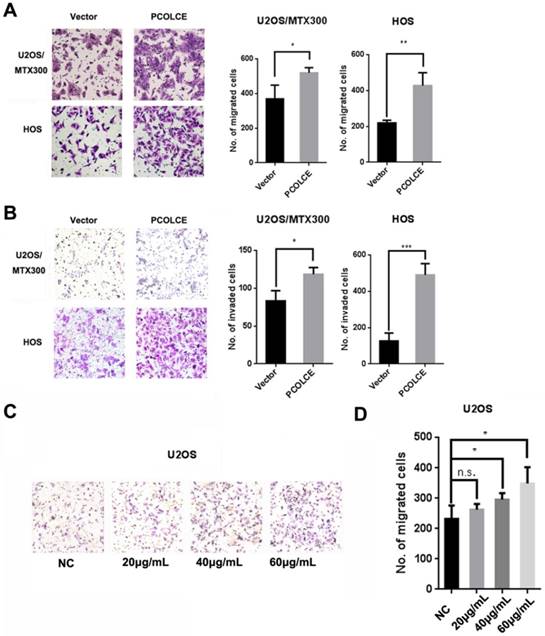
The stable overexpression of PCOLCE in U2OS, U2OS/MTX300 and HOS cells was also generated (Fig. S4A-C). Cell migration and invasion, but not cell viability, were dramatically promoted in these stable cells compared to those in cells stably overexpressing the empty vector (Fig. 3A and 3B, S4D-G). Furthermore, we purified His-tagged PCOLCE recombinant protein from HEK293T cells transiently expressing His-tagged PCOLCE (Fig. S4H) to test whether this recombinant protein could promote the migration of osteosarcoma cells. As shown in Fig. 3C and 3D, cell migration was significantly promoted in U2OS cells treated with the purified His-tagged PCOLCE recombinant protein in a concentration-dependent manner compared to the cell migration of the control cells. The results suggest that PCOLCE may play a critical role in promoting osteosarcoma metastasis.
Down-regulation of PCOLCE by shRNAs suppresses osteosarcoma metastasis. (A, B). Cell migration (A) and invasion (B) assays were performed and quantified in the indicated stable cells as described in the Materials and Methods. Representative images and quantification analyses are shown (mean ± SD, n=3, *p<0.05, **p<0.01, ***p<0.001). (C-E). The orthotopic osteosarcoma metastasis model in vivo using the indicated U2OS/MTX-Luc stable cells, as described in the Materials and Methods section. (C) Bioluminescence images for mice in the indicated groups. (D, E) Representative images of H&E staining (D) and quantification analyses (E) of the metastatic lung nodules from the indicated groups (mean ± SD, n=5, **p<0.01.
The N-glycosylation of PCOLCE is essential for its enhancement of metastasis
Glycosylation modifications are crucial for the ECM-mediated malignant phenotype during cancer [23], and PCOLCE may have N-linked oligosaccharides modified with sialic residues in human serum [24]; we explored which residue of PCOLCE could be glycosylated and the significance of this event. Two putative N-glycosylation sites of PCOLCE were predicted using the N-X-S/T consensus sequence [25] (Fig. 4A). Therefore, we generated two PCOLCE mutants by individually changing each Asn (N) into Gln (Q). As shown in Fig. 4B, the molecular weight of the N29Q mutant, but not that of the N431Q mutant, was slightly lower than its wild-type counterpart, whereas their molecular weights became identical under PNGase F treatment. As PNGase F specifically removes the N-linked glycosylation, the band shifts of both wild-type and its N431Q mutant, but not of its N29Q mutant, were observed after PNGase F treatment, indicating that PCOLCE has the N-linked glycosylation at N29. Notably, the protein level of the PCOLCE-N29Q mutant was markedly reduced compared to that of its wild-type protein (Fig. 4B), indicating that the N-linked glycosylation of PCOLCE at Asn29 may impact its protein stability. Indeed, as shown in Fig. 4C, the PCOLCE-N29Q mutant protein had a shorter half-life than that of its wild-type protein. In addition, as shown in Fig. 4D, wild-type PCOLCE was detected in both cell lysate and supernatant, while the N29Q mutant was hardly detected in supernatant, indicating that the secretion of the PCOLCE-N29Q mutant into media was much less compared to wild-type PCOLCE. The results suggest that PCOLCE is N-glycosylated at Asn29, which positively regulates its protein stability and its secretion into the ECM.
Next, we asked whether the Asn29 glycosylation of PCOLCE is required for its functions in osteosarcoma. As shown in Fig. 4E-4H and Fig. S5A-C, the enhancement of cell migration and metastasis induced by wild-type PCOLCE was not observed in the N29Q mutant both in vitro and in vivo. This finding indicates that the glycosylation of PCOLCE at Asn29 is necessary for its promotion of osteosarcoma metastasis.
PCOLCE promotes the migration and invasion of osteosarcoma cells. (A, B). Cell migration (A) and invasion (B) assays were performed and quantified in the indicated stable cells as described in the Materials and Methods section. Representative images and quantification analyses are shown (mean ± SD, n=3, *p<0.05, **p<0.01). (C, D) U2OS cells were treated with the indicated concentrations of PCOLCE-His tag protein from (Fig. S4H) for 24 h and were then subjected to the migration assay. Representative images and quantification analyses are shown (mean ± SD, n=3, *p<0.05).
PCOLCE is transcriptionally up-regulated by TWIST1
Since PCOLCE has been reported to be directly regulated by TWIST1 in mouse embryonic tissues[26], and two putative E-box binding sites for TWIST1, -542~-533 and -408~-399, were predicted in the PCOLCE promoter using JASPAR [27] as shown in Fig. 5A. In addition, the expression levels were positively correlated between TWIST1 and PCOLCE in osteosarcoma tissues (Fig. 5B). Therefore, we speculated that TWIST1 could transcriptionally up-regulate PCOLCE in osteosarcoma cells. This was the case, as the mRNA levels of PCOLCE were significantly increased in both U2OS and HOS cells stably overexpressing TWIST1 compared to those stably expressing the empty vector (Fig. 5C). To test whether PCOLCE is a direct target of TWIST1, the depletion of a single or double E-box in the dual-luciferase reporter of the PCOLCE promoter was generated. As shown in Fig. 5D, E-box1, but not E-box2, was crucial for the up-regulation of the PCOLCE promoter by TWIST1. Indeed, the association of TWIST1 with the PCOLCE promoter was further verified by a chromatin immunoprecipitation (ChIP) assay (Fig. 5E). Taken together, our results indicate that TWIST1 transcriptionally up-regulates PCOLCE by directly binding to E-box1 within the PCOLCE promoter.
Glycosylation of PCOLCE is essential for its function in osteosarcoma. (A) A schematic illustration of the predicted N-linked glycosylation sites in PCOLCE. (B) 293T cells were transiently transfected with the indicated plasmids for 48 h and were incubated with or without PNGase F as indicated; then, the cell lysates were harvested and subjected to Western blotting. (C) 293T cells were transiently transfected with PCOLCE wild-type or N29Q mutant plasmids for 48 h and were incubated with 20 mg/mL cycloheximide (CHX) for the indicated times. The cell lysates were harvested and analyzed by Western blotting. (D) The cell lysates and supernatant with (reduced) or without (nonreduced) β-mercaptoethanol from the indicated cells were analyzed by Western blotting. (E) A cell migration assay was performed and quantified in the indicated stable cells as described in the Materials and Methods section. The representative images and quantification analyses were shown (mean ± SD, n=3, **p<0.01, ***p<0.001). (F-H) The orthotopic osteosarcoma metastasis model in vivo using the indicated U2OS/MTX-Luc stable cells, as described in the Materials and Methods section. (F) Bioluminescence images for mice in the indicated groups. (G, H) Representative images of H&E staining (G) and quantification analyses (H) of the metastatic lung nodules from the indicated groups (mean ± SD, n=5, *p<0.05).
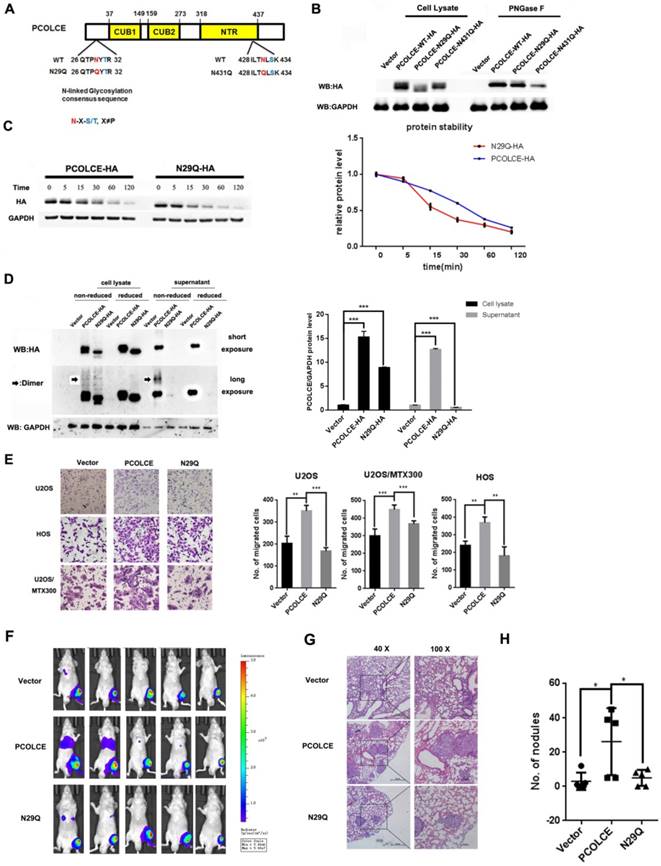
TWIST1 transcriptionally up-regulates PCOLCE in osteosarcoma cells. (A) A schematic illustration of putative TWIST1 binding sites (E-boxes) in the promoter of PCOLCE. (B) The correlation between TWIST1 mRNA levels and PCOLCE mRNA levels in the clinical samples (16 osteosarcoma and 4 normal control) shown in Fig. 1A. (C) The mRNA levels of PCOLCE were measured by qRT-PCR in the indicated cells stably transfected with the vector or TWIST1 (mean ± SD, n=3, ***p<0.001). (D) The ChIP-qPCR assay using an anti-Flag antibody was performed with U2OS cells expressing the vector or Flag-TWIST1 (mean ± SD, n=3, **p<0.01). (E) HEK293T cells were transfected with the indicated luciferase reporter for 48 h and were then subjected to the luciferase activity assay as described in the Materials and Methods section. (mean ± SD, n=3, **p<0.01).
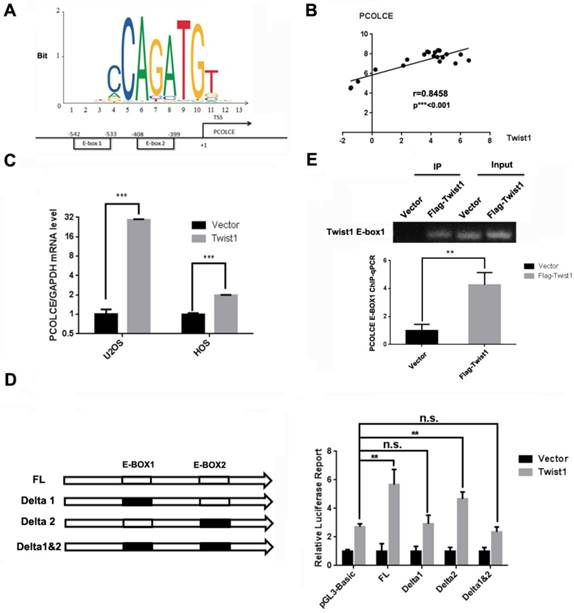
TWIST1 may also up-regulate some procollagens in osteosarcoma tissues
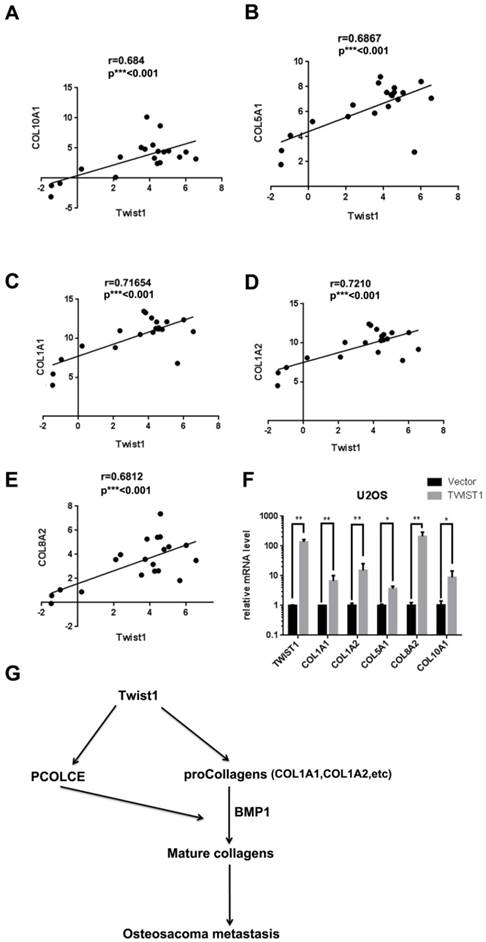
Since PCOLCE promotes the maturation of procollagens, we determined the status of several procollagens in osteosarcoma tissues. As shown in Fig. 6A-6E, the mRNA levels of several procollagens, such as COL1A1, COL1A2, COL5A1, COL8A2 and COL10A, were positively correlated with TWIST1 in our RNA-seq data (Fig. 1A). Indeed, the mRNA levels of these procollagens were significantly increased in U2OS cells stably expressing TWIST1 compared to those in cells stably expressing the vector (Fig. 6F). These results indicate that TWIST1 may also up-regulate some procollagens in osteosarcoma tissues.
Discussion
In this report, we revealed that PCOLCE and some procollagens were up-regulated by TWIST1 in osteosarcoma cell lines and tissues and that the N-glycosylation of PCOLCE at Asn29 was crucial for promoting osteosarcoma metastasis.
PCOLCE specifically enhances the activity of BMP-1, a zinc metalloproteinase that modulates collagen deposition in the extracellular matrix by removing C-propeptides from procollagen I, II and III [11]. Increased collagen deposition is the most well-recognized ECM alteration during cancer progression [28], suggesting that PCOLCE may be involved in cancer metastasis. Recently, Salza et al. reported that PCOLCE might be involved in tumor growth based on the interaction network of PCOLCE [29]. Our results showed that PCOLCE plays a critical role in promoting the metastasis of osteosarcoma, as the overexpression of PCOLCE promotes cell migration, invasion and lung metastasis (Figs. 3A, 3B, 4E-H, S4D, S4E) whereas the knockdown of PCOLCE had the opposite effects (Fig. 2, S3A, S3B) in osteosarcoma.
TWIST1 up-regulates Collagen expression. (A-E) The correlations between TWIST1 mRNA levels and mRNA levels of COL10A1 (A), COL15A1 (B), COL1A2 (C), COL5A1 (D) or COL8A2 (E) in the clinical samples (16 osteosarcoma and 4 normal controls) are shown in Fig. 1A. (F) The mRNA levels of COL1A1, COL1A2, COL5A1, COL8A2 and COL10A1 in the U2OS cells with the stable overexpression vector or Flag-TWIST1 were measured by qRT-PCR (mean ± SD, n=3, *p<0.05, **p<0.01). (G) The proposed model for the function and mechanism of PCOLCE in osteosarcoma. TWIST1 may up-regulate both PCOLCE and procollagens, and then PCOLCE helps BMP1 accelerate the maturation of collagen, which in turn promotes osteosarcoma metastasis.
A previous study has indicated that PCOLCE may have N-linked oligosaccharides decorated with sialic residues in human serum [24]. Here, we have demonstrated that PCOLCE is N-glycosylated at Asn29, which is essential for its protein stability, secretion and the enhancement of metastasis in osteosarcoma (Fig. 4E-H). Notably, serum PCOLCE may act as a biomarker of diseases, such as muscle [16], liver [30] and cardiac [31] fibrosis. PCOLCE could be detected in the supernatant of osteosarcoma cells (Fig. 4D), and the purified PCOLCE recombinant protein could promote cell migration in U2OS cells (Figs. 3C-3E); this indicates that PCOLCE may be a potential noninvasive biomarker to predict metastasis in osteosarcoma patients.
We proved that TWIST1 could up-regulate PCOLCE by directly binding to the E-box1 of the PCOLCE promoter (Fig. 5). In addition, TWIST1 also increased the mRNA levels of procollagens, such as COL1A1, COL1A2, COL5A1, COL8A2 and COL10A (Fig. 6A), which are potential substrates for PCOLCE. This may provide a new explanation that TWIST1 is a key regulator in cancer metastasis in addition to being a key regulator of EMT[32] [33] because the correlations between TWIST1 mRNA levels and PCOLCE mRNA levels were also observed in 18 different cancer types from the ChIPBase v2.0 database [34] (Supplementary Table 1).
In summary, we propose that TWIST1 may up-regulate both PCOLCE and procollagens and that PCOLCE helps BMP1 to accelerate the maturation of collagen, which in turn, promotes osteosarcoma metastasis; this is illustrated in Fig. 6G.
Supplementary Material
Supplementary figures and table 1.
Supplementary table S1.
Supplementary table S2.
Acknowledgements
This work was supported by grants from the National Key Research and Development Program of China [2016YFC0904601 and 2016YFA0500304 to T.K. ], the National Nature Science Foundation of China (NSFC) [81530081 and 31571395 to T.K., 81772922 to Y.W., 81702758 to J.W.], the Guangdong Natural Science Foundation Team Project [2014A030312015 to T.K.], the Special Key Project of Guangzhou Scientific Research [201607020038 to T.K.], the Natural Science Foundation of Guangdong Province [2016A030310218 to Y.W.], the Fundamental Research Funds for the Central Universities [16ykpy39 to Y.W.], the Sci-Tech Project Foundation of Guangzhou City [201607020038 to T.K.], and the Project funded by China Postdoctoral Science Foundation [to L.Z.].
Competing Interests
The authors have declared that no competing interest exists.
References
1. Luetke A, Meyers PA, Lewis I, Juergens H. Osteosarcoma treatment - where do we stand? A state of the art review. Cancer treatment reviews. 2014;40:523-32
2. Zambo I, Vesely K. [WHO classification of tumours of soft tissue and bone 2013: the main changes compared to the 3rd edition]. Ceskoslovenska patologie. 2014;50:64-70
3. Schwartz CL, Gorlick R, Teot L, Krailo M, Chen Z, Goorin A. et al. Multiple drug resistance in osteogenic sarcoma: INT0133 from the Children's Oncology Group. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2007;25:2057-62
4. Gill J, Ahluwalia MK, Geller D, Gorlick R. New targets and approaches in osteosarcoma. Pharmacology & therapeutics. 2013;137:89-99
5. Bonnans C, Chou J, Werb Z. Remodelling the extracellular matrix in development and disease. Nature reviews Molecular cell biology. 2014;15:786-801
6. Abedin M, King N. Diverse evolutionary paths to cell adhesion. Trends in cell biology. 2010;20:734-42
7. Liotta LA, Tryggvason K, Garbisa S, Hart I, Foltz CM, Shafie S. Metastatic potential correlates with enzymatic degradation of basement membrane collagen. Nature. 1980;284:67-8
8. Mook OR, Frederiks WM, Van Noorden CJ. The role of gelatinases in colorectal cancer progression and metastasis. Biochimica et biophysica acta. 2004;1705:69-89
9. Cox TR, Rumney RMH, Schoof EM, Perryman L, Hoye AM, Agrawal A. et al. The hypoxic cancer secretome induces pre-metastatic bone lesions through lysyl oxidase. Nature. 2015;522:106-10
10. Wu X, Liu T, Fang O, Leach LJ, Hu X, Luo Z. miR-194 suppresses metastasis of non-small cell lung cancer through regulating expression of BMP1 and p27(kip1). Oncogene. 2014;33:1506-14
11. Moali C, Font B, Ruggiero F, Eichenberger D, Rousselle P, Francois V. et al. Substrate-specific modulation of a multisubstrate proteinase. C-terminal processing of fibrillar procollagens is the only BMP-1-dependent activity to be enhanced by PCPE-1. The Journal of biological chemistry. 2005;280:24188-94
12. Pulido D, Sharma U, Vadon-Le Goff S, Hussain SA, Cordes S, Mariano N. et al. Structural Basis for the Acceleration of Procollagen Processing by Procollagen C-Proteinase Enhancer-1. Structure. 2018;26:1384-92 e3
13. Vadon-Le Goff S, Kronenberg D, Bourhis JM, Bijakowski C, Raynal N, Ruggiero F. et al. Procollagen C-proteinase enhancer stimulates procollagen processing by binding to the C-propeptide region only. The Journal of biological chemistry. 2011;286:38932-8
14. Weiss T, Ricard-Blum S, Moschcovich L, Wineman E, Mesilaty S, Kessler E. Binding of procollagen C-proteinase enhancer-1 (PCPE-1) to heparin/heparan sulfate: properties and role in PCPE-1 interaction with cells. The Journal of biological chemistry. 2010;285:33867-74
15. Bourhis JM, Vadon-Le Goff S, Afrache H, Mariano N, Kronenberg D, Thielens N. et al. Procollagen C-proteinase enhancer grasps the stalk of the C-propeptide trimer to boost collagen precursor maturation. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:6394-9
16. Hassoun E, Safrin M, Ziv H, Pri-Chen S, Kessler E. Procollagen C-Proteinase Enhancer 1 (PCPE-1) as a Plasma Marker of Muscle and Liver Fibrosis in Mice. PloS one. 2016;11:e0159606
17. Massoudi D, Germer CJ, Glisch JM, Greenspan DS. Procollagen C-proteinase enhancer 1 (PCPE-1) functions as an anti-angiogenic factor and enhances epithelial recovery in injured cornea. Cell and tissue research. 2017;370:461-76
18. Raz V, Sterrenburg E, Routledge S, Venema A, van der Sluijs BM, Trollet C. et al. Nuclear entrapment and extracellular depletion of PCOLCE is associated with muscle degeneration in oculopharyngeal muscular dystrophy. BMC neurology. 2013;13:70
19. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114-20
20. Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36
21. Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ. et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511-5
22. Wang X, Li L, Wu Y, Zhang R, Zhang M, Liao D. et al. CBX4 Suppresses Metastasis via Recruitment of HDAC3 to the Runx2 Promoter in Colorectal Carcinoma. Cancer research. 2016;76:7277-89
23. Marsico G, Russo L, Quondamatteo F, Pandit A. Glycosylation and Integrin Regulation in Cancer. Trends in cancer. 2018;4:537-52
24. Mesilaty-Gross S, Anikster Y, Vilensky B, Wolf I, Phillip M, Gat-Yablonski G. Different patterns of human serum procollagen C-proteinase enhancer1 (PCPE1). Clinica chimica acta; international journal of clinical chemistry. 2009;403:76-80
25. Nita-Lazar M, Wacker M, Schegg B, Amber S, Aebi M. The N-X-S/T consensus sequence is required but not sufficient for bacterial N-linked protein glycosylation. Glycobiology. 2005;15:361-7
26. Bildsoe H, Fan X, Wilkie EE, Ashoti A, Jones VJ, Power M. et al. Transcriptional targets of TWIST1 in the cranial mesoderm regulate cell-matrix interactions and mesenchyme maintenance. Developmental biology. 2016;418:189-203
27. Khan A, Fornes O, Stigliani A, Gheorghe M, Castro-Mondragon JA, van der Lee R. et al. JASPAR 2018: update of the open-access database of transcription factor binding profiles and its web framework. Nucleic acids research. 2018;46:D1284
28. Lu P, Weaver VM, Werb Z. The extracellular matrix: a dynamic niche in cancer progression. The Journal of cell biology. 2012;196:395-406
29. Salza R, Peysselon F, Chautard E, Faye C, Moschcovich L, Weiss T. et al. Extended interaction network of procollagen C-proteinase enhancer-1 in the extracellular matrix. The Biochemical journal. 2014;457:137-49
30. Gokce O, Ozenirler S, Atak Yucel A, Oruklu N, Yilmaz Esendagli G, Karahan S. Evaluation of serum procollagen C-proteinase enhancer 1 level as a fibrosis marker in patients with chronic hepatitis B. European journal of gastroenterology & hepatology. 2018;30:918-24
31. Kessler-Icekson G, Schlesinger H, Freimann S, Kessler E. Expression of procollagen C-proteinase enhancer-1 in the remodeling rat heart is stimulated by aldosterone. The international journal of biochemistry & cell biology. 2006;38:358-65
32. Kang Y, Massague J. Epithelial-mesenchymal transitions: twist in development and metastasis. Cell. 2004;118:277-9
33. Lyu P, Zhang SD, Yuen HF, McCrudden CM, Wen Q, Chan KW. et al. Identification of TWIST-interacting genes in prostate cancer. Science China Life sciences. 2017;60:386-96
34. Zhou KR, Liu S, Sun WJ, Zheng LL, Zhou H, Yang JH. et al. ChIPBase v2.0: decoding transcriptional regulatory networks of non-coding RNAs and protein-coding genes from ChIP-seq data. Nucleic acids research. 2017;45:D43-D50
Author contact
![]() Corresponding authors: Jing Chen, Ph.D., chenjingwhedu.cn, Yuanzhong Wu, Ph.D., wuyzorg.cn or Tiebang Kang, Ph.D., kangtborg.cn
Corresponding authors: Jing Chen, Ph.D., chenjingwhedu.cn, Yuanzhong Wu, Ph.D., wuyzorg.cn or Tiebang Kang, Ph.D., kangtborg.cn
 Global reach, higher impact
Global reach, higher impact