13.3
Impact Factor
Theranostics 2019; 9(14):4130-4140. doi:10.7150/thno.34692 This issue Cite
Review
Liquid biopsy in ovarian cancer: recent advances in circulating extracellular vesicle detection for early diagnosis and monitoring progression
1. Department of Obstetrics and Gynecology, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, Henan 450052, China
2. Cancer Care Centre, St George Hospital, Kogarah, NSW 2217, Australia.
3. St George and Sutherland Clinical School, Faculty of Medicine, UNSW Sydney, NSW 2052, Australia.
4. Institute of Precision Cancer Medicine and Pathology and Department of Pathology, Jinan University Medical College, Guangzhou 510630, China.
5. The Academy of Medical Sciences, Zhengzhou University, Henan 450001, China
6. School of Basic Medical Sciences, Zhengzhou University, Henan 450001, China
*Lei Chang and Jie Ni contributed equally to this work.
Received 2019-3-7; Accepted 2019-5-1; Published 2019-5-31
Abstract
The current biomarkers available in the clinic are not enough for early diagnosis or for monitoring disease progression of ovarian cancer. Liquid biopsy is a minimally invasive test and has the advantage of early diagnosis and real-time monitoring of treatment response. Although significant progress has been made in the usage of circulating tumor cells and cell-free DNA for ovarian cancer diagnosis, their potential for early detection or monitoring progression remains elusive. Extracellular vesicles (EVs) are a heterogeneous group of lipid membranous particles released from almost all cell types. EVs contain proteins, mRNA, DNA fragments, non-coding RNAs, and lipids and play a critical role in intercellular communication. Emerging evidence suggests that EVs have crucial roles in cancer development and metastasis, thus holding promise for liquid biopsy-based biomarker discovery for ovarian cancer diagnosis. In this review, we discuss the advantages of EV-based liquid biopsy, summarize the protein biomarkers identified from EVs in ovarian cancer, and highlight the utility of new technologies recently developed for EV detection with an emphasis on their use for diagnosing ovarian cancer, monitoring cancer progression, and developing personalized medicine.
Keywords: Extracellular vesicle, ovarian cancer, liquid biopsy, diagnosis
Introduction
Ovarian cancer (OC) is one of the most lethal gynecological malignancies. It is the fifth leading cause of cancer-related deaths among females, affecting over 290,000 women worldwide annually [1] with an estimated 22,240 new cases and 14,070 deaths in the United States in 2018 [2]. Due to the lack of early symptoms, physical signs, and effective screening approaches for early diagnosis, approximately 70% of OC cases are not diagnosed until they are in advanced stages, which only have a 20% 5-year survival rate. However, if diagnosed at early stages, the 5-year survival rate for Stage I and II OC is 89% and 71%, respectively [2]. Therefore, early diagnosis using effective biomarkers and screening approaches is of high importance and may improve the prognosis of a large number of OC patients.
The diagnosis of OC is mainly based on levels of biomarker CA-125 in blood and imaging [3, 4]. CA-125, also known as MUC16, is the most clinically utilized biomarker for monitoring the response to treatment and detecting disease recurrence in OC [5]. However, CA-125 levels are not always increased in the early stages of OC and not every OC patient shows elevated CA-125 levels. In addition, some other diseases such as endometriosis, inflammation, and other types of cancers [6-8] can also cause elevated CA-125 levels. Furthermore, even with using CA-125 as a screening marker, the overall survival rate in OC has not significantly changed in clinical trials [4, 9]. As a result, no professional group recommends screening ovarian cancer using CA-125 in the general population. Therefore, it is of great importance to find new approaches to detect early stage OC.
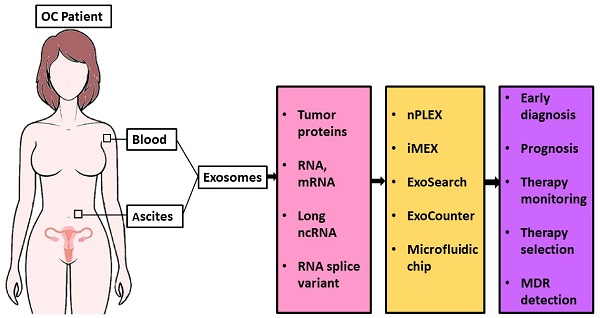
Extracellular vesicles (EVs) including exosomes, microvesicles, and other membranous structures are abundantly released into the extracellular space by almost all types of cells. EVs carry complex biological information from their original cells and are useful sources for cancer diagnosis in a non-invasive manner [10]. According to the International Society of Extracellular Vesicles (ISEV), the term “extracellular vesicles” is the appropriate terminology for heterogeneous populations of vesicles isolated from cell culture supernatants or physiological fluids [11]. Throughout this review, exosomes will be referred to as EVs.
Exosomes are cell-secreted membranous nanoscale vesicles with diameters of 50-150 nm that contain mRNA, microRNA, small interfering RNA, and proteins [12-15]. These exosomal contents are representative of its originating cell and contribute to intercellular communications [16]. Exosomes attract considerable interest in the research community due to their role in regulating multiple physiological processes and mediating systemic dissemination in various cancers [17]. Several reports have demonstrated that exosomes exist in blood and ascites of OC patients [18, 19]. In addition, exosomes and exosomal cargoes, such as microRNAs, were found to play a crucial role in disease progression and potentially facilitate chemoresistance in OC [20-22]. Therefore, OC-derived exosomes have the potential to be used as biomarkers for the early detection of cancer and follow-up monitoring.
Liquid biopsy, a recent and hot topic in cancer detection, has been considered for the early diagnosis of cancer [23]. Generally speaking, liquid biopsy involves the collection and analysis of circulating tumor cells (CTCs), circulating tumor DNA (ctDNA), circulating cell-free microRNAs (cfmiRNAs), and exosomes [24]. Liquid biopsy has already been used in OC research [24]. Both CTCs and ctDNAs in OC have been intensively studied for clinical significance in the last two decades and the advances in the field have been recently reviewed [25-27]. This review highlights the recent progress in new techniques for OC EV detection and mainly focuses on EV protein biomarkers for OC early detection, monitoring cancer progression, and personalized therapy.
EVs for liquid biopsy
Tissue biopsy versus liquid biopsy
Surgical tissue biopsies are invasive procedures and can be associated with complications such as bleeding and infection [28]. In addition, biopsies are often difficult to perform on organs that lie deep within the body and the use is limited as they can give false negative results due to sampling bias [29].
Compared to conventional tissue biopsy, liquid biopsy is growing in popularity because it is minimally invasive, easy to use, and can have high throughput. ctDNA in the plasma of OC patients can identify relapse or drug resistance well before clinical symptoms appear, enabling earlier intervention and better patient outcomes [30]. Liquid biopsies measure various tumor biomarkers such as proteins, nucleic acids, cells, and EVs in body fluid like blood. Thus, liquid biopsies are advantageous over traditional tissue biopsies as blood samples can be easily collected longitudinally and in large quantities, making it an attractive platform for large-scale screening of tumor-specific mutations [31, 32]. It also has the potential of providing new insights into prognosis, patient follow-up, treatment response, and more recently, early diagnosis and population screening [33].
Advantages of EVs for liquid biopsy
Liquid biopsies of CTCs, ctDNA, and EVs are promising for early-stage cancer detection and real-time monitoring the dynamics of cancer progression and metastasis [23, 34]. It has been shown that cancer cells release EVs containing cancer-specific contents that can be easily isolated from various body fluids [35].
However, using either CTCs or ctDNA as cancer biomarkers faces multiple technical and translational challenges. First, scarcity and heterogeneity of CTCs make the isolation and characterization of CTCs extremely hard [36]. Second, high fragmentation, low abundance, and low stability of ctDNA largely hampered the utility of ctDNA in routine clinical practice [37, 38].
Compared with CTCs and ctDNA, EVs possess advantages in terms of abundance, stability, and accessibility. First, EVs are abundant (108-13 exosomes/mL) in plasma and other body fluids. Secondly, EVs are very stable [39] and can be stored at -80C° for months and even years while maintaining protein and nucleic acid quality. Furthermore, the contents of EVs are tumor-specific and correlate with tumor staging and prognosis [40]. In addition, EVs are broadly distributed in body fluids and thus, can be easily obtained. From the same type of tissue, cancer cells were found to shed more EVs compared to normal cells, indicating EVs are a much more abundant biomarker source in liquid biopsy compared to CTCs [41]. Therefore, based on these merits, more emphasis has been put on EVs as a biomarker source for liquid biopsy of cancer in recent years [42, 43].
EV protein biomarkers in ovarian cancer
In addition to common exosomal proteins such as TSG101, CD9, CD81, and CD63, some other OC-induced proteins have also been investigated for screening and diagnosis. For example, claudin-4 protein is released from OC cells via exosomes. It was reported that positive expression of claudin-4 in exosomes in blood was shown in 32 of 63 OC patients, but in only 1 of 50 samples from healthy controls, with 51% sensitivity and 98% specificity [44], indicating its clinical significance for OC diagnosis. Additionally, the co-expression of exosomal claudin-4 and CA125 has also been suggested as a putative combination marker [44].
A recent proteomic analysis of exosomes from OC cell lines showed enrichment of OC-specific markers, such as mesothelin (MSLN), carcinoembryonic antigen (CEA), MUC16 (CA125), and WAP four-disulfide core domain protein 2 (WFDC2) [45]. Liang et al. demonstrated that OC-derived exosomes carried a protein expression signature that was also overexpressed in OC tissues including epithelial cell adhesion molecule (EpCAM), proliferation cell nuclear antigen (PCNA), tubulin beta-3 chain (TUBB3), epidermal growth factor receptor (EGFR), apolipoprotein E (APOE), claudin 3 (CLDN3), fatty acid synthase (FASN), ERBB2, and L1 cell adhesion molecule (L1CAM), suggesting that these proteins could be used as diagnostic markers or therapeutic targets for OC [46]. Peng et al. found 70 kilodalton heat shock protein (HSP70), major histocompatibility complex class I molecule (MHC-I), and CD81 molecular signatures on the ascites-derived exosomes of 85% OC patients. They also showed that these exosomes could decrease the cytotoxicity of peripheral blood mononuclear cells in the presence of dendritic cells [47], suggesting the immunotherapy potential of exosomes. Szajnik et al. found that exosomes from OC plasma contain distinguishable levels of TGF-β1 and melanoma-associated antigen (MAGE) 3/6 proteins compared with those from benign tumors, indicating a diagnostic value for these biomarkers [48]. In addition, a soluble activated leukocyte cell adhesion molecule (sALCAM) was identified in EVs of sera and ascites of OC patients [49], which correlated with more aggressive tumor types. In another study, soluble E-cadherin (sE-cad) was found to be released with EVs into the ascitic fluid and the levels were able to distinguish between OC and benign disease [50]. Altogether, these data suggest that the exosomal protein content may offer a novel approach to the diagnosis of OC.
CD24 is a glycosyl-phosphatidylinositol-linked glycoprotein at the cell surface and its expression has been correlated with shortened patient survival in OC [51]. EpCAM is a glycosylated 30-40 kDa transmembrane protein and is expressed in essentially all human adenocarcinomas, including OC [52, 53]. Overexpression of EpCAM was found in primary OC, as well as metastatic and recurrent/chemotherapy-resistant OC [54]. In one study, the exosomal proteins CD24 and EpCAM were isolated from ascites fluid of OC patients, suggesting their prognostic role in the clinic or value in treatment planning [18]. In another study, CD24 and EpCAM were selectively present on ascites exosomes of OC patients and proposed as putative biomarkers for OC detection [55]. Ketter et al. found that OC ascites-derived exosomes containing proteins such as L1CAM, CD24, ADAM metallopeptidase domain 10 (ADAM10), and extracellular matrix metalloproteinase inducer (EMMPRIN) were associated with an increased potential of tumor progression, which indicates that these exosomal protein markers are potential therapeutic targets for OC treatment [56]. A microfluidic “ExoSearch chip” has been designed for the non-invasive diagnosis of OC by multiplexed measurement of three exosomal tumor markers: CA125, EpCAM, and CD24 [57]. This new microfluidic technique will be detailed later in this review. Together, these studies indicate CD24 and EpCAM are useful exosome biomarkers for OC diagnosis and prognosis. Table 1 summarizes the EV protein markers in OC diagnosis, prognosis, and therapy for personalized medicine.
The potential EV protein biomarkers identified hold promise for screening for early diagnosis of OC. Due to tumor heterogeneity, instead of a single marker for OC detection, a panel of biomarkers will be more useful and reliable for OC early diagnosis and screening high-risk individuals, such as women with BRCA1 or BRCA2 mutation, OC patients' first-degree relatives, or those with history of early breast cancer. In addition, the panel of biomarkers could be used to distinguish low-grade OC from high-grade cases to predict the prognosis of OC patients and to better select an appropriate treatment. Furthermore, the panel of biomarkers could be used for longitudinal monitoring of therapy response (chemotherapy, immunotherapy, or combination therapy). Several multiplexed analysis platforms have recently been developed for exosome isolation and high-throughput screening of clinical samples [58-60]. On the non-protein side, a novel urine exosomal RNA-based test, ExoDx® Prostate (IntelliScore), is now available for clinical use as a Laboratory Developed Test (LDT) in the US. Using a 3-gene expression panel, this test facilitated the identification of high-grade prostate cancer patients among those with elevated levels of biomarker PSA [61]. These new platforms and assays are useful tools to expedite exosome-related research and the clinical translation of this research into OC detection.
In summary, EV protein biomarkers are an attractive source for providing clinically useful information for the management of OC. Therefore, using sensitive profiling methods like proteomic analysis to discriminate between body fluid EV proteins from OC patients and control patients will be very useful for identifying novel biomarkers for early diagnosis and real-time monitoring of cancer progression. Although the candidate exosomal protein biomarkers in Table 1 have been identified for OC, the role of these EV proteins in OC progression, occurrence, and treatment response is still unclear. Moreover, to further validate the EV biomarkers identified in OC liquid biopsies, these marker candidates should be validated in multicenter clinical trials.
EV isolation and detection
Isolation and detection are two important and indivisible parts of EV studies. It would be ideal if detection could be achieved with raw materials such as blood and urine. However, this is very challenging to achieve with these complex biofluids, as the presence of proteins may cause the actual targets to be hard to detect. Therefore, many isolation methods involve both purification and enrichment, which make the EV concentration higher for better detection. While most studies involve a pre-isolation step before the actual analysis, there have recently been some attempts to combine isolation and analysis into one system, especially with lab-on-chip devices [62, 63].
Physical isolation techniques are used to isolate EVs based on their physical properties like density, surface charge, or size. Conventional bulk methods based on physical isolation include ultracentrifugation, ultrafiltration, and size exclusive chromatography (SEC). Ultracentrifugation is considered the gold standard; however, it is time-consuming and has always been associated with additional issues, such as low recovery and low purity [64]. Recently, new separation technologies have been developed, mostly based on microfluidic platforms utilizing the physical properties of EVs. These new technologies include acoustic, membrane filtration, viscoelastic flow, nanowire trapping, and lateral displacement systems [35, 65]. Whereas physical separation techniques yield higher numbers of EVs without the need for labelling or modification, they usually co-isolate different types of EVs, protein aggregates, and other particle contaminants. For example, protein bound complexes co-exist with EVs when isolated using the polyethylene glycolebased precipitation method [66]. In addition, direct isolation of cell- or tissue-specific exosomes is not possible when using physical separation methods, as they do not target surface biomarkers.
Summary of potential EV protein biomarkers in OC diagnosis, prognosis, and therapy
| Putative biomarker | Source | Clinical sample number | Isolation method | Clinical significance | Reference |
|---|---|---|---|---|---|
| Claudin-4 | OVCAR2, OVCAR3, OVCA420, OVCA433, BG1, Hey and UCI101 cell lines; Plasma samples | HGSCOC patients (63), normal healthy controls (50) | Ultracentrifugation | Diagnosis, Prognosis | [44] |
| MSLN, CEA, MUC16 (CA125), WFDC2 | OVCAR3, OVCAR433, OVCAR5 and SKOV3 cell lines | N/A | Ultracentrifugation | Diagnosis | [45] |
| EpCAM, PCNA, TUBB3, EGFR, APOE, CLDN3, FASN, ERBB2, CD171 | OVCAR3, IGROV1 cell lines | N/A | Ultracentrifugation | Diagnosis | [46] |
| HSP70, MHC-I, CD81 | Ascites samples | OC patients (35) | Ultracentrifugation | Immunotherapy | [47] |
| TGF-β1, MAGE3, MAGE6, | Plasma samples | OC patients (22), Patients with benign tumors (10), normal healthy controls (10) | Ultracentrifugation | Predicting response to therapy, Prognosis | [48] |
| sALCAM | Sera and ascites samples | OC patients (61) | Ultracentrifugation | Diagnosis | [49] |
| CD24, EpCAM | Ascites samples | OC patients (16) | Ultracentrifugation | Predicting response to therapy, Prognosis | [18] |
| CD24, EpCAM | Ascites samples | OC patients (24) | Ultracentrifugation | Diagnosis | [55] |
| L1CAM, CD24, ADAM10, and EMMPRIN | Ascites samples | OC patients (20) | Ultracentrifugation | Therapeutic target | [56] |
| CA125, EpCAM, CD24 | Plasma samples | OC patients (15), normal healthy controls (5) | microfluidic ExoSearch chip | Diagnosis | [57] |
| sE-cad | OVCAR-3, Caov-3, OV-90, TOV21G, and TOV112D cell lines; Ascites samples | OC patients (35), Other cancer patients (15) | Ultracentrifugation | Diagnosis, Prognosis, Therapeutic target | [50] |
Notes: APOE: apolipoprotein E; CLDN3: claudin 3; EGFR: epidermal growth factor receptor; FASN: fatty acid synthase; HGSOC: high-grade serous ovarian cancer; MAGE: melanoma-associated antigen; N/A: not available; PCNA: proliferation cell nuclear antigen; sALCAM: soluble activated leukocyte cell adhesion molecule; sE-cad: soluble E-cadherin; TUBB3: tubulin beta-3 chain.
Unlike the physical isolation techniques, it has been demonstrated that biological- or affinity-based separation techniques are better at isolating specific subtypes of exosomes by targeting surface proteins mainly from the tetraspanin family (e.g., CD9, CD63, and CD81) [67]. These methods are able to directly characterize the captured exosomes or lyse the exosomes for downstream analysis. However, as it is difficult to remove EVs from the binding molecules, these isolated EVs cannot be used for the functional analysis of intact EVs. Magnetic bead kits are commercially available for biomarker-specific exosome isolation (e.g. beads from ThermoFisher Scientific and System Biosciences). However, these approaches are typically expensive and require multiple steps for washing and enrichment. Recently, microfluidic devices, which bring magnetic beads into lab-on-chip systems, have been developed. These lab-on-chip systems combine all necessary steps into one device: sample loading, mixing, incubation, washing, and downstream analysis for proteins and RNAs. The lab-on-chip systems make the clinical translation of EV analysis possible [57, 68].
Analysis of isolated exosomes is typically based on conventional detection approaches to measure the expression of exosomal proteins, such as western blot, enzyme-linked immunosorbent assays (ELISA), and flow cytometry (FCM), [69]. In ELISA, exosomes are immobilized onto a solid phase, followed by labelling with fluorescent- or enzyme-conjugated antibodies (Abs) for optical detection. In FCM, exosomes are bound to Ab-conjugated microbeads and then analyzed by measuring fluorescence of fluorescence-conjugated Abs. With these types of labelling methods, the detection signals such as absorbance (OD) or fluorescence intensity provide only the relative quantity of exosomes.
Because of the small size of EVs, most FCM-based analyses still rely on microbeads to capture EVs. Microbeads enable the analysis of EVs based on biomarkers on their surface. However, existing FCM methods have limited sensitivity and resolution to analyze EVs directly, as it tends to miss or underestimate small vesicles (< 200 nm) due to “Swarm Theory” [70]. Recently, highly sensitive FCMs are under development to distinguish particles as small as 100 nm [71] so that single EVs can be interrogated. Moreover, imaging-based technology has been developed to analyze single EVs in a multiplexed format [72]. Kibria et al. developed a microFCM platform that is capable of assessing the expression of CD47 in single circulating exosomes from breast cancer patients [73]. These new technologies provide opportunities for profiling single exosome and thus, differentiate different exosome subsets.
Physical analysis has been achieved for EVs as well. For physical analysis, pre-isolation to obtain a high purity EV population is particularly important. Particle size distribution and concentration are usually measured by nanoparticle tracking analysis (NTA), FCM, and tunable resistive pulse sensing [74]. NTA is a standard method for characterization and measurement of the concentration of exosomes or vesicles (< 200 nm). In NTA, a light beam illuminates the particles in the solution and the path of each particle is captured to determine its velocity and diffusivity, which will then be used to calculate the particle concentration and size distribution [75, 76]. NTA is a simple and quick analysis. However, the results regarding size and concentration are affected by different parameters during video capture and analysis, such as camera level and threshold. In addition, the linear range for NTA to provide an accurate measurement is around 108-109 particle/mL, which limit its application in measuring samples with low particle concentration. An alternative to NTA, tunable resistive pulse sensing (TRPS), is based on the ionic current change when a particle passes through a size-tunable nanopore. As TRPS measures individual particles, it has less strict requirements on the particle concentration. However, the particle size range that can be measured by TRPS is limited by the size of the nanopore. The nanopore may need to be changed when measuring particles in different size ranges. In addition, TRPS is not suitable for analyzing heterogeneous samples, such as plasma, as the nanopores tend to get clogged with large particles. These two techniques have recently been compared for EVs in clinical cerebrospinal fluids, suggesting that both methods are capable of assessing EVs derived from body fluids and that a multi-platform quantitation will be required to guide clinical studies [77]. Apart from multi-platform quantitation, the addition of pre-isolation procedures of exosomes such as ultracentrifugation and SEC, or precipitation reagents (such as polyethylene glycol) have also been suggested to better assess the size and distribution of EVs. Nevertheless, it has been shown that neither the total number of EVs nor the size of EVs is accurate in differentiating different status of cancers and healthy controls [43]. Thus, these physical parameters need to be combined with molecular information for clinical relevance.
Overall, bulk methods based on counting or labelling have limitations such as being time-consuming, labor-intensive, or insensitive. These limitations are greatly hindering the translation of current exosome analytical methods into clinical settings where real-time monitoring and high-throughput analysis is required for samples with low exosome abundance.
New technologies for EV analysis in ovarian cancer in clinical applications
Various new technologies have recently been developed to improve the sensitivity and throughput for EV analysis, such as microfluidic technology, which has previously been shown to have unique advantages in exosome separation, genomic and proteomic analysis, as well as quantitative biology. It also features low sample volume requirement and simple sample processing, which makes it feasible for point-of-care clinical utilities. The following approaches have been recently developed for OC exosome characterization and shown promise in the clinical setting for OC diagnosis and prognosis.
Profiling of OC patient exosomes with nPLEX. (A) An image showing an nPLEX chip integrated with a multichannel microfluidic cell for independent and parallel analyses. (B) Analysis of ascites-derived exosomes from OC and healthy patients by the nPLEX sensor. (C) Exosomal protein levels of EpCAM and CD24 in ascites samples from patients measured by nPLEX. (D) Longitudinal monitoring of treatment responses in ascites samples from OC patients before and after chemotherapy with nPLEX. Reprinted by permission from Springer Nature: BMC Springer Nature, Nature Biotechnology. Label-free detection and molecular profiling of exosomes with a nano-plasmonic sensor, H. Im et al., copyright 2014.
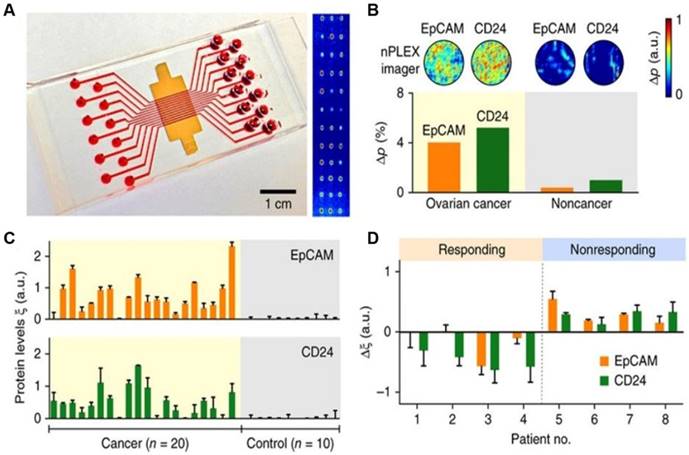
The nano-plasmonic exosome (nPLEX) assay
The nPLEX assay is a label-free, high-throughput approach for quantitative analysis of exosomes [58]. This method is based on transmission surface plasmon resonance to detect proteins on the surface or in the lysates of exosomes. This approach had improved sensitivity compared with conventional modalities and could be portably operated when integrated with miniaturized optics. Im et al. demonstrated that nPLEX could identify OC-derived exosomes from ascites in patients by detecting CD24 and EpCAM, suggesting its potential for diagnostics (Figure 1). Compared to conventional methods, the nPLEX technology has advantages such as high sensitivity, label-free exosome analyses, and continuous real-time monitoring of molecular markers.
The integrated magneto-electrochemical exosome (iMEX)
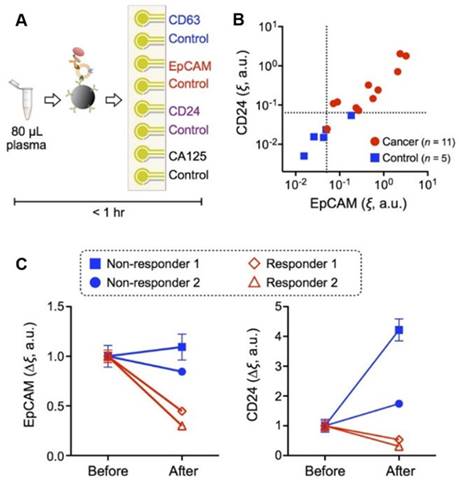
With iMEX assay exosomes were immunomagnetically captured from OC patient samples and assessed through an electrochemical reaction. Combining immunomagnetic enrichment and enzymatic amplification, the approach demonstrates high sensitivity, cell-specific detection, sensor miniaturization, and high-throughput ability for exosome measurements [59].
The iMEX is a portable exosome detection system with the capacity to perform measurements in parallel. The sensor can simultaneously detect multiple protein markers within an hour while consuming only 10 μL of plasma per marker, which outperforms conventional methods in terms of sensitivity and speed. This group found higher levels of EpCAM and CD24 in EVs from OC patients than those from healthy controls, and both metrics showed high correlation (Figure 2). In addition, they also examined iMEX's potential for real-time monitoring EV markers EpCAM and CD24 in plasma of OC patients before and after drug treatment. Their results suggested that the “nonresponding” patients had high expression levels of EpCAM and CD24 compared with the “responding” patients (Figure 2C).
Compared with nPLEX, iMEX has lower sensitivity and throughput, but is less complex and does not require nanofabrication, which makes it an affordable and miniaturized platform for on-site exosome detection.
iMEX for clinical applications. (A) iMEX assay for clinical OC plasma analysis with CD63, EpCAM, CD24, and CA125 markers. (B) EpCAM and CD24 levels analyzed by the iMEX assay were much higher in OC patients. (C) Longitudinal monitoring of drug treatment responses with the iMEX assay. EpCAM and CD24 levels in responders were decreased significantly, but their levels in nonresponders were stable (EpCAM) or increased (CD24) after treatment. Reprinted with permission from S. Jeong et al., Integrated Magneto-Electrochemical Sensor for Exosome Analysis, ACS Nano, 10 (2016) 1802-1809. Copyright (2016) American Chemical Society.
ExoSearch
ExoSearch is a simple microfluidic approach for the rapid preparation of blood plasma exosomes for in situ, multiplexed detection using immunomagnetic beads [57]. ExoSearch chip has been employed for plasma-based diagnosis of OC by multiplexed evaluation of the expression levels of CA-125, EpCAM, and CD24 on the surface of exosomes in 20 OC patients, which demonstrated superior diagnostic power (AUC = 1.0, p = 0.001). The ExoSearch chip has the capability to perform simultaneous and quantitative evaluation of a biomarker panel from the same exosome subpopulation with improved reproducibility. In addition, this assay can acquire different subpopulations of exosomes from a wide range of input volumes (10 μL to 10 mL), largely facilitating the downstream molecular analysis and profiling. However, given the small number of patients recruited in the study, future studies with a large-scale cohort is required to further validate the diagnostic value of the ExoSearch chip.
Later, this group developed another sensitive microfluidic platform based on a new graphene oxide/polydopamine (GO/PDA) nano-interface (nano-IMEX), which could discriminate OC patients from healthy controls by using 2 μL plasma without sample processing [60]. This suggests that this platform could provide a more robust assay to evaluate exosomes for non-invasive detection and precision treatment of OC.
ExoCounter
Kabe Y et al. recently designed a novel device, the ExoCounter, to quantify the number of exosomes in the sera of OC patients. In this system, exosomes can be captured in the groove of an Ab-coated optical disc, labeled with Ab-conjugated magnetic nanobeads, and then counted with an optical disc drive [78].
This team demonstrated that this new approach could detect specific exosomes derived from cell supernatants or human serum without any enrichment procedures. In addition, ExoCounter had high detection sensitivity and linearity compared with conventional detection methods such as ELISA or FCM. Using ExoCounter, the CD9/HER2-positive exosomes were shown to be significantly increased in patients with OC compared with healthy controls and noncancer disease patients. Therefore, this method is very suitable for liquid biopsies of OC exosome biomarkers for diagnosis and progression monitoring.
Microfluidic affinity separation chip
A herringbone-grooved microfluidic device has recently been developed for direct isolation of exosomes using biomarkers CD9 and EpCAM from small volumes of serum of high-grade serous ovarian cancer (HGSOC) patients. Using this device, they found that both total and EpCAM+ exosome numbers increase concurrently with disease progression in HGSOC [79]. This approach can be used to isolate intact and label-free biomarker specific exosomes for predicting HGSOC disease stages, as well as facilitating downstream functional studies.
Summary of the new technologies recently developed in EV detection of OC blood
| Assay | Tested marker | Sample source | Sample volume | Isolation method | Assay time | Level of Detection (LOD) | Advantage | Reference |
|---|---|---|---|---|---|---|---|---|
| nPLEX | CD24, EpCAM | OC patient and normal health ascites samples | n/a | transmission surface plasmon resonance through periodic nanohole arrays | < 30 min | 105 sensing elements | highly sensitive, label-free exosome analyses, portable operation, real-time monitoring of molecular binding. | [58] |
| ExoSearch | CA-125, EpCAM, CD24 | OC patient and normal health plasma samples | 20 μL | multiplexed detection using immunomagnetic beads | ~40 min | n/a | simultaneous quantitative evaluation of multiple markers, a wide range of preparation volumes, higher reproducibility. | [57] |
| nano-IMEX | CD9, CD63, CD81, EpCAM | OC patient and normal health plasma samples | 2 μL without sample processing | based on a new GO/PDA nano-interface | n/a | 103-fold higher than that of bench-top chemiluminescence ELISA | improving the detection sensitivity and dynamic range, small sample volume, effectively suppressing the effects of non-specific exosome adsorption. | [60] |
| iMEX | EpCAM, CD24, CD63, CD125 | OC patient and normal health plasma samples | 10 μL | combining magnetic enrichment and enzymatic amplification | readouts within 1 h; 10 μL/min rate. | detection sensitivity of <105 vesicles | highly sensitive, cell-specific exosome detection, sensor miniaturization, scale-up for high-throughput measurements. | [59] |
| ExoCounter | CD9, CD63, CD147, HER2 | OC patient and normal health serum samples | 0.39 μL | combining the properties of nanobeads with optical disc technology | 150 min | 800-fold higher than that of the ExoTest | The higher detection sensitivity and linearity with this system, high performance in the direct detection of exosomes. | [78] |
| Microfluidic affinity separation chip | CD9, EpCAM | healthy, benign, stage I, and stage IV HGSCOC patient serum samples | < 100 μL | herringbone-grooved microfluidic device | < 20 min for capture and release | n/a | higher yield, higher specificity, inexpensive, rapid, requiring minimal sample volume. | [79] |
Notes: GO: graphene oxide; HGSOC: high-grade serous ovarian cancer; iMEX: integrated magneto-electrochemical exosome; n/a: information not available; nano-IMEX: nano-interfaced microfluidic exosome; nPLEX: nano-plasmonic exosome; PDA: polydopamine.
In comparison with traditional isolation methods, this platform features a rapid (< 20 minutes for capture and release) and cost-effective method with a high yield and specificity and low sample volume requirement (< 100 μL) to distinguish significant differences in HGSOC disease stages, making itself suitable for clinical applications. In addition, as the exosomes captured by the platform are intact and label-free, this method allows further downstream characterization and experimentation, both on and off chip.
In summary, all new techniques recently developed for EV detection hold promise for OC early diagnosis and monitoring cancer progression. However, new OC exosomal markers should continue to be tested using these technologies and a large number of OC samples need to be used for validation studies to confirm their clinical significance.
The current challenges for EV biomarker detection and application in OC
While exosomes show great promise as biomarkers for OC diagnosis and real-time progression monitoring, there are still several limitations that need to be overcome prior to more widespread clinical application. First, standardized and consistent methods need to be established for the isolation and enrichment of tumor-derived exosomes from blood samples, as no clear consensus has been reached regarding the optimal method for isolation and quantification of exosomes [11]. Second, the identified existing exosome protein biomarkers in OC need to be validated in a large set of sample cohorts to find the impact on clinical outcomes such as improved early detection, progression-free survival, or overall survival rates. Some of the new technologies mentioned in the previous sections have follow-up studies that involve clinical sample cohorts. For instance, the nPLEX assay developed by Im et al. [58] has been applied to pancreatic ductal adenocarcinoma in more than 100 clinical samples [80]. Other new technologies need to be validated in clinical studies. Third, the time and costs for exosome processing and analysis should be significantly reduced for clinical application and these non-invasive detection methods should be accurate and fast [42]. Some of the new technologies described above have effectively addressed this challenge, aiming towards developing rapid and cost-effective tests. In addition, many other factors such as stress, hypoxia, tumor types, and growth patterns can influence the secretion of exosomes and should be taken into consideration during processing and analysis [81, 82]. Despite these challenges, exosomes have shown significant potential as future liquid biopsy biomarkers for OC and further research and development is warranted in this area.
Conclusions
Liquid biopsy is nearly ready to offer a robust, yet minimally invasive tool for the diagnosis and comprehensive management of OC. Apart from being able to provide valuable information for diagnosis when the tumor is less accessible, blood-based exosome tests may also allow for real-time monitoring of the tumor evolution and evaluation of treatment efficacy. Although most of the biomarkers available today require prospective validation, the development of non-invasive EV-based liquid biopsy has already emerged and paved the way to improve the early detection, evaluation of response to therapy, prognosis, and outcome in OC patients.
As surrogates of cancer cells, exosomes are promising for precise and personalized cancer diagnosis and real-time monitoring cancer progression. Using a panel of identified exosomal protein markers as a “cancer signature” may provide improved detection in screening OC for early diagnosis.
The EV cargo provides a promising source for the discovery of liquid biopsy biomarkers. The rapid advances in next-generation “omics” and EV capture platforms are the driving approaches for disease stratification, diagnosis, and monitoring. Further advancements in EV isolation methods that potentially prevent overestimation and contamination of EVs may allow the study of discrete EVs from body fluids, hence holding great promise for future diagnostic applications, where isolation and examination of individual EVs are paramount.
More rapid and defined EV isolation procedures have been recently developed. This should enable the seamless integration of EV isolation and analyses into clinical diagnostic pipelines. This is crucial since time-consuming isolation procedures that require expensive, specialized equipment (i.e. ultracentrifugation) are unlikely to be feasible for routine clinical practice. In addition, specific exosomal cargo molecules (proteins, RNAs, lipids, etc.) are likely to be identified/validated in the context of defined clinical questions.
Abbreviations
Abs: antibodies; ADAM10: ADAM metallopeptidase domain 10; APOE: apolipoprotein E; AUC: Area under the ROC curve; CEA: carcinoembryonic antigen; ctDNA: circulating tumour DNA; CTCs: circulating tumor cells; cfmiRNAs: circulating cell-free microRNAs; CLDN3: claudin 3; EGFR: epidermal growth factor receptor; ELISA: enzyme-linked immunosorbent assays; EpCAM: epithelial cell adhesion molecule; EMMPRIN: extracellular matrix metalloproteinase inducer; EV: extracellular vesicle; FASN: fatty acid synthase; FCM: flow cytometry; GO/PDA: graphene oxide/polydopamine; HGSOC: high-grade serous ovarian cancer; HSP70: 70 kilodalton heat shock protein; iMEX: integrated magneto-electrochemical exosome; ISEV: International Society of Extracellular Vesicles; MAGE: melanoma-associated antigen; MHC-I: major histocompatibility complex class I molecule; MSLN: mesothelin; NTA: nanoparticle tracking analysis; nPLEX: nano-plasmonic exosome; OC: ovarian cancer; PCNA: proliferation cell nuclear antigen; sALCAM: soluble activated leukocyte cell adhesion molecule; sE-cad: soluble E-cadherin; SEC: size exclusive chromatography; TUBB3: tubulin beta-3 chain; WFDC2: WAP four-disulfide core domain protein 2.
Acknowledgements
The authors thank funding support by Cancer Institute NSW Early Career Fellowship (Y.Z.); Cancer Research Trust Fund (Cancer Care Centre, St George Hospital); Prostate and Breast Cancer Foundation (PBCF) and University International Postgraduate Award (UIPA), UNSW Sydney (B.P.), and National Natural Science Foundation of China (81572876, 81773087, 81071736 and 30973508) (H.Z.).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424
2. Torre LA, Trabert B, DeSantis CE, Miller KD, Samimi G, Runowicz CD. et al. Ovarian cancer statistics, 2018. CA Cancer J Clin. 2018;68:284-96
3. van Nagell JR Jr, Hoff JT. Transvaginal ultrasonography in ovarian cancer screening: current perspectives. Int J Womens Health. 2013;6:25-33
4. Jacobs IJ, Menon U, Ryan A, Gentry-Maharaj A, Burnell M, Kalsi JK. et al. Ovarian cancer screening and mortality in the UK Collaborative Trial of Ovarian Cancer Screening (UKCTOCS): a randomised controlled trial. Lancet. 2016;387:945-56
5. Nolen BM, Lokshin AE. Multianalyte assay systems in the differential diagnosis of ovarian cancer. Expert Opin Med Diagn. 2012;6:131-8
6. Szekanecz E, Sandor Z, Antal-Szalmas P, Soos L, Lakos G, Besenyei T. et al. Increased production of the soluble tumor-associated antigens CA19-9, CA125, and CA15-3 in rheumatoid arthritis: potential adhesion molecules in synovial inflammation? Ann N Y Acad Sci. 2007;1108:359-71
7. Johnson CC, Kessel B, Riley TL, Ragard LR, Williams CR, Xu JL. et al. The epidemiology of CA-125 in women without evidence of ovarian cancer in the Prostate, Lung, Colorectal and Ovarian Cancer (PLCO) Screening Trial. Gynecol Oncol. 2008;110:383-9
8. Daher RMF, Rosa ESJC, Poli-Neto OB, Candido-Dos-Reis FJ, Nogueira AA. Diagnosis of endometriosis in women with chronic pelvic pain. Clin Exp Obstet Gynecol. 2016;43:512-5
9. Buys SS, Partridge E, Black A, Johnson CC, Lamerato L, Isaacs C. et al. Effect of screening on ovarian cancer mortality: the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Randomized Controlled Trial. JAMA. 2011;305:2295-303
10. Vaidyanathan R, Soon RH, Zhang P, Jiang K, Lim CT. Cancer diagnosis: from tumor to liquid biopsy and beyond. Lab Chip. 2018;19:11-34
11. Witwer KW, Buzas EI, Bemis LT, Bora A, Lasser C, Lotvall J. et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J Extracell Vesicles. 2013:2
12. Kalra H, Drummen GP, Mathivanan S. Focus on Extracellular Vesicles: Introducing the Next Small Big Thing. Int J Mol Sci. 2016;17:170
13. Colombo M, Raposo G, Thery C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255-89
14. Jabalee J, Towle R, Garnis C. The Role of Extracellular Vesicles in Cancer: Cargo, Function, and Therapeutic Implications. Cells. 2018;7:93
15. Lin Y, Dong H, Deng W, Lin W, Li K, Xiong X. et al. Evaluation of salivary exosomal chimeric GOLM1-NAA35 RNA as a potential biomarker in esophageal carcinoma. Clin Cancer Res. 2019 Feb 11. doi: 10.1158/1078-0432.CCR-18-3169. [Epub ahead of print]. PMID: 30745298
16. Ozawa PMM, Alkhilaiwi F, Cavalli IJ, Malheiros D, de Souza Fonseca Ribeiro EM, Cavalli LR. Extracellular vesicles from triple-negative breast cancer cells promote proliferation and drug resistance in non-tumorigenic breast cells. Breast Cancer Res Treat. 2018;172:713-23
17. Tkach M, Thery C. Communication by Extracellular Vesicles: Where We Are and Where We Need to Go. Cell. 2016;164:1226-32
18. Runz S, Keller S, Rupp C, Stoeck A, Issa Y, Koensgen D. et al. Malignant ascites-derived exosomes of ovarian carcinoma patients contain CD24 and EpCAM. Gynecol Oncol. 2007;107:563-71
19. Shender VO, Pavlyukov MS, Ziganshin RH, Arapidi GP, Kovalchuk SI, Anikanov NA. et al. Proteome-metabolome profiling of ovarian cancer ascites reveals novel components involved in intercellular communication. Mol Cell Proteomics. 2014;13:3558-71
20. Vaksman O, Trope C, Davidson B, Reich R. Exosome-derived miRNAs and ovarian carcinoma progression. Carcinogenesis. 2014;35:2113-20
21. Crow J, Atay S, Banskota S, Artale B, Schmitt S, Godwin AK. Exosomes as mediators of platinum resistance in ovarian cancer. Oncotarget. 2017;8:11917-36
22. Alharbi M, Zuniga F, Elfeky O, Guanzon D, Lai A, Rice GE. et al. The potential role of miRNAs and exosomes in chemotherapy in ovarian cancer. Endocr Relat Cancer. 2018;25:R663-r85
23. Mellby LD, Nyberg AP, Johansen JS, Wingren C, Nordestgaard BG, Bojesen SE. et al. Serum Biomarker Signature-Based Liquid Biopsy for Diagnosis of Early-Stage Pancreatic Cancer. J Clin Oncol. 2018;36:2887-94
24. Giannopoulou L, Zavridou M, Kasimir-Bauer S, Lianidou ES. Liquid biopsy in ovarian cancer: the potential of circulating miRNAs and exosomes. Transl Res. 2019;205:77-91
25. Van Berckelaer C, Brouwers AJ, Peeters DJ, Tjalma W, Trinh XB, van Dam PA. Current and future role of circulating tumor cells in patients with epithelial ovarian cancer. Eur J Surg Oncol. 2016;42:1772-9
26. Zhou Q, Li W, Leng B, Zheng W, He Z, Zuo M. et al. Circulating Cell Free DNA as the Diagnostic Marker for Ovarian Cancer: A Systematic Review and Meta-Analysis. PLoS One. 2016;11:e0155495
27. Cheng X, Zhang L, Chen Y, Qing C. Circulating cell-free DNA and circulating tumor cells, the "liquid biopsies" in ovarian cancer. J Ovarian Res. 2017;10:75
28. Anastasiadis A, Zapala L, Cordeiro E, Antoniewicz A, Dimitriadis G, De Reijke T. Complications of prostate biopsy. Expert Rev Anticancer Ther. 2013;13:829-37
29. Bedard PL, Hansen AR, Ratain MJ, Siu LL. Tumour heterogeneity in the clinic. Nature. 2013;501:355-64
30. Parkinson CA, Gale D, Piskorz AM, Biggs H, Hodgkin C, Addley H. et al. Exploratory Analysis of TP53 Mutations in Circulating Tumour DNA as Biomarkers of Treatment Response for Patients with Relapsed High-Grade Serous Ovarian Carcinoma: A Retrospective Study. PLoS Med. 2016;13:e1002198
31. Brock G, Castellanos-Rizaldos E, Hu L, Coticchia C, Skog J. Liquid biopsy for cancer screening, patient stratification and monitoring. Transl Cancer Res. 2015;4:10
32. Balaj L, Lessard R, Dai L, Cho YJ, Pomeroy SL, Breakefield XO. et al. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat Commun. 2011;2:180
33. Cohen JD, Li L, Wang Y, Thoburn C, Afsari B, Danilova L. et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. 2018;359:926-30
34. He J, Tan W, Ma J. Circulating tumor cells and DNA for real-time EGFR detection and monitoring of non-small-cell lung cancer. Future Oncol. 2017;13:787-97
35. Wang W, Luo J, Wang S. Recent Progress in Isolation and Detection of Extracellular Vesicles for Cancer Diagnostics. Adv Healthc Mater. 2018;7:e1800484
36. Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331:1559-64
37. Sedlackova T, Repiska G, Celec P, Szemes T, Minarik G. Fragmentation of DNA affects the accuracy of the DNA quantitation by the commonly used methods. Biol Proced Online. 2013;15:5
38. Ignatiadis M, Lee M, Jeffrey SS. Circulating Tumor Cells and Circulating Tumor DNA: Challenges and Opportunities on the Path to Clinical Utility. Clin Cancer Res. 2015;21:4786-800
39. Boukouris S, Mathivanan S. Exosomes in bodily fluids are a highly stable resource of disease biomarkers. Proteomics Clin Appl. 2015;9:358-67
40. Fan TWM, Zhang X, Wang C, Yang Y, Kang WY, Arnold S. et al. Exosomal lipids for classifying early and late stage non-small cell lung cancer. Anal Chim Acta. 2018;1037:256-64
41. Soung YH, Ford S, Zhang V, Chung J. Exosomes in Cancer Diagnostics. Cancers (Basel). 2017;9:8
42. Nawaz M, Camussi G, Valadi H, Nazarenko I, Ekstrom K, Wang X. et al. The emerging role of extracellular vesicles as biomarkers for urogenital cancers. Nat Rev Urol. 2014;11:688-701
43. Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, Kaye J. et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523:177-82
44. Li J, Sherman-Baust CA, Tsai-Turton M, Bristow RE, Roden RB, Morin PJ. Claudin-containing exosomes in the peripheral circulation of women with ovarian cancer. BMC Cancer. 2009;9:244
45. Sinha A, Ignatchenko V, Ignatchenko A, Mejia-Guerrero S, Kislinger T. In-depth proteomic analyses of ovarian cancer cell line exosomes reveals differential enrichment of functional categories compared to the NCI 60 proteome. Biochem Biophys Res Commun. 2014;445:694-701
46. Liang B, Peng P, Chen S, Li L, Zhang M, Cao D. et al. Characterization and proteomic analysis of ovarian cancer-derived exosomes. J Proteomics. 2013;80:171-82
47. Peng P, Yan Y, Keng S. Exosomes in the ascites of ovarian cancer patients: origin and effects on anti-tumor immunity. Oncol Rep. 2011;25:749-62
48. Szajnik M, Derbis M, Lach M, Patalas P, Michalak M, Drzewiecka H. et al. Exosomes in Plasma of Patients with Ovarian Carcinoma: Potential Biomarkers of Tumor Progression and Response to Therapy. Gynecol Obstet (Sunnyvale). 2013(Suppl 4):3
49. Carbotti G, Orengo AM, Mezzanzanica D, Bagnoli M, Brizzolara A, Emionite L. et al. Activated leukocyte cell adhesion molecule soluble form: a potential biomarker of epithelial ovarian cancer is increased in type II tumors. Int J Cancer. 2013;132:2597-605
50. Tang MKS, Yue PYK, Ip PP, Huang RL, Lai HC, Cheung ANY. et al. Soluble E-cadherin promotes tumor angiogenesis and localizes to exosome surface. Nat Commun. 2018;9:2270
51. Kristiansen G, Denkert C, Schluns K, Dahl E, Pilarsky C, Hauptmann S. CD24 is expressed in ovarian cancer and is a new independent prognostic marker of patient survival. Am J Pathol. 2002;161:1215-21
52. Went PT, Lugli A, Meier S, Bundi M, Mirlacher M, Sauter G. et al. Frequent EpCam protein expression in human carcinomas. Hum Pathol. 2004;35:122-8
53. Kloudova K, Hromadkova H, Partlova S, Brtnicky T, Rob L, Bartunkova J. et al. Expression of tumor antigens on primary ovarian cancer cells compared to established ovarian cancer cell lines. Oncotarget. 2016;7:46120-6
54. Bellone S, Siegel ER, Cocco E, Cargnelutti M, Silasi DA, Azodi M. et al. Overexpression of epithelial cell adhesion molecule in primary, metastatic, and recurrent/chemotherapy-resistant epithelial ovarian cancer: implications for epithelial cell adhesion molecule-specific immunotherapy. Int J Gynecol Cancer. 2009;19:860-6
55. Rupp AK, Rupp C, Keller S, Brase JC, Ehehalt R, Fogel M. et al. Loss of EpCAM expression in breast cancer derived serum exosomes: role of proteolytic cleavage. Gynecol Oncol. 2011;122:437-46
56. Keller S, Konig AK, Marme F, Runz S, Wolterink S, Koensgen D. et al. Systemic presence and tumor-growth promoting effect of ovarian carcinoma released exosomes. Cancer Lett. 2009;278:73-81
57. Zhao Z, Yang Y, Zeng Y, He M. A microfluidic ExoSearch chip for multiplexed exosome detection towards blood-based ovarian cancer diagnosis. Lab Chip. 2016;16:489-96
58. Im H, Shao H, Park YI, Peterson VM, Castro CM, Weissleder R. et al. Label-free detection and molecular profiling of exosomes with a nano-plasmonic sensor. Nat Biotechnol. 2014;32:490-5
59. Jeong S, Park J, Pathania D, Castro CM, Weissleder R, Lee H. Integrated Magneto-Electrochemical Sensor for Exosome Analysis. ACS Nano. 2016;10:1802-9
60. Zhang P, He M, Zeng Y. Ultrasensitive microfluidic analysis of circulating exosomes using a nanostructured graphene oxide/polydopamine coating. Lab Chip. 2016;16:3033-42
61. McKiernan J, Donovan MJ, O'Neill V, Bentink S, Noerholm M, Belzer S. et al. A Novel Urine Exosome Gene Expression Assay to Predict High-grade Prostate Cancer at Initial Biopsy. JAMA oncology. 2016;2:882-9
62. Kanwar SS, Dunlay CJ, Simeone DM, Nagrath S. Microfluidic device (ExoChip) for on-chip isolation, quantification and characterization of circulating exosomes. Lab Chip. 2014;14:1891-900
63. He M, Crow J, Roth M, Zeng Y, Godwin AK. Integrated immunoisolation and protein analysis of circulating exosomes using microfluidic technology. Lab Chip. 2014;14:3773-80
64. Tauro BJ, Greening DW, Mathias RA, Ji H, Mathivanan S, Scott AM. et al. Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes. Methods. 2012;56:293-304
65. Ramirez MI, Amorim MG, Gadelha C, Milic I, Welsh JA, Freitas VM. et al. Technical challenges of working with extracellular vesicles. Nanoscale. 2018;10:881-906
66. Gould SJ, Raposo G. As we wait: coping with an imperfect nomenclature for extracellular vesicles. J Extracell Vesicles. 2013;2:20389
67. Bu H, He D, He X, Wang K. Exosomes: Isolation, Analysis, and Applications in Cancer Detection and Therapy. Chembiochem: a European journal of chemical biology. 2019;20:451-61
68. Shao H, Chung J, Lee K, Balaj L, Min C, Carter BS. et al. Chip-based analysis of exosomal mRNA mediating drug resistance in glioblastoma. Nat Commun. 2015;6:6999
69. Liu C, Yang Y, Wu Y. Recent Advances in Exosomal Protein Detection Via Liquid Biopsy Biosensors for Cancer Screening, Diagnosis, and Prognosis. AAPS J. 2018;20:41
70. van der Pol E, van Gemert MJ, Sturk A, Nieuwland R, van Leeuwen TG. Single vs. swarm detection of microparticles and exosomes by flow cytometry. J Thromb Haemost. 2012;10:919-30
71. Tian Y, Ma L, Gong M, Su G, Zhu S, Zhang W. et al. Protein Profiling and Sizing of Extracellular Vesicles from Colorectal Cancer Patients via Flow Cytometry. ACS Nano. 2018;12:671-80
72. Lee K, Fraser K, Ghaddar B, Yang K, Kim E, Balaj L. et al. Multiplexed Profiling of Single Extracellular Vesicles. ACS Nano. 2018;12:494-503
73. Kibria G, Ramos EK, Lee KE, Bedoyan S, Huang S, Samaeekia R. et al. A rapid, automated surface protein profiling of single circulating exosomes in human blood. Sci Rep. 2016;6:36502
74. Momen-Heravi F, Balaj L, Alian S, Tigges J, Toxavidis V, Ericsson M. et al. Alternative methods for characterization of extracellular vesicles. Front Physiol. 2012;3:354
75. Gardiner C, Ferreira YJ, Dragovic RA, Redman CW, Sargent IL. Extracellular vesicle sizing and enumeration by nanoparticle tracking analysis. J Extracell Vesicles. 2013;2:19671
76. Ramteke A, Ting H, Agarwal C, Mateen S, Somasagara R, Hussain A. et al. Exosomes secreted under hypoxia enhance invasiveness and stemness of prostate cancer cells by targeting adherens junction molecules. Mol Carcinog. 2015;54:554-65
77. Akers JC, Ramakrishnan V, Nolan JP, Duggan E, Fu CC, Hochberg FH. et al. Comparative Analysis of Technologies for Quantifying Extracellular Vesicles (EVs) in Clinical Cerebrospinal Fluids (CSF). PLoS One. 2016;11:e0149866
78. Kabe Y, Suematsu M, Sakamoto S, Hirai M, Koike I, Hishiki T. et al. Development of a Highly Sensitive Device for Counting the Number of Disease-Specific Exosomes in Human Sera. Clin Chem. 2018;64:1463-73
79. Hisey CL, Dorayappan KDP, Cohn DE, Selvendiran K, Hansford DJ. Microfluidic affinity separation chip for selective capture and release of label-free ovarian cancer exosomes. Lab Chip. 2018;18:3144-53
80. Yang KS, Im H, Hong S, Pergolini I, Del Castillo AF, Wang R. et al. Multiparametric plasma EV profiling facilitates diagnosis of pancreatic malignancy. Sci Transl Med. 2017;9:aal3226
81. Shao C, Yang F, Miao S, Liu W, Wang C, Shu Y. et al. Role of hypoxia-induced exosomes in tumor biology. Mol Cancer. 2018;17:120
82. O'Neill CP, Gilligan KE, Dwyer RM. Role of Extracellular Vesicles (EVs) in Cell Stress Response and Resistance to Cancer Therapy. Cancers (Basel). 2019;11:136
Author contact
![]() Corresponding authors: Level 2, Research and Education Centre, St George Hospital, 4-10 South St, Kogarah, NSW 2217, Australia. Email addresses: y.liedu.au (Y. Li). or haozhangedu.cn (H. Zhang)
Corresponding authors: Level 2, Research and Education Centre, St George Hospital, 4-10 South St, Kogarah, NSW 2217, Australia. Email addresses: y.liedu.au (Y. Li). or haozhangedu.cn (H. Zhang)
 Global reach, higher impact
Global reach, higher impact