13.3
Impact Factor
Theranostics 2019; 9(13):3840-3852. doi:10.7150/thno.33370 This issue Cite
Research Paper
Suppression of YAP/TAZ-Notch1-NICD axis by bromodomain and extraterminal protein inhibition impairs liver regeneration
1. Department of Surgery, Second Affiliated Hospital of School of Medicine, Zhejiang University, Hangzhou 310058, China
2. Department of Biochemistry, and Department of Cardiology of Second Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou 310058, China.
3. College of Life Sciences and Medicine, Zhejiang Sci-Tech University, Hangzhou 310018, China
4. Department of Thyroid and Breast Surgery, Tongde Hospital of Zhejiang Province, Hangzhou, 310000, China
5. Department of general surgery, Zhejiang Hospital, Hangzhou, 310000, China.
* Equal contribution
Received 2019-1-21; Accepted 2019-5-6; Published 2019-5-31
Abstract
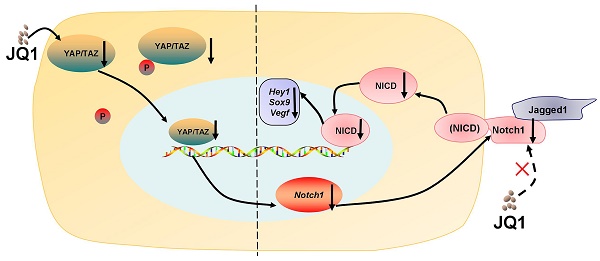
Background and aims: Biological mechanisms that control liver regeneration remain poorly defined. However, these mechanisms are remarkable issues in the clinic that affect management of hepatic loss caused by liver surgery, traumatic injury, chronic infection, or liver poisoning. Increasing evidence has shown that various growth factors, cytokines, and metabolic signaling pathways affect the liver regenerative process. Our aim is to study the effect of bromodomain and extraterminal (BET) protein inhibition on liver regeneration and its mechanism.
Methods: We studied the role of BET protein inhibitor, JQ1, in liver regeneration in a mouse model after 70% partial hepatectomy (PH). We evaluated yes-associated protein (YAP)/transcriptional co-activator with PDZ-binding motif (TAZ) and Notch signaling pathways, which were affected by BET protein inhibitor in mouse hepatic tissues and primary hepatocytes in vivo and AML12 cell lines in vitro. We evaluated the relationship of YAP/TAZ and Notch signaling pathway using YAP/TAZ pathway inhibitor in liver regeneration in vivo. Moreover, we analyzed the relationship of YAP/TAZ and Notch signaling pathways via overexpression or RNA silencing of Yap in AML12 cells. Furthermore, we used Yap overexpression mouse model to examine whether it can rescue liver regeneration damage caused by inhibition of BET proteins.
Results: In this study, we report that BET protein inhibitor JQ1 molecule impairs the early phase of liver regeneration in a mouse model after 70% PH. Mechanistically, YAP/TAZ and Notch1-NICD pathways were suppressed by BET protein inhibitor in mouse hepatic tissues and primary hepatocytes in vivo and mouse AML12 cell lines in vitro. By using YAP/TAZ pathway inhibitor, we confirmed that the liver regeneration and the activation of Notch pathway were impaired by the inhibition of YAP/TAZ pathway in vivo. Furthermore, the study showed that Yap knockdown by shRNA in normal mouse hepatic cell line downregulated Notch1 signal transduction, whereas Yap overexpression promoted Notch1-NICD signals. Specific overexpression of Yap in mouse liver could rescue the effect of BET protein inhibition on liver regeneration injury.
Conclusion: These results revealed the crucial role of the YAP/TAZ-Notch1-NICD axis in liver regeneration. Therefore, BET protein inhibitors must be used in caution in the treatment of hepatic diseases by reason of its suppressive roles in liver regeneration.
Keywords: liver regeneration, BET proteins, YAP/TAZ, Notch signaling pathway
Introduction
The liver is capable of strong regeneration. Many treatments for liver diseases, such as liver cancer, hepatic traumatic injury, chronic liver infection and liver poisoning are regulated by efficient liver regeneration. Moreover, liver regeneration is de facto a complex process, wherein multiple mechanical events and various signaling molecules are initiated at the early stage of liver injury to ensure an entire process of liver regeneration [1]. Therefore, determining the underlying mechanism of liver regeneration is crucial particularly for clinical scenarios.
Recently, bromodomain and extraterminal domain (BET) protein inhibitors have been considered as potential therapeutic drugs for the treatment of inflammatory diseases, metabolic disorders, cancer and autoimmune diseases [2]. The BET protein family in mammals includes BRD2, BRD3, BRD4 and BRDT. These proteins share conserved bromodomains, which bind to acetylated chromatin and functions as transcriptional regulators. In particular, BRD4 is a mediator component that recruits transcriptional regulatory complex to regulate the expression of a group of proteins, such as c-Myc [3]. (S)-tert-butyl2-(4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl)acetate (JQ1) has been shown to act specifically against BET proteins, and it competitively binds to BRD4 with high affinity as a first-in-class selective and potent BRD4 inhibitor [4-6]. Furthermore, JQ1 can displace BET bromodomains from chromatin and disturb BRD4 function, thereby leading to the promotion of apoptosis and cell cycle arrest [2, 7].
Previous studies have shown that BET proteins and their inhibitors are closely involved in hepatic cancer. Compared with normal hepatocyte, BRD4 is overexpressed in cell lines of hepatocellular carcinoma (HCC). Knockdown of BRD4 expression impairs proliferation, invasion and migration of HCC cell lines [8, 9]. Treatment with JQ1 has been shown to reduce cell growth in HCC cell lines and a xenograft tumor model [10]. Furthermore, JQ1 protects and reverses liver fibrotic responses in carbon tetrachloride-induced fibrosis in mouse model [11]. These studies imply that BET protein inhibitor might be a potential treatment option for liver diseases.
Hippo/Yes-associated protein (YAP) signaling pathway is a critical regulator of liver size [12, 13]. Moreover, YAP and its transcriptional co-activator with PDZ-binding motif (TAZ) have been identified as key regulators of cell proliferation and organ size [14]. Non-phosphorylated YAP and TAZ translocate into the nucleus as the activation of Hippo pathway is reduced, and bind with transcription factors, such as TEAD family proteins, and initiate their downstream gene expressions. YAP/TAZ target genes are closely associated with cell proliferation, anti-apoptosis and amplification of progenitor/stem cells [15]. Activation of YAP/TAZ in liver has been shown to result in liver injury and liver cancer formation in mouse models [12, 13, 16]. In addition, YAP/TAZ activation has been observed in bile acid-induced liver injury and nonalcoholic hepatosteatosis [17-19]. These data suggest that YAP/TAZ activation is a common event in liver disease.
A recent study has reported that BET proteins are crucial during liver regeneration in a zebrafish model [20], but specific mechanisms remain unclear. Our study showed that BET protein inhibitor JQ1 inhibited liver regeneration in a mouse model after 70% partial hepatectomy (PH) and culture cells by the inhibiting YAP/TAZ-Notch1-NICD axis. Our data provide insight into the function of YAP/TAZ-Notch1-NICD axis in liver regeneration and the risk of BET protein inhibitors, such as JQ1, in the treatment of liver diseases.
Methods
Chemical Materials
JQ1 was purchased from Selleck Chemicals Co. (Texas, USA), and dissolved in dimethylsulfoxide (DMSO) to a concentration of 50 mg/mL and subsequently diluted to a working concentration of 50 mg/kg in a solution of 10% hydroxypropyl β-cyclodextrin in sterile water (vehicle solution). JQ1 was dissolved in DMSO to a concentration of 500 nM for in vitro experiments. The YAP/TAZ signaling pathway inhibitor Verteporfin was purchased from Selleck Chemicals Co. (Texas, USA), and dissolved in DMSO to a concentration of 100 mg/mL. The working solution was prepared at 10 mg/mL in PBS.
Animal Studies
Male C57BL6/J mice (six-week-old) were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China). The animal studies were performed under the guidelines of the Institutional Animal Care and Use Committee of Zhejiang University. Animals were maintained pathogen-free under constant humidity and temperature in a 12 hours dark/ 12 hours light cycle.
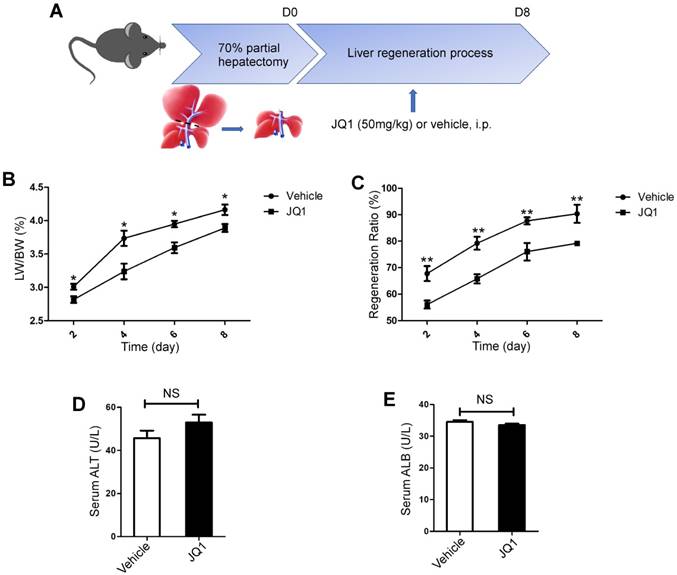
All surgeries were performed by one person under Isoflurane (Sigma, USA) anesthesia. 70% PH was carried out according to the method described by Higgins and Anderson [21]. In this model, two thirds of the liver (median and left lobes) was removed. One hour after surgery, animals were intraperitoneally (i.p.) injected with JQ1 (50 mg/kg body weight) or vehicle solution and daily intraperitoneally administered for consecutive five days at the same concentration of JQ1 after 70% PH. Mice were respectively sacrificed at day 2, 4, 6, and 8 after 70% PH for further analysis (Figure. 1A). The wet liver remnant weight and the total body weight of mice were utilized as hepatic regenerative index to evaluate progress of liver regeneration. One hour before liver harvest, the mice were intraperitoneally injected with 50 mg/kg 5-bromo-2'-deoxyuridine (BrdU) (Sigma, USA). A concentration of 5 mg/mL BrdU was dissolved in phosphate-buffered saline (PBS). At the indicated time-points, the mice were anesthetized for blood collection and livers harvest. Liver weight and body weight were measured, and liver tissues were collected in liquid nitrogen or fixed in 4% paraformalin. Serum concentrations of alanine aminotransferase (ALT) and albumin (ALB) were measured.
For in vivo YAP inhibition, six-week-old male C57BL6/J mice were performed 70% PH. One hour after surgery, mice were intraperitoneally injected with verteporfin (20 mg/kg body weight) or vehicle solution and every other day intraperitoneally administered the same concentration of JQ1 or vehicle solution.
For Yap overexpression mice model, six-week-old male C57BL/6J mice were administered AAV-YAP (1×1011 v.g.) (Vigene biosciences) in normal saline (intraperitoneal injection) for 4 weeks. Mice in control group were administered AAV-Vector (1×1011 v.g.) (Vigene biosciences) in normal saline (intraperitoneal injection) for 4 weeks. Then, mice were performed 70% PH. The JQ1 treatment after 70% PH was followed the method described above.
Statistical analysis
GraphPad Prism 7.0.4 software (GraphPad Software, La Jolla, CA, USA) was used for experimental data analysis. All experiments were independently repeated at least three times with triplicate samples. Statistical analysis was performed using the student T-test. Statistical significance was determined when p<0.05 (two-tailed). Values are expressed as the mean ± standard error of the mean (SEM).
Results
BET protein inhibition significantly suppressed liver regeneration after partial hepatectomy
To test whether BET protein inhibition could influence liver regeneration, JQ1, which is a specific inhibitor of BET proteins, was utilized in a mouse model of 70% PH. JQ1 treatment group manifested a significantly lower liver weight to body weight ratio (LW/BW) compared with the control group, which was injected with the vehicle solution at days 2, 4, 6 and 8 (day 2, p=0.0248; day 4, p=0.0152; day 6, p=0.0178; day 8, p=0.0392. Figure 1B). However, the peak of LW/BW difference was found at day 4 after 70% PH. To comprehensively evaluate the effect of BET protein inhibition on liver regeneration, we further utilized the ratio of liver weight post-resection to the total liver weight before resection, which was considered as the liver regenerative ratio between these two groups. Our findings showed that JQ1 administration resulted in significantly lower liver regenerative ratio compared with the control group at days 2, 4, 6, and 8 after PH (day 2, p=0.0059; day 4, p=0.0010; day 6, p=0.0079; day 8, p=0.0065. Figure 1C), thereby indicating that BET protein inhibition could evidently impair liver regeneration at the early stage. To further test whether BET protein inhibition could affect liver function, peripheral level of alanine aminotransferase (ALT) and albumin (ALB) was measured at day 2 after 70% PH, and no significant difference was observed (p>0.05, Figure 1D -E).
BET protein inhibition prevented hepatocyte proliferation in vivo
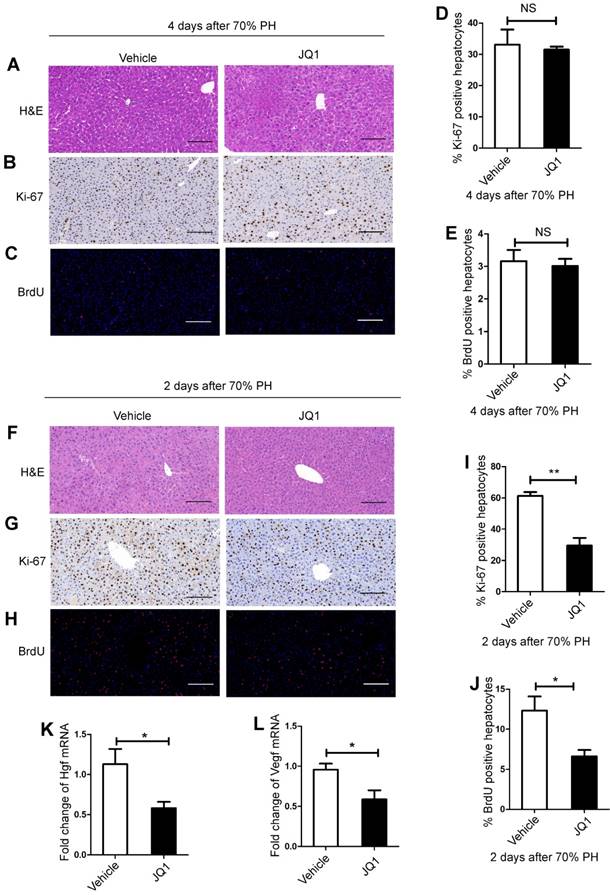
To investigate the underlying mechanism of the suppressive effect of JQ1 on liver regeneration, we quantitatively evaluated hepatic cellular proliferation after 70% PH. As liver regenerative difference peaked at day 4, histological HE staining and Ki-67 and BrdU staining were performed for those liver tissues harvested at day 4 after 70% PH. However, no statistical difference was observed between the two groups (Figure 2A-E). We speculated that earlier events at earlier time points might remarkably cause subsequent significant differences of hepatic cellular proliferation. Therefore, Ki-67 and BrdU staining were performed for those liver tissues harvested at day 2 after 70% PH. We found that the levels of Ki-67 and BrdU-positive hepatocytes in the JQ1 treatment group were significantly lower than those of control group at day 2 after 70% PH (Ki-67, p=0.0042; BrdU, p=0.0417. Figure 2F-J). These results illustrated that the effect of JQ1 on hepatocyte proliferation occurs at the early stage of the liver regeneration process. We examined the mRNA expression levels of hepatocyte growth (Hgf) and vascular endothelial growth factors (Vegf), which are two crucial factors in liver regeneration. Using liver tissues at day 2 after 70% PH, the expression levels of Hgf and Vegf were significantly lower in the JQ1 treatment group compared with those of the control group (Hgf, p=0.0183; Vegf, p=0.0283. Figure 2K-L). These data suggested that BET protein inhibition significantly impaired hepatocyte proliferation in vivo following 70% PH.
BET protein inhibition down-regulated YAP/TAZ expression during liver regeneration in vivo
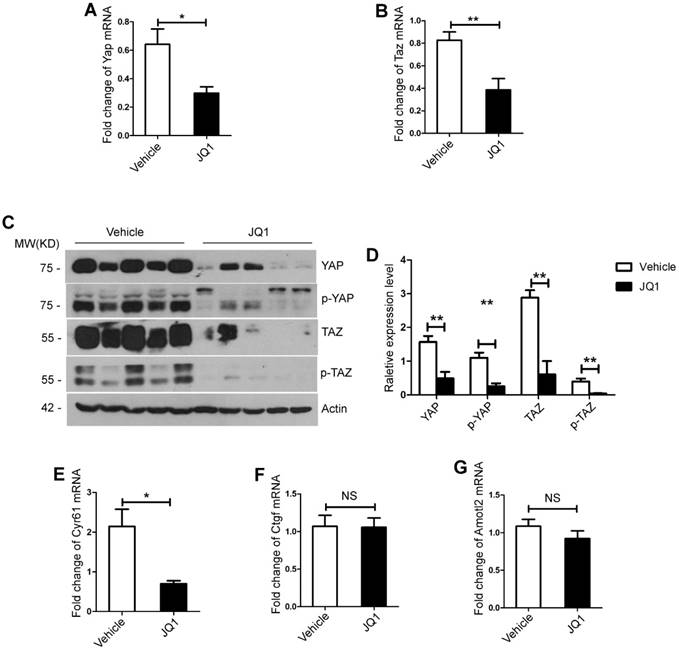
Previous studies have indicated that YAP/TAZ overexpression promotes proliferation of hepatocytes [22, 23]. We further tested whether the negative impact of BET protein inhibition on liver regeneration was mediated by YAP/TAZ activation. First, RT-PCR analysis exhibited that mRNA expression levels of Yap and Taz in the liver tissues and primary hepatocytes were significantly lower in the JQ1 treatment group compared with that of the control group at day 2 after 70% PH (the liver tissues, Yap, p=0.0179; Taz, p=0.0253, Figure S2A-B. the primary hepatocytes, Yap, p=0.0184; Taz, p=0.0072, Figure 3A-B). Afterwards, Western blot analysis was conducted on the liver tissues and primary hepatocytes at the time point that displayed protein levels of total YAP and TAZ, and their inactive forms of phosphorylated YAP (p-YAP) and phosphorylated TAZ (p-TAZ) were all significantly lower in the JQ1 treatment group than those of the control group (the liver tissues, Figure S2C; the primary hepatocytes, Figure 3C-D). Given our finding of decreased YAP/TAZ expression inhibited by JQ1 at levels of mRNA and protein, we further examined the mRNA levels of target genes of YAP/TAZ, Cyr61, Amotl2 and Ctgf in the liver tissues and primary hepatocytes respectively. Our results indicated that the expression of Cyr61 had a statistical difference between the two groups (the liver tissues, p=0.0307, Figure S2E; the primary hepatocytes, p=0.0106, Figure 3E), and no significant difference was observed in the expression levels of Amotl2 and Ctgf (p>0.05) (the liver tissues, Figure S2F-G; the primary hepatocytes, Figure 3F-G). These findings implied that YAP/TAZ activation was specifically impaired by BET protein inhibition after 70% PH in vivo.
BET proteins inhibition significantly suppressed liver regeneration after partial hepatectomy. (A) Schematic diagram of this present experimental study. (B) Liver regenerative index of LW/BW was analyzed for both experimental group (JQ1 intraperitoneal injection at the dose of 50 mg/kg, n=5) and control group (vehicle solution, n=5) after PH. (C) The liver regenerative ratio of JQ1 treatment group (n=5) and control group (n=5) after PH. (D and E) Peripheral level of ALT and albumin was measured at day 2 after PH. NS P≥0.05, *P<0.05, **P<0.01. Data are expressed as mean ± S.E.M.
BET proteins inhibition down-regulated the Notch signaling pathway in vivo
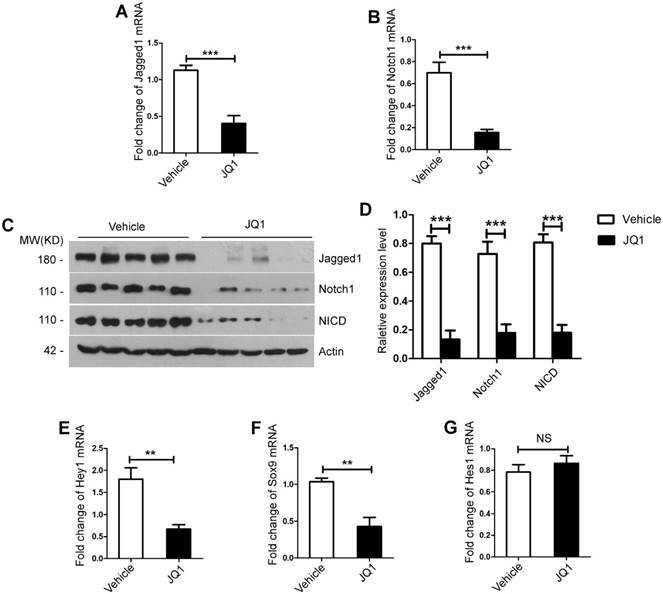
Notch signaling pathway is vital during liver regeneration. Notch1 and Jagged1 are crucial in cell proliferation during liver regeneration process [24]. We further investigated whether Notch signaling pathway was affected by BET protein inhibition during liver regeneration. RT-PCR analysis showed that mRNA expression levels of Notch signaling pathway ligand Jagged1 and receptor Notch1 in the liver tissues and primary hepatocytes were significantly lower in the JQ1 treatment group compared with those of the control group at day 2 (the liver tissues, Jagged1, p=0.0410; Notch1, p=0.0155, Figure S3A-B. the primary hepatocytes, Jagged1, p=0.0004; Notch1, p=0.0006, Figure 4A-B).
BET proteins inhibition prevented hepatocyte proliferation in vivo. (A) HE staining of liver tissues from JQ1 treatment group and control group mice at day 4 after 70% PH (200× original magnification). Scale bars, 200μm. (B) Ki-67 staining of liver tissues from JQ1 treatment group and control group mice at day 4 after 70% PH (200× original magnification). Scale bars, 200μm. (C) BrdU staining of liver tissues from JQ1 treatment group and control group mice at day 4 after 70% PH (200× original magnification). Scale bars, 200μm. (D) Ki-67-positive hepatocytes count at day 4 after 70% PH. (E) BrdU-positive hepatocytes count at day 4 after 70% PH. NS≥0.05. (F) HE staining of liver tissues from JQ1 treatment group and control group mice at day 2 after 70% PH (200× original magnification). Scale bars, 200μm. (G) Ki-67 staining of liver tissues from JQ1 treatment group and control group mice at day 2 after 70% PH (200× original magnification). Scale bars, 200μm. (H) BrdU staining of liver tissues from JQ1 treatment group and control group mice at day 2 after 70% PH (200× original magnification). Scale bars, 200μm. (I) Ki-67-positive hepatocytes count at day 2 after 70% PH. (J) BrdU-positive hepatocytes count at day 2 after 70% PH. (K and L) Fold change in Hgf and Vegf mRNA expression in liver tissues from JQ1 treatment group and control group mice at day 2 after 70% PH. NS P≥0.05, *P<0.05, **P<0.01. Data are expressed as mean ± S.E.M.
BET proteins inhibition down-regulated YAP/TAZ expression during liver regeneration in vivo. (A) Fold change in Yap mRNA expression in primary hepatocytes derived from JQ1 treatment group and control group mice at day 2 after 70% PH. (B) Fold change in Taz mRNA expression in primary hepatocytes derived from JQ1 treatment group and control group mice at day 2 after 70% PH. (C-D) Western blot analysis of YAP, p-YAP, TAZ and p-TAZ protein levels in primary hepatocytes derived from JQ1 treatment group and control group mice at day 2 after 70% PH. (E-G) Fold change in YAP/TAZ target genes, Cyr61, AmotL2, and Ctgf mRNA expression in primary hepatocytes derived from JQ1 treatment group and control group mice. NS P≥0.05, *P<0.05, **P<0.01. Data are expressed as mean ± S.E.M.
Moreover, Western blot analysis showed that the protein levels of Jagged1 and Notch1 and the nucleus translocation of NICD in both the liver tissues and primary hepatocytes significantly decreased in the JQ1 treatment group at day 2 (the liver tissues, Figure S3C-D; the primary hepatocytes, Figure 4C-D). Subsequently, we examined the expression of target genes of Notch signaling pathways, namely, Hey1, Hes1 and Sox9 in both the liver tissues and primary hepatocytes. The results showed that mRNA expression levels of Hey1 and Sox9 significantly downregulated by JQ1 treatment (Hey1, p=0.0004; Sox9, p=0.0064, Figure S3E-F. Hey1, p=0.0018; Sox9, p=0.0019, Figure 4E-F), whereas no significant difference was observed in the expression of has1 gene (the liver tissues, Figure S3G; the primary hepatocytes, Figure 4G). Overall, these findings confirmed that BET protein inhibition impaired the Notch signaling pathway during liver regeneration after 70% PH in vivo.
YAP/TAZ inhibition significantly suppressed liver regeneration after partial hepatectomy and down-regulated the Notch signaling pathway in vivo
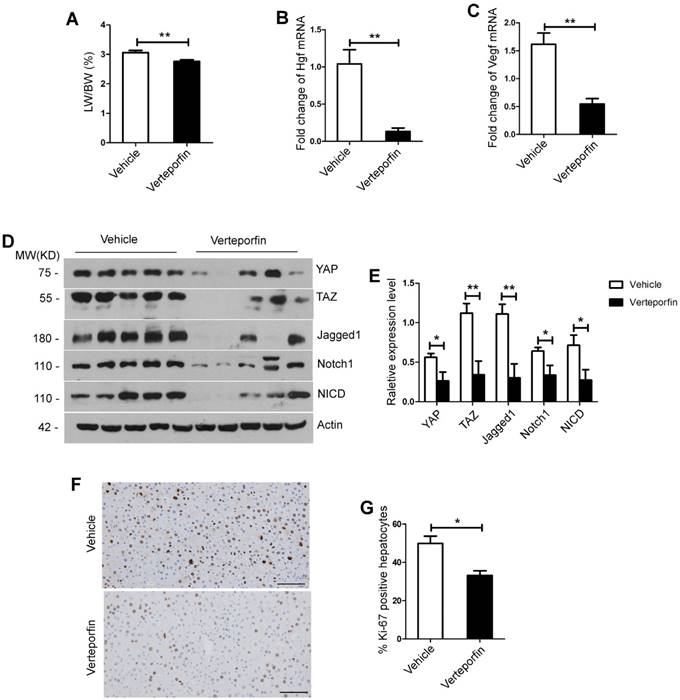
To further validate the effects of inhibition of YAP/TAZ signaling pathway on liver regeneration, we performed YAP/TAZ pathway inhibitor (verteporfin) after 70% PH in mice. The results showed that the verteporfin treatment group exhibited a significant decrease in LW/BW ratio compared with its counterpart control group, which was injected with the vehicle solution at day 2 and day 4 after 70% PH (day 2, p=0.0095, Figure 5A; day 4, p=0.0039, Figure S4A). Using primary hepatocytes at day 2 after 70% PH, the expression levels of Hgf and Vegf were significantly lower in the verteporfin treatment group compared with those of the control group (Hgf, p=0.0017; Vegf, p=0.0014, Figure 5B-C). Afterwards, Western blot analysis was performed on the primary hepatocytes at day 2 after 70% PH and the liver tissues at day 4 after 70% PH showed that the protein levels of YAP, TAZ, Jagged1, Notch1 and NICD were all significantly lower in the verteporfin treatment group compared with those of the control group (the primary hepatocytes, Figure 5D-E; the liver samples, Figure S4B-C). Moreover, the results showed that the numbers of Ki-67 positive hepatocytes in the verteporfin treatment group were significantly lower compared with those of the control group at day 2 and day 4 after 70% PH (day 2, p=0.0203, Figure 5F-G; day 4, p=0.0032, Figure S4D-E). These data suggest that YAP/TAZ pathway inhibition significantly impaired hepatocyte proliferation after 70% PH, and the activity of Notch signaling pathway was downregulated following the inhibition of YAP/TAZ signaling pathway in vivo.
BET proteins inhibition down-regulated the Notch signaling pathway in vivo. (A) Fold change in Notch1 mRNA expression in primary hepatocytes derived from JQ1 treatment group and control group mice at day 2 after 70% PH. (B) Fold change in Jagged1 mRNA expression in primary hepatocytes derived from JQ1 treatment group and control group mice at day 2 after 70% PH. (C-D) Western blot analysis of Notch1, Jagged1 and NICD protein levels in primary hepatocytes derived from JQ1 treatment group and control group mice at day 2 after 70% PH. (E-G) Fold change in Notch signaling pathway target genes, Hey1, Sox9 and Hes1 mRNA expression in primary hepatocytes derived from JQ1 treatment group and control group mice at day 2 after 70% PH. NS P≥0.05, *P<0.05, **P<0.01, ***P<0.001. Data are expressed as mean ± S.E.M.
Both YAP/TAZ expression and Notch signaling pathway were impaired by BET protein inhibition in vitro
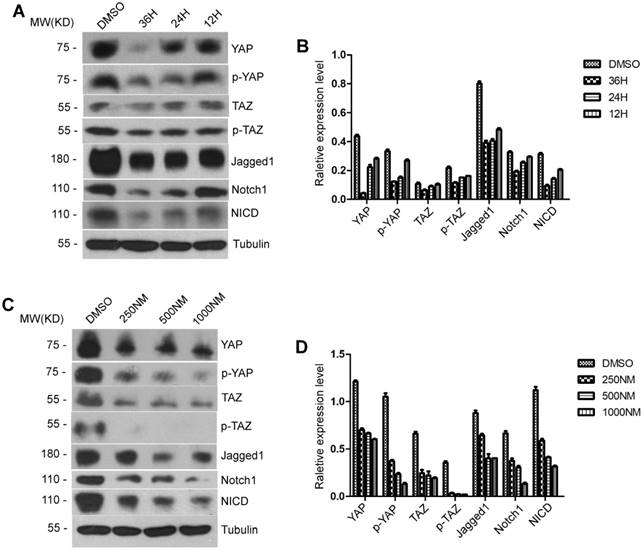
To test the effect of BET protein inhibition on YAP/TAZ and Notch signaling pathways in mouse hepatic cell lines, AML12 cells were treated with 500 ng/ml of JQ1 for 12, 24 and 36 hours. Western blot analysis showed a significant decrease in the extension time of JQ1 treatment in the total YAP, TAZ, p-YAP and p-TAZ proteins. By contrast, significant decreases in the protein levels of Jagged1, Notch1 and NICD were found with the extension time of JQ1 treatment (Figure 6A-B). Afterwards, we performed experiments on a concentration gradient of JQ1 in AML12 cells for 24 hours. AML12 cells were treated with JQ1 at concentrations of 250, 500 and 1000 nM for 24 hours. Significant reductions in the protein expression levels of total YAP, TAZ, p-YAP and p-TAZ were observed as the concentration of JQ1 treatment was increased. By contrast, a significant difference was observed in the protein levels of Jagged1, Notch1 and NICD as the JQ1 treatment concentration was increased (Figure 6C-D). These findings indicated that BET protein inhibition suppressed the activation of YAP/TAZ and Notch signaling pathways in normal hepatocytes. Furthermore, this suppressive effect of BET protein inhibition was time- and dose-dependent.
YAP/TAZ inhibition significantly suppressed liver regeneration after partial hepatectomy and down-regulated the Notch signaling pathway in vivo. (A) Liver regenerative index of LW/BW was analyzed for both experimental group (Verteporfin intraperitoneal injection at the dose of 50 mg/kg, n=5) and control group (vehicle solution, n=5) at day 2 after 70% PH. (B-C) Fold change in Hgf and Vegf mRNA expression in primary hepatocytes derived from Verteporfin treatment group and control group mice at day 2 after 70% PH. (D-E) Western blot analysis of YAP, TAZ, Notch1, Jagged1 and NICD protein levels in primary hepatocytes derived from Verteporfin treatment group and control group mice at day 2 after 70% PH. (F) Ki-67 staining of liver tissues from Verteporfin treatment group and control group mice at day 2 after 70% PH (200× original magnification). Scale bars, 200μm. (G) Ki-67-positive cell count at day 2 after 70% PH. *P<0.05, **P<0.01. Data are expressed as mean ± S.E.M.
The Notch receptor Notch1 was a Functional YAP Target
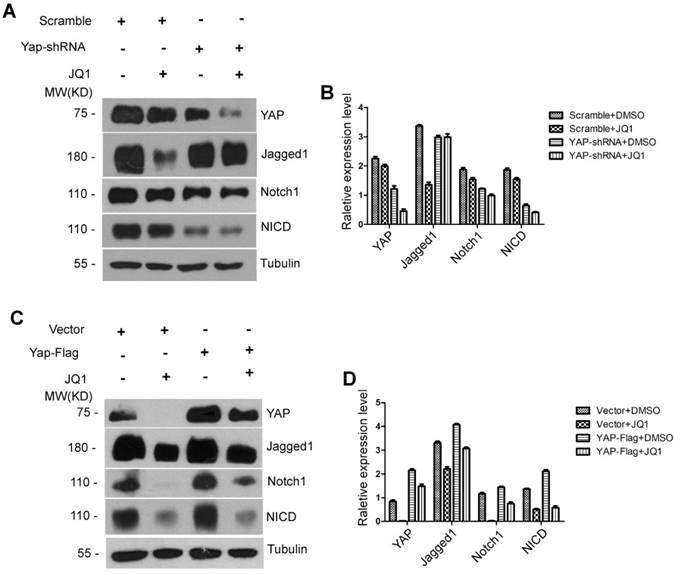
In tumor research models, the YAP/TAZ signaling pathway interacts with the Notch signaling pathway, which is a functional target of YAP/TAZ [25-27]. However, no studies have been conducted on liver regeneration to investigate whether the Notch signaling pathway is downstream of YAP/TAZ or whether BET protein inhibition is a direct effect on the expression of the Notch signaling pathway. We hypothesized that the inhibitory effect of JQ1 on liver regeneration is obtained by YAP/TAZ regulation of the Notch signaling pathway (YAP/TAZ-Notch axis). To prove this hypothesis, we constructed AML12 Yap shRNA knockdown and AML12 Yap flag cell lines. First, AML12 Yap shRNA knockdown and AML12 scrambled shRNA cells were treated with JQ1 or DMSO for 24 hours, respectively. The results showed that the protein level of YAP significantly decreased in AML12 Yap shRNA knockdown cells compared with that of AML12 scrambled shRNA cells irrespective of cells treated with JQ1 or DMSO. The protein levels of Notch1 and NICD were concurrently altered with the protein level of YAP. However, the level of Jagged1 did not change with the Yap expression (Figure 7A-B). Accordingly, we implemented a remediation assay. AML12 Yap flag and AML12 vector cells were treated with JQ1 or DMSO for 24 hours, respectively. We found that the protein level of YAP was significantly increased in AML12 Yap flag cells compared with that of AML12 vector cells regardless of these cells being treated with JQ1 or DMSO. The protein levels of Notch1 and NICD were concurrently altered with the protein level of YAP. However, the level of Jagged1 was not altered with Yap (Figure 7C-D). These results indicated that JQ1 indirectly downregulates the activity of the Notch signaling pathway by inhibiting the YAP expression in BET protein inhibitors in AML12 cells. Furthermore, downstream molecule Notch1 was shown to be a functional target of YAP/TAZ.
Both YAP/TAZ expression and Notch signaling pathway were impaired by BET proteins inhibition in vitro. (A-B) Western blot analysis of YAP/TAZ pathway signaling and Notch pathway signaling protein levels with time gradients treated by JQ1 in AML12 cells. (C-D) Western blot analysis of YAP/TAZ pathway signaling and Notch pathway signaling protein levels with concentration gradients treated by JQ1 for 24 hours in AML12 cells.
The liver regeneration injury caused by the inhibition of BET protein can be rescued by Yap overexpression in mouse liver
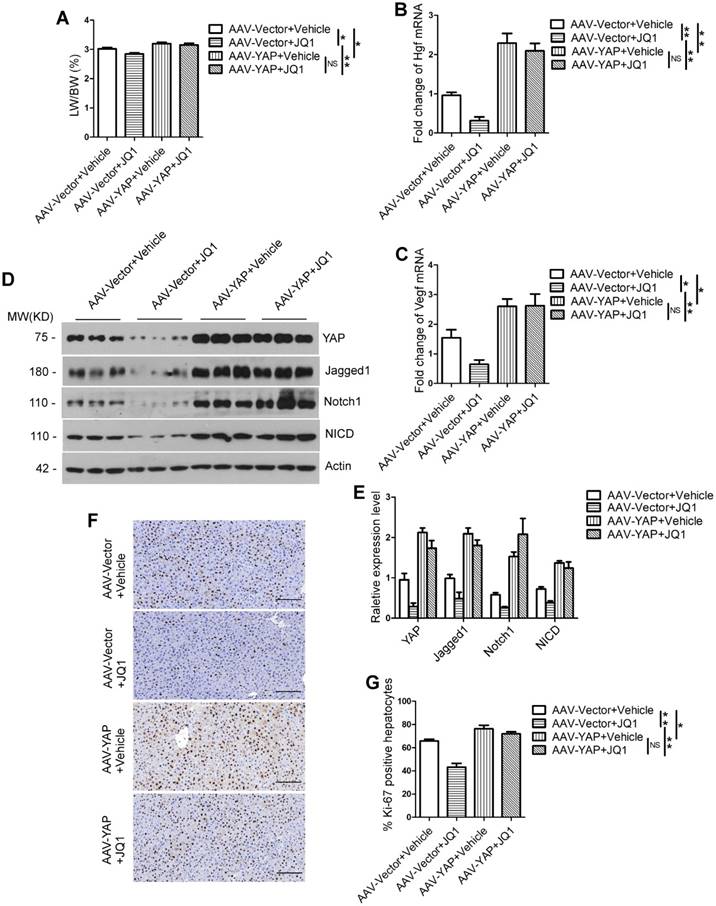
To further prove that BET protein inhibition impairs liver regeneration through YAP/TAZ-Notch1-NICD axis, we constructed a mouse model of Yap overexpression in liver by infecting the mice with adeno-associated virus (AAV). We performed JQ1 after 70% PH in AAV-vector group and AAV-Yap group mice, and harvested liver at day 2 and day 4 after 70% PH. The results showed that mice treated with JQ1 had no significant difference in LW/BW ratio from those treated with the vehicle solution in the AAV-Yap group (day 2, p=0.5633; day 4, p=0.5727). However, the LW/BW ratio was significantly lower in mice treated with JQ1 compared with those treated with the vehicle solution in the AAV-vector group (day 2, p=0.0260; day 4, p=0.0365). In addition, the LW/BW ratio of the AAV-Yap group treated with JQ1 significantly increased than that of the AAV-vector group treated with JQ1 (day 2, p=0.0027; day 4, p=0.0365) (day 2, Figure 8A; day 4, Figure S5A). Using primary hepatocytes at day 2 after 70% PH, the expression levels of Hgf and Vegf were significantly higher in the AAV-Yap group compared with those of the AAV-Vector group; however, there were no significant difference between the mice treated with JQ1 and vehicle solution in the AAV-Yap group (Figure 5B-C). Western blot analysis was performed on the primary hepatocytes at day 2 after 70% PH and the liver tissues at day 4 after 70% PH showed that the protein levels of Jagged1, Notch1 and NICD were significantly higher in the AAV-Yap group than those of the AAV-Vector group. However, there was no significant difference between the mice treated with JQ1 and vehicle solution in the AAV-Yap group (day 2, Figure 8D-E; day 4, Figure S5B-C). Moreover, the numbers of Ki-67 positive hepatocytes in the AAV-Vector group were significantly lower compared with those of the AAV-Yap group in the JQ1 treatment mice (day 2, p=0.0013; day 4, p=0.0128). Furthermore, the numbers of Ki-67 positive hepatocytes in the AAV-Vector group were significantly lower than those of the AAV-Yap group in the vehicle solution treatment mice at day 2 and day 4 after 70% PH (day 2, p=0.0338; day 4, p=0.0164). However, no statistical difference was observed in AAV-Yap group between the mice treated with JQ1 and vehicle solution respectively (day 2, p=0.2663; day 4, p=0.0164) (day 2, Figure 8F-G; day 4, Figure S5D-E). These results indicated that specific overexpression of Yap in mouse liver could rescue the effect of BET proteins inhibition on liver regeneration injury and the activity of Notch signaling pathway was upregulated following the overexpression of Yap in vivo.
Discussion
Increasing evidence supports the importance of BET protein inhibitors, such as JQ1, in the treatment of a series of liver pathologies due to their ability to protect against liver fibrosis [11], and repress cell growth in tumor xenograft HCC models [10]. The majority of HCC requires surgical resection of liver tissues, and liver fibrosis occurs in patients with chronic liver diseases. Both are in need of subsequent liver regeneration. However, our findings revealed that inhibiting the BET protein might result in impaired liver regeneration, thereby implying an adverse impact of BET protein inhibition on the treatment of HCC and liver fibrosis. Recently, lineage-tracing studies in mice demonstrate that preexisting hepatocytes are the main sources of regenerated hepatocytes in current oval cell activation models [28, 29]. Our results showed that BET proteins inhibition prevents hepatocyte proliferation in the mouse model of hepatocyte-driven liver regeneration.
The Notch receptor Notch1 was a Functional YAP Target. (A) Western blot analysis of YAP, Jagged1, Notch1 and NICD protein levels treated by JQ1 or DMSO after infection by Yap shRNA adenovirus in AML12 cells. (B) Western blot analysis of YAP, Jagged1, Notch1 and NICD protein levels treated by JQ1 or DMSO after transfection of PAMD-Yap-Flag plasmids in AML12 cells.
The liver regeneration injury caused by the inhibition of BET proteins can be rescued by Yap overexpression in mouse liver. (A) LW/BW ratio was analyzed for AAV-Vector+Vehicle group, AAV-Vector+JQ1 group, AAV-Yap+Vector group and AAV-Yap+JQ1 group mice at day 2 after 70% PH. (B-C) Fold change in Hgf and Vegf mRNA expression in primary hepatocytes derived from AAV-Vector+Vehicle group, AAV-Vector+JQ1 group, AAV-Yap+Vector group and AAV-Yap+JQ1 group mice at day 2 after 70% PH. (D-E) Western blot analysis of YAP, Notch1, Jagged1 and NICD protein levels in primary hepatocytes derived from AAV-Vector+Vehicle group, AAV-Vector+JQ1 group, AAV-Yap+Vector group and AAV-Yap+JQ1 group mice at day 2 after 70% PH. (F) Ki-67 staining of liver tissues from AAV-Vector+Vehicle group, AAV-Vector+JQ1 group, AAV-Yap+Vector group and AAV-Yap+JQ1 group mice at day 2 after 70% PH (200× original magnification). Scale bars, 200μm. (G) Ki-67-positive cell count at day 2 after 70% PH. NS≥0.05, *P<0.05, **P<0.01. Data are expressed as mean ± S.E.M.
Although previous studies have described the role of YAP/TAZ overexpression in enhanced proliferation of hepatocytes, their underlying mechanisms remain unclear in liver regeneration [22, 23]. Our findings revealed a novel mechanism of YAP/TAZ-Notch1-NICD axis in hepatocytes and liver regeneration, wherein Notch receptor Notch1 was positively regulated by YAP in hepatocytes. The suppression of YAP/TAZ-Notch1-NICD axis by BET protein inhibition being able to impair liver regeneration has not been previously described. Furthermore, we applied the specific inhibitor of the YAP/TAZ signaling pathway in vivo, thereby confirming that inhibition of this signaling pathway affects liver regeneration. Additionally, we found that inhibiting the YAP/TAZ signaling pathway decreases the expression of Notch signaling pathway-associated proteins, thereby indicating that the YAP/TAZ signaling pathway regulates the Notch signaling pathway during liver regeneration.
Association among YAP, Notch1, and Jagged1 has been reported in a tumor model (Drosophila) [25, 26], but further details regarding their interactions has been unclear. In normal hepatocytes, the regulation pattern of YAP on the Notch pathway is different from that in liver cancer cells. The Notch signaling pathway regulates several important cellular processes, including cell survival, differentiation, proliferation, and apoptosis [30-33]. In mammals, two types of ligands (Jagged1-2 and DLL1, DLL3-4) and four Notch receptors (Notch1-4) exist in the Notch signaling pathway [34]. In hepatocytes, Notch1 is predominantly expressed and is crucial in regulating cell proliferation [24]. The Notch signaling pathway is activated as the ligand Jagged1 binds to the receptor Notch1, which is released by γ-secretase complex, thereby resulting in the release of NICD. NICD can translocate into the nucleus and lead to the transcription of Notch target genes, including proliferation-related genes [35].
Numerous studies have indicated that Jagged1 and Notch1 are crucial in hepatocyte proliferation during liver regeneration [24, 36, 37]. Our findings further revealed that the Notch receptor Notch1 was a Functional YAP target in liver. Previous study showed an increased nuclear YAP, elevated levels of Jagged1 and NICD, and activation of Notch target genes in mammals' Mst1/2-deficient intestinal cells [27]. Our mouse model with 70% PH showed that BET protein inhibition suppressed YAP/TAZ expression, subsequently leading to remarkable reductions in Jagged1 and Notch1. Then, nucleus translocation of NICD was significantly decreased, which significantly downregulated its downstream gene expressions of Hey1, Hes1, and Sox9. Notch signaling target genes, such as Hes1, Hey1, Vegf and Sox9, have been documented for their roles in cell proliferation and differentiation in livers [31, 32]. Cyr61, as a target gene of Yap, was downregulated by BET protein inhibition, whereas the expressions of two other target genes (Ctgf and Amotl2) remain unchanged. This effector mechanism requires further investigation for additional signaling pathways in the future.
In vitro experimental study using AML12 cell line exhibited that the Notch signaling pathway might be directly interacted by regulating the expression of YAP, and knockdown or overexpression of Yap. Furthermore, the suppression of liver regeneration caused by JQ1 was rescued in the liver specificity Yap overexpression mouse model in the rescue experiment. The protein expression levels of Notch signaling pathway increased follow the overexpression of Yap. These findings support our hypothesis that YAP/TAZ-Notch1-NICD axis regulated by BET protein inhibition plays a major role in regulating hepatocyte proliferation during liver regeneration. Moreover, the BET protein inhibitor could indirectly affect the expression of Notch1.
A recent study showed that BET protein inhibition inhibits liver regeneration by inhibiting the Wnt signaling pathway [38]. Our data demonstrate that the BET proteins inhibition affect the liver regeneration by inhibiting the YAP/TAZ-Notch1-NICD axis, and uncovered the role of YAP/TAZ-Notch1-NICD in liver regeneration. It would be of significance in shedding light on further understanding of the mechanism of action of BET proteins during liver regeneration. YAP/TAZ and Notch signaling as well as Wnt signaling pathway are the downstream effectors of BET proteins in liver regeneration settings. Moreover, in the study mentioned above, the serum AST levels were increased in JQ1-injected mice at 40 hours after 70% PH. However, our study showed that the serum ALT or albumin levels were not increased in JQ1-injected mice at 2 days after 70% PH. The effect of JQ1 on liver function requires further investigation in the future.
In summary, our study revealed a crucial role of YAP/TAZ-Notch1-NICD axis in hepatocytes, which is downregulated by BET protein inhibition, wherein this downregulation impairs hepatocyte-driven liver regeneration. Therefore, utilizing BET protein inhibitors must be cautiously used to treat liver diseases, such as HCC and liver fibrosis. Our findings are significant in further understanding the biological behaviors of liver regeneration.
Abbreviations
BET: bromo- and extra-terminal domain; YAP: Yes-associated protein; TAZ: transcriptional co-activator with PDZ-binding motif; NICD: Notch intracellular domain; HGF: hepatocyte growth factor; VEGF: vascular endothelial growth factor; ALT: alanine aminotransferase; ALB: albumin; BrdU: 5-bromo-2'-deoxyuridine.
Supplementary Material
Supplementary methods, figures and table.
Acknowledgements
We thank Professor Lanfen Chen (Xiamen University) for providing the PAMD-YAP-Flag plasmids. The project was supported by the National Science Foundation for Outstanding Young Scholars of China (No. 81522006), the Fundamental Research Funds for the Central Universities (2015XZZX004-21), the National Natural Science Foundation of China (No. 81470527, No. 81870306), Zhejiang Provincial 151 Talent Project, and Zhejiang Provincial Outstanding Youth Foundation (No. LR13H020001).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Diehl AM, Chute J. Underlying potential: cellular and molecular determinants of adult liver repair. J Clin Invest. 2013;123:1858-60
2. Alghamdi S, Khan I, Beeravolu N, McKee C, Thibodeau B, Wilson G. et al. BET protein inhibitor JQ1 inhibits growth and modulates WNT signaling in mesenchymal stem cells. Stem Cell Res Ther. 2016;7:22
3. Zhang L, Tong Y, Zhang X, Pan M, Chen S. Arsenic sulfide combined with JQ1, chemotherapy agents, or celecoxib inhibit gastric and colon cancer cell growth. Drug Des Devel Ther. 2015;9:5851-62
4. Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA. et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524-8
5. Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM. et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904-17
6. Baud MGJ, Lin-Shiao E, Cardote T, Tallant C, Pschibul A, Chan KH. et al. Chemical biology. A bump-and-hole approach to engineer controlled selectivity of BET bromodomain chemical probes. Science. 2014;346:638-41
7. Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O. et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067-73
8. Wang YH, Sui XM, Sui YN, Zhu QW, Yan K, Wang LS. et al. BRD4 induces cell migration and invasion in HCC cells through MMP-2 and MMP-9 activation mediated by the Sonic hedgehog signaling pathway. Oncol Lett. 2015;10:2227-32
9. Zhang P, Dong Z, Cai J, Zhang C, Shen Z, Ke A. et al. BRD4 promotes tumor growth and epithelial-mesenchymal transition in hepatocellular carcinoma. Int J Immunopathol Pharmacol. 2015;28:36-44
10. Li GQ, Guo WZ, Zhang Y, Seng JJ, Zhang HP, Ma XX. et al. Suppression of BRD4 inhibits human hepatocellular carcinoma by repressing MYC and enhancing BIM expression. Oncotarget. 2016;7:2462-74
11. Ding N, Hah N, Yu RT, Sherman MH, Benner C, Leblanc M. et al. BRD4 is a novel therapeutic target for liver fibrosis. Proc Natl Acad Sci U S A. 2015;112:15713-8
12. Camargo FD, Gokhale S, Johnnidis JB, Fu D, Bell GW, Jaenisch R. et al. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr Biol. 2007;17:2054-60
13. Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA. et al. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120-33
14. Zhao B, Tumaneng K, Guan KL. The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nat Cell Biol. 2011;13:877-83
15. Zhao B, Ye X, Yu J, Li L, Li W, Li S. et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962-71
16. Zhou D, Conrad C, Xia F, Park JS, Payer B, Yin Y. et al. Mst1 and Mst2 maintain hepatocyte quiescence and suppress hepatocellular carcinoma development through inactivation of the Yap1 oncogene. Cancer Cell. 2009;16:425-38
17. Bai H, Zhang N, Xu Y, Chen Q, Khan M, Potter JJ. et al. Yes-associated protein regulates the hepatic response after bile duct ligation. Hepatology. 2012;56:1097-107
18. Machado MV, Michelotti GA, Pereira TA, Xie G, Premont R, Cortez-Pinto H. et al. Accumulation of duct cells with activated YAP parallels fibrosis progression in non-alcoholic fatty liver disease. J Hepatol. 2015;63:962-70
19. Wang X, Zheng Z, Caviglia JM, Corey KE, Herfel TM, Cai B. et al. Hepatocyte TAZ/WWTR1 Promotes Inflammation and Fibrosis in Nonalcoholic Steatohepatitis. Cell Metab. 2016;24:848-62
20. Ko S, Choi TY, Russell JO, So J, Monga SPS, Shin D. Bromodomain and extraterminal (BET) proteins regulate biliary-driven liver regeneration. J Hepatol. 2016;64:316-25
21. Higgins GK, Johnson D Jr. Detection of Antihypertensive Agents in Urine. Trans Assoc Life Insur Med Dir Am. 1964;48:186-90
22. Grijalva JL, Huizenga M, Mueller K, Rodriguez S, Brazzo J, Camargo F. et al. Dynamic alterations in Hippo signaling pathway and YAP activation during liver regeneration. Am J Physiol Gastrointest Liver Physiol. 2014;307:G196-204
23. Lu L, Finegold MJ, Johnson RL. Hippo pathway coactivators Yap and Taz are required to coordinate mammalian liver regeneration. Exp Mol Med. 2018;50:e423
24. Kohler C, Bell AW, Bowen WC, Monga SP, Fleig W, Michalopoulos GK. Expression of Notch-1 and its ligand Jagged-1 in rat liver during liver regeneration. Hepatology. 2004;39:1056-65
25. Tschaharganeh DF, Chen X, Latzko P, Malz M, Gaida MM, Felix K. et al. Yes-associated protein up-regulates Jagged-1 and activates the Notch pathway in human hepatocellular carcinoma. Gastroenterology. 2013;144:1530-42 e12
26. Cho E, Feng Y, Rauskolb C, Maitra S, Fehon R, Irvine KD. Delineation of a Fat tumor suppressor pathway. Nat Genet. 2006;38:1142-50
27. Zhou D, Zhang Y, Wu H, Barry E, Yin Y, Lawrence E. et al. Mst1 and Mst2 protein kinases restrain intestinal stem cell proliferation and colonic tumorigenesis by inhibition of Yes-associated protein (Yap) overabundance. Proc Natl Acad Sci U S A. 2011;108:E1312-20
28. Yanger K, Knigin D, Zong Y, Maggs L, Gu G, Akiyama H. et al. Adult hepatocytes are generated by self-duplication rather than stem cell differentiation. Cell Stem Cell. 2014;15:340-9
29. Schaub JR, Malato Y, Gormond C, Willenbring H. Evidence against a stem cell origin of new hepatocytes in a common mouse model of chronic liver injury. Cell Rep. 2014;8:933-9
30. Zhou W, He Q, Zhang C, He X, Cui Z, Liu F. et al. BLOS2 negatively regulates Notch signaling during neural and hematopoietic stem and progenitor cell development. Elife. 2016;5:e18108
31. Kessler M, Hoffmann K, Brinkmann V, Thieck O, Jackisch S, Toelle B. et al. The Notch and Wnt pathways regulate stemness and differentiation in human fallopian tube organoids. Nat. Commun. 2015;6:8989
32. Kozlovskaja-Gumbrienė A, Yi R, Alexander R, Aman A, Jiskra R, Nagelberg D. et al. Proliferation-independent regulation of organ size by Fgf/Notch signaling. Elife. 2017;6:e21049
33. Zeng C, Xing R, Liu J, Xing F. Role of CSL-dependent and independent Notch signaling pathways in cell apoptosis. Apoptosis. 2016;21:1-12
34. Morell CM, Strazzabosco M. Notch signaling and new therapeutic options in liver disease. J Hepatol. 2014;60:885-90
35. Geisler F, Strazzabosco M. Emerging roles of Notch signaling in liver disease. Hepatology. 2015;61:382-92
36. Xu L, Gu L, Tao X, Xu Y, Qi Y, Yin L. et al. Effect of dioscin on promoting liver regeneration via activating Notch1/Jagged1 signal pathway. Phytomedicine. 2018;38:107-17
37. Zhang F, Zhang J, Li X, Li B, Tao K, Yue S. Notch signaling pathway regulates cell cycle in proliferating hepatocytes involved in liver regeneration. J Gastroenterol Hepatol. 2018;33:1538-47
38. Russell JO, Ko S, Saggi HS, Singh S, Poddar M, Shin D. et al. Bromodomain and Extraterminal (BET) Proteins Regulate Hepatocyte Proliferation in Hepatocyte-Driven Liver Regeneration. Am J Pathol. 2018;188:1389-405
Author contact
![]() Corresponding authors: Weihua Gong, Department of Surgery, Second Affiliated Hospital of School of Medicine, Zhejiang University, Hangzhou 310058, China, E-mail: weihuagongedu.cn, Tel: +86 571-87783580, Fax: +86 571-87783581 or Qiming Sun, Department of Biochemistry, and Department of Cardiology of Second Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou 310058, China, E-mail: qmsunedu.cn, Tel: +86 571-88208505, Fax: +86 571-88208505. Weihua Gong is the Lead Contact.
Corresponding authors: Weihua Gong, Department of Surgery, Second Affiliated Hospital of School of Medicine, Zhejiang University, Hangzhou 310058, China, E-mail: weihuagongedu.cn, Tel: +86 571-87783580, Fax: +86 571-87783581 or Qiming Sun, Department of Biochemistry, and Department of Cardiology of Second Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou 310058, China, E-mail: qmsunedu.cn, Tel: +86 571-88208505, Fax: +86 571-88208505. Weihua Gong is the Lead Contact.
 Global reach, higher impact
Global reach, higher impact