13.3
Impact Factor
Theranostics 2019; 9(13):3723-3731. doi:10.7150/thno.33980 This issue Cite
Research Paper
One-step colorimetric genotyping of single nucleotide polymorphism using probe-enhanced loop-mediated isothermal amplification (PE-LAMP)
1. Natural Products Research Center, Chengdu Institute of Biology, Chinese Academy of Sciences, Chengdu 610041, P. R. China
2. University of Chinese Academy of Sciences, Beijing 100049, P. R. China
3. Chengdu University of Traditional Chinese Medicine, Chengdu 611137, P. R. China.
Received 2019-2-11; Accepted 2019-4-10; Published 2019-5-31
Abstract
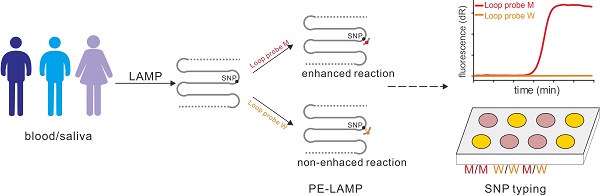
Single nucleotide polymorphism (SNP) is the most abundant molecular marker associated with many physiologic and pathologic phenotypes. An isothermal, accurate and cost-effective SNP detection could make a great difference in point-of-care testing (POCT) or on-site diagnosis. However, there are two challenges, the expensive instrument and labor-intensive process, faced by the development of on-site SNP detection. We reported a novel SNP typing method based on the probe-enhanced loop-mediated isothermal amplification (PE-LAMP), which combines the oligonucleotide probe with a conventional LAMP to realize the SNP discrimination by analyzing the great discrepancy in amplification efficiency.
Methods: We firstly constructed the genotyping method by combining the hybridization of the specific probe with the powerful amplification of LAMP. Then we validated the method by genotyping the SNP rs3741219 and we sought to realize one-step visualized typing. Finally, we applied the method to pharmacogenomic testing by genotyping CYP2C19*2 and MDR1 C3435T.
Results: The PE-LAMP was successfully constructed to detect SNP and the sensitivity of our method is as low as 1000 copies of target DNA, which is sufficient to routine diagnosis. The high specificity in detecting mutant in the presence of excess wild-type allele could be achieved. It has shown good performance in helping predict the individual response of antiplatelet drug Clopidogrel through typing simply treated saliva samples.
Conclusions: The proposed method is one-step, colorimetric, specific and sensitive enough to detect crudely treated samples, showing great potential in the pharmacogenomic study and POCT use.
Keywords: SNP, LAMP, colorimetric detection, pharmacogenomic
Introduction
Single nucleotide polymorphism (SNP) is the most common genetic variation, with the amount of more than 9 million reported in public databases [1]. With the progression of post-genome projects, such as HapMap (http://www.hapmap.org) and GWAS (http://www.ebi.ac.uk), abundant SNPs linked with phenotypic changes have been found to be useful in understanding human physiology. From this aspect, researchers and clinicians are seeking for using SNP as a new biomarker for diagnosis and prediction in the era of individualized medicine. For instance, many SNPs associated with the susceptibility of cancers are able to help make precise diagnostic [2]. Moreover, many SNPs related to drug response are available to predict the therapy efficacy [3].
Intensive efforts have been devoted to SNP detection. Currently, the prevailing SNP detection methods mainly depend on the polymerase chain reaction (PCR), such as TaqMan assay [4], restriction fragment length polymorphism (RFLP) [5] and sequencing. However, the laborious operation as well as the need for auxiliary equipment limit these methods to the laboratory with specialized facilities. The emergence of isothermal amplification strategies for nucleic acids has been revolutionizing the development of genetic detection, which eliminates the need for non-portable thermal cycler [6, 7]. Loop-mediated isothermal amplification (LAMP) is one of the most practical methods which was developed by Notomi and his colleagues in 2000 [8]. Conventional LAMP uses two primer pairs targeting to six regions on a target sequence and a mesophilic DNA polymerase with strand-displacement activity to realize specific amplification. LAMP shows great potential in clinical detection due to its good tolerance to crudely processed samples [9, 10]. Furthermore, the loop primer has been introduced to LAMP, leading to a significant enhancement of reaction rate [11]. In the last few years, several LAMP-based SNP detection methods have been reported, such as allele-specific LAMP (AS-LAMP) [12-15], the one-step strand displacement coupled LAMP (LAMP-OSD) [16], and the LAMP combined with allele-selective oligonucleotide hybridization (LAMP-ASO) [17]. The strategy of AS-LAMP is placing the site of interest at the 3' or 5' end of the initial primers such that the specific amplification could be realized by mesophilic DNA polymerases with the ability of mismatch discrimination. However, many DNA polymerases used in isothermal amplification may initial extension regardless of the 3'-terminal mismatches [18]. The addition of unique mismatch-binding protein Taq MutS was able to overcome this problem [19], but the binding activity of MutS depends on the composition of each mismatched base pair [20]. Additionally, the primer used to realize AS-LAMP strictly depends on the position of the SNP in target, making these methods inflexible in primer design. The LAMP-OSD assay, as well as the LAMP-ASO reaction, were more complex, which required more time on signal readout. Apart from the advance in isothermal amplification, the great progress in isothermal genetic detection is attributed to the development of reporting methods. The reporting system is vital to genetic detection because it directly presents the result of a whole assay. Among the reporting systems, colorimetric sensor could be a promising tool in the applications of point-of-care (POC) diagnosis and field test due to the portability, low cost, and no need for complicated instrumentation. More important, the signal change can be directly read out by naked eyes. Recently, many colorimetric reporting systems have been reported to detect isothermal amplification such as the DNAzyme [21] and modified nanozyme [22, 23]. The methods used for monitoring LAMP reaction have been drawing attention from diverse fields of science, but a suitable colorimetric tool for SNP detection is still a challenge. Except for the real-time fluorescence assay that depends on the bulky instrument, currently described reporting systems in LAMP-based SNP detection include several visualized methods such as enzyme-linked immunosorbent assay [17], nanoparticles [24] and lateral flow dipstick [13]. These visualized reporting systems, though costly and laborious, are promising in POCT. Nevertheless, these measurements require tube opening at the endpoint, which is an unwise choice in LAMP reaction due to the high risk of carry-over contamination. The one-step colorimetric reporting way is, therefore, more attractive. Some colorimetric indicators are capable of reflecting the results of isothermal reaction in a single-step way, such as metal ion indicators [25, 26] and pH-sensitive dyes [27]. However, none of them has been reported to be used for LAMP-based SNP discrimination, which might be ascribed to the narrow time gap between specific and unspecific amplification.
Herein, we presented a novel strategy to genotype SNP based on the PE-LAMP, in which the loop primer was conferred a new role of SNP detection. In the meantime, we sought to realize the visualized genotyping without uncapping, making the new method a user-friendly tool in typing SNP.
Materials and methods
Plasmid construction and preparation of the real sample
To obtain the standard template, two plasmids containing the rs3741219 specific sequence which differing in a single nucleotide were constructed by using pEASY®-T1 Cloning Kit (TransGen Biotech Company Limited, China). Then the recombined plasmids were purified with AxyPrep Plasmid purification kit (Corning Life Science, USA) and verified via sequencing by Sangon Biotech Company Limited (Shanghai, China). The final DNA quality and concentration were measured by the NanoDrop UV-Vis spectrophotometer (Thermo Fisher Scientific Corporation, USA). The genomic samples were collected from saliva samples provided by the informed individuals. After collection, 1 mL saliva sample was centrifuged at 5400 g for 5 min at room temperature. Then the precipitate was simply processed by the commercial DNA extraction kit (Sigma-Aldrich Corporation, USA). Finally, 1 μL lysed saliva sample was directly added to the LAMP or PCR condition without purification.
Primer and probe design
Two sets of primers named outer and inner primers were designed according to the principle of the LAMP. Additionally, loop probes (LPs) were designed according to the allelic type of SNP. The primers for direct sequencing were also designed. All sequences were given in supplemental Tables S1 and S2.
PCR reaction and verification of SNP
For PCR reaction, the reaction was performed in 50 μL condition containing 1 U phanta Max Super-fidelity DNA polymerase, 25 μL 2x phanta Max buffer (Vazyme Biotech Company Limited, China), 0.2 mM deoxyribonucleoside triphosphates (dNTP), and 0.4 μM of forward and reverse primers. The PCR reaction was incubated in a C1000TM Thermal Cycler PCR (Bio-Rad Laboratories, USA) for total of 35 cycles (95 °C for 3 min, followed by 35 cycles of 30 s at 95 °C, 45 s at 60 °C and 30 s at 72 °C. A final elongation step at 72 °C for 5 min). The amplification products were verified by 2% agarose gel and subjected to direct sequencing.
Probe-enhanced LAMP reaction
For PE-LAMP reaction, the whole reaction was performed in 25 μL mixture containing 1.6 μM each of inner primers, 0.4 μM each of outer primers, 0.8 μM LP, 1 M betaine (Sigma-Aldrich), 2.5 μL 10x LAMP buffer (200 mM Tris-HCl (pH 8.8), 100 mM (NH4)2SO4, 40 mM MgCl2, and 600 mM KCl), 0.8 mM dNTP, 4 U Bsm DNA polymerase large fragment (Thermo Fisher Scientific), and 1 μL DNA template. The reaction solution was incubated at an optimized temperature for different SNP sites.
Amplification analysis
For real-time fluorescent LAMP, 1/50,000 diluted original SYBR Green I (Thermo Fisher Scientific, USA) was preadded to the reaction condition. As for visualized detection, 100 μM Neutral red (N-red) was added to the LAMP solution before the amplification. The amplification products were verified by 2% agarose gel.
The sensitivity and specificity of the method
The sensitivity of PE-LAMP reaction was evaluated by using 10-fold serial dilutions of the plasmid. The specificity of PE-LAMP reaction was measured by using the mixed templates containing different ratios (100%, 10%, 1%, 0.1% and 0%, regards to total 106 copies of targets) of mutant rs3741219 plasmids.
Application of the method using saliva samples
The real-life samples with defined genotype (CYP2C19*2 and MDR1 C3435T) were used to evaluate the method. When using colorimetric genotyping, the whole assay was carried out by pre-adding N-red to the reaction tubes.
Results and Discussion
The mechanism of SNP detection based on PE-LAMP
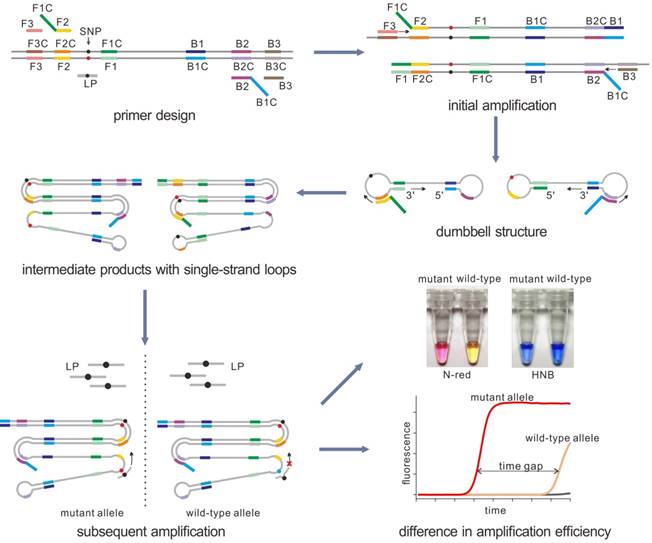
The principle of our strategy was shown in Figure 1A, a set of LAMP primers including inner and outer primers were designed according to the target sequence. The SNP site was flanked by the two regions of the inner primer (F1C and F2C) so as to make sure that the SNP allele sits in the loop domain of the LAMP products. Then with the strand displacement ability of mesophilic DNA polymerase, the initial amplification could produce the dumbbell-shaped products. After several rounds of amplification by using dumbbell structure as the template, a few intermediate products with single strand loop containing the SNP allele could be generated. Then the loop probe (LP), designed to detect the mutant allele, was used to hybrid with the mutant-containing loop domain. When the LP was perfectly complementary to the target region, the subsequent LAMP reaction could be significantly enhanced. On the contrary, the reaction containing the wild-type template could not be enhanced efficiently due to the single-base mismatch that leads to unstable hybridization between LP and the loop under the reaction temperature. Due to the difference in amplification rate, the mutant may be detected by the time gap between these two reactions. Based on the PE-LAMP, SNP genotyping could be realized through proceeding two independent reactions, each comprising one kind of LP that corresponds to the allelic type of the SNP. In our strategy, the SNP detection mainly depends on the probe hybridization, rather than the discrimination ability of isothermal DNA polymerase. The detection result could be reported in a fluorescent real-time way by using a double-stranded DNA binding dye or in a visual way with the aid of the colorimetric indicators.
Construction of the method
To investigate the feasibility of the strategy, we designed a set of LAMP primers according to the SNP (rs3741219) (Figure S1), which is known as a tag SNP in the human genome [28]. We firstly constructed two plasmids that contain mutant allele (C) and wild-type allele (T), respectively.
The schematic of detection of SNP by using the PE-LAMP. The outer primers (F3 and B3), the inner primers (F1C-F2 and B1C-B2) as well as the loop probe (LP) for the mutant target are designed. Then LAMP amplifies the target, yielding a few intermediate products with single-strand loops. When the specific LP hybrid to the target loop, the subsequent amplification will be accelerated. While the reaction containing the wild-type target remains low efficiency. The real-time fluorescence tracing is realized by SYBR Green I. For the colorimetric analysis, Neutral red (N-red) or Hydroxy naphthol blue (HNB) is pre-added to the reaction tubes.
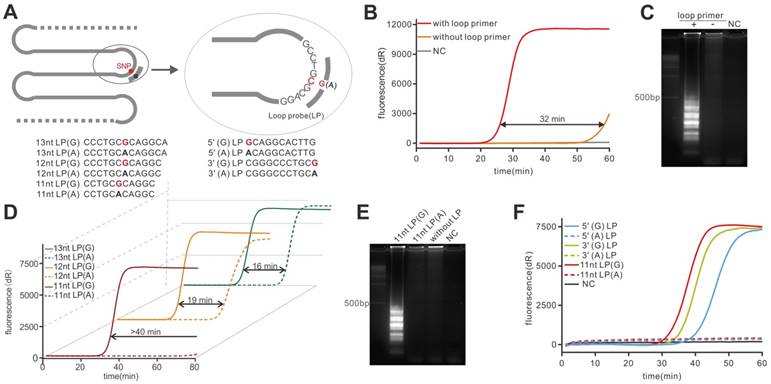
Then the LAMP reaction was carried out using the plasmid template and we found that the reaction could be performed under the temperature ranging from 57 °C to 62 °C (Figure S1). To validate the enhancement of loop primer to LAMP rate, we conducted two real-time LAMP reactions with and without a loop primer, respectively. The real-time fluorescent LAMP (Figure 2B) showed that the amplification rate of the loop primer-involving reaction was greatly enhanced compared to that lacking loop primer. Then the LAMP products were subjected to agarose gel electrophoresis and the typical ladder on agarose gel (Figure 2C) also indicated that the amplification rate could be significantly enhanced by the loop primer, which was consistent with the previous report [11]. In virtue of the role that the loop primer played in the LAMP reaction, there could be a possibility of making the loop primer a specific probe which links the amplification efficiency to the allelic constitution. According to our strategy, the specific hybridization of the LP should be the core part to realize the amplification difference which is the basis of SNP discrimination. The hybridization is a thermodynamic process, thus, the LP should be designed to destabilize the non-specific hybridization but stabilize the specific one. To obtain the ideal discrimination, it is critical to select the LP with a proper melting temperature (Tm). Thus, a series of LPs with different length were tested under the maximum reaction temperature (Figure 2A). As can be seen from Figure 2D, all the reactions that comprise mismatching LPs showed poorer amplification efficiency compared to the reaction containing matched LPs. Unexpectedly, however, among these mismatching LPs, the LPs with 11-nucleotide (nt) in length could provide no enhancement to the reaction in 80 min so that the biggest time gap (more than 40 min) between the two reactions (11nt LP(G) vs 11nt LP(A), Figure 2D) could be obtained. By contrast, the non-specific amplification signal from the others mismatched LPs could be detected soon after the specific amplification signal, thus showing the relatively narrow time gap. The agarose gel pattern in Figure 2E also showed that the reaction containing 11nt LP(A) was similar to the reaction without LP, demonstrating the 11nt mismatched LP could not stably bind to the target under the reaction temperature. Thus, the 11nt LPs presented better discrimination ability in our model. Additionally, like the design of a hybridization probe in DNA microarrays [29], the SNP position in probe should also be considered. Therefore, three pairs of LPs, with the allele at 5' end, 3' end and middle, respectively, were used to investigate the positional effect of the mutant in LP (Figure 2A). As shown in Figure 2F, we found that both three sets of LPs fulfilled the requirement of discrimination in an hour, however, the specific LP (11 nt LP(G)) showed a higher amplification efficiency, indicating the faster discrimination. Additionally, when extending the reaction time, the LP with the SNP at the center could provide a maximum time gap (49.9 min, Table S3) that represented optimal discrimination. The result was repeated by using the wild-type plasmid (Figure S2). This result was similar to previously reported studies [30, 31], which might be explained by the assumption that a mismatch at the center of a probe was most disruptive to hybridization. Taken together, we chose the 11nt LP with the allele at the center as the optimal probe for the following study and the results showed that the specific probe could successfully detect different targets (Figure S3).
Demonstration of PE-LAMP and selection of the LP. (A) Schematic of the hybridization of LP and all LPs used. The mutation in LP was highlighted in red. (B) Validation of accelerated LAMP by using 15nt loop primer. (C) Gel electrophoresis analysis of the loop primer effect on LAMP reaction. (D) Selection of the LP with proper length using mutant plasmid. (E) Gel electrophoresis analysis of probe-enhanced LAMP using 11nt LPs. (F) Investigating the positional effect of the mutant in LP.
Genotyping of SNP and visualized analysis
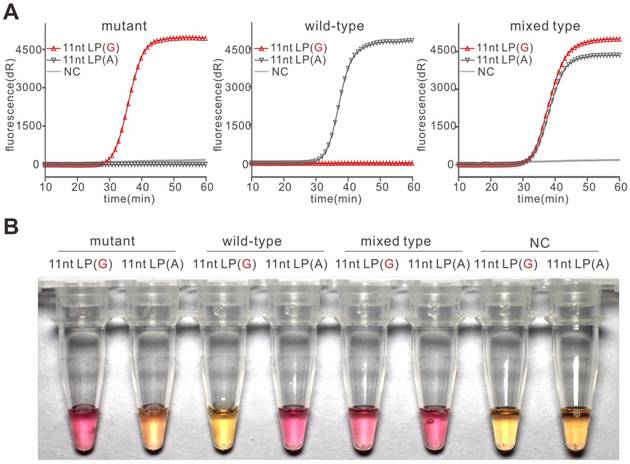
After the demonstration of PE-LAMP in SNP detection, we explored its ability in SNP genotyping. We assigned 11nt LP(G) and 11nt LP(A) to two parallel reactions containing the same template such that we could determine the genotype of the sample by analyzing the difference between these reactions (Figure S4). In experiments with plasmid templates, the results in Figure 3A showed that only the reaction containing matching LP had a rapid increase in fluorescence in an hour, demonstrating the PE-LAMP assay could accurately discriminate homozygotes and heterozygote. Then we sought to realize the colorimetric genotyping. Although there are many ways to achieve visualization, the one-step visualized detection is far more attractive to LAMP reaction. Neutral red (N-red) is a pH indicator that manifests color change from orange to pink over pH 8.0 to 6.8 [27]. We explored the feasibility of visualized detection by pre-adding N-red. As presented in Figure 3B, the color of the tubes with matching LP turned pink while the tubes with mismatching LP remained orange when using N-red as the indicator. The color change of the N-red could be explained by the pH drop which was caused by the released hydrogen ion as the DNA polymerase incorporates dNTP into the nascent DNA. In the meantime, we also obtained similar results by using the classic indicator, Hydroxy naphthol blue (HNB) (Figure S5). All the colorimetric results were consistent with the discrimination results using fluorescence, suggesting that the PE-LAMP assay was capable of colorimetric SNP typing. As far as we know, neither of these two indicators was mentioned in LAMP-based SNP detection. It might due to that the indicators need a wider time window to realize distinctly single-nucleotide mutation discrimination, which was hardly obtained by the existing LAMP-based SNP detection method. According to our results, the PE-LAMP was able to provide a sufficient time gap to obtain an optimal colorimetric difference. Meanwhile, the indicator was pre-added to the reaction, thus reducing the possible carry-over contamination.
Genotyping of SNP rs3741219 using the PE-LAMP. (A) The real-time fluorescent typing of mutant type plasmid (rs3741219 C), wild-type plasmid (rs3741219 T) and mixed type plasmid (rs3741219 C and T). Each experiment was repeated three times. (B) Using pre-added N-red to colorimetrically type plasmid templates.
Analysis of the sensitivity and specificity
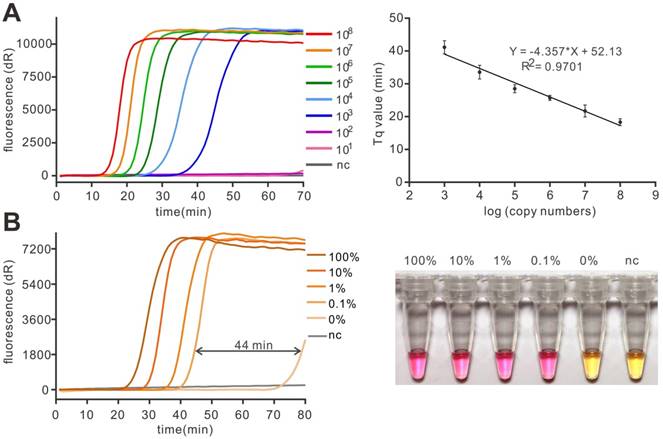
Although SNP detection has been thought to be a qualitative detection in most cases, the crudely treated samples may challenge the application of a method. Thus, a method with high sensitivity is more favorable in SNP detection. To determine the sensitivity of the whole assay, the gradient dilution plasmid templates (101 to 108) were used for PE-LAMP. In this experiment, the Tq value, which is similar to the Cq in real-time PCR, was defined as the reaction time but not the cycle number when fluorescence intensity exceeds an arbitrary threshold. According to the result, as few as 103 copies of the target could achieve efficient amplification (Figure 4A and Figure S6). The amplification Tq values were in linear correlation to the concentration of target (Figure 4B), indicating a potential of the method in quantification through real-time assay. Notably, it is commonly believed that 1 μL of extracted clinical sample, about 104-105 copies, is enough for routine detection [24]. Thus, the sensitivity of this strategy is enough to give a rapid and accurate genotyping result when applied for routine detection in clinical use. In order to investigate the specificity of our method, the probe specific to the mutant template was used to detect the total 106 copies of mixed templates harboring mutant templates at different ratios (100%, 10%, 1%, 0.1 and 0%, in percentage). According to the results, the time gaps between the mutant-harbouring reaction and the background (100% wild-type) were substantial enough to give a significant discrimination even in the presence of 0.1% mutant targets (1 mutant in 999 wild-type, 44 min, Figure 4C) and the distinct colorimetric discrimination (Figure 4D), implying the method was specific to detect the low abundant mutant target. The high repeatability was obtained in Figure S7. The experiments, in this case, were similar to the routine detection of acquired somatic mutation, in which the amount of total genomic DNA was fixed but the mutant was present in low abundance. The proposed method was competent to deal with such a situation with high specificity, indicating a great potential in the liquid biopsy.
Evaluation of the strategy using the real-life sample
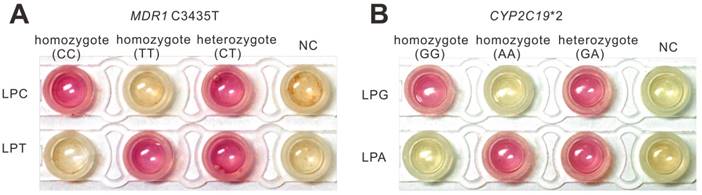
To explore the feasibility of this method in practical use, the strategy was further applied to pharmacogenomic test using human genomic DNA samples. The pharmacogenomic test is a useful method to predict the drug response using human genetic makeup. We verified the feasibility of the method in genotyping genome using saliva lysates (Figure S8). Then we investigated the further use of this strategy to genotype two SNPs (CYP2C19*2 (G681A) and MDR1 C3435T) that relate to the antiplatelet drug Clopidogrel. CYP2C19*2 is the most common allele which accounts for about 75-85% of loss-of-function mutation in the CYP2C19 gene [32] that affects the metabolism of Clopidogrel. P-glycoprotein, encoded by the MDR1 gene, is involved in drug transport and MDR1 polymorphisms, especially C3435T, was found to affect the function of P-glycoprotein [33]. There are many clinical trials have demonstrated that CYP2C19*2 and MDR1 C3435T played an important role in predicting the clinical outcome of Clopidogrel [34-36]. According to the pharmacogenomic test, as illustrated in Figure S9A, we devised a workflow to realize pharmacogenomic test using the PE-LAMP. The saliva sample was simply treated by the fast extraction kit that is commercially available and then the saliva lysate was subjected to genotyping by PE-LAMP. To evaluate our method, we sequenced several candidates and confirmed their genotypes at these two SNP sites (Figure S9B). The fluorescence curves presented in Figure S10 and S11 demonstrated that the probes designed here were capable of discriminating different types of these two SNPs. The colorimetric discrimination is shown in Figure 5, which was in accordance with the sequencing results, indicated that the proposed method could be an effective tool in the development of on-site SNP genotyping. As we expected, the colorimetric genotyping could be finished in an hour. Apart from the saliva sample, the whole blood samples were also conducted to evaluate the method (Figure S12). When compared to some currently reported methods, the performance of the PE-LAMP was comparable to some of the existing methods such as the high-resolution melting analysis (HRM) and the reported LAMP-based methods (Figure S13 and Table S4).
The sensitivity and specificity of PE-LAMP. (A) The sensitivity of the real-time fluorescent PE-LAMP (left) and the linear relationship between the Tq value and the logarithm of the copy number (right) in the range of 108 copies to 103 copies. The error bars are the standard deviation of three repetitive measurements. (B) Real-time fluorescent PE-LAMP (left) and colorimetric detection (right) using 11nt LP(G) (specific to mutant) to detect mutant templates at varying percentages (100%, 10%, 1%, 0.1%, 0%).
Application of PE-LAMP to genotype real-life samples. (A) The colorimetric typing of homozygous and heterozygous MDR1 C3435T allele by using saliva samples. (B) The colorimetric typing of homozygous and heterozygous CYP2C19*2 allele by using saliva samples.
In particular, the one-step operation, as well as less requirement of instrument in comparison with these methods, make the proposed method more user-friendly. Furthermore, as an isothermal method, the PE-LAMP could be improved in the future by simplifying the DNA extraction, using improved DNA polymerase or the highly sensitive signal reporter. These improvements could make a difference in shortening the assay time, which may make the proposed method a fast diagnosis tool in the future.
Conclusions
We reported here a simple and cost-effective strategy for sensitive and colorimetric genotyping of SNP at a constant temperature. The proposed technique takes advantage of the hybridization of oligonucleotide probe and the powerful amplification of LAMP, resulting in a dramatic discrepancy in amplification efficiency. Unlike the recently reported LAMP-based SNP detections, our strategy principally depends on the hybridization of specific probe, whereby the influence of mismatch extension caused by mesophilic DNA polymerase is minimized. Besides, the primer design in our strategy is more flexible, making this method easier in selecting the proper probe for an allele. In summary, this method comprises several merits: (i) the whole assay is based on the PE-LAMP, which is competent to genotype a real-life sample in an isothermal manner; (ii) the method can be coupled with colorimetric indicator to realize the one-step visualized detection, which can be observed by naked eyes and eliminates the risk of carry-over contamination; (iii) the sensitivity and specificity of the method are comparable to the routine detection platform, supporting its application in clinical test; (iv) the hybridization probe is sensitive enough to discriminate the single-base mutation, thus the strategy may have a great potential in detecting other forms of genetic variation including deletions, insertions and duplications; (v) the method uses the unmodified probe and there is no additional protein in need except for the DNA polymerase, making this method cost-effective than many existing methods. With these features, the proposed method needs only a simple temperature controller, much suitable for decentralization healthcare in source-limited regions, which is expected to provide a robust platform for SNP genotyping.
Abbreviations
LAMP: loop-mediated isothermal amplification; POCT: point-of-care testing; HapMap: haplotype map; GWAS: genome-wide association study; RFLP: restriction fragment length polymorphism; AS-LAMP: allele-specific LAMP; CYP2C19: cytochrome P450 family 2 subfamily C member 19; MDR1: multidrug resistance protein 1, also known as P-glycoprotein 1.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
This work was supported by the National Natural Science Foundation of China [grant numbers 21572222, 21708037, 21877108]; the Innovative Team of Sichuan Province [grant number 2017TD0021]; Chengdu Municipal Bureau of Science and Technology [grant number 2015-HM02-00099-SF, 2016-HM01-00371-SF]; China Postdoctoral Science Foundation [grant number 2017M612998].
Contributions
Conceptualization: Tang Z, Deng Y, Ding S; Formal analysis and Investigation: Chen R, Chen GY, Li M, Wang JY, Zou JW, Du F, Dong J, Cui X, Huang X; Supervision: Tang Z and Deng Y.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Kim S, Misra A. SNP genotyping: technologies and biomedical applications. Annu Rev Biomed Eng. 2007;9:289-320
2. Sparano JA, Gray RJ, Makower DF, Pritchard KI, Albain KS, Hayes DF. et al. Prospective validation of a 21-gene expression assay in breast cancer. N Engl J Med. 2015;373:2005-14
3. Wiechec E, Hansen LL. The effect of genetic variability on drug response in conventional breast cancer treatment. Eur J Pharmacol. 2009;625:122-30
4. Schleinitz D, Distefano JK, Kovacs P. Targeted SNP genotyping using the TaqMan® assay. Methods Mol Biol. 2011;700:77-87
5. Zhang R, Zhu Z, Zhu H, Nguyen T, Yao F, Xia K. et al. SNP cutter: a comprehensive tool for SNP PCR-RFLP assay design. Nucleic Acids Res. 2005;33:W489-92
6. Yan L, Zhou J, Zheng Y, Gamson AS, Roembke BT, Nakayama S. et al. Isothermal amplified detection of DNA and RNA. Mol Biosyst. 2014;10:970-1003
7. Zhao Y, Chen F, Li Q, Wang L, Fan C. Isothermal amplification of nucleic acids. Chem Rev. 2015;115:12491-545
8. Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N. et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:E63
9. Karanis P, Ongerth J. LAMP-a powerful and flexible tool for monitoring microbial pathogens. Trends Parasitol. 2009;25:498-9
10. Kaneko H, Kawana T, Fukushima E, Suzutani T. Tolerance of loop-mediated isothermal amplification to a culture medium and biological substances. J Biochem Biophys Methods. 2007;70:499-501
11. Nagamine K, Hase T, Notomi T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol Cell Probes. 2002;16:223-9
12. Badolo A, Okado K, Guelbeogo WM, Aonuma H, Bando H, Fukumoto S. et al. Development of an allele-specific, loop-mediated, isothermal amplification method (AS-LAMP) to detect the L1014F kdr-w mutation in Anopheles gambiae s. l. Malar J. 2012;11:227
13. Yongkiettrakul S, Kampeera J, Chareanchim W, Rattanajak R, Pornthanakasem W, Kiatpathomchai W. et al. Simple detection of single nucleotide polymorphism in Plasmodium falciparum by SNP-LAMP assay combined with lateral flow dipstick. Parasitol Int. 2017;66:964-71
14. Liu X, Zhang C, Zhao M, Liu K, Li H, Li N. et al. A direct isothermal amplification system adapted for rapid SNP genotyping of multifarious sample types. Biosens Bioelectron. 2018;115:70-6
15. Zhang L, Zhang Y, Wang C, Feng Q, Fan F, Zhang G. et al. Integrated microcapillary for sample-to-answer nucleic acid pretreatment, amplification, and detection. Anal Chem. 2014;86:10461-6
16. Jiang YS, Bhadra S, Li B, Wu YR, Milligan JN, Ellington AD. Robust strand exchange reactions for the sequence-specific, real-time detection of nucleic acid amplicons. Anal Chem. 2015;87:3314-20
17. Yamanaka ES, Tortajada-Genaro LA, Pastor N, Maquieira A. Polymorphism genotyping based on loop-mediated isothermal amplification and smartphone detection. Biosens Bioelectron. 2018;109:177-83
18. Chen F, Zhao Y, Fan C, Zhao Y. Mismatch extension of DNA polymerases and high-accuracy single nucleotide polymorphism diagnostics by gold nanoparticle-improved isothermal amplification. Anal Chem. 2015;87:8718-23
19. Mitani Y, Lezhava A, Kawai Y, Kikuchi T, Oguchi-Katayama A, Kogo Y. et al. Rapid SNP diagnostics using asymmetric isothermal amplification and a new mismatch-suppression technology. Nat Methods. 2007;4:257-62
20. Whitehouse A, Deeble J, Parmar R, Taylor GR, Markham AF, Meredith DM. Analysis of the mismatch and insertion/deletion binding properties of Thermus thermophilus, HB8, MutS. Biochem Biophys Res Commun. 1997;233:834-7
21. Xu H, Wu D, Li CQ, Lu Z, Liao XY, Huang J. et al. Label-free colorimetric detection of cancer related gene based on two-step amplification of molecular machine. Biosens Bioelectron. 2017;90:314-20
22. Zeng C, Lu N, Wen Y, Liu G, Zhang R, Zhang J. et al. Engineering nanozymes using DNA for catalytic regulation. ACS Appl Mater Interfaces. 2019;11:1790-9
23. Lu N, Zhang M, Ding L, Zheng J, Zeng C, Wen Y. et al. Yolk-shell nanostructured Fe3O4@C magnetic nanoparticles with enhanced peroxidase-like activity for label-free colorimetric detection of H2O2 and glucose. Nanoscale. 2017;9:4508-15
24. Lu Y, Ma X, Wang J, Sheng N, Dong T, Song Q. et al. Visualized detection of single-base difference in multiplexed loop-mediated isothermal amplification amplicons by invasive reaction coupled with oligonucleotide probe-modified gold nanoparticles. Biosens Bioelectron. 2017;90:388-93
25. Tomita N, Mori Y, Kanda H, Notomi T. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat Protoc. 2008;3:877-82
26. Goto M, Honda E, Ogura A, Nomoto A, Hanaki K. Colorimetric detection of loop-mediated isothermal amplification reaction by using hydroxy naphthol blue. Biotechniques. 2009;46:167-72
27. Tanner NA, Zhang Y, Evans TC Jr. Visual detection of isothermal nucleic acid amplification using pH-sensitive dyes. Biotechniques. 2015;58:59-68
28. Xia Z, Yan R, Duan F, Song C, Wang P, Wang K. Genetic polymorphisms in long noncoding RNA H19 are associated with susceptibility to breast cancer in Chinese population. Medicine. 2016;95:e2771
29. Gardner SN, Thissen JB, McLoughlin KS, Slezak T, Jaing CJ. Optimizing SNP microarray probe design for high accuracy microbial genotyping. J Microbiol Methods. 2013;94:303-10
30. Duan F, Pauley MA, Spindel ER, Zhang L, Norgren RB. Large scale analysis of positional effects of single-base mismatches on microarray gene expression data. BioData Min. 2010;3:2
31. Tan JC, Patel JJ, Tan A, Blain JC, Albert TJ, Lobo NF. et al. Optimizing comparative genomic hybridization probes for genotyping and SNP detection in Plasmodium falciparum. Genomics. 2009;93:543-50
32. Chen L, Qin S, Xie J, Tang J, Yang L, Shen W. et al. Genetic polymorphism analysis of CYP2C19 in Chinese Han populations from different geographic areas of mainland China. Pharmacogenomics. 2008;9:691-702
33. Hoffmeyer S, Burk O, von Richter O, Arnold HP, Brockmoller J, Johne A. et al. Functional polymorphisms of the human multidrug-resistance gene: multiple sequence variations and correlation of one allele with P-glycoprotein expression and activity in vivo. Proc Natl Acad Sci U S A. 2000;97:3473-8
34. Mega JL, Close SL, Wiviott SD, Shen L, Walker JR, Simon T. et al. Genetic variants in ABCB1 and CYP2C19 and cardiovascular outcomes after treatment with clopidogrel and prasugrel in the TRITON-TIMI 38 trial: a pharmacogenetic analysis. Lancet. 2010;376:1312-9
35. Hou X, Han W, Gan Q, Liu Y, Fang W. CYP2C19 and ABCB1 genetic polymorphisms correlate with the recurrence of ischemic cardiovascular adverse events after clopidogrel treatment. J Clin Lab Anal. 2018;32:e22369
36. Wang XQ, Shen CL, Wang BN, Huang XH, Hu ZL, Li J. Genetic polymorphisms of CYP2C19 2 and ABCB1 C3435T affect the pharmacokinetic and pharmacodynamic responses to clopidogrel in 401 patients with acute coronary syndrome. Gene. 2015;558:200-7
Author contact
![]() Corresponding authors: Zhuo Tang, Chengdu Institute of Biology, Chinese Academy of Sciences, Chengdu, Sichuan, China. Fax: +86-28-82890648; E-mail: tangzhuoac.cn; Yun Deng, Chengdu University of Traditional Chinese Medicine, Chengdu, Sichuan, China. Fax: +86-28-85243250; E-mail: dengyun2000com.
Corresponding authors: Zhuo Tang, Chengdu Institute of Biology, Chinese Academy of Sciences, Chengdu, Sichuan, China. Fax: +86-28-82890648; E-mail: tangzhuoac.cn; Yun Deng, Chengdu University of Traditional Chinese Medicine, Chengdu, Sichuan, China. Fax: +86-28-85243250; E-mail: dengyun2000com.
 Global reach, higher impact
Global reach, higher impact