13.3
Impact Factor
Theranostics 2019; 9(12):3565-3579. doi:10.7150/thno.33046 This issue Cite
Research Paper
Regulated Delayed Shigella flexneri 2a O-antigen Synthesis in Live Recombinant Salmonella enterica Serovar Typhimurium Induces Comparable Levels of Protective Immune Responses with Constitutive Antigen Synthesis System
1. Department of Infectious Diseases and Immunology, College of Veterinary Medicine, University of Florida, Gainesville, FL, USA
2. College of Animal Science and Technology, Southwest University, Chongqing, China
Received 2019-1-11; Accepted 2019-4-21; Published 2019-5-26
Abstract
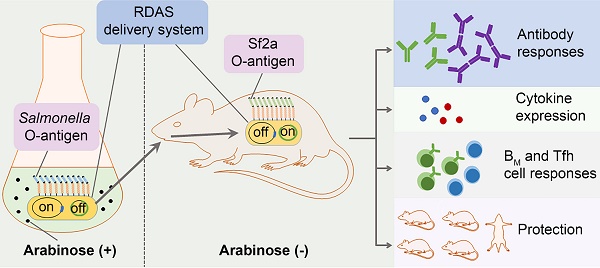
Shigella flexneri (S. flexneri), a leading cause of bacillary dysentery, is a major public health concern particularly affecting children in developing nations. We have constructed a novel attenuated Salmonella vaccine system based on the regulated delayed antigen synthesis (RDAS) and regulated delayed expression of attenuating phenotype (RDEAP) systems for delivering the S. flexneri 2a (Sf2a) O-antigen.
Methods: The new Salmonella vaccine platform was constructed through chromosomal integration of the araC PBAD lacI and araC PBAD wbaP cassettes, resulting in a gradual depletion of WbaP enzyme. An expression vector, encoding Sf2a O-antigen biosynthesis under the control of the LacI-repressible Ptrc promoter, was maintained in the Salmonella vaccine strain through antibiotic-independent selection. Mice immunized with the vaccine candidates were evaluated for cell-mediate and humoral immune responses.
Results: In the presence of exogenous arabinose, the Salmonella vaccine strain synthesized native Salmonella LPS as a consequence of WbaP expression. Moreover, arabinose supported LacI expression, thereby repressing Sf2a O-antigen production. In the absence of arabinose in vivo, native Salmonella LPS synthesis is repressed whilst the synthesis of the Sf2a O-antigen is induced. Murine immunization with the Salmonella vaccine strain elicited robust Sf2a-specific protective immune responses together with long term immunity.
Conclusion: These findings demonstrate the protective efficacy of recombinant Sf2a O-antigen delivered by a Salmonella vaccine platform.
Keywords: Shigella flexneri 2a, regulated delayed antigen synthesis, O-antigen, attenuated Salmonella vaccine.
Introduction
Recombinant attenuated Salmonella strains are attractive vaccine platforms that provide needle-free, low-cost, and highly-versatile antigen delivery systems [1]. Salmonella, one of the most extensively studied bacterial genera, is readily manipulated to construct recombinant attenuated Salmonella vaccines (RASVs). Through modification of RASVs and antigen expression systems, host immune responses can be tailored to elicit humoral, cell-mediated, or mucosal immunity bias. One of the key features of current RASV vaccine candidates is the ability to undergo regulated delayed attenuation. Upon oral immunization, the RASV synthesizes virulent factors essential for host colonization and invasion. Due to the absence of carbohydrate inducers in vivo, the RASV strain gradually experienced controlled attenuation. Conversely, the production of antigens maintained by antibiotic-independent balanced lethal plasmids is enhanced in the mammalian system. The combined attributes of delayed attenuation and antigen expression enable productive colonization of the internal effector lymphoid tissues and the robust stimulation of mucosal and systemic humoral and cell-mediated immune responses [2].
The efficiency of recombinant vector vaccines largely depends on the capability to deliver foreign antigens to induce a protective immune response. Many strategies, such as high-copy number expression vectors or strong promoters, have been previously investigated to maximize heterologous antigen expression [3, 4]. Increased energy expenditure dedicating to plasmid replication and maintenance imparts substantial metabolic burden. Furthermore, over-expression of antigens may be detrimental or toxic to the vaccine strain. Therefore, conventional strategies to enhance antigen expression often impair vaccine efficacy [5]. Many alternative approaches, including chromosomal integration of heterologous genes [6], construction of stabilized low-copy-number expression plasmids [7], surface display of heterologous antigen [8], and the use of runaway vectors [9], have been investigated to support antigen synthesis while maintaining vaccine strain immunogenicity. One of the most popular approaches to improve RASV immune responses is the use of in vivo inducible promoters, including pagC [10], nirB [11], spv and dps [12]. In previous work, in order to reduce the metabolic burden imposed by constitutive antigen gene expression, we utilized the Ptrc promoter to develop the regulated delayed antigen synthesis (RDAS) system [13]. In this system, the Ptrc promoter is repressed by LacI, which is produced from an araC PBAD lacI cassette inserted into the chromosome of the RASV. When a vaccine strain is grown in vitro in the presence of arabinose, LacI is synthesized to preclude the Ptrc-controlled expression of the antigen gene. When the Salmonella vaccine reaches tissues in the host, in the absence of arabinose, LacI is no longer produced and the antigen synthesis is initiated [13]. In order to achieve an effective balance of attenuation and immunogenicity of RASVs, we also developed systems of regulated delayed expression of attenuating phenotypes (RDEAP) to maintain the ability of RASVs to colonize internal lymphoid tissues. One of these systems involved native RASV LPS synthesis upon in vitro growth media supplementation with arabinose [14]. The expression of Salmonella LPS at the time of immunization supported invasion of intestinal mucosal cells. Due to the absence of arabinose in the mammalian system, in vivo synthesis of the Salmonella O-antigen was ceased. The regulated delayed LPS deficiency enabled vaccine strain tissue invasion while sparing energy for the synthesis of heterologous antigens. Additionally, the in vivo impairment of native Salmonella O-antigen diminished undesired immune responses generated against the RASV vector.
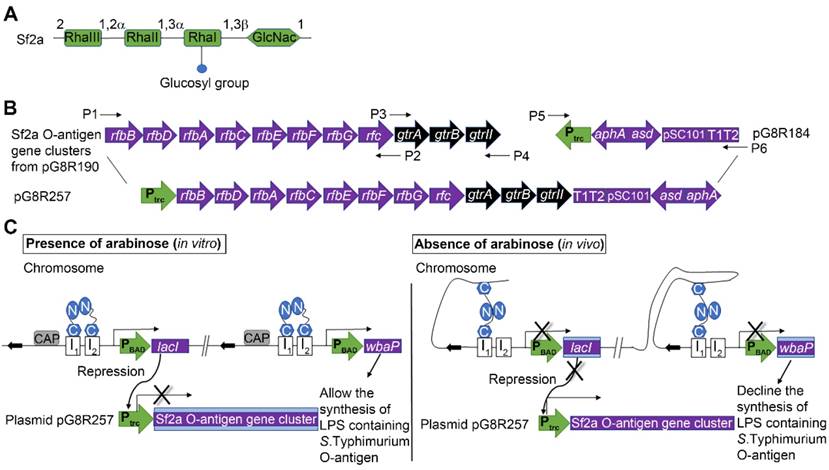
Shigella, the etiological agent of shigellosis or bacillary dysentery, is Gram-negative enteroinvasive bacterial pathogens [15]. Largely as a consequence of poor sanitation conditions, S. flexneri is a leading cause of disease in the developing world. Shigellosis continues to be a major public health concern among children less than 5 years old in many developing countries [16]. Treatment strategies are limited to post-exposure antibiotics which are compromised by the emergence of antibiotic resistant strains. Therefore, the development of equitable and cost-effective vaccines against S. flexneri has been viewed as a high priority by the World Health Organization [17]. Significant efforts have been made in the development of Shigella vaccines, including attenuated or inactivated whole-cell bacterial vaccines, subunit or conjugated vaccines, and outer membrane vesicle vaccines (OMV) [18]. However, all these vaccine candidates suffer from poor immunogenicity, are insufficiently attenuated, or require complicated purification procedures. Currently, no licensed vaccine is available against Shigella infection in humans. S. flexneri, which causes the highest mortality rate among the Shigella species [19], encompasses at least 14 serotypes based on the structure of the lipopolysaccharide O-antigen repeats. All S. flexneri serotypes, aside from serotype 6, are variants of a conserved, repeated structural unit referred to as serotype Y. The serotype Y O-antigen is comprised of repeating tetrasaccharide N-acetylglucosamine-rhamnose-rhamnose-rhamnose units [20]. Differences between classical S. flexneri serotypes are generated by temperate bacteriophages through addition of either glucosyl or O-acetyl residues at specific positions on the repeating units. For instance, S. flexneri 2a (Sf2a) O-antigen has glycosylation on a specific rhamnose residue of the repeating units (Fig. 1A) [21]. As the host immune responses against S. flexneri infection are thought to be serotype specific, which means that immunity generated against a specific S. flexneri serotype only provides protection against strains of the same serotype [22], O-antigen structures are believed to be the primary targets of the immune response. Moreover, there is a substantial body of evidence indicating that antibodies generated against Shigella O-antigen play a key role in affording protection [23]. As pure polysaccharides are mostly T-cell independent (TI) antigens and are poorly immunogenic, glycoconjugate vaccines generated by chemical conjugation of polysaccharides to immunogenic proteins were developed in order to stimulate polysaccharide-specific T-cell dependent (TD) immune responses [24]. However, the production of glycoconjugates requires a complicated purification processes, substantially increasing the cost of a vaccine. Strategies have been employed to construct live vector vaccines to synthesize Shigella O-antigen in rough E. coli or attenuated Salmonella strains. However, a hybrid E.coli vaccine failed to protect Shigella infection in humans because it had no capacity to invade epithelial cells and showed only weak protection even with introduced epithelial invasiveness [25]. A Salmonella vaccine strain, using the attenuated Salmonella vaccine vector Ty21a expressing the Sf2a O-antigen, required laborious manipulation and maintained all the genes required for native Salmonella O-antigen synthesis [26]. The synthesis of both the Shigella and Salmonella O-antigens throughout the bacterial infectious cycle is believed to result in the excessive consumption of nutrients and energy, thereby reducing immunogenicity.
In this study, a new RASV system was developed which combines the RDAS and RDEAP features for delivering of exogenous O-antigens. In this system, the araC PBAD lacI and araC PBAD wbaP cassettes were chromosomally integrated in the parental RASV (with a native wbaP deletion). The LacI-repressible Ptrc promoter was used to control expression of Sf2a O-antigen biosynthesis genes. During in vitro propagation in the presence of arabinose, WbaP was induced to support native Salmonella O-antigen synthesis; whereas, LacI was induced to repress Sf2a O-antigen production. In the absence of arabinose in vivo, native Salmonella LPS was repressed while LacI induction was relieved, thereby Sf2a O-antigen production started. Oral immunization of mice with the new RASV strains delivering Sf2a O-antigen elicited protective immune responses against Sf2a infection together with long term immunity. Taken together, the regulated delayed expression of the Sf2a O-antigen reduced potential toxicity of foreign antigen expression and alleviated RASV metabolic burden. Moreover, the expression of native S. Typhimurium O-antigen upon immunization mimicked the early stages of colonization, thereby enhancing RASV immunogenicity.
Bacterial strains and plasmids used in this study
| Strains or Plasmids | Description | Source |
|---|---|---|
| Suicide and cloning vectors | ||
| pYA4278 | Suicide vector, sacB mobRP4 R6K ori Cm+, pRE112-T-vector | [28] |
| pYA5457 | Suicide vector, pagL7 deletion | pYA4278 [34] |
| pYA5470 | Suicide vector, pagL7 deletion and TT araC PBAD wbaP insertion | This study |
| pYA3700 | Cloning vector containing TT araC PBAD cassette | [29] |
| Recombinant plasmids | ||
| pG8R184 | Asd+ vector, pSC101 ori, Kan+ | Lab stock |
| pG8R190 | Asd+ vector carrying genes involved in the biosynthesis of Sf2a O-antigen (galF-rfc fused with gtrA-gtrII), pSC101 ori | Lab stock |
| pG8R257 | Asd+ vector carrying genes involved in the biosynthesis of Sf2a O-antign (rfbB-rfc fused with gtrA-gtrII) driven by the Ptrc promoter, pSC101 ori | This study |
| E. coli strains | ||
| χ6097 | ∆(lac-pro) rpsL ∆asdA4 ∆ (zhf-2::Tn10)thi 80dlacZ ∆M15 | K-12/F- |
| χ7213 | thi-1 thr-1 leuB6 glnV44 fhuA21 lacY1 recA1 RP4-2-Tc::Mu λpir ∆asdA4 ∆zhf-2::Tn10 | [30] |
| S. enterica serovar Typhimurium strains | ||
| χ3761 | Wild-type UK-1 | Lab collection |
| χ9241 | ∆pabA1516 ∆pabB232 ∆asdA16 ∆araBAD23 ∆relA198::araC PBADlacI TT | [13] |
| χ11316 | ∆pabA1516 ∆pabB232 ∆asdA16 ∆araBAD23 ∆relA198::araC PBADlacI TT ∆wbaP45 | χ9241 [28] |
| χ12491 | ∆pabA1516 ∆pabB232 ∆asdA16 ∆araBAD23 ∆relA198::araC PBADlacI TT ∆wbaP45 ΔpagL7 | χ11316 This study |
| χ12514 | ∆pabA1516 ∆pabB232 ∆asdA16 ∆araBAD23 ∆relA198::araC PBADlacI TT ∆wbaP45 ΔpagL7::TT araC PBAD wbaP | χ12491 This study |
| Strains for immunization | ||
| Vaccine strain 1 (Vac1) | χ12514 electroplated with pG8R257 | |
| Vac2 | χ9241 electroplated with pG8R257 | |
| Vac3 | χ11316 electroplated with pG8R190 | |
| Vac4 | χ9241 electroplated with pG8R190 | |
| Vac5 | χ9241 electroplated with pG8R184 | |
| Shigella flexneri 2a (Sf2a) Strain | ||
| 2457T | Wild-type Sf2a strain | Lab collection |
Methods
Bacterial strains, plasmids, media, and growth conditions
Strains and plasmids used in this work are listed in Table 1. The wild type S. Typhimurium strain and transformants of S. Typhimurium and E. coli strains containing the asdA gene were grown at 37°C in Luria-Bertani (LB) broth or on LB agar. In order to propagate strains with the asdA gene deletion, growth medium was supplemented with 50 μg/ml diaminopimelic acid (DAP) [27]. S. Typhimurium strains carrying Ptrc promoter sequence used for Sf2a O-antigen detection and animal immunizations were grown at 37°C on LB agar or in LB broth containing 0.1% arabinose. The wild-type Sf2a (2457T) was grown at 37 °C in trypticase soy broth (TSB) or on TSB agar containing 0.01% Congo red.
Plasmid and mutant strain construction
Primers used in this study are described in Table 2. Genes involved in the biosynthesis of the Sf2a O-antigen were amplified from plasmid pG8R190 and integrated into the plasmid pG8R184 backbone. Amplicons containing 18-20 base pair overlapping regions (Fig. 1B) were assembled via the NEBuilder HiFi DNA Assembly Master Mix (New England Biolabs (NEB), Ipswich, MA, USA) to generate plasmid pG8R257. The vector control plasmid pG8R184 as well as the pG8R190 and pG8R257 plasmids encoding Sf2a O-antigen biosynthesis genes were transferred to asd-deficient S. Typhimurium by electroporation. The resulting transformants were selected on LB agar plates lacking DAP.
For generating the pagL deletion, approximately 350-bp fragments from upstream and downstream of the pagL gene were amplified from the χ3761 genome, using primers P7/P8 and P9/P10 (Table 2), respectively. Following agarose gel purification, the two PCR fragments were combined through overlapping PCR. The addition of terminal adenine residues at both ends of the joined PCR products was catalyzed by the GoTaq enzyme (Promega, Madison, WI). The PCR product was ligated to the AhdI treated plasmid pYA4278 [28] to generate plasmid pYA5457.
For construction of the ΔpagL ::TT araC PBAD wbaP deletion-insertion mutation, the wbaP gene was amplified by P11 and P12 (Table 2) from the χ3761 genome and inserted into pYA3700 [29]. The fragment TT araC PBAD wbaP was then amplified (with primers P12 and P13) and digested with NotI and SbfI. The gel purified product was ligated to the NotI-SbfI-digested plasmid pYA5457 to form suicide vector pYA5470.
E. coli strain χ7213 [30] harboring the suicide vector of pYA5457 was conjugated with S. Typhimurium strain χ11316 [28] in order to generate the ΔpagL mutation through allelic exchange, resulting in strain χ12491. The ΔpagL::TT araC PBAD wbaP mutation was similarly introduced into χ12491 to yield χ12514. The genotypes of ΔpagL and ΔpagL::TT araC PBAD wbaP mutations were verified by PCR. LPS profiles of S. Typhimurium strains were examined as described by Hitchcock and Brown [31].
Primers used in this study
| Primer ID | Sequence (5'-3') |
|---|---|
| P1 | CAGGAAACAGACCATGAAGATACTTGTTACTGGTGGC |
| P2 | CATTTCTTCTCCTTTATTTTGCTCCAGAAGTGAGG |
| P3 | GCAAAATAAAGGAGAAGAAATGTTAAAGTTATTTGTAAAGTA |
| P4 | TCTCATCCGCCAAAACAGCCTTAAATATTAAATGGAAGCCACC |
| P5 | AAGTATCTTCATGGTCTGTTTCCTGTGTGAAATTG |
| P6 | TAAGGCTGTTTTGGCGGATGAGAGAAG |
| P7 | AGACTATCTTTACTGGCAGG |
| P8 | CACCTGCAGGGCATGCGGCCGCTCCACCACCATTTCAATGTC |
| P9 | GAGCGGCCGCATGCCCTGCAGGTGAAGTTGAATAACAATTAGC |
| P10 | TGCGGATGAAGCTGCCGACC |
| P11 | GCAGTCGACAAGGCTCTATATGGATAATATTGATAATAAG |
| P12 | ACGGTACCTGCAGGCTTAATACGCACCATCTCGC |
| P13 | ATGCGGCCGCAGATCTTTTATTATTCTATCC |
Western blotting
Wild-type S. Typhimurium χ3761, wild-type Sf2a 2457T, and the mutant strains of S. Typhimurium harboring recombinant plasmids were grown overnight at 37 °C in LB broth supplemented with 0.1% arabinose when needed. LPS were extracted and fractionated by Tris-glycine PAGE as described previously [31]. The separated LPS were transferred to a nitrocellulose membrane and immunologically detected with 1:1000 dilutions of rabbit polyclonal antisera against S. flexneri type 2a O-antigen (Tianjin Biochip Corporation, Tianjin, China) and Salmonella O Group B Antiserum (BD, Sparks, MD, USA) for the identification Sf2a O-antigen and S. Typhimurium O-antigen profiles, respectively.
Animal experiments
All animal experimentation was carried out in compliance with the recommendations of the Animal Welfare Act and US Government Principles for the Utilization and Care of Vertebrate Animals Used in Testing, Research, and Training. The protocol was reviewed and approved by the University of Florida Institutional Animal Care and Use Committee.
For immunization, the RASV strains were grown overnight in LB broth at 37 °C. LB broth supplemented with 0.1% arabinose was used for growth of RDAS strains. The overnight cultures of vaccine strains were diluted 1:100 and grown at 37°C to an OD600 of 0.8 to 0.9. Each group consisting of 16 seven-week-old female BALB/c mice (Charles River Laboratories, Wilmington, MA) were acclimated for 7 days upon arrival before initiating experiments. Groups of mice were orally inoculated with 20 μl of buffered saline with gelatin (BSG) containing approximately 1×109 CFU of each vaccine strain on day 0 and boosted on day 14 with an identical dose of the same strain. Mice administrated with 1×109 CFU of χ9241 (pG8R184) or with 20 μl of plain BSG were included as control groups. Three mice in each group were euthanized on day 7 and day 75 for detection of cell-mediated immune responses and memory immune responses, respectively. Blood samples collected on day 28 through mandibular vein puncture were allowed to coagulate at 37 °C for 2 h. The serum samples were collected and stored at -80 °C, after centrifugation. Vaginal-wash samples were collected and stored as described previously [13].
The overnight culture of a Congo red positive Sf2a 2457T colony was diluted 1:100 in fresh TSB and grown at 37 °C (up to OD600 0.8-0.9). Two weeks after the secondary immunization (day 28), 10 mice in each group were challenged with 100 μl of BSG containing 2457T (5 × 107 CFU per mouse) through the intraperitoneal route as described previously [32].
Antigen preparation and ELISA
S. Typhimurium LPS and Sf2a LPS were extracted using the LPS extraction kit (Boca Scientific, Westwood, MA) from strains of χ3761 and 2457T, respectively. An enzyme-linked immunosorbent assay (ELISA) was used to assay antibodies in serum and vaginal washes against Sf2a and S. Typhimurium LPS. The 96-well flat-bottom microtiter plates were coated overnight with 100 μl/well of poly-L-lysine (MW.260000, 10 μg/ml) (Sigma Aldrich, St. Louis, MO) in 0.01 M phosphate-buffered saline (PBS) at pH7.2 at room temperature. The LPS samples suspended in PBS were applied with 100 μl volumes in each well. The coated plates were incubated for 1 h at 37°C and blocked with the SEA BLOCK Blocking Buffer (Thermo Fisher Scientific, Waltham, MA) for 1 h at room temperature. A 100 μl/well of diluted serum sample (1:100) or vaginal wash (1:10) was added to individual wells in triplicate and incubated for 1 h at 37°C. Plates were then treated with biotinylated goat anti-mouse IgG, IgG1, IgG2a or IgA (Southern Biotechnology Associates, Birmingham, AL). After incubation of wells with a streptavidin-alkaline phosphatase conjugate (Southern Biotechnology) for 1 h at 37°C, the p-nitrophenyl phosphate (PNPP, Thermo Fisher Scientific) was added for color development. The optical density (OD) unit was read at 405 nm using an automated ELISA plate reader (model EL311SX; Biotek, Winooski, VT). Unconjugated IgG (IgG-UNLB), IgG1(IgG1-UNLB), IgG2a (IgG2a-UNLB) and IgA (IgA-UNLB) derived from normal mice (Southern Biotechnology) were serially diluted in 2-fold steps in PBS (IgG-UNLB, from 5 μg/ml to 40 ng/ml; IgG1-UNLB and IgG2a-UNLB, from 1 μg/ml to 8 ng/ml; IgA-UNLB, from 62.5 ng/ml to 0.488 ng/ml). The OD values at 405nm were plotted against the representative concentrations of the diluted unconjugated antibody solutions to create the standard curves using linear regression in Excel (R2≥0.98). The antibody concentrations were calculated from the corresponding curve.
Splenic lymphocytes proliferation
Mice were euthanized 7 days after primary immunization (3 mice per group) and spleens were removed aseptically. Spleen cell suspensions were obtained by passing through a 70 μm sterile cell strainer (Fisher brand, Houston, TX; 1 spleen/strainer) and the erythrocytes were lysed with RBC lysis buffer (eBioscience, San Diego, CA). Splenocytes were resuspended in RPMI 1640 supplemented with 5% fetal calf serum (FCS). The suspended cells were inoculated in a 96-well cell culture plate (3 × 105 cells/well) and incubated in a 5% CO2 incubator at 37 °C. For experimental test groups, cells were cultured with the Sf2a LPS or S. Typhimurium LPS (30 ng/well). As a positive control, cells were cultured with phytohemagglutinin (PHA, 10 μg/ml). For the negative control, cells were incubated with sterile culture medium. After 72 h of incubation, 20 μl of MTT (5 mg/ml in PBS) was added to each well. After 4 h of incubation at 37 °C in the 5% CO2 incubator, cells were centrifuged at 200 g for 5 min and the supernatants were removed. The colored crystals were dissolved with 100 μL per well of DMSO by agitation at 37 °C for 10 min. The OD was read at 490 nm using the microplate reader (ELISA plate reader). Cell proliferation was calculated as stimulation index (SI), which is the ratio of the mean absorbance of triplicate wells with PHA or LPS stimulation to the mean absorbance of triplicate wells of the negative control.
Cell Stimulation and Flow Cytometry
Single cell suspensions of splenic lymphocytes were stimulated ex vivo with the Sf2a LPS or S. Typhimurium LPS (50 ng / 2 × 106 cells) for 8 h in the presence of Protein Transport Inhibitor Cocktail (eBioscience). Cell surface staining was performed using fluorophore-conjugated antibodies at 4 °C for 30 min. Intracellular staining was performed by eBioscience™ Intracellular Fixation and Permeabilization Buffer Set. All the cells were run on the BD LSRFortessaTM cell analyzer. Dead cells were excluded by Zombie RedTM (Biolegend, San Diego, CA) for detection of cytokines and IgG+ memory B (BM) cells, or Helix NPTM Blue (Biolegend) for detection of T follicular helper (Tfh) Cells. Cells were stained with the rat anti-mouse CD16/CD32 monoclonal antibody (eBioscience) to block Fc receptors.
Cytokine expression of CD4+ T cells was detected 7 days after primary immunization with rat anti-mouse CD3-Alexa Fluor 700, CD4-FITC, IFNγ-APC/Cy7 and IL-4-Alexa Fluor 647, using APC/Cy7 conjugated rat IgG1κ and Alexa Fluor 647 conjugated rat IgG1κ (Biolegend) as isotype controls. To determine the percentage of antigen-specific cells, the ratio of cytokine-positive cells of unstimulated sample was subtracted from the antigen-stimulated value of the same mouse.
On day 75 (61 days after secondary immunization), Tfh cells were identified as CD3+CD4+CD44+CD62L+CXCR5+PD-1+IL-21+ lymphocytic cells after staining with anti-mouse CD3-PE, CD4-FITC, CD44-APC/Cy7, CD62L-Alexa Fluor 700, CXCR5-Brilliant Violet 421TM, PD-1-PE/DazzleTM 594, and IL-21-APC / isotype APC conjugated Rat IgG2aκ (Biolegend). IgG+ BM cells were identified as IgG+B220low/intCD80+IgD- or IgG+B220highCD80+IgD- live lymphocytes by staining with anti-mouse B220-Pacific Blue, CD80-FITC, IgD-Alexa Fluor 700, and IgG-APC (Biolegend) / isotype APC conjugated Goat IgG (R&D Systems, Minneapolis, MN). Data from both of the Tfh cells and IgG+ BM assays were further displayed as antigen-specific cells which were calculated by subtracting the number of unstimulated Tfh or BM cells from antigen-stimulated values.
Shigella-specific serum bactericidal antibody (SBA) assay
Serum samples in each group were heated at 56 °C for 30 min and pooled. Inactivated serum samples (with an initial dilution of 1:10) were added to 96-well U-bottom plates and serially diluted 2-fold in Muller Hinton broth. The final volume of the reaction mixture was made up to 100 μl/well, containing 10 μL of diluted serum, 10 μL of the Sf2a 2457T cells (1000 CFU), 25 μl of baby rabbit complement (BRC, Sigma). The plates containing the reaction mixture were incubated for 1 h at 37°C in a shaker at 200 rpm. At the end of incubation, 10 μl of each reaction mixture was plated on the TSB agar and grown overnight. Colonies were counted and the percentage of killed bacteria was calculated using the equation [1- (the average CFU of the tested control / the average CFU of negative control]. For each sample, the testing and plating were performed in duplicate (four independent CFUs per sample), the four CFU counts were averaged for each sample to calculate the percentage of killed bacteria.
Statistical analyses
The GraphPad Prism 5 software package (Graph Software, San Diego, CA) was used for statistical analyses. The one-way analysis of variance (ANOVA) followed by Bonferroni's multiple comparison test was used to evaluate differences in antibody concentrations and cell numbers of antigen-specific BM and Tfh cells. Bonferroni's multiple comparison test in a two-way ANOVA was used to analyze the differences in percentages of antigen-specific cells and stimulation indexes of proliferation. All data mentioned above were expressed as means with deviations. The Kaplan-Meier method was applied to analyze the survival data. The survival differences were assessed by the log-rank (Mantel-Cox) test. The significance levels were set at 0.05, 0.01 and 0.001.
Results
RDAS system was constructed for delivering Sf2a O-antigen by S. Typhimurium
In order to reduce the adverse effects of RASV strains caused by antigen overexpression, an RDAS system was previously established [13]. Moreover, this RDAS system was utilized to facilitate the controlled synthesis of LPS containing exogenous O-antigens in a ΔwbaP mutant [28]. Previous studies demonstrated that the ΔwbaP deletion impaired synthesis of O-antigen O-unit (undecaprenyl-phosphate-O-unit), thereby preventing S. Typhimurium LPS production.
In this study, in order to achieve the regulated delayed synthesis of Sf2a O-antigen in RASV strains while maintaining the cell invasive capacity as wild-type Salmonella, we developed a new RDAS system by chromosomally integrating the TT araC PBAD wbaP cassette in place of the pagL gene of χ11316 [28] (Fig. 1C) to generate RDAS strain χ12514. In this system, both lacI and wbaP are placed under the control of the arabinose-inducible araC PBAD promoter. When RDAS strain χ12514 is cultured in vitro in the presence of arabinose, LacI is stimulated to repress the transcription of Ptrc-driven exogenous O-antigen genes and WbaP is expressed to facilitate the synthesis of S. Typhimurium native LPS. Upon in vivo inoculation, due to the absence of free arabinose, the expression of LacI and WbaP are repressed in strain χ12514. Therefore, S. Typhimurium native LPS is gradually suppressed in a mammalian system. PagL is capable of modifying the lipid A structure of S. Typhimurium through removal of the acyl chain [33]. Here, pagL was chosen as the integration site for the ΔpagL::TT araC PBAD wbaP deletion/insertion mutation, because deletion of pagL did not alter the virulence of the Salmonella and maintaining the immunogenicity and capability of colonization [34].
The Asd+ recombinant plasmid pG8R257 (Fig. 1B) was constructed by integrating genes involved in the biosynthesis of the basic O-antigen (rfbB-rfc) and the Sf2a serotype-specific modification genes (gtrA-gtrII) under the control of Ptrc promoter. Plasmid pG8R257 was introduced into RDAS strains χ12514 (with araC PBAD-regulated wbaP) and χ9241 (without mutation in wbaP), yielding Vac1 and Vac2, respectively. The pG8R190 plasmid carrying original Sf2a O-antigen gene cluster was also introduced into strains χ11316 and χ9241 respectively, referred to as Vac3 and Vac4. Vac5 reflects strain χ9241 containing the pG8R184 empty control vector.
The RDAS system for delivering Sf2a O-antigen by attenuated Salmonella strains. (A) Structure of the Sf2a O-antigen. (B) Construction of plasmid pG8R257. Plasmid sequences include genes involved in the biosynthesis of Sf2a O-antigen (rfbB-rfc fused with gtrA-gtrII) under the control of Ptrc promoter; the aphA and asdA genes. (C) Diagram of the RDAS system, modified from Shifeng et al, 2010. The RDAS strain χ12514 could express LacI and WbaP under the control of the arabinose-inducible araC PBAD promoter in the presence of arabinose in vitro. Therefore, the transcription of Ptrc-driven Sf2a O-antigen genes was repressed by LacI and synthesis of S. Typhimurium native LPS initiated at the participation of WbaP. In animal tissues, in the absence of free arabinose, LacI and WbaP could not be produced. Thus, the transcription of Sf2a O-antigen genes started and the S. Typhimurium native LPS could no longer be synthesized. Rha, rhamnose; GlcNac, N-acetylglucosamine.
LPS synthesis in RDAS strains and constitutive antigen synthesis strains. (A) LPSs from transfected strains grown in nutrient broth with (+) or without (-) 0.1% arabinose were separated by 12% SDS-PAGE, and detected by silver staining method. (B) Detection of Sf2a O-antigen synthesis with the antisera against S. flexneri type 2a O-antigen by western blot. (C) Identification of S. Typhimurium O-antigen with the rabbit polyclonal Salmonella group B antisera by western blot.
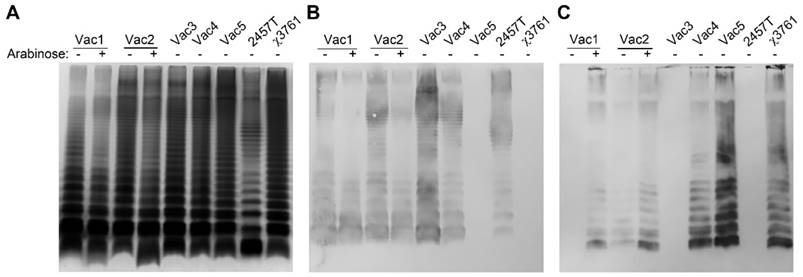
When cultured in the presence of 0.1% arabinose, both Vac1 and Vac2 indicated unique LPS banding patterns compared with the synthesis pattern when cultured in the absence of arabinose (Fig. 2A). As expected, Vac1 (χ12514 with pG8R257) primarily synthesized LPS recognized by antisera generated against S. flexneri type 2a O-antigen (Fig. 2B) in the absence of arabinose. The detection of Sf2a O-antigen indicated that all the genes from Ptrc to T1T2 in the pG8R257 were transcribed and expressed successfully. When 0.1% arabinose was supplemented in the culture medium, S. Typhimurium LPS was detected in the Vac1 (χ12514 with pG8R257) strain (Fig. 2C). The Vac2 strain (χ9241 with pG8R257), which retained all the genes required for native S. Typhimurium LPS synthesis, produced a combination of Sf2a O-antigen and S. Typhimurium O-antigen when cultured in the absence of arabinose. When 0.1% arabinose was added to the growth medium, the native Salmonella LPS was primarily produced by Vac2. Only the Sf2a O-antigen could be detected in strains Vac3 (χ11316 with pG8R190) and both Sf2a O-antigen and Salmonella O-antigen could be detected in Vac4 (χ9241 with pG8R190). Conversely, strain Vac5 (χ9241 with pG8R184) solely synthesized LPS recognized by anti-S. Typhimurium O-antigen antibody.
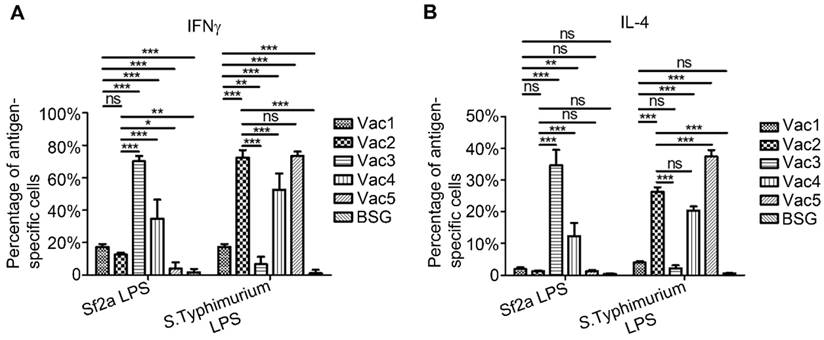
RDAS strains elicited lower Sf2a LPS-specific CD4+-T-Cell responses compared to strains that constitutively produce Sf2a O-antigen
T cell activation is believed to be a key element for the development of efficient humoral and cell-mediated immune responses, along with the priming of memory immunity. However, natural infection of Shigella has been described to inhibit CD4+-T-cell migration, therefore compromising host immune responses [35]. In our previous study, it was demonstrated that immunization of mice with RASV strains can increase CD4+ cytokine responses, thereby stimulating a Th1-type response [36]. In this study, the Sf2a LPS-specific cytokine expression of CD4+ T cells was detected using intracellular staining combined with flow cytometry 7 days after primary immunization. The RDAS strains of Vac1 (χ12514 with pG8R257) and Vac2 (χ9241 with pG8R257) significantly increased IFNγ stimulation relative to the Vac5 (empty plasmid control) and BSG treated groups. However, the levels of IFNγ produced in response to RDAS strains Vac1 and Vac2 were significantly less pronounced compared to strains Vac3 (χ11316 with pG8R190) and Vac4 (χ9241 with pG8R190), which constitutively synthesized Sf2a O-antigen (Fig. 3A). Both RDAS strains Vac1 and Vac2 failed to elicit robust Sf2a LPS-specific IL-4 on day 7 post primary immunization. To the contrary, the Vac3 and Vac4 strains which constitutively produced Sf2a O-antigens induced significantly increased levels of IL-4 production relative to the RDAS strains as well as the Vac5 and BSG controls (Fig. 3B). S. Typhimurium LPS-specific cytokines production was significantly lower in the groups immunized with strains Vac1 (synthesizing Sf2a O-antigen in vivo and S. Typhimurium O-antigen in vitro) and Vac3 (constitutively producing Sf2a O-antigen only) compared to the Vac2, Vac4 and Vac5 strains which synthesized S. Typhimurium O-antigen constitutively.
RDAS strains delivering Sf2a O-antigen induced robust humoral immune responses
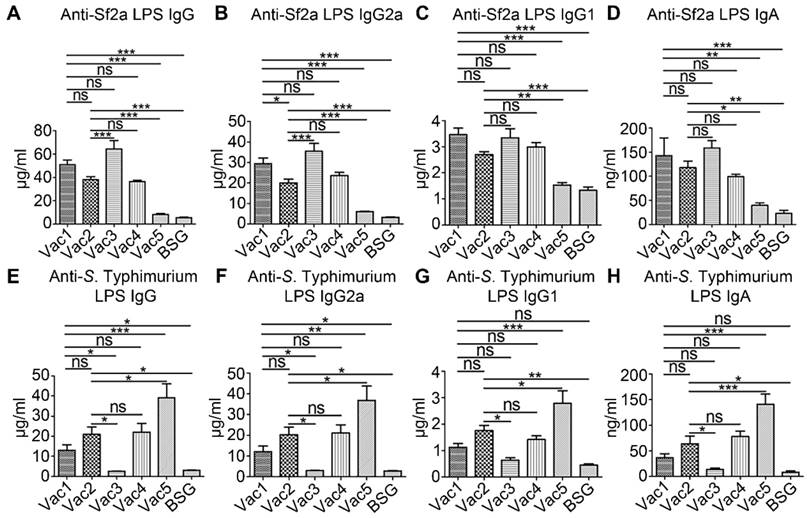
Serum IgG and mucosal IgA responses against Sf2a LPS and S. Typhimurium LPS were measured by ELISA 2 weeks after secondary immunization. Both of the Vac1 and Vac2 RDAS strains induced significantly increased levels of Sf2a LPS-specific IgG relative to the Vac5 and BSG control groups (Fig. 4A). Moreover, the RDAS strain Vac1 (χ12514 with pG8R257), which synthesized native S. Typhimurium LPS in the presence of arabinose in vitro and produced LPS containing Sf2a O-antigen in vivo, triggered Sf2a LPS-specific IgG production comparable to the Vac3 strain (χ11316 with pG8R190) constitutively producing Sf2a O-antigen. The Vac2 strain (χ9241 with pG8R257), which synthesized mainly Salmonella native LPS in vitro but produced LPS containing both of S. Typhimurium O-antigen and Sf2a O-antigen in vivo, induced similar levels of IgG compared to Vac4, which constitutively produced both native S. Typhimurium and foreign Sf2a O-antigens. The levels of S. Typhimurium LPS-specific IgG induced by the Vac1 and Vac2 RDAS strains were significantly enhanced relative to Vac3 (producing only Sf2a O-antigen) immunized and BSG treated groups, but lower than Vac5 (producing only S. Typhimurium O-antigen) immunized group (Fig. 4E). Further analysis revealed that IgG2a was the predominant subtype of LPS-specific IgG elicited by the RASV strains (Fig. 4B, 4C, 4F and 4G). Both of the Vac1 and Vac2 RDAS strains stimulated similar levels of Sf2a LPS-specific vaginal IgA compared to the Vac3 and Vac4 strains that constitutively synthesized Sf2a O-antigen (Fig. 4D). The Vac2 strain (with Sf2a O-antigen in vivo and constitutively synthesizing S. Typhimurium LPS) stimulated the production of S. Typhimurium LPS-specific vaginal IgA; whereas, Vac1 (synthesizing Sf2a O-antigen in vivo and S. Typhimurium O-antigen in vitro) failed to elicit vaginal mucosal IgA responses (Fig. 4H). The level of vaginal IgA generated against S. Typhimurium LPS in Vac2 immunized group was comparable to that detected in the Vac4 (constitutively producing both of Sf2a and S. Typhimurium O-antigen) immunized group. The Vac5 (producing only S. Typhimurium O-antigen) group indicated significantly increased vaginal IgA against S. Typhimurium LPS relative to all other treatment groups.
Cytokine expression of CD4+ T cells stimulated with Sf2a or S. Typhimurium LPS ex vivo at 7 days after primary immunization. Bar graphs depicted the proportions of antigen-specific cells expressing IFNγ (A) and IL-4 (B), which were calculated by deducting the percentages of cytokine-positive cells of unstimulated samples from the antigen-stimulated values of corresponding samples. Data were summarized from two (n=6) independent experiments. * P < 0.05, ** P <0.01, *** P <0.001.
Systemic and mucosal antibody concentrations against Sf2a and S. Typhimurium LPS. Serum IgG, IgG2a, IgG1and mucosal IgA against Sf2a LPS (A-D) and S. Typhimurium LPS (E-H) were measured by ELISA at 14 days after secondary immunization. The RDAS strains (Vac1 and Vac2) could stimulate comparable levels of Sf2a LPS-specific antibody responses with the constitutive antigen synthesis strains (Vac3 and Vac4). * P < 0.05, ** P <0.01, *** P <0.001.
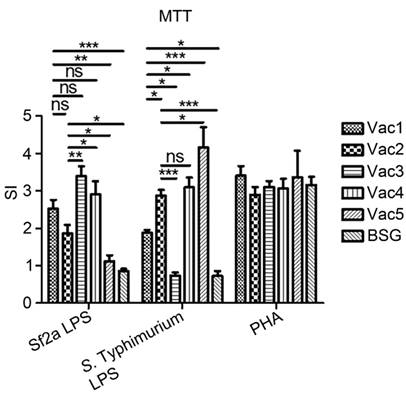
Spleen cell proliferation in response to Sf2a and S. Typhimurium LPS. Spleen cell suspensions were prepared from mice immunized with indicated Salmonella strains or BSG at 7 days after primary immunization. Cells (3 × 105 cells/well) were stimulated by Sf2a LPS, S. Typhimurium LPS, PHA (positive control), or culture medium (negative control) for 72 h. Cell proliferation was measured with the MTT based colorimetric assay. * P < 0.05, ** P <0.01, *** P <0.001.
RDAS strains stimulated Sf2a LPS-specific splenic lymphocyte proliferation
Priming of cell-mediated immune responses plays important roles in inducing protective immunity against S. flexneri infections [35]. The proliferative responses of spleen cells in response to Sf2a LPS and S. Typhimurium LPS stimulation were measured 7 days after primary immunization (Fig. 5). Both of the Vac1 and Vac2 RDAS strains stimulated significantly elevated Sf2a LPS-specific proliferation compared to the Vac5 and the BSG control groups. The Vac1 strain, synthesizing Sf2a O-antigen in vivo and S. Typhimurium O-antigen in vitro, demonstrated comparable capability in stimulating Sf2a LPS-specific splenocyte proliferation to the strains constitutively producing Sf2a O-antigen (Vac3 and Vac4). The Vac2 strain, which produced Sf2a O-antigen in vivo and constitutively synthesized S. Typhimurium LPS, induced equivalent splenocyte proliferation upon Sf2a LPS stimulation compared to Vac1. However, the splenocyte proliferation of the Vac2 groups was significantly lower than that of the Vac 3 (constitutively producing Sf2a O-antigen only) and Vac4 (constitutively producing both of Sf2a and S. Typhimurium O-antigen) groups. When stimulated with S. Typhimurium LPS, the splenocyte proliferation in Vac1 group was significantly higher than the Vac 3 immunized and the BSG treated groups. The S. Typhimurium LPS-specific splenocyte proliferation in the Vac2 group was greater than Vac1, but markedly lower than that of Vac5 (empty plasmid control, producing only S. Typhimurium LPS).
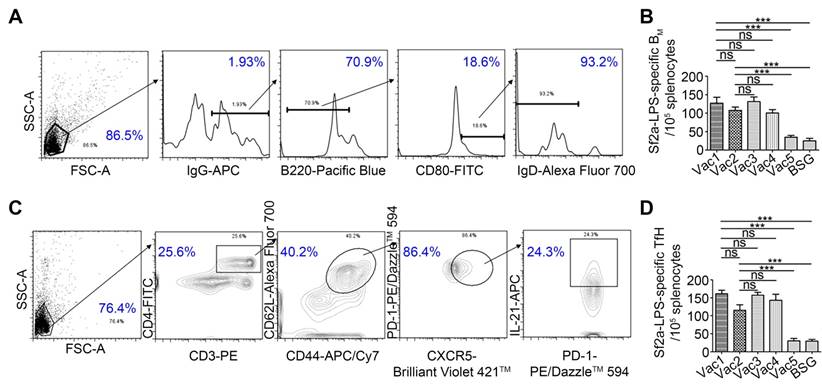
RDAS strains elicited Sf2a LPS-specific BM and Tfh cell responses
Memory B cell (BM) responses to Shigella antigens, including LPS, have been indicated to be a suitable surrogate of protection in shigellosis [37]. Memory B cells are believed to be differentiated from germinal center (GC) B cells, with the help of Tfh cells, which may provide support for GC induction, B cell affinity maturation, and generation of memory B cells [38]. Memory Tfh cells may also provide rapid assistance to BM cells in the spleen and lymph nodes (LNs) [39]. In this study, we measured both the Sf2a LPS-specific IgG+ BM and Tfh cell responses by flow cytometry on Day 75 (61 days after secondary immunization) (Fig. 6). Upon stimulation with Sf2a LPS, the ratio of IgG+B220low/intCD80+IgD- BM cells [40, 41] in groups immunized with the RDAS strains (Vac1 and Vac2) was significantly increased relative to the Vac5 (empty plasmid control) and BSG treated groups. Moreover, the IgG+B220low/intCD80+IgD- BM cell responses in the RDAS treated groups were comparable to the groups treated with RASV strains constitutively producing Sf2a O-antigen (Fig. 6A and 6B). The ratio of IgG+B220highCD80+IgD- cells showed no difference among groups (data not shown). Numbers of Tfh cells (CD3+CD4+CD44+CD62L+CXCR5+PD-1+IL-21+) with or without Sf2a LPS stimulation were also detected and analyzed on Day 75. These findings indicated that Sf2a LPS-specific Tfh cell responses were significantly increased in the RDAS treated groups relative to the empty vector and negative control groups. The level of Tfh cell responses were comparable among the groups immunized with the RDAS strains (Vac1 and Vac2) and the groups administered with strains constitutively synthesizing Sf2a O-antigen (Vac 3 and Vac 4) (Fig. 6C and 6D).
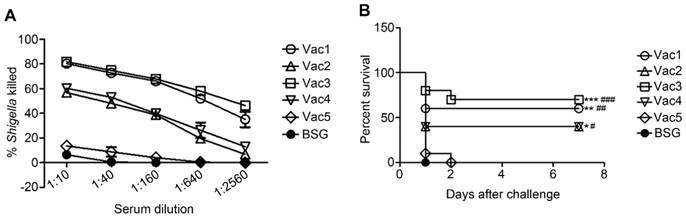
Serum bactericidal assay
Serum bactericidal assays were performed to determine the functional capability of Sf2a-specific antibodies in killing 2457T through the complement-mediated pathway (Fig. 7A). Pooled serum samples collected from groups, which received the Vac1 strain (synthesizing Sf2a O-antigen in vivo and S. Typhimurium O-antigen in vitro) and the Vac3 strain (constitutively producing only Sf2a O-antigen) showed similar levels of killing capacities, which appeared to be more efficient than the other four groups in killing 2457T. The level of serum killing activity of the group immunized with Vac2 (producing Sf2a in vivo and constitutively synthesizing S. Typhimurium LPS) was equivalent to that of Vac4 (constitutively producing both of Sf2a and S. Typhimurium O-antigen) immunized group. The serum killing capacity of all experimental groups surpassed that of Vac5 (empty plasmid control) and BSG treated groups.
Protection conferred by RDAS strains against Sf2a challenge
A murine model system was utilized in order to evaluate the protective efficacy conferred by Salmonella strains synthesizing Sf2a O-antigen against virulent Sf2a challenge. BALB/c mice were orally immunized with Sf2a O-antigen-synthesizing strains administered in two identical doses, with the booster delivered two weeks following the primary immunization event. Two weeks after receiving the second dose, mice were intraperitoneally challenged with 5 × 107 CFU of virulent Sf2a strain 2457T. All RASV strains producing Sf2a O-antigen provided significant protection against Sf2a challenge compared to the empty-plasmid and BSG controls (Fig. 7B). Immunizations with the Vac1 and Vac3 strains (synthesizing only Sf2a O-antigen in vivo) demonstrated slightly enhanced levels of protection relative to the Vac2 and Vac4 strains (producing both of Sf2a O-antigen and S. Typhimurium O-antigen in vivo), although this trend was not statistically significant. The RDAS strain Vac1 (synthesizing Sf2a O-antigen in vivo and S. Typhimurium O-antigen in vitro) provided a comparable level of protection to the Vac3 strain (constitutively producing Sf2a O-antigen only).
BM cells and Tfh cells were determined by flow cytometry ex vivo on day 75 (61 days after secondary immunization). (A) Gating strategy used to identify BM cells. (B) The antigen-specific BM cells were calculated by subtracting the number of IgG+B220low/intCD80+IgD- B cells in the sample without stimulation from that stimulated with Sf2a LPS. (C) Tfh cells were gated as the CD3+CD4+CD44+CD62L+CXCR5+PD-1+IL-21+ cells. (D) The Sf2a-LPS-specific Tfh cells were counted by subtracting the number of CD3+CD4+CD44+CD62L+CXCR5+PD-1+IL-21+ cells in the unstimulated sample from that stimulated by Sf2a LPS. Data from two independent experiments (n=6) were statistically analyzed. * P < 0.05, ** P <0.01, *** P <0.001.
Sf2a-specific serum bactericidal assays (A) and protection of mice against the challenge of virulent Sf2a strain 2457T (B). (A) Serially diluted serum from each group collected on Day 28 (two weeks after secondary immunization) were used to determine and assay the percentages of killed 2457T. (B) Female BALB/c mice immunized with 2 doses of vaccine strains (1 × 109 CFU per dose) expressing Sf2a O-antigen showed significantly greater protection against the challenge of 2457T (5 × 107 CFU per mouse) through the intraperitoneal route two weeks after the secondary immunization, compared to the empty-plasmid and BSG controls. * P < 0.05 compared to BSG, ** P<0.01 compared to BSG, *** P <0.001 compared to BSG, # P < 0.05 compared to Vac5, ## P <0.01 compared to Vac5, ### P <0.001 compared to Vac5.
Discussion
The key aspects of an ideal RASV system include complete attenuation of the live vaccine strain whilst retaining robust invasive capability, thereby supporting a robust immune response without compromising safety [1]. To achieve this objective, we combined the RDAS and RDEAP attributes to develop vaccine strains which exhibit controlled expression of the Sf2a O-antigen. In this system, during in vitro propagation in the presence of arabinose, the vaccine strain synthesizes native Salmonella LPS but represses the production of exogenous Sf2a O-antigen, and after invasion of the host tissues, in the absence of arabinose, the synthesis of native LPS is repressed and the transcription of exogenous Sf2a O-antigen biosynthesis is induced.
Both the Vac1 and Vac2 strains, which have the araC PBAD lacI cassette inserted into the chromosome, efficiently synthesize the Sf2a O-antigen when cultured in the absence of arabinose. It is important to note that the Sf2a O-antigen was still detected in Vac1 and Vac2 when cultured in the presence of 0.1% arabinose, indicating that the expression of a single copy of lacI partially represses the transcription of the Sf2a O-antigen genes under the control of the Ptrc promoter (Fig. 2B). Increased copy numbers of araC PBAD lacI may suppress the expression of exogenous antigen more efficiently. However, overexpression of lacI was reported to disrupt S. Typhimurium colonization in host tissues [42]. In this study, the level of LacI expressed from the single copy in our new RDAS system induced an efficient humoral immune response. The other approach to achieve the tight regulation of transcription of the Sf2a O-antigen gene cluster is via modification of a Shine-Dalgarno (SD) sequence and/or start codon of Ptrc promoter [14]. As this promoter needs to drive a 13kb gene cluster in our construct, we did not investigate the effects of alteration of SD sequence or start codon of Ptrc on the transcription of the Sf2a O-antigen gene cluster. Therefore, the use of single copy of araC PBAD lacI and a Ptrc carrying an appropriate SD sequence and/or start codon may be pursued in other RASV vaccine candidates in order to maintain a balance between attenuation and immunogenicity.
In this study, immunization with the RDAS strains (Vac1 and Vac2) induced significantly decreased production of IFNγ and IL-4 in response to Sf2a LPS-stimulation compared to strains synthesizing Sf2a O-antigen constitutively (Vac3 and Vac4). This result is consistent with a previous study in which impaired cytokine immune responses were attributed to the expression level of LacI in the RDAS system [43]. Our findings suggest that the decreased cytokine expression in this study may result from a delay in Sf2a O-antigen production at the early stage of bacteria colonization. The O-antigen of Gram-negative bacteria is synthesized by three groups of proteins, including proteins for nucleotide sugar biosynthesis, sugar transferases and proteins involved in O-antigen assembly and export [44]. Genes involved in nucleotide sugar biosynthesis and sugar transfer were cloned under the control of the regulated promoter in our system. As O-antigen biosynthesis is a multifactorial process, the controlled stimulation of O-antigen production may not be as efficient as regulated protein antigen expression. Thus, when colonizing the mammalian system, the induction of O-antigen biosynthesis by the RDAS candidates may suffer a detrimental lag phase. The potential lapse in O-antigen production may be responsible for the reduced stimulation of host immune responses relative to the constitutive antigen synthesis systems. This reminds us of the importance to consider about the lag phase in the initiation of adaptive immunity, when the RDAS vaccine strains are applied for delivering O-antigens in the future.
The synthesis of native Salmonella LPS O-antigen was controlled in this study through arabinose-regulated expression of WbaP. This RDAS system (Vac1) solely produced Salmonella native LPS in vitro in the presence of 0.1% arabinose, and gradually suppressed the synthesis of Salmonella native LPS upon host tissue invasion where free arabinose was no longer available. Moreover, due to the absence of arabinose in the mammalian system, inducible LacI expression was alleviated, thereby enabling synthesis of LPS containing exogenous Sf2a O-antigen. The Vac1 strain elicited increased anti-Sf2a-LPS IgG2a and serum bactericidal activity relative to the Vac2 strain, which lacked modified WbaP regulation and produced LPS containing both Salmonella native O-antigen and Sf2a O-antigen in vivo. This result indicated that depletion of native LPS synthesis in vivo may benefit the RASV by sparing nutrients and energy for producing exogenous antigens, thereby increasing antigen-specific immune responses. The Vac3 strain carrying a ∆wbaP mutation (producing only Sf2a O-antigen constitutively) induced comparable protective immune responses to the Vac1 strain (synthesizing S. Typhimurium O-antigen in vitro and Sf2a O-antigen in vivo) upon 14 days after secondary immunization. This result suggests that constitutive expression of LPS containing the Sf2a O-antigen does not negatively influence the ability of RASV strain of Vac3 to invade the host and induce an efficacious immune response, indicating that smooth LPS of RASV is essential for initiating the colonization and invasion of host cells and later inducing immune responses regardless of its mixed or solely O-antigen presence on the surface of RASV [45].
Natural protective immunity against Shigella infection is developed after several rounds of re-infection and is notably short-lasting [35]. Moreover, protective immunity elicited by natural infection is poorly established in young patients [46]. In this study, the RDAS strains producing LPS containing Sf2a O-antigen induced efficient IgG+ BM and memory Tfh cell responses, of which the levels were comparable to those of the constitutive antigen synthesis strains. Furthermore, the RDAS strains induced robust humoral immune responses and provided significant protection against the challenge of wild-type Sf2a 2457T in a murine model system.
Although the RDAS system developed in this study shows promise for prevention of Shigella infection, additional refinement should be pursued before progressing to human trials. One major issue is the lack of clinically relevant non-primate animal models strictly mimicking human infection with Shigella. Previous studies have attempted to develop Shigella infection models using rabbits, guinea pigs and mice. As infection of the rabbit ileal loop can induce acute inflammation and tissue destruction, this model is more suitable for studying Shigella interactions with the intestinal barrier and subsequent induction of an innate immune response [47, 48]. In the guinea pig keratoconjunctivitis model, Shigella invades the corneal epithelia and spreads to adjacent cells, causing ulcerative conjunctivitis. However, due to the limited availability of immunological tools, characterizing the immune response induced in this model remains a challenging limitation [49]. Thus, mouse models are widely used due to the low cost, ease of handling, and availability of immunological research tools. Since oral infection with Shigella in adult mice does not cause diarrheal disease, the mouse pulmonary pneumonia infection model is commonly used to examine protective efficacy [50, 51]. However, the clinical irrelevance of the pulmonary pneumonia infection model remains a serious concern. A mouse model of shigellosis by intraperitoneal (IP) infection was successfully developed by Jin-Young Yang et al. to produce clinical symptoms that mimic human shigellosis [32] and this model was applied to predict protective efficacy of vaccine candidates targeting Shigella flexneri 2a and 3a infection [26]. In this study, we applied the IP infection model. We acknowledge that the immune responses elicited by this IP infection model do not entirely reflect natural human infection. Therefore, further studies are necessary in higher-level organisms, such as application of the intragastrically infected macaque monkey model, in order to fully characterize the protective efficacy and immune responses elicited by the RASV candidates [52].
Another concern with live vaccine candidates pertains to the inherent risk of insufficient attenuation or virulence reversion, thereby causing adverse events. For the RDAS strains used in this study, attenuation was achieved through complete deletion of virulence genes, as opposed to introducing inactivating point mutations, which minimizes the likelihood of reversion. Furthermore, there are two well-established attenuating deletions (ΔpabA1516 and ΔpabB232) introduced to the candidate strains to further reduce the probability of reversion. It is important to note that wild-type S. Typhimurium generally causes a self-limited infection in humans [53], and prior studies have shown that an attenuated S. Typhimurium strain (WT05) elicits negligible clinical symptoms in a clinical trial [54]. Thus, the RDAS system has great potential for development of a safe, live-attenuated vaccine platform. The system we developed in this study to reduce the side effects of overexpressed foreign antigens in the vaccine strains provides an alternative to the constitutive antigen synthesis system in Shigella vaccine development based on the attenuated Salmonella strains producing exogenous O-antigen.
Abbreviations
Sf2a: Shigella flexneri 2a; RDAS: regulated delayed antigen synthesis; RDEAP: regulated delayed expression of attenuating phenotype; RASV(s): recombinant attenuated Salmonella vaccine(s); WbaP: the Und-P galactose phosphotransferase; BSG: buffered saline with gelatin; Tfh cells: T follicular helper cells; BM cells: memory B cells; Rha: rhamnose; GlcNac: N-acetylglucosamine.
Acknowledgements
The graphical abstract was prepared using the Scientific Illustration Toolkits from Motifolio (Motifolio Inc, Ellicott City, MD, USA).
We thank Dr Stephan Willias (University of Florida, Gainesville, Florida, USA) for editing the manuscript.
Funding
This work was supported by NIH R01 grant AI112680 and the fundamental research funds for the Central Universities (SWU117061 and 117062).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Clark-Curtiss JE, Curtiss R 3rd. Salmonella vaccines: conduits for protective antigens. J Immunol. 2018;200:39-48
2. Curtiss R 3rd, Xin W, Li Y, Kong W, Wanda SY, Gunn B. et al. New technologies in using recombinant attenuated Salmonella vaccine vectors. Crit Rev Immunol. 2010;30:255-70
3. Smith MA, Bidochka MJ. Bacterial fitness and plasmid loss: the importance of culture conditions and plasmid size. Can J Microbiol. 1998;44:351-5
4. Cheah UE, Weigand WA, Stark BC. Effects of recombinant plasmid size on cellular processes in Escherichia coli. Plasmid. 1987;18:127-34
5. Galen JE, Levine MM. Can a 'flawless' live vector vaccine strain be engineered? Trends Microbiol. 2001;9:372-6
6. Stratford R, McKelvie ND, Hughes NJ, Aldred E, Wiseman C, Curtis J. et al. Optimization of Salmonella enterica serovar Typhi ΔaroC ΔssaV derivatives as vehicles for delivering heterologous antigens by chromosomal integration and in vivo inducible promoters. Infect Immun. 2005;73:362-8
7. Galen JE, Wang JY, Chinchilla M, Vindurampulle C, Vogel JE, Levy H. et al. A new generation of stable, nonantibiotic, low-copy-number plasmids improves immune responses to foreign antigens in Salmonella enterica serovar Typhi live vectors. Infect Immun. 2010;78:337-47
8. Lee JS, Shin KS, Pan JG, Kim CJ. Surface-displayed viral antigens on Salmonella carrier vaccine. Nat Biotechnol. 2000;18:645-8
9. Torres-Escobar A, Juarez-Rodriguez MD, Gunn BM, Branger CG, Tinge SA, Curtiss R 3rd. Fine-tuning synthesis of Yersinia pestis LcrV from runaway-like replication balanced-lethal plasmid in a Salmonella enterica serovar Typhimurium vaccine induces protection against a lethal Y. pestis challenge in mice. Infect Immun. 2010;78:2529-43
10. Hohmann EL, Oletta CA, Loomis WP, Miller SI. Macrophage-inducible expression of a model antigen in Salmonella typhimurium enhances immunogenicity. Proc Natl Acad Sci U S A. 1995;92:2904-8
11. Chatfield SN, Charles IG, Makoff AJ, Oxer MD, Dougan G, Pickard D. et al. Use of the nirB promoter to direct the stable expression of heterologous antigens in Salmonella oral vaccine strains: development of a single-dose oral tetanus vaccine. Biotechnology (N Y). 1992;10:888-92
12. Marshall DG, Haque A, Fowler R, Del Guidice G, Dorman CJ, Dougan G. et al. Use of the stationary phase inducible promoters, spv and dps, to drive heterologous antigen expression in Salmonella vaccine strains. Vaccine. 2000;18:1298-306
13. Wang S, Li Y, Scarpellini G, Kong W, Shi H, Baek CH. et al. Salmonella vaccine vectors displaying delayed antigen synthesis in vivo to enhance immunogenicity. Infect Immun. 2010;78:3969-80
14. Kong Q, Liu Q, Roland KL, Curtiss R 3rd. Regulated delayed expression of rfaH in an attenuated Salmonella enterica serovar Typhimurium vaccine enhances immunogenicity of outer membrane proteins and a heterologous antigen. Infect Immun. 2009;77:5572-82
15. Kotloff KL, Winickoff JP, Ivanoff B, Clemens JD, Swerdlow DL, Sansonetti PJ. et al. Global burden of Shigella infections: implications for vaccine development and implementation of control strategies. Bull World Health Organ. 1999;77:651-66
16. Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S. et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet. 2013;382:209-22
17. Research priorities for diarrhoeal disease vaccines. Memorandum from a WHO meeting. Bull World Health Organ. 1991;69:667-76
18. Mani S, Wierzba T, Walker RI. Status of vaccine research and development for Shigella. Vaccine. 2016;34:2887-94
19. Roberts F, Jennison AV, Verma NK. The Shigella flexneri serotype Y vaccine candidate SFL124 originated from a serotype 2a background. FEMS Immunol Med Microbiol. 2005;45:285-9
20. Simmons DA, Romanowska E. Structure and biology of Shigella flexneri O antigens. J Med Microbiol. 1987;23:289-302
21. Allison GE, Verma NK. Serotype-converting bacteriophages and O-antigen modification in Shigella flexneri. Trends Microbiol. 2000;8:17-23
22. Jennison AV, Verma NK. Shigella flexneri infection: pathogenesis and vaccine development. FEMS Microbiol Rev. 2004;28:43-58
23. Levine MM, Kotloff KL, Barry EM, Pasetti MF, Sztein MB. Clinical trials of Shigella vaccines: two steps forward and one step back on a long, hard road. Nat Rev Microbiol. 2007;5:540-53
24. Passwell JH, Harlev E, Ashkenazi S, Chu C, Miron D, Ramon R. et al. Safety and immunogenicity of improved Shigella O-specific polysaccharide-protein conjugate vaccines in adults in Israel. Infect Immun. 2001;69:1351-7
25. Kotloff KL, Herrington DA, Hale TL, Newland JW, Van De Verg L, Cogan JP. et al. Safety, immunogenicity, and efficacy in monkeys and humans of invasive Escherichia coli K-12 hybrid vaccine candidates expressing Shigella flexneri 2a somatic antigen. Infect Immun. 1992;60:2218-24
26. Dharmasena MN, Osorio M, Takeda K, Stibitz S, Kopecko DJ. Stable chromosomal expression of Shigella flexneri 2a and 3a O-antigens in the live Salmonella oral vaccine vector Ty21a. Clin Vaccine Immunol. 2017:24
27. Curtiss R 3rd, Nakayama K, Kelly SM. Recombinant avirulent Salmonella vaccine strains with stable maintenance and high level expression of cloned genes in vivo. Immunol Invest. 1989;18:583-96
28. Kong Q, Yang J, Liu Q, Alamuri P, Roland KL, Curtiss R 3rd. Effect of deletion of genes involved in lipopolysaccharide core and O-antigen synthesis on virulence and immunogenicity of Salmonella enterica serovar Typhimurium. Infect Immun. 2011;79:4227-39
29. Santander J, Wanda SY, Nickerson CA, Curtiss R 3rd. Role of RpoS in fine-tuning the synthesis of Vi capsular polysaccharide in Salmonella enterica serotype Typhi. Infect Immun. 2007;75:1382-92
30. Roland K, Curtiss R 3rd, Sizemore D. Construction and evaluation of a Δcya Δcrp Salmonella typhimurium strain expressing avian pathogenic Escherichia coli O78 LPS as a vaccine to prevent airsacculitis in chickens. Avian Dis. 1999;43:429-41
31. Hitchcock PJ, Brown TM. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J Bacteriol. 1983;154:269-77
32. Yang JY, Lee SN, Chang SY, Ko HJ, Ryu S, Kweon MN. A mouse model of shigellosis by intraperitoneal infection. J Infect Dis. 2014;209:203-15
33. Raetz CR, Reynolds CM, Trent MS, Bishop RE. Lipid A modification systems in gram-negative bacteria. Annu Rev Biochem. 2007;76:295-329
34. Kong Q, Six DA, Roland KL, Liu Q, Gu L, Reynolds CM. et al. Salmonella synthesizing 1-dephosphorylated [corrected] lipopolysaccharide exhibits low endotoxic activity while retaining its immunogenicity. J Immunol. 2011;187:412-23
35. Salgado-Pabon W, Celli S, Arena ET, Nothelfer K, Roux P, Sellge G. et al. Shigella impairs T lymphocyte dynamics in vivo. Proc Natl Acad Sci U S A. 2013;110:4458-63
36. Li Y, Wang S, Xin W, Scarpellini G, Shi Z, Gunn B. et al. A sopB deletion mutation enhances the immunogenicity and protective efficacy of a heterologous antigen delivered by live attenuated Salmonella enterica vaccines. Infect Immun. 2008;76:5238-46
37. Simon JK, Wahid R, Maciel M Jr, Picking WL, Kotloff KL, Levine MM. et al. Antigen-specific B memory cell responses to lipopolysaccharide (LPS) and invasion plasmid antigen (Ipa) B elicited in volunteers vaccinated with live-attenuated Shigella flexneri 2a vaccine candidates. Vaccine. 2009;27:565-72
38. Ma CS, Suryani S, Avery DT, Chan A, Nanan R, Santner-Nanan B. et al. Early commitment of naive human CD4(+) T cells to the T follicular helper (T(FH)) cell lineage is induced by IL-12. Immunol Cell Biol. 2009;87:590-600
39. Ise W, Inoue T, McLachlan JB, Kometani K, Kubo M, Okada T. et al. Memory B cells contribute to rapid Bcl6 expression by memory follicular helper T cells. Proc Natl Acad Sci U S A. 2014;111:11792-7
40. Anderson SM, Tomayko MM, Ahuja A, Haberman AM, Shlomchik MJ. New markers for murine memory B cells that define mutated and unmutated subsets. J Exp Med. 2007;204:2103-14
41. McHeyzer-Williams LJ, Cool M, McHeyzer-Williams MG. Antigen-specific B cell memory: expression and replenishment of a novel b220(-) memory b cell compartment. J Exp Med. 2000;191:1149-66
42. Eriksson S, Lucchini S, Thompson A, Rhen M, Hinton JC. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol Microbiol. 2003;47:103-18
43. Wang S, Li Y, Shi H, Sun W, Roland KL, Curtiss R 3rd. Comparison of a regulated delayed antigen synthesis system with in vivo-inducible promoters for antigen delivery by live attenuated Salmonella vaccines. Infect Immun. 2011;79:937-49
44. Samuel G, Reeves P. Biosynthesis of O-antigens: genes and pathways involved in nucleotide sugar precursor synthesis and O-antigen assembly. Carbohydr Res. 2003;338:2503-19
45. Han Y, Liu Q, Willias S, Liang K, Li P, Cheng A. et al. A bivalent vaccine derived from attenuated Salmonella expressing O-antigen polysaccharide provides protection against avian pathogenic Escherichia coli O1 and O2 infection. Vaccine. 2018;36:1038-46
46. Raqib R, Qadri F, SarkEr P, Mia SM, Sansonnetti PJ, Albert MJ. et al. Delayed and reduced adaptive humoral immune responses in children with shigellosis compared with in adults. Scand J Immunol. 2002;55:414-23
47. Wassef JS, Keren DF, Mailloux JL. Role of M cells in initial antigen uptake and in ulcer formation in the rabbit intestinal loop model of shigellosis. Infect Immun. 1989;57:858-63
48. Schnupf P, Sansonetti PJ. Quantitative RT-PCR profiling of the rabbit immune response: assessment of acute Shigella flexneri infection. PLoS One. 2012;7:e36446
49. Phalipon A, Sansonetti PJ. Shigella's ways of manipulating the host intestinal innate and adaptive immune system: a tool box for survival? Immunol Cell Biol. 2007;85:119-29
50. Voino-Yasenetsky MV, Voino-Yasenetskaya MK. Experimental pneumonia caused by bacteria of the Shigella group. Acta Morphol Acad Sci Hung. 1962;11:439-54
51. Shim DH, Chang SY, Park SM, Jang H, Carbis R, Czerkinsky C. et al. Immunogenicity and protective efficacy offered by a ribosomal-based vaccine from Shigella flexneri 2a. Vaccine. 2007;25:4828-36
52. Rout WR, Formal SB, Giannella RA, Dammin GJ. Pathophysiology of Shigella diarrhea in the rhesus monkey: intestinal transport, morphological, and bacteriological studies. Gastroenterology. 1975;68:270-8
53. Galen JE, Curtiss R 3rd. The delicate balance in genetically engineering live vaccines. Vaccine. 2014;32:4376-85
54. Hindle Z, Chatfield SN, Phillimore J, Bentley M, Johnson J, Cosgrove CA. et al. Characterization of Salmonella enterica derivatives harboring defined aroC and Salmonella pathogenicity island 2 type III secretion system (ssaV) mutations by immunization of healthy volunteers. Infect Immun. 2002;70:3457-67
Author contact
![]() Corresponding author: Qingke Kong, qingke.kongedu or kongqikicom, Tel/Fax: 352-294-4140
Corresponding author: Qingke Kong, qingke.kongedu or kongqikicom, Tel/Fax: 352-294-4140
 Global reach, higher impact
Global reach, higher impact