13.3
Impact Factor
Theranostics 2019; 9(9):2606-2617. doi:10.7150/thno.32344 This issue Cite
Research Paper
Targeting PRMT5 Activity Inhibits the Malignancy of Hepatocellular Carcinoma by Promoting the Transcription of HNF4α
1. Department of Gastroenterology, Changzheng Hospital, Second Military Medical University, Shanghai, 200003, China
2. Drug Discovery and Design Center, CAS Key Laboratory of Receptor Research, State Key Laboratory of Drug Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences (CAS), Shanghai 201203, China
3. School of Biological Science and Technology, University of Jinan, Jinan 250022, China
4. Departments of Gastroenterology, Shanghai East Hospital, Tongji University, Shanghai, 200120, China
5. International Cooperation Laboratory on Signal Transduction of Eastern Hepatobiliary Surgery Institute, Second Military Medical University, Shanghai, China
6. Department of Pharmacy, Changzheng Hospital, Second Military Medical University, 415 Fengyang Road, Shanghai 200003, China
7. Open Studio for Druggability Research of Marine Natural Products, Pilot National Laboratory for Marine Science and Technology, Qingdao, 266237, China
*These authors contributed equally
Received 2018-12-17; Accepted 2019-3-19; Published 2019-4-13
Abstract
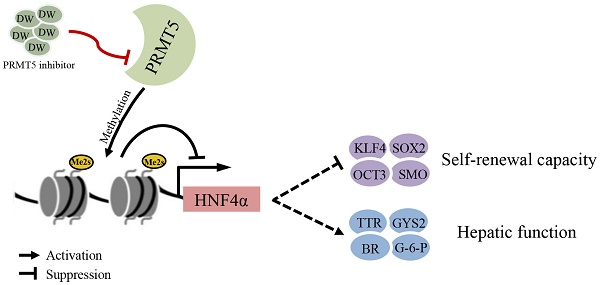
Background: Liver cancer stem cells (LCSCs) are responsible for the initiation, progression and chemoresistance of liver cancer. However, no agent targeting LCSC is available in the clinic to date. Here, we investigated the effects of targeting protein arginine methyltransferase 5 (PRMT5), an epigenetic regulator, on LCSCs and HCC using a novel PRMT5 inhibitor DW14800.
Methods: Tumor spheroid formation culture was used to enrich LCSCs and assess their self-renewal capability. Human alpha-1-antitrypsin (A1AT) ELISA, acetylated low-density lipoprotein (ac-LDL) uptake, periodic acid-Schiff (PAS) reactions and senescence associated β-galactosidase (SA-β-gal) activity assays were performed to examine the differentiation status of HCC cells. The effects of DW14800 on HCC malignancy were assessed in HCC cell lines and on an HCC xenograft model in mice. Chromatin immunoprecipitation was applied to clarify the transcriptional regulation of HNF4α by PRMT5-mediated Histone H4 arginine-3 symmetrical dimethylation (H4R3me2s).
Results: Quantitative real-time PCR revealed that the expression of PRMT5 was upregulated in LCSCs. DW14800 specifically decreased the symmetrical dimethylation of arginine residues in HCC cells. Treatment of DW14800 suppressed the self-renewal capacity of LCSCs while re-establishing hepatocyte-specific characteristics in HCC cells. DW14800 displayed antitumor effects in HCC cells in vitro and in xenograft HCC in vivo. Importantly, ChIP assay showed that PRMT5 and H4R3me2s bound to the promoter region of HNF4α gene, and DW14800 increased the expression of HNF4α via reducing the H4R3me2s levels and enhancing the transcription of HNF4α.
Conclusions: Our data revealed the significance of targeting PRMT5 activity in LCSC elimination and HCC differentiation, and proposed that DW14800 may represent a promising therapeutic agent for HCC in the clinic.
Keywords: PRMT5, HNF4α, hepatocellular carcinoma, cancer stem cells
Introduction
Liver cancer is the sixth most common cancer and the fourth leading cause of cancer death worldwide in 2018 [1-3]. The mortality of liver cancer rose by 1.6% per year in men and by 2.7% per year in women from 2011 to 2015 while the death rates for the other 4 major cancers (lung, breast, prostate, and colorectal) declined [1]. Hepatocellular carcinoma (HCC), comprising 75%-85% cases of liver cancer, is one of the most common cancer with an extremely poor prognosis [2]. Sorafenib, a multikinase inhibitor, was licensed as the first FDA approved first-line targeted drug for advanced HCC [4]. However, the efficacy of sorafenib is far from satisfactory in clinical practice [5, 6]. Thus, the identification of novel targets for HCC treatment is urgently needed.
Cancer stem cells (CSCs) are a small subset in cancer and thought to be responsible for tumor initiation, maintenance, heterogeneity, and chemoresistance [7, 8]. Identifying targets that are critical for CSC function is important for the development of novel cancer therapies. Epigenetics is crucial for the regulation of stem cells, progenitor cells and CSCs. It has been hypothesized that histone methylation and DNA methylation could “reset” CSCs toward differentiation [8-11]. Recent studies have indicated that epigenetic modulators could be anticipated to serve as new targets for cancer prevention, diagnosis and treatment.
Protein arginine methyltransferase 5 (PRMT5), the predominant type II PRMT, catalyzes the formation of ω-NG-monomethyl arginine (MMA) and symmetric ω-NG,NG-dimethylarginine (SDMA) and specifically methylates a wide spectrum of substrates, including histone substrates [12-16]. Symmetric dimethylations in histones H4 residue Arg3 (H4R3) and H3 residue Arg8 (H3R8) catalyzed by PRMT5 remodel the chromatin structure to repress transcription and play important roles in epigenetic control of gene expression [17-19]. PRMT5 is associated with the development of leukemia, lymphoma, glioblastoma, lung and breast cancer and has attracted attention as a novel drug target [20-24]. Two types of PRMT5 inhibitors, SAM competitive inhibitors and SAM uncompetitive inhibitors, have been developed over the past few years. Among these inhibitors, the SAM-uncompetitive PRMT5 inhibitor EPZ015666 has displayed effective anti-tumor activity in MCL xenograft animal models [25]. Recent studies suggested that PRMT5 is upregulated in patient HCCs and may play a role in hepatocarcinogenesis [26-28], suggesting that targeting PRMT5 may have therapeutic effects on HCC. Nevertheless, effect of PRMT5 inhibitors on HCC remains unclear.
Hepatocyte nuclear factor 4α (HNF4α) is a liver-enriched transcription factor and plays a key role in hepatocyte differentiation. HNF4α expression is inversely associated with HCC differentiation status and HNF4α reduction is a critical molecular event during hepatocarcinogenesis [29]. Our previous studies suggested that HNF4α delivery could reverse the malignant phenotypes of HCC by inducing the redifferentiation of HCC cells toward hepatocytes [30]. Moreover, HNF4α can suppress hepatocyte epithelial-mesenchymal transition (EMT) and cancer stem cell generation to block hepatocarcinogenesis [31]. Recent studies have successfully converted hepatoma cells to hepatocyte-like cells with forced expression of HNF4α, HNF1α, and FOXA3 [32, 33]. These studies have suggested the differentiation therapy with HNFs is an effective strategy for HCC treatment.
In this study, we found that PRMT5 is upregulated in LCSCs and involved in the maintenance of LCSCs. Targeting PRMT5 activity with a novel PRMT5 inhibitor DW14800 significantly eliminated the HCC malignancy and re-established the hepatocyte features in HCC cells via upregulating the transcription of HNF4α.
Methods
Cell lines and cell cultures
The human HCC cell lines Huh-7 and Hep3B were obtained from Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). Cell lines were routinely tested for mycoplasma contamination using a Mycoalert detection kit (Lonza) and authenticated by short tandem repeat analysis. Huh-7 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% heat-inactivated fetal bovine serum (FBS). Hep3B cells were cultured in Eagle's minimum essential medium (MEM) supplemented with 10% FBS and nonessential amino acid (NEAA). Primary human hepatocytes were obtained from Xeno Tech and cultured in William's E medium (Xeno Tech) containing 50 μg/ml penicillin-streptomycin and 6 μg/ml insulin.
Reagents
The compound EPZ015666 was purchased from Selleck. The compound DW14800 was designed by structure-based drug design and the structural optimization according to EPZ015666, and synthesized by Drug Discovery and Design Center, Shanghai Institute of Materia Medica [34].
RNA interference
Small interfering RNA for PRMT5 (5′-GCCCAGUUUGAGAUGCCUUAU-3′), HNF4α (5′-CCACAUGUACUCCUGCAGA-3′) and the negative control (NC) (5′-UUCUCCGAACGUGUCACGU-3′) were purchased from GenePharma (Shanghai GenePharma Co., Ltd, Shanghai, China).
Real-time PCR
Total RNA was isolated from cells or tissues following the standard Trizol (Takara) protocol. First-strand cDNA was synthesized from total RNA using the RT Master Mix (Takara). Transcript levels were detected using SYBR Green-based real-time PCR performed by the ABI StepOne Real-time PCR Detection System (Life Technologies). The mRNA levels were normalized to those of β-actin mRNA. At least three independent experiments were performed using each condition. Primer sequences are shown in Table S1.
Western blotting analysis
Proteins were extracted using lysis buffer (125 mM Tris-HCl pH 6.8, 25% glycerol, 5% SDS) supplemented with protease inhibitor (Roche), separated using sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and then transferred onto nitrocellulose membranes (HAHY00010, Millipore). The membranes were blocked in PBST containing 5% skim milk and then incubated with primary antibody. After incubation with a secondary antibody (donkey-anti-mouse or donkey-anti-rabbit, IRDye 700 or IRDye 800, respectively), signals were quantified using an Odyssey infrared imaging system (LI-COR) at 700 nm or 800 nm. The following primary antibodies were used: SDMA (Cell Signaling), PRMT5 (Abcam), HNF4α (Santa Cruz), H4R3me2s (Active Motif), and GAPDH (Boster).
Tumor spheroid formation culture
Huh-7 and Hep3B cells were seeded into ultra-low-attachment dishes and cultured in serum-free DMEM/F12 supplemented with 2% B-27, 20 ng/ml epidermal growth factor (EGF), 20 ng/ml basic fibroblast growth factor (bFGF) and 4 mg/ ml glutamine. The tumor spheres formed in 2 weeks. To detect the effect of DW14800 on the primary tumor spheroid formation, Huh-7 and Hep3B cells pretreated with 1.0 μM DW14800 or dimethyl sulfoxide (DMSO) for 5 days were digested and cultured with spheroid formation medium in ultra-low-attachment 35 mm-dish at 5000 cells/dish. After 2 weeks, each dish was examined using a light microscope by choosing 10 fields of cells, and the total field number of spheres was counted. To detect the effect of DW14800 on the secondary tumor spheroid formation, the spheres formed by HCC cells were resuspended and counted and then were inoculated in ultra-low-attachment 96-well plates at 3 cells per well and cultured with spheroid formation medium containing DMSO or 1.0 μM DW14800. After 10 days, each well was examined using a light microscope, and the total well numbers and volumes of spheres were recorded. To further detect the effect of PRMT5 knockdown on the spheroid formation, Huh-7 and Hep3B cells were transfected with control siNC and siPRMT5 for 12 h and inoculated in ultra-low-attachment 35 mm-dish at 5000 cells/dish for primary spheroid formation assay. After 2 weeks, each dish was examined using a light microscope and the total field number of spheres was counted. Relative sphere volumes were calculated at the same time points using the following equation: volume=length×(width)2×π/6.
Enzymatic selectivity test assays
The inhibition activity of DW14800 (compound 39) against PRMT1, PRMT3, PRMT4, PRMT6, PRMT7, and PRMT8 were evaluated as previously described [34, 35].
Alpha-1-antitrypsin (A1AT) ELISA
Human A1AT was measured by the Human alpha-1-Antitrypsin ELISA Kit (Assay Pro) according to the manufacturer's instructions. For in vitro analysis, HCC cells were treated with 1.0 μM DW14800 in 6-well plates for 4 days, and then the medium was replaced by fresh medium without DW14800 for another 24 h. The supernatant was acquired and measured by the kit.
Periodic acid-Schiff (PAS) reaction
To assess stored glycogen, Huh-7 and Hep3B cells treated with 1.0 μM DW14800 for 5 days were stained with a PAS Reaction Kit (Shanghai Hongqiao Lexiang Medical Reagent Technology Co., Ltd., China) according to the manufacturer's instructions. In brief, the cells were fixed in 4% paraformaldehyde for 5 min and then oxidized in periodic acid for 15 min. After washing with ddH2O, the cells were incubated with Schiff's reagent for 5-20 min. The stored glycogen was visualized under a light microscope. Image analysis software (Image-Pro Plus 6.0, Media Cybernetics) was used to measure the stained area.
Acetylated low-density lipoprotein (ac-LDL) uptake assay
HCC cells were seeded in 12-well plates and treated with 1.0 μM DW14800 for 5 days, and then the medium was replaced by fresh medium containing Dil-Ac-LDL (Invitrogen) at a concentration of 10 μg/ml for 4 h at 37°C. Each well was observed and photographed using the fluorescence microscopy.
Senescence associated β-galactosidase (SA-β-gal) activity assay
Huh-7 and Hep3B cells were treated with DMSO, 1.0 μM or 4.0 μM DW14800 for five days. The senescence of the HCC cells was detected with a Senescence β-Galactosidase Staining Kit (Beyotime, China). The cells were fixed for 15 min with 4% paraformaldehyde in PBS and then incubated at 37 °C overnight with fresh senescence associated β-gal staining solution. The senescent cells were photographed under a microscope. Image analysis software (Image-Pro Plus 6.0, Media Cybernetics) was used to count the senescent cells.
Apoptosis assay
Huh-7 and Hep3B cells were treated with DMSO, 1.0 μM or 4.0 μM DW14800 for five days. The apoptotic cells were detected with an Annexin V-FITC/PI Apoptosis Kit (Multi Sciences) according to the manufacturer's instructions. The cells were harvested using 0.25% trypsin and washed twice by phosphate buffer saline (PBS), then resuspended in Binding Buffer at a concentration of 1 × 106 cells/ml. Cells were incubated with 300 μl PBS containing 5 μl Annexin V-FITC and 5 μl of propidium iodide for 15 min at room temperature in the dark. Cells were then evaluated by FACS (Navios, Beckman Coulter, CA, USA).
Cell proliferation assay, migration and invasion assay
Huh-7 and Hep3B cells were plated into 96-well plates at 3 × 103/well and cultured in medium containing DW14800. The number of metabolically active cells was determined using the Cell Counting Kit-8 (CCK8, Dojindo, Tokyo, Japan) every 2 days for 1 week. In the rescue experiments, HCC cells transfected with siNC or siHNF4α were plated into 96-well plates at 8 × 103/well and cultured in medium containing DW14800. Then, the cell viability was detected on day 3.
In vitro migration and invasion assays were performed using Transwell chambers (BD Bioscience), without or with Matrigel, according to the manufacturer's instructions. In brief, 4 × 104 HCC cells for migration assay and 6 × 104 HCC cells for invasion assay in serum-free medium containing DW14800 were seeded in the upper chamber. Medium supplemented with FBS and the same concentration of DW14800 was added into the lower chamber. After incubation for 48 h at 37 °C, cells remaining on the upper membrane were removed with cotton swabs. Cells on the lower surface of the membrane were fixed and stained with 0.1% crystal violet, 20% methanol. Five fields of cells on the lower membrane were photographed and counted to estimate cell density. Image analysis software (Image-Pro Plus 6.0, Media Cybernetics) was used to measure the stained area. At least three independent experiments were performed for each condition.
Human HCC xenograft model
Male athymic BALB/c nude mice (5 weeks old) were purchased and maintained under specific pathogen-free conditions with a 12 h on/off light cycle. One million Huh-7 cells were inoculated subcutaneously in the right flank. When tumors reached an average volume of approximately 60 mm3, the mice were randomly divided into two groups (8 animals per group) and injected with DMSO or 0.5 mg DW14800 (0.5 mg in 100 μl DMSO) intratumorally (i.t). Mice were sacrificed, and tumors were excised and weighed. Tumor volumes were calculated according to the following equation: volume=length×(width)2×π/6.
Immunohistochemical staining and TUNEL staining
Formalin-fixed paraffin-embedded sections were deparaffinized in xylene and rehydrated in graded alcohols. Endogenous peroxidase was blocked by 3% H2O2 followed by antigen retrieval. Slides were incubated with primary antibodies overnight at 4 °C and incubated with a secondary antibody at room temperature for 30 min. The following primary antibodies were used: Ki67 (Santa Cruz), SDMA (Cell Signaling), and HNF4α (Abcam). The staining was developed using an EnVision Detection Rabbit/Mouse Kit (GK500710, GeneTech, Shanghai, China). Immunohistofluorescence of TUNEL staining was performed on paraffin-embedded tumor sections according to the manufacturer's instruction (Beyotime).
Chromatin immunoprecipitation (ChIP) assay
Huh-7 cells were harvested and cross-linked in 1% formaldehyde for 10 min at room temperature. Then, cells were added with 125 mM glycine and incubated for 5 min at room temperature. Cells were washed with precooled PBS and centrifuged. Then, we discarded the supernatant and resuspended the cells in FA lysis buffer (50 mM HEPES-KOH pH 7.5, 140 mM NaCl, 1 mM EDTA pH 8.0, 1% Triton-X-100, 0.1% SDS, protease inhibitor). The samples were sonicated and 50 μl of products were removed to assess the DNA fragment size. The remainder was stored at -80 °C. Antibodies for control IgG (Cell Signaling), H4R3me2s (Active Motif) and PRMT5 (Santa Cruz) were used for immunoprecipitation. DNA extracted from 10 μl pre-immunoprecipitated samples was used as input controls. Beads were washed with low-salt ChIP wash buffer (0.1% SDS, 1% Triton-X-100, 2 mM EDTA pH 8.0, 150 mM NaCl, 20 mM Tris-HCl pH 8.0) 3 times and then high-salt ChIP wash buffer (0.1% SDS, 1% Triton-X-100, 2 mM EDTA pH 8.0, 500 mM NaCl, 20 mM Tris-HCl pH 8.0) once. The immune-precipitated chromatins were eluted with ChIP elution buffer (1% SDS, 100 mM NaHCO3) and incubated at 37 °C for 30 min, followed by 6 h at 65 °C to reverse the cross-links. DNA was amplified by quantitative real-time PCR and normalized to the input. The primer in the eIF4E promoter was used as a positive control [36]. The primer for a nonspecific site was used as the negative control [37]. Primer sequences are shown in Table S2.
Statistical analyses
Data analyses were performed with Prism 5 (GraphPad Software, La Jolla, CA). For experiments involving only two groups, data were analyzed with Student's t-tests. The data for animals are presented as the mean ± SEM. Other data are presented as the mean ± SD. Statistical significance was set at * P < 0.05, ** P < 0.01, and *** P < 0.001. P < 0.05 was considered statistically significant.
Results
Targeting PRMT5 activity suppresses the self-renewal capability of LCSCs
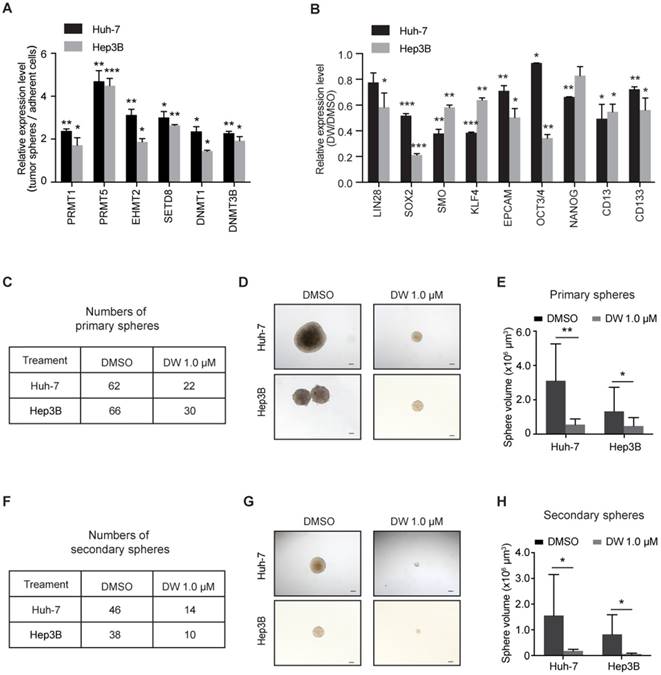
To identify the key epigenetic regulators in LCSC maintenance, we examined the expression of various epigenetic modulator enzymes, including histone arginine methyltransferase (PRMT1 and PRMT5), histone lysine methyltransferase (EHMT2 and SETD8) and DNA methyltransferase (DNMT1 and DNMT3B), in LCSCs enriched by spheroid culture. As shown in Figure S1, the levels of stemness-associated genes, including LIN28, SOX2, SMO, OCT3/4, KLF4, NANOG and CD13, as well as the LCSC marker CD133 were evidently increased in the LCSC-enriched spheres. As expected, the expression of some epigenetic modulators was increased in LCSCs. Interestingly, the levels of PRMT5 were increased more robustly in the stem-like tumor spheres compared to other enzymes (Figure 1A). Importantly, knockdown of PRMT5 dramatically decreased the expression of stemness genes in HCC cells (Figure S2A-D), indicating that PRMT5 may play an important role in maintaining the pluripotency of LCSCs.
Our recent study reported a new class of potent PRMT5 inhibitors designed by a structure-based strategy. These inhibitors bound to the substrate binding site of PRMT5 and presented high selectivity over other PRMTs [34]. We then evaluated the effect of this class of inhibitors on targeting PRMT5 and LCSCs in HCC cells. One of these inhibitors, DW14800 (also called Compound 39) with high activity against PRMT5 (IC50 = 17 nM) (Figure S3A, B), reduced the cellular symmetric dimethylarginine in HCC cells and the PRMT5-mediated methylation marks, H4R3me2s and H3R8me2s, in a dose-dependent manner, whereas no obvious decrease of asymmetrical dimethylarginine was detected (Figure S3C-E). More importantly, inhibition of PRMT5 with DW14800 suppressed the expression of stemness genes in HCC cells (Figure 1B).
We next investigated the effect of DW14800 on the sphere formation of HCC cells. Huh-7 and Hep3B cells were pretreated with DMSO or DW14800 and cultured in sphere-forming medium for 2 weeks. About 62 and 66 tumor spheres were observed in the DMSO group, whereas only 22 and 30 spheres formed in the DW14800-treated group in Huh-7 and Hep3B cells, respectively (Figure 1C). Moreover, the size of the tumor spheres formed from HCC cells pretreated with DW14800 was markedly smaller than those from the control cells (Figure 1D, E). We also performed a secondary spheroid formation experiment with Huh-7 and Hep3B cells. The result showed that DW14800 treatment significantly reduced the formation of spheres from passaged primary spheres (Figure 1F-H). Consistently, knockdown of PRMT5 also remarkably suppressed the sphere formation of HCC cells (Figure S2E-G). Collectively, these data suggested that targeting PRMT5 activity reduced the self-renewal capability of LCSCs.
Targeting PRMT5 activity re-establishes the expression profile of hepatocyte in HCC cells
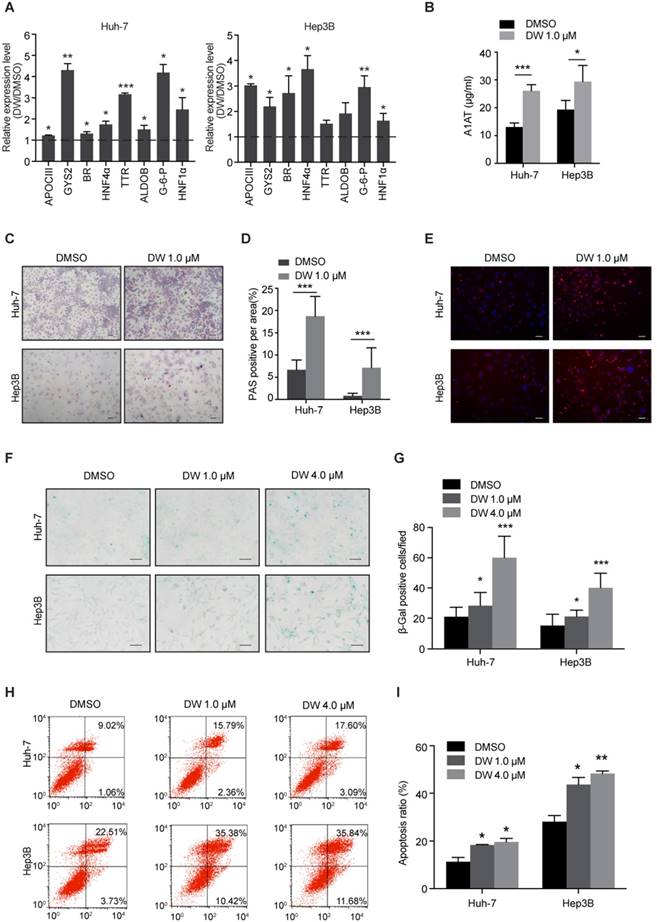
Since DW14800 decreased the expression of stemness genes in HCC cells, we then examined whether targeting PRMT5 activity affects the hepatic functions of HCC cells. Interestingly, real-time PCR showed that DW14800 treatment upregulated the expression of hepatocyte-specific markers, including glycogen synthetase 2 (GYS2), glucose-6-phosphatase (G-6-P), biliverdin reductase (BR), aldolase B (ALDOB), apolipoprotein C3 (APOCIII), transthyretin (TTR), and hepatocyte nuclear factor 1α (HNF1α) in both Huh-7 and Hep3B cells (Figure 2A). Knockdown of PRMT5 in HCC cells also confirmed the effect of targeting PRMT5 on the expression of hepatic function genes (Figure S2H, I). In addition, ELISA assay showed that DW14800-treated HCC cells displayed a stronger capability for secreting the protein alpha-1-antitrypsin (A1AT) (Figure 2B). Periodic acid-Schiff (PAS) reactions showed that the typical hepatocyte function of storing glycogen was increased in HCC cells treated with DW14800 (Figure 2C, D). Moreover, DW14800 treatment enhanced the hepatic function of acetylated low density lipoprotein (ac-LDL) intake in HCC cells (Figure 2E). Furthermore, senescence associated β-galactosidase (SA-β-gal) staining revealed that DW14800 treatment induced the cellular senescence of HCC cells (Figure 2F, G). The proportion of apoptotic cells was significantly increased in HCC cells treated with DW14800 (Figure 2H, I). Taken together, we concluded that targeting PRMT5 activity results in the recovery of hepatic function in HCC cells and might induce the redifferentiation of HCC cells toward hepatocytes.
Targeting PRMT5 activity reduces the self-renewal capacity of HCC cells. (A) Real-time PCR analysis of the expression levels of epigenetic enzymes. (B) The expression of stemness markers was decreased in Huh-7 and Hep3B cells treated with DW14800. (C) DW14800 reduced the frequency of primary spheroid formation of Huh-7 and Hep3B cells. (D) Representative photographs of primary tumor spheres. Scale bars = 100 μm. (E) Quantification analysis of the size of primary tumor spheres formed from HCC cells. (F) The frequency of secondary spheroid formation of HCC cells. (G) Representative photographs of secondary tumor spheres. Scale bars = 100 μm. (H) Quantification analysis of the size of secondary tumor spheres formed. The data are presented as the mean ± SD. * P < 0.05, ** P < 0.01, *** P < 0.001.
DW14800 exhibits robust therapeutic effect on HCC cells
Previous studies have reported that the downregulation of PRMT5 led to the impaired proliferation and invasion of HCC cells [26]. Therefore, we assessed the therapeutic effect of DW14800 on HCC cells. As shown in Figure 3A, DW14800 significantly inhibited the proliferation of HCC cells in a dose-dependent manner. The transwell assays revealed that DW14800 remarkably suppressed the migration and invasion of HCC cells (Figure 3B, C). EPZ015666 is the first PRMT5 inhibitor that bound to the substrate binding site with a hydrogen bond and hydrophobic interactions, the antitumor activity of which has been confirmed in MCL xenograft models [25]. Herein, we found that the IC50 of DW14800 against the proliferation of Huh-7 and Hep3B cells was 2.7 μM and 1.4 μM, respectively, which is much lower than EPZ015666 (Figure S4). We also confirmed that DW14800 could inhibit the malignancy of HCC cells in a superior manner than EPZ015666 (Figure S5). The last but not the least, DW14800 treatment (1.0 μM), which dramatically impaired the survival of Huh-7 cells, had no obvious toxicity on primary human hepatocytes (Figure S6), suggesting its translational value in cancer therapy.
DW14800 increases the characteristic hepatocyte markers of HCC cells. (A) The expression of characteristic hepatocyte markers was increased in Huh-7 and Hep3B cells treated with DW14800. (B) Human alpha-1-antitrypsin (A1AT) secretion after DW14800 treatment in HCC cells was measured by ELISA. (C) Periodic acid-Schiff (PAS) staining was used to detect the stored glycogen in Huh-7 and Hep3B cells. Representative images of PAS staining of Huh-7 and Hep3B cells treated with DW14800 or vehicle were shown. Scale bars = 100 μm. (D) DW14800 increased the stored glycogen of HCC cells. (E) Acetylated low-density lipoprotein (ac-LDL) uptake ability of HCC cells was analyzed using the Dil-ac-LDL fluorescent substrate (red). Scale bars = 50 μm. (F) Representative images of senescence-associated β-galactosidase activity staining of Huh-7 and Hep3B cells. Scale bars = 50 μm. (G) Quantification of the β-gal positive cells. (H, I) The apoptosis of HCC cells was detected by Annexin V-FITC/PI staining. The data are presented as the mean ± SD. * P < 0.05, ** P < 0.01, *** P < 0.001.
DW14800 suppresses malignant phenotypes of HCC in vitro and in HCC xenograft models. (A) The proliferation of Huh-7 and Hep3B cells treated with DW14800 was measured using the Cell Counting Kit-8 (CCK8). (B, C) Migration (B) and invasion (C) of Huh-7 and Hep3B cells were significantly inhibited by DW14800. The data are presented as the mean ± SD. * P < 0.05, ** P < 0.01, *** P < 0.001. (D) Growth curves of Huh-7 xenografts treated with DW14800 or vehicle. (E) Images of tumor nodules from the subcutaneous mouse xenograft models (n = 8 in each group). (F) Tumor weight of Huh-7 xenografts. (G) Western blotting analysis of the levels of SDMA in tumor nodules. (H) Immunohistochemical staining and TUNEL assay showed the reduction of Ki67 expression and levels of SDMA along with an increase in DNA strand breaks in the DW14800 group compared to those in the control group. Scale bars = 50 μm. The data for animals are presented as the mean ± SEM. * P < 0.05, ** P < 0.01.
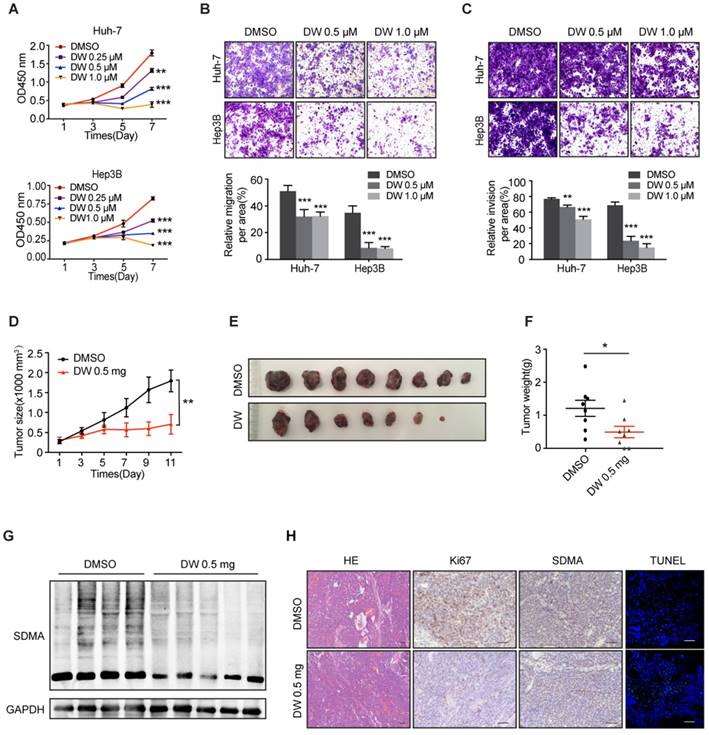
We next explored the potential therapeutic effects of DW14800 on HCC xenografts in mice. The size and weight of HCC xenografts treated with DW14800 were significantly less than the control xenografts (Figure 3D-F). Moreover, the reduced levels of symmetric dimethylarginine were confirmed in DW14800-treated HCC xenografts (Figure 3G, H). Furthermore, immunohistochemical staining and TUNEL assay of tumors also showed that DW14800 treatment resulted in a remarkable decrease of Ki67 positive cells in parallel with an increase in apoptotic cells (Figure 3H). Taken together, our data suggested that DW14800 is a selective and active PRMT5 inhibitor with potent anti-HCC effect.
Targeting PRMT5 activity gives rise to HNF4α induction in HCC cells
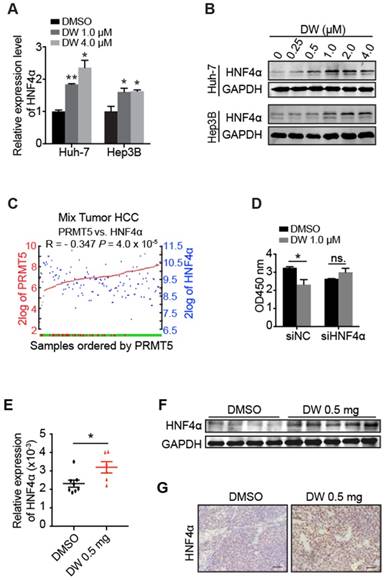
Our previous studies demonstrated that HNF4α delivery inhibited HCC growth through the induction of HCC cells differentiation toward hepatocytes [30, 31]. Interestingly, we found that DW14800 treatment markedly increased the mRNA and protein levels of HNF4α in Huh-7 and Hep3B cells (Figure 4A, B). Analysis of human HCC microarray data sets (R2: Genomics Analysis and Visualization Platform) revealed that HNF4α and PRMT5 are inversely correlated at the level of mRNA expression in human HCC samples (P < 0.001; N = 134) (Figure 4C). Importantly, the suppression of HCC cell proliferation by DW14800 was rescued upon HNF4α knockdown (Figure 4D), suggesting that DW14800 repressed HCC growth through, at least partially, targeting HNF4α. Consistently, the increased expression of HNF4α was detected in DW14800-treated HCC xenografts by real-time PCR, western blotting and immunohistochemical staining (Figure 4E-G).
DW14800 increases the expression of HNF4α. (A) The mRNA levels of HNF4α in Huh-7 and Hep3B cells treated with DW14800 for 5 days were analyzed by RT-PCR. (B) Western blotting analysis of HNF4α in HCC cells. (C) Human HCC microarray datasets reveal that the mRNA expression level of PRMT5 was negatively correlated with HNF4α in HCC specimens (P < 0.001) (N = 134). The red and green samples represent the normal tissues and tumor tissues, respectively. The red and blue blots separately represent the mRNA level of PRMT5 and HNF4α. The data were from R2: genomics analysis and visualization platform. (D) Knockdown of HNF4α attenuated the inhibitory effect of DW14800 in Huh-7 cells. The data are presented as the mean ± SD. * P < 0.05, ** P < 0.01, ns: no statistical significance. (E) The expression levels of HNF4α in the tumor nodules were detected by real-time PCR. The data are presented as the mean ± SEM. * P < 0.05. (F) Western blotting analysis of the levels of HNF4α in tumor nodules. (G) Immunohistochemical staining showed the increase of HNF4α in the DW14800 group compared to those in the control group. Scale bars = 50 μm.
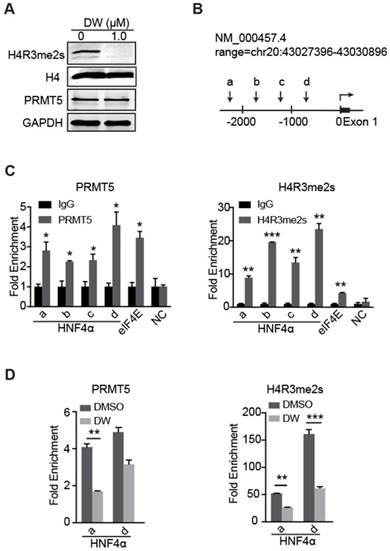
Targeting PRMT5 activity releases the transcription of HNF4α gene repressed by H4R3me2s
Previous studies have reported that PRMT5-mediated H4R3me2s marks specific repressed loci in mammals and facilitates gene repression [17-19]. Herein, our data showed that DW14800 also reduced the symmetrical dimethylation of H4R3 in HCC cells, whereas no obvious alteration of PRMT5 expression was observed (Figure 5A, S3D). To understand how DW14800 upregulates HNF4α levels, we examined whether H4R3me2s occupied the promoter region of the HNF4α gene. As shown in Figure 5B, four pairs of primers in different regions of HNF4α promoter were designed (Figure 5B). Quantitative ChIP experiments showed that both PRMT5 and H4R3me2s mark bound to the promoter region of HNF4α (Figure 5C). Moreover, ChIP assays revealed that the enrichment of PRMT5 and H4R3me2s at the HNF4α promoter was reduced by the treatment of DW14800 (Figure 5D). These results indicated that DW14800 promoted HNF4α transcription by regulating the binding of H4R3me2s to the HNF4α promoter.
DW14800 decreases the occupation of PRMT5 and H4R3me2s on the promoter region of HNF4α. (A) The arginine symmetric dimethylation of histone H4 levels was significantly reduced in Huh-7 cells treated with DW14800 for 5 days. (B) Schematic representation of the HNF4α gene. A set of arrowheads indicate ChIP PCR amplicons. (C) A ChIP assay was performed to analyze PRMT5 and H4R3me2s occupancy at the HNF4α promoter in Huh-7 cells. The eIf4E promoter was used as a positive control. DNA samples from immunoprecipitation with normal rabbit IgG (IgG) were used as controls. (D) DW14800 treatment reduced the binding of PRMT5 and H4R3me2s to the HNF4α promoter. The data are presented as the mean ± SD. * P < 0.05, ** P < 0.01, *** P < 0.001.
Discussion
Cancer stem cells, a minor proportion of cells in tumor bulk with distinct self-renewal ability, give rise to cancer initiation and facilitate cancer progression [38-40]. One previous study demonstrated that Prmt5 plays an essential role in the maintenance of pluripotency of mouse embryonic stem cells (ESCs) by repressing certain differentiation-associated genes [41]. Moreover, PRMT5 has been reported to be involved in the progression of multiple cancers, including leukemia, lymphoma, lung cancer, liver cancer, colorectal cancer and breast cancer [9, 19, 21, 23, 42-46]. Chiang et al. showed that PRMT5 exerted as a critical regulator of breast CSC survival via the epigenetic regulation of FOXP1 [9]. Additionally, Banasavadi-Siddegowda et al. reported that PRMT5 played an important role in the self-renewal of mature and undifferentiated GBM cells [22]. These studies imply that PRMT5 inhibitors might potentially eradicate CSCs and thus suppress tumor growth. Here, we found that PRMT5 plays an important role in maintaining the pluripotency of LCSCs and targeting PRMT5 activity with DW14800 significantly reduced the self-renewal capability of LCSC. Moreover, DW14800 treatment resulted in the recovery of hepatic function and increased cell senescence and apoptosis of HCC cells, indicating that targeting undifferentiated cancer cells (especially CSCs) by PRMT5 may be a promising approach for HCC therapy.
HNF4α is a key regulator in the hepatocyte differentiation during embryonic development. It is also important in the maintenance of the differentiated phenotype of mature hepatocytes in adult liver. Other groups and we have elucidated that HNF4α plays a critical role during the hepatocarcinogenesis [29, 31, 32] and HNF4α alone is sufficient to induce the differentiation of HCC [30]. It has been well accepted that symmetric dimethylation of H4R3 catalyzed by PRMT5 could repress the transcription of certain genes. In the current study, we found that targeting PRMT5 activity with DW14800 significantly increased HNF4α expression both in vitro and in vivo. In the further study, we found that targeting PRMT5 activity decreased the levels of H4R3me2s and thus facilitated the transcription of HNF4α. As expected, knockdown of HNF4α abrogated the decrease of cell proliferation triggered by DW14800. These data suggested that HNF4α functions as a downstream effector of PRMT5 and DW14800 treatment inhibited HCC growth at least partially through the upregulation of HNF4α. From a clinical standpoint, targeting PRMT5 might present as a promising strategy for differentiation therapy of HCC.
As an oncogene, PRMT5 has been suggested as a therapeutic target in a variety of cancers [15, 19, 25]. Various PRMT5 inhibitors have been discovered in recent years. Most of existed PRMT5 inhibitors were aimed for SAM-binding site and designed to disturb the interaction of SAM and PRMT5 [35, 47]. However, due to their native binding state, it is difficult to identify small molecules with inhibitory activity stronger than SAM. Fortunately, the discovery of EPZ015666 and its new binding site provides a new clue for developing non-SAM competitive inhibitors [25]. Most recently, we have identified DW14800 as a non-SAM competitive PRMT5 inhibitor based on EPZ015666 by structure-based drug design and structural optimization [34]. The identification of DW14800 offered us a tool to explore the biological effects driven by type II PRMT-mediated substrate methylation in cancer and other diseases. Herein, we revealed that DW14800 specifically targeted PRMT5 activity in HCC cells and eliminated the malignancy of HCC cells. Through applying DW14800, we achieved a better understanding of the PRMT5 function in HCC.
In conclusion, our study highlights the importance of PRMT5 in LCSC maintenance and proposes its significance in drug development targeting LCSCs. As anticipated, targeting PRMT5 activity with DW14800 suppressed the self-renewal capacity of LCSCs and induced the differentiation of HCC cells. Mechanistically, DW14800 increased the expression of HNF4α by decreasing PRMT5-mediated symmetric methylation of H4R3. These findings indicate that PRMT5 could be a novel therapeutic target in LCSCs and DW14800 may represent as a novel therapeutic agent for human HCC.
Abbreviations
ALDOB: aldolase B; APOCIII: apolipoprotein C3; BR: biliverdin reductase; CD13: alanyl aminopeptidase membrane; CSC: cancer stem cell; DNMT1: DNA methyltransferase 1; DNMT3B: DNA methyltransferase 3B; EMHT2: euchromatic histone lysine methyltransferase 2; EMT: epithelial-mesenchymal transition; FOXA3: forkhead box transcription factor A3; GYS2: glycogen synthetase 2; G-6-P: glucose-6-phosphatase; HCC: hepatocellular carcinoma; HNF: hepatocyte nuclear factor; HNF1α: hepatocyte nuclear factor 1α; HNF4α: hepatocyte nuclear factor 4α; KLF4: Kruppel like factor 4; LCSC: liver cancer stem cell; LIN28: lin-28 homolog A; MCL: mantle cell lymphoma; MMA: monomethyl arginine; NANOG: nanog homeobox; OCT3/4: POU class 5 homeobox 1; PRMT: protein arginine methyltransferase; PRMT5: protein arginine methyltransferase 5; SAM: S-adenosyl-l-methionine; SDMA: symmetric dimethylarginine; SMO: smoothened frizzled class receptor; SOX2: SRY-box 2; TTR: transthyretin.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81530019 and 81572377 to W.F.X, 81625022, 91853205 and 81821005 to C.L, 81572377 and 81772523 to X.Z., 81703415 to S.C. and 81803438 to K.Z), the Shanghai Science and Technology Committee (15431907100 to W.F.X.), the National Science and Technology Major Project (2018ZX09711002 to C.L.), the K. C. Wong Education Foundation to C.L. and Shanghai Sailing Program (17YF1423100 to S.C.).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7-30
2. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424
3. Fact sheets by Population-Globocan-IARC. September, 2018. http://gco.iarc.fr/today/data/factsheets/cancers/11-Liver-fact-sheet.pdf
4. Tong M, Che N, Zhou L, Luk ST, Kau PW, Chai S. et al. Efficacy of annexin A3 blockade in sensitizing hepatocellular carcinoma to sorafenib and regorafenib. J Hepatol. 2018;69:826-39
5. Zhu AX, Kudo M, Assenat E, Cattan S, Kang Y-K, Lim HY. et al. Effect of Everolimus on Survival in Advanced Hepatocellular Carcinoma After Failure of Sorafenib. Jama. 2014;312:57-67
6. Llovet JM, Montal R, Sia D, Finn RS. Molecular therapies and precision medicine for hepatocellular carcinoma. Nat Rev Clin Oncol. 2018;15:599-616
7. Wang Y, Cardenas H, Fang F, Condello S, Taverna P, Segar M. et al. Epigenetic targeting of ovarian cancer stem cells. Cancer Res. 2014;74:4922-36
8. Matteucci C, Balestrieri E, Argaw-Denboba A, Sinibaldi-Vallebona P. Human endogenous retroviruses role in cancer cell stemness. Semin Cancer Biol. 2018;53:17-30
9. Chiang K, Zielinska AE, Shaaban AM, Sanchez-Bailon MP, Jarrold J, Clarke TL. et al. PRMT5 Is a Critical Regulator of Breast Cancer Stem Cell Function via Histone Methylation and FOXP1 Expression. Cell Rep. 2017;21:3498-3513
10. Toh TB, Lim JJ, Chow EK. Epigenetics in cancer stem cells. Mol Cancer. 2017;16:29
11. Hua F, Shang S, Yang YW, Zhang HZ, Xu TL, Yu JJ. et al. TRIB3 Interacts with Betacatenin and TCF4 to Increase Stem Cell Features of Colorectal Cancer Stem Cells and Tumorigenesis. Gastroenterology. 2018
12. Ren J, Wang Y, Liang Y, Zhang Y, Bao S, Xu Z. Methylation of Ribosomal Protein S10 by Protein-arginine Methyltransferase 5 Regulates Ribosome Biogenesis. J Biol Chem. 2010;285:12695
13. Hsu JM, Chen CT, Chou CK, Kuo HP, Li LY, Lin CY. et al. Crosstalk between Arg 1175 methylation and Tyr 1173 phosphorylation negatively modulates EGFR-mediated ERK activation. Nat Cell Biol. 2011;13:174-81
14. Burgos ES, Wilczek C, Onikubo T, Bonanno JB, Jansong J, Reimer U. et al. Histone H2A and H4 N-Terminal Tails are Positioned by the MEP50 WD-Repeat Protein for Efficient Methylation by the PRMT5 Arginine Methyltransferase. J Biol Chem. 2015;290:9674-89
15. Blanc RS, Richard S. Arginine Methylation: The Coming of Age. Mol Cell. 2017;65:8-24
16. Jansson M, Durant ST, Cho EC, Sheahan S, Edelmann M, Kessler B. et al. Arginine methylation regulates the p53 response. Nat Cell Biol. 2008;10:1431-9
17. Tsutsui T, Fukasawa R, Shinmyouzu K, Nakagawa R, Tobe K, Tanaka A. et al. Mediator complex recruits epigenetic regulators via its two cyclin-dependent kinase subunits to repress transcription of immune response genes. J Biol Chem. 2013;288:20955-65
18. Xu X, Hoang S, Mayo MW, Bekiranov S. Application of machine learning methods to histone methylation ChIP-Seq data reveals H4R3me2 globally represses gene expression. Bmc Bioinformatics. 2010;11:396
19. Tarighat SS, Santhanam R, Frankhouser D, Radomska HS, Lai H, Anghelina M. et al. The dual epigenetic role of PRMT5 in acute myeloid leukemia: gene activation and repression via histone arginine methylation. Leukemia. 2015;30:1388-95
20. Jin Y, Zhou J, Xu F, Jin B, Cui L, Wang Y. et al. Targeting methyltransferase PRMT5 eliminates leukemia stem cells in chronic myelogenous leukemia. J Clin Invest. 2016;126:3961-80
21. Serio J, Ropa J, Chen W, Mysliwski M, Saha N, Chen L. et al. The PAF complex regulation of Prmt5 facilitates the progression and maintenance of MLL fusion leukemia. Oncogene. 2018;37:450-60
22. Banasavadi-Siddegowda YK, Welker AM, An M, Yang X, Zhou W, Shi G. et al. PRMT5 as a druggable target for glioblastoma therapy. Neuro-oncology. 2018;20:753-63
23. Ibrahim R, Matsubara D, Osman W, Morikawa T, Goto A, Morita S. et al. Expression of PRMT5 in lung adenocarcinoma and its significance in epithelial-mesenchymal transition. Hum Pathol. 2014;45:1397-405
24. Huang S, Chi Y, Qin Y, Wang Z, Xiu B, Su Y. et al. CAPG enhances breast cancer metastasis by competing with PRMT5 to modulate STC-1 transcription. Theranostics. 2018;8:2549-64
25. Chan-Penebre E, Kuplast KG, Majer CR, Boriack-Sjodin PA, Wigle TJ, Johnston LD. et al. A selective inhibitor of PRMT5 with in vivo and in vitro potency in MCL models. Nat Chem Biol. 2015;11:432-7
26. Zhang B, Dong S, Li Z, Lu L, Zhang S, Chen X. et al. Targeting protein arginine methyltransferase 5 inhibits human hepatocellular carcinoma growth via the downregulation of beta-catenin. J Transl Med. 2015;13:349
27. Li Z, Zhang J, Liu X, Li S, Wang Q, Chen D. et al. The LINC01138 drives malignancies via activating arginine methyltransferase 5 in hepatocellular carcinoma. Nat Commun. 2018;9:1572
28. Kanda M, Murotani K, Sugimoto H, Miwa T, Umeda S, Suenaga M. et al. An integrated multigene expression panel to predict long-term survival after curative hepatectomy in patients with hepatocellular carcinoma. Oncotarget. 2017;8:71070-9
29. Hatziapostolou M, Polytarchou C, Aggelidou E, Drakaki A, Poultsides GA, Jaeger SA. et al. An HNF4alpha-miRNA inflammatory feedback circuit regulates hepatocellular oncogenesis. Cell. 2011;147:1233-47
30. Yin C, Lin Y, Zhang X, Chen YX, Zeng X, Yue HY. et al. Differentiation therapy of hepatocellular carcinoma in mice with recombinant adenovirus carrying hepatocyte nuclear factor-4alpha gene. Hepatology. 2008;48:1528-39
31. Ning BF, Ding J, Yin C, Zhong W, Wu K, Zeng X. et al. Hepatocyte nuclear factor 4 alpha suppresses the development of hepatocellular carcinoma. Cancer Res. 2010;70:7640-51
32. Takashima Y, Horisawa K, Udono M, Ohkawa Y, Suzuki A. Prolonged inhibition of hepatocellular carcinoma cell proliferation by combinatorial expression of defined transcription factors. Cancer Sci. 2018;109:3543-53
33. Cheng Z, He Z, Cai Y, Zhang C, Fu G, Li H. et al. Conversion of hepatoma cells to hepatocyte-like cells by defined hepatocyte nuclear factors. Cell Res. 2019;29:124-35
34. Shao J, Zhu K, Du D, Zhang Y, Tao H, Chen Z. et al. Discovery of 2-substituted-N-(3-(3,4-dihydroisoquinolin-2(1H)-yl)-2-hydroxypropyl)-1,2,3,4-tet rahydroisoquinoline-6-carboxamide as potent and selective protein arginine methyltransferases 5 inhibitors: Design, synthesis and biological evaluation. Eur J Med Chem. 2019;164:317-33
35. Mao R, Shao J, Zhu K, Zhang Y, Ding H, Zhang C. et al. Potent, Selective, and Cell Active Protein Arginine Methyltransferase 5 (PRMT5) Inhibitor Developed by Structure-Based Virtual Screening and Hit Optimization. J Med Chem. 2017;60:6289-304
36. Zhang B, Dong S, Zhu R, Hu C, Hou J, Li Y. et al. Targeting protein arginine methyltransferase 5 inhibits colorectal cancer growth by decreasing arginine methylation of eIF4E and FGFR3. Oncotarget. 2015;6:22799-811
37. Yin C, Wang PQ, Xu WP, Yang Y, Zhang Q, Ning BF. et al. Hepatocyte nuclear factor-4α reverses malignancy of hepatocellular carcinoma through regulating miR-134 in the DLK1-DIO3 region. Hepatology. 2013;58:1964-76
38. Bhartiya D, Patel H, Ganguly R, Shaikh A, Shukla Y, Sharma D. et al. Novel Insights into Adult and Cancer Stem Cell Biology. Stem Cells Dev. 2018;27:1527-39
39. Glinka Y, Mohammed N, Subramaniam V, Jothy S, Prud'homme GJ. Neuropilin-1 is expressed by breast cancer stem-like cells and is linked to NF-kappaB activation and tumor sphere formation. Biochem Biophys Res Commun. 2012;425:775-80
40. Bhatlekar S, Fields JZ, Boman BM. Role of HOX Genes in Stem Cell Differentiation and Cancer. Stem Cells Int. 2018;2018:3569493
41. Tee WW, Pardo M, Theunissen TW, Yu L, Choudhary JS, Hajkova P. et al. Prmt5 is essential for early mouse development and acts in the cytoplasm to maintain ES cell pluripotency. Genes Dev. 2010;24:2772-7
42. Koh CM, Bezzi M, Low DH, Ang WX, Teo SX, Gay FP. et al. MYC regulates the core pre-mRNA splicing machinery as an essential step in lymphomagenesis. Nature. 2015;523:96-100
43. Li Y, Chitnis N, Nakagawa H, Kita Y, Natsugoe S, Yang Y. et al. PRMT5 is required for lymphomagenesis triggered by multiple oncogenic drivers. Cancer Discov. 2015;5:288-303
44. Jing P, Zhao N, Ye M, Zhang Y, Zhang Z, Sun J. et al. Protein arginine methyltransferase 5 promotes lung cancer metastasis via the epigenetic regulation of miR-99 family/FGFR3 signaling. Cancer Lett. 2018;427:38-48
45. Wei L, Chiu DK, Tsang FH, Law DC, Cheng CL, Au SL. et al. Histone methyltransferase G9a promotes liver cancer development by epigenetic silencing of tumor suppressor gene RARRES3. J Hepatol. 2017;67:758-769
46. Wang Z, Kong J, Wu Y, Zhang J, Wang T, Li N. et al. PRMT5 determines the sensitivity to chemotherapeutics by governing stemness in breast cancer. Breast Cancer Res Treat. 2017;168:1-12
47. Bonday ZQ, Cortez GS, Grogan MJ, Antonysamy S, Weichert K, Bocchinfuso WP. et al. LLY-283, a Potent and Selective Inhibitor of Arginine Methyltransferase 5, PRMT5, with Antitumor Activity. ACS Med Chem Lett. 2018;9:612-7
Author contact
![]() Corresponding authors: Wei-Fen Xie, Department of Gastroenterology, Changzheng Hospital, Second Military Medical University, 415 Fengyang Road, Shanghai 200003, China. Tel.: (86-21) 8188-5341; Fax: (86-21) 8188-9624; E-mail: weifenxiecom.cn. Cheng Luo, CAS Key Laboratory of Receptor Research, State Key Laboratory of Drug Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai 201203, China. Tel.: (86-21) 5080-6918; Fax: (86-21) 5080-7188; E-mail: cluoac.cn. Xin Zhang, Department of Pharmacy, Changzheng Hospital, Second Military Medical University Shanghai, 200003, China. Tel.: (86-21) 8187-1330; E-mail: zhang68com
Corresponding authors: Wei-Fen Xie, Department of Gastroenterology, Changzheng Hospital, Second Military Medical University, 415 Fengyang Road, Shanghai 200003, China. Tel.: (86-21) 8188-5341; Fax: (86-21) 8188-9624; E-mail: weifenxiecom.cn. Cheng Luo, CAS Key Laboratory of Receptor Research, State Key Laboratory of Drug Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai 201203, China. Tel.: (86-21) 5080-6918; Fax: (86-21) 5080-7188; E-mail: cluoac.cn. Xin Zhang, Department of Pharmacy, Changzheng Hospital, Second Military Medical University Shanghai, 200003, China. Tel.: (86-21) 8187-1330; E-mail: zhang68com
 Global reach, higher impact
Global reach, higher impact