13.3
Impact Factor
Theranostics 2019; 9(9):2475-2488. doi:10.7150/thno.31144 This issue Cite
Research Paper
In-depth serum proteomics reveals biomarkers of psoriasis severity and response to traditional Chinese medicine
1. Guangdong Provincial Hospital of Chinese Medicine, Guangdong Provincial Academy of Chinese Medical Sciences, Guangdong Provincial Key Laboratory of Clinical Research on Traditional Chinese Medicine Syndrome, Guangzhou 510120, China.
2. State Key Laboratory of Analytical Chemistry for Life Science, School of Chemistry and Chemical Engineering, Nanjing University, Nanjing, China.
3. State Key Laboratory of Proteomics, Beijing Proteome Research Center, National Center for Protein Sciences, Beijing Institute of Lifeomics, Beijing, China.
4. School of Life Sciences, Westlake University; Institute for Basic Medical Sciences, Westlake Institute for Advanced Study, Hangzhou, Zhejiang, China.
5. Department of Rheumatology and Clinical Immunology, University Medical Center Utrecht, Utrecht, The Netherlands.
6. Department of Dermatology and Allergology, University Medical Center Utrecht, Utrecht, the Netherlands.
*Contributed equally to this manuscript
Abstract
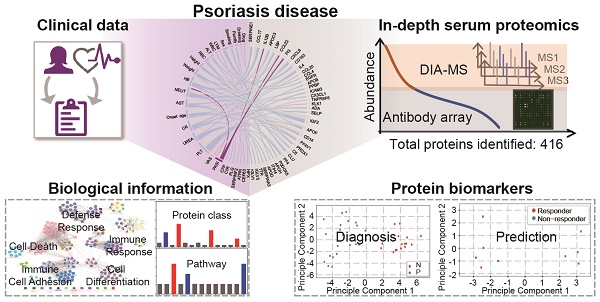
Serum and plasma contain abundant biological information that reflect the body's physiological and pathological conditions and are therefore a valuable sample type for disease biomarkers. However, comprehensive profiling of the serological proteome is challenging due to the wide range of protein concentrations in serum.
Methods: To address this challenge, we developed a novel in-depth serum proteomics platform capable of analyzing the serum proteome across ~10 orders or magnitude by combining data obtained from Data Independent Acquisition Mass Spectrometry (DIA-MS) and customizable antibody microarrays.
Results: Using psoriasis as a proof-of-concept disease model, we screened 50 serum proteomes from healthy controls and psoriasis patients before and after treatment with traditional Chinese medicine (YinXieLing) on our in-depth serum proteomics platform. We identified 106 differentially-expressed proteins in psoriasis patients involved in psoriasis-relevant biological processes, such as blood coagulation, inflammation, apoptosis and angiogenesis signaling pathways. In addition, unbiased clustering and principle component analysis revealed 58 proteins discriminating healthy volunteers from psoriasis patients and 12 proteins distinguishing responders from non-responders to YinXieLing. To further demonstrate the clinical utility of our platform, we performed correlation analyses between serum proteomes and psoriasis activity and found a positive association between the psoriasis area and severity index (PASI) score with three serum proteins (PI3, CCL22, IL-12B).
Conclusion: Taken together, these results demonstrate the clinical utility of our in-depth serum proteomics platform to identify specific diagnostic and predictive biomarkers of psoriasis and other immune-mediated diseases.
Keywords: Proteomics, Data-Independent Acquisition Mass Spectrometry, Antibody Microarray, Biomarker, Psoriasis
 Global reach, higher impact
Global reach, higher impact