13.3
Impact Factor
Theranostics 2019; 9(8):2198-2208. doi:10.7150/thno.30798 This issue Cite
Research Paper
Long noncoding RNA TUG1 inhibits osteogenesis of bone marrow mesenchymal stem cells via Smad5 after irradiation
Department of Blood Transfusion, Lab of Radiation biology, The Second Affiliated Hospital, Third Military Medical University, Chongqing, 400037, China.
*These authors contributed equally to this work and should be considered co-first authors.
Received 2018-10-19; Accepted 2019-2-18; Published 2019-4-12
Abstract
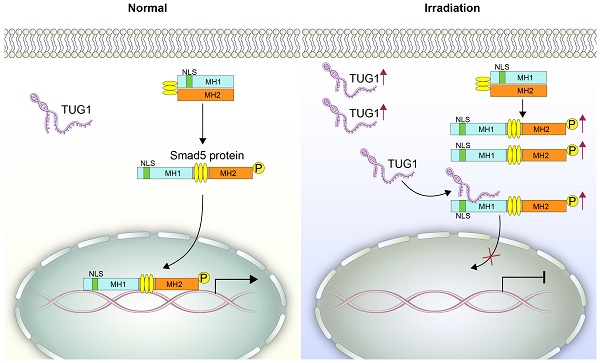
Irradiation can greatly inhibit osteogenesis of bone marrow mesenchymal stem cells (BM-MSCs). However, the mechanism remains unclear.
Methods: We analyzed the expression profile of long noncoding RNAs (lncRNAs) in BM-MSCs using microarray data. LncRNA TUG1 (Taurine Upregulated Gene 1) was selected and tested in radiated BM-MSCs and non-radiated BM-MSCs. Functional analyses (in vitro) were performed to confirm the role of TUG1 in the osteogenic inhibition induced by irradiation. A RIP (RNA immunoprecipitation) assay was performed to detect the interaction of TUG1 and Smad5. Smad5 and the phosphorylated Smad5 (p-Smad5) were tested by western blot. The nuclear translocation of p-Smad5 were tested by immunofluorescence analysis. Furthermore, a series of Smad5 deletions was constructed to identify the TUG1 binding site of Smad5.
Results: We found that numerous lncRNAs, including TUG1, exhibit significant expression differences after irradiation. After irradiation TUG1 was significantly increased in BM-MSCs and inhibited osteogenesis. Furthermore, TUG1 directly bound to Smad5, an osteogenic enhancer. Although the phosphorylation level of Smad5 was increased following irradiation, osteogenesis of BM-MSCs was decreased. Mechanistically, TUG1 interacting with the 50-90 aa region of Smad5 and blocks the nuclear translocation of p-Smad5, abolishing osteogenic signalling after irradiation.
Conclusion: These results indicate that TUG1 is a negative regulator of Smad5 signalling and suppresses osteogenesis of BM-MSCs after irradiation.
Keywords: TUG1, Osteogenesis, Bone Marrow Mesenchymal Stem Cells, Smad5, Irradiation
Introduction
Haematopoietic acute radiation sickness is the most common disease caused by high-dose irradiation [1]. The abnormal haematopoietic environment caused by irradiation is an important barrier to haematopoietic recovery [2].
BM-MSCs play important roles in haematopoietic environment, supporting and regulating bone haematopoiesis. Most of the non-haematopoietic cells in bone marrow are derived from BM-MSCs, including adipocytes and osteoblasts [3]. The differentiation of BM-MSCs, especially the osteoblastic differentiation of BM-MSCs, is crucial to the structure and function of haematopoietic environment [4]. However, BM-MSC osteogenic differentiation can be greatly suppressed by irradiation. As a result, the structure and function of haematopoietic environment become abnormal, which hinders haematopoietic recovery [5].Thus, it is essential to study how irradiation inhibits the osteogenic differentiation of BM-MSCs.
The osteogenesis of BM-MSCs is regulated by a complex osteogenic signalling network, including TGF-β signalling [6], Wnt signalling [7], and MAPK signalling [8]. TGF-β signalling is a key signalling pathway in the osteogenic differentiation of BM-MSCs. This signalling pathway relies on a series of Smad proteins including R-Smads (receptor-regulated Smads), Co-Smads (common-partner Smads), and I-Smads (Inhibitory Smads) [9]. Smad5 is an R-Smad protein. It participates in the osteogenic differentiation of BM-MSCs by acting as a transcription factor [10]. When osteogenic signalling is transducted into the cytoplasm of BM-MSCs, Smad5 is phosphorylated and then is directed into the nucleus, regulating the expression of target genes to induce osteogenic differentiation [9]. In this process, the nuclear translocation of Smad5 is critical for the osteogenic signalling transduction. However, we found that although the phosphorylation level of Smad5 significantly increased after irradiation, the osteogenesis of BM-MSCs was significantly inhibited, suggesting that unknown inhibitors induced by irradiation suppress Smad5 signalling and osteogenesis of BM-MSCs.
Long noncoding RNAs (lncRNAs) are defined as transcripts longer than 200 nucleotides and were originally regarded as transcriptional “noise” [11, 12]. In recent years, many studies have reported that lncRNAs are involved in cell osteogenic differentiation, such as lncRNA BORG [13], lncRNA ANCR [14], and lncRNA DANCR [15]. Our previous microarray data revealed that numerous lncRNAs exhibit significant expression differences after irradiation. Further analysis of the results of the lncRNA and co-expression networks revealed that the expression of lncRNA TUG1 (Taurine Upregulated Gene 1) (FC>2) was significantly increased after irradiation and may interact with Smad5, a key molecule in the osteogenic differentiation of BM-MSCs.
LncRNA TUG1 is a 7.1 kb lncRNA and was firstly identified in retinal cells. It can be upregulated by taurine, and it regulate photoreceptor differentiation [16]. It is involved in many diseases, including neurodegeneration and cancers. However, the role of TUG1 in the osteogenic differentiation of human BM-MSCs and its mechanism remains unclear.
In this study, we explored the regulatory roles of TUG1 in Smad5 signalling during the osteogenic differentiation of BM-MSCs after irradiation. We propose that TUG1 is a key regulator of osteogenic differentiation after the irradiation of BM-MSCs.
Materials and methods
Cells Lines and Culture
Human BM-MSCs were obtained from ScienCell Research Laboratories (No. 7500, ScienCell) and were cultured in a complete medium (No. HUCMX-90011, Cyagen Biosciences, Inc.) at 37 °C and 5 % CO2. All media were changed every other day. The cells were passaged to a secondary culture when dense colonies of spindle-shaped cells covered approximately 80 %-90 % of the culture dish. BM-MSCs from passage 10 (P10) were used in this study. For the radiation treatment, when cultured cells reached 80 % confluence, they were irradiated with Co-60 for 13min at a rate of 0.69 Gy/min.
Plasmids Constructs
The flag-tagged Smad5 deletion constructs (WT, ∆25, ∆50, ∆90) were created using forward primers containing the BamH I site and a common reverse primer containing the SaL I site. Primer sequences are summarized in Table 1. The PCR products were sequentially digested with BamH I and SaL I and ligated into similarly digested pCMV-N-Flag-Smad5 vector. The in-frame ligation of all plasmids was confirmed by automated sequencing. Then, the vectors were transferred into BM-MSCs for protein expression by Lipofectamine® 3000(L3000015, Invitrogen).
The primers of Smad5 deletion constructs
| Gene name | Sequence | Restriction site |
|---|---|---|
| WT | Forward primer: 5′-CGCGGATCCACGTCAATGGCCAGCTTGTTT-3′ | BamH I |
| ∆25 | Forward primer: 5′-CGCGGATCCGAGGAGGAGAAATGGGCAGAA-3′ | BamH I |
| ∆50 | Forward primer: 5′-CGCGGATCCGAACTGGAGAAAGCCTTGAGC-3′ | BamH I |
| ∆90 | Forward primer: 5′-CGCGGATCCCGTGTTTGGCGCTGGCCGGA-3′ | BamH I |
| WT, ∆25, ∆50, ∆90 | Reverse primer: 5′-ACGCGTCGACTTATGAAACAGAAGATATGGGG-3′ | SaL I |
Cell transfection
TUG1 knockdown (or overexpression) was conducted via lentiviral transfection. Briefly, three lentiviral (si-TUG1 or ov-TUG1) constructs were generated based on different regions of the human TUG1 sequence (NCBI accession NR_002323.2). The same lentiviral vector containing an insert of nonspecific RNA oligonucleotide (si-NC or ov-NC) was used as a negative control. The si-TUG1 sequence was 5′-GGATATAGCCAGAGAACAA-3′, the ov-TUG1 sequence was 5′-CACTATCGGAGACAAAGCGG-3′, and the negative control sequence was 5′-GTTCTCCGAACGTGTCACGT-3′. Lentiviruses containing these plasmids were generated by transfecting the 293T packaging cell line with the siRNA vector (pGreen-Puro-TUG1) and overexpression vector (dCAS-VP64-blast, MS2-P65-HSF1-Hygro, H-TUG1 gRNA-Puro). Lentiviral vectors were added into BM-MSCs culture medium. After 72 h, BM-MSCs harbouring the constructs were selected using the corresponding antibiotics (puromycin: selected, 3 µg/mL; maintained, 1 µg/mL; blast: selected, 6 µg/mL; maintained, 1 µg/mL; Hygromycin B: selected, 150 µg/mL; maintained, 50 µg/mL).
Polymerase chain reaction
Total RNA was extracted using the Trizol Reagent according to the manufacturer's protocol (No. 10296010, Invitrogen). Next, 1 µg of the extracted RNA was reverse-transcribed using PrimeScript RT reagent Kit with gDNA Eraser (No. RR047A, Takara). qRT-PCR was performed in triplicate in 20 µl reactions using SYBR® Premix Ex Taq™ II (Tli RNaseH Plus) (No. RR820A, Takara). The reaction protocol was as follows: heating for 10 min at 95 °C, followed by 40 cycles of amplification (15 s at 95 °C and 1 min at 60 °C). Data were analysed using the delta-delta Ct method. The primer sequences are summarized in Table 2.
PCR Primer sequences
| Gene name | Sequence |
|---|---|
| TUG1 | Forward primer: 5′-TAGCAGTTCCCCAATCCTTG-3′ |
| Reverse primer: 5′-CACAAATTCCCATCATTCCC-3′ | |
| β-actin | Forward primer: 5′-ACCCCGTGCTGCTGACCGAG-3′ |
| Reverse primer: 5′-TCCCGGCCAGCCAGGTCCA-3′ | |
| Runx2 | Forward primer: 5′-TGCCACCTCTGACTTCTGC-3′ |
| Reverse primer: 5′-GATGAAATGCCTGGGAACTG-3′ | |
| OGN | Forward primer: 5′-TGCCTTGATAGGAGGAAAACA-3′ |
| Reverse primer: 5′-GATCCCCAAAAGCATTTAAGG-3′ | |
| ALP | Forward primer: 5′-ACGTGGCTAAGAATGTCATC-3′ |
| Reverse primer: 5′-CTGGTAGGCGATGTCCTTA-3′ | |
| OCN | Forward primer: 5′-TGAGAGCCCTCACACTCCTC-3′ |
| Reverse primer: 5′-CGCCTGGGTCTCTTCACTAC-3′ | |
| Osterix | Forward primer: 5′-CCCACCTCAGGCTATGCTAA-3′ |
| Reverse primer: 5′-CACTGGGCAGACAGTCAGAA-3′ | |
| TUG1-Mouse | Forward primer: 5′-CATAGTATCATCTTCGGGTTAC-3′ |
| Reverse primer: 5′-CACAAAATGCATGTAGGTTC-3′ | |
| β-actin-Mouse | Forward primer: 5′-AGCCATGTACGTAGCCATCC-3′ |
| Reverse primer: 5′-CTCAGCTGTGGTGGTGAA-3′ |
Osteogenic differentiation
To detect osteogenic differentiation, cells were treated in human mesenchymal stem cell osteogenic differentiation medium (No. HUXMA-90021, Cyagen) for 2 weeks. Cells were plated at a density of 10,000 cells/cm2 in 6 well cell culture clusters and treated every other day with the medium. Of note, because of the limited cell number, TUG1-overpression BM-MSCs were plated at a density of 2,000 cells/cm2 in 24 well cell culture clusters and treated every other day with the medium. After differentiation, we removed the osteogenic differentiation medium from the wells and fixed the cells with 4 % formaldehyde solution in PBS for 30 min. Then, we stained the cells with alizarin red S solution (No. S0141, Cyagen) for 10 min, followed by 3 extensive washes with PBS. The images were visualized under a light microscope (Leica DMIRB, Germany). For quantitation, the cells were stained with alizarin red S and destained with ethylpyridinium chloride (Wako Pure Chemical Industries, Ltd). Then, the extracted stain was transferred to a 96-well plate, and absorbance was measured at 562 nm using a Varioskan FLASH microplate reader (Thermo).
Western Blotting and Antibodies
Protein expression in the samples was analysed by western blotting. The total protein lysate was extracted with cell lysis buffer for western blotting and immunoprecipitation (No. P0013, Beyotime) and denatured by boiling. Protein samples were resolved on 12 % SDS-polyacrylamide gels and transferred to polyvinylidene fluoride membranes (PVDF Western Blotting Membranes, Roche). Membranes were blocked in PBS containing 5 % (w/v) non-fat dry milk and 0.1 % Tween-20 for 2 h, incubated with the appropriate antibodies overnight, washed with TBST buffer, and then incubated with secondary antibodies for 2 h. Blots were developed using a horseradish peroxidase-linked secondary antibody and developed with a chemiluminescent detection system (Phototope-HRP Western blot detection kit, New England Biolab).
The primary antibodies used for blotting were as follows: anti-Smad5 (1:1000, 12534, Cell Signaling Technology), anti-p-Smad5 (1:1000, 9516S, Cell Signaling Technology), anti-OGN (1:1000, sc-374463, Santa Cruz), anti-Runx2 (1:1000, sc-390351; Santa Cruz), and anti-β-actin (1:1000, sc-47778, Santa Cruz).
Immunofluorescence
After treatment, cells were fixed with 4 % formaldehyde for 30 min at room temperature and then incubated in blocking solution containing 5 % bovine serum albumin in PBS. For permeabilization, cells were incubated in 0.2 % Triton X-100. Then, the cells were incubated with anti-p-Smad5 (1:50) overnight at 4 °C. The next day, the cells were incubated with secondary antibodies conjugated to Alexa Fluor 647 and Cy3 for 30 min at 37 °C. For nuclear staining, the cells were incubated with 4,6-diamidino-2-phenylindole (DAPI) for 3 min. Fluorescence was visualized using laser confocal microscopy (Leica SP5, Heidelberg, Germany).
RNA-binding protein immunoprecipitation (RIP) assay
RIP experiments were conducted using the EZ-Magna RIP Kit (17-700, Millipore Corporation). Briefly, cells were harvested; lysed; and then reacted with RIP buffer containing magnetic beads conjugated with anti-Smad5 antibody, anti-Flag antibody (14793, Cell Signaling Technology) or the corresponding negative control IgG (Millipore). After the antibody was recovered using protein A/G beads, precipitates were evaluated by PCR analysis.
Isolation of nuclear and cytoplasmic protein
The nuclear and cytoplasmic fractions were separated with a Nuclear and Cytoplasmic Protein Extraction Kit (P0028, Beyotime, China). Approximately 4.0 × 106 cells were lysed in the cytoplasmic fraction buffer and separated from the sediment, then, the nuclear fraction was collected from the supernatant. Protein expression in the samples was analysed by western blot.
RNA fluorescence in situ hybridization (FISH) and immunofluorescence
The 48 fluorescently labeled oligonucleotides targeted to different regions of TUG1 (Table S2) were designed and hybridized to fixed BM-MSCs according to the protocol of the manufacturer (Biosearch Technologie). The BM-MSCs were rinsed briefly in PBS and then fixed in 3.7% formaldehyde in PBS (pH 7.4) for 10 min at room temperature and permeabilized in 70% ethyl alcohol at 4°C at least 1 h. Then Hybridization was carried out using probe sets and p-Smad5 antibody in a moist chamber at 37°C for 12-16 h according to the protocol (Biosearch Technologies).The image sections was acquired using a laser confocal microscopy (Leica SP5, Heidelberg, Germany).
Statistical analysis
The mRNA levels of TUG1, Runx2, OGN, OCN, Osterix in the tested samples are expressed as cycle threshold (CT) levels. Normalized copy numbers (relative quantification) were calculated using the ΔΔCT equation. Data were presented as the mean ± standard error of mean (SEM). Statistical analysis was performed using GraphPad Prism version 6.0. An independent t-test was used to compare data obtained from the experimental group with those obtained from the control group. The results are considered significant at *P< 0.05, **P < 0.01, and ***P < 0.001.
Results
The expression level of TUG1 increases in BM-MSCs after irradiation
In vivo and in vitro studies have shown that irradiation can strongly inhibit the osteogenic differentiation of BM-MSCs [17, 18]. Consistent with previous studies, our data show that the osteogenic differentiation of BM-MSCs is significantly decreased after irradiation (Figure 1A, 1B, 1C).
TUG1 increases after irradiation in BM-MSCs. (A-C) The effect of irradiation on osteogenesis of BM-MSCs. (A) Representative images of alizarin red staining of non-irradiated BM-MSCs (control) and irradiated BM-MSCs (IR) in osteogenesis. (B) qRT-PCR analysis of mRNA levels of osteogenic markers, and (C) Western blot analysis of protein expression levels of osteogenic markers in osteogenesis.of BM-MSCs. (D) Heat map of differentially expressed lncRNAs (53 upregulated lncRNAs and 4 downregulated lncRNAs) between non-irradiated BM-MSCs and irradiated BM-MSCs. (E) The expression levels of TUG1 in BM-MSCs within 14 days after irradiation. Relative expressions of genes were normalized by β-actin in qRT-PCR. All experiments were performed in triplicate, and the results are expressed as the means ±SEM. *P < 0.05; **P < 0.01; ***P < 0.001; ns: not significant. Abbreviations: IR: irradiated BM-MSCs; Runx2: runt related transcription factor 2; OGN: osteoglycin.
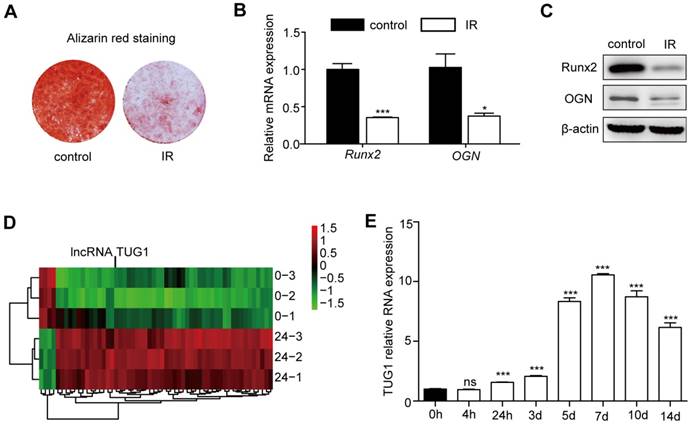
To explore the role of lncRNAs in BM-MSCs after irradiation, we designed a customized microarray to probe the expression profiles of 27,984 human transcripts that have been annotated as potential noncoding RNAs. The expression profiles were probed for the control and 24 h after irradiation. A total of 57 potential noncoding RNAs were upregulated or downregulated by > 2-fold after irradiation (Figure 1D, Table S1). Of these transcripts, 53 lncRNAs were highly induced after irradiation, while 4 lncRNAs showed reduced expression. We focused on a highly upregulated lncRNA transcript, TUG1 (Figure 1D).
Then, we evaluated the expression level of TUG1 at different time points after irradiation by qRT-PCR. TUG1 expression level significantly increased after irradiation in 14 days (Figure 1E). In addition, the expression levels of TUG1 in mice bone marrow were significantly increased after irradiation (Figure S3A).
TUG1 suppresses the osteogenic differentiation of BM-MSCs after irradiation
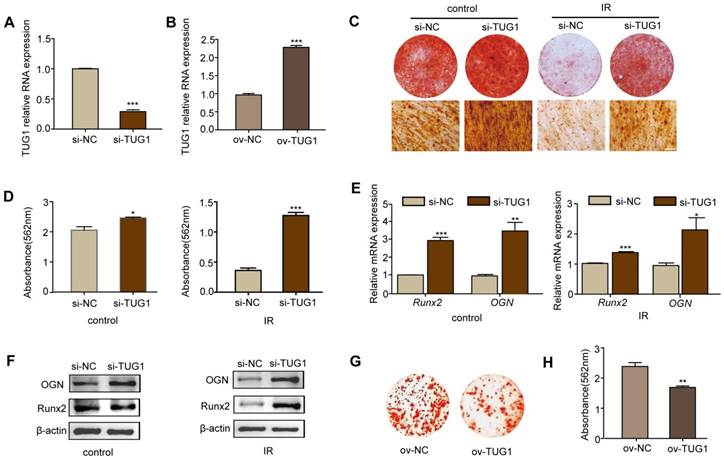
To study the role of TUG1 in the osteogenic differentiation of BM-MSCs after irradiation, we knocked down the expression of TUG1 with siRNA vector (pGreen-Puro-TUG1) and overexpressed TUG1 by CRISPR/CAS9 (Figure 2A, 2B).
After 7 days of osteogenic induction, alkaline phosphatase (ALP) staining (Figure S4A), ALP quantification of osteogenic induction (Figure S4B) and the mRNA expression levels of the osteogenic-related markers, including ALP (Figure S4C), Runx2 (runt related transcription factor 2) (Figure S4D), OCN (osteocalcin) (Figure S4E) and osterix (Figure S4F), were increased in TUG1-knockdown BM-MSCs at day 7 both after irradiation and under non-irradiated conditions.
After 14 days of osteogenic induction, we determined the effect of TUG1 on matrix mineralization by performing alizarin red S staining and the expression of the key osteogenic markers Runx2 and OGN by performing qRT-PCR and western blot. Irradiated BM-MSCs showed lower rates of calcium deposition than non-irradiated BM-MSCs. Moreover, TUG1-knockdown BM-MSCs showed higher rates of calcium deposition than control BM-MSCs both after irradiation and under non-irradiated conditions (Figure 2C-2F). The mRNA and protein expression levels of the osteogenic-related markers Runx2 and OGN were higher in TUG1-knockdown BM-MSCs (Figure 2E, 2F). However, the TUG1-overexpression BM-MSCs showed lower rates of calcium deposition than control BM-MSCs in non-irradiated condition (Figure 2G, 2H). These results show that TUG1 may inhibit the osteogenic differentiation of BM-MSCs.
TUG1 inhibits the osteogenic differentiation of BM-MSCs. (A-B) qRT-PCR analysis of RNA expression levels of TUG1 after BM-MSCs were transfected with TUG1-siRNA vector (si-TUG1), empty vector (si-NC) (A), TUG1-overexpression vectors (ov-TUG1) and empty CRISPR/CAS9 vectors (ov-NC) (B). (C-D) Representative images of alizarin red staining of alizarin red staining (C) with quantification of the dye extracted from alizarin red S staining (D) in osteogenesis.of BM-MSCs. Control: non-irradiated BM-MSCs; IR: irradiated BM-MSCs; Scale bars, 100 μm. (E-F) qRT-PCR analysis of mRNA expression (E) and western blot analysis of protein (F) levels of osteogenic markers in osteogenesis of BM-MSCs. (G-H) Representative images of alizarin red staining of alizarin red staining (G) with quantification of the dye extracted from alizarin red S staining (H) in osteogenesis.of ov-NC and ov-TUG1 BM-MSCs. All experiments were performed in triplicate, and the results are expressed as the means ±SEM. *P < 0.05; **P < 0.01; ***P < 0.001; ns: not significant. Abbreviations: IR: irradiated BM-MSCs; si-NC: BM-MSCs transfected with empty vector, si-TUG1: BM-MSCs transfected with TUG1-siRNA vector, ov-TUG1: BM-MSCs transfected with TUG1-overexpression vectors, ov-NC: BM-MSCs transfected with empty CRISPR/CAS9 vectors.
TUG1 directly interacts with Smad5
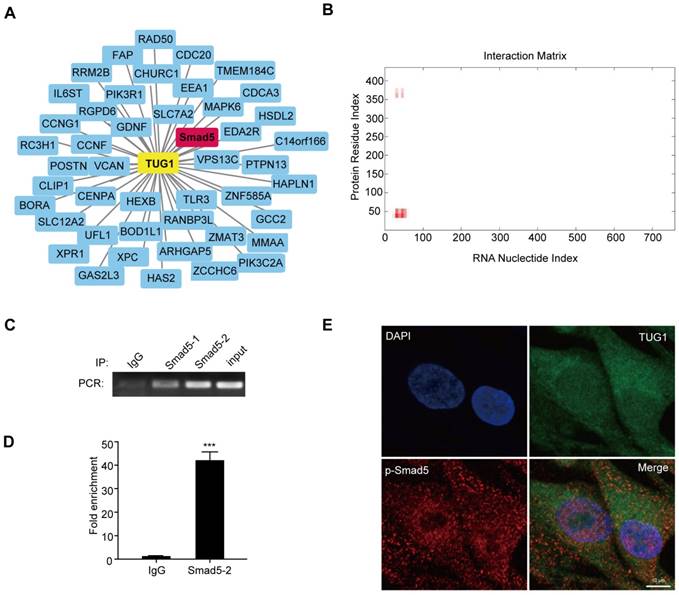
To clarify how TUG1 modulates the osteogenic differentiation of BM-MSCs after irradiation, we analysed the co-expression network maps of TUG1 with the microarray data. There are 46 mRNAs related to TUG1, including Smad5 (Figure 3A), which is an important positive transcription factor in the osteogenic differentiation of BM-MSCs. Smad5 consists of three regions: a conserved N-terminal domain (MH1 domain), a conserved C-terminal domain (MH2 domain) and a more divergent linker region [19]. Then, we used the online RNA-protein binding prediction site catRAPID to predict the interaction between TUG1 and Smad5. The result showed a high possibility that TUG1 binds to the MH1 domain of Smad5 (Figure 3B). To test whether TUG1 binds to Smad5, we performed a RIP assay and found that TUG1 RNA levels were detected in Smad5 immunoprecipitates relative to those of the control IgG immunoprecipitates (Figure 3C, 3D, Figure S1). These results support bioinformatics predictions indicating that TUG1 can directly bind to Smad5.
Moreover, RNA FISH assay of TUG1 combined with immunofluorescence detection of p-Smad5 in BM-MSCs showed that TUG1 was colocalized with p-Smad5 mainly in the cytoplasm of BM-MSCs (Figure 3E), indicating that TUG1 may regulate cytoplasmic p-Smad5 activity.
TUG1 regulates the subcellular localization of p-Smad5
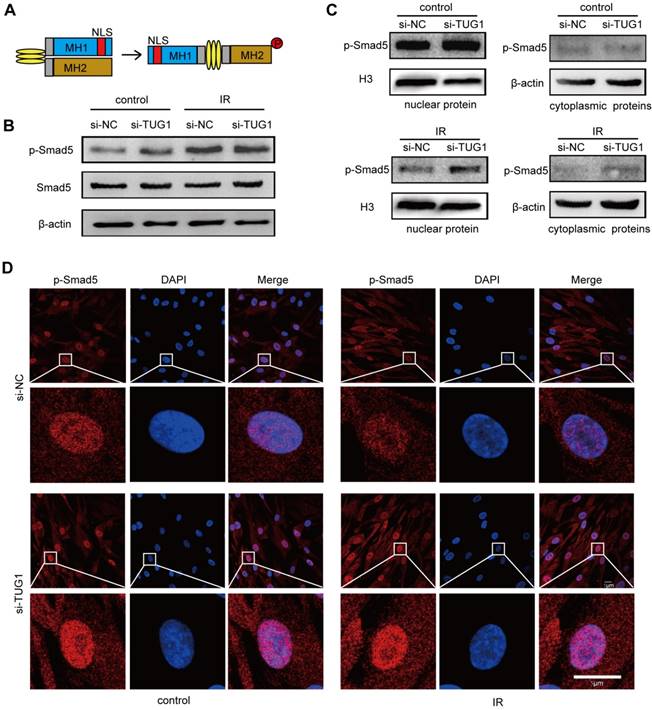
Smad5 is a key transcription factor in the osteogenic differentiation of BM-MSCs [10]. Under physiological conditions, Smad5 is mainly located in the cytoplasm. When Smad5 is phosphorylated, it is directed into the nucleus [20], where it regulates the expression of osteogenic genes and thus induces the osteogenic differentiation of BM-MSCs [21, 22] (Figure 4A). During this process, the nuclear translocation of Smad5 is crucial to osteogenic modulation.
TUG1 directly binds to Smad5. (A) Co-expression network maps showing 46 putative RNA binding proteins (RBPs) related to TUG1. (B) Online RNA-protein binding prediction site catRAPID predicted the interaction region between TUG1 and Smad5. (C) RIP assay was performed to detect the interaction between TUG1 and Smad5. PCR detection of the indicated RNAs retrieved by immunoglobulin G (IgG) in BM-MSCs (lgG), Smad5-specific antibody in non-radiated BM-MSCs (Smad5-1), Smad5-specific antibody in radiated BM-MSCs (Smad5-2), and positive control(Input). (D) qRT-PCR detection of the indicated RNAs retrieved by Smad5-specific antibody compared with IgG in the RIP assay within BM-MSCs. (E) Colocalization analysis: RNA FISH assay of TUG1 combined with immunofluorescence detection of p-Smad5 in BM-MSCs. Scale bars, 10 um. DAPI, 4′,6-diamidino-2-phenylindole. All experiments were performed in triplicate, and the results are expressed as the means ±SEM. ***P < 0.001. Abbreviations: DAPI, 4′,6-diamidino-2-phenylindole, IgG: immunoglobulin G, RNA FISH: RNA Fluorescence In Situ Hybridization; RBPs: RNA binding proteins.
To further elucidate the relationship between TUG1 and Smad5, we analysed the effect of TUG1-knockdown on Smad5 expression, phosphorylation and nuclear translocation in irradiated and non-irradiated BM-MSCs. Western blot results showed that the protein level of smad5 and its phosphorylation level was not significantly changed in TUG1-knockdown BM-MSCs (Figure 4B). Interestingly, although irradiation could not affect the protein level of Smad5, the phosphorylation level of Smad5 increased after irradiation (Figure 4B). The phosphorylation level of Smad5 also increased in mice after irradiation (Figure S3). According to previous studies, Smad5 phosphorylation promotes the nuclear translocation of Smad5. However, although irradiation treatment promoted the phosphorylation of Smad5, the nuclear translocation of p-Smad5 in BM-MSCs was significantly inhibited, as the protein level of p-Smad5 in the nucleus was markedly decreased after irradiation. Surprisingly, TUG1 knockdown, rescued the nuclear translocation of p-Smad5 after irradiation (Figure 4C, 4D). TUG1 might act as a negative regulator of Smad5 by blocking the nuclear translocation of p-Smad5.
TUG1 regulates the subcellular localization of p-Smad5. (A)The structure of Smad5. NLS: nuclear localization sequence. (B) Western blot analysis of the Smad5 expression and phosphorylation level in BM-MSCs. Control: non-irradiated BM-MSCs; IR: irradiated BM-MSCs; si-NC: BM-MSCs transfected with empty vector, si-TUG1: BM-MSCs transfected with TUG1-siRNA vector. (C) Western blot analysis of the cytoplasmic and nuclear phosphorylated Smad5 levels in BM-MSCs. (D) Immunofluorescence analysis of the nuclear translocation of phosphorylated Smad5 in BM-MSCs. Scale bars, 5 μm. Abbreviations: DAPI, 4′,6-diamidino-2-phenylindole; IR: irradiated BM-MSCs; NLS: nuclear localization sequence. p-Smad5: phosphorylated Smad5; si-NC: BM-MSCs transfected with empty vector, si-TUG1: BM-MSCs transfected with TUG1-siRNA vector.
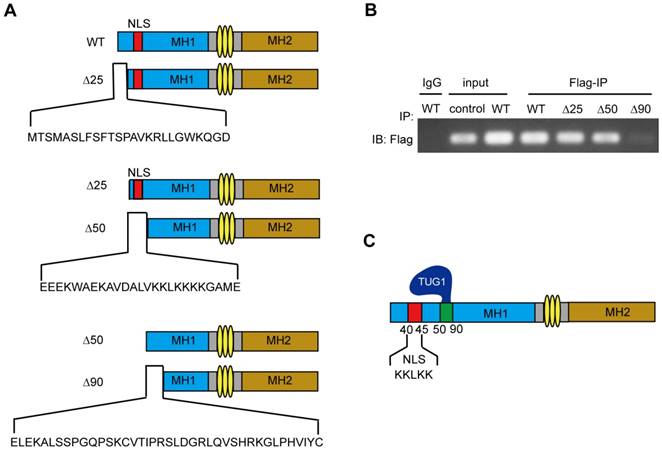
The 50-90 aa region of Smad5 is required for interaction with TUG1
To further identify the TUG1 binding site of Smad5, we constructed flag-tagged Smad5 deletions (WT: wild-type Smad5, ∆25: the 25-435 aa of Smad5, ∆50: the 50-435 aa of Smad5, ∆90: the 90-435 aa of Smad5) (Figure 5A) and transferred the constructed plasmids into BM-MSCs to express the corresponding proteins. Then, we extracted the cellular proteins for the RIP experiments to determine whether TUG1 interacts with these Smad5 deletion constructs.
We found that the Smad5 WT, ∆25, ∆50 deletion constructs all physically interact with TUG1 (Figure 5B). However, the ∆90 deletion constructs failed to interact with TUG1, suggesting that the region between 50-90 of Smad5 is important for TUG1 binding (Figure 5B, Figure S2). The schematic of TUG1 bound to Smad5 is shown in Figure 5C. This result shown that the 50-90 aa region of Smad5 is required for its interaction with TUG1.
The main domain of Smad5 interacting with TUG1 is near to NLS of Smad5. (A)The schematic illustration of Smad5 deletion constructs. WT: wild-type Smad5, ∆25: ∆25 deletion constructs, ∆50: ∆50 deletion constructs, ∆90: ∆90 deletion constructs. (B) RIP assay to detect the interaction between TUG1 and the Smad5 deletion constructs. IgG: negative control; Input: positive control; Flag-IP (Anti-flag immunoprecipitation): PCR detection of the indicated RNAs retrieved by Flag-specific antibody within BM-MSCs. Control: BM-MSCs, WT: BM-MSCs transfected with flag-tagged wild-type Smad5, ∆25: BM-MSCs transfected with the flag-tagged ∆25 deletion constructs, ∆50: BM-MSCs transfected with the flag-tagged ∆50 deletion constructs, ∆90: BM-MSCs transfected with the flag-tagged ∆90 deletion constructs. (C) The schematic diagram of TUG1 bound to Smad5. Abbreviations: Flag-IP: Anti-flag immunoprecipitation, NLS: nuclear localization sequence, IgG: immunoglobulin G, WT: wild type.
Discussion
Bone marrow mesenchymal stem cells (BM-MSCs), the important stromal cells in bone marrow, can differentiate into different stromal cells including osteoblasts, to form the haematopoietic environment. During this process, the osteoblastic differentiation of BM-MSCs is a critical step in the formation of the haematopoietic niche [4]. At different stages of osteogenic differentiation, the cell population, including osteoprogenitors, osteoblasts and osteocytes, may form distinct niches for different haematopoietic progenitors. By secreting IL-7 and IGF-1, osteoprogenitors can support early-stage B lineage differentiation [23]. Osteoblasts support HSC (hematopoietic stem cell) maintenance [24, 25]. Mature osteoblasts are thought to support T cell differentiation [26]. Although osteocytes are embedded in the bone, they are indispensable for HSPC (hematopoietic stem and progenitor cell) mobilization and B and T lymphopoiesis [5, 27]. A deficiency of these cell populations can lead to different haematopoietic deficiencies. Thus, the osteogenic differentiation of BM-MSCs is crucial to bone haematopoiesis.
Irradiation can cause damage to the hematopoietic system. The major targets of irradiation in bone marrow include two types of stem cells: BM-MSCs and HSCs. After irradiation, numerous HSCs are apoptotic. Although BM-MSCs partially survive, their osteogenic differentiation function is significantly inhibited [17, 18, 28, 29].
LncRNA plays an important role in regulating the osteogenic differentiation of BM-MSCs [13-15]. Thousands of lncRNAs are differentially expressed in BM-MSCs under normal in vitro osteogenic induction [30]. Several important lncRNAs of BM-MSC osteogenesis, including lncRNA-H19 [31, 32], and lncRNA-ANCR [14], had been found. Our study found a novel regulator of BM-MSC osteogenic differentiation, lncRNA TUG1, which inhibits osteogenic differentiation of BM-MSCs after radiation injury.
In our microarray study, we found that TUG1 increase in BM-MSCs after irradiation. Consistent with previous studies, our data showed that the osteogenic differentiation of BM-MSCs was markedly inhibited after irradiation. Knocking down of TUG1 rescues the osteogenic differentiation of BM-MSCs after irradiation. Over-expression of TUG1 exhibits an inhibitory effect. These results indicated that TUG1 acts as a negative regulator in osteogenesis of BM-MSCs after radiation (Figure 2). Many studies indicate that TUG1 has important pathological and physiological functions. Early study reported that TUG1 is an important molecule involved in the development of the mouse retina [16]. TUG1 can co-ordinately promote the self-renewal of glioma stem cells [33]. Recent studies explored the roles of TUG1 on osteogenic differentiation. Yu et al demonstrated that TUG1 knockdown induced inhibition of osteoblast differentiation of human aortic valve interstitial cells (VICs) in aortic valve calcification [34]. In addition, He et al demonstrated that TUG1 facilitates osteogenic differentiation of periodontal ligament stem cells (PDLSCs) [35]. However, the role of TUG1 in osteogenic differentiation of BM-MSCs remain unclear.
Co-expression networks revealed that TUG1 may interact with Smad5 to regulate osteogenic differentiation of BM-MSCs (Figure 3A).
Smad5 is an important transcription factor in osteogenic differentiation and promotes the osteogenic differentiation of BM-MSCs [10]. As a receptor-regulated Smad (R-Smad) protein, Smad5 contains MH1 domain and MH2 domain. The MH1 domain of Smad5 contains a typical nuclear localization sequence "KKLKK" (NLS sequence), which is necessary for transport of Smad molecules from the cytoplasm to the nucleus [19]. The MH2 domain contains the receptor phosphorylation site SSXS and is a functional regulatory region of the Smad protein [36]. Under physiological conditions, Smad5 is mainly located in the cytoplasm, MH1 and MH2 bind to each other to inhibit activity. When the MH2 domain is phosphorylated, its conformation changes, and the NLS sequence in the MH1 domain is exposed, thereby directing the Smad5 protein into the nucleus [20], and then regulates the expression of target genes (Runx2, OGN) to induce osteogenic differentiation. In this process, the phosphorylation and the nuclear translocation of Smad5 is very important. Here, we found that irradiation significantly increased the phosphorylation level of Smad5. However, although irradiation treatment promoted the phosphorylation of Smad5, the nuclear translocation of p-Smad5 and osteogenesis in BM-MSCs was significantly inhibited, suggesting that unknown inhibitors induced by irradiation suppress nuclear translocation of p-Smad5.
Inhibitors of Smad5 include Smad6 [37], Smad7 [38], I-Smads (Inhibitory Smads), Smurf1 [39], SCP4/CTDSPL2 and PPM1A/PP2Ca [40], MicroRNA-222-3p [10], and so on. Smad6 and Smad7 were originally identified as inhibitory proteins for TGF-β family signalling [37] [38] and they stably bind to TGF-β type I receptors and inhibits phosphorylation of Smad5, resulting in inhibition of TGF-β signalling [41]. In the nucleus, I-Smads (Inhibitory Smads) can bind Smad5 complex to inhibit its transcription. Smurf1 can reduce the activity of Smad5, through I-Smads binding Smad5 by Smad5-mediated proteasome pathway [39]. The phosphatases, including SCP4/CTDSPL2 and PPM1A/PP2Ca, bind to p-Smad5 to mediate Smad5 dephosphorylation inactivation [40]. MicroRNA-222-3p can inhibit the expression of Smad5 then inhibit the osteogenic differentiation of BM-MSCs [10]. Dysregulation of regulatory molecules upstream of Smad5 can lead to the occurrence of multiple diseases [42].
However, mechanism of negative modulation of p-Smad5 nuclear translocation is not clear. We found that TUG1 is a novel inhibitor of p-Smad5 by suppressing nuclear translocation of p-Smad5. Interestingly, knocking down TUG1 could not significantly change the phosphorylation of Smad5 in BM-MSCs but rescue nuclear localization of p-Smad5 after irradiation (Figure 4). Hence, TUG1 might act as an inhibitor by suppressing nuclear translocation of p-Smad5.
RIP data showed that TUG1 can bind to Smad5 directly. On-line prediction demonstrated that TUG1 could bind to the N-terminal region including NLS sequence of Smad5, probably regulating the nuclear translocation of p-Smad5. Subsequently, a series of flag-tagged Smad5 deletions were constructed. We identified that the 50-90 aa of Smad5 is required for its interaction with TUG1. This region is right near the NLS (40-45 aa) of Smad5. These indicated that TUG1 may obscure the NLS of Smad5 to block the nuclear translocation of p-Smad5 and suppresses the osteogenic differentiation of BM-MSCs.
Conclusion
We propose that TUG1 act as a negative regulator of osteogenic differentiation by modulateing the nuclear translocation of p-Smad5 after irradiation. Our findings might provide a new target for controlling osteogenesis in BM-MSCs, which are crucial to the structure and function of the hematopoietic microenvironment after irradiation.
Abbreviations
ALP: Alkaline phosphatase; BM-MSC: bone marrow mesenchymal stem cell; Co-Smad: common-partner Smad; DAPI: 4',6-diamidino2-phenylindole; FISH: fluorescence in situ hybridization; HSC: hematopoietic stem cell; HSPC: hematopoietic stem and progenitor cell; I-Smad: Inhibitory Smad; lncRNA: long noncoding RNA; NLS: nuclear localization sequence; OCN: osteocalcin; OGN: osteoglycin; p-Smad5: phosphorylated Smad5; RBPs: RNA binding proteins; RIP: RNA immunoprecipitation; R-Smad: receptor-regulated Smad; Runx2: runt related transcription factor 2; siRNA: small interfering RNA; TUG1: Taurine Upregulated Gene 1.
Supplementary Material
Supplementary figures and table.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant 81472915), the National Natural Youth Science Foundation of China (Grant 81402634), the National Science and Technology Major Project of the Ministry of Science and Technology of China (Grant 2018ZX09J18103-004).
Author Contributions
Q.R. and Z.L. designed experiments. W.Z, L.C and J.W. performed experiments. J.L. and X.Z performed bioinformatics analyses. Y.X. and C.W. provided expert advice. W.Z, L.C and J.W. wrote the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Fliedner TM, Chao NJ, Bader JL. et al. Stem cells, multiorgan failure in radiation emergency medical preparedness: a U.S./European Consultation Workshop. Stem Cells. 2009;27:1205-1211
2. Cao X, Wu X, Frassica D. et al. Irradiation induces bone injury by damaging bone marrow microenvironment for stem cells. Proc Natl Acad Sci U S A. 2011;108:1609-1614
3. Bianco P. "Mesenchymal" stem cells. Annu Rev Cell Dev Biol. 2014;30:677-704
4. Wu J, Zhang W, Ran Q. et al. The Differentiation Balance of Bone Marrow Mesenchymal Stem Cells Is Crucial to Hematopoiesis. Stem Cells Int. 2018;2018:1540148
5. Asada N, Katayama Y, Sato M. et al. Matrix-embedded osteocytes regulate mobilization of hematopoietic stem/progenitor cells. Cell Stem Cell. 2013;12:737-747
6. Song Q, Zhong L, Chen C. et al. miR-21 synergizes with BMP9 in osteogenic differentiation by activating the BMP9/Smad signaling pathway in murine multilineage cells. Int J Mol Med. 2015;36:1497-1506
7. Taipaleenmaki H, Abdallah BM, AlDahmash A. et al. Wnt signalling mediates the cross-talk between bone marrow derived pre-adipocytic and pre-osteoblastic cell populations. Exp Cell Res. 2011;317:745-756
8. Yu Y, Wang L, Yu J. et al. Dentin matrix proteins (DMPs) enhance differentiation of BMMSCs via ERK and P38 MAPK pathways. Cell Tissue Res. 2014;356:171-182
9. Miyazono K, Kamiya Y, Morikawa M. Bone morphogenetic protein receptors and signal transduction. J Biochem. 2010;147:35-51
10. Yan J, Guo D, Yang S. et al. Inhibition of miR-222-3p activity promoted osteogenic differentiation of hBMSCs by regulating Smad5-RUNX2 signal axis. Biochem Biophys Res Commun. 2016;470:498-503
11. Ponjavic J, Ponting CP, Lunter G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 2007;17:556-565
12. Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629-641
13. Takeda K, Ichijo H, Fujii M. et al. Identification of a novel bone morphogenetic protein-responsive gene that may function as a noncoding RNA. J Biol Chem. 1998;273:17079-17085
14. Zhu L, Xu PC. Downregulated LncRNA-ANCR promotes osteoblast differentiation by targeting EZH2 and regulating Runx2 expression. Biochem Biophys Res Commun. 2013;432:612-617
15. Zhang J, Tao Z, Wang Y. Long noncoding RNA DANCR regulates the proliferation and osteogenic differentiation of human bone-derived marrow mesenchymal stem cells via the p38 MAPK pathway. Int J Mol Med. 2018;41:213-219
16. Young TL, Matsuda T, Cepko CL. The noncoding RNA taurine upregulated gene 1 is required for differentiation of the murine retina. Curr Biol. 2005;15:501-512
17. Islam MS, Stemig ME, Takahashi Y. et al. Radiation response of mesenchymal stem cells derived from bone marrow and human pluripotent stem cells. J Radiat Res. 2015;56:269-277
18. Xu X, Li R, Zhou Y. et al. Dysregulated systemic lymphocytes affect the balance of osteogenic/adipogenic differentiation of bone mesenchymal stem cells after local irradiation. Stem Cell Res Ther. 2017;8:71
19. Xiao Z, Liu X, Lodish HF. Importin beta mediates nuclear translocation of Smad 3. J Biol Chem. 2000;275:23425-23428
20. Xiao Z, Watson N, Rodriguez C. et al. Nucleocytoplasmic shuttling of Smad1 conferred by its nuclear localization and nuclear export signals. J Biol Chem. 2001;276:39404-39410
21. Lee KS, Kim HJ, Li QL. et al. Runx2 is a common target of transforming growth factor beta1 and bone morphogenetic protein 2, and cooperation between Runx2 and Smad5 induces osteoblast-specific gene expression in the pluripotent mesenchymal precursor cell line C2C12. Mol Cell Biol. 2000;20:8783-8792
22. Javed A, Bae JS, Afzal F. et al. Structural coupling of Smad and Runx2 for execution of the BMP2 osteogenic signal. J Biol Chem. 2008;283:8412-8422
23. Yu VW, Lymperi S, Oki T. et al. Distinctive Mesenchymal-Parenchymal Cell Pairings Govern B Cell Differentiation in the Bone Marrow. Stem Cell Rep. 2016;7:220-235
24. Calvi LM, Adams GB, Weibrecht KW. et al. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature. 2003;425:841-846
25. Visnjic D, Kalajzic Z, Rowe DW. et al. Hematopoiesis is severely altered in mice with an induced osteoblast deficiency. Blood. 2004;103:3258-3264
26. Yu VW, Saez B, Cook C. et al. Specific bone cells produce DLL4 to generate thymus-seeding progenitors from bone marrow. J Exp Med. 2015;212:759-774
27. Sato M, Asada N, Kawano Y. et al. Osteocytes regulate primary lymphoid organs and fat metabolism. Cell Metab. 2013;18:749-758
28. Zhang X, Xiang L, Ran Q. et al. Crif1 Promotes Adipogenic Differentiation of Bone Marrow Mesenchymal Stem Cells After Irradiation by Modulating the PKA/CREB Signaling Pathway. Stem Cells. 2015;33:1915-1926
29. Nicolay NH, Lopez PR, Saffrich R. et al. Radio-resistant mesenchymal stem cells: mechanisms of resistance and potential implications for the clinic. Oncotarget. 2015;6:19366-19380
30. Wang L, Wang Y, Li Z. et al. Differential expression of long noncoding ribonucleic acids during osteogenic differentiation of human bone marrow mesenchymal stem cells. Int Orthop. 2015;39:1013-1019
31. Huang Y, Zheng Y, Jia L. et al. Long Noncoding RNA H19 Promotes Osteoblast Differentiation Via TGF-beta1/Smad3/HDAC Signaling Pathway by Deriving miR-675. Stem Cells. 2015;33:3481-3492
32. Liang WC, Fu WM, Wang YB. et al. H19 activates Wnt signaling and promotes osteoblast differentiation by functioning as a competing endogenous RNA. Sci Rep. 2016;6:20121
33. Katsushima K, Natsume A, Ohka F. et al. Targeting the Notch-regulated non-coding RNA TUG1 for glioma treatment. Nat Commun. 2016;7:13616
34. Yu C, Li L, Xie F. et al. LncRNA TUG1 sponges miR-204-5p to promote osteoblast differentiation through upregulating Runx2 in aortic valve calcification. Cardiovasc Res. 2018;114:168-179
35. He Q, Yang S, Gu X. et al. Long noncoding RNA TUG1 facilitates osteogenic differentiation of periodontal ligament stem cells via interacting with Lin28A. Cell Death Dis. 2018;9:455
36. Lo RS, Chen YG, Shi Y. et al. The L3 loop: a structural motif determining specific interactions between SMAD proteins and TGF-beta receptors. Embo J. 1998;17:996-1005
37. Imamura T, Takase M, Nishihara A. et al. Smad6 inhibits signalling by the TGF-beta superfamily. Nature. 1997;389:622-626
38. Hayashi H, Abdollah S, Qiu Y. et al. The MAD-related protein Smad7 associates with the TGF beta receptor and functions as an antagonist of TGF beta signaling. Cell. 1997;89:1165-1173
39. Murakami G, Watabe T, Takaoka K. et al. Cooperative inhibition of bone morphogenetic protein signaling by Smurf1 and inhibitory Smads. Mol Biol Cell. 2003;14:2809-2817
40. Zhao Y, Xiao M, Sun B. et al. C-terminal domain (CTD) small phosphatase-like 2 modulates the canonical bone morphogenetic protein (BMP) signaling and mesenchymal differentiation via Smad dephosphorylation. J Biol Chem. 2014;289:26441-26450
41. Goto K, Kamiya Y, Imamura T. et al. Selective inhibitory effects of Smad6 on bone morphogenetic protein type I receptors. J Biol Chem. 2007;282:20603-20611
42. MacFarlane EG, Haupt J, Dietz HC. et al. TGF-beta Family Signaling in Connective Tissue and Skeletal Diseases. Cold Spring Harb Perspect Biol. 2017:9
Author contact
![]() Corresponding authors: louise-r-qcom, johnneyusccom
Corresponding authors: louise-r-qcom, johnneyusccom
 Global reach, higher impact
Global reach, higher impact