13.3
Impact Factor
Theranostics 2019; 9(8):2143-2157. doi:10.7150/thno.29552 This issue Cite
Research Paper
Engineering stem cell cardiac patch with microvascular features representative of native myocardium
1. Department of Biomedical Engineering, Michigan Technological University, Houghton, MI 49931, USA;
2. Stem Cell and Regenerative Medicine Center, University of Wisconsin, Madison, WI 53705, USA.
Received 2018-8-28; Accepted 2019-2-14; Published 2019-4-9
Abstract
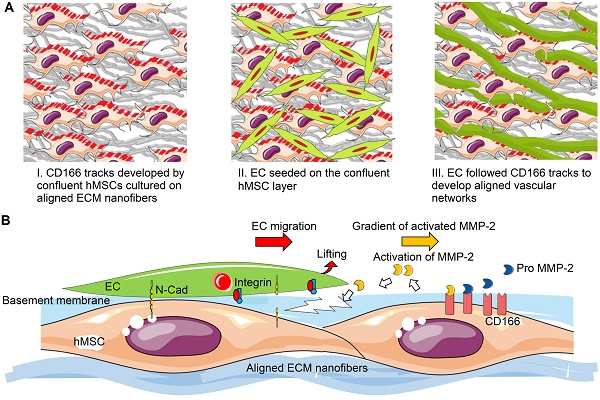
The natural myocardium is a highly aligned tissue with an oriented vasculature. Its characteristic cellular as well as nanoscale extracellular matrix (ECM) organization along with an oriented vascular network ensures appropriate blood supply and functional performance. Although significant efforts have been made to develop anisotropic cardiac structure, currently neither an ideal biomaterial nor an effective vascularization strategy to engineer oriented and high-density capillary-like microvessels has been achieved for clinical cardiovascular therapies. A naturally derived oriented ECM nanofibrous scaffold mimics the physiological structure and components of tissue ECM and guides neovascular network formation. The objective of this study was to create an oriented and dense microvessel network with physiological myocardial microvascular features.
Methods: Highly aligned decellularized human dermal fibroblast sheets were used as ECM scaffold to regulate physiological alignment of microvascular networks by co-culturing human mesenchymal stem cells (hMSCs) and endothelial cells (ECs). The influence of topographical features on hMSC and EC interaction was investigated to understand underlying mechanisms of neovasculature formation.
Results: Results demonstrate that the ECM topography can be translated to ECs via CD166 tracks and significantly improved hMSC-EC crosstalk and vascular network formation. The aligned ECM nanofibers enhanced structure, length, and density of microvascular networks compared to randomly organized nanofibrous ECM. Moreover, hMSC-EC co-culture promoted secretion of pro-angiogenic growth factors and matrix remodeling via metalloprotease-2 (MMP-2) activation, which resulted in highly dense vascular network formation with intercapillary distance (20 μm) similar to the native myocardium.
Conclusion: HMSC-EC co-culture on the highly aligned ECM generates physiologically oriented and dense microvascular network, which holds great potential for cardiac tissue engineering.
Keywords: Extracellular matrix nanofibers, prevascularization, vascular density, vascular maturation, vascular orientation
Introduction
The natural myocardium is a highly aligned tissue with an oriented vasculature [1, 2]. The specific cellular and nanoscale extracellular matrix (ECM) organization of the myocardium make it mechanically strong, while the orientation of microvessels ensure its effective blood supply and functional performance. In the past decade, significant efforts have been focused on developing synthetic or naturally aligned nanofibrous scaffolds to form an anisotropic cardiac structure, stimulate angiogenesis, or incorporate engineered microvessels to maintain tissue viability after implantation. However, neither an ideal biomaterial nor an effective vascularization strategy to engineer oriented capillary-like microvessels has yet been achieved for clinical cardiovascular therapies.
Most capillaries in the heart muscle run along and tend to wrap around the cardiomyocytes in a parallel arrangement with respect to the myocardial fibers. Such an orientation determines the direction of blood flow along a particular fiber (or a group of fibers) [1, 2]. Therefore, it is important to create aligned microvessels for efficient oxygen and nutrient transport, and also incorporate natural inlet and outlet sides, facilitating the anastomosis of the cardiac patch to the host vasculature post-implantation [3]. Advanced microfabrication [4-7] and 3D printing [8, 9] show great promise in fabricating tissue constructs with defined microvessel structure. Unfortunately, these microvessels are markedly larger in diameter (100 μm-1mm) than native capillaries (< 10 μm) [10], and undoubtedly will significantly lower the microvessel density and constrain the blood supply and functional performance of the engineered cardiac patch after implantation. Several groups reported that alignment of engineered microvessels has been achieved via mechanical strain applied to the cellular constructs [3, 11-13]. Although the cardiac patch engraftment was significantly improved due to the rapid microvessel inosculation on injured myocardium, the dynamic cultures dramatically complicated the fabrication process. Moreover, the microvessel density was low and the patch failed in replicating the natural myocardium structure. Additionally, the materials utilized in these applications, as in microfabrication and 3D printing, are normally limited to hydrogels. Unfortunately, hydrogels normally cannot attain the complex structure, composition, and mechanical strength needed to match the aligned nanofibrous ECM found in natural myocardium.
A variety of synthetic and natural materials have been developed as cardiovascular tissue substitutes. However, they are limited in real applications because their degradation products can cause adverse tissue reactions. Decellularized natural ECMs from native tissues are composed of multiple proteins interlaced with proteoglycans. The ECM not only serves as a supporting scaffold with receptor sites to orient cells, but also acts as a communication portal that provides a means for cells to interact via signaling factors to regulate a variety of molecular functions. Nevertheless, natural ECM suffers from problems of autologous tissue/organ scarcity, host responses, and pathogen transfer when allogenic and xenogenic tissues and organs are used [14]. Human cell-derived ECM scaffolds can be screened for pathogens to eliminate the provocation of undesirable inflammatory and immunological reactions from natural tissues and organs [14, 15]. It thus offers a promising alternative to ECM derived from natural tissues. Human dermal fibroblasts can be easily isolated and used to a high passage without a decrease in growth rate [16-18]. They also secrete abundant ECM biomacromolecules such as fibronectin, proteoglycans, collagen and elastin. Therefore, ECM scaffolding derived from human dermal fibroblast cell sheets is an attractive candidate for engineering a completely biological cardiac patch.
When using a suitable substrate, such as Matrigel® or fibrin gel, ECs can self-assemble microvessels at the same order of magnitude density as native capillaries. Nevertheless, their structure experiences a post-implant regression [19]. Native microvessels are surrounded and stabilized by sparsely distributed pericytes. Similarly, supporting cells are required to stabilize engineered microvessels [20]. Mesenchymal stem cells (MSCs) can function as pericytes and secrete proangiogenic factors during tissue regeneration [21]. Moreover, MSCs are easily obtainable, extensively proliferative in vitro, immunomodulatory [22-30], and can stimulate tissue regeneration [23, 28, 31, 32]. Although MSCs do not differentiate into cardiomyocytes, the beneficial effect of MSCs in promoting myocardial regeneration is well documented. It has been shown, for example, that intramyocardial injection of MSCs in small animal models of myocardial infarction (MI) can reduce fibrosis and infarct size, form new cardiac tissue, increase capillary density, and improve cardiac contractility and left ventricular ejection fraction [33-35]. Moreover, injection of MSC sheet fragments, a cluster form of MSCs mixed with self-assembled ECM, can further improve cell retention and deliver cell therapy [36]. These unique properties enable hMSCs to combine with ECM scaffolds and ECs to biofabricate a prevascularized patch, in which hMSCs play a dual role of stabilizing vasculature formed by ECs in vitro and regenerating the infarcted cardiac tissue after implantation.
It is important to create aligned microvessels in an engineered hMSC patch to biomimic the natural aligned myocardial capillaries and promote its functional performance after implantation. Although the role of aligned fibers in guiding cell alignment is well established, their effect on microvessel alignment is not well understood. Our group has previously derived a highly aligned ECM nanofibrous scaffold by decellularizing an aligned human dermal fibroblast sheet. The nanofibers are around 80 nm in their diameter, similar to the size of collagen nanofibers in vivo. Moreover, the aligned nanofibers elicited a reduced immune response compared their randomly organized nanofibrous counterpart, and could be repopulated with hMSCs and then stacked or rolled into a thick 3D structure for engineering diverse tissues [37, 38]. In this study, we used this aligned ECM nanofibrous scaffold to guide the interaction of hMSCs and ECs to form highly oriented microvessels under our previously optimized oxygen (O2) conditions: 7 day hMSC culture at 2% O2 followed by 7 day hMSC-EC co-culture at 20% O2. Such a combination of O2 culture conditions facilitated hMSCs to maintain their angiogenic property, which further improved the microvessels' development and maturation [39-41]. Moreover, we elucidated the role of nanotopographical ECM organization on microvessel alignment, density and maturation, which are important considerations in the development of prevascularized 3D tissues. The prevascularized ECM scaffolds could serve as building blocks to fabricate a 3D prevascularized hMSC patch through multi-layering approaches.
Materials and Methods
Fabrication of aligned ECM scaffolds
The ECM scaffolds derived from human dermal fibroblast sheets (ATCC, Manasass, VA) were prepared as previously described [42]. Briefly, aligned polydimethylsiloxane (PDMS) substrates were casted from nano-grated masters with 350 nm grating width and 130 nm grating depth, and coated with type I collage (Sigma-Aldrich, St Louis, MO). Human dermal fibroblasts were seeded on top of the material surface and cultured in Dulbecco's modified eagle medium (DMEM) with 20% fetal bovine serum (FBS) for 3 weeks with a medium change every 72 hours (h). The obtained cell sheets were gently peeled off from the PDMS layer and decellularized with 0.5% sodium dodecyl sulfate, 10 mM Tris (Bio-rad, Hercules, CA), and 25 Mm Ethylenediaminetetraacetic acid (EDTA, Sigma-Aldrich) solution for 15 minutes (min). After the decellularization process, the ECM sheets were thoroughly washed with PBS, followed by incubation in DMEM with 20% FBS, and 1% penicillin/streptomycin for 48 h at room temperature to further decompose the remaining DNA in the ECM sheets. The structure of ECM nanofibers is shown in Supplemental Figure 1.
ECM prevascularization
The hMSCs were obtained from Texas A&M University Health Sciences Center. Passage 3 hMSCs at a density of 10,000 cells/cm2 were seeded on the decellularized ECM sheets and cultured in α-minimum essential medium (α-MEM) with 20% FBS, 1% Pen/strep, and 1% L-glutamine (Thermo Fisher Scientific, Waltham, MA). Following the optimal culture conditions we have achieved in our previous studies [39, 40], hMSCs were maintained under hypoxia (2% O2) condition for 7 days. The hMSCs grown on aligned ECM nanofibrous scaffold is shown in Supplemental Figure 1. Subsequently, the passage 3 human umbilical vein endothelial cells (HUVECs) (Lonza, Walkersville, MD), which have been typically used as model ECs, were seeded at a density of 20,000 cells/cm2 on top of the hMSCs/ECM constructs. The co-cultures were maintained in normoxia (20% O2) up to 7 days in endothelial cell growth media (EGM, Lonza), which was changed every 48 h.
Vasculature staining and imaging
The co-cultured prevascularized constructs were washed with PBS, fixed and stained with mouse polyclonal antibody against human CD31 (Abcam, Cambridge, MA), and followed by DyLight 488 horse anti-mouse IgG secondary antibody (Vector laboratories, Burlingame, CA). The samples were washed and incubated in 4′, 6-diamidino-2-phenylindole (DAPI, Sigma-Aldrich) solution to label the cell nuclei. Images were captured under an Olympus FV-1000 confocal microscope. The vascular networks were quantified by the ImageJ software, using methods previously described [43]. We firstly chose one vessel that aligned along the main direction of the nanogratings and set the angle of it as 0 degree, the angles of other vessels were measured and defined as Δ Angle. Experiments were triplicated, and 5 non-overlapping panels were randomly analyzed from each individual sample for unbiased statistical analysis. The same immunofluorescent staining was performed with anti-CD146 and anti-CD166 primary antibody staining. CD146 is known as the melanoma cell adhesion molecule (MCAM), which is regarded as a pericyte marker [44]. CD166 stains activated leukocyte cell adhesion molecule, and it was used to detect hMSC-EC interaction [45]. Images were captured under an Olympus FV-1000 confocal microscope by depth scan every 1 µm. The obtained images were reconstructed into 3-D views by a Fiji software.
Enzyme-linked immunosorbent assay (ELISA) of soluble angiogenic growth factors
An ELISA assay was employed to quantify angiogenic growth factors. The cell culture medium was collected from different groups at different time points. Growth factors in the hMSC sheet were extracted as previously described, and 1 mL extraction buffer was used for each sample [10]. The concentration of vascular endothelial growth factor (VEGF), basic fibroblast growth factor (bFGF), transforming growth factor beta 1 (TGF-β1), and angiopoietin 1 (Ang-1) were determined using ELISA kits (R&D Systems, Minneapolis, MN) following the manufacture's protocol. Absorbance was measured with a plate reader (Molecular Devices, USA) at 450 nm with correction wavelength set at 540 nm. Experiments were triplicated and each sample and standard was measured in duplicates.
Real-time reverse transcription polymerase chain reaction (RT-qPCR)
Ribonucleic acid (RNA) was extracted using an RNeasy® extraction kit (Qiagen, Valencia, CA). Extracted RNA was reverse transcribed into cDNA using a reverse transcription kit (Life Technologies, Rockville, MD) and amplified by the StepOnePlus™ Real-Time PCR System (Applied Biosystems, Waltham, MA) using SYBR® Green Real Time PCR Master Mixes (Life Technology). Customized KiCqStart® SYBR® Green Primers (Sigma-Aldrich) were used, with the sequences listed in Table 1. The GAPDH gene was used as an endogenous control. Data was analyzed using the ∆∆Ct method. Amplification measurements were triplicated and three individual samples were tested for statistical analysis.
List of primer sequences
| Common name | Symbol/ Gene ID | Forward sequence | Reverse sequence |
|---|---|---|---|
| GAPDH | GAPDH/2597 | 5′ - ACAGTTGCCATGTAGACC | 5′ - TTTTTGGTTGAGCACAGG |
| VEGFA | VEGFA/7422 | 5′ - GACCAAAGAAAGATAGAGCAAG | 5′ - ATACGCTCCAGGACTTATAC |
| bFGF | FGF2/2247 | 5′ - CAAGCAGAAGAGAGAGGAG | 5′ - CACTCATCCGTAACACATTTAG |
| Paxillin | PXN/5829 | 5' - ATCCTGGATAAAGTGGTGAC | 5' - AAAGAAGGCTCCACACTG |
| N-Cadherin | CDH2/1000 | 5' - ACATATGTGATGACCGTAAC | 5' - TTTTTCTCGATCAAGTCCAG |
| Integrin α2 | ITGA2/3673 | 5' - GGTGGGGTTAATTCAGTATG | 5' - ATATTGGGATGTCTGGGATG |
| Integrin β1 | ITGB1/3688 | 5' - ATTCCCTTTCCTCAGAAGTC | 5' - TTTTCTTCCATTTTCCCCTG |
| MMP-2 | MMP2/4313 | 5' - TTCTGGAGATACAATGAGGTG | 5' - CTTGAAGAAGTAGCTGTGAC |
Gelatin zymography for MMP-2 assessment
Gelatin zymography was performed to assess matrix metalloprotease (MMP) enzymatic activity. Briefly, the medium was collected from different time points during co-culture. The collected medium from different groups was diluted to the same total protein concentration as measured by the bicinchoninic acid assay (BCA, Thermo Fisher Scientific). Polyacrylamide gels containing 4 mg / mL gelatin were used to separate the protein samples through electrophoresis. Non-reducing (beta mercaptoethanol free) sample buffer was used to dilute protein samples, and 10 µL of each sample was loaded into each well. Gels containing separated proteins were incubated for 24 h at 37 oC in incubation buffer (1% Triton X-100, 50 mM Tris, 5 mM CaCl2, and 1µM ZnCl2) to allow gelatinase reaction. Gels were then stained with coomassie blue (Sigma-Aldrich) staining solution for 30 min, followed by destaining until clear bands could be observed. The density of the bands was analyzed by imageJ. The experiment was triplicated for statistical analysis.
MMP inhibition
The MMP inhibition was performed to determine MMP inhibitor's influence on the structural properties of microvessels. A MMP inhibitor, APR 100 (Tocris Bioscience, Bristol, UK), was dissolved in dimethyl sulfoxide (DMSO) (Corning, NY) to prepare a 1mM stock solution. APR 100 is a selective inhibitor of MMP-2 (IC50 = 12 nM), which also displays selectivity over MMP-9, MMP-3, MMP-1 and MMP-7 (IC50 = 200, 4500, > 50000 and > 50000 nM respectively). Three different working concentrations (1 µM, 5 µM, 10 µM) of APR 100 were prepared in endothelial cell growth medium (EGM), which was used during the 7-day co-culture. No APR 100 was added in the control group. After 7 days of co-culture, prevascularized constructs were washed with PBS, fixed and stained with mouse polyclonal antibody against human CD31 (Abcam), followed by DyLight 488 horse anti-mouse IgG secondary antibody (Vector laboratories). Samples were mounted, and images were captured under an Olympus FV-1000 confocal microscope. Experiments were triplicated, and at least 5 non-overlapping areas from each sample were randomly observed.
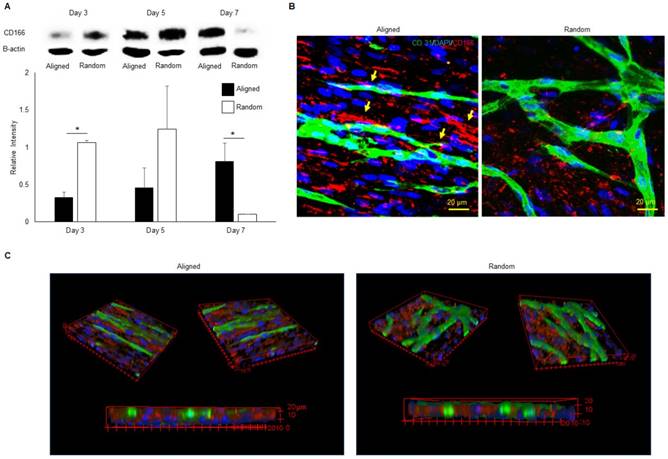
Western blot of activated leukocyte cell adhesion molecule (CD166)
Western blotting was performed on both aligned and random co-cultured constructs to assess the protein expression of CD166. Construct lysates were prepared using radioimmunoprecipitation assay (RIPA) buffer with 1% protease inhibitors (Thermo Fisher). Denatured cell lysates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and transferred onto a polyvinylidene fluoride (PVDF) membrane. Blocking was performed with Tris-buffered saline (TBS) supplemented with 0.1% Tween-20 (TBST) (Thermo Fisher Scientific, Waltham, MA) and 5% dry milk (Bio-rad). The blocked membrane was incubated with the primary anti-CD166 antibody (Abcam) overnight at 4 °C. Horseradish peroxidase-conjugated secondary antibody (1:3000, Bio-rad) and enhanced luminol-based chemiluminescent (ECL) substrate (Bio-rad) was used to develop the bands. The developed membrane was imaged using a FluorChem Imaging System (Alpha InnoTech, San Leandro, CA).
Statistical analysis
All statistical results were obtained from triplicated experiments. The results were reported as mean ± standard deviation. Statistical comparisons between experimental conditions were performed using one-way ANOVA and Tukey's post hoc test. Results were considered statistically significant for p < 0.05.
Results
Morphology of microvascular networks
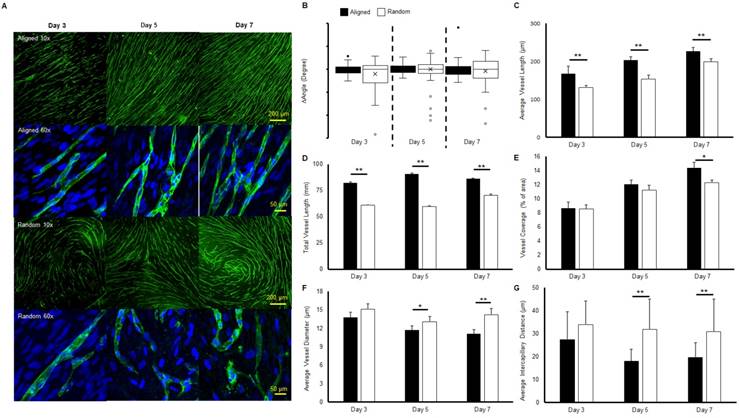
Immunofluorescent staining of microvessels from aligned and random groups at different time points are presented in Figure 1a. The microvessel alignment was observed in the aligned groups throughout the entire co-culture period, while curves and swirls of vascular networks were observed in the random groups. Higher magnification imaging revealed the single vessel morphology. Similar structures were observed from both aligned and random groups at day 3. At days 5 and 7, distinct boundaries and tubular morphologies were observed in the aligned groups, while the vascular structure was relatively impaired in the random groups. The characterization of microvessels was organized in Figure 1(b-e). The alignment of the vascular networks was displayed as a boxplot (Figure 1b). The interquartile range (IQR), which showed the range of variation, was computed from the boxplot. The IQR values were significantly lower in the aligned groups (2.7 fold at day 3, 1.5 fold at day 5, and 2.2 fold at day 7, p < 0.01), as aligned groups exhibited less variation in their organization compared with the wider and full range variation found in random groups. The average vessel length in aligned groups was significantly higher than in the random groups (1.3 fold at day 3 and day 5, 1.2-fold at day 7, p < 0.01), and reached 0.23 mm at day 7 (Figure 1c). A similar trend was observed in total microvessel length, which was significantly higher in the aligned groups than their random counterparts (1.3 fold at day 3, 1.5 fold at day 5, and 1.1 fold at day 7, p < 0.01, Figure 1d). The vessel coverage percentage was significantly higher in the aligned groups at day 7 (p < 0.05), with a total of 14.4% area covered by the vascular networks (Figure 1e). However, the average vessel diameter of the random groups was 1.3 fold wider at day 7 (p < 0.01), with wider vessels also observed at days 3 and 5 (p < 0.05) (Figure 1f). The intercapillary distance was significantly lower at day 5 (± 13.8 μm) and day 7 (± 11.2 μm) (1.7 fold, p < 0.01) for the aligned groups, in accordance with the higher vessel density and coverage percentage.
Morphologies and characteristics of co-cultured microvascular network. (A) Immunofluorescent images of co-cultured microvascular networks at different time points. (B) Boxplot analysis of vessel alignment for aligned and random co-cultured groups. (C) Average vessel length. (D) Total vessel length. (E) Vessel coverage percentage. (F) Average vessel diameter. (G) Average intercapillary distance. The reduced interquartile measurement deviation of aligned groups indicates less vessel alignment variation compared to the random groups. The microvascular characterization demonstrated a denser and enhanced vascular structure in the aligned group. (*p < 0.05, **p < 0.01)
Assessment of pro-angiogenic growth factor in co-culture medium
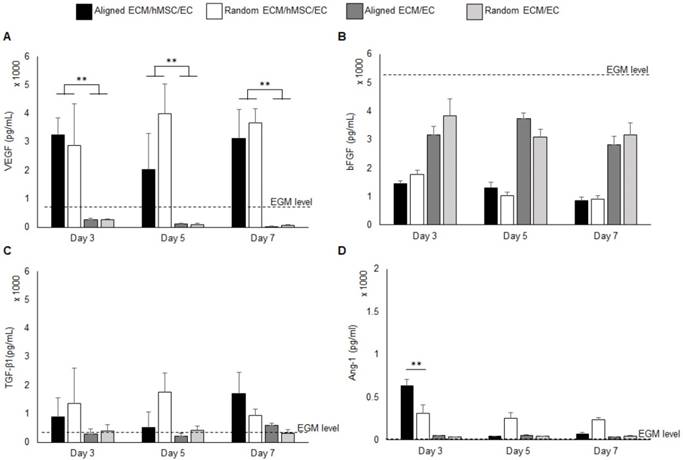
The pro-angiogenic growth factors in the medium from the co-culture system for both aligned and random groups were quantified by ELISA assay. Monocultures of ECs on both aligned and random ECM were used as controls to evaluate the pro-angiogenic growth factor release mediated by hMSCs. The background EGM concentration was also measured for reference. The VEGF concentration was significantly elevated (10 fold, p < 0.01) in the co-culture groups throughout the entire period, as it was below the EGM concentration in the monoculture groups (Figure 2a). Interestingly, at day 5, the VEGF concentration in the aligned culture medium decreased compared to the random co-culture group. The VEGF consumption at day 5 explains the increased total vascular length in the aligned co-culture group, as indicated in Figure 1d. Both co-cultures and monocultures consumed bFGF. Compared with monocultures, the co-cultures consumed additional bFGF, which was 2 fold reduced in the system. Aligned and random co-cultures consumed similar amounts of bFGF over the cell culture period (Figure 2b). The TGF-β1 concentration was slightly higher in co-cultured groups than monoculture groups and EGM level (Figure 2c), and no significant differences were identified between the aligned and random groups. The Ang-1 concentration was robustly increased to 634 pg / mL at day 3 in the aligned co-culture groups (2 fold higher than random groups, p < 0.01), and significantly dropped at days 5 and 7 to less than 100 pg/mL. In contrast, the Ang-1 level held steady in random groups throughout the entire culture period (250-300 pg/mL). It was also observed that the monocultures had slightly higher Ang-1 concentration (less than 50 pg/mL) compared to EGM (Figure 2d).
Pro-angiogenic growth factors in co-cultured constructs
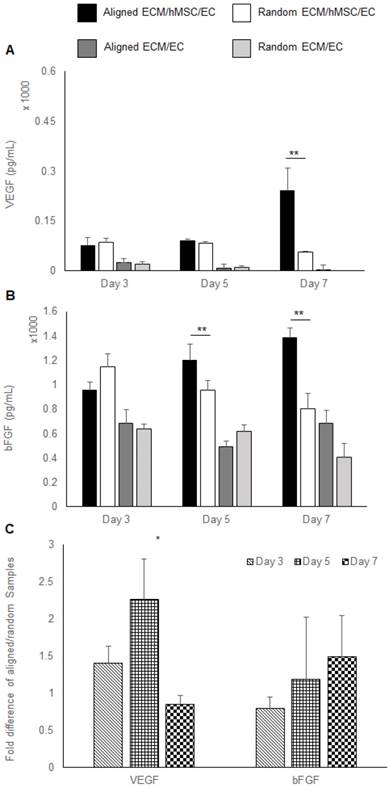
Growth factors extracted from co-cultured constructs were also measured by ELISA. A higher concentration of VEGF was localized to the co-cultured groups (3 folds, p < 0.01). At day 7, only the aligned groups showed significant increase of VEGF concentration (5 fold higher than random groups, p < 0.01). All the other samples had a low localized VEGF concentration, which was lower than 100 pg/mL. No VEGF was detected in the day 7 random monoculture group (Figure 3a). A higher concentration of bFGF was found in co-cultured constructs relative to monocultured groups. Importantly, a higher bFGF concentration was observed in aligned co-culture groups than random co-cultured groups at day 5 (1.3 fold, p < 0.01) and day 7 (1.8 fold, p < 0.01) (Figure 3b). Results of the qPCR analysis of VEGF and bFGF mRNA expression of aligned co-cultured groups were normalized to the random co-cultured groups. The mRNA expression levels of VEGF were 1.5 fold higher at day 3 and 2.3 fold (p <0.05) higher at day 5, indicating the accelerated angiogenesis process before day 7. Following that, the VEGF mRNA was elevated in random co-cultured groups at day 7, denoting its delayed angiogenesis until day 7. The bFGF mRNA expression was lower for the aligned co-cultured groups at day 3, but was 1.2 fold higher at day 5 and 1.5 fold higher at day 7. However, no statistical differences were detected for bFGF mRNA expression level (Figure 3c).
Pro-angiogenic growth factors VEGF (A), bFGF (B), TGF-β1 (C), and Ang-1 (D) concentration in the growth medium. The background concentration of each factor was measured and marked as “EGM level” on each graph. The co-cultures with hMSCs secreted high level of VEGF than EC monocultures. (*p < 0.05, **p < 0.01)
Pro-angiogenic growth factors expression. (A) VEGF and (B) bFGF localized in the constructs assessed by ELISA. (C) The mRNA expression of VEGF and bFGF was measured by qRT-PCR. The aligned ECM/hMSC/EC has significantly more localized VEGF (day 7) and bFGF (day 5 and day 7) than the random ECM/MSC/EC group. (*p < 0.05, **p < 0.01)
Maturation assessment through pericyte identification
Immunofluorescent staining of CD146/CD31 was performed to assess the maturation of the vascular structures at day 7 (Figure 4). Representative images demonstrated clear lumen formation in microvessels of aligned groups. Leading cells of the sprout known as 'tip cells' were observed in both groups, as shown in left images. Tip cells guide other ECs by sensing its surrounding environment for guidance cues. Aligned hMSC nuclei were observed at the base of the tip cell in the aligned group. In contrast, the tip cell in the random group followed the underlying hMSCs without a pre-defined direction. These images provided important evidence regarding the role of hMSCs in guiding ECs by transferring topographical features of the ECM. Moreover, the CD31 positive microvessels covered with CD146 positive cells could be clearly identified from the cross-sectional view. In contrast, a flatter lumen was observed in the random group with less CD146 staining around the microvessel lumen.
Adhesion molecule mRNA expression and mapping
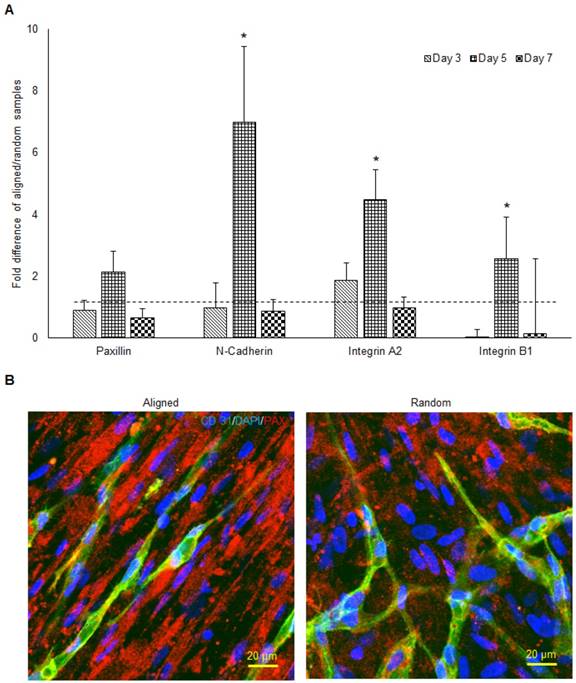
The qPCR analysis of paxillin, N-cadherin, integrin α2, and integrin β1 mRNA expression of aligned co-cultured groups were performed and normalized to the random co-cultured groups (Figure 5a). While mRNA expression was similar in paxillin at day 3 and day 7, day 5 paxillin expression was up-regulated 2 fold (no significance) in the aligned groups. N-cadherin mRNA expression followed the same trend, as the day 5 N-cadherin expression was enhanced 7 fold (p < 0.05) in the aligned groups. Integrin α2 mRNA expression was up-regulated in the aligned groups at day 3 (2 fold, p < 0.05) and day 5 (4.5 fold, p < 0.05), and returned to the same level as random at day 7. Immunofluorescent staining of paxillin is presented in Figure 5b. Although red fluorescent paxillin positive staining was visualized for both aligned and random co-cultured groups, the paxillin expression pattern was highly aligned, complementing the aligned morphology of the vascular networks. In contrast, the vascular networks on the random co-cultured samples showed a random paxillin pattern, which followed the ECM nanofibers' topography.
Matrix remodeling activity and MMP-2 activation
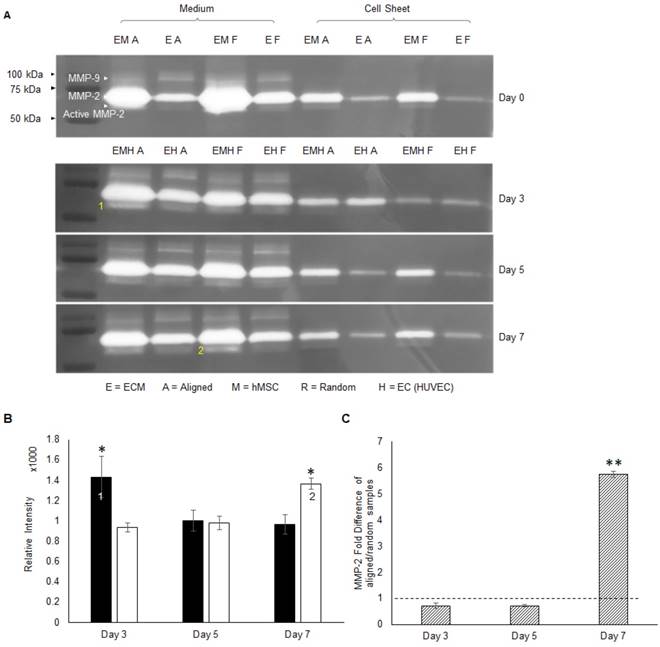
The hMSC-secreted ECM consists of collagen I, collagen IV, laminin and fibronectin etc. [41]. The migration of ECs on top of the hMSC layer requires degradation of ECM components. The MMP family is responsible for ECM remodeling and vessel development, in which gelatinase MMP-2 and MMP-9 can degrade gelatins, collagens, laminin and aggrecan [46]. Gelatin zymography analysis was performed to screen the identity of MMP during angiogenesis. From the gel images in Figure 6a, MMP-2 at 75 kDa was extensively expressed compared to MMP-9 at 92 kDa. Whereas MMP-9 was absent from the co-cultured constructs. Thus, MMP-2 was identified as the major player, and activated MMP-2 at 68 kDa bands were detected as well. Activated MMP-2 expression was quantitatively analyzed by the band intensity. These results indicated that aligned co-cultured groups had higher activated MMP-2 concentration at day 3 (1.5 fold, p < 0.05), which gradually decreased by day 7, whereas random co-culture groups showed gradual and significantly higher MMP-2 concentration (1.4 fold, p < 0.05) at day 7 (Figure 6b). Moreover, co-culture groups showed higher MMP-2 concentration compared to monoculture groups (only ECs) in both culture medium and cellular constructs (Figure 6a), which indicated a possible role of hMSCs in MMP-2 activation via CD166. The qPCR analysis of MMP-2 mRNA expression of aligned co-cultured groups were performed and normalized to the random co-cultured groups. Total MMP-2 mRNA expression was similar between aligned and random co-cultured groups at days 3 and 5, but drastically increased (5.7 fold, p < 0.01) at day 7 for the aligned co-cultured groups (Figure 6c), which indicated highly active matrix remodeling.
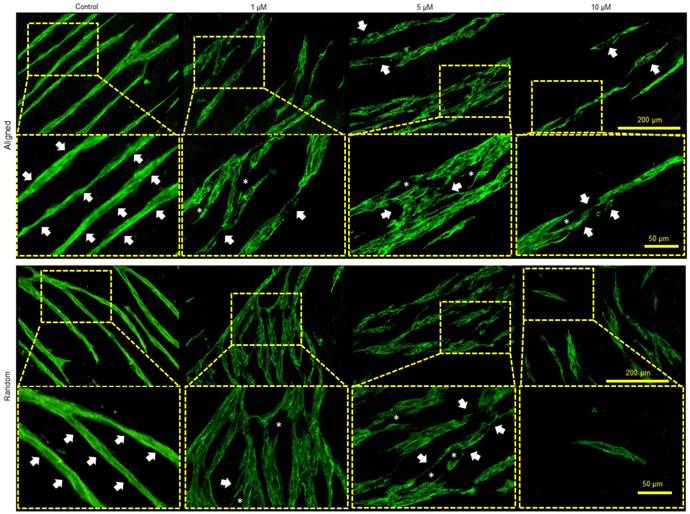
To further verify the function of MMP in the co-culture system, a MMP inhibitor APR100 was introduced in the culture medium with three different concentrations (1 µM, 5 µM and 10 µM). It was observed that in the presence of APR100, especially at its high concentrations (5 µM and 10 µM), large “structural gaps” were formed in the branches of the microvessels, as indicated by white asterisks in Figure 7. Whereas, in control groups, microvessels showed overall intact structures without such large gaps. Moreover, when exposed to MMP inhibition, microvessels showed discontinued and/or compromised vessel wall structure, which looked more like flat cellular patches rather than tubular structure, as indicated by white arrows. By increasing APR100 concentration from 1 µM to 10 µM, it was observed that the majority of the microvessels were shortened in their length along with reduced vascular density.
CD166 protein expression and organization
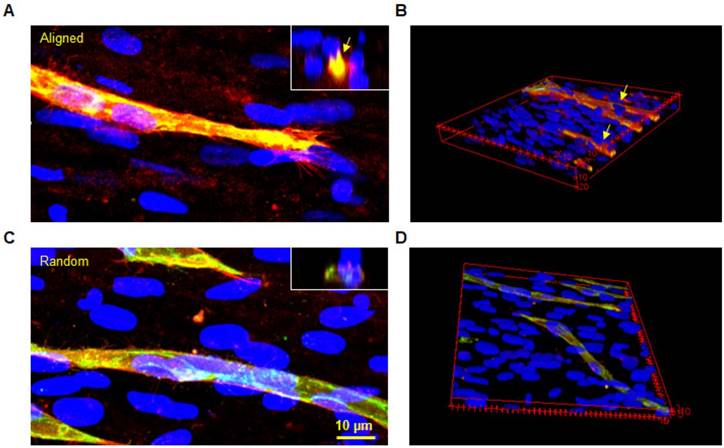
The western blot analysis of CD166 was performed on aligned and random co-cultured groups at different time points. Up-regulation of CD166 was observed at day 3 (3 fold, p < 0.05) and day 5 (3 fold, no significance) in random co-cultured groups (Figure 8a). However, up-regulation of CD166 was found in aligned co-cultured groups at day 7 (9 fold, p < 0.05). Confocal Z-stack images displayed CD166 tracks on aligned co-cultured groups, which guided the orientation of vascular networks (Figure 8b, arrows). No clear tracks of CD166 were observed on the random groups. 3D confocal imaging suggested that CD166 tracks localized to the bottom of the vascular networks, guiding the progression of the vessels in aligned groups. In comparison, the random co-cultured groups showed scattered CD166 expression, providing no guidance to the vascular progression (Figure 8c).
Co-immunofluorescent staining of CD146 (red) and CD31 (green) for both aligned and random groups. The microvessel was well insulated by a CD146 positive cell layer in the aligned group (arrows). Similarly, the cross-sectional view revealed a complete lumen by CD31 positive cells, which were covered with CD146 positive cells. The 3D image further proved this finding in the 3D structure.
The mRNA expression of cell adhesion molecules responsible for cell-ECM interaction (A) and the morphology of paxillin (B) in both aligned and random samples. The aligned samples presented significantly higher mRNA expression at day 5. The paxillin were organized along the direction of hMSCs and microvessels. (*p < 0.05)
Discussion
Native myocardium is anisotropic, consisting not only of aligned cardiomyocytes, but also similarly aligned capillaries. It has been shown that most cardiomyocytes are aligned at an angle of less than 45 degrees [47]. The capillaries between muscle cells are organized in the direction of muscle fibers, in order to fulfill interstitial oxygen and nutrient diffusion requirements for cell metabolism [48, 49]. The orientation of cells and capillaries gives rise to the robust density of microvasculature. In the native heart, the average intercapillary distance is 20 µm [50]. Moreover, the capillaries are wrapped by mural cells to ensure effective support during mass transport and blood flow. Therefore, an engineered myocardium should not only mimic the cell alignment, but also the capillary orientation, high density and maturity, to more closely mimic the architecture and support the functional performance of engineered tissues post-implantation. Although attempts have been made by some groups to create aligned vessels using micropatterning [51, 52], interstitial flow [53], and tensile loading [54], none of the resulting vascular networks are comparable with the physiological high alignment and density of native tissues. Aligned ECM nanofibers not only preserve the biomimetic nanofibrous topography, but also provides a biological niche and bioactivities of native ECM of target tissues [55, 56]. Such a biomimetic scaffold offers great opportunity for engineering an anisotropic cardiac patch with highly organized cells and capillary geometries, while generating insight into the mechanism of capillary formation, progression, and maturation.
The activity of MMPs was assessed by gelatin zymography (A), and MMP-2 was identified as the major membrane cleaving enzyme in this co-cultured angiogenesis system. Note that 1) the early activation of MMP-2 in aligned ECM/hMSC/EC groups and 2) the delayed MMP-2 activation in the random counterparts (B). (C) The total MMP-2 mRNA expression was up-regulated in the aligned ECM/hMSC/EC groups at day 7. (*p < 0.05, **p < 0.01)
Alignment promoted vascular network density and maturation
It has been reported that an aligned topography provides surface guidance for EC elongation and motility, which can promote vascular formation in implantable materials [57, 58]. Several studies have also shown that MSC differentiation could be regulated by provision of topographical cues [59, 60], and that growth factor release profiles varied under different topography[61]. Those topographic cues included nano-scale fibers, ridges, and pores of the basement membrane, which were in direct contact with the attached cells [62]. Our results on the density of newly formed vasculature prove that nanofibrous alignment enhances vascular formation (Figure 1). Compared with other groups' prevascularization strategies through EC/MC co-culture, our construct generated superior microvascular alignment, length and density [3, 51, 52, 61, 63]. Importantly, the average intercapillary distance of our aligned vascular networks after 5 days co-culture was below 20 μm, which matched the physiological intercapillary distance (~20 μm) that is observed in native myocardium [50]. Whereas in other cardiac tissue engineering strategies that have generated aligned and physiologically sized microvessels, their intercapillary space ranges from 50 μm to 100 μm [3, 52]. MSCs can be recruited by neovasculature as pericytes to wrap the microvessels and promote their lumen formation and stabilization [64]. This theory explains the enhanced maturation of our prevascularized aligned groups in their 3D morphologies (Figure 6c), which was further evidenced by a layer of pericytes wrapping around the aligned vessels as shown by the positive CD146 staining (Figure 4).
Influence of MMP-2/9 inhibitor APR100 on microvascular network formation. Aligned and randomly organized microvascular networks formed under 1 µM, 5 µM and 10 µM of APR100 after 7 days of EC culture. No APR100 in the control groups. White arrows and asterisks (*) indicate the vessel walls and the structural gaps within microvascular networks, respectively. Both aligned and randomly organized control groups showed intact vessel wall, whereas fewer microvessels and large structural gaps appeared within branches of microvascular networks in MMP-inhibited samples, especially those with higher APR100 concentration.
Western blot analysis (A), 2D (B) and 3D (C) morphologies of CD166 expression at different time points for both aligned and random co-culture groups. The CD166 tracks were identified in aligned ECM/hMSC/EC groups, as identified by yellow arrows (B) The CD166 was concentrated at the interface between hMSCs and ECs in the aligned groups and guided the microvessels progression, while scattered CD166 expression was observed in random groups. (*p < 0.05, **p < 0.01)
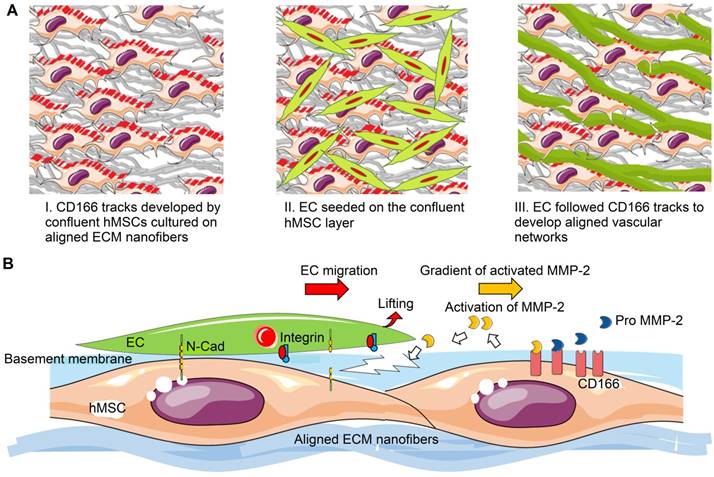
Schematic representation of aligned vascular network development by hMSC-EC co-culture on decellularized nanofibrous aligned ECM construct. (A) hMSCs transferred topographical features of aligned nanofibrous ECM to ECs in order to develop aligned and dense vascular network. (B) CD166-expressing hMSCs modulated MMP-2 activation, which further regulated directional ECM remodeling. Intercellular crosstalk between hMSCs and ECs resulted in vascular network formation.
Topography regulated angiogenic growth factors in co-culture environment
Angiogenic growth factors in cell cultures play a significant role in vascular network formation [65, 66]. hMSCs provide essential paracrine secretions including VEGF, bFGF, IGF-1, TGF-β, Ang-1 and HGF to promote EC-mediated angiogenesis [67, 68], in which TGF-β is a factor that promotes pericyte proliferation [69, 70]. In the present study, the activity of hMSCs could be responsible for enhanced expression of VEGF in both culture medium and co-cultured constructs compared to the EC/ECM monocultures (Figure 2a and 3a). The increased genotypic and phenotypic expression of VEGF in aligned co-cultures (Figure 3a, c) compared to random groups was consistent with the significantly improved microvessel density, as shown by their increased vessel length and vessel coverage (Figure 1). The bFGF concentration in cell culture medium of co-cultured constructs was reduced compared to monocultured constructs (Figure 2b). This increased consumption of bFGF could have contributed to the development of high-density capillary networks. However, the opposite phenomenon was observed in aligned cellular constructs, where bFGF expression was significantly increased compared to all other groups (Figure 3b). This significant increase in VEGF and bFGF expression in aligned cellular constructs compared to random ones was attributed to the topographical features. Studies have shown that TGF-β promotes EC quiescence and MSC differentiation towards pericytes during the vascular maturation process [21, 71-74]. Increased levels of TGF-β were observed in both aligned and random co-cultured constructs compared to monocultures, but no significant difference was shown (p > 0.01). The pericyte-like cells surrounding the microvessels could be attributed to the differentiation of hMSCs, causing by the synergized effect of bFGF and TGF-β [69], which resulted in higher expression of pericyte protein CD146 at day 7 (Figure 4). Besides TGF-β, angiopoietins are a major family of angiogenic growth factors. MSCs secrete Ang-1, especially during wound healing, which recruits macrophages and endothelial lineage cells [75-77]. Ang-1 phosphorylates the Tie-2 receptor and this Ang-1/Tie-2 interaction regulates maturation of newly formed vasculature, which ultimately results in complex functional vascular network formation [78, 79]. Our previous research indicated that hMSCs cultured on the ECM in hypoxic environments promoted expression of Ang-1[39]. Similarly, in this study a hypoxic environment during hMSCs culture on aligned ECM could be responsible for the initial increased expression of Ang-1 at day 3, which decreased gradually during normoxic co-culture (Figure 2d). Significantly increased levels of Ang-1 at day 3 in aligned co-cultures (Figure 2d) compared to all other groups might have led to development of an aligned and more matured vascular network. In summary, enhanced expression of these angiogenic factors in aligned co-cultured constructs compared to random constructs shows the effects of topographical features on microvascular network alignment and maturation.
Topography guided anisotropic microvascular progression
Various studies have demonstrated that matrix proteolytic activities can result in development of topographical cues in order to facilitate EC migration, vessel stabilization, and vessel maturation [80, 81]. Adhesion molecules are involved in matrix proteolytic events, which regulate construct remodeling during vascular formation. One of the focal adhesion molecules, paxillin, which was present at the ventral surface of cells, formed aligned tracks along the direction of aligned nanofibers and hMSCs (Figure 5). In addition, the mRNA of focal adhesion proteins was highly expressed in aligned groups. These organized and more intense focal adhesion sites could modulate and direct the migration of tip cells (Figure 4), which is the first important step for angiogenic sprouting [82]. Moreover, significantly higher MMP-2 was activated in aligned samples at early stages of angiogenic development (Figure 6a). CD-166 has been identified as the modulator of MMP-2 activation during ECM remodeling [83]. Investigation of matrix remodeling during angiogenesis indicated that the activation of MMP-2 was dependent on the density of CD166 positive cells, including MSCs [84, 85]. Clear CD166 tracks were observed in the aligned co-culture group along with aligned vascular network (Figure 8b). In contrast, the vascular network was more aberrant on random nanofibrous ECM due to scattered CD166 expression (Figure 8b & 8c), which resulted in randomly oriented and less dense vascular network formation. It is thus possible that through CD166, the aligned topographical feature upregulated hMSCs' capabilities for MMP-2 activation, which might have played a role in the increased vascular network density. This was verified by MMP inhibition in the co-culture system, where the microvessels showed a deteriorated vessel wall, along with shortened vessel length and reduced vascular density (Figure 7).
Based on our results, potential mechanism of nano-topography guided angiogenesis can be concluded in the following steps: 1) hMSCs followed contact guidance that was provided by aligned ECM nanofibers, and formed aligned confluent hMSC layer in the same direction 2) CD166-expressing hMSCs formed vascular guiding tracks. Importantly, CD166 mediated MMP-2 activation, and MMP-2 in turn mediated ECM remodeling. Activated MMP-2 possibly helped EC migration along the aligned ECM nanofibers. 3) Intercellular interaction between ECs and MSCs promoted the pro-angiogenic factor release and deposition, which resulted in aligned, stable and mature vascular network formation, as illustrated in Figure 9.
Conclusions
In this study, we demonstrated successful engineering of highly oriented, dense and matured vascular networks on an aligned nanofibrous ECM construct by hMSC/EC co-culture. The ECM topography greatly influenced the vascular network development by enhancing vascular alignment, angiogenic growth factor secretion, and ECM remodeling. Importantly, our study indicated that hMSCs successfully translated the alignment of nanofibrous ECM scaffold to ECs and guided the microvascular structure. These prevascularized aligned ECM constructs with highly dense and mature microvascular networks could be stacked on each other to construct completely biological 3D tissue to accelerate cardiac regeneration.
Acknowledgements
This study was supported by the National Institutes of Health (1R15CA202656 and 1R15HL115521-01A1) and the National Science Foundation (1703570) to FZ. It was also supported by NIH 1U01HL134764-01 to TJK.
Supplementary Material
Supplementary figures.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Kaneko N, Matsuda R, Toda M, Shimamoto K. Three-dimensional reconstruction of the human capillary network and the intramyocardial micronecrosis. Am J Physiol Heart Circ Physiol. 2011;300:H754-H61
2. Emerson GG, Segal SS. Alignment of microvascular units along skeletal muscle fibers of hamster retractor; 1997.
3. Morin KT, Dries-Devlin JL, Tranquillo RT. Engineered microvessels with strong alignment and high lumen density via cell-induced fibrin gel compaction and interstitial flow. Tissue Eng Part A. 2013;20:553-65
4. Zheng Y, Chen JM, Craven M, Choi NW, Totorica S, Diaz-Santana A. et al. In vitro microvessels for the study of angiogenesis and thrombosis. Proc Natl Acad Sci U S A. 2012;109:9342-7
5. Li X, Xu SL, He PN, Liu YX. In Vitro Recapitulation of Functional Microvessels for the Study of Endothelial Shear Response, Nitric Oxide and Ca2+ (i). PLoS One. 2015:10
6. Ye XF, Lu L, Kolewe ME, Park H, Larson BL, Kim ES. et al. A biodegradable microvessel scaffold as a framework to enable vascular support of engineered tissues. Biomaterials. 2013;34:10007-15
7. Chiu LLY, Montgomery M, Liang Y, Liu HJ, Radisic M. Perfusable branching microvessel bed for vascularization of engineered tissues. Proc Natl Acad Sci U S A. 2012;109:E3414-E23
8. Lee VK, Kim DY, Ngo HG, Lee Y, Seo L, Yoo SS. et al. Creating perfused functional vascular channels using 3D bio-printing technology. Biomaterials. 2014;35:8092-102
9. Kolesky DB, Truby RL, Gladman AS, Busbee TA, Homan KA, Lewis JA. 3D Bioprinting of Vascularized, Heterogeneous Cell-Laden Tissue Constructs. Adv Mater. 2014;26:3124-30
10. Tsuji T, Sasaki Y, Tanaka M, Hanabata N, Hada R, Munakata A. Microvessel morphology and vascular endothelial growth factor expression in human colonic carcinoma with or without metastasis. Laboratory Investigation. 2002;82:555-62
11. Schaefer JA, Guzman PA, Riemenschneider SB, Kamp TJ, Tranquillo RT. A cardiac patch from aligned microvessel and cardiomyocyte patches. J Tissue Eng Regen Med. 2018;12:546-56
12. Krishnan L, Underwood CJ, Maas S, Ellis BJ, Kode TC, Hoying JB. et al. Effect of mechanical boundary conditions on orientation of angiogenic microvessels. Cardiovasc Res. 2008;78:324-32
13. Riemenschneider SB, Mattia DJ, Wendel JS, Schaefer JA, Ye L, Guzman PA. et al. Inosculation and perfusion of pre-vascularized tissue patches containing aligned human microvessels after myocardial infarction. Biomaterials. 2016;97:51-61
14. Lu HX, Hoshiba T, Kawazoe N, Koda I, Song MH, Chen GP. Cultured cell-derived extracellular matrix scaffolds for tissue engineering. Biomaterials. 2011;32:9658-66
15. Lu HX, Hoshiba T, Kawazoe N, Chen GP. Autologous extracellular matrix scaffolds for tissue engineering. Biomaterials. 2011;32:2489-99
16. Ahlfors JEW, Billiar KL. Biomechanical and biochemical characteristics of a human fibroblast-produced and remodeled matrix. Biomaterials. 2007;28:2183-91
17. Grinnell F, Fukamizu H, Pawelek P, Nakagawa S. Collagen Processing, Crosslinking, and Fibril Bundle Assembly in Matrix Produced by Fibroblasts in Long-Term Cultures Supplemented with Ascorbic-Acid. Exp Cell Res. 1989;181:483-91
18. Ishikawa O, Kondo A, Okada K, Miyachi Y, Furumura M. Morphological and biochemical analyses on fibroblasts and self-produced collagens in a novel three-dimensional culture. Brit J Dermatol. 1997;136:6-11
19. Baffert F, Le T, Sennino B, Thurston G, Kuo C, Hu Lowe D. et al. Cellular changes in normal blood capillaries undergoing regression after inhibition of VEGF signaling. Am J Physiol Heart Circ Physiol. 2006;290:H547-H59
20. Costa Almeida R, Granja PL, Soares R, Guerreiro SG. Cellular strategies to promote vascularisation in tissue engineering applications. Eur Cell Mater. 2014;28:51-67
21. Mills SJ, Cowin AJ, Kaur P. Pericytes, mesenchymal stem cells and the wound healing process. Cells. 2013;2:621-34
22. Hashi CK, Zhu YQ, Yang GY, Young WL, Hsiao BS, Wang K. et al. Antithrombogenic property of bone marrow mesenchymal stem cells in nanofibrous vascular grafts. Proc Natl Acad Sci U S A. 2007;104:11915-20
23. Caplan AI, Wagner J, Kean T, Young R, Dennis JE. Optimizing mesenchymal stem cell-based therapeutics. Curr Opin Biotech. 2009;20:531-6
24. Nemeth K, Leelahavanichkul A, Yuen PST, Mayer B, Parmelee A, Doi K. et al. Bone marrow stromal cells attenuate sepsis via prostaglandin E(2)-dependent reprogramming of host macrophages to increase their interleukin-10 production. Nat Med. 2009;15:42-9
25. Groh ME, Maitra B, Szekely E, Koc ON. Human mesenchymal stem cells require monocyte-mediated activation to suppress alloreactive T cells. Exp Hematol. 2005;33:928-34
26. Ren GW, Xu GW, Zhang LY, Zhang YY, Zhao X, Roberts A. et al. The interaction between mesenchymal stem cells and the immune system. Cell Res. 2008:18
27. Ren GW, Zhang LY, Zhao X, Xu GW, Zhang YY, Roberts AI. et al. Mesenchymal stem cell-mediated immunosuppression occurs via concerted action of chemokines and nitric oxide. Cell stem cell. 2008;2:141-50
28. Caplan AI, Correa D. The MSC: An Injury Drugstore. Cell stem cell. 2011;9:11-5
29. Salem HK, Thiemermann C. Mesenchymal Stromal Cells: Current Understanding and Clinical Status. Stem Cells. 2010;28:585-96
30. Tuan RS, Aronin CEP. Therapeutic Potential of the Immunomodulatory Activities of Adult Mesenchymal Stem Cells. Birth Defects Res C. 2010;90:67-74
31. Caplan AI, Dennis JE. Mesenchymal stem cells as trophic mediators. J Cell Biochem. 2006;98:1076-84
32. Haynesworth SE, Baber MA, Caplan AI. Cytokine expression by human marrow-derived mesenchymal progenitor cells in vitro: Effects of dexamethasone and IL-1 alpha. J Cell Physiol. 1996;166:585-92
33. Orlic D, Kajstura J, Chimenti S, Jakoniuk I, Anderson SM, Li BS. et al. Bone marrow cells regenerate infarcted myocardium. Nature. 2001;410:701-5
34. Kudo M, Wang YG, Wani MA, Xu MF, Ayub A, Ashraf M. Implantation of bone marrow stem cells reduces the infarction and fibrosis in ischemic mouse heart. J Mol Cell Cardiol. 2003;35:1113-9
35. Zhang SH, Guo JX, Zhang P, Liu YG, Jia ZQ, Ma KT. et al. Long-term effects of bone marrow mononuclear cell transplantation on left ventricular function and remodeling in rats. Life Sciences. 2004;74:2853-64
36. Wang CC, Chen CH, Lin WW, Hwang SM, Hsieh PCH, Lai PH. et al. Direct intramyocardial injection of mesenchymal stem cell sheet fragments improves cardiac functions after infarction. Cardiovasc Res. 2008;77:515-24
37. Xing Q, Qian Z, Kannan B, Tahtinen M, Zhao F. Osteogenic Differentiation Evaluation of an Engineered Extracellular Matrix Based Tissue Sheet for Potential Periosteum Replacement. ACS Appl Mater Interfaces. 2015;7:23239-47
38. Xing Q, Qian Z, Tahtinen M, Yap AH, Yates K, Zhao F. Aligned Nanofibrous Cell-Derived Extracellular Matrix for Anisotropic Vascular Graft Construction. Adv Healthc Mater. 2017:1601333 -n/a
39. Zhang L, Qian Z, Tahtinen M, Qi S, Zhao F. Prevascularization of natural nanofibrous extracellular matrix for engineering completely biological three-dimensional prevascularized tissues for diverse applications. J Tissue Eng Regen Med. 2018;12:e1325-e36
40. Zhang L, Xing Q, Qian Z, Tahtinen M, Zhang Z, Shearier E. et al. Hypoxia created human mesenchymal stem cell sheet for prevascularized 3D tissue construction. Adv Healthc Mater. 2016;5:342-52
41. Zhao F, Veldhuis JJ, Duan Y, Yang Y, Christoforou N, Ma T. et al. Low oxygen tension and synthetic nanogratings improve the uniformity and stemness of human mesenchymal stem cell layer. Mol Ther. 2010;18:1010-8
42. Xing Q, Yates K, Tahtinen M, Shearier E, Qian Z, Zhao F. Decellularization of fibroblast cell sheets for natural extracellular matrix scaffold preparation. Tissue Eng Part C Methods. 2014;21:77-87
43. Chen X, Aledia AS, Ghajar CM, Griffith CK, Putnam AJ, Hughes CC. et al. Prevascularization of a fibrin-based tissue construct accelerates the formation of functional anastomosis with host vasculature. Tissue Eng Part A. 2008;15:1363-71
44. Li Q, Yu Y, Bischoff J, Mulliken JB, Olsen BR. Differential expression of CD146 in tissues and endothelial cells derived from infantile haemangioma and normal human skin. J Pathol. 2003;201:296-302
45. Bühring HJ, Treml S, Cerabona F, De Zwart P, Kanz L, Sobiesiak M. Phenotypic characterization of distinct human bone marrow-derived MSC subsets. Ann N Y Acad Sci. 2009;1176:124-34
46. Galis ZS, Khatri JJ. Matrix metalloproteinases in vascular remodeling and atherogenesis: the good, the bad, and the ugly. Circ Res. 2002;90:251-62
47. Lunkenheimer PP, Redmann K, Kling N, Jiang X, Rothaus K, Cryer CW. et al. Three-dimensional architecture of the left ventricular myocardium. Anat Rec A Discov Mol Cell Evol Biol. 2006;288A:565-78
48. Emerson GG, Segal SS. Alignment of microvascular units along skeletal muscle fibers of hamster retractor. J Appl Physiol. 1997;82:42-8
49. Oosthoek PW, Moorman AFM, Sauer U, Gittenberger-de Groot AC. Capillary Distribution in the Ventricles of Hearts With Pulmonary Atresia and Intact Ventricular Septum. Circulation. 1995;91:1790-8
50. Schmidt RF, Biederman-Thorson MA, Thews G. Human Physiology: Springer Berlin Heidelberg; 2013.
51. Baranski JD, Chaturvedi RR, Stevens KR, Eyckmans J, Carvalho B, Solorzano RD. et al. Geometric control of vascular networks to enhance engineered tissue integration and function. Proc Natl Acad Sci U S A. 2013;110:7586-91
52. Thomson KS, Korte FS, Giachelli CM, Ratner BD, Regnier M, Scatena M. Prevascularized microtemplated fibrin scaffolds for cardiac tissue engineering applications. Tissue Eng Part A. 2013;19:967-77
53. Steward R, Tambe D, Hardin CC, Krishnan R, Fredberg JJ. Fluid shear, intercellular stress, and endothelial cell alignment. Am J Physiol Cell Physiol. 2015;308:C657-C64
54. Greiner AM, Chen H, Spatz JP, Kemkemer R. Cyclic Tensile Strain Controls Cell Shape and Directs Actin Stress Fiber Formation and Focal Adhesion Alignment in Spreading Cells. PLoS One. 2013;8:e77328
55. Hutmacher DW. Scaffold design and fabrication technologies for engineering tissues-state of the art and future perspectives. J Biomater Sci Polym Ed. 2001;12:107-24
56. Chan B, Leong K. Scaffolding in tissue engineering: general approaches and tissue-specific considerations. Eur Spine J. 2008;17:467-79
57. Huang NF, Okogbaa J, Lee JC, Jha A, Zaitseva TS, Paukshto MV. et al. The modulation of endothelial cell morphology, function, and survival using anisotropic nanofibrillar collagen scaffolds. Biomaterials. 2013;34:4038-47
58. Qutub AA, Popel AS. Elongation, proliferation & migration differentiate endothelial cell phenotypes and determine capillary sprouting. BMC Syst Biol. 2009;3:13
59. Dalby MJ, Gadegaard N, Tare R, Andar A, Riehle MO, Herzyk P. et al. The control of human mesenchymal cell differentiation using nanoscale symmetry and disorder. Nat Mater. 2007;6:997
60. Wu Y, Yang Z, Law JBK, He AY, Abbas AA, Denslin V. et al. The combined effect of substrate stiffness and surface topography on chondrogenic differentiation of mesenchymal stem cells. Tissue Eng Part A. 2017;23:43-54
61. Hofer HR, Tuan RS. Secreted trophic factors of mesenchymal stem cells support neurovascular and musculoskeletal therapies. Stem Cell Res Ther. 2016;7:131
62. Abrams G, Goodman S, Nealey P, Franco M, Murphy C. Nanoscale topography of the basement membrane underlying the corneal epithelium of the rhesus macaque. Cell Tissue Res. 2000;299:39-46
63. Costa-Almeida R, Gomez-Lazaro M, Ramalho C, Granja PL, Soares R, Guerreiro SG. Fibroblast-Endothelial Partners for Vascularization Strategies in Tissue Engineering. Tissue Eng Part A. 2015;21:1055-65
64. Peppiatt CM, Howarth C, Mobbs P, Attwell D. Bidirectional control of CNS capillary diameter by pericytes. Nature. 2006;443:700
65. Armulik A, Abramsson A, Betsholtz C. Endothelial/pericyte interactions. Circ Res. 2005;97:512-23
66. Zisch AH, Lutolf MP, Ehrbar M, Raeber GP, Rizzi SC, Davies N. et al. Cell-demanded release of VEGF from synthetic, biointeractive cell-ingrowth matrices for vascularized tissue growth. FASEB J. 2003
67. Mayer H, Bertram H, Lindenmaier W, Korff T, Weber H, Weich H. Vascular endothelial growth factor (VEGF-A) expression in human mesenchymal stem cells: autocrine and paracrine role on osteoblastic and endothelial differentiation. J Cell Biochem. 2005;95:827-39
68. Sopko NA, Turturice BA, Becker ME, Brown CR, Dong F, Popovic ZB. et al. Bone marrow support of the heart in pressure overload is lost with aging. PLoS One. 2010;5:e15187
69. Papetti M, Shujath J, Riley KN, Herman IM. FGF-2 antagonizes the TGF-β1-mediated induction of pericyte α-smooth muscle actin expression: a role for myf-5 and Smad-mediated signaling pathways. Invest Ophthalmol Vis Sci. 2003;44:4994-5005
70. Nico B, Mangieri D, Corsi P, De Giorgis M, Vacca A, Roncali L. et al. Vascular endothelial growth factor-A, vascular endothelial growth factor receptor-2 and angiopoietin-2 expression in the mouse choroid plexuses. Brain Res. 2004;1013:256-9
71. Kurpinski K, Lam H, Chu J, Wang A, Kim A, Tsay E. et al. Transforming growth factor-β and notch signaling mediate stem cell differentiation into smooth muscle cells. Stem Cells. 2010;28:734-42
72. Antonelli-Orlidge A, Saunders KB, Smith SR, D'Amore PA. An activated form of transforming growth factor beta is produced by cocultures of endothelial cells and pericytes. Proc Natl Acad Sci U S A. 1989;86:4544-8
73. Darland DC, D'Amore PA. TGF beta is required for the formation of capillary-like structures in three-dimensional cocultures of 10T1/2 and endothelial cells. Angiogenesis. 2001;4:11-20
74. Orlidge A, D'Amore PA. Inhibition of capillary endothelial cell growth by pericytes and smooth muscle cells. J Cell Biol. 1987;105:1455-62
75. Rustad KC, Wong VW, Sorkin M, Glotzbach JP, Major MR, Rajadas J. et al. Enhancement of mesenchymal stem cell angiogenic capacity and stemness by a biomimetic hydrogel scaffold. Biomaterials. 2012;33:80-90
76. Wu Y, Chen L, Scott PG, Tredget EE. Mesenchymal stem cells enhance wound healing through differentiation and angiogenesis. Stem Cells. 2007;25:2648-59
77. Chen L, Tredget EE, Wu PY, Wu Y. Paracrine factors of mesenchymal stem cells recruit macrophages and endothelial lineage cells and enhance wound healing. PLoS One. 2008;3:e1886
78. Cho CH, Kim KE, Byun J, Jang HS, Kim DK, Baluk P. et al. Long-term and sustained COMP-Ang1 induces long-lasting vascular enlargement and enhanced blood flow. Circ Res. 2005;97:86-94
79. Babaei S, Teichert-Kuliszewska K, Zhang Q, Jones N, Dumont DJ, Stewart DJ. Angiogenic actions of angiopoietin-1 require endothelium-derived nitric oxide. Am J Pathol. 2003;162:1927-36
80. Stratman AN, Malotte KM, Mahan RD, Davis MJ, Davis GE. Pericyte recruitment during vasculogenic tube assembly stimulates endothelial basement membrane matrix formation. Blood. 2009;114:5091-101
81. Stratman AN, Saunders WB, Sacharidou A, Koh W, Fisher KE, Zawieja DC. et al. Endothelial cell lumen and vascular guidance tunnel formation requires MT1-MMP-dependent proteolysis in 3-dimensional collagen matrices. Blood. 2009;114:237-47
82. Sauteur L, Krudewig A, Herwig L, Ehrenfeuchter N, Lenard A, Affolter M. et al. Cdh5/VE-cadherin promotes endothelial cell interface elongation via cortical actin polymerization during angiogenic sprouting. Cell Rep. 2014;9:504-13
83. Lunter PC, van Kilsdonk JWJ, van Beek H, Cornelissen IMHA, Bergers M, Willems PHGM. et al. Activated Leukocyte Cell Adhesion Molecule (ALCAM/CD166/MEMD), a Novel Actor in Invasive Growth, Controls Matrix Metalloproteinase Activity. Cancer Res. 2005;65:8801-8
84. Kijima N, Hosen N, Kagawa N, Hashimoto N, Nakano A, Fujimoto Y. et al. CD166/Activated leukocyte cell adhesion molecule is expressed on glioblastoma progenitor cells and involved in the regulation of tumor cell invasion. Neuro Oncol. 2012;14:1254-64
85. Maleki M, Ghanbarvand F, Reza Behvarz M, Ejtemaei M, Ghadirkhomi E. Comparison of Mesenchymal Stem Cell Markers in Multiple Human Adult Stem Cells. Int J Stem Cells. 2014;7:118-26
Author contact
![]() Corresponding author: Feng Zhao, Department of Biomedical Engineering, Michigan Technological University, 1400 Townsend Drive, Houghton, MI 49931, U.S. Tel: 906-487-2852 Fax: 906-487-1717 Email: fengzhaoedu
Corresponding author: Feng Zhao, Department of Biomedical Engineering, Michigan Technological University, 1400 Townsend Drive, Houghton, MI 49931, U.S. Tel: 906-487-2852 Fax: 906-487-1717 Email: fengzhaoedu
 Global reach, higher impact
Global reach, higher impact