13.3
Impact Factor
Theranostics 2019; 9(5):1303-1322. doi:10.7150/thno.30309 This issue Cite
Review
The Applications of Radiomics in Precision Diagnosis and Treatment of Oncology: Opportunities and Challenges
1. CAS Key Laboratory of Molecular Imaging, Institute of Automation, Beijing, 100190, China
2. Department of Radiology, Henan Provincial People's Hospital & the People's Hospital of Zhengzhou University, Zhengzhou, Henan, 450003, China
3. Department of Hepatobiliary Surgery, The Affiliated Hospital of Southwest Medical University, Luzhou, Sichuan, 646000, China
4. Engineering Research Center of Molecular and Neuro Imaging of Ministry of Education, School of Life Science and Technology, Xidian University, Xi'an, Shaanxi, 710126, China
5. University of Chinese Academy of Sciences, Beijing, 100080, China
6. Collaborative Innovation Center for Internet Healthcare, Zhengzhou University, Zhengzhou, Henan, 450052, China
7. Beijing Advanced Innovation Center for Big Data-Based Precision Medicine, Beihang University, Beijing, 100191, China
*These authors contributed equally to this work.
Received 2018-9-30; Accepted 2019-1-10; Published 2019-2-12
Abstract
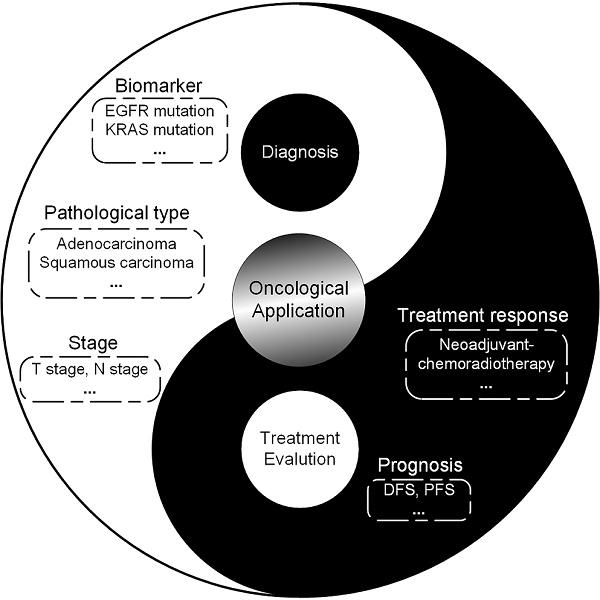
Medical imaging can assess the tumor and its environment in their entirety, which makes it suitable for monitoring the temporal and spatial characteristics of the tumor. Progress in computational methods, especially in artificial intelligence for medical image process and analysis, has converted these images into quantitative and minable data associated with clinical events in oncology management. This concept was first described as radiomics in 2012. Since then, computer scientists, radiologists, and oncologists have gravitated towards this new tool and exploited advanced methodologies to mine the information behind medical images. On the basis of a great quantity of radiographic images and novel computational technologies, researchers developed and validated radiomic models that may improve the accuracy of diagnoses and therapy response assessments. Here, we review the recent methodological developments in radiomics, including data acquisition, tumor segmentation, feature extraction, and modelling, as well as the rapidly developing deep learning technology. Moreover, we outline the main applications of radiomics in diagnosis, treatment planning and evaluations in the field of oncology with the aim of developing quantitative and personalized medicine. Finally, we discuss the challenges in the field of radiomics and the scope and clinical applicability of these methods.
Keywords: radiomics, medical imaging, precision diagnosis and treatment, oncology
Introduction
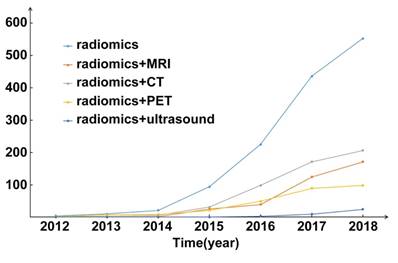
Multimodality medical images contain a great deal of valuable information reflecting the development and progression of cancer. Advancements in data mining and machine learning make it possible to extract many quantitative features and convert the rapidly increasing number of medical images into minable data. This comprehensive method used to analyze medical images is known as radiomics. The concept of radiomics was first proposed in 2012 [1, 2], and has since attracted the attention of researchers throughout the world (Figure 1).
Initially, radiomics refers to extracting large amounts of high-dimensional quantitative features from multimodality medical images such as those of computed tomography (CT), magnetic resonance imaging (MRI), positron emission tomography (PET) and ultrasonography (US), then the mining of correlations between these features and the diagnosis/ prognosis of cancer [3, 4]. These correlations are revealed with imaging analyses, and tumors can then be decoded into different imaging phenotypes [5]. With complimentary information from clinical reports, treatment responses, and genomic/proteomic assays, radiomics may reflect the global outlook of cancer [6]. Compared with previous ways that process medical images as pictures for visual inspection, radiomics introduces a new way to mine the information contained in the medical images. Radiomics has been rapidly developed toward clinical application [7-9] in the hope that it will advance precision diagnostics and cancer treatment.
Radiomics may provide quantitative and objective support for decisions surrounding cancer detection and treatment [10]. This is accomplished with obtaining quantitative information from medical images, combining imaging features with clinical information, genomic information and other information, and mining these data to detect radiomic biomarkers. Radiomics incorporates a series of computational technologies, and the methodologies used in radiomics are usually oriented for clinical problems. Great progress has been made within the field of radiomics regarding technology that meets clinical requirements and benefits diagnostics and cancer treatments.
Here, we will review recent developments in the methodologies in radiomics, mainly involving feature detection and model construction. We will also discuss the scope and the challenges associated with radiomics for precision diagnostics and cancer treatment. Finally, we will offer our opinion on the future steps required for better acceptance of radiomics.
Publication statistics of radiomics since 2012. The number of publications is going straight up. Abbreviations: CT, Computed Tomography; MRI, Magnetic Resonance Imaging; PET, Positron Emission Tomography
Development of Methodology and Technology in Radiomics
Radiomics involves several essential steps (see pipeline of radiomics in Figure 2 and Figure 3), including data acquisition and preprocessing, tumor segmentation, feature extraction, knowledge discovery, and modeling. A series of advancements have been made in each of these areas.
Data acquisition and preprocessing
Radiomics usually begins with data acquisition. It relies on a large volume of medical images and corresponding clinical data to reveal the correlation that exists between them. Radiomics was first proposed using CT images [1] and soon after was applied in the analysis of MR images [2]. There were also studies based on PET [11, 12] and US [13] images utilizing similar strategy. Medical images used for radiomic analysis are collected from different hospitals or data centers; thus, these images are usually obtained using different parameters and protocols and reconstructed with different software. The differences may bring unexpected influences on the radiomic model [14]. The influence of different image acquisition parameters on the reproducibility and quality of the extracted features in different modalities including CT, PET, MRI and US as well as the preprocessing of multi-modality images have been discussed in several recent literatures and a systematic review published by Larue et al. in 2017 [15].
For CT images, differences in CT scanners, voxel size and reconstruction kernels are the main source of influence [16-24]. Shafiq-ul-Hassan et al. [16] suggested that resampling the pixel size could minimize the dependency of radiomic features on voxel size. In addition, applying normalization on the gray level and voxel size in radiomic studies could improve the robustness of features [17]. Zhao et al. [19] found that smooth and sharp reconstruction kernels should not co-occur in a study, as combining data that includes both two kernels could lead to uncontrollable discrepancies. Another recent study by Mackin et al. found that variations in tube current do not significantly affect the radiomic features [23]. A most recent study comprehensively demonstrated that using a controlled protocol can reduce variability in radiomic features, while resampling image thickness cannot reduce the adverse effect derived from thickness variations [24].
There are also several studies focused on the influence of image acquisition on features extraction for PET images. In 2010, Galavis et al. has shown that most of the quantitative textural features presented large variations due to the changes in the grid size, reconstruction algorithm and number of iterations [25]. Recent results from Yan et al. [26] and Veld et al. [27] further confirmed this finding. Recently, Gallivanone et al [28] found that reconstruction settings largely affects the stability of radiomic features, and only 26% were stable when different parameters were considered in this anthropomorphic phantom study. Respiratory motion is another factor that should be concerned. Oliver et al. [29] and Grootjans et al. [30] showed that respiratory motion significantly affects the quantitative textural features reflecting tumor heterogeneity with PET. In addition, they found that standard uptake value (SUV) discretization could also impact the results of textural features detecting. Compare to three-dimensional (3D)-PET, respiratory-gated (4D) PET can reduce motion blurring and generate more robust features [29, 31, 32].
The effects of MRI field strength, imaging protocols, and manufacturers on radiomic features have been investigated by several studies using either living samples or physical phantoms [33-38]. For instance, Jirak et al [34] and Mayerhoefer et al [35] suggested that some features were sensitive to MR acquisition parameters including TR, TE and bandwidth, which was confirmed by a recent simulation study [39]. Savio et al. [36] investigated the effect of slice thickness on texture-based model in differentiating plaque from normal tissue, and showed that the moderate differences caused by thickness does not affect the classification results. The research conducted by Waugh et al. [40] showed that differences of acquisition parameters may not significantly affect the ability of texture analysis in classification task. Moreover, some mature algorithms and tools could help to remove unwanted low-frequency intensity nonuniformity [41, 42] and perform image intensity normalization [43-45] in order to reduce the difference caused by different acquisition parameters of brain MRI.
US-derived quantitative features have been proved to be strongly correlated with breast biologic characteristics [46], gestational age [47], neonatal respiratory morbidity [48]. They can also be used to detecting malignant tumors from benign tumors in thyroid [49, 50] and breast [51, 52]. In a review in terms of ultrasound image processing methods for carotid plaque morphology analysis [53], Kyriacou et al. demonstrated that a standard acquisition protocol, image normalizing method helps to generate reproducible measurements. So far, there are still few studies concerning on the reproducibility and stability of quantitative features obtained from US in oncology.
Considering the influence of different image acquisition parameters, researchers should pay more attention on the differences of imaging protocols and provide essential parameters to achieve reproducebility and comparability with other radiomic studies [10]. As the importance of using standard imaging protocols should be taken into account [6], researchers should develop specific methods aiming at one disease compared with others.
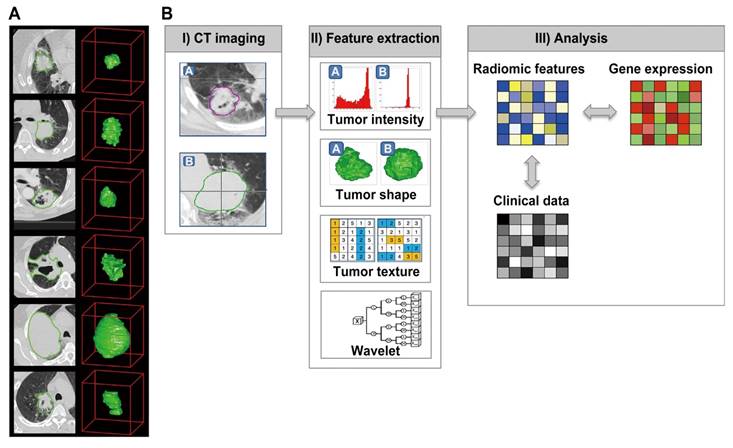
The initially radiomics pipeline with medical images. Reproduced with permission from [4]. (a) Example CT images of patients with lung cancer. (b) Strategy of radiomic analysis.
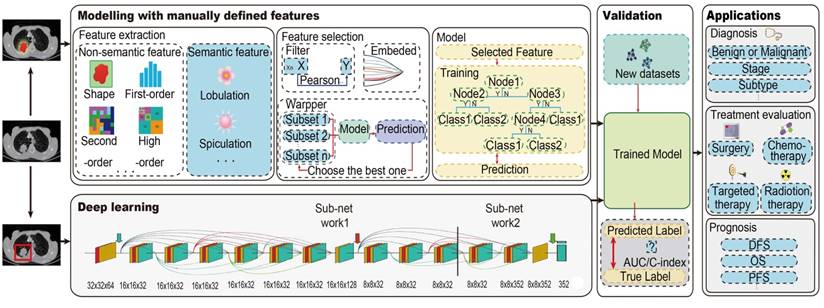
The radiomics pipeline of Modelling with manually defined features and Deep learning. For Modelling with manually defined features, it includes the main steps: data acquisition and preprocessing, tumor segmentation, feature extraction and selection, and modeling. For deep learning, it is an end-to-end method without separate steps of feature extraction, feature selection and modelling. Trained model from both two methods should be validated with new dataset, and then could be applied. Abbreviations: AUC, area under the receiver operating characteristic curve; C-index, concordance index; DFS, disease-free survival; PFS, progression-free survival; OS, overall survival
Tumor segmentation
In radiomic analysis, it is the entire primary tumor that is usually analyzed. Tumor segmentation determines which region will be analyzed further, so this becomes a fundamental step in radiomics. Segmentation includes manual, semiautomatic, and automatic segmentation methods. However, regardless of the method used, there will be certain challenges. First, there is no golden standard for tumor segmentation [2], and the manual segmentation is quite boring and time consuming. Radiologists have to go through every slice of the imaging sequence to delineate the regions of interest, and a mask for a large tumor may contain dozens of slices. Second, there exist numerous morphological variations as tumors are very different from geometric objects, and the variations are usually difficult to model. Third, tumor margins could be blurred by the partial volume effect and not well defined in medical images. Furthermore, segmentation methods especially their reproducibility and reliability are very important. For instance, inter-reader variability of the interpretation for in-plane nodule boundaries could be very large [54]. Several researches have provided that features can be changed with different delineation. For example, van Velden et al. found 25 features were sensitive to the change in delineation using two baseline whole-body PET/CT scans from eleven NSCLC patients [27]. Also Leijenaar et al. found that 71% of radiomic features were stable in a test-retest cohort of 11 NSCLC patients using a semiautomatic segmentation, and 91% of radiomic features were stable in an inter-observer cohort of 23 NSCLC patients with manual segmentation [12]. Since the features are extracted based on the segmented tumors, segmentation methods could ensure that the radiomic features are reproducible and reliable.
Intra-class correlation coefficients (ICC) are usually used to evaluate the inter- and intra-reader agreement. Several studies suggested that ROIs should be detected with an acceptable ICC, so that the further extracted features can be used [7, 8]. Several automatic or semi-automatic segmentation methods have been developed to minimize manual cost and increase reproducibility of tumor segmentation. Parmar et al. [55] compared radiomic features computed by a semiautomatic region growing volumetric segmentation algorithm and manual segmentation with NSCLC tumors in twenty patients; they found that the radiomic features generated with the semiautomatic segmentation had significantly higher reproducibility and more robustness compared with the manual segmentation. Heye et al. [56] compared the inter- and intra-observer variability with manual segmentation versus that with semiautomatic segmentation (“random walker”), preformed with MR Oncotreat software using 15 DCE MR studies. They concluded that the semiautomatic segmentation can provide a significant reduction in inter-observer variability compared with the manual method. These studies demonstrated the potential of semiautomatic segmentation in consistency and repeatability. In addition, Christ et al. [57] proposed a method for medical image segmentation integrating marker controlled watershed segmentation and clustering algorithm, which achieved better segmentation than the conservative watershed algorithm in MR images was achieved. Hatt et al. [58] summarized some PET automatic segmentation algorithms and concluded that the segmentation with manual and visual verification show higher reproducibility and reliability than simple threshold-based approaches.
Until now, in morphological imaging (CT, US or MRI), most radiomic studies have used manually segmented ROIs for analysis even though it is quite time consuming and entails large inter-reader variability. In PET or SPECT imaging, several automatic or semiautomatic segmentation algorithms were performed. Although segmentation methods for medical images have been a hot spot of concern for a long time, there is still a long way to detect a fully automated application [59-61].
Feature extraction
Radiomics was initially defined as the extraction of high-throughput features from images. Quantitative imaging features are an important characteristic in radiomics because they bridge medical images and the clinical end point. Typically, there are two main categories in imaging features: manually defined features (including semantics and non-semantics) and deep learning features (Figure 4).
Manually defined Features
Semantic features could be defined as the qualitatively described empirical features proposed by radiologists. These features cannot be described with efficient mathematical expression, but they are useful in clinical settings and in radiomic analysis. For example, Liu et al. suggested that semantic features of CT images were correlated with epidermal-growth-factor-receptor (EGFR) status in patients with lung cancer [62].
The main features we used for radiomic analysis could be divided into three parts: Empirical features, Statistical features and Deep learning features. All these features could be visualized and interpreted with physical meanings. However, what we should do further is to unravel their physiological significance.
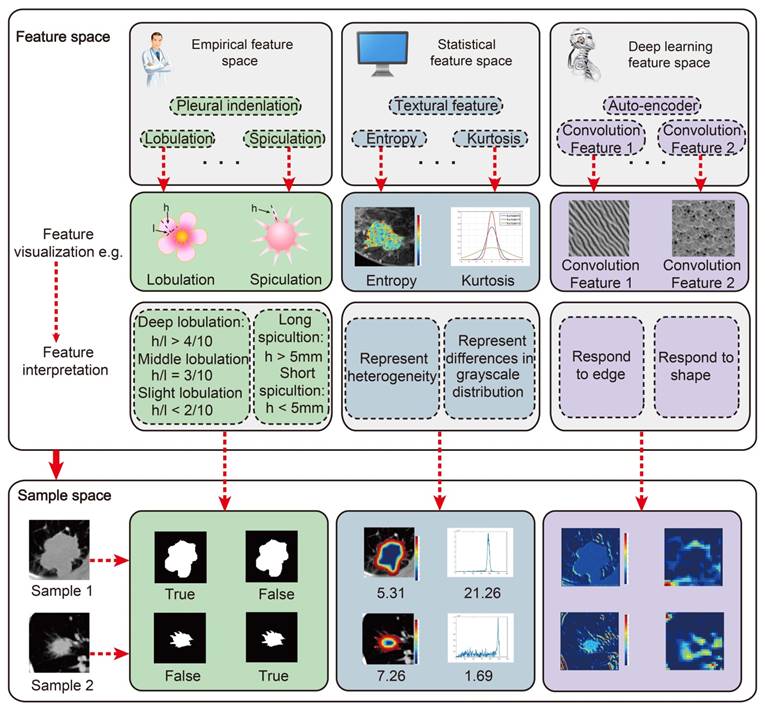
Non-semantic features could be defined as quantitatively described imaging features with mathematical expressions. For the current radiomic studies, the mostly used non-semantic features are usually obtained from predefined ROIs. First group contains the shape features describing the tumors, including volume, surface area, compactness, and etc. Second group contains a large number of first-, second-, and high-order features constructed from the intensity of each voxel in the ROIs [6]. First-order features describe the distribution of the intensity within the ROI, including maximum, median, minimum, and entropy. Second-order features describe the statistical relationships between voxels, such as textural features related with the representative of tumor heterogeneity. High-order features usually referred to statistical features calculated on matrices that consider relationships between three or more pixels. In addition, wavelet features, referred to as multiresolution image scaling features, and fractals, referred to as model-based features, were also used a lot in radiomic studies [63-65]. Other computational imaging features such as local binary patterns (LBP) [66] and scale-invariant feature transform (SIFT) [67] can also be used in radiomic analysis.
Deep Learning Features
Deep learning represents a class of algorithms that use the stacked neural network structure [68]. A deep learning model generally includes the following computational components: convolution, pooling, activation, full connection and batch normalization. All the computational components are defined as layers, and they are stacked layer-by-layer. The most important components of the deep-learning model are the convolutional layer and fully connected layer that can adaptively learn features from data. Therefore, the outputs of the convolutional layer or the fully connected layer can be defined as deep-learning features.
The typical manner of extracting deep-learning features is to train a deep learning model using the clinical outcomes, and then extract the outputs of several layers as deep learning features [69]. For example, after training a convolutional neural network (CNN) for EGFR mutation prediction in lung cancer, the results of the final fully connected layer was extracted to obtain deep learning features for EGFR status prediction [70]. Another method of extracting deep-learning features is to use an unsupervised auto-encoder. When there is not enough data to train a supervised deep learning model, an auto-encoder provides an alternative method to extract deep learning features [71-73]. A typical auto-encoder includes an encoder network and a decoder network. The result of the final convolutional layer is defined as a deep learning feature.
When compared with manually defined features, deep learning features are more specific to clinical outcomes and data. Most manually defined features describe general information about a tumor (e.g., shape, intensity, and texture), but may lack specificity regarding clinical outcomes. For example, when designing manually defined features, clinical outcomes are not considered in most situations, therefore, the manually defined features may not be suitable for clinical labels. In contrast, deep learning features are learned from data directly, and therefore, the deep learning features can be adapted to specific datasets and related to clinical outcomes more naturally. Generally, deep learning features are extracted from convolutional layers in the deep learning model. Each convolutional layer includes hundreds of convolutional filters that are defined as deep learning features. The deep learning features can describe multi-level tumor information from low-level visual characteristics to high-level abstract features. For example, in recurrence analysis of high-grade ovarian cancer, a convolutional auto encoder was designed to extract deep learning features from CT image. As illustrated in Fig. 3, features in the first convolutional layer describe simple tumor intensity information, and features in the second layer describe tumor edge information. Features in a deeper layer (the 3rd layer) describe complex tumor shape information. As the convolutional layer goes deeper, high-level abstract features are extracted, which are not interpretable visually, but tend to be associated with the recurrence status of the tumor. Without the need to explicitly specify feature formulas, deep learning features naturally include much of the same information as in manually defined features such as the intensity information (the 1st layer), edge information (the 2nd feature) and shape information (the 3rd). Since deep learning features are learned from data directly, they have a relationship with the recurrence status of the tumor (Fig. 3b). In addition, deep learning features can be designed flexibly since different CNN structures provide different deep learning features, and there is no need carefully consider design feature formulas.
Knowledge discovery and modeling
Generally, thousands of quantitative imaging features could be obtained for the tumors. However, too many features may include redundant information and cause overfitting. Thus, feature selection should be performed to preserve the relevant features. A prediction model can then be constructed with the selected key features according to the clinical endpoint.
Feature Selection
The most commonly used feature selection methods in radiomic studies can be divided into three broad categories: filter, wrapper, and embedding [74].
Filter methods usually evaluate the features without involving the model; therefore, these are also called independent methods. There two main kinds of filter methods: univariate and multivariate methods. Univariate filters mostly rank features based on quality (i.e., on the Chi-squared test or Mann-Whitney U test). Multivariate filters are composed of rankers and subset selectors like those used in correlation-based feature selection. With wrapper methods, features are generated first and then evaluated using the model. This means all wrapper methods are considered multivariate and subset-selector methods. With embedded methods, a feature subset is proposed and evaluated during the construction of the model.
Most radiomic studies use filters for coarse selection. For example, several studies used ICCs to detect stable features [4, 73, 75], and others also ranked the features using Student's t-test [8] or the Mann-Whitney U test [76]. Another commonly used feature-selection method in radiomic studies is the least absolute shrinkage and selection operator (LASSO), which is an embedded method that can produce selected features and a prediction model together [77-81].
Modelling
The goal of radiomics is to construct a prediction model for clinical outcomes with selected features. Machine learning provides several modelling methods to achieve this goal. Typically, supervised, semi-supervised, and unsupervised learning are fundamental strategies used according to the different levels of available clinical outcomes.
Supervised learning is a learning structure that requires clinical labels to train the model. A supervised learning model includes two steps: training and testing. In the process of model training, training samples with corresponding clinical labels are paired to train the model. Through a pre-defined loss function, the model learns the relationship between the feature and the clinical labels. In the testing phase, the well-trained model is used directly to test its predictive performance. There are many supervised learning methods that have shown great performance in radiomic analysis, such as the support vector machine [8], LASSO-logistic regression [7] and random forest [73]. Despite the good performance of many supervised-learning methods, they usually require large amounts of training samples to avoid overfitting.
In many situations, clinical labels may not be enough to train a supervised model, and unsupervised learning can be an alternative. Unsupervised learning methods group data into several clusters according to the similarity between samples. In this process, the clinical label is not necessary for model training. The unsupervised model utilizes a distance measurement to calculate the similarity between samples. The similar training samples are grouped together, while the dissimilar samples are assigned into different groups. These models include many clustering algorithms such as k-means clustering [82], fuzzy clustering [83] and consensus clustering [84]. For example, a consensus clustering model was used in a previous study to detect glioblastoma phenotypic subtypes [85], and patients were classified into three groups, each with different molecular pathway activities and a different prognosis.
Generally, supervised learning shows stronger performance than unsupervised learning since the clinical label is considered during the supervised model training. However, insufficiently labelled data limits the performance of supervised learning. As a trade-off between better performance and enough training data, semi-supervised learning could be a good choice. Semi-supervised learning uses a large amount of unlabeled data to mine tumor information and utilizes small amounts of labelled data to build the relationship between features and clinical labels. For predicting the recurrence for high-grade serous ovarian cancer, a semi-supervised deep learning framework was proposed [86]. The semi-supervised framework can be divided into an unsupervised feature learning phase and a supervised model training phase. In the unsupervised learning phase, a convolutional auto encoder was built to compress tumor images into low-dimensional features. In this process, the auto encoder tried to extract the intrinsic characteristics of the tumor without a clinical label. Afterwards, a supervised Cox model was built to model the relationships between the features and clinical labels.
Validation
A radiomic model must be validated to show its potential value for clinical application. An independent, external, validated model is considered to be more credible than an internally validated model, as results from independently obtained data are usually more robust. Furthermore, prospectively validated models have the most credibility for radiomic studies. There are dozens of tools to measure the performance of radiomic models. For discrimination analysis, the receiver operating characteristic (ROC) curve is the mostly used method to report the performance of proposed models. Moreover, the area under the curve (AUC), sensitivity, and specificity of the model can also be used to evaluate if the model can predictive the clinical outcome. For survival analysis, the concordance index (C-index) and time-dependent ROC curve are usually used for validation. In addition, calibration is a useful tool for both discrimination analysis and survival analysis, as it can obtain the agreement between the observed clinical outcomes and model predictions [87].
Applications of Radiomics in Diagnosis and Treatment of Oncology
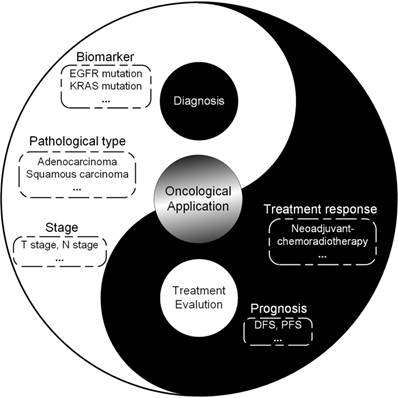
Since radiomics was proposed, rapidly increasing radiomic studies have been published to improve the diagnosis and treatment of cancer. More and more studies demonstrate the value of radiomic features as a complimentary tool to decision making of clinicians. Here, we will provide a brief introduction to radiomic studies, focusing on the clinical problems during the whole process of diagnosis and treatment in several typical cancers including brain tumors, head-and-neck cancer, breast cancer, lung cancer, prostate cancer, colorectal cancer, liver cancer and gastric cancer (Figure 5).
Scope of radiomics for diagnosis and treatment evaluation, suggesting the potential directions radiomics could be applied for. Abbreviations: EGFR: epidermal growth factor receptor; IDH: isocitrate dehydrogenase; KRAS: Kirsten rat sarcoma viral oncogene homolog; MGMT: O-6-methylguanine-DNA methyltransferase
Brain Tumors
Malignant brain tumors can be divided into primary tumors which started within the brain (mainly referred to as gliomas, meningiomas, and chordomas), and secondary tumors that spread from elsewhere, known as brain-metastasis tumors [88]. Current brain-tumor imaging protocols incorporate CT, multi-parametric MRI, and sometimes PET. As use of radiomics could extract large amounts of quantitative imaging features and capture intratumoral and intertumoral heterogeneity, it makes a radiomic analysis could assess imaging phenotypes that may influence the diagnosis and treatment evaluation of brain tumors.
Diagnosis
Although standard brain imaging can provide results enough for tumor grading, improve was needed for newly proposed imaging protocols. Bai et al. first utilized diffusion kurtosis MRI for the grading of gliomas and detected good performance in the grading of gliomas [89]. Moreover, radiomics also attracted attention for the prediction of molecular subtypes of brain tumors, as the precision diagnosis of gene-expression patterns could potentially enhance decision making for targeted therapies [90]. Isocitrate dehydrogenase (IDH) [91], O6-methylguanine-DNA methyltransferase (MGMT) [92, 93], 1p/19q co-deletion [94], EGFR expression level [95], Ki-67 expression level [96], p53 status [97], and ATRX mutation [98] have been the main focus in prediction studies of molecular subtype in brain tumors. Radiomic analysis based on multi-parametric MRI aided the prediction of molecular characteristics. More importantly, imaging phenotypes were shown to be associated with molecular pathway activities that may determine the type of targeted therapy [85].
Treatment evaluation and Prognosis
There has been considerable interest in treatment evaluation and prognosis for brain tumors to identify imaging phenotypes that may predict the treatment response in patients with glioblastoma. Two recent studies investigating the responses to bevacizumab treatment in recurrent glioblastoma patients suggested the potential of radiomics to predict different response to the treatment [99, 100]. These studies both detected the potential of radiomics to aid in cancer treatment decision-making at a low cost. However, a bevacizumab-naïve control group was needed for these studies to confirm the predictive value. In addition, radiomics provided a new option for determining prognosis for brain tumors. Recent studies have suggested that features detected from MRI and PET were significantly associated with the survival of patients with gliomas [101, 102]. In the future, a model could be constructed using radiomics to improve both treatment planning and prognosis.
Head- and-neck cancer
Head-and-neck cancer is one of the cancers that radiomics has also been widely applied [103].
Diagnosis
Ren et al. built a MRI based radiomic signature to predict the stage of head and neck cancer preoperatively [104]. They found that the radiomic signature from contrast-enhanced T1-weighted MR images and T2-weighted MR images had good performance in discriminating different stages. Leijenaar et al found that CT radiomic signature could predict HPV (p16) status in oropharyngeal squamous cell carcinoma [105]. Zhou et al proposed a 3-dimensional deep learning model to predict lymph node metastasis in nasopharyngeal cancer (NPC) [106]. Chen et al. evaluated the association between Tumor PD-1 expression and Immunohistochemical biomarkers or radiomic features from PET imaging in NPC [107]. Crispin-Ortuzar et al predicted hypoxia status using PET/CT radiomics in NPC patients [108].
Treatment evaluation and Prognosis
Zhang et al. proposed that multi-parametric MRI based radiomics could be a novel prognostic factor in advanced NPC [109]. They collected 118 advanced NPC patients and found that radiomic signature achieved significantly improved performance evaluating PFS compared with the TNM staging system. Furthermore, Zhang et al. evaluated 6 different feature selection methods and 9 different classifiers for prediction of local failure and distant failure in advanced NPC [110]. They found that Random Forest performed best among the 9 methods. Wang et al. [111] investigated the value of radiomic signatures in prediction of early response to induction chemotherapy in NPC patients. They found that radiomic signature had a good performance in predicting early response to induction chemotherapy. Lu and Lv et al. evaluated the robustness of radiomic features obtained from different PET images in NPC patients [112, 113]. Wu et al. performed survival prediction in high-grade Osteosarcoma using radiomics of diagnostic CT [114]. They found that radiomic nomogram, which combines a radiomic signature and clinical factors, showed better calibration and classification capacity than a model with only clinical factors. Elhalawani et al investigated radiomic signatures for local recurrence in oropharyngeal cancer [115]. A radiomic signature consisting of 2 radiomic features showed robust discrimination ability of recurrence. Vallières et al used radiomics for assessing tumor failure in patients with NPC from 4 centers [116]. Gabryś et al predicted the xerostomia in NPC patients after radiotherapy [117]. They compared the performance of 6 feature selection, 7 classification, and 10 data balancing methods.
Breast cancer
Breast cancer is the cancer with the highest incidence in women worldwide. As it is a known heterogeneous disease, the precise diagnosis and early prediction of response to the treatment are hotspots of the clinical practice and research in breast cancer. Radiomics, combining multi-modality imaging data and clinical information, is now used widely in breast cancer research.
Diagnosis
There are several diagnostic imaging modalities for breast cancer, including US, PET/CT, mammography, and MRI. Thus, radiomic researches in breast cancer also covered all these modalities. Several previous studies utilized radiomics for the prediction of breast cancer subtype or the status of ER, PR, Ki67 and HER2 with US [46], mammography [118], PET/CT [119, 120], and MRI [121, 122]. Specifically, Antropova and his colleague proposed a feature fusion method that could combine US, MRI and mammography together for better diagnosis of breast cancer [118]. In addition of the characterization of breast cancer, radiomics could also provide a non-invasive approach for prediction of sentinel lymph node metastasis [123].
Treatment evaluation and Prognosis
Most radiomic studies in breast cancer have focused on the evaluation of response to therapy. Chan et al. [124] developed an automated method to predict treatment failure in early breast cancer patients with a pretreatment MRI scan. Most other studies attempted to obtain a radiomic biomarker for pathological complete response (pCR) to neoadjuvant chemotherapy, which is a hot point in breast cancer research. Braman et al found that intra- and peri-tumoral features of DCE-MRI could contribute to pretreatment prediction of pCR [125]. Other studies also showed that T1WI, T2WI and DWI could also help to detect pCR [76, 126]. While several studies approved the potential of MRI as a biomarker for detecting pCR, Tran et al pointed out that diffuse optical spectroscopic could also be a useful tool for pCR prediction [127].
There are also radiomic studies focusing on the prognosis of breast cancer. Park et al [128] have developed a radiomic signature combining MRI features and clinical information for individualized estimation of disease free survival in breast cancer patients.
Lung cancer
Lung cancer is the most harmful cancer, and its prevalence continues to increase worldwide [129]. Radiomics has been widely used in the diagnosis, treatment evaluation, and prognosis in lung cancer [130, 131].
Diagnosis
One of the most important diagnostic applications of radiomics is screening of lung cancer. Kumar et al performed a classification between malignant and benign lesions with diagnostic data from the LIDC-IDRI dataset [132]. The proposed method achieved a sensitivity of 79.06% and a specificity of 76.11%. Shen et al. proposed a deep learning model based on CT images and achieved better prediction results for malignant lung nodule compared with previous methods [133]. Maldonado et al. developed a noninvasive radiomic method to discriminate solid and ground glass opacities using high resolution CT [134]. Besides diagnostic CT, researches on low-dose CT were also performed for screening malignant lung nodules. Carter et al. [135] performed a screening study on patients with low-dose CT screening detected lung cancer in National Lung Cancer Screening Trial (NLST) dataset. Accuracies of 80% and 79% were found for predicting nodules that will develop cancerous in one or two years, respectively. Liu et al. also utilized a low-dose CT cohort from the United States and radiological image traits of lung nodule to predict malignancy and achieved impressing results [136].
Accurate preoperative TNM staging is also important for treatment decisions of lung cancer. Aerts et al. extracted 440 CT radiomic features from 1,019 patients and found that many of the radiomic features were correlated with the overall stage, T stage, N stage, and M stage of lung cancer and head-and-neck cancer [4, 137]. Coroller et al. used CT radiomic signature to predict distant metastasis (M staging) on a United States cohort of 182 pathologically confirmed lung adenocarcinoma [138] and Zhou et al. combined CT radiomic features and clinical risk factors to identify the distant metastasis based on a Chinese cohort including 348 lung cancer patients [139]. The results from all of these studies showed good performance of radiomics on preoperative M staging. Wu et al. used PET/CT radiomic features to predict distant metastasis with 101 early-stage NSCLC patients and they found additional value of PET in M staging [140].
Besides screening and staging, radiomics has been applied to predict the gene mutation or pathological type in lung cancer. Liu et al. and Zhang et al. all found that EGFR mutation was associated with CT radiomic features [141-143]. Rios et al. developed CT radiomic signatures to distinguish somatic mutations on a dataset of 763 lung adenocarcinoma patients [73]. They found that a CT radiomic signature that is related with radiographic heterogeneity can successfully predict EGFR status. Wu et al. used radiomics to predict the histologic type of lung cancer, adenocarcinoma or squamous carcinoma, on two NSCLC cohorts from Netherlands [144]. Zhu et al. performed a similar study on a Chinese cohort and also found that radiomic signature could serve as a diagnostic factor for histologic subtype classification of NSCLC [145]. Fan et al. [146] developed a radiomic signature for the preoperative discrimination of lung invasive adenocarcinoma manifesting as a ground-glass nodule.
Treatment evaluation and Prognosis
Treatment response evaluation is essential in lung cancer due to its value in treatment decision. Mattonen et al. found that radiomic signatures could predict the recurrence after Stereotactic Ablative Radiation Therapy (SART) in Lung Cancer [147, 148]. Fave et al. used delta-radiomic features to predict outcomes in Stage III NSCLC patients during the radiation therapy [149]. Their results suggest the change of radiomic features due to radiation therapy would be indicators of tumor response. Coroller et al. found that pretreatment CT radiomic features could predict pathological response after neoadjuvant chemoradiation in patients with advanced NSCLC [150]. Aerts et al. found that CT radiomic feature before treatment is able to predict EGFR mutation status in NSCLC and associated with gefitinib response [151]. Cook et al. used heterogeneity features in PET images to evaluate the treatment response of Erlotinib in NSCLC [152]. Song et al. found that CT radiomic signature could predict progression-free survival after TKI therapy [153].
Besides treatment response, many radiomic studies have focused on prognosis. Aerts et al. found the clusters of radiomic features were associated with the prognosis of lung cancer [4]. Song et al. also demonstrated a relationship between CT radiomic features and overall survival in patients with NSCLC [154]. Balagurunathan et al. tested the reproducibility and prognosis of quantitative radiomic features from CT images, and many features were associated with the prognosis of lung cancer [75]. Coroller et al. found that radiomic features are prognostic for both distant metastasis and survival [138]. Huang et al. found that the radiomic signature was associated with DFS [155].
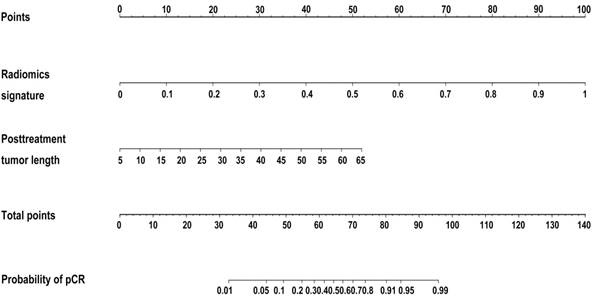
Developed radiomics nomogram for prediction of pCR to NCRT in rectal cancer. Reproduced with permission from [8].
Colorectal cancer
Colorectal cancer is a common and fatal type of cancer. The incidence and deaths of colorectal cancer are increasing every year, and many questions remain regarding the three different stages of diagnosis, treatment evaluation, and prognosis.
Diagnosis
Accurate identification of the extent of lymph-node metastasis in colorectal cancer patients is critical for the determination of treatment strategies. If it is confirmed that the patient has no lymph-node metastasis before surgery, there is no need to perform lymph-node dissection during surgery [156-158]. Huang et al. performed radiomic analysis on 526 patients with colorectal cancer [7]. Based on preoperative CT images of the portal vein, the radiomic features were extracted, and a nomogram was established. The C-index was 0.736 on the training set and 0.778 on the validation set. Moreover, the authors proposed an easy-to-use nomogram based on the radiomic model to help its application (Figure 6). Another relevant application of radiomics in colorectal cancer is the prediction of KRAS/NRAS/ BRAF mutation. The National Comprehensive Cancer Network (NCCN) guidelines for colorectal cancer recommend that patients should screen for KRAS/ NRAS/BRAF mutations, because these mutations are indicative of poor response to cetuximab and panitumumab [159-162]. Therefore, determination of the KRAS/NRAS/BRAF mutation status before or during treatment is essential to predict treatment efficacy and achieve a personalized diagnosis. Yang et al.[163] extracted 346 radiomic features from the preoperative CT images of 117 patients with colorectal cancer for a radiomic analysis, achieving an AUC of 0.869.
Treatment evaluation and Prognosis
Patients with rectal cancer usually undergo neoadjuvant chemoradiotherapy before surgery [164, 165], but it is not known whether pCR is achieved before neoadjuvant chemoradiotherapy. This information is only obtained after surgery. Approximately 15-27% of locally advanced patients will achieve pCR [166, 167], and studies have shown that these patients do not need surgery [168-170]. Based on information from 48 patients, Ke et al. [171] constructed an artificial neural network based model, and the AUC was 0.71-0.79. Liu et al. applied radiomics to identify patients with locally advanced rectal cancer who achieved PCR before surgery [8]. The study enrolled 222 patients with locally advanced rectal cancer. T2WI and DWI were used to extract 2252 radiomic features. The AUC was 0.976 (95% confidence interval [CI], 0.9185-0.9711), and the predicted PCR accuracy was 94.3% (95% CI, 91.9%-97.1%). Based on 114 patients (18 PCR) Natally et al. extracted radiomic features on T2WI and made predictions using random forest classification; the AUC was 0.93, sensitivity 100%, and specificity 91% [172].
Although treatment may improve survival, it has not proven to improve the DFS of patients with locally advanced rectal cancer [13, 173, 174]. Distant metastasis is the main cause of patient treatment failure. For high-risk patients, increased systemic therapy reduces the risk of metastasis and improves survival. Therefore, pre-operative risk stratification may help to select individualized treatment strategies and improve the prognosis of patients. Based on 108 patients with locally advanced rectal cancer who underwent total mesorectal excision [175], Meng et al. extracted 485 radiomic features; the C- index of the radiomic signature was 0.767 (95% CI, 0.72-0.86), and the 3-year time-dependent AUC was 0.827. Combined with clinical features, the C-index and AUC of the Cox model were improved: 0.788 (95% CI, 0.72-0.86) and 0.837, respectively.
Identification of patients who need more aggressive treatment and follow-up is critical. Lovinfosse et al. explored the significance of PET/CT-based radiomic features for prognosis. The study extracted the SUVmax, SUVmean, metabolic tumor volume, total lesion glycolysis, and texture features, established univariate and multivariate Cox models, and analyzed the prognostic performance of the features [176].
Prostate cancer
Prostate cancer (PCa) is the most common cancer affecting men in Western societies [177]. Multi-parametric MRI (mp-MRI) plays important role in diagnosis and treatment of PCa because of the advantage of high soft tissue contrast.
Diagnosis
The non-PCa condition such as prostatitis and benign prostatic hyperplasia is easily confused with PCa. Ginsburg SB et al utilized mp-MRI based radiomic model for differentiation between PCa and non-PCa [178]. The result showed that the validation model achieved relatively good performance with high AUC using T2 WI, ADC, and T2 WI and ADC features for PCa detection. PCa exhibits an extraordinary variable biological behavior. According to NCCN Prostate Cancer, version 3, the identification of aggressive PCa is vital for clinic decision. Some radiomics models based on mp-MRI were developed and validated to identify PCa aggressiveness in patients [179, 180]. The results showed that radiomic models could be helpful for identifying the presence and absence of PCa aggressiveness compared with PIRADS v2.0 assessment, an mp-MRI based subjective assessment method. The pathological Gleason score (GS) of PCa has clinical implications for the treatment and prognosis. Chaddad et al. achieved the best performance for GS = 3 + 4, and 64.76% for GS ≥ 4 + 3 to predict the GS of PCa patients via radiomic features based on mp-MRI[181].
Treatment evaluation and Prognosis
In addition, radiomics is also used for treatment evaluation of PCa. Many researchers successfully utilized radiomic features derived from pretreatment mp-MRI to predict biochemical recurrence of PCa after radical prostatectomy, radical radiotherapy or external beam radiotherapy [182, 183].
Liver and Gastric cancer
As medical imaging is playing a more and more important role in the management of liver cancer and gastric cancer, there are more opportunities to utilize quantitative imaging analysis in the clinical setting for diagnosis and treatment evaluation.
Diagnosis
Radiomics is mainly used in cases of liver cancer for noninvasive prediction of microvascular invasion (MiVI). Two research teams have verified the validity of radiomics for preoperative MiVI prediction [184, 185]. Bakr et al. extracted quantitative radiomic and semantic imaging features from the triphasic CT scans of 28 treatment-naïve hepatocellular carcinoma (HCC) patients [184]. Delta radiomic features were designed as the absolute difference and ratio computed from all pairs of the three imaging phases. The proposed imaging features from arterial and venous phases were regarded as potential biomarkers for MiVI prediction in HCC. Jie et al. further used a cohort of 304 patients with hepatitis B virus-related (HBV) HCCs to validate the usefulness of imaging biomarkers for MiVI prediction [185]. The radiomics-based nomogram showed satisfactory predictive accuracy for MiVI in HBV-related HCC.
For gastric cancer (GC), Ma et al. evaluated the value of CT-based radiomic signature to discriminate Borrmann type IV GC from primary gastric lymphoma (PGL) [186]. Ba-Ssalamah et al. used CT texture features to discriminate GC, lymphoma, and gastrointestinal stromal tumors [187]. Liu et al. explored the application of radiomics in predicting pathologic differentiation for the type of GC [188]. Liu et al. used radiomic analysis to predict T stage, N stage, overall stage, and perineural invasion in patients with GC [189-191].
Treatment evaluation and Prognosis
Radiomics-based methods for treatment evaluation and prognostic prediction in liver cancer rely on two aspects: overall survival (OS) and recurrence predictions. Zhou et al. proved the prognostic power of radiomic signatures for early recurrence of HCC with partial liver resection [192]. The LASSO was used to build the radiomic signature. The AUC of the combined model was 0.836, which shows it performed better than clinical predictors. Akai et al. investigated the use of random forest along with radiomic analysis to predict DFS and OS for HCCs with resection as treatment [193]. Furthermore, Cozzi et al. proved the radiomic signature could be used as a robust biomarker for local control and survival prediction [194]. Compacity and Barcelona Clinic Liver Cancer (BCLC) stage were finally chosen as effective covariates for prognosis prediction. All these studies indicated that radiomic signatures are potential biomarkers for prognostic prediction in liver cancer research.
Radiomics is also a useful tool for treatment evaluation and prognosis in GC. Giganti et al. found that radiomic texture features had predictive ability for treatment response for neoadjuvant therapy in patients with GC [195]. Hyun et al. found that textural features on CT images were associated with better prognosis in patients with HER2-positive gastric cancer who received target therapy [196]. Giganti et al. investigated the association between CT based radiomic features and overall survival in patients with GC [197]. Texture features were significantly associated with the prognosis of GC.
Challenges and Prospects for Radiomics
In recent years, we have witnessed the progress of radiomics in methodologies and clinical applications. Its potential has been revealed in helping clinical experts to uncover cancer characteristics that fail to be appreciated by naked eyes. In particular, radiomics not only helps radiologists to make precision diagnoses, but also provides oncologists a useful tool for treatment planning and response evaluation [131, 198]. Based on the progress outlined here, further improvements in radiomics will advance clinical care for cancer patients. Next, we will discuss the challenges and opportunities for further radiomics studies.
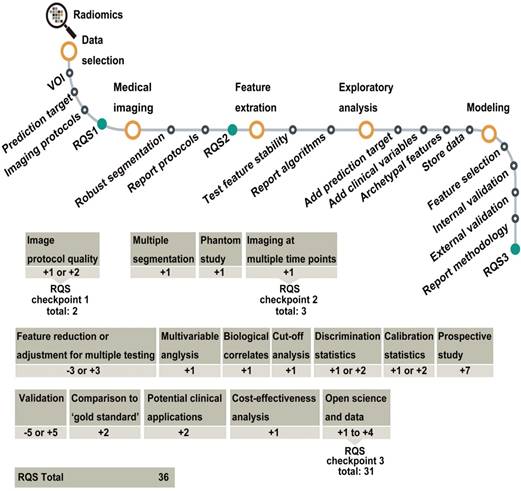
Flowchart depicting the workflow of radiomics and the application of the RQS. Reproduced with permission from [10]. Abbreviations: RQS, radiomics quality score.
Big clinical data and data sharing
A reliable conclusion is based on enough data, as is radiomics. Big and standardized clinical data will make radiomics clinically applicable. However, the radiomic researchers need better access to big data, as medical images are dispersed in different hospitals or data centers, and the size of the patient population is the basis of large amounts of medical images. Another challenge is that the collection of medical images is time consuming. Thus, data sharing among institutes and hospitals around the country or even around the world is important for radiomics, although it presents complex logistical problems. The Cancer Imaging Archive (TCIA) provides a good example of data sharing with a large portion of clinical information [199], and it is still growing with contributions from different institutes and hospitals. Researchers could develop new methods and tools to validate their hypotheses with this large dataset [59, 79]. The Quantitative Imaging Network (QIN) is another useful project for data integration that may promote radiomic studies [200]. However, high quality datasets are still needed for radiomic researches [201].
Reproducibility and quality control
Reproducibility and quality control are involved in the entire radiomic process, including data acquisition, feature extraction and selection, modelling, and validation. Every decision in this process affects the result, its reproducibility, and the quality of the research. Moreover, the inclusion and exclusion criteria of patients in radiomic studies are other important factors related that affect reproducibility. Different criteria may result in the availability of different cohorts for analysis, especially for small cohort studies. Therefore, developing unique inclusion and exclusion criteria for similar clinical applications of radiomics should be considered seriously. Disclosure of the details in every step is important; with the development of data sharing and unique criteria of patients' inclusion, models may be validated using an open dataset that could become the standard for comparison of future studies. Recently, the image biomarker standardization initiative (IBSI) towards seeking standardization for improved reproducibility of high-throughput imaging analyses mainly focused on radiomics was proposed [69]. This was a highly valuable step towards the improvement of radiomic studies. In addition, the radiomics quality score (RQS) (Figure 7) was proposed to aid in the assessment of radiomics studies [10]. Improving research quality will increase the influence of radiomics.
Interpretability of radiomic features and models
Clinical experts often think of the radiomic model as a black box that can provide good prediction results for particular clinical outcomes [131, 198], which tends to make radiomics a less accepted approach. Improvement of the interpretability of radiomic features and models is urgently needed. For manually defined imaging features, it is easier to correlate them using pathophysiology; for deep learning, it is a little more difficult. Visualization of deep learning features and prediction models could potentially help to solve this problem. Fortunately, there are many researchers investigating methods to accomplish this goal and several tools have been proposed to diminish the black box perception [202].
Conclusions
Radiomics is still a newly proposed and rapidly progressing field that integrates radiology, oncology, and machine learning. With the growth of clinical data and advanced machine-learning methodologies, it is playing an increasingly important role in precision diagnostics and oncology. Further research should focus on the reproducibility and interpretability to further make radiomics more acceptable in the field.
Specifications of radiomic studies in different cancers
| Studies | Study design | No. of patients (training + testing) | No. and type of radiomic features | Statistical analysis | Image Modality | Clinical Characteristics |
|---|---|---|---|---|---|---|
| Brain tumor | ||||||
| Kickingereder et al [88] | Retrospective, single center study | 79 + 40 | 6095 (first-order, volume and shape, texture) | Cox regression analysis | MRI | Prognosis |
| Xi et al [90] | Retrospective, single center study | 98 + 20 | 1665 (first-order, size and shape, texture, wavelet) | LASSO, SVM | MRI | Diagnosis |
| Kickingereder et al [91] | Retrospective, single center study | 121 + 60 | 1043 (first-order, size and shape, texture) | Chi-square and Wilcoxon test, LASSO, Cox regression | MRI | Prognosis |
| Han et al [92] | Retrospective, single center study | 184 + 93 | 79 (size and shape, intensity, textural, wavelet) | random forest, U test | MRI | Diagnosis |
| Li et al [93] | Retrospective, single center study | 200 + 70 | 431 (first-order, size and shape, texture, wavelet) | logistic regression, t test, Chi-square test | MRI | Diagnosis |
| Li et al [95] | Retrospective, single center study | 180 + 92 | 431 (first-order, size and shape, texture, wavelet) | t test, Chi-square test, LASSO, SVM | MRI | Diagnosis |
| Kickingereder et al [97] | Retrospective, single center study | 112 + 60 | 4842 (first-order, size and shape, texture, wavelet) | PCA, Cox regression model | MRI | Treatment evaluation |
| Grossmann et al [98] | Retrospective, single center study | 57 + 56 | 65 (first order statistics, size and shape, texture) | PCA, Spearman rank correlation, Wilcoxon test, Cox | MRI | Treatment evaluation and Prognosis |
| Papp et al [99] | Retrospective, single center study | 70 | 48 (Histogram, shape, texture) | Geometrical Probability Covering Algorithms, | PET | Prognosis |
| Head and neck cancer | ||||||
| Ren et al [102] | Retrospective, single center study | 85 + 42 | 970 (shape, intensity, textural and wavelet) | LASSO | MRI | Diagnosis |
| Leijenaar et al [103] | Retrospective, multi-center study | 628 + 150 | 902 (intensity, shape, texture, Wavelet and Laplacian) | LASSO and multivariable logistic regression | CT | Diagnosis |
| Zhou et al [104] | Retrospective, single center study | 122 + 39 | 257 (intensity, texture, and geometric) | 3D CNN | PET/CT | Diagnosis |
| Chen et al [105] | Retrospective, single center study | 53 | 41 (textural features or histograms) | logistic regression analysis | PET/CT | Diagnosis |
| Zhang et al [107] | Retrospective, single center study | 88 + 30 | 970 (shape, intensity, textural and wavelet) | LASSO | MRI | Prognosis |
| Wang et al [109] | Retrospective, single center study | 120 | 591 (texture) | LASSO | MRI | Treatment evaluation |
| Wu et al [111] | Retrospective, single center study | 102 + 48 | 474 (shape, intensity, textural and wavelet) | LASSO | CT | Prognosis |
| Elhalawani et al [113] | Retrospective, single center study | 420 + 45 | 134 (intensity direct, histogram, shape and texture) | multivariate Cox | CT | Prognosis |
| Vallières et al [114] | Retrospective, multi-center study | 194 + 106 | 1615 (intensity, shape, texture) | random forest | PET/CT | Prognosis |
| Breast cancer | ||||||
| Guo et al [46] | Retrospective, single center study | 215 | 463 (morphology, intensity, texture, wavelet) | t test, ICC, LASSO, and SVM | US | Diagnosis |
| Antropova et al [116] | Retrospective, single center study | 2060 | Deep learning features | CNN, SVM | FFDM, US, MRI | Diagnosis |
| Antunovic et al [117] | Retrospective, single center study | 43 | 20 (size and shape, first-order) | Univariate analysis, hierarchical clustering, exact Fisher's test | PET/CT | Diagnosis |
| Ha et al [118] | Retrospective, single center study | 73 | 109 (texture) | Hierarchical clustering, logistic regression, Cox | PET | Diagnosis and Prognosis |
| Saha et al [120] | Retrospective, single center study | 461 + 461 | 529 (first order, size and shape, texture) | correlation, RF, logistic regression | MRI | Diagnosis |
| Chan et al [122] | Retrospective, single center study | 563 | 8192 (washin and washout intensity values) | PCA, LASSO, logistic regression, K-M survival | MRI | Treatment Evaluation |
| Braman et al [123] | Retrospective, single center study | 117 | 99 (texture) | mRMR, consensus clustering, LDA, DLDA, QDA, naïve Bayes, SVM | MRI | Treatment Evaluation |
| Chamming et al [76] | Retrospective, single center study | 85 | texture | Mann-Whitney U test, logistic regression | MRI | Treatment Evaluation |
| Partridge et al [124] | Prospective, multi-center study | 272 |  ADC, ADC,  FTV FTV | Z-test, logistic regression | MRI | Treatment Evaluation |
| Tran et al [125] | Retrospective, single center study | 37 | 40 (texture) | Logistic regression, naïve Bayes, k-NN | DOS | Treatment Evaluation |
| Park et al [126] | Retrospective, single center study | 194+100 | 156 (texture) | Elastic net, Cox Model, K-M analysis | MRI | Prognosis |
| Lung cancer | ||||||
| Kumar et al [130] | Retrospective, multi-center study | 38106 + 4234 | 500 (Deep learning features) | 3D CNN | CT | Diagnosis. |
| Shen et al [131] | Retrospective, multi-center study | 825 + 275 | Deep learning features + 319 (first order, shape and size, textural, and wavelet) | mrmr, SVM, MC-CNN | CT | Diagnosis |
| Aerts et al [4] | Retrospective, multi-center study | 474 + 575 | 440 (intensity, shape, texture and wavelet) | Cluster and Cox | CT | Prognosis and Diagnosis |
| Parmar et al [135] | Retrospective, multi-center study | 558 + 320 | 440 (intensity, shape, texture and wavelet) | Cluster and Cox | CT | Prognosis |
| Coroller et al [136] | Retrospective, single center study | 98 + 84 | 635 (intensity, shape, texture, LoG and Wavelet) | mrmr and Cox | CT | Prognosis |
| Zhou et al [137] | Retrospective, multi-center study | 241 + 107 | 485 (intensity, shape, texture, and wavelet) | concave minimization and SVM | CT | Prognosis |
| Jia et al [138] | Retrospective, single center study | 70 + 31 | 70 (statistical, histogram, morphologic, and texture) | c-means, ICC, linear correlation coefficient, LASSO and Cox | PET | Prognosis |
| Liu et al [139] | Retrospective, single center study | 298 | 219 (size, shape, location, air space, histogram, laws texture and wavelet) | Multiple logistic regression analysis, and SVM | CT | Diagnosis |
| Zhang et al [141] | Retrospective, single center study | 140 + 40 | 485 (shape, intensity, textural and wavelet) | LASSO | CT | Diagnosis |
| Rios et al [56] | Retrospective, multi-center study | 353 + 352 | 440 (intensity, shape and texture) | MRMR, Random forest classifier | CT | Diagnosis |
| Wu et al [142] | Retrospective, multi-center study | 198 + 152 | 440 (intensity, shape and texture) | 24 Feature Selection Methods, 3 Classifiers | CT | Diagnosis |
| Zhu et al [143] | Retrospective, single center study | 81 + 48 | 485 (shape, intensity, textural and wavelet) | LASSO | CT | Diagnosis |
| Fan et al [144] | Retrospective, multi-center study | 160 + 235 | 355 (shape, intensity, textural and wavelet) | LASSO | CT | Diagnosis |
| Fave et al [147] | Retrospective, single center study | 107 | 212 (texture, shape) | Cox, AIC | CT | Prognosis |
| Coroller et al [148] | Retrospective, single center study | 101 + 26 | 440 (intensity, shape and texture) | PCA | CT | Treatment evaluation |
| Aerts et al [149] | Retrospective, multi-center study | 47 | 183 (shape, intensity, textural and wavelet) | Coefficient of Variation | CT | Treatment evaluation |
| Song et al [151] | Retrospective, multi-center study | 117 + 197 | 1032 (shape, intensity, textural and wavelet) | LASSO and COX | CT | Treatment evaluation |
| Song et al [152] | Retrospective, single center study | 661 + 61 | 592 (shape, intensity, textural and wavelet) | SVM and COX | CT | Prognosis |
| Balagurunathan et al [55] | Retrospective, single center study | 32 + 59 | 329 (shape, size, and texture) | concordance correlation coefficient, and K-M | CT | Prognosis |
| Huang et al [155] | Retrospective, single center study | 141 + 141 | 132 (textural) | LASSO and COX | CT | Prognosis |
| Colorectal cancer | ||||||
| Huang et al [7] | Retrospective, single center study | 326 + 200 | 150 (histogram and GLCM) | LASSO | CT | Diagnosis |
| Yang et al [161] | Retrospective, single center study | 61 + 56 | 346 (histogram, shape, and texture) | ICC, SVM | CT | Diagnosis |
| Ke et al [169] | Retrospective, single center study | 48 | 103 (histogram, GLCM, shape) | Mann-Whitney test, T test, ANN | MRI | Treatment evaluation |
| Liu et al [8] | Retrospective, single center study | 152 + 70 | 2252 (intensity, shape, wavelet features) | T test, LASSO | MRI | Treatment evaluation |
| Natally et al [170] | Retrospective, single center study | 93 + 21 | 34 (texture, wavelet features) | Wilcoxon rank-sum test, random forest | MRI | Treatment evaluation |
| Meng et al [173] | Retrospective, single center study | 54 + 54 | 485 (Shape, Intensity Texture) | log-rank test, LASSO Cox regression | MRI | Prognosis |
| Lovinfosze et al [174] | Retrospective, single center study | 86 | (Histogram, texture) | Cox regression analysis | PET/CT | Prognosis |
| Prostate cancer | ||||||
| Chen et al [176] | Retrospective, single center study | 266 + 155 | 781 (first- and second-order, histogram, texture, and Form Factor Parameters) | LASSO | T2WI, ADC | Diagnosis |
| Wang et al [177] | Retrospective, single center study | 54 | 40 (texture features) | RFE-SVM | DCE T1WI DWI T2WI | Diagnosis |
| Algohary et al [178] | Retrospective, single center study | 30 + 15 | 308 (first-order statistics, Gabor, Laws, and Haralick) | QDA ,RF,SVM | T2WI, ADC | Diagnosis |
| Chaddad et al [179] | Retrospective, single center study | 99 | 57 (GLCM,JIM) | Random Forest | ADC T2WI | Diagnosis |
| Shiradkar et al [181] | Retrospective, multi-center study | 70 + 50 | 150 (first-order statistics, Gabor, Laws texture) | A machine-learning classifier | T2WI, ADC | Prognosis |
| Liver and Gastric cancer | ||||||
| Bakr et al [182] | Retrospective, single center study | 28 | 4176 (Intensity, Texture, Shape, Edge) | ICC Lasso | CT | Diagnosis |
| Peng et al [183] | Retrospective, single center study | 184 + 120 | 980 (Shape, Intensity, texture) | Lasso | CT | Diagnosis |
| Ma et al [184] | Retrospective, single center study | 70 | 485 (shape, intensity, textural and wavelet) | LASSO | CT | Diagnosis |
| Ba-Ssalamah et al [185] | Retrospective, single center study | 47 | 30 (histogram, texture) | LDA, K-NN | CT | Diagnosis |
| Zhou et al [190] | Retrospective, single center study | 215 | 300 (histogram, GLCM) | ICC, LASSO | CT | Prognosis |
| Akai et al [191] | Retrospective, single center study | 127 | 96 (filtration, histogram) | Multivariate Cox Random Forest | CT | Prognosis |
| Cozzi et al [192] | Retrospective, single center study | 138 | 35 (shape and size, histogram, second and high order) | Cox | CT | Prognosis |
| Francesco et al [195] | Retrospective, single center study | 56 | 107 (first- and second-order Textural, shape and size) | Cox | CT | Prognosis |
Abbreviations
ADC: apparent diffusion coefficient; AUC: area under receiver operating characteristic curve; C-index: concordance index; CNN: convolutional neural network; CT: Computed Tomography; DFS: Disease-free survival; DWI: diffusion weighted imaging; EGFR: epidermal growth factor receptor; HCC: hepatocellular carcinoma; ICC: Intra-class correlation coefficients; IDH: isocitrate dehydrogenase; KRAS: Kirsten rat sarcoma viral oncogene homolog; LASSO: least absolute shrinkage and selection operator; LBP: local binary pattern; MGMT: O-6-methylguanine-DNA methyltransferase; MiVI: microvascular invasion; MRI: Magnetic Resonance Imaging; NSCLC: non-small cell lung cancer; OS: Overall survival; PET: Positron Emission Tomography; PFS: Progression-free survival; ROC: receiver operating characteristic; ROI: region of interest; RQS: radiomics quality score; SIFT: scale-invariant feature transform; SUV: standard uptake value; TKI: tyrosine kinase inhibitor; US: ultrasound.
Acknowledgements
This paper is supported by the Beijing Natural Science Foundation under Grant No. 7182109, the National Natural Science Foundation of China (Grant Nos. 81772012 and 81227901), the National Key Research and Development Plan of China under Grant Nos. 2017YFA0205200, Chinese Academy of Sciences under Grant No. GJJSTD20170004.The authors would like to acknowledge the instrumental and technical support of Multi-modal biomedical imaging experimental platform, Institute of Automation, Chinese Academy of Sciences.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RG, Granton P. et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48:441-6
2. Kumar V, Gu Y, Basu S, Berglund A, Eschrich SA, Schabath MB. et al. Radiomics: the process and the challenges. Magn Reson Imaging. 2012;30:1234-48
3. Gatenby RA, Grove O, Gillies RJ. Quantitative Imaging in Cancer Evolution and Ecology. Radiology. 2013;269:8-15
4. Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Carvalho S. et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5:4006
5. Aerts HJ. The potential of radiomic-based phenotyping in precision medicine: a review. JAMA Oncol. 2016;2:1636-42
6. Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images Are More than Pictures, They Are Data. Radiology. 2016;278:563-77
7. Huang YQ, Liang CH, He L, Tian J, Liang CS, Chen X. et al. Development and Validation of a Radiomics Nomogram for Preoperative Prediction of Lymph Node Metastasis in Colorectal Cancer. Journal of Clinical Oncology. 2016;34:2157
8. Liu ZY, Zhang XY, Shi YJ, Wang L, Zhu HT, Tang ZC. et al. Radiomics Analysis for Evaluation of Pathological Complete Response to Neoadjuvant Chemoradiotherapy in Locally Advanced Rectal Cancer. Clin Cancer Res. 2017;23:7253-62
9. Sun R, Limkin EJ, Vakalopoulou M, Dercle L, Champiat S, Han SR. et al. A radiomics approach to assess tumour-infiltrating CD8 cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: an imaging biomarker, retrospective multicohort study. Lancet Oncol. 2018;19:1180-91
10. Lambin P, Leijenaar RTH, Deist TM, Peerlings J, de Jong EEC, van Timmeren J. et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14:749-62
11. El Naqa I, Grigsby P, Apte A, Kidd E, Donnelly E, Khullar D. et al. Exploring feature-based approaches in PET images for predicting cancer treatment outcomes. Pattern Recognit. 2009;42:1162-71
12. Leijenaar RT, Carvalho S, Velazquez ER, Van Elmpt WJ, Parmar C, Hoekstra OS. et al. Stability of FDG-PET radiomics features: an integrated analysis of test-retest and inter-observer variability. Acta oncologica. 2013;52:1391-7
13. Sauer R, Becker H, Hohenberger W, Rödel C, Wittekind C, Fietkau R. et al. Preoperative versus postoperative chemoradiotherapy for rectal cancer. New Engl J Med. 2004;351:1731-40
14. He L, Huang Y, Ma Z, Liang C, Liang C, Liu Z. Effects of contrast-enhancement, reconstruction slice thickness and convolution kernel on the diagnostic performance of radiomics signature in solitary pulmonary nodule. Sci Rep. 2016;6:34921
15. Larue RT, Defraene G, De RD, Lambin P, Van EW. Quantitative radiomics studies for tissue characterization: a review of technology and methodological procedures. British Journal of Radiology. 2017;90:20160665
16. Shafiq-Ul-Hassan M, Zhang GG, Latifi K, Ullah G, Hunt DC, Balagurunathan Y. et al. Intrinsic dependencies of CT radiomic features on voxel size and number of gray levels. Medical Physics. 2017;44:1050
17. Shafiqulhassan M, Latifi K, Zhang G, Ullah G, Gillies R, Moros E. Voxel size and gray level normalization of CT radiomic features in lung cancer. Scientific Reports. 2018:8
18. Lu L, Ehmke RC, Schwartz LH, Zhao B. Assessing Agreement between Radiomic Features Computed for Multiple CT Imaging Settings. Plos One. 2016;11:e0166550
19. Zhao B, Tan Y, Tsai WY, Qi J, Xie C, Lu L. et al. Reproducibility of radiomics for deciphering tumor phenotype with imaging. Sci Rep. 2016;6:23428
20. Zhao B, Tan Y, Wei YT, Schwartz LH, Lin L. Exploring Variability in CT Characterization of Tumors: A Preliminary Phantom Study 1. Translational Oncology. 2014;7:88-93
21. Rthm L, van Timmeren JE, Eec DJ, Feliciani G, Rth L, Wmj S. et al. Influence of gray level discretization on radiomic feature stability for different CT scanners, tube currents and slice thicknesses: a comprehensive phantom study. Acta Oncologica. 2017;56:1
22. Mackin D, Fave X, Zhang L, Yang J, Jones AK, Ng CS. et al. Harmonizing the pixel size in retrospective computed tomography radiomics studies. Plos One. 2017;12:e0178524
23. Mackin D, Ger R, Dodge C, Fave X, Chi PC, Zhang L. et al. Effect of tube current on computed tomography radiomic features. Sci Rep. 2018;8:2354
24. Ger RB, Zhou S, Chi P-CM, Lee HJ, Layman RR, Jones AK. et al. Comprehensive investigation on controlling for CT imaging variabilities in radiomics studies. Scientific reports. 2018;8:13047
25. Galavis PE, Hollensen C, Jallow N, Paliwal B, Jeraj R. Variability of textural features in FDG PET images due to different acquisition modes and reconstruction parameters. Acta Oncologica. 2010;49:1012-6
26. Jianhua Y, Jason Lim CS, Hoi Yin L, Lih Kin K, Sinha AK, Swee Tian Q. et al. Impact of Image Reconstruction Settings on Texture Features in 18F-FDG PET. Journal of Nuclear Medicine Official Publication Society of Nuclear Medicine. 2015;56:1667
27. Velden FHPV, Kramer GM, Frings V, Nissen IA, Mulder ER, Langen AJD. et al. Repeatability of Radiomic Features in Non-Small-Cell Lung Cancer [ 18 F]FDG-PET/CT Studies: Impact of Reconstruction and Delineation. Molecular Imaging & Biology. 2016;18:788-95
28. Gallivanone F, Interlenghi M, D'Ambrosio D, Trifirò G, Castiglioni I. Parameters Influencing PET Imaging Features: A Phantom Study with Irregular and Heterogeneous Synthetic Lesions. Contrast media & molecular imaging. 2018. 2018
29. Oliver JA, Budzevich M, Zhang GG, Dilling TJ, Latifi K, Moros EG. Variability of Image Features Computed from Conventional and Respiratory-Gated PET/CT Images of Lung Cancer. Translational Oncology. 2015;8:524-34
30. Grootjans W, Tixier F, Cs VDV, Vriens D, Le RC, Bussink J. et al. The Impact of Optimal Respiratory Gating and Image Noise on Evaluation of Intratumor Heterogeneity on 18F-FDG PET Imaging of Lung Cancer. Journal of Nuclear Medicine Official Publication Society of Nuclear Medicine. 2017;57:1692
31. Yip S, Mccall K, Aristophanous M, Chen AB, Aerts HJ, Berbeco R. Comparison of Texture Features Derived from Static and Respiratory-Gated PET Images in Non-Small Cell Lung Cancer. Plos One. 2015;9:e115510
32. Carles M, Bach T, Torresespallardo I, Baltas D, Nestle U, Martíbonmatí L. Significance of the impact of motion compensation on the variability of PET image features. Physics in Medicine & Biology. 2018:63
33. Herlidou-Même S, Constans JM, Carsin B, Olivie D, Eliat PA, Nadal-Desbarats L. et al. MRI texture analysis on texture test objects, normal brain and intracranial tumors. Magnetic Resonance Imaging. 2003;21:989-93
34. Jirak D, Dezortova MM. Phantoms for texture analysis of MR images. Long-term and multi-center study. Medical Physics. 2004;31:616
35. Mayerhoefer ME, Szomolanyi P, Jirak D, Materka A, Trattnig S. Effects of MRI acquisition parameter variations and protocol heterogeneity on the results of texture analysis and pattern discrimination: an application-oriented study. Medical Physics. 2009;36:1236
36. Savio SJ, Harrison LC, Luukkaala T, Heinonen T, Dastidar P, Soimakallio S. et al. Effect of slice thickness on brain magnetic resonance image texture analysis. Biomedical engineering online. 2010;9:60
37. Waugh SA, Lerski RA, Bidaut L, Thompson AM. The influence of field strength and different clinical breast MRI protocols on the outcome of texture analysis using foam phantoms. Medical Physics. 2011;38:5058
38. Yang F, Dogan N, Stoyanova R, Ford JC. Evaluation of radiomic texture feature error due to MRI acquisition and reconstruction: A simulation study utilizing ground truth. Physica Medica. 2018;50:26-36
39. John F, Nesrin D, Lori Y, Fei Y. Quantitative Radiomics: Impact of Pulse Sequence Parameter Selection on MRI-Based Textural Features of the Brain. Contrast Media Mol Imaging. 2018;2018:1-9
40. Waugh SA, Purdie CA, Jordan LB, Vinnicombe S, Lerski RA, Martin P. et al. Magnetic resonance imaging texture analysis classification of primary breast cancer. European Radiology. 2016;26:322-30
41. Sled JG, Zijdenbos AP, Evans AC. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Transactions on Medical Imaging. 1998;17:87-97
42. Tustison NJ, Avants BB, Cook PA, Zheng Y, Egan A, Yushkevich PA. et al. N4ITK: improved N3 bias correction. IEEE transactions on medical imaging. 2010;29:1310-20
43. Avants BB, Tustison NJ, Song G, Cook PA, Klein A, Gee JC. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage. 2011;54:2033-44
44. Shinohara RT, Sweeney EM, Goldsmith J, Shiee N, Mateen FJ, Calabresi PA. et al. Normalization Techniques for Statistical Inference from Magnetic Resonance Imaging. Bepress. 2013
45. Shinohara RT, Sweeney EM, Goldsmith J, Shiee N, Mateen FJ, Calabresi PA. et al. Statistical normalization techniques for magnetic resonance imaging. NeuroImage: Clinical. 2014;6:9-19
46. Guo Y, Hu Y, Qiao M, Wang Y, Yu J, Li J. et al. Radiomics analysis on ultrasound for prediction of biologic behavior in breast invasive ductal carcinoma. Clin Breast Cancer. 2018;18:e335-e44
47. Cobo T, Bonet-Carne E, Martínez-Terrón M, Perez-Moreno A, Elías N, Luque J. et al. Feasibility and reproducibility of fetal lung texture analysis by automatic quantitative ultrasound analysis and correlation with gestational age. Fetal diagnosis and therapy. 2012;31:230-6
48. Bonet-Carne E, Palacio M, Cobo T, Perez-Moreno A, Lopez M, Piraquive JP. et al. Quantitative ultrasound texture analysis of fetal lungs to predict neonatal respiratory morbidity. Ultrasound Obstet Gynecol. 2015;45:427-33
49. Wu MH, Chen CN, Chen KY, Ho MC, Tai HC, Wang YH. et al. Quantitative analysis of echogenicity for patients with thyroid nodules. Scientific Reports. 2016;6:35632
50. Gesheng S, Fuzhong X, Chengqi Z. A Model Using Texture Features to Differentiate the Nature of Thyroid Nodules on Sonography. Journal of Ultrasound in Medicine Official Journal of the American Institute of Ultrasound in Medicine. 2015;34:1753-60
51. Woo Kyung M, Yao-Sian H, Chung-Ming L, Chiun-Sheng H, Sun BM, Won Hwa K. et al. Computer-aided diagnosis for distinguishing between triple-negative breast cancer and fibroadenomas based on ultrasound texture features. Medical Physics. 2015;42:3024-35
52. Ali Abbasian A, Akbar G, Afshin M. Classification of breast tumors using sonographic texture analysis. Journal of Ultrasound in Medicine Official Journal of the American Institute of Ultrasound in Medicine. 2015;34:225-31
53. Kyriacou EC, Constantinos P, Marios P, Christos L, Christodoulos C, Kakkos SK. et al. A review of noninvasive ultrasound image processing methods in the analysis of carotid plaque morphology for the assessment of stroke risk. IEEE Transactions on Information Technology in Biomedicine A Publication of the IEEE Engineering in Medicine & Biology Society. 2010;14:1027
54. Armato SG, McLennan G, Bidaut L, McNitt-Gray MF, Meyer CR, Reeves AP. et al. The Lung Image Database Consortium, (LIDC) and Image Database Resource Initiative (IDRI): A Completed Reference Database of Lung Nodules on CT Scans. Medical Physics. 2011;38:915-31
55. Parmar C, Velazquez ER, Leijenaar R, Jermoumi M, Carvalho S, Mak RH. et al. Robust radiomics feature quantification using semiautomatic volumetric segmentation. PloS one. 2014;9:e102107
56. Tobias H, Merkle EM, Reiner CS, Davenport MS, Horvath JJ, Sebastian F. et al. Reproducibility of dynamic contrast-enhanced MR imaging. Part II. Comparison of intra- and interobserver variability with manual region of interest placement versus semiautomatic lesion segmentation and histogram analysis. Radiology. 2013;266:812-21
57. Christ MJ, Parvathi R. Segmentation of medical image using clustering and watershed algorithms. American Journal of Applied Sciences. 2011;8:1349
58. Hatt M, Lee JA, Schmidtlein CR, Naqa IE, Caldwell C, De Bernardi E. et al. Classification and evaluation strategies of auto-segmentation approaches for PET: Report of AAPM task group No. 211. Medical physics. 2017:44
59. Song JD, Yang CY, Fan L, Wang K, Yang F, Liu SY. et al. Lung Lesion Extraction Using a Toboggan Based Growing Automatic Segmentation Approach. Ieee Transactions on Medical Imaging. 2016;35:337-53
60. Polan D, Brady S, Kaufman R. Tissue Segmentation of Computed Tomography Images Using a Random Forest Algorithm: A Feasibility Study. Medical Physics. 2016;43:3330-1
61. Wang S, Zhou M, Liu ZY, Liu ZY, Gu DS, Zang YL. et al. Central focused convolutional neural networks: Developing a data-driven model for lung nodule segmentation. Med Image Anal. 2017;40:172-83
62. Liu Y, Kim J, Qu F, Liu S, Wang H, Balagurunathan Y. et al. CT Features Associated with Epidermal Growth Factor Receptor Mutation Status in Patients with Lung Adenocarcinoma. Radiology. 2016;280:271-80
63. Chicklore S, Goh V, Siddique M, Roy A, Marsden PK, Cook GJ. Quantifying tumour heterogeneity in 18F-FDG PET/CT imaging by texture analysis. Eur J Nucl Med Mol Imaging. 2013;40:133-40
64. Parekh V, Jacobs MA. Radiomics: a new application from established techniques. Expert Rev Precis Med Drug Dev. 2016;1:207-26
65. Scalco E, Rizzo G. Texture analysis of medical images for radiotherapy applications. Br J Radiol. 2017;90:20160642
66. Ojala T, Pietikainen M, Maenpaa T. Multiresolution gray-scale and rotation invariant texture classification with local binary patterns. Ieee T Pattern Anal. 2002;24:971-87
67. Ambrosini RD, Wang P, O'Dell WG. Computer-Aided Detection of Metastatic Brain Tumors Using Automated Three-Dimensional Template Matching. Journal of Magnetic Resonance Imaging. 2010;31:85-93
68. LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436-44
69. Hu Z, Tang J, Wang Z, Kai Z, Lin Z, Sun Q. Deep Learning for Image-based Cancer Detection and Diagnosis — A Survey. Pattern Recogn. 2018;83:134-49
70. Yu DD, Zhou M, Yang F, Dong D, Gevaert O, Liu ZY. et al. Convolutional Neural Networks for Predicting Molecular Profiles of Non-Small Cell Lung Cancer. 2017 Ieee 14th International Symposium on Biomedical Imaging (Isbi 2017). 2017:569-72
71. Chaudhary K, Poirion OB, Lu L, Garmire LX. Deep Learning-Based Multi-Omics Integration Robustly Predicts Survival in Liver Cancer. Clin Cancer Res. 2018;24:1248-59
72. Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M. et al. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42:60-88
73. Rios VE, Parmar C, Liu Y, Coroller TP, Cruz G, Stringfield O. et al. Somatic Mutations Drive Distinct Imaging Phenotypes in Lung Cancer. Cancer research. 2017;77:3922-30
74. Bagherzadeh-Khiabani F, Ramezankhani A, Azizi F, Hadaegh F, Steyerberg EW, Khalili D. A tutorial on variable selection for clinical prediction models: feature selection methods in data mining could improve the results. J Clin Epidemiol. 2016;71:76-85
75. Balagurunathan Y, Gu Y, Wang H, Kumar V, Grove O, Hawkins S. et al. Reproducibility and Prognosis of Quantitative Features Extracted from CT Images. Transl Oncol. 2014;7:72-87
76. Chamming's F, Ueno Y, Ferre R, Kao E, Jannot AS, Chong J. et al. Features from Computerized Texture Analysis of Breast Cancers at Pretreatment MR Imaging Are Associated with Response to Neoadjuvant Chemotherapy. Radiology. 2018;286:412-20
77. Liu Z, Wang Y, Liu X, Du Y, Tang Z, Wang K. et al. Radiomics analysis allows for precise prediction of epilepsy in patients with low-grade gliomas. Neuroimage Clin. 2018;19:271-8
78. Guo J, Liu Z, Shen C, Li Z, Yan F, Tian J. et al. MR-based radiomics signature in differentiating ocular adnexal lymphoma from idiopathic orbital inflammation. Eur Radiol. 2018;28:3872-81
79. Tang Z, Liu Z, Li R, Yang X, Cui X, Wang S. et al. Identifying the white matter impairments among ART-naive HIV patients: a multivariate pattern analysis of DTI data. Eur Radiol. 2017;27:4153-62
80. Shen C, Liu Z, Guan M, Song J, Lian Y, Wang S. et al. 2D and 3D CT Radiomics Features Prognostic Performance Comparison in Non-Small Cell Lung Cancer. Transl Oncol. 2017;10:886-94
81. Shen C, Liu Z, Wang Z, Guo J, Zhang H, Wang Y. et al. Building CT Radiomics Based Nomogram for Preoperative Esophageal Cancer Patients Lymph Node Metastasis Prediction. Transl Oncol. 2018;11:815-24
82. Arthur D, Vassilvitskii S. k-means plus plus: The Advantages of Careful Seeding. Proceedings of the Eighteenth Annual Acm-Siam Symposium on Discrete Algorithms. 2007:1027-35
83. Clark MC, Hall LO, Goldgof DB, Velthuizen R, Murtagh FR, Silbiger MS. Automatic tumor segmentation using knowledge-based techniques. Ieee Transactions on Medical Imaging. 1998;17:187-201
84. Monti S, Tamayo P, Mesirov J, Golub T. Consensus clustering: A resampling-based method for class discovery and visualization of gene expression microarray data. Mach Learn. 2003;52:91-118
85. Itakura H, Achrol AS, Mitchell LA, Loya JJ, Liu T, Westbroek EM. et al. Magnetic resonance image features identify glioblastoma phenotypic subtypes with distinct molecular pathway activities. Sci Transl Med. 2015:7
86. Wang S, Liu Z, Rong Y, Zhou B, Bai Y, Wei W. et al. Deep learning provides a new computed tomography-based prognostic biomarker for recurrence prediction in high-grade serous ovarian cancer. Radiother Oncol. 2018
87. Kramer AA, Zimmerman JE. Assessing the calibration of mortality benchmarks in critical care: The Hosmer-Lemeshow test revisited. Crit Care Med. 2007;35:2052-6
88. Brandao LA. Adult Brain Tumors: Imaging Characterization of Primary, Secondary, and Extraparenchymal Tumors in the Central Nervous System, Including Findings on Advanced MR Imaging Techniques as well as Treatment-Related Abnormalities. Neuroimaging Clin N Am. 2016;26:xiii-xiv
89. Bai Y, Lin YS, Tian J, Shi DP, Cheng JL, Haacke EM. et al. Grading of Gliomas by Using Monoexponential, Biexponential, and Stretched Exponential Diffusion-weighted MR Imaging and Diffusion Kurtosis MR Imaging. Radiology. 2016;278:496-504
90. Kickingereder P, Burth S, Wick A, Gotz M, Eidel O, Schlemmer HP. et al. Radiomic Profiling of Glioblastoma: Identifying an Imaging Predictor of Patient Survival with Improved Performance over Established Clinical and Radiologic Risk Models. Radiology. 2016;280:880-9
91. Zhou H, Vallieres M, Bai HX, Su C, Tang H, Oldridge D. et al. MRI features predict survival and molecular markers in diffuse lower-grade gliomas. Neuro Oncol. 2017;19:862-70
92. Xi YB, Guo F, Xu ZL, Li C, Wei W, Tian P. et al. Radiomics signature: A potential biomarker for the prediction of MGMT promoter methylation in glioblastoma. J Magn Reson Imaging. 2018;47:1380-7
93. Kickingereder P, Neuberger U, Bonekamp D, Piechotta PL, Gotz M, Wick A. et al. Radiomic subtyping improves disease stratification beyond key molecular, clinical, and standard imaging characteristics in patients with glioblastoma. Neuro Oncol. 2018;20:848-57
94. Han Y, Xie Z, Zang Y, Zhang S, Gu D, Zhou M. et al. Non-invasive genotype prediction of chromosome 1p/19q co-deletion by development and validation of an MRI-based radiomics signature in lower-grade gliomas. J Neuro-Oncol. 2018;140:297-306
95. Li YM, Liu X, Xu KB, Qian ZH, Wang K, Fan X. et al. MRI features can predict EGFR expression in lower grade gliomas: A voxel-based radiomic analysis. European Radiology. 2018;28:356-62
96. Li YM, Qian ZH, Xu KB, Wang K, Fan X, Li SW. et al. Radiomic features predict Ki-67 expression level and survival in lower grade gliomas. J Neuro-Oncol. 2017;135:317-24
97. Li Y, Qian Z, Xu K, Wang K, Fan X, Li S. et al. MRI features predict p53 status in lower-grade gliomas via a machine-learning approach. Neuroimage Clin. 2018;17:306-11
98. Li YM, Liu X, Qian ZH, Sun ZY, Xu KB, Wang K. et al. Genotype prediction of ATRX mutation in lower-grade gliomas using an MRI radiomics signature. European Radiology. 2018;28:2960-8
99. Kickingereder P, Gotz M, Muschelli J, Wick A, Neuberger U, Shinohara RT. et al. Large-scale Radiomic Profiling of Recurrent Glioblastoma Identifies an Imaging Predictor for Stratifying Anti-Angiogenic Treatment Response. Clinical Cancer Research. 2016;22:5765-71
100. Grossmann P, Narayan V, Chang K, Rahman R, Abrey L, Reardon DA. et al. Quantitative imaging biomarkers for risk stratification of patients with recurrent glioblastoma treated with bevacizumab. Neuro Oncol. 2017;19:1688-97
101. Papp L, Potsch N, Grahovac M, Schmidbauer V, Woehrer A, Preusser M. et al. Glioma Survival Prediction with Combined Analysis of In Vivo C-11-MET PET Features, Ex Vivo Features, and Patient Features by Supervised Machine Learning. J Nucl Med. 2018;59:892-9
102. Perez-Beteta J, Molina-Garcia D, Ortiz-Alhambra JA, Fernandez-Romero A, Luque B, Arregui E. et al. Tumor Surface Regularity at MR Imaging Predicts Survival and Response to Surgery in Patients with Glioblastoma. Radiology. 2018;288:218-25
103. Jethanandani A, Lin TA, Volpe S, Elhalawani H, Mohamed ASR, Yang P. et al. Exploring Applications of Radiomics in Magnetic Resonance Imaging of Head and Neck Cancer: A Systematic Review. Frontiers in Oncology. 2018:8
104. Ren J, Tian J, Yuan Y, Dong D, Li X, Shi Y. et al. Magnetic resonance imaging based radiomics signature for the preoperative discrimination of stage I-II and III-IV head and neck squamous cell carcinoma. European journal of radiology. 2018;106:1-6
105. Rth L, Bogowicz M, Jochems A, Fjp H, Fwr F, Huang SH. et al. Development and validation of a radiomic signature to predict HPV (p16) status from standard CT imaging: a multicenter study. British Journal of Radiology. 2018;91:20170498
106. Zhou Z, Chen L, Sher D, Zhang Q, Shah J, Pham N-L. et al. Predicting Lymph Node Metastasis in Head and Neck Cancer by Combining Many-objective Radiomics and 3-dimensioal Convolutional Neural Network through Evidential Reasoning. Conf Proc IEEE Eng Med Biol Soc. 2018:1-4
107. Chen RY, Lin YC, Shen WC, Hsieh TC, Yen KY, Chen SW. et al. Associations of Tumor PD-1 Ligands, Immunohistochemical Studies, and Textural Features in 18 F-FDG PET in Squamous Cell Carcinoma of the Head and Neck. Sci Rep. 2018;8:105
108. Crispin-Ortuzar M, Apte A, Grkovski M, Oh JH, Lee NY, Schöder H. et al. Predicting hypoxia status using a combination of contrast-enhanced computed tomography and [18F]-Fluorodeoxyglucose positron emission tomography radiomics features. Radiother Oncol. 2018;127:36-42
109. Zhang B, Tian J, Dong D, Gu D, Dong Y, Zhang L. et al. Radiomics Features of Multiparametric MRI as Novel Prognostic Factors in Advanced Nasopharyngeal Carcinoma. Clinical Cancer Research An Official Journal of the American Association for Cancer Research. 2017;23:4259
110. Zhang B, He X, Ouyang F, Gu D, Dong Y, Zhang L. et al. Radiomic machine-learning classifiers for prognostic biomarkers of advanced nasopharyngeal carcinoma. Cancer Letters. 2017;403:21
111. Wang G, He L, Yuan C, Huang Y, Liu Z, Liang C. Pretreatment MR imaging radiomics signatures for response prediction to induction chemotherapy in patients with nasopharyngeal carcinoma. European Journal of Radiology. 2018;98:100-6
112. Lu L, Lv W, Jiang J, Ma J, Feng Q, Rahmim A. et al. Robustness of Radiomic Features in [ 11 C]Choline and [ 18 F]FDG PET/CT Imaging of Nasopharyngeal Carcinoma: Impact of Segmentation and Discretization. Molecular Imaging and Biology. 2016;18:935-45
113. Lv W, Yuan Q, Wang Q, Ma J, Jiang J, Wei Y. et al. Robustness versus disease differentiation when varying parameter settings in radiomics features: application to nasopharyngeal PET/CT. European Radiology. 2018;28:1-10
114. Wu Y, Xu L, Yang P, Lin N, Huang X, Pan W. et al. Survival Prediction in High-grade Osteosarcoma Using Radiomics of Diagnostic Computed Tomography. EBioMedicine. 2018;34:27-34
115. Elhalawani H, Kanwar A, Mohamed ASR, White A, Zafereo J, Wong A. et al. Investigation of radiomic signatures for local recurrence using primary tumor texture analysis in oropharyngeal head and neck cancer patients. Scientific Reports. 2018:8
116. Vallières M, Kayrivest E, Perrin LJ, Liem X, Furstoss C, Aerts HJWL. et al. Radiomics strategies for risk assessment of tumour failure in head-and-neck cancer. Scientific Reports. 2017;7:10117
117. Gabryś HS, Buettner F, Sterzing F. Design and Selection of Machine Learning Methods Using Radiomics and Dosiomics for Normal Tissue Complication Probability Modeling of Xerostomia. Frontiers in Oncology. 2018:8
118. Antropova N, Huynh BQ, Giger ML. A Deep Feature Fusion Methodology for Breast Cancer Diagnosis Demonstrated on Three Imaging Modality Datasets. Medical Physics. 2017;44:5162
119. Antunovic L, Gallivanone F, Sollini M, Sagona A, Invento A, Manfrinato G. et al. [ 18 F]FDG PET/CT features for the molecular characterization of primary breast tumors. European Journal of Nuclear Medicine & Molecular Imaging. 2017;44:1945-54
120. Ha S, Park S, Bang JI, Kim EK, Lee HY. Metabolic Radiomics for Pretreatment 18 F-FDG PET/CT to Characterize Locally Advanced Breast Cancer: Histopathologic Characteristics, Response to Neoadjuvant Chemotherapy, and Prognosis. Scientific Reports. 2017;7:1556
121. Guo W, Li H, Zhu Y, Lan L, Yang S, Drukker K. et al. Prediction of clinical phenotypes in invasive breast carcinomas from the integration of radiomics and genomics data. Journal of Medical Imaging. 2015;2:041007
122. Saha A, Harowicz MR, Grimm LJ, Kim CE, Ghate SV, Walsh R. et al. A machine learning approach to radiogenomics of breast cancer: a study of 922 subjects and 529 DCE-MRI features. Br J Cancer. 2018;119:508
123. Dong Y, Feng Q, Yang W, Lu Z, Deng C, Zhang L. et al. Preoperative prediction of sentinel lymph node metastasis in breast cancer based on radiomics of T2-weighted fat-suppression and diffusion-weighted MRI. European Radiology. 2018;28:582-91
124. Chan H, Bhm VDV, Loo CE, Kga G. Eigentumors for prediction of treatment failure in patients with early-stage breast cancer using dynamic contrast-enhanced MRI: a feasibility study. Physics in Medicine & Biology. 2017:62
125. Braman NM, Etesami M, Prasanna P, Dubchuk C, Gilmore H, Tiwari P. et al. Intratumoral and peritumoral radiomics for the pretreatment prediction of pathological complete response to neoadjuvant chemotherapy based on breast DCE-MRI. Breast Cancer Research. 2017;19:57
126. Partridge SC, Zhang Z, Newitt DC, Gibbs JE, Chenevert TL, Rosen MA. et al. Diffusion-weighted MRI findings predict pathologic response in neoadjuvant treatment of breast cancer: the ACRIN 6698 Multicenter Trial. Radiology. 2018. 1802 73
127. Tran WT, Gangeh MJ, Sannachi L, Chin L, Watkins E, Bruni SG. et al. Predicting breast cancer response to neoadjuvant chemotherapy using pretreatment diffuse optical spectroscopic texture analysis. British Journal of Cancer. 2017;116:1329-39
128. Park H, Lim Y, Ko ES, Cho H-h, Lee JE, Han B-K. et al. Radiomics Signature on Magnetic Resonance Imaging: Association with Disease-Free Survival in Patients with Invasive Breast Cancer. Clinical Cancer Research. 2018 clincanres. 3783.2017
129. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA: A Cancer Journal for Clinicians. 2017;67:7-30
130. Shaikh FA, Kolowitz BJ, Awan O, Aerts HJ, Reden Av, Halabi S. et al. Technical Challenges in the Clinical Application of Radiomics. JCO Clinical Cancer Informatics. 2017:1-8
131. Limkin EJ, Sun R, Dercle L, Zacharaki EI, Robert C, Reuze S. et al. Promises and challenges for the implementation of computational medical imaging (radiomics) in oncology. Ann Oncol. 2017;28:1191-206
132. Kumar D, Shafiee MJ, Chung AG, Khalvati F, Haider MA, Wong A. Discovery radiomics for computed tomography cancer detection. Cornell University Library. 2015
133. Shen W, Zhou M, Yang F, Yu D, Dong D, Yang C. et al. Multi-crop Convolutional Neural Networks for lung nodule malignancy suspiciousness classification. Pattern Recognition. 2017;61:663-73
134. Maldonado F, Boland JM, Raghunath S, Aubry MC, Bartholmai BJ, Deandrade M. et al. Non-invasive Characterization of the Histopathologic Features of Pulmonary Nodules of the Lung Adenocarcinoma Spectrum using Computer Aided Nodule Assessment and Risk Yield (CANARY) - a Pilot Study. Journal of Thoracic Oncology. 2013;8:452-60
135. Carter BW, Godoy MC, Erasmus JJ. Predicting Malignant Nodules from Screening CTs. Journal of Thoracic Oncology. 2016;11:2045-7
136. Liu Y, Balagurunathan Y, Atwater T, Antic S, Li Q, Walker RC. et al. Radiological Image traits Predictive of Cancer Status in Pulmonary Nodules. Clinical Cancer Research. 2016;23:1442-9
137. Parmar C, Leijenaar RT, Grossmann P, Rios VE, Bussink J, Rietveld D. et al. Radiomic feature clusters and prognostic signatures specific for Lung and Head & Neck cancer. Scientific Reports. 2015;5:11044
138. Coroller TP, Grossmann P, Hou Y, Velazquez ER, Leijenaar RTH, Hermann G. et al. CT-based radiomic signature predicts distant metastasis in lung adenocarcinoma. Radiotherapy & Oncology. 2015;114:345-50
139. Zhou H, Dong D, Chen B, Fang M, Cheng Y, Gan Y. et al. Diagnosis of Distant Metastasis of Lung Cancer: Based on Clinical and Radiomic Features. Translational Oncology. 2018;11:31-6
140. Jia W, Aguilera T, Shultz D, Gudur M, Rubin DL Jr BWL. et al. Early-Stage Non-Small Cell Lung Cancer: Quantitative Imaging Characteristics of 18F Fluorodeoxyglucose PET/CT Allow Prediction of Distant Metastasis. Radiology. 2016;281:151829
141. Liu Y, Kim J, Balagurunathan Y, Li Q, Garcia AL, Stringfield O. et al. Radiomic Features Are Associated With EGFR Mutation Status in Lung Adenocarcinomas. Clinical Lung Cancer. 2016;17:441-8.e6
142. Liu Y, Kim J, Qu F, Liu S, Wang H, Balagurunathan Y. et al. CT Features Associated with Epidermal Growth Factor Receptor Mutation Status in Patients with Lung Adenocarcinoma. Radiology. 2016;280:151455
143. Zhang L, Chen B, Liu X, Song J, Fang M, Hu C. et al. Quantitative Biomarkers for Prediction of Epidermal Growth Factor Receptor Mutation in Non-Small Cell Lung Cancer. Translational Oncology. 2018;11:94-101
144. Wu W, Parmar C, Grossmann P, Quackenbush J, Lambin P, Bussink J. et al. Exploratory Study to Identify Radiomics Classifiers for Lung Cancer Histology. Frontiers in Oncology. 2016;6:71
145. Zhu X, Dong D, Chen Z, Fang M, Zhang L, Song J. et al. Radiomic signature as a diagnostic factor for histologic subtype classification of non-small cell lung cancer. European Radiology. 2018:1-7
146. Fan L, Fang MJ, Li ZB, Tu WT, Wang SP, Chen WF. et al. Radiomics signature: a biomarker for the preoperative discrimination of lung invasive adenocarcinoma manifesting as a ground-glass nodule. European Radiology. 2018:1-9
147. Mattonen SA, Tetar S, Palma DA, Senan S, Ward AD. Automated Texture Analysis for Prediction of Recurrence After Stereotactic Ablative Radiation Therapy for Lung Cancer. International Journal of Radiation Oncology Biology Physics. 2015;93:S5-S6
148. Mattonen SA, Palma DA, Haasbeek CJ, Senan S, Ward AD. Early prediction of tumor recurrence based on CT texture changes after stereotactic ablative radiotherapy (SABR) for lung cancer. Medical Physics. 2014;41:033502
149. Fave X, Zhang L, Yang J, Mackin D, Balter P, Gomez D. et al. Delta-radiomics features for the prediction of patient outcomes in non-small cell lung cancer. Scientific Reports. 2017;7:588
150. Coroller TP, Agrawal V, Narayan V, Hou Y, Grossmann P, Lee SW. et al. Radiomic phenotype features predict pathological response in non-small cell lung cancer. Radiother Oncol. 2016;119:480-6
151. Aerts HJWL, Patrick G, Tan Y, Oxnard GG, Naiyer R, Schwartz LH. et al. Defining a Radiomic Response Phenotype: A Pilot Study using targeted therapy in NSCLC. Scientific Reports. 2016;6:33860
152. Cook GJ, O'Brien ME, Siddique M, Chicklore S, Loi HY, Sharma B. et al. Non-Small Cell Lung Cancer Treated with Erlotinib: Heterogeneity of (18)F-FDG Uptake at PET-Association with Treatment Response and Prognosis. Radiology. 2015;276:883-93
153. Song J, Shi J, Dong D, Fang M, Zhong W, Wang K. et al. A new approach to predict progression-free survival in stage IV EGFR-mutant NSCLC patients with EGFR-TKI therapy. Clinical Cancer Research. 2018
154. Song J, Liu Z, Zhong W, Huang Y, Ma Z, Dong D. et al. Non-small cell lung cancer: quantitative phenotypic analysis of CT images as a potential marker of prognosis. Scientific Reports. 2016;6:38282
155. Huang Y, Liu Z, He L, Chen X, Pan D, Ma Z. et al. Radiomics Signature: A Potential Biomarker for the Prediction of Disease-Free Survival in Early-Stage (I or II) Non-Small Cell Lung Cancer. Radiology. 2016;281:947
156. Chang GJ, Rodriguezbigas MA, Skibber JM, Moyer VA. Lymph node evaluation and survival after curative resection of colon cancer: systematic review. J Natl Cancer Inst. 2004;99:433-41
157. Glasgow SC, Bleier JI, Burgart LJ, Finne CO, Lowry AC. Meta-analysis of histopathological features of primary colorectal cancers that predict lymph node metastases. Journal of Gastrointestinal Surgery Official Journal of the Society for Surgery of the Alimentary Tract. 2012;16:1019
158. Toiyama Y, Inoue Y, Shimura T, Fujikawa H, Saigusa S, Hiro J. et al. Serum Angiopoietin-like Protein 2 Improves Preoperative Detection of Lymph Node Metastasis in Colorectal Cancer. Anticancer Research. 2015;35:2849-56
159. Van CE, Lenz HJ, Köhne CH, Heinemann V, Tejpar S, Melezínek I. et al. Fluorouracil, leucovorin, and irinotecan plus cetuximab treatment and RAS mutations in colorectal cancer. Journal of Clinical Oncology. 2015;33:692-700
160. Peeters M, Oliner KS, Price TJ, Cervantes A, Sobrero AF, Ducreux M. et al. Analysis of KRAS/NRAS Mutations in a Phase III Study of Panitumumab with FOLFIRI Compared with FOLFIRI Alone as Second-line Treatment for Metastatic Colorectal Cancer. Clin Cancer Res. 2015;21:5469-79
161. Barras D, Missiaglia E, Wirapati P, Sieber OM, Jorissen RN, Love C. et al. BRAF V600E Mutant Colorectal Cancer Subtypes Based on Gene Expression. Clinical Cancer Research An Official Journal of the American Association for Cancer Research. 2017;23:104
162. Douillard JY, Oliner KS, Siena S, Tabernero J, Burkes R, Barugel M. et al. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. New Engl J Med. 2013;369:1023-34
163. Yang L, Dong D, Fang M, Zhu Y, Zang Y, Liu Z. et al. Can CT-based radiomics signature predict KRAS/NRAS/BRAF mutations in colorectal cancer? European radiology. 2018;28:2058-67
164. Cj VDV, Boelens PG, Borras JM, Coebergh JW, Cervantes A, Blomqvist L. et al. EURECCA colorectal: multidisciplinary management: European consensus conference colon & rectum. European Journal of Cancer. 2014;50:1.e-e34
165. Van GW, Marijnen CA, Nagtegaal ID, Kranenbarg EM, Putter H, Wiggers T. et al. Preoperative radiotherapy combined with total mesorectal excision for resectable rectal cancer: 12-year follow-up of the multicentre, randomised controlled TME trial. Lancet Oncol. 2011;12:575-82
166. Sanghera P, Wong DW, Mcconkey CC, Geh JI, Hartley A. Chemoradiotherapy for rectal cancer: an updated analysis of factors affecting pathological response. Clinical Oncology. 2008;20:176-83
167. Maas M, Nelemans PV, Das P, Rodel C, Kuo LJ, Calvo FA. et al. Long-term outcome in patients with a pathological complete response after chemoradiation for rectal cancer: a pooled analysis of individual patient data. Lancet Oncology. 2010;11:835-44
168. Maas M, Beets-Tan RG, Lambregts DM, Lammering G, Nelemans PJ, Engelen SM. et al. Wait-and-see policy for clinical complete responders after chemoradiation for rectal cancer. Coloproctology. 2012;34:375-6
169. Renehan AG, Malcomson L, Emsley R, Gollins S, Maw A, Myint AS. et al. Watch-and-wait approach versus surgical resection after chemoradiotherapy for patients with rectal cancer (the OnCoRe project): a propensity-score matched cohort analysis. Lancet Oncology. 2016;17:174-83
170. Marijnen CA. Organ preservation in rectal cancer: have all questions been answered? Lancet Oncology. 2015;16:e13-e22
171. Nie K, Shi L, Chen Q, Hu X, Jabbour SK, Yue N. et al. Rectal Cancer: Assessment of Neoadjuvant Chemoradiation Outcome based on Radiomics of Multiparametric MRI. Clinical Cancer Research. 2016;22:5256-64
172. Horvat N, Veeraraghavan H, Khan M, Blazic I, Zheng J, Capanu M. et al. MR Imaging of Rectal Cancer: Radiomics Analysis to Assess Treatment Response after Neoadjuvant Therapy. Radiology. 2018. 1723 00
173. Rodel C, Liersch T, Becker H, Fietkau R, Hohenberger W, Hothorn T. et al. Preoperative chemoradiotherapy and postoperative chemotherapy with fluorouracil and oxaliplatin versus fluorouracil alone in locally advanced rectal cancer: initial results of the German CAO/ARO/AIO-04 randomised phase 3 trial. Lancet Oncology. 2012;13:679-87
174. Bosset JF, Collette L, Calais G, Mineur L, Maingon P, Radosevic-Jelic L. et al. Chemotherapy with preoperative radiotherapy in rectal cancer. N Engl J Med. 2006;355:1114-23
175. Meng Y, Zhang Y, Dong D, Li C, Liang X, Zhang C. et al. Novel radiomic signature as a prognostic biomarker for locally advanced rectal cancer. Journal of Magnetic Resonance Imaging. 2018
176. Lovinfosse P, Polus M, Van DD, Martinive P, Daenen F, Hatt M. et al. FDG PET/CT radiomics for predicting the outcome of locally advanced rectal cancer. Eur J Nucl Med Mol Imaging. 2018;45:365-75
177. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7-30
178. Chen T, Li M, Gu Y, Zhang Y, Yang S, Wei C. et al. Prostate Cancer Differentiation and Aggressiveness: Assessment With a Radiomic-Based Model vs. PI-RADS v2. J Magn Reson Imaging. 2018
179. Wang J, Wu CJ, Bao ML, Zhang J, Wang XN, Zhang YD. Machine learning-based analysis of MR radiomics can help to improve the diagnostic performance of PI-RADS v2 in clinically relevant prostate cancer. European Radiology. 2017;27:4082-90
180. Algohary A, Viswanath S, Shiradkar R, Ghose S, Pahwa S, Moses D. et al. Radiomic features on MRI enable risk categorization of prostate cancer patients on active surveillance: Preliminary findings. J Magn Reson Imaging. 2018;48:818-28
181. Chaddad A, Kucharczyk MJ, Niazi T. Multimodal Radiomic Features for the Predicting Gleason Score of Prostate Cancer. Cancers. 2018:10
182. Lin YC, Lin G, Hong JH, Lin YP, Chen FH, Ng SH. et al. Diffusion radiomics analysis of intratumoral heterogeneity in a murine prostate cancer model following radiotherapy: Pixelwise correlation with histology. Journal of Magnetic Resonance Imaging. 2017;46:483-9
183. Shiradkar R, Ghose S, Jambor I, Taimen P, Ettala O, Purysko AS. et al. Radiomic features from pretreatment biparametric MRI predict prostate cancer biochemical recurrence: Preliminary findings. J Magn Reson Imaging. 2018;48:1626-36
184. Bakr S, Echegaray S, Shah R, Kamaya A, Louie J, Napel S. et al. Noninvasive radiomics signature based on quantitative analysis of computed tomography images as a surrogate for microvascular invasion in hepatocellular carcinoma: a pilot study. Journal of Medical Imaging. 2017;4:041303
185. Peng J, Zhang J, Zhang Q, Xu Y, Zhou J, Liu L. A radiomics nomogram for preoperative prediction of microvascular invasion risk in hepatitis B virus-related hepatocellular carcinoma. Diagnostic & Interventional Radiology. 2018;24:121-7
186. Ma Z, Fang M, Huang Y, He L, Chen X, Liang C. et al. CT-based radiomics signature for differentiating Borrmann type IV gastric cancer from primary gastric lymphoma. European Journal of Radiology. 2017;91:142
187. Ba-Ssalamah A, Muin D, Schernthaner R, Kulinna-Cosentini C, Bastati N, Stift J. et al. Texture-based classification of different gastric tumors at contrast-enhanced CT. European Journal of Radiology. 2013;82:E537-E43
188. Liu S, Song L, Ji C, Zheng H, Xia P, Zhang Y. et al. Application of CT texture analysis in predicting histopathological characteristics of gastric cancers. European Radiology. 2017;27:1-9
189. Liu S, Zhang Y, Chen L, Guan W, Guan Y, Ge Y. et al. Whole-lesion apparent diffusion coefficient histogram analysis: significance in T and N staging of gastric cancers. Bmc Cancer. 2017;17:665
190. Liu S, Zhang Y, Xia J, Chen L, Guan W, Guan Y. et al. Predicting the nodal status in gastric cancers: The role of apparent diffusion coefficient histogram characteristic analysis. Magnetic Resonance Imaging. 2017;42:144
191. Liu S, Zheng H, Zhang Y, Chen L, Guan W, Guan Y. et al. Whole-volume apparent diffusion coefficient-based entropy parameters for assessment of gastric cancer aggressiveness. Journal of Magnetic Resonance Imaging. 2017;47:168
192. Zhou Y, He L, Huang Y, Chen S, Wu P, Ye W. et al. CT-based radiomics signature: a potential biomarker for preoperative prediction of early recurrence in hepatocellular carcinoma. Abdominal Radiology. 2017;42:1695-704
193. Akai H, Yasaka K, Kunimatsu A, Nojima M, Kokudo T, Kokudo N. et al. Predicting prognosis of resected hepatocellular carcinoma by radiomics analysis with random survival forest. Diagnostic & Interventional Imaging. 2018
194. Cozzi L, Dinapoli N, Fogliata A, Hsu WC, Reggiori G, Lobefalo F. et al. Radiomics based analysis to predict local control and survival in hepatocellular carcinoma patients treated with volumetric modulated arc therapy. Bmc Cancer. 2017;17:829
195. Giganti F, Marra P, Ambrosi A, Salerno A, Antunes S, Chiari D. et al. Pre-treatment MDCT-based texture analysis for therapy response prediction in gastric cancer: Comparison with tumour regression grade at final histology. European Journal of Radiology. 2017;90:129
196. Hyun YS, Hoon KY, Jin LY, Jihoon P, Won KJ, Seung LH. et al. Tumor Heterogeneity in Human Epidermal Growth Factor Receptor 2 (HER2)-Positive Advanced Gastric Cancer Assessed by CT Texture Analysis: Association with Survival after Trastuzumab Treatment. Plos One. 2016;11:e0161278
197. Giganti F, Antunes S, Salerno A, Ambrosi A, Marra P, Nicoletti R. et al. Gastric cancer: texture analysis from multidetector computed tomography as a potential preoperative prognostic biomarker. European Radiology. 2016;27:1-9
198. Verma V, Simone CB 2nd, Krishnan S, Lin SH, Yang J, Hahn SM. The Rise of Radiomics and Implications for Oncologic Management. J Natl Cancer Inst. 2017:109
199. Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P. et al. The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J Digit Imaging. 2013;26:1045-57
200. Kalpathy-Cramer J, Freymann JB, Kirby JS, Kinahan PE, Prior FW. Quantitative Imaging Network: Data Sharing and Competitive Algorithm Validation Leveraging The Cancer Imaging Archive. Translational Oncology. 2014;7:147-52
201. O'Connor JPB, Aboagye EO, Adams JE, Aerts HJWL, Barrington SF, Beer AJ. et al. Imaging biomarker roadmap for cancer studies. Nat Rev Clin Oncol. 2017;14:169-86
202. Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D. Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization. ICCV. 2017;3:618-26
Author contact
![]() Corresponding authors: Jie Tian, Address: Institute of Automation, Chinese Academy of Sciences, Beijing, 100190, China. E-mail: jie.tianac.cn or Bo Li, Address: Department of Hepatobiliary Surgery, the Affiliated Hospital of Southwest Medical University, Luzhou, Sichuan, 646000, China. E-mail: liboer2002com or Meiyun Wang, Address: Henan Provincial People's Hospital & the People's Hospital of Zhengzhou University, Zhengzhou, Henan, 450003, China. E-mail: marian9999com
Corresponding authors: Jie Tian, Address: Institute of Automation, Chinese Academy of Sciences, Beijing, 100190, China. E-mail: jie.tianac.cn or Bo Li, Address: Department of Hepatobiliary Surgery, the Affiliated Hospital of Southwest Medical University, Luzhou, Sichuan, 646000, China. E-mail: liboer2002com or Meiyun Wang, Address: Henan Provincial People's Hospital & the People's Hospital of Zhengzhou University, Zhengzhou, Henan, 450003, China. E-mail: marian9999com
 Global reach, higher impact
Global reach, higher impact