13.3
Impact Factor
Theranostics 2019; 9(4):1015-1028. doi:10.7150/thno.30853 This issue Cite
Review
Design strategies and application progress of therapeutic exosomes
Department of Molecular Oncology, Eastern Hepatobiliary Surgical Hospital & National Center for Liver Cancer, Second Military Medical University, Shanghai 200438, China
Received 2018-10-22; Accepted 2019-1-17; Published 2019-1-30
Abstract
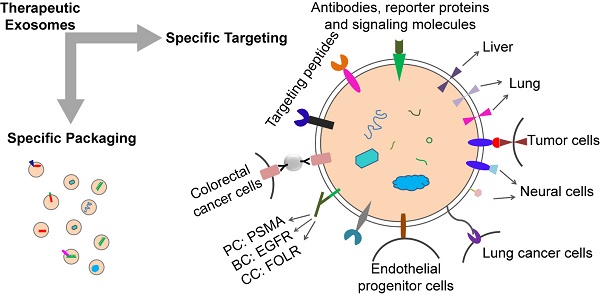
Exosomes have great potential to be drug delivery vehicles due to their natural material transportation properties, intrinsic long-term circulatory capability, and excellent biocompatibility, which are suitable for delivering a variety of chemicals, proteins, nucleic acids, and gene therapeutic agents. However, an effective method of loading specific protein agents into exosomes for absorption by target cells is still lacking. The application potential of exosome is still limited. In this review, we discussed the methods for loading specific treating molecules (proteins, nucleic acids and small chemicals) into exosomes, the design strategies for cell and tissue targeting, and the factors for exosome formation. This review can be used as a reference for further research as well as for the development of therapeutic exosomes.
Keywords: exosome, drug carrier, therapeutic strategy
Introduction
Exosomes are specialized membranous 50-150 nm sized vesicles released from living cells to the extracellular microenvironment. According to the proposal of the international society of extracellular vesicles (ISEV) in 2018, exosome identification methods include transmission electron microscope (TEM), nanosight particle size analyzer, and protein markers. Exosomes observed by transmission electron microscopy are usually saucer-shaped or hemispherical with a concave side. Specific marker molecules were found on the surface of exosomes, such as CD9, CD81 and CD63 etc. [1]. Théry and Marina et al. reviewed the biogenesis, secretion, composition, function, and intercellular interactions of exosomes and other extracellular vesicles successively [2, 3]. Recently, van Niel et al. more systematically reviewed the cellular biological characteristics of extracellular vesicles [4]. With a deeper understanding of exosomes, its clinical application has attracted more and more attention. Tumor-derived exosomes or TDEx (tumor-derived extracellular vesicles) regulate tumor microenvironment and assist in distant metastasis. The molecular markers on exosomes or TDEx make it an effective liquid biopsy indicator for cancer diagnosis and prognostic monitoring [5, 6].
One of the most useful properties of exosomes is their ability to cross barriers such as the cytoplasmic membrane and the blood/brain barrier. This makes them ideal as therapeutic delivery molecules. Exosomes have great potential of being a drug carrier due to its natural material transportation properties, intrinsic long-term circulatory capability, and excellent biocompatibility, which are suitable to deliver a variety of chemicals, proteins, nucleic acids and gene therapeutic agents. For example, the modified exosomes could be used to treat pancreatic cancer by delivering siRNA, and the CD47 protein on the exosomes will prevent them from being phagocytosed by immune cells, making the exosomes more efficient than synthetic analogiliposomes. By using the targeting effect of exosomes, drugs such as adriamycin can be directly applied to tumor cells, limiting toxicity to normal cells [7]. Studies have found that the cellular immunity, angiogenesis, and regenerative effects in cell therapy mediated by mesenchymal stem cells (MSCs) and some other cells are implemented by the exosomes released from these cells [8, 9], which provides the possibility of “cell-free cell therapy”. Therefore, exosome therapy has shown great application potential from oncology to regenerative medicine.
Much remains to be done before exosomes can be transformed into new technologies and treatments. Side effects need to be considered to ensure that the exosomes do not play a deleterious role, such as transferring drug resistance or inhibiting immune responses. Standardized methods for the separation, characterization, and storage of exosomes need to be developed. More importantly, as a nano-vesicle carrier from cell source, the potential of exosomes is weakened due to the shortage of good methods to load specific proteins with therapeutic effects into exosomes and to promote their absorption by the specific cells. Targeted modification of exosomes through genetic engineering or chemical methods may increase the carrying capacity of the exosomes and improve their specificity. This review will discuss load-specific and targeted design strategies for engineered exosomes and the factors that promote exosome formation. This article can be used as a reference for further research and development on engineered exosomes.
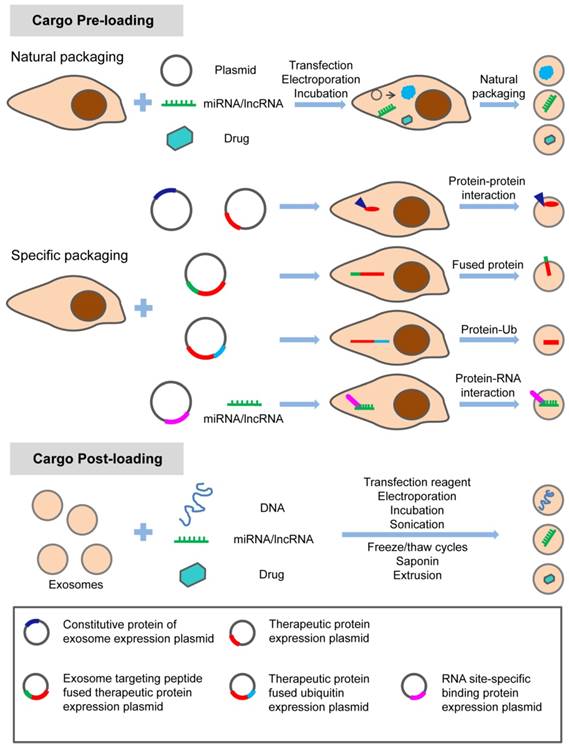
Methods of loading engineered exosomes with specific proteins
Protein deficiency and dysfunction are important causes of many diseases. For example, the occurrence of malignant tumors is associated with the loss and dysfunction of certain tumor suppressors. Therefore, increasing the corresponding protein level is one of the ways to treat diseases. Given the risks of genomic changes, it is more desirable to deliver therapeutic protein molecules to the target through effective drug delivery vectors. The biological characteristics of exosomes have attracted the attention and exploration of more and more researchers. At present, there are several strategies to load specific proteins into the exosomes and increase the protein contents in the exosomes to exert the therapeutic effect. The strategies include:
Overexpression of cytoplasmic proteins in donor cells
To load proteins into exosomes by genetic engineering of donor cells, the vectors containing the gene of interest are transfected into the donor cells. Proteins encoded by the inserted genes are produced by donor cells and then secreted into extracellular vesicles through the natural packaging processes. Exosomes full of proteins can be obtained by isolating and purifying the extracellular vesicles in the cell culture supernatant [10]. Although this approach seems simple and feasible, there are many problems to be solved. Cytotoxicity is the first consideration. The expression of cytotoxic protein can inhibit the proliferation of donor cells and even induce their apoptosis. Promiscuous interactions and impaired biological responses also need to be considered. This will provide a great obstacle for the production of exosomes. On the other hand, there should be a complex sorting system to determine which molecules can enter exosomes. This method of expressing target proteins in donor cells makes the target proteins nonspecific and randomly packaged into exosomes with low loading efficiency. Therefore, several attempts have been made to specifically load target proteins into exosomes, such as the fusion of the target protein with the constitutive protein of exosomes and the specific modification of the target protein.
Fusion of target protein with constitutive protein of exosomes
By fusing the target protein with the constitutive protein of exosomes (e.g., CD63), the protein of interest can be delivered into the exosomes as mediated by the constitutive protein expression. This can improve the specificity of target protein loading into exosomes. Di Bonito et al. designed a Nef/E7 DNA vector expressing Nef exosome-anchoring protein fused with HPV-E7 and generated large amounts of exosomes containing the Nef-E7 fusion protein [11]. The property of the Nef (mut), a nonfunctional mutant of the HIV-1 Nef protein, was utilized to assemble the proteins into exosomes. By fusing the target proteins to the C-terminal of Nef (mut), the fusion proteins can be delivered into exosomes without external neutralization or degradation [12].
Enabling the target protein to be recognized by the exosome sorting machine ESCRT can also specifically improve its exosome content. By constructing a chimeric vector which contained the ectodomain of CD8 and the cytosolic domain of the transmembrane subunit of Env protein (from the bovine leukemia virus) and transfecting it into K562 cells, the chimera was effectively packaged into the released exosomes. It was found that some motifs in Env cytosolic domain could be recognized by the ESCRT [13]. These peptides, which could be recognized by the ESCRT machinery, are capable of being fused with specific proteins and loaded into exosomes for the treatment of disease. The Nedd4 family-interacting protein 1 (Ndfip1) plays an important role in loading proteins, such as the Nedd4 family protein, in exosomes. It was reported that Ndfip1could increase the secretion of exosomes. Under normal circumstances, Nedd4, Nedd4-2, and Itch were not present in exosomes, but the expression of Ndfip1 caused recruitment of three Nedd4 proteins into exosomes [14].
There is a close relationship between exosomes and viruses. Exosomes are involved in virus replication and diffusion, antiviral molecular transport, infection immunity and immune regulation, and microenvironment modulation. The target protein can be loaded into exosomes through a virus pseudotype packaging mechanism and its transport capacity can be improved. The exogenous protein was fused with vesicular stomatitis virus glycoprotein (VSVG) and transfected into HEK293 cells. Exosomes containing VSVG fusion protein were released. VSVG without the extracellular domain was able to deliver exogenous proteins into the exosomes, but the full-length VSVG was better internalized by recipient cells, indicating that the extracellular domain is associated with the cell tropism [15].
In addition, many newly discovered exosome proteins may also be applied in this field. Neoplastic tissues uptake high amount of Transferrin (Tf) and Lactoferrin (Lf). GAPDH is able to capture Tf and Lf to the exosomes, and a study provided a significant clue for delivery of these iron-carrying proteins by exosomes [16]. The tumor-associated macrophages (TAMs) of M2 phenotype were enriched in gastric cancer, and the exosomes secreted by these macrophages were rich in the apolipoprotein E (ApoE), whose expression level is much higher than ones of any other factors [17]. ApoE protein may have potential of delivering molecules into tumor exosomes. Exosome secretion is also a mechanism of selective toxin elimination, such as alpha-Synuclein. Researchers demonstrated that the amount of SCAMP5 (a secretory membrane protein) was obviously increased in the brains of Huntington's disease patients. This protein could promote the unconventional secretion of the co-localized alpha-Synuclein [18]. SCAMP protein family plays an important role in exosome protein sorting and targeting.
Additionally, a new method to load proteins into exosomes, named “exosomes for protein loading via optically reversible protein-protein interactions (EXPLORs)”, was described. The principle of this method is coupling the membrane protein CD9 with CIBN, the CRY2 with the cargo protein. When activated by light, CIBN interacts with CRY2, leading to the loading of cargo proteins into the exosomes. Only blue light is required by the EXPLOR method to load proteins into the exosomes; no protein purification steps are needed [19].
In summary, fusing the target protein with the constitutive protein of the exosome did improve the concentration of the target protein in the exosome. However, whether the fusion protein affects the function of the target protein needs to be verified. Therefore, researchers have tried to select peptides as guides to fuse with the target protein to minimize the impact on protein function. Myoferlin, a new exosome protein which contains several C2-domains, is conserved in vesicle fusion and endocytosis. In tumor cells, Myoferlin depletion causes a dysregulation of exosome protein loading, indicating that Myoferlin or the C2 domain could be involved in exosome protein loading [20]. The late-domain (L-domain) pathway was used to load proteins into exosomes, and a study demonstrated that labeling a WW tag to the Cre recombinase could help the L-domain-containing protein Ndfip1 recognize the fusion protein, which allowed the target protein to be ubiquitinated and loaded into exosomes. The function of these delivered proteins was also intact [21]. In addition, most of these guiding proteins or peptides are transmembrane proteins or peptides (as shown in Table 1). They target proteins to the exosome membrane, and whether this affects the therapeutic function of the target protein in recipient cells requires further study.
Proteins and peptides with the potential of packaging specific protein molecules into exosomes or carrying targeting molecules on the surface of exosomes
| Proteins or Peptides | Descriptions | References |
|---|---|---|
| CD63 | Tetraspanin | [22] |
| TSG101 | Component of the ESCRT-I complex | [23] |
| ARRDC | Arrestin family of protein | |
| Palmitoylated tdTomato | Tandem dimer Tomato fused at NH2-termini with a palmitoylation signal for EV membrane labelling. | [24] |
| Lactadherin C1C2 domain | Membrane glycoprotein | [25] |
| EGF VIII | Transmembrane glycoprotein | [26] |
| PDGFR TM domain | Cell surface tyrosine kinase receptor | [27] |
| HIV-1 Nef (mut) | Released in extracellular vesicles | [12] |
| VSVG | Vesicular stomatitis virus glycoprotein | [15] |
| LAMP2B | Lysosome-Associated Membrane Glycoprotein 2 | [28] |
| LAMP1 | Lysosome-Associated Membrane Glycoprotein 1 | |
| ALIX-1 | Cytosolic protein that associates with MVB by interacting with ESCRT-III subunit SNF7 | |
| CD9 | Tetraspanin | [29] |
| CD81 | Tetraspanin | |
| HSP70 | Heat Shock Protein | |
| HSP90 | Heat Shock Protein | |
| MHC | Anchored in the membrane | [30] |
| SCAMPs | Secretory Carrier-Associated Membrane Protein | [18] |
| ApoE | Apolipoprotein E | [17] |
| WW tag | Recognized by the L-domain-containing protein Ndfip1, resulting in ubiquitination and loading into exosomes. | [21] |
List of proteins commonly found in all exosomes studied to date [28]
| Protein category | Gene symbols |
|---|---|
| Antigen-presentation | HLA |
| Cell adhesion | MFGE8, THBS1, ITG, CLDN1 |
| Cell structure and motility | ACT, ACTN, CFL1, VIL2, MSN, MYH, MYL, RDX, TUB |
| Heat shock proteins and chaperones | HSPA8, SP90AB1, CCT |
| Metabolic enzymes | ALDOA, FASN, GAPDH, PGK1, PGAM1, PKM2 |
| MVB biogenesis | PDCD6IP, TSG101, VPS28, VPS37, VPS25, VPS36, SNF8, CHMP |
| Signaling proteins | YWHA, HRAS, ARHGDIA, RHOC, RAP1B, RAP2B, RRAS2, IQGAP1, SDCBP, RHOA, GNB, GNG, GNAS, GNA, GNAI, GNAO, GNAQ |
| Tetraspanins | CD9, CD63, CD81, CD82 |
| Transcription and protein synthesis | HIST, RPS, RPS27A, EEF1A1 |
| Trafficking and membrane fusion | ANXA, ARF, AP2A1, AP2B1, CLTC, GDI2, RAB, SNAP23, STX3 |
According to existing studies, proteins and peptides with the potential of packaging specific protein molecules into exosomes or carrying targeting molecules on the surface of exosomes are summarized (Table 1). In addition, a variety of proteins involved in the formation of exosomes also have the potential to package specific molecules into exosomes or carrying targeting molecules (Table 2). Application strategies utilizing this need to be further explored. It may be possible to obtain more applicable guiding peptides directed to exosomes.
Specific modifications of target protein
Exosomes are natural antigen transport carriers and are increasingly considered to be potential new vaccine carriers. A fusion of ubiquitin to target proteins at the C-terminus was developed, and the fusion protein was expressed in a human embryonic kidney (HEK 293) cell line. The concentration of target proteins in exosomes was increased about tenfold when they were fused to ubiquitin. Also, these exosomes which contained recombinant antigen could induce the T cell response [31]. Similarly, this method can be used to load protein molecules with therapeutic effect into engineered exosomes to play a role in therapies. The mono-ubiquitinated Syntaxin 3 (Stx3) was rapidly endocytosed and excreted into exosomes. However, the mutant of Stx3 (Stx3-5R) which was not ubiquitinated failed to enter this pathway [32]. Major histocompatibility complex class II (MHC-II) molecules are rich in exosomes secreted by the APCs (antigen presenting cells). It has been reported that ubiquitination of the MHC-II β-chain cytoplasmic tail in many cell types contributes to the sorting of MHC-II molecules into exosomes. However, the ubiquitination of MHC-II was not indispensable for its entering into exosomes [33]. Therefore, protein ubiquitination could be one of the signals for loading the protein into exosomes specifically. In addition, fusion of the N-terminus of target proteins with a myristoylation or palmitoylation tag [34] or a transmembrane motif could make fusion proteins target to the inner surface of the plasma membrane [27]. However, it is still unknown whether the modification of the target protein, especially the ubiquitination modification, will lead to the degradation of the target protein by the proteasome and affect its function in the recipient cells.
Mechanical methods of producing exosomes loaded with target proteins
Overexpression of a cytoplasmic protein in cell lines, combined with mechanical selection of different pore sizes, can produce vesicles containing the target protein. Haney et al. tried several methods including: incubation at room temperature, saponin permeabilization, repeated freeze-thaw, sonication, and mechanical extrusion to load the catalase into exosomes to treat Parkinson's disease (PD). The loading of catalase into exosomes by sonication, mechanical extrusion, or saponin permeabilization was of high efficiency, which maintained a sustained release and protected the catalase from degradation by proteases. In vitro, these exosomes were taken up by neuronal cells, indicating that they have significant neuroprotective effect [35].
There are two main problems with engineered exosomes obtained by mechanical methods. One is the high technical requirement of exosome purification, and the others are the integrity of exosomes and the maintenance of biological activity. The composition of the cell-generated exosomes is different from that of the mechanically generated exosomes (or “vesicles”, to be precise). This difference may affect the efficacy of engineered exosomes. In the future, these two problems need to be solved and proved.
Methods of loading specific nucleic acids into engineered exosomes
There are several methods to load nucleic acids into exosomes, including transfection with commercialized reagents [36], electroporation, guidance of proteins and signature sequences, transfecting donor cells and producing hybrid exosomes with liposomes.
Transfection with commercialized reagents
By isolating exosomes from HeLa cells and ascites through ultracentrifugation, the short interfering RNA (siRNA) AF488 was mixed with lipofectamine and the exosomes were incubated with this mixture at RT for 30 min. After transfection, the mixture was washed 3-5 times and ultra-filtered. The exosomes loaded with siRNA were cultured together with the receptor cells for 24 hours. The exogenous siRNA could then be transferred to recipient cells [37, 38].
Electroporation
The most common method for loading miRNA or siRNA into exosomes is electroporation. The exosome membrane protein Lamp2b was fused with the RVG peptide, which is neuron-specific. The siRNA was then loaded into purified exosomes by electroporation to make these exosomes target neuron cells. The BACE1-targeted siRNA can be delivering to the brain to treat Alzheimer's disease [39].
Transfection of exosomes directly with nucleic acid by transfection reagents or electroporation is straightforward, but it is not efficient. It requires separation and purification of exosomes before and after transfection. Currently, methods for the preparation and purification of exosomes in large quantities include differential ultracentrifugation and sucrose density gradient ultracentrifugation [40], size-based separation (ultrafiltration) [40, 41], and exosome precipitation [42]. High-speed centrifugation may destroy vesicles and reduce sample quality. Repeated purification is bound to result in loss of exosomes and loss of mass.
Guidance of proteins, peptides or signature sequences
Based on the disadvantages of these two methods, researchers have turned their attention to the method of actively packaging nucleic acids into exosomes. One is active packaging using proteins that bind to specific RNA sequences. A method was reported to produce customized engineered exosomes in engineered mammalian cells. The RNA packaging device in the exosome producing cells packaged specific mRNAs into exosomes and delivered the mRNA to the cytoplasm of target cells. This delivery was of high efficiency and concentration of exosomes was not required. Additionally, importing the engineered cells into the PD mice models successfully delivered mRNA to the brain. The mRNA of catalase which has therapeutic effect could significantly reduce the neuroinflammation and neurotoxicity [43].
The new technology includes an active packaging device for specific RNA and a cytosolic delivery helper for RNA delivery. Researchers applied archaeal ribosomal protein L7Ae, which binds to the C/D box RNA structure, to RNA packaging devices, [44-46]. Binding of L7Ae to the C-terminus of CD 63 and insertion of C/D box into the reporter gene's 3'-UTR can well package the mRNA of the reporter gene into the exosomes through the interaction of L7Ae and the C/D box in the 3'-UTR. It has been reported that Connexin 43 (Cx43) can be accumulated in exosomes, and the S368A mutation of Cx43 can enhance the information delivery from exosomes to target cells [47]. Therefore, the gap junction protein, Cx43, is used as a potential cytoplasmic transfer assistant. The RNA packaging device and the cytoplasmic transfer assistant were co-transfected into mammalian cells for the production of exosomes, and these cells were then prepared into engineered cells. C/D box was then inserted into the 3'-UTR of the target gene expression vector, and the vector was used to transfect the engineered cells. In this way, the exosomes produced by the engineered cells are packed with specific mRNAs and can efficiently transfer the mRNAs to the cytoplasm of the target cells.
The other method is to use the conserved sequence of exosome-enriched RNAs (eRNAs) to carry out active packaging. The researchers speculated that eRNAs may contain a specific common sequence which target RNAs into exosome as a cis-element. By analyzing RNAs in exosomes based on this hypothesis, it was found that only three motifs were potential signature sequences. These results are helpful to our understanding on the selective targeting of candidate RNAs to these exosomes for therapeutic purposes [48].
In addition, the tumor suppressive miRNAs present in the exosomes of advanced cancers were examined. The miR-193a could interact with major vault protein (MVP). Lack of MVP caused the miR-193a to accumulate in the donor cells instead of exosomes, and this inhibited cancer development [49]. GW182 and AGO2 are two important components of the RNA-induced silencing complex (RISC). They are related to the MVBs and may be involved in the process of miRNA distribution to exosomes [50, 51]. The exosome protein Myoferlin is usually highly expressed in tumor cells. The transportation of nuclei acids into exosomes lacking Myoferlin is significantly decreased [20]. Whether protein molecules like MVP, GW182, AGO2 and Myoferlin can carry specific nucleic acid sequences into the exosomes remains to be explored; these proteins might be a tool for packaging specific nucleic acids into the exosomes. This is a promising direction. In the future, bioinformatics analysis will be required to obtain more specific RNA-sequence binding proteins and conserved sequences of eRNAs.
Transfecting donor cells
Similar to protein packaging methods described above, the parental cells can also be transfected with miRNA or siRNA which are then secreted naturally into the exosomes by these parental cells. The release of exosomes from fat stem cells overexpressing miR-126 plays a therapeutic role in alleviating acute myocardial ischemic injury [52]. HEK293T cells were transfected with the inhibitor of miR-214, and the exosomes Exo-anti-214 secreted by these cells can reverse the DDP resistance of gastric cancer [53]. When HEK293T cells were transfected with HGF siRNA, the obtained exosomes containing HGF siRNAs could inhibit the proliferation and migration of cancer cells and vascular cells in in vivo and in vitro experiments [54]. However, this approach is limited by cytotoxicity, poor specificity, and inefficient packaging.
Producing hybrid exosomes with liposomes
Targeting of the CRISPR/Cas9 to the receptor cells is crucial for in vivo gene editing. However, the efficiency of packaging macromolecular nucleic acids by exosomes is quite low. A type of hybrid exosome containing liposomes through simple incubation was developed, and these mixed nano-particles were able to package large plasmids, including the CRISPR-Cas9 expression vector, efficiently. Additionally, these hybrid nanoparticles could be endocytosed and expressed by the mesenchymal stem cells (MSCs). The liposomes cannot transfect MSCs alone. To sum up, the exosome-liposome hybrid nanoparticles can deliver CRISPR-Cas9 system in MSCs, which offers a promising application prospect in gene manipulation [55].
In short, despite the wide use of electroporation, their clinical application has been limited to some, but not all, RNA cargo. The modified miRNAs, shRNAs, mRNAs, or RNAs are unable to be loaded into exosomes by electroporation, indicating that this method is not suitable for some RNAs with special structures [56]. The EXOtic devices developed by Kojima group enable the specific and highly efficient delivery of specific mRNA. The conserved sequences of exosome-enriched RNAs also have good application prospect and the methods for loading RNAs into exosomes need to be further studied.
Methods of loading small molecules into exosomes
There is growing interest in using exosomes as carriers of chemotherapy drugs. Methods of loading small molecule drugs into exosomes include direct mixing, incubation, ultrasonic treatment, eddy oscillation, etc.
The hydrophobic drug curcumin can be loaded into exosomes by direct mixing [57]. Paclitaxel can be introduced into exosomes by direct mixing and ultrasonic treatment [58, 59]. Different methods, including incubation at room temperature, electrical perforation, and ultrasonic treatment, were developed to load paclitaxel (PTX) into exosomes released by macrophages, and their size, drug release, stability, and antitumor effect in vitro were evaluated. It was found that the ultrasonic treatment increased the load of drug and sustained release of exosomes [60]. Small molecules can also be naturally secreted into extracellular vesicles (EVs) by incubating with parental cells. Microvesicles containing paclitaxel were produced by incubating MSC with paclitaxel [61]. In addition, several different ways of loading catalase into exosomes ex vivo such as incubation, saponin permeabilization, freeze and thaw, sonication, and extrusion were used to attempt to treat Parkinson's disease (PD) [62]. These methods can also be used to load exosomes with small molecule drugs. Currently, most of these small molecule drugs are loaded into exosomes by these passive loading methods. The disadvantages of these approaches have always been the loss and degradation of exosomes from multiple purification steps. In addition, prolonged in vitro treatment and the inherent physicochemical properties of drug molecules also affect the bioactivity and stability of exosomes. Therefore, before exosomes can be widely used for treatment, the stability of exosomes and proper storage conditions are also worth studying. The methods for loading the specific treating molecules (proteins, nucleic acids and small chemicals) into exosomes are summarized in the schematic diagram (Figure 1).
Design strategy for therapeutic exosome targeting
Loading and targeting are the keys to the therapeutic potential of exosomes. Systemically delivered exosomes mainly accumulate in kidney, liver, and spleen [63]. However, in the application of therapeutic exosomes, it is more desirable to accurately target a specific type of cells (e.g., tumor cells) or a certain type of tissue (e.g., brain tissue) than broad distribution to the liver, kidney and spleen. Therefore, researchers have tried to improve the targeting of exosomes through the study of exosome donor cells and the modification of surface molecules on exosomes.
Donor cells determine the targeting of exosomes
Because exosomes are vesicles secreted by cells, they express certain lipid and cellular adhesion molecules which have specificity for certain types of receptor cells. Some studies have shown that exosomes have a natural targeting capability based on donor cells.
Researchers have found that WJ-MSCs (mesenchymal stem cells, MSCs) are able to deliver miRNAs into glioblastoma (GBM) cells. The marked miR-124 were delivered into the U87 GBM together with WJ-MSCs by exosome-dependent or independent ways. These results indicated that the delivery of miRNA by exosomes from WJ-MSC may provide a new method for miRNA replacement therapy in GBM [64].
Methods of loading specific proteins, nucleic acids and small molecular drugs into engineered exosomes. The specific treating molecules can be loaded into exosomes before or after exosome production through natural or specific packaging.
Exosomes derived from prostate cancer cells MDA-PCa2b can target osteoblasts. In vivo and in vitro studies showed that MDA-PCa2b exosomes could transfer mir-141-3p to osteoblasts [65]. In 2016, Zhao et al found that tumor-associated fibroblast derived exosomes (CDEs) can be consumed by prostate cancer cells in patients with prostate cancer [66]. Several clues about how exosomes influence organ-specific metastasis were provided, and the exosomes, which target different organs, had different cell-binding receptor proteins, or integrins, on the surface. Different types of exosomes tend to enter organs with a large number of ligands corresponding to their surface integrins. Integrin αVβ5 targets exosomes exhibit tropism for the liver while integrin α6β4 and integrin α6β1 target exosomes to lung [67, 68]. Exosome targeting therapy mediated by different integrins also has good application prospects. This suggests that tumor-associated fibroblast derived exosomes can be used to target drug delivery to cancer cells. Exosomes from different tumor sources can deliver drugs to different organs. There are also potential risks. Tumor-derived or tumor-associated fibroblast-derived exosomes carry not only the targeted molecules we need, but also other molecules on the surface or inside of these cells, which may lead to the occurrence, metastasis and progression of tumors. So researchers have considered the effect of removing these molecules. Targeted protein or chemical modification of the exosome surface is a good choice.
Assembly of targeting proteins or chemical modifications on exosome surface
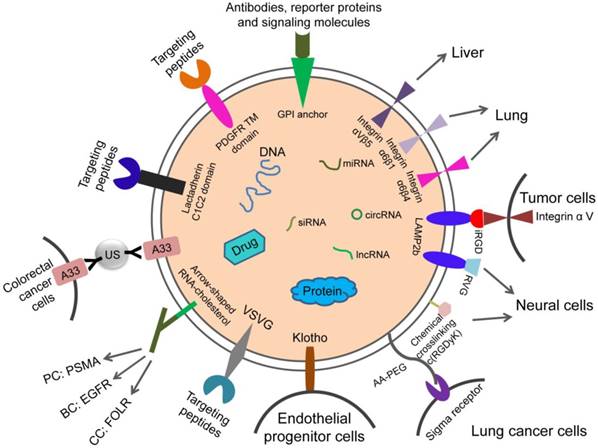
Targeting exosomes can be acquired by constructing targeting molecules such as antibody fragments or peptides which can recognize the target antigens on the surface of exosomes. Targeting ligands are added to the surface of exosomes. The most common technique is to insert targeting proteins into donor cells, which will then secrete exosomes containing targeting proteins. In recent years, the most common means to achieve accurate targeting of exosomes is to use the Lactadherin C1C2 domain with synthetic tag and Lamp2b with synthetic tag.
The iRGD specifically recognizes αV integrins on the surface of tumor cells. This can be used for exosome therapy for malignant tumors. The fusion protein of iRGD and Lamp2b was overexpressed in immature dendritic cells (imDCs), and large amounts of exosomes carrying iRGD-targeting proteins were obtained by purification. The imDCs-exosomes were loaded with doxorubicin by electroporation and were found to have a specific effect on breast cancer cells which express the αV integrin, indicating a targeted therapeutic role of these exosomes in breast cancer mouse models [69].
Rabies virus glycoprotein (RVG) and c(RGDyK) peptides target brain tissue. RVG- exosomes can be used for targeted delivery of gene therapy to the brain [70]. The fusion of exosome protein Lamp2b with RVG could modify the exosomes, which allows for efficient delivery of miR-124 by exosomes to brain infarctions [70]. Researchers have employed modified exosomes expressing the neuron-specific RVG peptide on the membrane surface to deliver opioid receptor (MOR) siRNA into the brain to treat morphine addiction. They found that these exosomes efficiently and specifically delivered MOR siRNA into Neuro2A cells and the mouse brain. Researchers have also delivered MOR-siRNA into the brain by targeted RVG exosomes to treat morphine relapse. These exosomes efficiently reduced MOR expression and inhibited morphine relapse [71]. By chemically conjugating the c(RGDyK) on the surface of exosomes, the engineered c(RGDyK)-conjugated exosomes (cRGD-Exo) are able to target ischemic brain lesions following intravenous administration [72].
Klotho protein has the property of binding to circulating endothelial progenitor cells (EPCs) and can be used for exosome therapy targeting EPCs. Researchers transfected MSCs with Klotho expression plasmids and loaded the adenosine kinase (ADK) siRNA into exosomes (Klotho/ADK siRNA-exosomes) by electroporation. Flow cytometry results showed that the Klotho/ADK siRNA-exosomes captured the circulating endothelial progenitor cells (EPCs) and were endocytosed by these EPCs effectively. Then the exosomes were decomposed into Klotho protein and ADK siRNA [73].
The combination of exosomes and various nanoparticles by chemically editing donor membranes or exosome membranes is a hot topic in recent years. This method provides a new method for precise targeting of exosomes. For example, nanobodies which anchor the glycosylphos-phatidylinositol (GPI) on surfaces of EVs can display various proteins such as reporter proteins, antibodies, and signaling molecules [63].
Attaching antibody-like (Y-shaped) RNA nanoparticles to microvesicles can specifically deliver effective RNA therapeutic agents (siRNA) to cancer cells. Extracellular microvesicles were prepared to carry antibody-like RNA molecules called “aptamers” that bind to highly expressed surface marker in three tumor types: prostate cancer (targeting PSMA), breast cancer (targeting EGFR), and colorectal cancer (targeting folic acid receptor). All the vesicles were loaded with siRNA which suppresses expression of the survivin gene. As the test therapy, they inhibited the development of cancer cells without obvious toxicity [74].
Exosomes coated with high-density antibodies which target specific ligands may be a new delivery system for targeted drugs against human cancers [75]. The isolated exosomes (A33-Exo) from A33 positive LIM1215 cells were loaded with doxorubicin (Dox), then the surface-carboxyl superparamagnetic iron oxide nanoparticles (US) were coated with A33 antibody to bind to A33-positive exosomes, so as to target A33-positive colon cancer cells. The results suggested that A33Ab-US-Exo/Dox had good tumor-targeted ability and inhibited tumor growth. By combining the exosomes with nano-materials through chemical modification of the donor cell membrane, the exosomes were designed to response to the infrared light. The designed exosomes effectively accumulated at target sites through dual ligand-mediated endocytosis. The gold nanorods at the exosome surfaces transferred the near-infrared light into heat energy, affected the permeability of exosome membrane to improve the release of drug from exosomes, and inhibited the tumor development. This technique provided a new strategy for individualized precision cancer therapy in the future [76]. The aminoethyl anisamide-polyethylene glycol (AA-PEG) can target lung cancer cells with sigma receptor overexpression. It can be used for exosome therapy of lung cancer. The targeting exosomes loaded with PTX and AA-PEG had a high loading capacity and accumulated in cancer cells upon systemic administration [77].
Adhesion molecules (glycoproteins) on the surface of platelets play a significant role in targeting and retention in damaged tissue. Cheng's team used this principle to create platelet nanoparticles and modified them on the surface of cardiac stem cells. Platelet nanoparticles act as a “mantle” for platelets, with binding molecules that target but do not activate thrombosis. The targeting function of platelets can be used to mediate other types of stem cells and exosomes and guide them to repair damaged heart muscle [78].
Although researchers have made impressive progress in the modification of engineered exosomes, most of the above-mentioned techniques are based on the modification of exosome donor cells. These changes may affect the protein composition and function of exosomes. Therefore, it is necessary to explore new techniques for targeted modification of exosomes while minimizing changes in their composition. For example, nanobody-PEG-micelles were prepared by conjugating nanobodies targeting the epidermal growth factor receptor (EGFR) to phospholipid (DMPE)-PEG derivatives. When micelles were mixed with EVs derived from Neuro2A cells or platelets, the transfer of nanobody-PEG-lipids to the EV membranes was temperature-dependent. This process had no effect on EV size distribution, protein composition, or morphology. This research proposed post-insertion as a new technique to give these isolated EVs targeting capacity, which would avoid modification of EVs cells. More importantly, the insertion of ligand-conjugated PEG-derivatized phospholipids in EV membranes improved the cell specificity and prolonged circulation duration of EVs, which increased the accumulation of EVs in targeted tissues [79]. The design strategies for cell and tissue targeting are summarized in the schematic diagram (Figure 2).
Design strategies for therapeutic exosome targeting. Targeting is the key to the therapeutic potential of exosomes. It can be improved by selecting the exosome donor cells and the modification of surface molecules on exosomes. AA-PEG: aminoethylanisamide-polyethylene glycol; GPI: glycosylphos-phatidylinositol; RVG: rabies virus glycoprotein; VSVG: vesicular stomatitis virus glycoprotein; A33Ab-US: the surface-carboxyl superparamagnetic iron oxide nanoparticles (US) were coated with A33 antibody, to bind to A33-positive exosomes; iRGD: an αⅤ integrin-specific internalizing peptide; PDGFR TM domain: PDGFR transmembrane domain.
Origins of engineered exosomes and factors promoting their formation
Sources of engineered exosomes
A potential source of therapeutic exosome is immature dendritic cells (imDCs). The exosomes secreted by imDCs lack surface markers such as MHC-I, MHC-II, CD40, CD86, etc., resulting in low immunogenicity. They can be produced from CD34+ cells isolated from the patient's peripheral blood and are one of the preferred sources of therapeutic exosomes. MSCs can be extracted from a patient's bone marrow, fat, and other tissues. The exosomes from MSCs are attractive because they mediate the immunosuppressive effect of treatment based on bone marrow MSCs [80].
Use of cancer cell-derived exosomes to deliver drugs and vaccines for immunotherapy is promising. Cancer exosomes are able to transmit antigens to dendritic cells and induce T-cell-mediated immune responses to cancer cells [30]. Due to the specific expression of Tetraspanin in exosomes from tumor sources, they tend to interact with ligands of different tissues. This makes it possible to use cancer-derived exosomes for cancer-targeting and selective drug delivery [81]. However, cancer derived exosomes also have many potential risks, because they have Tetraspanins, Urokinase plasminogen activator, Cathepsin D, Vimentin, and other cancer cell surface derived molecules [82, 83], which may promote tumor proliferation and metastasis or cause immunosuppression [84-86].
Based on reliable sources and safety, plants and fruits can be used as another source for isolating exosomes for clinical use [87]. Agricultural products like milk and fruits are economical and practical sources for exosomes. Isolated exosomes from grapefruit were decorated with targeting modifications and were then loaded with the antitumor drugs doxorubicin and curcumin. These modified exosomes targeted inflammatory cancers and had anti-inflammatory effects in mice [88]. Exosomes were also isolated from milk and attempts were made to load various drugs into them for the treatment of lung cancer. These exosomes not only have high yield and safety, but also do not activate host immune response [89].
Factors affecting exosome formation
Exosomes are tiny EVs, usually obtained in small amounts. This small yield has been hampering the expansion of basic research on exogenous analysis and drug delivery applications [90]. To improve the production efficiency of exosomes, several methods to enhance the total yield of exosomes are reviewed. This review will focus on five key factors: intracellular calcium level, external stress, cytoskeletal blocking, drug effects, and gene expression factors.
Intracellular calcium level
Several studies have shown that increased intracellular calcium can lead to the formation of extracellular vesicles [91-95]. It was reported that when intracellular calcium levels were increased through tannin stimulation, the production of exosomes increased [91].
External stress
Thermal stress [96], anoxia [97], radiation [98], and pH of the microenvironment [99] can increase EV production. Although stress leads to an increase in EV production, the composition of EVs produced in this case is significantly different [100]. Therefore, the production conditions of the exosomes used for treatment may directly influence the efficiency and therapeutic effect of the engineered exosomes. The optimal conditions for the mass production of exosomes have yet to be explored.
Cytoskeletal blocking
A protocol to stimulate vesicle production and separate microvesicles using sulfhydryl-blocking agent has been published [101]. Incubation with mercapto blockers which hinders cytoskeletal function is a way to induce rapid EV generation [102-104]. In addition to chemical agents, bio actin/myosin inhibitors and chemical crosslinking agents are also used to induce vesicular formation by blocking actin function and stopping vesicular contraction [105, 106].
Drug effects
Chemical compounds which regulate the biogenesis and release of exosomes were screened using the aggressive prostate cancer (PCa) CD63-GFP-expressing C4-2B cells, including 4580 compounds from the NPC library and LOPAC library. The results showed that the lead compounds tipifarnib, neticonazole, climbazole, ketoconazole, and triadimenol were effective inhibitors, while the sitafloxacin, forskolin, SB218795, fenoterol, nitrefazole, and pentetrazol were activators for biogenesis and/or release of PC cell exosomes [107].
Other studies have shown that PA treatment of hepatocytes significantly enhanced exosome production and changed the expression profile of exosome miRNA. In addition, the exosomes from PA-treated hepatocytes induced an increase in the expression of genes related to fibrosis in hepatic stellate cells (HSCs) [108]. The ketotifen, a mast cell stabilizer, inhibited the release of exosome from cancer cells [109].
Gene expression factors
Hypoxia treatment improves the paracrine effect of MSCs. Hypoxia treatment increases the neutral Sphingomyelinase 2 (nSMase2)'s expression level, which is important for exosome secretion [110]. Neutral Sphingomyelinase (nSMase) inhibitors (also called SMPD2 and SMPD3) inhibit the release of exosome from cells [111]. Therefore, overexpression of nSMase can promote the production of exosomes. Other researchers have increased extracellular vesicular productivity by CD9 overexpression. Increased expression of the tetraspanin CD9 led to a higher production of extracellular vesicles by four different human cell lines [112]. In addition, the secretion of exosomes released from the donor MSCs with the overexpression of HIF-1α was enhanced [113].
Other factors
To increase the yield of exosomes, Emam et al. tried to stimulate cancer cells by adding liposomes in vitro. The fluid but not the solid cationic liposomes provided the strongest stimulation. This research indicated that the physicochemical property determined whether it behaved as an inhibitor or a stimulant. Liposomal stimulation may be an effective way to improve the yield of exosomes. We can fine-tune the biological properties of exosome by controlling stimulant liposomes' physicochemical property [114].
Stimulus of triggering exosome release
| Stimulus | Details | References |
|---|---|---|
| Adjustment of physical and chemical factors | Thermal stress | [96] |
| Hypoxia | [97] | |
| Radiation | [98] | |
| pH | [99] | |
| Nutrient deficiency | [115, 116] | |
| Increase intracellular calcium concentration | Tannin | [91] |
| Cytoskeletal fixation | Sulfhydryl-blocking | [101] |
| Actin/Myosin inhibitor | [106] | |
| Chemical crosslinking agent | [105] | |
| Drug stimulation | Sitafloxacin | [107] |
| Forskolin | ||
| SB218795 | ||
| Fenoterol | ||
| Nitrefazole | ||
| Pentetrazol | ||
| Palmitic acid | [108] | |
| Gene overexpression | SMPD2 | [111] |
| SMPD3 | ||
| CD9 | [112] | |
| HIF-1α | [113] |
In view of all the ways to promote EV formation (Table 3), the sulfhydryl-blocking reagent which can inhibit cytoskeleton has superiority. It can produce EVs rapidly and increase the output within only one hour [105] whereas EV production by calcium induction, stress induction, and other drug-induction takes 12 hours or even several days to significantly increase the output [91, 99]. Engineering cells need to be prepared by overexpressing a molecule to promote the production of exosomes. Most importantly, the influence of external factors may lead to changes in the expression profile of internal and surface molecules of exosomes, which may affect the packaging and delivery efficiency of drugs into exosomes. Therefore, it is imperative to find a way to improve the production of exosomes while maintaining the same biological activity as naturally formed EVs.
Conclusions and perspective
Exosomes are of great clinical value due to their strong biocompatibility, offering a new frontier in drug delivery. However, its application also has some risks, such as immunosuppression and tumorigenesis. Since exosomes can be secreted from many types of cells, the use of tumor exosomes can be avoided by selecting the source of donor cells. Using immune cells as the producer of exosomes, the modified exosomes were designed for clinical use by artificially optimizing the integration of specific loadings.
Although the study of exosome application has made great progress, many challenges still remain. During the processing and modification of engineered exosomes, changes to donor cells and exosomes may affect exosome content or protein composition. This may affect the bioactivity and efficacy of engineered exosomes. Therefore, it is necessary to select the transformation method with little influence on the morphology and composition of exosomes. Cytotoxicity, promiscuous interactions, and impaired biological responses during modification should also be considered. Another challenge is to achieve large scale production of exosomes for clinical use. There have been reports of rapid purification of therapeutic exosomes on a large-scale. However, this technique still requires expansion to different cell types. It remains to be studied which cells are most suitable for the production of exosomes. Nevertheless, the idea of using exosomes as a vehicle is promising and inspiring. More in vivo studies are needed for the application of exosomes in clinical practice. Further studies on the efficacy and toxicity of exosomes are still needed.
Abbreviations
AA-PEG: aminoethylanisamide-polyethylene glycol; ADK: adenosine kinase; APCs: antigen presenting cells; ApoE: apolipo protein E; CRISPR: clustered regularly interspaced short palindromic repeats; Cx43: connexin 43; Dox: doxorubicin; EGFR: epidermal growth factor receptor; EPCs: endothelial progenitor cells; eRNAs: exosome-enriched RNAs; ESCRT: endosomal sorting complex required for transport; EVs: extracellular vesicles; EXPLORs: exosomes for protein loading via optically reversible protein-protein interactions; GBM: glioblastoma; GPI: glycosylphos-phatidylinositol; HSCs: hepatic stellate cells; imDCs: immature dendritic cells; MHC-II: major histocompatibility complex class II; MSCs: mesenchymal stem cells; MVP: major vault protein; Ndfip1: Nedd4 family-interacting protein 1; nSMase: neutral sphingomyelinase; PCa: prostate cancer; PD: parkinson's disease; PTX: paclitaxel; RISC: RNA-induced silencing complex; RVG: rabies virus glycoprotein; SCAMP5: secretory membrane protein 5; Stx3: syntaxin 3; TAMs: tumor-associated macrophages; TDEx: tumor-derived extracellular vesicles; VSVG: vesicular stomatitis virus glycoprotein.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81572863 & 81773251 to C. S.).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Tkach M, Théry C. Communication by extracellular vesicles: where we are and where we need to go. Cell. 2016;164:1226-32
2. Théry C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2:569-79
3. Marina C, Graça R, Clotilde T. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255-89
4. van Niel G, D'Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Bio. 2018;19:213-28
5. Aman S, Zamila K, Anjali S. Tumor exosomes: cellular postmen of cancer diagnosis and personalized therapy. Nanomedicine. 2016;11:421-37
6. Boriachek K, Islam MN, Möller A, Salomon C, Nguyen N-T, Hossain MSA. et al. Biological functions and current advances in isolation and detection strategies for exosome nanovesicles. Small. 2018;14:1702153
7. Kamerkar S, LeBleu VS, Sugimoto H, Yang S, Ruivo CF, Melo SA. et al. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature. 2017;546:498-503
8. Baglio SR, Pegtel DM, Baldini N. Mesenchymal stem cell secreted vesicles provide novel opportunities in (stem) cell-free therapy. Front Physiol. 2012;3:359
9. Barile L, Lionetti V, Cervio E, Matteucci M, Gherghiceanu M, Popescu LM. et al. Extracellular vesicles from human cardiac progenitor cells inhibit cardiomyocyte apoptosis and improve cardiac function after myocardial infarction. Cardiovasc Res. 2014;103:530-41
10. Hall J, Prabhakar S, Balaj L, Lai CP, Cerione RA, Breakefield XO. Delivery of therapeutic proteins via extracellular vesicles: review and potential treatments for Parkinson's disease, glioma, and schwannoma. Cell Mol Neurobiol. 2016;36:417-27
11. Di Bonito P, Chiozzini C, Arenaccio C, Anticoli S, Manfredi F, Olivetta E. et al. Antitumor HPV E7-specific CTL activity elicited by in vivo engineered exosomes produced through DNA inoculation. Int J Nanomedicine. 2017;12:4579-91
12. Manfredi F, Di Bonito P, Arenaccio C, Anticoli S, Federico M. Incorporation of heterologous proteins in engineered exosomes. Methods Mol Biol. 2016;1448:249-60
13. De Gassart A, Trentin B, Martin M, Hocquellet A, Bette-Bobillo P, Mamoun R. et al. Exosomal sorting of the cytoplasmic domain of bovine leukemia virus TM Env protein. Cell Biol Int. 2009;33:36-48
14. Putz U, Howitt J, Lackovic J, Foot N, Kumar S, Silke J. et al. Nedd4 family-interacting protein 1 (Ndfip1) is required for the exosomal secretion of Nedd4 family proteins. J Biol Chem. 2008;283:32621-7
15. Meyer C, Losacco J, Stickney Z, Li L, Marriott G, Lu B. Pseudotyping exosomes for enhanced protein delivery in mammalian cells. Int J Nanomedicine. 2017;12:3153-70
16. Malhotra H, Sheokand N, Kumar S, Chauhan AS, Kumar M, Jakhar P. et al. Exosomes: tunable nano vehicles for macromolecular delivery of transferrin and lactoferrin to specific intracellular compartment. J Biomed Nanotechnol. 2016;12:1101-14
17. Zheng P, Luo Q, Wang W, Li J, Wang T, Wang P. et al. Tumor-associated macrophages-derived exosomes promote the migration of gastric cancer cells by transfer of functional Apolipoprotein E. Cell Death Dis. 2018;9:434
18. Yang Y, Qin M, Bao P, Xu W, Xu J. Secretory carrier membrane protein 5 is an autophagy inhibitor that promotes the secretion of α-Synuclein via exosome. PLoS One. 2017;12:e0180892
19. Yim N, Choi C. Extracellular vesicles as novel carriers for therapeutic molecules. BMB Rep. 2016;49:585-6
20. Blomme A, Fahmy K, Peulen O, Costanza B, Fontaine M, Struman I. et al. Myoferlin is a novel exosomal protein and functional regulator of cancer-derived exosomes. Oncotarget. 2016;7:83669-83
21. Sterzenbach U, Putz U, Low LH, Silke J, Tan SS, Howitt J. Engineered exosomes as vehicles for biologically active proteins. Mol Ther. 2017;25:1269-78
22. Amano T, Furuno T, Hirashima N, Ohyama N, Nakanishi M. Dynamics of intracellular granules with CD63-GFP in rat basophilic leukemia cells. J Biochem. 2001;129:739-44
23. Nabhan JF, Hu R, Oh RS, Cohen SN, Lu Q. Formation and release of arrestin domain-containing protein 1-mediated microvesicles (ARMMs) at plasma membrane by recruitment of TSG101 protein. Proc Natl Acad Sci USA. 2012;109:4146-51
24. Lai CP, Kim EY, Badr CE, Weissleder R, Mempel TR, Tannous BA. et al. Visualization and tracking of tumour extracellular vesicle delivery and RNA translation using multiplexed reporters. Nat Commun. 2015;6:7029
25. Hartman ZC, Wei J, Glass OK, Guo H, Lei G, Yang XY. et al. Increasing vaccine potency through exosome antigen targeting. Vaccine. 2011;29:9361-7
26. Zeelenberg IS, Ostrowski M, Krumeich S, Bobrie A, Jancic C, Boissonnas A. et al. Targeting tumor antigens to secreted membrane vesicles in vivo induces efficient antitumor immune responses. Cancer Res. 2008;68:1228-35
27. Lai CP, Mardini O, Ericsson M, Prabhakar S, Maguire C, Chen JW. et al. Dynamic biodistribution of extracellular vesicles in vivo using a multimodal imaging reporter. ACS Nano. 2014;8:483-94
28. Simpson RJ, Jensen SS, Lim JW. Proteomic profiling of exosomes: current perspectives. Proteomics. 2008;8:4083-99
29. Blanchard N, Lankar D, Faure F, Regnault A, Dumont C, Raposo G. et al. TCR activation of human T cells induces the production of exosomes bearing the TCR/CD3/zeta complex. J Immunol. 2002;168:3235-41
30. Wolfers J, Lozier A, Raposo G, Regnault A, Théry C, Masurier C. et al. Tumor-derived exosomes are a source of shared tumor rejection antigens for CTL cross-priming. Nat Med. 2001;7:297-303
31. Cheng Y, Schorey JS. Targeting soluble proteins to exosomes using a ubiquitin tag. Biotechnol Bioeng. 2016;113:1315-24
32. Giovannone AJ, Reales E, Bhattaram P, Fraile-Ramos A, Weimbs T. Monoubiquitination of syntaxin 3 leads to retrieval from the basolateral plasma membrane and facilitates cargo recruitment to exosomes. Mol Biol Cell. 2017;28:2843-53
33. Gauvreau ME, Côté MH, Bourgeois-Daigneault MC, Rivard LD, Xiu F, Brunet A. et al. Sorting of MHC class II molecules into exosomes through a ubiquitin-independent pathway. Traffic. 2009;10:1518-27
34. Shen B, Wu N, Yang JM, Gould SJ. Protein targeting to exosomes/microvesicles by plasma membrane anchors. J Biol Chem. 2011;286:14383-95
35. Haney MJ, Klyachko NL, Zhao Y, Gupta R, Plotnikova EG, He Z. et al. Exosomes as drug delivery vehicles for Parkinson's disease therapy. J Control Release. 2015;207:18-30
36. Yang T, Fogarty B, LaForge B, Aziz S, Pham T, Lai L. et al. Delivery of small interfering RNA to inhibit vascular endothelial growth factor in zebrafish using natural brain endothelia cell-secreted exosome nanovesicles for the treatment of brain cancer. AAPS J. 2017;19:475-86
37. Shtam TA, Kovalev RA, Varfolomeeva EY, Makarov EM, Kil YV, Filatov MV. Exosomes are natural carriers of exogenous siRNA to human cells in vitro. Cell Commun Signal. 2013;11:88
38. Li SP, Lin ZX, Jiang XY, Yu XY. Exosomal cargo-loading and synthetic exosome-mimics as potential therapeutic tools. Acta Pharmacol Sin. 2018;39:542-51
39. Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29:341-5
40. Théry C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006 Chapter 3: Unit 3.22
41. Heinemann ML, Vykoukal J. Sequential filtration: a gentle method for the isolation of functional extracellular vesicles. Methods Mol Biol. 2017;1660:33-41
42. Niu Z, Pang RTK, Liu W, Li Q, Cheng R, Yeung WSB. Polymer-based precipitation preserves biological activities of extracellular vesicles from an endometrial cell line. PLoS One. 2017;12:e0186534
43. Kojima R, Bojar D, Rizzi G, Hamri GC-E, El-Baba MD, Saxena P. et al. Designer exosomes produced by implanted cells intracerebrally deliver therapeutic cargo for Parkinson's disease treatment. Nat Commun. 2018;9:1305
44. Ausländer S, Ausländer D, Müller M, Wieland M, Fussenegger M. Programmable single-cell mammalian biocomputers. Nature. 2012;487:123-7
45. Saito H, Fujita Y, Kashida S, Hayashi K, Inoue T. Synthetic human cell fate regulation by protein-driven RNA switches. Nat Commun. 2011;2:160
46. Saito H, Kobayashi T, Hara T, Fujita Y, Hayashi K, Furushima R. et al. Synthetic translational regulation by an L7Ae-kink-turn RNP switch. Nat Chem Biol. 2010;6:71-8
47. Soares AR, Martins-Marques T, Ribeiro-Rodrigues T, Ferreira JV, Catarino S, Pinho MJ. et al. Gap junctional protein Cx43 is involved in the communication between extracellular vesicles and mammalian cells. Sci Rep. 2015;5:13243
48. Batagov AO, Kuznetsov VA, Kurochkin IV. Identification of nucleotide patterns enriched in secreted RNAs as putative cis-acting elements targeting them to exosome nano-vesicles. BMC Genomics. 2011;12:S18
49. Teng Y, Ren Y, Hu X, Mu J, Samykutty A, Zhuang X. et al. MVP-mediated exosomal sorting of miR-193a promotes colon cancer progression. Nat Commun. 2017;8:14448
50. Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654-9
51. Gibbings DJ, Ciaudo C, Erhardt M, Voinnet O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat Cell Biol. 2009;11:1143-9
52. Luo Q, Guo D, Liu G, Chen G, Hang M, Jin M. Exosomes from miR-126-overexpressing Adscs are therapeutic in relieving acute myocardial ischaemic injury. Cell Physiol Biochem. 2017;44:2105-16
53. Wang X, Zhang H, Bai M, Ning T, Ge S, Deng T. et al. Exosomes serve as nanoparticles to deliver anti-miR-214 to reverse chemoresistance to cisplatin in gastric cancer. Mol Ther. 2018;26:774-83
54. Zhang H, Wang Y, Bai M, Wang J, Zhu K, Liu R. et al. Exosomes serve as nanoparticles to suppress tumor growth and angiogenesis in gastric cancer by delivering hepatocyte growth factor siRNA. Cancer Sci. 2018;109:629-41
55. Lin Y, Wu J, Gu W, Huang Y, Tong Z, Huang L. et al. Exosome-liposome hybrid nanoparticles deliver CRISPR/Cas9 system in MSCs. Adv Sci (Weinh). 2018;5:1700611
56. Ohno S, Takanashi M, Sudo K, Ueda S, Ishikawa A, Matsuyama N. et al. Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells. Mol Ther. 2013;21:185-91
57. Zarovni N, Corrado A, Guazzi P, Zocco D, Lari E, Radano G. et al. Integrated isolation and quantitative analysis of exosome shuttled proteins and nucleic acids using immunocapture approaches. Methods. 2015;87:46-58
58. Davies RT, Kim J, Jang SC, Choi EJ, Gho YS, Park J. Microfluidic filtration system to isolate extracellular vesicles from blood. Lab Chip. 2012;12:5202-10
59. Dragovic RA, Gardiner C, Brooks AS, Tannetta DS, Ferguson DJ, Hole P. et al. Sizing and phenotyping of cellular vesicles using nanoparticle tracking analysis. Nanomedicine. 2011;7:780-8
60. Kim MS, Haney MJ, Zhao Y, Mahajan V, Deygen I, Klyachko NL. et al. Development of exosome-encapsulated paclitaxel to overcome MDR in cancer cells. Nanomedicine. 2016;12:655-64
61. Pascucci L, Coccè V, Bonomi A, Ami D, Ceccarelli P, Ciusani E. et al. Paclitaxel is incorporated by mesenchymal stromal cells and released in exosomes that inhibit in vitro tumor growth: a new approach for drug delivery. J Control Release. 2014;192:262-70
62. Haney MJ, Klyachko NL, Zhao Y, Gupta R, Plotnikova EG, He Z. et al. Exosomes as drug delivery vehicles for Parkinson's disease therapy. J Control Release. 2015;207:18-30
63. Barile L, Vassalli G. Exosomes: therapy delivery tools and biomarkers of diseases. Pharmacol Ther. 2017;174:63-78
64. Sharif S, Ghahremani MH, Soleimani M. Delivery of exogenous miR-124 to glioblastoma multiform cells by Wharton's Jelly mesenchymal stem cells decreases cell proliferation and migration, and confers chemosensitivity. Stem Cell Rev. 2018;14:236-46
65. Ye Y, Li SL, Ma YY, Diao YJ, Yang L, Su MQ. et al. Exosomal miR-141-3p regulates osteoblast activity to promote the osteoblastic metastasis of prostate cancer. Oncotarget. 2017;8:94834-49
66. Zhao H, Yang L, Baddour J, Achreja A, Bernard V, Moss T. et al. Tumor microenvironment derived exosomes pleiotropically modulate cancer cell metabolism. eLife. 2016;5:e10250
67. Hoshino A, Costa-Silva B, Shen TL, Rodrigues G, Hashimoto A, Tesic Mark M. et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527:329-35
68. Rak J. Cancer: organ-seeking vesicles. Nature. 2015;527:312-4
69. Tian Y, Li S, Song J, Ji T, Zhu M, Anderson GJ. et al. A doxorubicin delivery platform using engineered natural membrane vesicle exosomes for targeted tumor therapy. Biomaterials. 2014;35:2383-90
70. Yang J, Zhang X, Chen X, Wang L, Yang G. Exosome mediated delivery of miR-124 promotes neurogenesis after ischemia. Mol Ther Nucleic Acids. 2017;7:278-87
71. Liu Y, Li D, Liu Z, Zhou Y, Chu D, Li X. et al. Targeted exosome-mediated delivery of opioid receptor Mu siRNA for the treatment of morphine relapse. Sci Rep. 2015;5:17543
72. Tian T, Zhang HX, He CP, Fan S, Zhu YL, Qi C. et al. Surface functionalized exosomes as targeted drug delivery vehicles for cerebral ischemia therapy. Biomaterials. 2018;150:137-49
73. Chen W, Yang M, Bai J, Li X, Kong X, Gao Y. et al. Exosome-modified tissue engineered blood vessel for endothelial progenitor cell capture and targeted siRNA delivery. Macromol Biosci. 2018;18:201700242
74. Pi F, Binzel DW, Lee TJ, Li Z, Sun M, Rychahou P. et al. Nanoparticle orientation to control RNA loading and ligand display on extracellular vesicles for cancer regression. Nat Nanotechnol. 2018;13:82-9
75. Li Y, Gao Y, Gong C, Wang Z, Xia Q, Gu F. et al. A33 antibody-functionalized exosomes for targeted delivery of doxorubicin against colorectal cancer. Nanomedicine. 2018;14:1973-85
76. Wang J, Dong Y, Li Y, Li W, Cheng K, Qian Y. et al. Designer exosomes for active targeted chemo-photothermal synergistic tumor therapy. Adv Funct Mater. 2018;18:201707360
77. Kim MS, Haney MJ, Zhao Y, Yuan D, Deygen I, Klyachko NL. et al. Engineering macrophage-derived exosomes for targeted paclitaxel delivery to pulmonary metastases: in vitro and in vivo evaluations. Nanomedicine. 2018;14:195-204
78. Tang J, Su T, Huang K, Dinh PU, Wang Z, Vandergriff A. et al. Targeted repair of heart injury by stem cells fused with platelet nanovesicles. Nat Biomed Eng. 2018;2:17-26
79. Kooijmans SAA, Fliervoet LAL, van der Meel R, Fens M, Heijnen HFG, van Bergen En Henegouwen PMP. et al. PEGylated and targeted extracellular vesicles display enhanced cell specificity and circulation time. J Control Release. 2016;224:77-85
80. Yeo RW, Lai RC, Zhang B, Tan SS, Yin Y, Teh BJ. et al. Mesenchymal stem cell: an efficient mass producer of exosomes for drug delivery. Adv Drug Deliv Rev. 2013;65:336-41
81. Rana S, Yue S, Stadel D, Zöller M. Toward tailored exosomes: the exosomal tetraspanin web contributes to target cell selection. Int J Biochem Cell Biol. 2012;44:1574-84
82. Zomer A, Maynard C, Verweij FJ, Kamermans A, Schäfer R, Beerling E. et al. In vivo imaging reveals extracellular vesicle-mediated phenocopying of metastatic behavior. Cell. 2015;161:1046-57
83. Harris DA, Patel SH, Gucek M, Hendrix A, Westbroek W, Taraska JW. Exosomes released from breast cancer carcinomas stimulate cell movement. PLoS One. 2015;10:e0117495
84. Tomihari M, Chung JS, Akiyoshi H, Cruz PD, Ariizumi K. DC-HIL/glycoprotein Nmb promotes growth of melanoma in mice by inhibiting the activation of tumor-reactive T cells. Cancer Res. 2010;70:5778-87
85. Kim JW, Wieckowski E, Taylor DD, Reichert TE, Watkins S, Whiteside TL. Fas ligand-positive membranous vesicles isolated from sera of patients with oral cancer induce apoptosis of activated T lymphocytes. Clin Cancer Res. 2005;11:1010-20
86. Luan X, Sansanaphongpricha K, Myers I, Chen H, Yuan H, Sun D. Engineering exosomes as refined biological nanoplatforms for drug delivery. Acta Pharmacol Sin. 2017;38:754-63
87. Ju S, Mu J, Dokland T, Zhuang X, Wang Q, Jiang H. et al. Grape exosome-like nanoparticles induce intestinal stem cells and protect mice from DSS-induced colitis. Mol Ther. 2013;21:1345-57
88. Wang Q, Ren Y, Mu J, Egilmez NK, Zhuang X, Deng Z. et al. Grapefruit-derived nanovectors use an activated leukocyte trafficking pathway to deliver therapeutic agents to inflammatory tumor sites. Cancer Res. 2015;75:2520-9
89. Munagala R, Aqil F, Jeyabalan J, Gupta RC. Bovine milk-derived exosomes for drug delivery. Cancer Lett. 2016;371:48-61
90. Ingato D, Lee JU, Sim SJ, Kwon YJ. Good things come in small packages: overcoming challenges to harness extracellular vesicles for therapeutic delivery. J Control Release. 2016;241:174-85
91. Savina A, Furlán M, Vidal M, Colombo MI. Exosome release is regulated by a calcium-dependent mechanism in K562 cells. J Biol Chem. 2003;278:20083-90
92. Distler JH, Pisetsky DS, Huber LC, Kalden JR, Gay S, Distler O. Microparticles as regulators of inflammation: novel players of cellular crosstalk in the rheumatic diseases. Arthritis Rheum. 2005;52:3337-48
93. Fauré J, Lachenal G, Court M, Hirrlinger J, Chatellard-Causse C, Blot B. et al. Exosomes are released by cultured cortical neurones. Mol Cell Neurosci. 2006;31:642-8
94. Krämer-Albers EM, Bretz N, Tenzer S, Winterstein C, Möbius W, Berger H. et al. Oligodendrocytes secrete exosomes containing major myelin and stress-protective proteins: trophic support for axons? Proteomics Clin Appl. 2007;1:1446-61
95. Miyoshi H, Umeshita K, Sakon M, Imajoh-Ohmi S, Fujitani K, Gotoh M. et al. Calpain activation in plasma membrane bleb formation during tert-butyl hydroperoxide-induced rat hepatocyte injury. Gastroenterology. 1996;110:1897-904
96. Hedlund M, Nagaeva O, Kargl D, Baranov V, Mincheva-Nilsson L. Thermal- and oxidative stress causes enhanced release of NKG2D ligand-bearing immunosuppressive exosomes in leukemia/lymphoma T and B cells. PLoS One. 2011;6:e16899
97. King HW, Michael MZ, Gleadle JM. Hypoxic enhancement of exosome release by breast cancer cells. BMC Cancer. 2012;12:421
98. Wysoczynski M, Ratajczak MZ. Lung cancer secreted microvesicles: underappreciated modulators of microenvironment in expanding tumors. Int J Cancer. 2009;125:1595-603
99. Parolini I, Federici C, Raggi C, Lugini L, Palleschi S, De Milito A. et al. Microenvironmental pH is a key factor for exosome traffic in tumor cells. J Biol Chem. 2009;284:34211-22
100. de Jong OG, Verhaar MC, Chen Y, Vader P, Gremmels H, Posthuma G. et al. Cellular stress conditions are reflected in the protein and RNA content of endothelial cell-derived exosomes. J Extracell Vesicles. 2012;1:18396
101. Zaborowski MP, Balaj L, Breakefield XO, Lai CP. Extracellular vesicles: composition, biological relevance, and methods of study. Bioscience. 2015;65:783-97
102. Scott RE, Perkins RG, Zschunke MA, Hoerl BJ, Maercklein PB. Plasma membrane vesiculation in 3T3 and SV3T3 cells. I. Morphological and biochemical characterization. J Cell Sci. 1979;35:229-43
103. Scott RE. Plasma membrane vesiculation: a new technique for isolation of plasma membranes. Science. 1976;194:743-5
104. Dalle-Donne I, Rossi R, Milzani A, Di Simplicio P, Colombo R. The actin cytoskeleton response to oxidants: from small heat shock protein phosphorylation to changes in the redox state of actin itself. Free Radic Biol Med. 2001;31:1624-32
105. Sezgin E, Kaiser HJ, Baumgart T, Schwille P, Simons K, Levental I. Elucidating membrane structure and protein behavior using giant plasma membrane vesicles. Nat Protoc. 2012;7:1042-51
106. Charras GT, Hu CK, Coughlin M, Mitchison TJ. Reassembly of contractile actin cortex in cell blebs. J Cell Biol. 2006;175:477-90
107. Datta A, Kim H, McGee L, Johnson AE, Talwar S, Marugan J. et al. High-throughput screening identified selective inhibitors of exosome biogenesis and secretion: a drug repurposing strategy for advanced cancer. Sci Rep. 2018;8:8161
108. Lee YS, Kim SY, Ko E, Lee JH, Yi HS, Yoo YJ. et al. Exosomes derived from palmitic acid-treated hepatocytes induce fibrotic activation of hepatic stellate cells. Sci Rep. 2017;7:3710
109. Khan FM, Saleh E, Alawadhi H, Harati R, Zimmermann WH, El-Awady R. Inhibition of exosome release by ketotifen enhances sensitivity of cancer cells to doxorubicin. Cancer Biol Ther. 2018;19:25-33
110. Zhu J, Lu K, Zhang N, Zhao Y, Ma Q, Shen J. et al. Myocardial reparative functions of exosomes from mesenchymal stem cells are enhanced by hypoxia treatment of the cells via transferring microRNA-210 in an nSMase2-dependent way. Artif Cells Nanomed Biotechnol. 2017:1-12
111. Menck K, Sönmezer C, Worst TS, Schulz M, Dihazi GH, Streit F. et al. Neutral sphingomyelinases control extracellular vesicles budding from the plasma membrane. J Extracell Vesicles. 2017;6:1378056
112. Böker KO, Lemus-Diaz N, Rinaldi Ferreira R, Schiller L, Schneider S, Gruber J. The impact of the CD9 tetraspanin on lentivirus infectivity and exosome secretion. Mol Ther. 2018;26:634-47
113. Gonzalez-King H, García NA, Ontoria-Oviedo I, Ciria M, Montero JA, Sepúlveda P. Hypoxia inducible factor-1alpha potentiates Jagged 1-mediated angiogenesis by mesenchymal stem cell-derived exosomes. Stem Cells. 2017;35:1747-59
114. Emam SE, Ando H, Abu Lila AS, Shimizu T, Ukawa M, Okuhira K. et al. A novel strategy to increase the yield of exosomes (extracellular vesicles) for an expansion of basic research. Biol Pharm Bull. 2018;41:733-42
115. Gao T, Guo W, Chen M, Huang J, Yuan Z, Zhang Y. et al. Extracellular vesicles and autophagy in osteoarthritis. Biomed Res Int. 2016;2016:2428915
116. Aubertin K, Silva AK, Luciani N, Espinosa A, Djemat A, Charue D. et al. Massive release of extracellular vesicles from cancer cells after photodynamic treatment or chemotherapy. Sci Rep. 2016;6:35376
Author contact
![]() Corresponding author: Changqing Su, MD, PhD, Department of Molecular Oncology, Eastern Hepatobiliary Surgical Hospital & National Center for Liver Cancer, Second Military Medical University, Shanghai 200438, China. Tel.: 0086-021-81875351; Email: suchangqingcom
Corresponding author: Changqing Su, MD, PhD, Department of Molecular Oncology, Eastern Hepatobiliary Surgical Hospital & National Center for Liver Cancer, Second Military Medical University, Shanghai 200438, China. Tel.: 0086-021-81875351; Email: suchangqingcom
 Global reach, higher impact
Global reach, higher impact