13.3
Impact Factor
Theranostics 2019; 9(2):573-587. doi:10.7150/thno.27654 This issue Cite
Research Paper
USP5 promotes epithelial-mesenchymal transition by stabilizing SLUG in hepatocellular carcinoma
1. State Key Laboratory of Medicinal Chemical Biology and College of Pharmacy, Nankai University, Tianjin, China
2. Tianjin Key Laboratory of Molecular Drug Research, Tianjin International Joint Academy of Biomedicine, Tianjin, China
3. Department of Gastroenterology and Hepatology, Tianjin Key Laboratory of Artificial Cells, Tianjin Institute of Hepatobiliary Disease, Tianjin Third Central Hospital, Tianjin, China
4. College of Life Science, Nankai University, Tianjin, China
* These authors contributed equally to this work.
Received 2018-6-4; Accepted 2018-11-3; Published 2019-1-1
Abstract
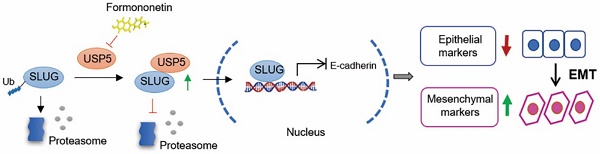
Rationale: The role of SLUG in epithelial-mesenchymal transition during tumor progression has been thoroughly studied, but its precise regulation remains poorly explored.
Methods: The affinity purification, mass spectrometry and CO-IP were performed to identify the interaction between SLUG and ubiquitin-specific protease 5 (USP5). Cycloheximide chase assays and deubiquitination assays confirmed that the effect of USP5 on the deubiquitin of SLUG. The dual-luciferase reporter and chromatin immunoprecipitation assays were employed to observe the direct transcriptional regulation of E-cadherin by SLUG effected by USP5. EMT related markers was detected by western blotting and immunofluorescence. Molecular docking, SPR sensor (biacore) and co-location were detected to prove Formononetin targets USP5.
Bioinformatics analysis was used to study the relation of USP5 and SLUG to malignancy degree of HCC. Cell migration, invasion in HCC cells and xenografts model in nude mouse were conducted to detect the promotion of USP5 and the inhibition of Formononetin on EMT.
Results: USP5 interacts with and stabilizes SLUG to regulate its abundance through USP5 deubiquitination activities in epithelial-mesenchymal transition (EMT) of hepatocellular carcinoma (HCC). USP5 is highly expressed and positively correlated with SLUG expression in HCC with high malignancy. Knockdown of USP5 inhibits SLUG deubiquitination and inhibits HCC cells proliferation, metastasis, and invasion, while overexpression of USP5 promotes SLUG stability and EMT in vitro and in vivo. Through virtual screening, we found that Formononetin exhibits excellent binding to USP5. Moreover, Formononetin inhibits deubiquitinating activities of USP5 to SLUG and consequently impedes the EMT and malignant progression of HCC.
Conclusion: Our findings reveal that USP5 serve as a potential target for tumor intervention and provide a preliminary antitumor therapy for inhibit EMT by targeting USP5 or its interaction with SLUG in HCC.
Keywords: USP5, SLUG, EMT, Formononetin, hepatocellular carcinoma
Introduction
Hepatocellular carcinoma is the most common malignant liver tumor, and epithelial-mesenchymal transition (EMT) plays a critical role in tumor progression [1, 2]. Tumor cells undergo EMT change to a mesenchymal phenotype with invasive capacities, thereby increasing the migratory capacity [3]. Therefore, inhibiting of EMT is an essential way to suppress tumor metastasis [2].
SLUG, a zinc-finger transcriptional factor, represses E-cadherin transcription via the E-box elements in the proximal E-cadherin promoter in EMT progression [4]. In addition, SLUG is aberrantly expressed in various cancers where it regulates diverse processes ranging from tumor cell invasion and metastasis to cell survival and proliferation [5-12]. The SLUG protein is quickly turned over by the ubiquitin-proteasome system. In tumor cells, high SLUG expression indicates the presence of a SLUG stabilizing factor [13].
Ubiquitination plays an important role in the post-translational modification of cellular proteins that are involved in multiple diseases, including cancers [14]. Many oncogenes and tumor suppressors are short-lived proteins targeted by the ubiquitin-proteasome system, and their deregulated proteolysis is involved in cancer [15]. USP5, a member of the ubiquitin-specific protease (USP) family of deubiquitinating enzymes (DUBs), removes ubiquitin from the proximal end of unanchored polyubiquitin chains [16-18]. USP5 targets several cytosolic proteins, such as p53 and Cav3.2 proteins, and subsequently regulates multiple cellular activities including the repair of DNA double-strand breaks, transmission of inflammatory pain and neural pain signals, immune response, and tumor cell proliferation [19-24]. However, the mechanism of USP5 in hepatocellular carcinoma progression remains unclear.
We reported that USP5 interacts with SLUG and inhibits its ubiquitination, thereby stabilizing SLUG expression in HCC. Knockdown of USP5 inhibits EMT and metastasis due to SLUG degradation in HCC. Formononetin, an O-methylated isoflavone phytoestrogen extracted from plants and herbs, targets USP5 and suppresses EMT by inhibiting the USP5-mediated deubiquitination of SLUG. This study revealed the enhancing effect of USP5 on the malignant progression of HCC and provided a potential antitumor therapy of HCC by inhibiting USP5 deubiquitination activities.
Results
SLUG is physically associated with USP5
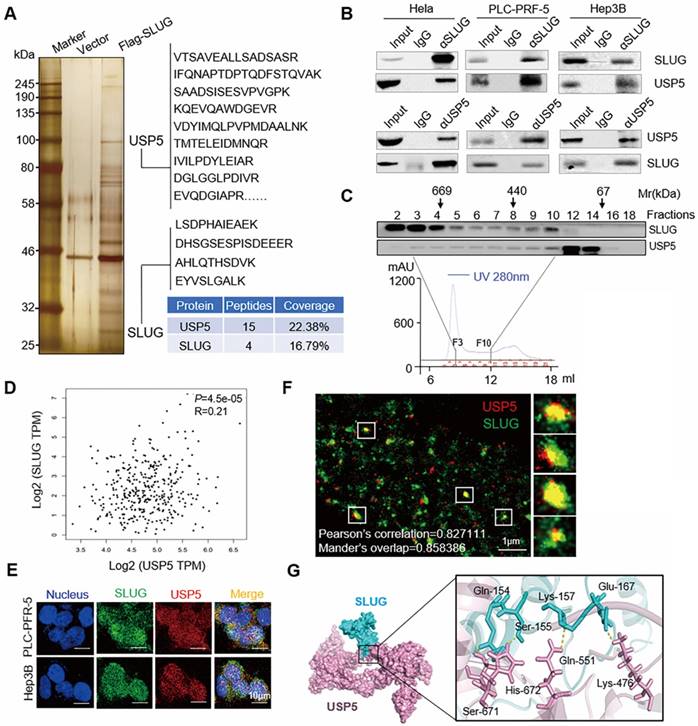
Affinity purification and mass spectrometry were performed to identify the interaction proteins of SLUG to comprehensively understand the function of SLUG. Whole-cell extracts from Hela cells with or without Flag-SLUG expression was applied to affinity purification and mass spectrometry analysis (Figure 1A). SLUG protein was associated with several protein interactions (Supplementary data 1). USP5, a DUB, was also found in the SLUG-containing protein complex.
To explore the relationship between SLUG and USP5, co-immunoprecipitation experiments were performed in Hela cell extracts and the results showed that SLUG efficiently co-immunoprecipitated with USP5. Meanwhile, USP5 was efficiently co-immunoprecipitated with SLUG. Similar observations were also detected in PLC-PRF-5 cells and Hep3B cells (Figure 1B).
To further confirm the interaction between SLUG and USP5, we performed fast protein liquid chromatography (FPLC) experiments with a Superdex 200 10/300 GL. The results indicated that native SLUG from Hela cells was eluted with an apparent molecular mass much greater than that of the monomeric protein. The majority of SLUG co-existed with USP5 in a multiprotein complex. The overlap peaked in fraction 10 (Figure 1C).
Correlation analysis of expression of USP5 and SLUG in TCGA database revealed that the expression level of USP5 and SLUG was positively correlated of TCGA HCC samples from GEPIA database (http://gepia.cancer-pku.cn) (P<0.05, R=0.21, Figure 1D). We examined the protein expression levels of USP5 and SLUG in multiple HCC cell lines and two normal liver cells. There was a correlation between the expression of USP5 and SLUG in PLC-PRF-5, MHCC-97L, MHCC-97H, SK-Hep-1, Hep3B cells, and the expression in normal liver cells was lower than that in most HCC cells (Figure S1). SLUG and USP5 were found to be co-located according to the immunofluorescence experiments in PLC-PRF-5 and Hep3B cells (Figure 1E). The multidirectional colocalization of the two proteins was accurately analyzed by immunostaining with antibodies against SLUG and USP5 via N-STORM (Figure 1F). Docking of protein-protein interface interaction of SLUG and USP5 were performed (Figure 1G).
USP5 stabilizes SLUG through its deubiquitination activity
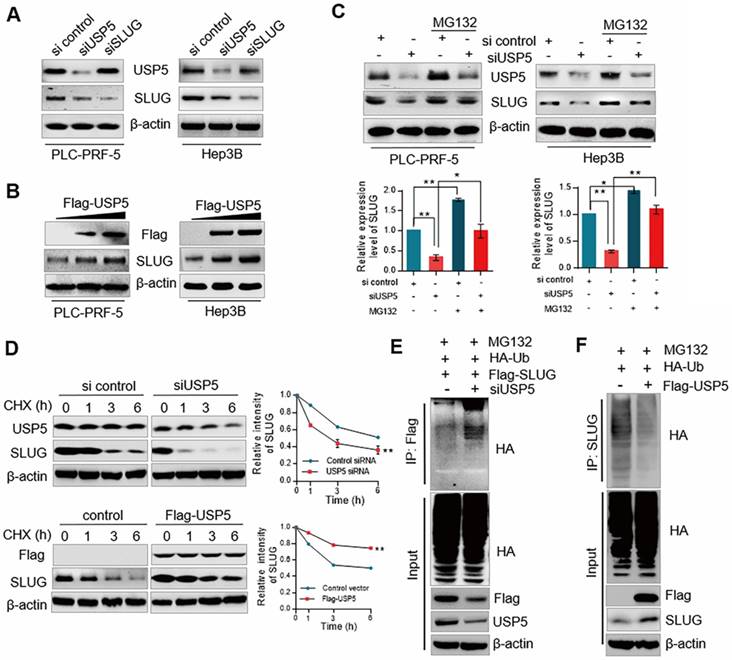
To understand the functional significance of the physical interaction and spatial co-localization between USP5 and SLUG, PLC-PRF-5 and Hep3B cells knocked down or overexpressed USP5 and SLUG. Knocked down USP5 decreased the expression of SLUG, while the expression of USP5 was nearly unchanged when SLUG was knocked down (Figure 2A). Similar observation was obtained in immunofluorescence analysis (Figure S2). USP5 overexpression increased SLUG expression in PLC-PRF-5 and Hep3B cells (Figure 2B). Moreover, the reduction in protein level of SLUG due to USP5 depletion could be rescued by MG132, a proteasome inhibitor (Figure 2C). Thus, we suspected that USP5 stabilized SLUG by inhibiting SLUG degradation through proteasome. For confirmation, cycloheximide (CHX) chase assays were conducted to detect the half-life of SLUG with USP5 knockdown or overexpressed treatments (Figure 2D). The results showed that SLUG half-life was decreased in USP5-deficient cell. Consistently, USP5 overexpression prolonged the half-life of SLUG. All the results indicate that USP5 regulates the stability of SLUG.
SLUG is physically associated with USP5. (A) Immunoaffinity purification and mass spectrometry analysis of SLUG-associated proteins in Hela cells. Representative peptide fragments and peptide coverage of SLUG and USP5 are shown. (B) Whole-cell lysates from HeLa, PLC-PRF-5 and Hep3B cells were immunoprecipitated followed by IB with antibodies against the indicated proteins. (C) Whole Hela cell extracts were fractionated on Superdex 200 10/300 GL with PBS. Chromatographic elution profiling and IB analysis of the chromatographic fractions with antibodies against USP5 and SLUG were conducted. (D) Statistical analysis of the correlation analysis between USP5 and SLUG expression of TCGA HCC samples from GEPIA database (P<0.05, R=0.21). (E) Co-localization of USP5 and SLUG was analyzed by immunostaining of PLC-PRF-5 cells with anti- USP5 and SLUG via confocal microscopy. Scale bars, 10 μm. (F) Accurate multidirectional co-localization of USP5 and SLUG was analyzed by immunostaining of PLC-PRF-5 cells with anti- USP5 and SLUG via N-STROM. Red, USP5; Green, SLUG. Pearson's correlation=0.827, Mander's overlap=0.858. (G)Molecular docking of USP5 and SLUG.
Considering the function of USP5 as a DUB, we speculated that USP5 may affect the stability of SLUG by deubiquitinating SLUG. The deubiquitination assays confirmed that ubiquitin on SLUG protein was significantly reduced in USP5 overexpressed cells and accordingly increased when USP5 was knocked down (Figs. 2E and 2F). Collectively, these results indicated that USP5 promotes SLUG stabilization by deubiquitinating SLUG.
USP5 promotes EMT and metastasis in hepatocellular carcinoma
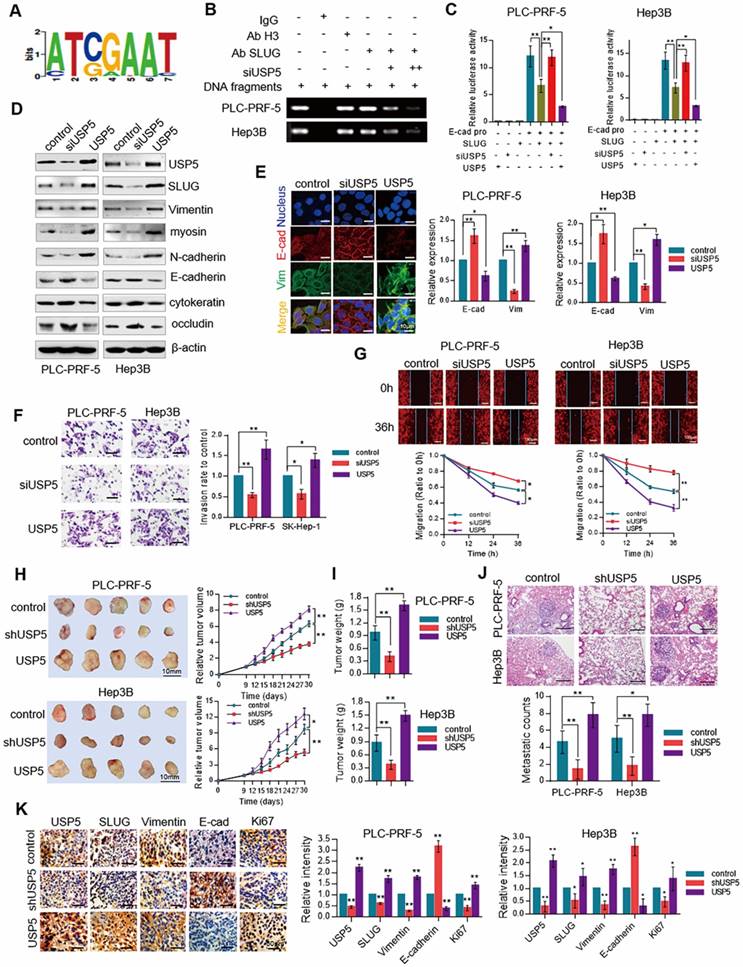
SLUG, a critical protein in EMT, promotes EMT progression by repressing E-cadherin expression [4]. To understand the biological significance of USP5-mediated stabilization of SLUG, we investigated the effect of USP5 on EMT. ChIPBASE motif analysis revealed the binding base sequence of SLUG (Figure 3A). Chromatin IP (ChIP) assay showed that the binding of SLUG to E-cadherin promoter was significantly inhibited in USP5-deficient cells (Figure 3B). Luciferase reporter gene assay showed that knockdown USP5 interfered with the transcriptional inhibition of SLUG on E-cadherin, and over-expression of USP5 promoted the transcriptional inhibition of SLUG on E-cadherin (Figure 3C). Western blot analysis further affirmed that the expression level of epithelial markers (E-cadherin, cytokeratin, and occludin) increased, and the expression level of mesenchymal markers (Vimentin, N-cadherin, and myosin) decreased in PLC-PRF-5 and Hep3B cells knocked down USP5, while overexpressed USP5, the EMT related markers had corresponding changes (Figure 3D). Similar observation was obtained in immunofluorescence analysis of E-cadherin and Vimentin in PLC-PRF-5 and Hep3B cells under USP5 siRNA or overexpressed treatment. Knockdown USP5 increased the fluorescence intensity of E-cadherin but reduced that of Vimentin, and the results was opposite in USP5 overexpressed cells (Figure 3E). Transwell assay and wound healing assay results also showed that knockdown of USP5 inhibited cell invasion and migration and overexpression of USP5 promoted cell invasion and migration (Figure 3F and 3G).
USP5 stabilizes SLUG through its deubiquitination activity. (A) PLC-PRF-5 and Hep3B cells were transfected with USP5 siRNA, SLUG siRNA. Cellular extracts were collected for WB analysis. (B) PLC-PRF-5 and Hep3B cells were transfected with different amounts of Flag-USP5 for WB analysis with antibodies against USP5 and SLUG. (C) PLC-PRF-5 and Hep3B cells were transfected with control siRNA or USP5 siRNA with or without 5 μM MG132 and followed by immunoblotting using anti-USP5 and SLUG. Intensity of each band from biological triplicate experiments was quantified. Each bar represents the mean ± SD for biological triplicate experiments. *P<0.05, **P<0.01, one-way ANOVA. (D) PLC-PRF-5 cells transfected with USP5 siRNA or FLAG-USP5 were treated with CHX and harvested at the indicated time followed by WB. Intensity of each band from biological triplicate experiments was quantified. Each point represents the mean ± SD for biological triplicate experiments. **P<0.01, Student's t-test. (E) PLC-PRF-5 cells were treated with USP5 siRNA together with HA-Ub and FLAG-SLUG followed by treatment with MG132. Cellular extracts were prepared for co-immunoprecipitation assays with anti-FLAG followed by IB with anti-HA. (F) PLC-PRF-5 cells were transfected with Flag-USP5 together with HA-Ub followed by treatment with MG132. Cellular extracts were prepared for co-immunoprecipitation assays with anti-SLUG followed by IB with anti-HA.
USP5 promotes EMT in hepatocellular carcinoma. (A) Motif analysis of SLUG ChIP-Seq cited from ChIPBASE. (B) PLC-PRF-5 and Hep3B cells were treated with different amounts of USP5 siRNA. Cellular extracts were prepared for ChIP assays with anti-SLUG. (C) PLC-PRF-5 and Hep3B cells were transfected with E-cadherin - dependent reporter gene plasmids. Luciferase activity was measured when cells overexpressed or knocked down USP5. (D) WB analysis of USP5, SLUG and EMT related markers in PLC-PRF-5 and Hep3B cells under USP5 knocked down or overexpressed treatment. (E) Immunofluorescence assay of PLC-PRF-5 and Hep3B cells treated with USP5 siRNA or overexpression vectors. The relative intensity of E-cadherin and Vimentin was analyzed by the Image J software. Scale bar, 10 μm. *P<0.05, **P<0.01, Student's t-test. (F) Transwell assay of PLC-PRF-5 and Hep3B cells treated with USP5 siRNA or USP5 overexpression vectors. Scale bar, 20 μm. *P<0.05, **P<0.01, Student's t-test. (G) Scratch assay of PLC-PRF-5 and Hep3B cells treated with USP5 siRNA or overexpression vectors and stained with DiD. Representative images from biological triplicate experiments are shown. Scale bar, 100 μm. *P<0.05, **P<0.01, Student's t-test. (H) PLC-PRF-5 and Hep3B cells knocked down or overexpressed USP5 was transplanted on nude mice, and tumor volumes were measured every 3 days. (I) Tumor weight of control, USP5 knockdown and overexpression groups. (J) Representative images and number of pulmonary metastasis nodules in control, knockdown and overexpressed USP5 groups (Scale bar, 200μm). (K) IHC analysis of USP5, SLUG, E-cadherin, Vimentin and Ki67 in tumors of control, shUSP5, and USP5 groups. scale bar, 50 μm. *P<0.05, **P<0.01. Student's t-test. Each bar represents the mean ± SD for biological triplicate experiments or different animal measurements (n = 5).
To establish the role of USP5/SLUG in the malignant progression of HCC in vivo, USP5-deficient or overexpressed PLC-PRF-5 and Hep3B cells were transplanted subcutaneously into nude mice to establish xenograft models. Tumor growth was suppressed in nude mice receiving tumor transplants with depletion of USP5 and was promoted in mice receiving tumor transplants with USP5 overexpression (Figure 3H). The tumors weight was decreased in knocked down USP5 groups and increased in USP5 overexpression groups (Figure 3I). Moreover, metastatic nodules of lung were decreased in USP5-deficient cells burdened mice and increased in USP5 overexpressed cells burdened mice (Figure 3J). We then examined the expression level of USP5, SLUG, E-cadherin, Vimentin, and Ki67 in xenografts with immunohistochemistry (IHC) staining assays. The results in Figure 3K showed that the expression of USP5, SLUG, and Vimentin reduced, whereas the expression of E-cadherin increased. The results were detected in USP5 overexpressed tumors. All the results indicated that USP5 promotes the growth and metastasis of hepatocellular carcinoma.
Expression level of USP5 and SLUG is related to malignancy degree of hepatocellular carcinoma
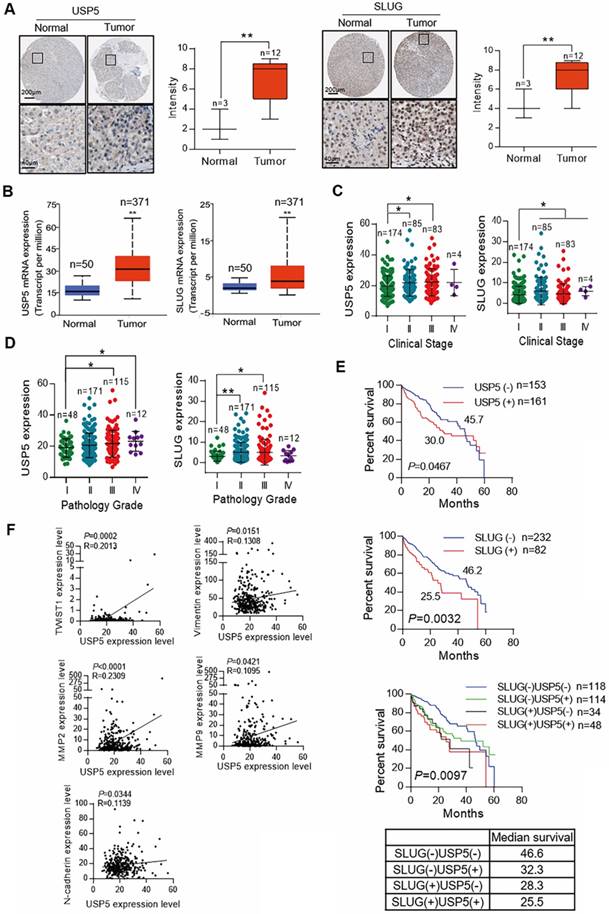
Clinical data were analyzed to confirm the role of USP5/SLUG axis in HCC. Representative image of immunohistochemical assay from the Human Protein Atlas and score is shown in Figure 4a. The results revealed that the expression of USP5 and SLUG in tumors was higher than that in normal liver tissues (Figure 4A). Cancer transcription analysis of TCGA samples from UALCAN database (http://ualcan.path.uab.edu/index.html) also showed that USP5 and SLUG were highly expressed in hepatocellular carcinoma tissues compared with normal liver tissue (Figure 4B). At the same time, the expression of USP5 and SLUG was positively correlated with clinical stage and pathological grade in TCGA hepatocellular carcinoma samples (Figs. 4C and 4D). Survival analysis also revealed that the high expression of USP5 and SLUG indicated poor prognosis (Figure 4E). The Correlation analysis of USP5 and key transcription factors and EMT related markers (TWIST1, Vimentin, MMP2, MMP9 and N-cadherin) revealed that the expression of USP5 was closely related to EMT and tumor metastasis. Our findings reveal that USP5/SLUG axis promotes the malignant progression of patients with HCC, and thus support the possibility of using USP5/SLUG as potential targets for HCC therapies.
Formononetin targets USP5 to inhibit the deubiquitination of SLUG
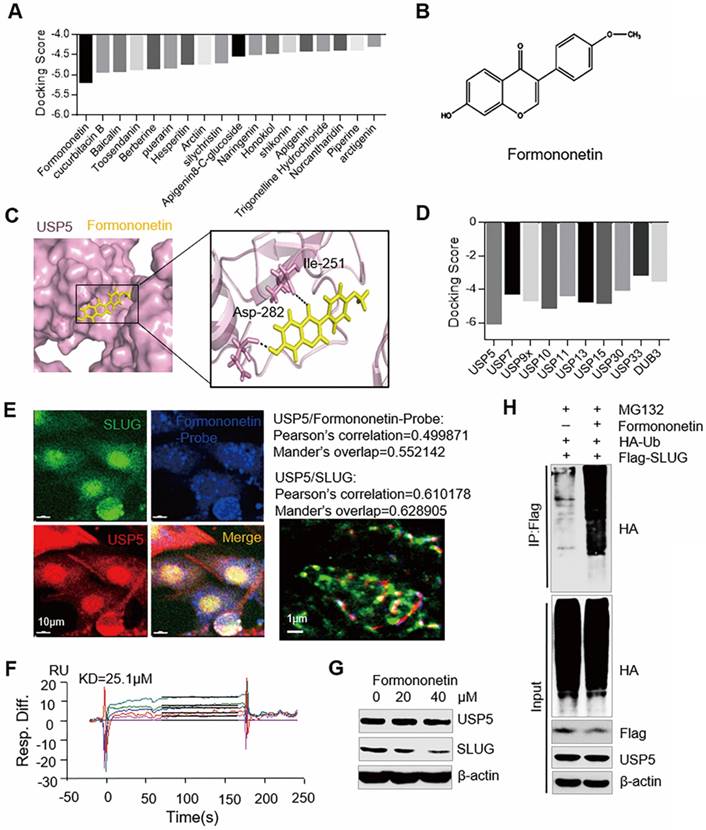
Based on the structure of USP5, lead compounds with highest docking score to USP5 were selected from the traditional Chinese medicine database by virtual screening (Figure 5A and 5B). Molecular dynamics simulation displayed the dynamic interactions that Formononetin binds to USP5 in 100 ns (Figure 5C). Molecular docking results also showed that Formononetin exhibited the highest docking score to USP5 among USP family members. This finding reveals the specificity of Formononetin (Figure 5D). According to the immunofluorescence probe co-localization images, we found that Formononetin-probe and USP5 were co-located (Pearson's correlation = 0.499871, Mander's overlap = 0.552142) (Figure 5E). The synthesis method of Formononetin-probe is shown in figure S3. The expression of USP5 and SLUG were co-location (Pearson's correlation = 0.610178, Mander's overlap = 0.628905) (Figure 5E). Biacore experiment was performed to further verify whether USP5 interacts with Formononetin. The dissociation constant (Kd value) was 25.1 μM (Figure 5F). All the results showed that Formononetin directly targets USP5.
To explore the influence of Formononetin on USP5/SLUG axis, PLC-PRF-5 cells treated with Formononetin at different concentrations were used for WB analysis (Figure 5G). The results indicated that the expression level of SLUG was decreased, whereas that of USP5 showed no noticeable change. We subsequently conducted deubiquitination assay to detect the ubiquitin of SLUG in PLC-PRF-5 cells treated with 40 μM of Formononetin (Figure 5H). The ubiquitin of SLUG was increased in Formononetin-treated group. These results indicate that Formononetin targets USP5 to inhibit the deubiquitination of SLUG.
Formononetin inhibits EMT of hepatocellular carcinoma
To investigate the influence of Formononetin on inhibiting USP5, ChIP assay was conducted in cells treated with Formononetin at different concentrations. The result showed that Formononetin treatment inhibited the binding of SLUG to the promoter of E-cadherin (Figure 6A). Luciferase reporter gene assay showed that Formononetin increased E-cadherin transcriptional activity in a dose-dependent manner (Figure 6B). Scanning electron microscopy (SEM) assay showed that Formononetin treatment stimulated the cell phenotype to change from mesenchymal phenotype to epithelial phenotype (Figure 6C). This result also supports by Western blot analysis. Western blot analysis further revealed that Formononetin treatment upregulated the expression level of the epithelial markers (E-cadherin, cytokeratin, and occludin) and downregulated the expression level of mesenchymal markers (Vimentin, N-cadherin, and myosin) in PLC-PRF-5 and Hep3B cells (Figure 6D). In addition, colony formation (Figure 6E), transwell (Figure 6F), and wound healing assay (Figure 6G) in PLC-PRF-5 and Hep3B cells showed that Formononetin inhibited the colony formation, migration, and invasion of HCC cells.
Expression of USP5 and SLUG is related to malignancy degree of hepatocellular carcinoma. (A) IHC analysis of the expression levels of USP5 and SLUG in tumors and normal liver tissues cited from the Human Protein Atlas. Representative images are shown. **P<0.01, Student's t-test. (B) Expression level of USP5 and SLUG in primary tumors (n=371) and normal liver tissues (n=50) based on TCGA dataset. **P<0.01. (C) Analysis of the expression levels of USP5 and SLUG in TCGA LIHC samples based on clinical stages (n=346). *P<0.05, one-way ANOVA. (D) Analysis of the expression levels of USP5 and SLUG in TCGA LIHC samples based on pathology grade (n=346). *P<0.05, **P<0.01, one-way ANOVA. (E) Kaplan-Meier curve showing the five-year survival rate of TCGA LIHC samples classified by USP5 or SLUG expression (n=346). (F) Correlation analysis of USP5 with TWIST1, Vimentin, MMP2, MMP9 and N-cadherin in TCGA LIHC samples (n=314). The correlation coefficient and P values are shown.
Formononetin targets USP5 to inhibit the deubiquitination of SLUG. (A) Molecular docking results of USP5 with the lead compounds were chosen from the traditional Chinese medicine. (B) Chemical structure of Formononetin. (C) Molecular dynamics (MD) simulation picture of the combination of Formononetin with USP5. (D) Molecular docking results of Formononetin and other USP family members. (E) Immunofluorescence probe co-localization of Formononetin probe (Blue), USP5 (Red), and SLUG (Green). Scale bar, 10 μm. Pearson's correlation and Mander's overlap values are shown. (F) Sensorgrams (Biacore analysis) of the interaction between Formononetin and USP5. (G) PLC-PRF-5 cells treated with Formononetin at different concentrations were collected for WB analysis. (H) PLC-PRF-5 cells transfected with Flag-SLUG and HA-Ub were treated with Formononetin (40 μM). Cellular extracts were prepared for co-immunoprecipitation assays with anti-Flag followed by IB with anti-HA.
To further investigate the influence of Formononetin on inhibiting EMT, the differential genes expression in PLC-PRF-5 cells treated with Formononetin were analyzed. GO analysis revealed that Formononetin treatments inhibited the activity of USPs, tissue migration, promoted the development of epithelial cell and the adhesion between cells (Figure 6H). KEGG analysis showed that Formononetin treatment contributed to ubiquitin-mediated proteolysis, inhibited the pathway in cancer including TGF-β and Toll-like receptor signaling pathway (Figure 6I) (Supplementary data 2).
Formononetin inhibits EMT of hepatocellular carcinoma. (A) PLC-PRF-5 and Hep3B cells were treated with different concentrations of Formononetin. Cellular extracts were prepared for CHIP assays with anti-SLUG. (B) PLC-PRF-5 and Hep3B cells were transfected with E-cadherin - dependent reporter gene plasmids. Luciferase activity was measured under 20μM, 40μM Formononetin treatment. *P<0.05, **P<0.01, one-way ANOVA. (C) Cell phenotype changes of PLC-PRF-5 cell treated with 20μM Formononetin. Scale bar, 10 μm. (D) Western blot analysis USP5, SLUG and EMT related markers in PLC-PRF-5 and Hep3B cells treated with different concentrations of Formononetin. (E) Colony formation assay of PLC-PRF-5 and Hep3B cells treated with Formononetin at different concentrations. **P<0.01, one-way ANOVA. (F) Transwell assay of PLC-PRF-5 and Hep3B cells treated with different concentrations of Formononetin. **P<0.01, one-way ANOVA. (G) Scratch assay of PLC-PRF-5 and Hep3B cells treated with Formononetin at different concentrations. **P<0.01, one-way ANOVA. (H) GO analysis of differentially expressed proteins in cells treated with Formononetin. (I) KEGG analysis of differentially expressed proteins in cells treated with Formononetin. (J) PLC-PRF-5 and Hep3B cells were subjected to nude mice by subcutaneous injection and treated with Formononetin. (K)Tumor volumes were measured every 3 days. *P<0.05, **P<0.01, one-way ANOVA. (L) Bioluminescent quantitation of tumors in control, Formononetin and shUSP5+Formononetin groups. *P<0.05, **P<0.01, one-way ANOVA. (M) Tumor weight in control, Formononetin and shUSP5+Formononetin groups. *P<0.05, **P<0.01, one-way ANOVA. (N) Representative lung metastasis specimens were sectioned and stained with H&E (scale bar, 200μm), and metastasis nodules were analyzed. *P<0.05, **P<0.01, one-way ANOVA. (O) IHC analysis of USP5, SLUG, E-cadherin, Vimentin and Ki67 in tumors tissues. Representative images and staining scores are shown. Scale bar, 50 μm. (P) USP5, SLUG, Vimentin and E-cadherin were detected by Western blotting in PLC-PRF-5 and Hep3B tumor tissues of each group. Error bars represent mean ± SD, *P<0.05, **P<0.01.
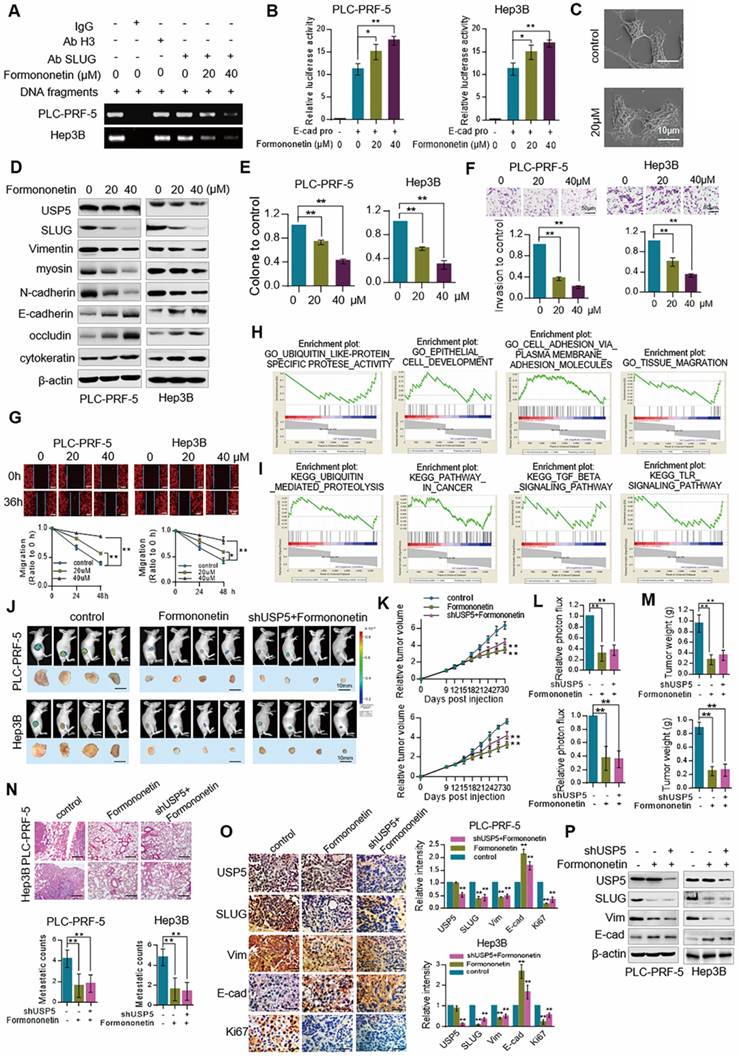
To verify the effect of Formononetin inhibits hepatocellular carcinoma in vivo, nude mice injected subcutaneously with GFP-labeled PLC-PRF-5 and Hep3B cells were given Formononetin treatment and/or combined with knocked down USP5 (Figure 6J). Obviously, tumor volume curve (Figure 6K), fluorescence intensity (Figure 6L), tumor weight (Figure 6M), and metastatic nodules (Figure 6N) in lung were inhibited in Formononetin treated group and there was no obvious difference in Formononetin treated and shUSP5 + Formononetin treated groups. IHC assay also revealed that the expression of SLUG, Vimentin and Ki67 decreased under Formononetin treatment whereas that of E-cadherin increased and there was no obvious difference in Formononetin treated and shUSP5 + Formononetin treated groups (Figure 6O). Western blot results are consistent with the IHC results (Figure 6P). All these results demonstrate that Formononetin inhibits EMT of hepatocellular carcinoma, and the effect of Formononetin depended on USP5.
Discussion
HCC is the second leading cause of death from cancer worldwide. The prognosis of HCC is extremely poor with an overall ratio of mortality to incidence of 0.95 (http://gco.iarc.fr/today/home). Tumor recurrence or metastasis is always detected in many patients with HCC after their initial response to standard cancer therapy [25]. EMT plays a crucial role in tumor malignant progression by facilitating tumor cell invasion and dissemination to distant organs, thereby resulting in cancer metastasis [26, 27]. Therefore, the underlying molecular mechanisms of EMT should be investigated to develop novel therapeutic strategies for suppressing metastasis and thus improving treatment outcome.
SLUG, a member of the SNAIL family of transcriptional repressors and is a key EMT regulator, triggers EMT by repressing E-cadherin expression [4]. SLUG is involved in various developmental and cellular processes, including the induction of cell motility and EMT [5]. SLUG also plays an important role in mesoderm formation, neural crest migration, and re-epithelialization of adult wounds [28]. However, SLUG is a highly unstable protein with short half-life, and the mechanism of controlling SLUG activity remains unclear [29].
In this study, we employed pulldown and LC-MS/MS experiment and found that SLUG interacted with the deubiquitinase USP5, a member of USP family that can cleave ubiquitin-ubiquitin and ubiquitin-protein bonds through its deubiquitinase activity [14]. CHX and in vivo deubiquitination assay revealed that USP5 removed ubiquitin on SLUG and prevented SLUG from being ubiquitin-degraded. The ubiquitin-proteasome system regulates core EMT-inducing transcriptional factors. USP5 was recently found to stabilize c-Maf protein and promote myeloma cell proliferation and survival [30]. However, the importance of USP5-promoted SLUG stabilization in EMT must be examined.
ChIP analysis showed that knockdown of USP5 decreased the binding of SLUG to the promoter of E-cadherin. In vitro experimental results further proved that the depletion of USP5 reduced the expression of E-cadherin and inhibited the migration of HCC cells. In vivo experiments showed that the tumor proliferation and metastasis in USP5- or SLUG-deficient nude mice were all repressed. Clinical data analysis confirmed that the expression of USP5 was positively correlated with clinical stage and pathological grade, and its high expression indicated a poor prognosis. In addition, the expression level of USP5 was closely associated with the metastasis and EMT-related protein expression. All the results suggest that USP5 can serve as a potential target for cancer treatment.
Chinese medicine has a long-standing history, and many traditional Chinese medicines exert anti-tumor effects. Formononetin of a high docking score with USP5 was filtered out through virtual screening. The Formononetin is a novel herbal isoflavonoid isolated from Astragalus membranaceus, a medicinal plant with antitumorigenic properties [31]. Deubiquitinating enzymes play key roles in many cell activities, such as tumorigenesis, cell cycle, apoptosis, receptor signaling, endocytosis, drug resistance and so on [32, 33]. The increasing evidences demonstrate multiple USP-involved regulation mechanisms in tumors, it may be developed as a novel therapy for this devastating disease [34]. Formononetin, as an inhibitor of USP5 activity, may regulate tumors through multiple pathways. Formononetin induces the apoptosis of human osteosarcoma cell line U2OS [35] and to inhibit angiogenesis and tumor cell invasion by down-regulated the expression of the key pro-angiogenic factors, including vascular endothelial growth factor and matrix metalloproteinases [31]. However, the direct target of Formononetin remains unknown. In this study, we show that Formononetin directly targets USP5 and inhibits its deubiquitinase activity, thereby destabilizing SLUG and repressing EMT and HCC progression. Bortezomib, also known as Velcade, is the first and currently the only proteasome inhibitor that is approved for clinical cancer therapy and shows excellent therapeutic efficacy in the treatment of multiple myeloma. However, the efficacy of Velcade is limited and is yet to be developed for targeted ubiquitination drugs [36, 37].
In conclusion, we reveal for the first time that USP5 is physically associated with and stabilizes SLUG by its deubiquitinase activity. Formononetin displays excellent anti-tumor activity through suppresses the EMT by inhibiting USP5 deubiquition for SLUG in HCC. Structural modification should be conducted to obtain compounds with improved performance, which is significant to the development of clinical anti-tumor drugs that target USP5/SLUG.
Methods
Cell culture and transfection
Hela, PLC-PRF-5, HepG2, MHCC-97L, MHCC-97H, LO2, THLE2, Hep3B, and SK-Hep-1 cells were purchased from KeyGEN BioTECH and OBIO, Shanghai. Cells are maintained in RPMI 1640 or DMEM medium (Neuronbc) with 10%FBS and 1%PS. All the cell lines were kept at 37 °C under a humidified atmosphere of 5% CO2 in an incubator, trypsinized, and passaged every 2 days. Plasmid DNA was transfected using Lipofectamine 2000 transfection reagent (Invitrogen).
Antibodies
For western blot: anti-USP5 (abcam, ab154170, 1:1000), anti-SLUG (Affinity, DF6202, 1:1000), N-cadherin (abcam, ab18203, 1:1000), Occlaudin-3 (Affinity, AF0129, 1:1000) Cytokeratin (Affinity, BF0197, 1:1000), myosin (Affinity, AF4725, 1:1000). For IF: anti-E-cadherin (CST, 14472, 1:50), anti-Vimentin (CST, 5741, 1:100), anti-SLUG (Affinity, DF6202, 1:100) and anti-USP5 antibodies (Abcam, ab154170, 1:100). For IHC: anti-E-cadherin (CST, 14472, 1:100), anti-Vimentin (CST, 5741,1:100), anti-SLUG (CST, 9585, 1:100), anti-USP5 (abcam, ab154170, 1:50), anti-Ki67 (Abcam, ab15580, 1:500).
Pull down and silver staining
Lysates from Hela cells expressing Flag-SLUG was prepared using 0.3% Nonidet P-40 lysis buffer (0.2 mM EDTA; 50 mM Tris-HCl, PH 7.4; 150 mM NaCl; 0.3% Nonidet P-40) containing protease inhibitor cocktail (Roche). Anti-Flag tag (L5) affinity beads (Biolegend) was used to incubate with the cell extracts for 12 h at 4 °C. After binding, the beads were washed with cold 0.1% Nonidet P-40 lysis buffer (0.2 mM EDTA, 50 mM Tris-HCl (PH 7.4), 150 mM NaCl, 0.1% Nonidet P-40). Flag peptide (Sigma) was then applied to the beads to elute the Flag protein complex as described by the manufacturer. The eluents were collected and visualized on 10% SDS-PAGE followed by silver staining with Fast Silver Stain Kit (Beyotime). Distinct protein bands were retrieved and analyzed by LC-MS/MS [38].
Immunopurification (IP)
For IP, 50 μl of 50% protein A/G agarose (Pierce) was incubated with control or specific antibodies (1-2 μg) for 8 h at 4 °C with constant rotation. Hela, PLC-PRF-5 and Hep3B cell lysates were prepared by incubating the cells in 0.3% Nonidet P-40 lysis buffer in the presence of protease inhibitor cocktails. The lysates were centrifuged at 12000 rpm for 10 min at 4°C and then incubated with antibody-conjugated beads for an additional 12 h. After incubation, beads were washed five times using cold 0.1% Nonidet P-40 lysis buffer. The precipitated proteins were eluted from the beads by re-suspending the beads in 2 × SDS-PAGE loading buffer and boiling for 10 min at 99 °C. The boiled immune complexes were subjected to SDS-PAGE followed by IB with USP5 (abcam, ab154170, 1:1000) and SLUG (Affinity, DF6202, 1:1000).
FPLC
Hela cell extracts were applied to a Superdex 200 10/300 GL (GE Healthcare) that was equilibrated with 1 × phosphate-buffered saline (PBS). The column was eluted at a flow rate of 0.5 mL/min, and fractions were collected. Western blotting analysis was conducted to detect USP5 (abcam, ab154170, 1:1000) and SLUG (Affinity, DF6202, 1:1000).
Immunofluorescence assay
For the immunofluorescence assay, PLC-PRF-5 and Hep3B cells subjected to different treatments were washed three times with 1 × PBS, fixed in 4% paraformaldehyde (pre-cooled at 4 °C, Solarbio) for 20 min, and blocked with 5% bovine serum albumin (BSA, KeyGEN BioTECH) containing 0.1% TritonX-100 (Sigma) for 30 min at room temperature. Then, the resultants were incubated with anti-E-cadherin (CST, 14472, 1:50) and anti-Vimentin (CST, 5741, 1:100) or anti-SLUG (Affinity, DF6202, 1:100) and anti-USP5 antibodies (Abcam, ab154170, 1:100). After washing with 1 × PBS again, and then incubated with fluorescently conjugated secondary antibodies (1:200, KeyGEN BioTECH) diluted in 5% BSA for about 50 min at room temperature. Finally, the cells were washed with 1 × PBS and mounted with the DAPI-containing mounting medium (Solarbio). The images of cells were taken with a laser scanning confocal microscope (Leica).
Deubiquitination assay
PLC-PRF-5 cells with different treatments were lysed by 0.3% Nonidet P-40 lysis buffer and centrifuged at 12000 rpm for 10 min. Then, anti-FLAG affinity gel or anti-SLUG antibody-conjugated protein A/G agarose were used to incubate with cellular extracts for 12 h at 4 °C. Thereafter, the resultants were washed five times with cold 0.1% Nonidet P-40 lysis buffer, boiled in SDS loading buffer, and subjected to SDS-PAGE followed by IB[39].
ChIP
PLC-PRF-5 cells treated with USP5 siRNA or Formononetin were cross-linked using 1% formaldehyde (Sigma-Aldrich). Then, the cells were lysed, and chromatin was sheared by sonication. Chromatin fraction was incubated with SLUG antibody overnight at 4 °C, and DNA was extracted and used for polymerase chain reaction amplification with E-cadherin specific primers (Forward: 5'-AGCCTCGGCAACATAGT-3', Reverse: 5'-CACCACACCGGCTAATT-3'). Histone H3 (PTM Bio) was used as a positive control and IgG was used as a negative control (abcam, ab190475).
Synthesize of Formononetin-Probe
A solution of Formononetin (100 mg, 0.37 mmol, meilunbio) and 4-Pentynoic acid (44 mg, 0.45 mmol) in CH2Cl2 (8 mL) was added with DCC (152 mg, 0.74 mmol) and DMAP (90 mg, 0.74 mmol). The reaction mixture was then stirred under nitrogen at 20 °C for 8 h. The aqueous solution was extracted with CH2Cl2 (3 × 10 mL). The combined organic layer was washed with H2O (2×10 mL), dried over anhydrous MgSO4, and filtered. The solvent was removed under reduced pressure. The crude product was purified by column chromatography (silica gel, PE:EA = 2:1) to facilitate Formononetin probe in 75% yield (97 mg): 1H-NMR (400 MHz, CDCl3) δ 8.33 (d, J = 8.7 Hz, 1H), 7.98 (s, 1H), 7.52 - 7.48 (m, 2H), 7.32 (d, J = 2.2 Hz, 1H), 7.18 (dd, J = 8.7, 2.2 Hz, 1H), 7.00 - 6.97 (m, 2H), 3.85 (s, 3H), 2.87 (t, J = 7.3 Hz, 2H), 2.66 (m, J = 7.3, 2.6 Hz, 2H), 2.07 (t, J = 2.6 Hz, 1H).
RNA interference
All siRNAs were transfected using Lipofectamine RNAi MAX following the recommendations of the manufacturer. The final concentration of the siRNA was 10 nM, and the cells were collected after 72 or 96 h for the experiments. SLUG siRNA sequence: 5'-CCCAUUCUGAUGUAAAGAAAU-3'. USP5 siRNA: 5'-GAUAGACAUGAACCAGCGGAU-3'.
Lentiviral production
The shRNAs targeting USP5 in the pLKO-U6-shRNA, carried by pLP1, pLP2, pLP VSV-G assistant vectors transfected into HEK293T cells. Viral supernatants were collected 48 h later, clarified by filtration, and concentrated by ultracentrifugation. The plasmids were purchased from OBIO, Shanghai. USP5 shRNA sequence: 5'-CCGGGACCACACGATTTGCCTCATTCTCGAGAATGAGGCAAATCGTGTGGTCTTTTT-3'.
Immunofluorescence co-localization
PLC-PRF-5 cells treated with or without Formononetin probe were fixed by 3.7% formaldehyde and blocked with 5% FBS containing 0.1% TritonX-100. Samples were then stained with anti-SLUG (Affinity, DF6202, 1:100) and anti-USP5 antibodies (Abcam, ab154170, 1:100) and secondary antibodies coupled to AlexaFluor 488 or 594 (Invitrogen). Formononetin probe was connected to 647 dyes by click reaction. Pictures were taken with the N-stochastic optical reconstruction microscopy system (N-STORM, Nikon, Tokyo, Japan).
Biacore
The experiments were carried out using Biacore T200 SPR sensor (Biacore, GE Healthcare) with control software version 3.0 and Sensor Chip CM5 (carboxymethylated dextran surface). All the assays were performed at 25 °C. USP5 was immobilized via amine groups in all of the four available flow cells. To this end, the chip surface was first activated following a standard EDC/NHS protocol11 with Biacore PBS-EP buffer used as the running buffer. USP5 at a concentration of 0.4 mg/mL in 10 mM phosphate buffers (pH 7.4) was then injected for 12 min followed by a 7-min injection of 1 M of ethanolamine (pH 8.5) to inactivate the residual active groups. Typically, approximately 4000 RU of USP5 was immobilized per flow cell. Formononetin was diluted at 3, 6, 12, 25, and 50 μM. The insoluble residue was pelleted by centrifugation and discarded. The supernatant of 200 μL was injected at a flow rate of 20 μL/min. Protein binding time was set to 3 min, and the dissociation time was 300 s. The chip was regenerated with glycine-HCl (pH 2.5, 10 mM) [40].
SEM
Cells were seeded on climbing films and treated with Formononetin. Then, the cells were fixed with 4% paraformaldehyde (pre-cooled at 4 °C), dehydrated in acetone/isoamyl acetate (1:1), dried with a gradient concentration of acetonitrile, and coated with gold. Subsequently, images of the cells were taken with a scanning electron microscope (LEO 1530 VP, Germany) [41].
Invasion assay
For the invasion assays, PLC-PRF-5 and Hep3B cells subjected to different treatments were added in top-chamber inserts coated with matrigel (BD Biosciences). The bottom chamber was filled with 500 μL of medium containing 10% FBS. After being cultured at 37 °C for 24 h, the cells transferred through the filter membrane at the bottom of the chambers were washed three times with 1 × PBS, fixed in 4% paraformaldehyde (pre-cooled at 4 °C), and stained with crystal violet staining solution (KeyGEN BioTECH). The passed cells were counted under a microscope (Nikon, Japan).
Colony formation
PLC-PRF-5 and Hep3B cells with different treatments were maintained in culture media for 14 days, and this procedure was followed by staining with crystal violet.
Migration assay
For the migration assays, PLC-PRF-5 and Hep3B cells with different treatments were scratched in the center of the well. Wound images were photographed every 12 h using a light microscope (Nikon, Japan).
Luciferase activity assay
PLC-PRF-5 and Hep3B cells were seeded in 96-well plates. After 24 h, the plasmids of SLUG, USP5 or siRNA were transfected separately into the cells or co-transfected with E-cadherin promoter. After 48 h, Gaussia luciferase and secreted alkaline phosphatase (SEAP) luciferase activities were measured consecutively by using Dual-Luciferase Reporter Assay System (GeneCopoeia Inc., USA). Gaussia luciferase was normalized to SEAP activity. The experiment was performed in triplicate [38].
Tumor xenograft
PLC-PRF-5 and Hep3B cells were infected with USP5 shRNA or USP5 overexpressed plasmids with lentiviruses in vitro. After forty-eight hours, 2 × 106 cells in PBS were injected into BALB/c nude mice (6-8 weeks old, Charles River, Beijing, China) by subcutaneous injection. For testing the effect of Formononetin on inhibiting liver cancer, 2 × 106 PLC-PRF-5 and Hep3B cells or cells knocked down USP5 suspended in PBS were subjected to BALB/c nude mice (5-6 weeks old) by subcutaneous injection. From the 15th to the 30th day since injection, the mice in experimental groups were treated with Formononetin at a concentration of 100 mg/kg every 2 days. By contrast, the mice in the control group were treated with the same volume of saline. Tumors were measured every 3 days using a Vernier calliper, and the volume was determined using the formula V=ab2/2 (a = length of tumor, b = width of tumor). After the mice were sacrificed, the tumors and lungs were harvested and fixed in 4% paraformaldehyde and embedded in paraffin for histologic examination or hematoxylin and eosin staining.
IHC
The tissues were deparaffinized with xylene and dehydrated with ethanol of decreasing concentrations. Endogenous peroxidase was blocked by incubating with 3% hydrogen peroxide for 15 min. The antigen retrieval was done in a steam pressure cooker with citrate buffered saline (pH 6.0) for 15 min at 95 °C. After incubation with normal goat serum for 20 min at room temperature to block unspecific labeling, the tissues were incubated with primary antibodies including anti-E-cadherin (CST, 14472, 1:100,), anti-Vimentin (CST, 5741, 1:100,), anti-SLUG (CST, 9585, 1:100), anti-USP5 (abcam, ab154170, 1:50), anti-Ki67 (Abcam, ab15580 1:500) antibodies in a humidified chamber overnight at 4 °C. Diaminobenzidine was utilized for color development and hematoxylin as counterstain. Expression levels of E-cadherin, Vimentin, USP5, and SLUG were independently evaluated by two investigators.
Molecular docking
The crystal structure of USP5 was downloaded from the Protein Data Bank (PDBID: 3IHP). The crystal structure of SLUG was modeled by SWISS-MODEL (https://www.swissmodel.expasy.org/). Then, the protein structure was prepared with by adding hydrogen, optimizing the H-bond assignment, assigning bond order, treating disulfides, and performing energy minimization to relax the structure. The HEX software was used to perform protein-protein docking. Schrodinger software was used to perform molecule screening. The ligand in the crystal structure was used to define the center site of a docking grid box, and the xyz dimensions of docking grid box were 60×60×60. The 3D structures of the Traditional Chinese medicine molecule were generated with LigPrep and were minimized with OPLS-2005 force field. Docking score was used to sort small molecules [42].
Bioinformatics analysis
PLC-PRF-5 cells treated with Formononetin (20 μM) or DMSO were lysed with TRIZOL reagent (Invitrogen) and underwent microarray-based high throughput gene expression profiling. The genes underwent GO and KEGG analysis by using gene set enrichment analysis (GSEA) soft. GO and KEGG gene sets was analyzed from MSigDB v2.5 gene set database (http://software.broadinstitute.org/gsea/msigdb/index.jsp). In generally, the analysis consists of the following steps. The list of all the gene names and corresponding quantitative value were loaded. Then, minimum number of gene sets matches to 15 and maximum number of gene matches to 500. Given a gene set, GSEA soft distinguishes whether the gene is belongs to the top or bottom of the list. And enrichment score indicates the degree of gene enrichment.
Statistical analysis
Data from biological triplicate experiments were presented with error bar as mean ± SD. Two-tailed unpaired Student's t-test was used for comparing two groups of data. One-ANOVA was used to compare multiple groups of data. P value of less than 0.05 was considered significant.
Supplementary Material
Supplementary figures.
Supplementary data 1 - SLUG 1 protein.
Supplementary data 1 - SLUG 2 protein.
Supplementary data 1 - SLUG 3 protein.
Supplementary data 1 - SLUG 4 protein.
Supplementary data 1 - SLUG 5 protein.
Supplementary data 1 - SLUG 6 protein.
Supplementary data 1 - SLUG 7 protein.
Supplementary data 1 - SLUG 8 protein.
Supplementary data 2 - NB C vs NB D signal.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (Grant nos. 81572838, 81402973, and 81703581), the National Science and Technology Major Project (Grant no. 2017ZX09306007), and Postdoctoral support scheme for innovative talents (Grant no. BX20180150).
Author contributions
TS SC and JM conceived and designed the projects. JM, YYL and WLZ wrote the manuscript. JM, YYL, JXH, KLQ, LCH, YL, BJZ performed the experiments. CY, TS, SC, BXQ, XYA, HGZ provided technical and material support. XRW, HZW, XYZ and YYX performed the clinical analysis.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. Clin Cancer Investig J. 2015;65:87-108
2. Lee GA, Hwang KA, Choi KC. Roles of dietary phytoestrogens on the regulation of epithelial-mesenchymal transition in diverse cancer metastasis. Toxins (Basel). 2016;8:6
3. Serrano-Gomez SJ, Maziveyi M, Alahari SK. Regulation of epithelial-mesenchymal transition through epigenetic and post-translational modifications. Mol Cancer. 2016;15:18
4. Hajra KM, Chen DY, Fearon ER. The SLUG zinc-finger protein represses E-cadherin in breast cancer. Cancer Res. 2002;62:1613-8
5. Nieto MA. The snail superfamily of zinc-finger transcription factors. Nat Rev Mol Cell Biol. 2002;3:155-66
6. Kajita M, McClinic KN, Wade PA. Aberrant expression of the transcription factors snail and slug alters the response to genotoxic stress. Mol Cell Biol. 2004;24:7559-66
7. Barrallo-Gimeno A, Nieto MA. The Snail genes as inducers of cell movement and survival: implications in development and cancer. Development. 2005;132:3151-61
8. Dhasarathy A, Phadke D, Mav D, Shah RR, Wade PA. The transcription factors Snail and Slug activate the transforming growth factor-beta signaling pathway in breast cancer. PloS one. 2011;6:e26514
9. Kurrey NK, K A, Bapat SA. Snail and Slug are major determinants of ovarian cancer invasiveness at the transcription level. Gynecol Oncol. 2005;97:155-65
10. Uygur B, Wu WS. SLUG promotes prostate cancer cell migration and invasion via CXCR4/CXCL12 axis. Mol Cancer. 2011;10:139
11. Turner FE, Broad S, Khanim FL, Jeanes A, Talma S, Hughes S. et al. Slug regulates integrin expression and cell proliferation in human epidermal keratinocytes. J Biol Chem. 2006;281:21321-31
12. Emadi Baygi M, Soheili ZS, Essmann F, Deezagi A, Engers R, Goering W. et al. Slug/SNAI2 regulates cell proliferation and invasiveness of metastatic prostate cancer cell lines. Tumour Biol. 2010;31:297-307
13. Wu ZQ, Li XY, Hu CY, Ford M, Kleer CG, Weiss SJ. Canonical Wnt signaling regulates Slug activity and links epithelial-mesenchymal transition with epigenetic breast cancer 1, Early Onset (BRCA1) repression. Proc Natl Acad Sci U S A. 2012;109:16654-9
14. Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK. et al. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773-86
15. Diaz VM, de Herreros AG. F-box proteins: Keeping the epithelial-to-mesenchymal transition (EMT) in check. Semin Cancer Biol. 2016;36:71-9
16. Tsai CH, Cheng HC, Wang YS, Lin P, Jen J, Kuo IY. et al. Small GTPase Rab37 targets tissue inhibitor of metalloproteinase 1 for exocytosis and thus suppresses tumour metastasis. Nat Commun. 2014;5:4804
17. Wilkinson KD, Tashayev VL, O'Connor LB, Larsen CN, Kasperek E, Pickart CM. Metabolism of the polyubiquitin degradation signal: structure, mechanism, and role of isopeptidase T. Biochem. 1995;34:14535-46
18. Stein RL, Chen Z, Melandri F. Kinetic studies of isopeptidase T: modulation of peptidase activity by ubiquitin. Biochem. 1995;34:12616-23
19. Dayal S, Sparks A, Jacob J, Allende-Vega N, Lane DP, Saville MK. Suppression of the deubiquitinating enzyme USP5 causes the accumulation of unanchored polyubiquitin and the activation of p53. J Biol Chem. 2009;284:5030-41
20. Gadotti VM, Caballero AG, Berger ND, Gladding CM, Chen L, Pfeifer TA. et al. Small organic molecule disruptors of Cav3.2 - USP5 interactions reverse inflammatory and neuropathic pain. Mol Pain. 2015;11:s12990-015 -0011
21. Shiomi N, Mori M, Tsuji H, Imai T, Inoue H, Tateishi S. et al. Human RAD18 is involved in S phase-specific single-strand break repair without PCNA monoubiquitination. Nucleic Acids Res. 2007;35:e9
22. Marger F, Gelot A, Alloui A, Matricon J, Ferrer JF, Barrere C. et al. T-type calcium channels contribute to colonic hypersensitivity in a rat model of irritable bowel syndrome. Proc Natl Acad Sci U S A. 2011;108:11268-73
23. Kummari E, Alugubelly N, Hsu CY, Dong B, Nanduri B, Edelmann MJ. Activity-based proteomic profiling of deubiquitinating enzymes in salmonella-infected macrophages leads to identification of putative function of UCH-L5 in inflammasome regulation. PloS one. 2015;10:e0135531
24. Potu H, Peterson LF, Pal A, Verhaegen M, Cao J, Talpaz M. et al. Usp5 links suppression of p53 and FAS levels in melanoma to the BRAF pathway. Oncotarget. 2014;5:5559-69
25. Nguyen LT, Song YW, Cho SK. Baicalein inhibits epithelial to mesenchymal transition via downregulation of Cyr61 and LOXL-2 in MDA-MB231 breast cancer cells. Mol Cells. 2016;39:909-14
26. Santamaria PG, Moreno-Bueno G, Portillo F, Cano A. EMT: Present and future in clinical oncology. Mol Oncol. 2017;11:718-38
27. Heerboth S, Housman G, Leary M, Longacre M, Byler S, Lapinska K. et al. EMT and tumor metastasis. Clin Transl Med. 2015;4:6
28. Boutet A, De Frutos CA, Maxwell PH, Mayol MJ, Romero J, Nieto MA. Snail activation disrupts tissue homeostasis and induces fibrosis in the adult kidney. EMBO J. 2006;25:5603-13
29. Dominguez D, Montserrat-Sentis B, Virgos-Soler A, Guaita S, Grueso J, Porta M. et al. Phosphorylation regulates the subcellular location and activity of the Snail transcriptional repressor. Mol Cell Biol. 2003;23:5078-89
30. Wang S, Juan J, Zhang Z, Du Y, Xu Y, Tong J. et al. Inhibition of the deubiquitinase USP5 leads to c-Maf protein degradation and myeloma cell apoptosis. Cell Death Dis. 2017;8:e3058
31. Auyeung KK, Law PC, Ko JK. Novel anti-angiogenic effects of formononetin in human colon cancer cells and tumor xenograft. Oncol Rep. 2012;28:2188-94
32. Liu CH, Goldberg AL, Qiu XB. New insights into the role of the ubiquitin-proteasome pathway in the regulation of apoptosis. Chang Gung Med J. 2007;30:469-79
33. Kaistha BP, Krattenmacher A, Fredebohm J, Schmidt H, Behrens D, Widder M. et al. The deubiquitinating enzyme USP5 promotes pancreatic cancer via modulating cell cycle regulators. Oncotarget. 2017;8:66215-25
34. Liu Y, Wang WM, Lu YF, Feng L, Li L, Pan MZ. et al. Usp5 functions as an oncogene for stimulating tumorigenesis in hepatocellular carcinoma. Oncotarget. 2017;8:50655-64
35. Hu W, Xiao Z. Formononetin induces apoptosis of human osteosarcoma cell line U2OS by regulating the expression of Bcl-2, Bax and MiR-375 in vitro and in vivo. Cell Physiol Biochem. 2015;37:933-9
36. Dicato M, Boccadoro M, Cavenagh J, Harousseau JL, Ludwig H, San Miguel J. et al. Management of multiple myeloma with bortezomib: experts review the data and debate the issues. Oncology. 2006;70:474-82
37. Orlowski RZ, Kuhn DJ. Proteasome inhibitors in cancer therapy: lessons from the first decade. Clin Cancer Res. 2008;14:1649-57
38. Meng J, Chen S, Lei YY, Han JX, Zhong WL, Wang XR. et al. Hsp90beta promotes aggressive vasculogenic mimicry via epithelial-mesenchymal transition in hepatocellular carcinoma. Oncogene. 2018
39. Lee MS, Jeong MH, Lee HW, Han HJ, Ko A, Hewitt SM. et al. PI3K/AKT activation induces PTEN ubiquitination and destabilization accelerating tumourigenesis. Nat Commun. 2015;6:7769
40. Wang S, Dong Y, Liang X. Development of a SPR aptasensor containing oriented aptamer for direct capture and detection of tetracycline in multiple honey samples. Biosens Bioelectron. 2018;109:1-7
41. Meng J, Chen S, Han JX, Qian B, Wang XR, Zhong WL. et al. Twist1 Regulates Vimentin through Cul2 Circular RNA to Promote EMT in Hepatocellular Carcinoma. Cancer Res. 2018;78:4150-62
42. Fatchiyah F, Hardiyanti F, Widodo N. Selective inhibition on RAGE-binding AGEs required by bioactive peptide Alpha-S2 case in protein from goat ethawah breed milk: study of biological modeling. Acta Inform Med. 2015;23:90-6
Author contact
![]() Corresponding authors: State Key Laboratory of Medicinal Chemical Biology and College of Pharmacy, Nankai University, Haihe Education Park, 38 Tongyan Road, Tianjin 300350, China. E-mail: tao.sunedu.cn; cheng.yangedu.cn; shuang7332com; honggang.zhou126.com.
Corresponding authors: State Key Laboratory of Medicinal Chemical Biology and College of Pharmacy, Nankai University, Haihe Education Park, 38 Tongyan Road, Tianjin 300350, China. E-mail: tao.sunedu.cn; cheng.yangedu.cn; shuang7332com; honggang.zhou126.com.
 Global reach, higher impact
Global reach, higher impact