13.3
Impact Factor
Theranostics 2019; 9(2):491-525. doi:10.7150/thno.29875 This issue Cite
Review
Recent Progress on Liquid Biopsy Analysis using Surface-Enhanced Raman Spectroscopy
School of Medicine, State Key Laboratory of Medicinal Chemical Biology, Key Laboratory of Bioactive Materials for Ministry of Education, Nankai University, 300071 Tianjin, China
Received 2018-9-11; Accepted 2018-11-30; Published 2019-1-1
Abstract
Traditional tissue biopsy is limited in understanding heterogeneity and dynamic evolution of tumors. Instead, analyzing circulating cancer markers in various body fluids, commonly referred to as “liquid biopsy”, has recently attracted remarkable interest for their great potential to be applied in non-invasive early cancer screening, tumor progression monitoring and therapy response assessment. Among the various approaches developed for liquid biopsy analysis, surface-enhanced Raman spectroscopy (SERS) has emerged as one of the most powerful techniques based on its high sensitivity, specificity, tremendous spectral multiplexing capacity for simultaneous target detection, as well as its unique capability for obtaining intrinsic fingerprint spectra of biomolecules. In this review, we will first briefly explain the mechanism of SERS, and then introduce recently reported SERS-based techniques for detection of circulating cancer markers including circulating tumor cells, exosomes, circulating tumor DNAs, microRNAs and cancer-related proteins. Cancer diagnosis based on SERS analysis of bulk body fluids will also be included. In the end, we will summarize the “state of the art” technologies of SERS-based platforms and discuss the challenges of translating them into clinical settings.
Keywords: liquid biopsy, surface-enhanced Raman spectroscopy, circulating tumor cell, exosome, circulating tumor DNA
Introduction
Molecular and cellular heterogeneity represent one of the greatest challenges in cancer diagnosis and therapy. The molecular landscapes of metastatic lesions might be varied from the primary tumor tissue, and the genome profile within a tumor also evolves dynamically over time [1]. Traditional tissue biopsy obtained by collecting a portion of cells from the tumor site reflects only a single snapshot of a small region, and therefore is inadequate for comprehensive characterization and tracking of a patient's tumor [2]. In the last few years, detection of tumor-derived components in body fluids (blood, urine, saliva, ascites, cerebrospinal fluid, etc.), commonly referred to as “liquid biopsy”, has been getting enormous attention in both medical research and clinic applications [3-5]. Compared to tissue biopsy, which is invasive and cannot be repeatedly performed, liquid biopsy enables physicians to non-invasively interrogate the dynamic evolution of tumors and monitor patients' response to therapeutic treatments in real time [6].
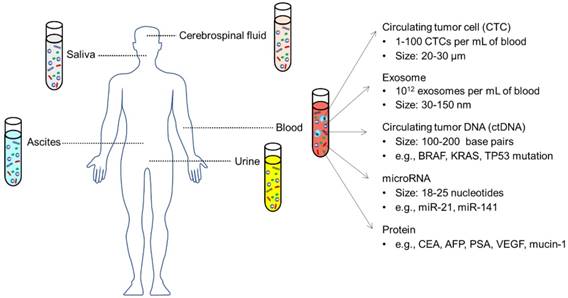
Plenty of substances from liquid biopsies have been found to be closely related to tumor stage and might serve as new biomarkers for cancer diagnosis/ prognosis (Figure 1), including circulating tumor cells (CTCs), exosomes, circulating tumor DNAs (ctDNAs), and non-coding microRNAs (miRNAs), alongside a series of cancer-related proteins [6, 7]. Tumor cells that shed from solid tumor sites and enter the circulatory system are called CTCs. In the past two decades, many studies have confirmed that CTCs are present in the blood stream at very early stages of cancer and play a major role in tumor metastasis [8]. CTCs represent the molecular characteristics of the corresponding tumor tissue and have been validated as prognostic markers for patients with metastatic breast, colorectal and prostate cancer in the clinic [9]. More recent studies have found that tumor cells excrete a large number of extracellular vesicles (EVs) into their mircoenvironment and use them for cell-cell communication [10]. EVs in body fluids, in particular exosomes (a subset of EVs with a size of 30-150 nm), contain proteins, lipids and nucleic acids inherited from the parental tumors cells and hold great promise to be used as novel diagnostic/prognostic markers [11-13]. In addition to complete cells or cell fragments, the tumor's genetic content can also be found in the circulating cell-free nucleic acids. Circulating tumor DNA (ctDNA) that derive from apoptotic/necrotic neoplastic cells or lysed CTCs represent genomic profiles closely related to tumor burden, intratumoral heterogeneity, therapy responses and resistance [14]. Non-coding microRNAs (miRNAs), which regulate gene expression, also exist in various biological fluids and distinct levels of circulating miRNAs might reflect the progress of cancer [15, 16]. Additionally, increased amounts of several kinds of proteins in the blood, such as prostate-specific antigen (PSA), carcino-embryonic antigen (CEA) and a series of carbohydrate antigens, have been found to be positively related to tumor stages [17].
Circulating biomarkers in different body fluids, including blood, urine, saliva, ascites, cerebrospinal fluid, etc. Adapted with permission from [7], copyright 2017 BMC.
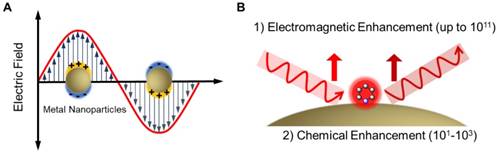
Despite the growing interest in biomarkers in liquid biopsy, establishing sensitive and reliable detection methods for clinical applications remains challenging due to the complexity of biological samples as well as the extremely low concentration of the analytes [18, 19]. In recent years, optical sensors based on surface-enhanced Raman spectroscopy (SERS) have emerged as one of the most powerful techniques in biological and biomedical analysis [20-22]. Raman spectroscopy provides fingerprint vibrational spectra of molecules and allows individual components in a mixture to be identified. However, Raman signals are usually weak due to the very low ratio of inelastic scattering events (approximately one in 106 photons being inelastically scattered) [23]. In the 1970s, Fleischmann et al. [24] and Van Duyne et al. [25] found that Raman signals can be significantly enhanced when molecules are adsorbed on a roughened metal surface. The enhancement factor can be a million-fold and enables detection of a monolayer species on a metal surface, leading to the initial establishment of the SERS technique. In 1997, two research groups independently reported single molecule/single nanoparticle SERS, where the enhancement factors were estimated to be as high as 1014 [26, 27]. Among the various mechanisms proposed to explain SERS in recent years, electromagnetic enhancement (EM) and chemical enhancement (CM) are the most widely accepted [20, 28] (Figure 2). In the EM mechanism, the interaction between electromagnetic waves with plasmonic (typically Au and Ag) nanostructures with dimensions much smaller than the wavelength induces collective oscillation of free electrons on the metal surface. When the frequency of the incident light matches the inherent oscillation frequency of free electrons in the metal, localized surface plasmons resonance (LSPR) occurs and leads to an enhancement in the incident optical field (Figure 2A). The CM arises from the charge transfer resonance between the reporter molecule and the nanostructure, requiring direct adsorption or chemical binding of the molecule on the metal surface, and is usually weaker than the EM [20]. In SERS (Figure 2B), the high local optical field induced by resonances between incident /scattered light and surface plasmons on the metal nanostructures provides more than 10 orders of magnitude signal enhancement (by EM); in addition, an enhancement factor of 101-103 is contributed by the CM. As a result, SERS combines the structural specificity and experimental flexibility of conventional Raman spectroscopy with the ultrahigh sensitivity provided by the plasmonic nanostructure- mediated signal amplification, making it a desirable technique for analysis of biomedical samples [29].
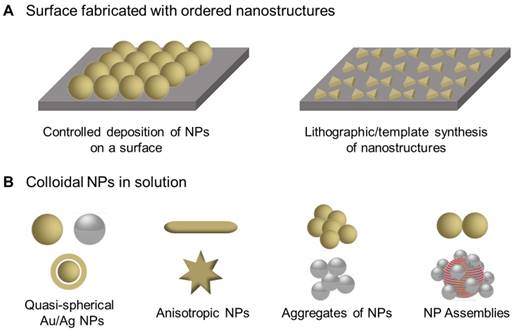
The rational design of plasmonic nanostructures with defined physicochemical properties is essential for SERS. In general, two platforms are commonly used as SERS-active substrates: periodic 2D nanostructures (Figure 3A), and colloidal nanoparticles (NPs) in solution (Figure 3B) [20]. Controlled deposition and lithographic/ template synthesis techniques are routinely used in the fabrication of nanostructured 2D surfaces due to the high level of reproducibility. For colloidal NPs, quasi-spherical NPs, anisotropic NPs, and NP clusters including aggregates and assemblies of NPs have been extensively synthesized and employed as SERS substrates [30]. Their optical properties strongly depend on the size, shape and composition of the NPs, and adjusting the LSPR peak into resonance with the incident laser wavelength is usually beneficial for enhanced optical field and optimized SERS signals.
(A) Illustration of the collective oscillation of free electrons in metal nanoparticles upon excitation by an electromagnetic wave. (B) Illustration of electromagnetic enhancement and chemical enhancement in SERS.
Representative SERS substrates. (A) Nanostructured surfaces prepared by controlled deposition of NPs or lithographic/template synthesis methods. (B) Colloidal NPs-based SERS substrates, including quasi-spherical NPs, anisotropic NPs, aggregates of NPs and NP assemblies.
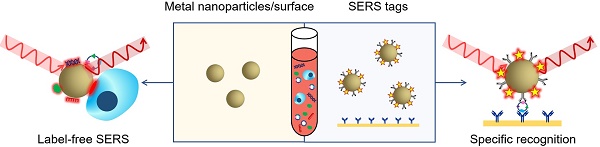
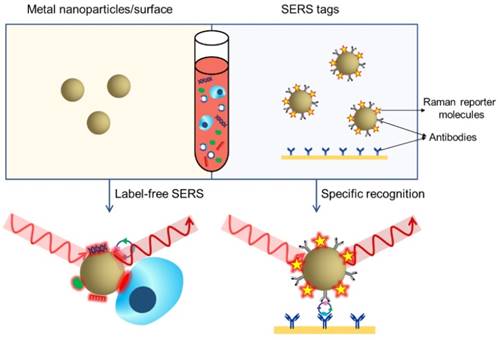
Two methodologies are mainly adopted in biomedical applications of SERS: label-free detection and indirect detection using SERS tags [20]. In label-free SERS detection (Figure 4, left), samples for analysis directly adsorb to the surface of metallic nanostructures and intrinsic fingerprint spectra of the biomolecules are obtained. Complex spectral analysis is usually a prerequisite to interpret the spectral information for discriminating biomolecules at different status or cells/microorganisms of different species [31, 32]. SERS tags (Figure 4, right) are typically composed of metallic NPs coated with Raman reporter molecules emitting strong and distinct Raman signals. By conjugating specific recognition molecules such as antibodies or aptamers, SERS tags can be used as optical labelling tools for indirect sensing/imaging of the target biomolecules in vitro and in vivo [30, 33-35]. Compared to traditional external labelling reagents like organic dyes or fluorophores, SERS tags offer advantages such as ultrasensitivity, tremendous multiplexing capacity, high photostability, the need of only a single laser to excite all SERS labels and minimized interference by autofluorescence from cells/tissues [20, 23]. In the past decade, both label-free SERS detection and SERS tags have been increasingly applied to liquid biopsy analysis, providing qualitative and quantitative information for cancer diagnosis, prognosis, and real-time monitoring of therapy response. In this review, we will summarize recent progress in the use of SERS for detection of CTCs, extracellular EVs, ctDNAs, miRNAs, cancer-related proteins, as well as bulk body fluids, and discuss the opportunities for developing new generation SERS-based platforms and the challenges of translating them to clinical settings. About the physical fundamentals of SERS, the rational design and optimization of SERS substrates, and the other broad applications of SERS-based techniques, we highly recommend several excellent reviews on the corresponding topics [20, 23, 28-30, 36, 37].
CTCs
The presence of CTCs in the blood of cancer patients was first discovered by Dr. Ashworth in 1869. However, little research was performed in the following years on CTCs due to their extraordinary rarity in blood vessels (1-100 CTCs amongst 5 billion erythrocytes and 10 million leukocytes) [8, 38]. With the development of new enrichment and detection techniques, CTCs have been increasingly studied since the mid-1990s and gained enormous attention for their clinical value to predict disease progression and survival in metastases [39, 40]. Numerous studies have confirmed that CTCs might be used as prognostic markers for breast, colorectal and prostate cancer; i.e., higher CTCs numbers correlate with increased metastasis and lower survival rates. In addition, recent studies have found that CTCs enter the blood stream at an unexpectedly early stage, even before malignancy could be detected, indicating the potential of CTCs to be used as noninvasive markers for early cancer diagnosis [41]. Furthermore, phenotype identification and molecular analysis of CTCs are expected to offer more insights into the understanding of tumor metastasis and guide therapeutic management.
SERS-based liquid biopsy analysis using a label-free SERS approach (left) or SERS tags (right). In label-free SERS, the spectroscopic signal results from analyte adsorption onto the SERS substrate, whereas in SERS tags-based specific recognition assays, the spectroscopic signal results from the reporter molecules on the SERS tags.
Current techniques for CTCs detection
In the past twenty years, a variety of technologies have been set up to separate and identify CTCs based on their unique physical and/or biological properties that are distinct from hematological cells. For example, ISET (isolation by size of epithelial tumor cells, Rarecells Inc.) discriminates CTCs by filtration through an 8 μm porous membrane due to the larger size of CTCs (20-30 μm) relative to hematologic cells. ApoStream (ApoCell) isolates CTCs based on the assumption that cancer cells have a more negative surface charge than hematologic cells. CellSearch (Janssen Diagnostics) is the first and currently only FDA-approved system for clinical automated CTC detection in breast, colorectal and prostate cancer patients. In this assay, putative CTCs are selected using magnetic beads modified with anti-epithelial cell adhesion molecule (EpCAM) antibodies, and then enumerated based on subsequent immunostaining results (cytokeratin+/DAPI+/ CD45-). Compared to the separation methods based on physical properties, the isolation purity is largely improved by using ligands for specific recognition; however, the labor-intensive and time-consuming procedures as well as high instrument costs remain a major obstacle and hinder their use in clinical diagnostics [38, 39, 42].
SERS detection of CTCs after enrichment
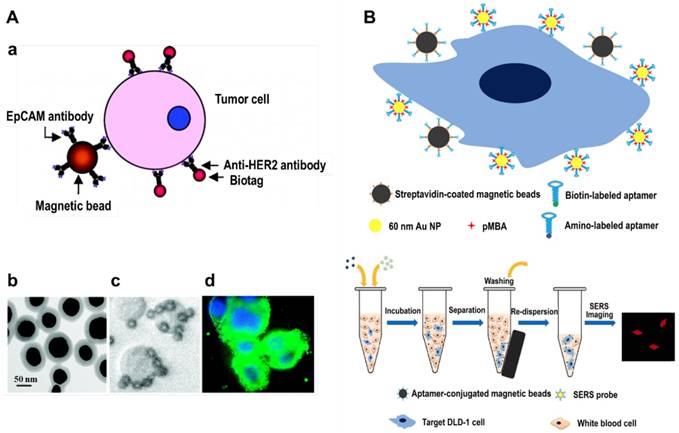
Sha et al. were the first to combine magnetic beads enrichment with SERS rapid detection and developed a no-wash assay for CTCs enumeration (in 2008) [43]. They modified magnetic beads with anti-EpCAM antibody and SERS tags with anti-human epidermal growth factor receptor-2 (HER2) antibody, respectively, thereby both NPs can specifically recognize breast cancer cells (Figure 5A). By adding the magnetic bead-EpCAM antibody and SERS-HER2 antibody conjugates to a blood sample, a sandwich structure (magnetic beads-CTC-SERS tags) will be formed and CTCs can be rapidly detected in the presence of whole blood, with a limit of detection (LOD) less than 50 cells/mL. Using the same strategy, Shi et al. fabricated folic acid-modified magnetic beads and SERS tags, which can specifically recognize CTCs since folate receptors are overexpressed in a variety of cancer cells [44]. In order to promote the in vivo application of the NPs, they built up a magnetic trapping system that can capture CTCs flowing inside a Tygon tube with different flow velocities ranging from 0.12 to 12 cm/s (mimicking the physiological situation in peripheral vessels), and were able to detect CTCs at a concentration of ∼300 cells/mL in a total sample volume of 10 mL within 7 min. Bhana et al. fabricated magnetic- SERS dual functional NPs by coating a gold shell on iron oxide NPs, and then conjugated anti-EpCAM or anti-HER2 antibodies on the surface. The highly integrated magnetic-SERS NPs cocktail allowed on-line magnetic separation and SERS detection of CTC-mimic SK-BR-3 cells in whole blood, with a detection sensitivity down to 1-2 cells/mL [45]. Aptamers are in vitro-selected single-stranded oligonucleotides that can bind to particular targets with high affinities and specificities, providing similar functions as antibodies but are easier to prepare and cost less. Sun et al. used a KDED2a-3 aptamer against DLD-1 cells (a colorectal adenocarcinoma cell line) to label magnetic NPs and SERS tags (Figure 5B), and the target cells were captured from buffer and whole blood with an efficiency of 73% and 55%, respectively [46].
SERS detection of CTCs after magnetic beads enrichment. (A) a, Schematic of the ternary immuno-complex formed by SERS tags and magnetic bead conjugates binding to the model CTC. b, TEM image of the SERS tags. c, Magnetic beads binding to SKBR3 cells. d, HER2 antibody-conjugated SERS tags (green) labeling of the SKBR3 membrane (Hoechst dye-labeled nuclei are in blue). Adapted with permission from [43], copyright 2008 American Chemical Society. (B) Schematic representation of CTCs capture and identification using aptamer-modified magnetic beads and SERS tags. Adapted with permission from [46], copyright 2015 Springer Nature.
Schematic of the ASGPR antibody-conjugated silver‐coated magnetic NPs, GPC3 antibody-conjugated SERS tags, and the operating principle for CTCs detection in human peripheral blood using dual-enhanced SERS.
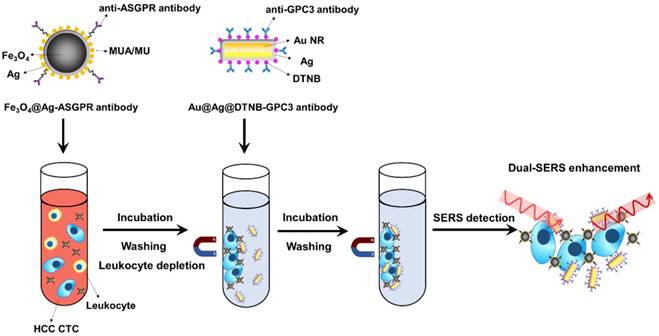
In order to improve the detection sensitivity, Pang et al. fabricated silver shell-coated magnetic beads and conjugated them with anti-asialoglyco protein receptor (ASGPR) antibody to recognize hepatocytes and used anti-glypican 3 (GPC3) antibody to decorate Au@Ag nanorods-based SERS tags (Figure 6). Aggregation of the magnetic beads and SERS tags on the surface of hepatocellular carcinoma (HCC) CTCs induced further plasmonic coupling between the Ag shell on the magnetic NPs and the Au@Ag nanorods apart from the inherent enhanced scattering of the SERS tags. The dual selective and dual-enhanced SERS signals enabled a LOD of 1 cell/mL for HCC CTC in human peripheral blood samples with a linear relationship from 1 to 100 cells/mL [47]. Besides magnetic NPs, nitrocellulose membrane was also utilized as one kind of cost-efficient and easily-prepared CTC-capture substrate. In a study performed by Zhang et al., 100 nonsmall-cell lung cancer (NSCLC) cells were spiked into 1 mL of human whole blood, among which 34 cells were captured by anti-EpCAM antibody-adsorbed nitrocellulose membrane and detected by SERS imaging [48].
SERS detection of CTCs without enrichment
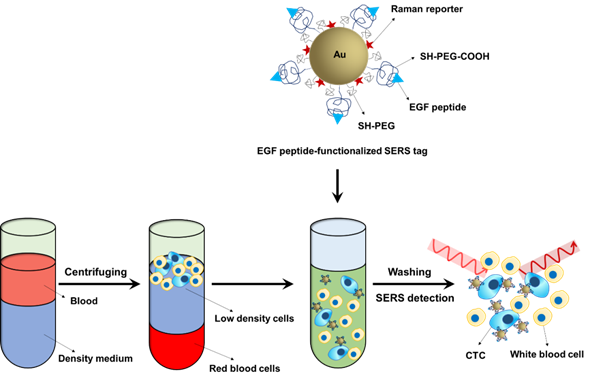
The majority of CTC detection techniques require an enrichment step before detection due to the rarity of CTCs in the peripheral blood and the presence of a large number of hematocytes. SERS is a highly sensitive technique that can detect even a single particle. Another key advantage of SERS is that it provides a sharp fingerprint-like spectrum, which is distinct from other interferences within the complex biological milieu [49]. Based on the super sensitivity and spectral specificity of SERS technology, Wang and coworkers developed a direct assay to detect CTCs in peripheral blood (Figure 7). In the direct assay, ~60 nm Au NPs were coated with QSY reporter molecules and then encapsulated with a mixed layer of thiolated polyethylene glycol (SH-PEG:SH-PEG-COOH=85:15). The closely packed PEG protection layer helped to stabilize the NPs and reduce non-specific interactions with blood cells, while the carboxy functional group was used for conjugation with epidermal growth factor (EGF) peptide as recognition groups to capture CTCs. The peripheral blood was mixed with mononuclear cell separation medium and centrifuged, and the obtained low-density cell layer containing white blood cells and CTCs was incubated with the SERS probes and then measured using a Raman system. This assay was able to detect as low as 5-50 tumor cells in 1 mL of blood and successfully identified CTCs in 19 patients with squamous cell carcinoma of the head and neck (SCCHN) [49].
Schematic of the Raman-encoded, PEG-stabilized, and EGF peptide-functionalized SERS tags and the assay principle for their use in CTCs detection in human peripheral blood without enrichment.
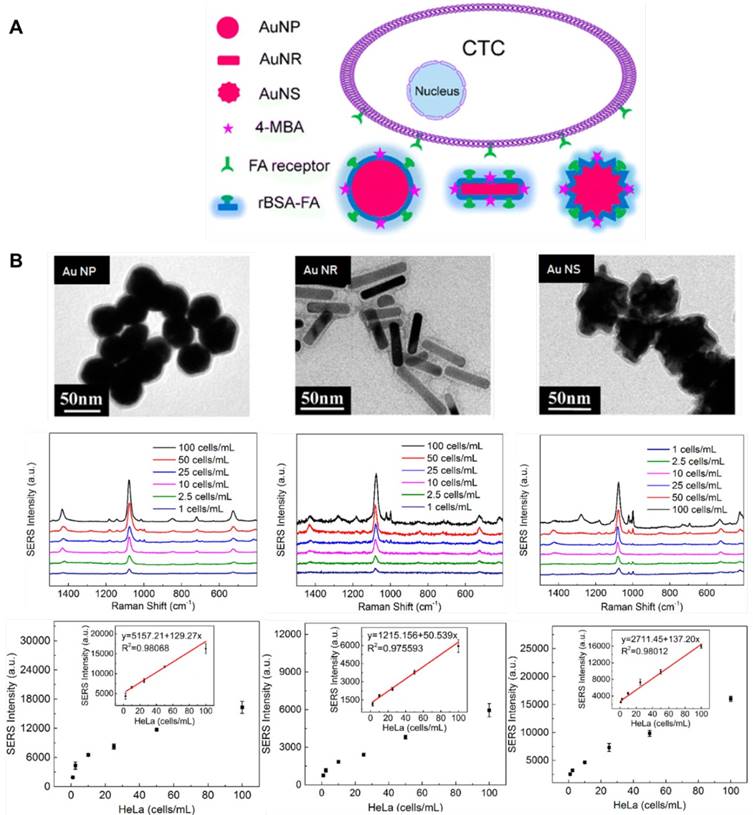
In order to reduce the thickness of the protection layer and improve SERS signal, Wu et al. used reductive bovine serum albumin (rBSA) instead of PEG to encapsulate the Au NPs, and folic acid was conjugated to the surface of the NPs to recognize tumor cells. They observed a linear relationship between SERS signal and the number of tumor cells in the range of 5-500 cells/mL, and the LOD was 5 cells/mL [50]. In their subsequent work, they fabricated SERS probes using spherical Au NPs, Au nanorods and Au nanostars as metal substrates, and then encapsulated the NPs with rBSA and conjugated them with folic acid (Figure 8). They optimized the modification conditions and proved that all three SERS probes can be utilized for CTCs detection in the blood without initial enrichment, among which Au nanostar was the most sensitive with a LOD of 1 cell/mL [51].
Multiplexed detection of CTCs based on SERS
EpCAM is highly expressed on normal epithelia and epithelial tumors but is absent on blood cells and has therefore been frequently used for positive enrichment of epithelial CTCs. However, it has recently been reported that circulating epithelial cells were detected in patients with benign colon diseases and using EpCAM as the single CTC marker might lead to false positive results [52]. Other studies have found that carcinoma cells can undergo an epithelial-to-mesenchymal transition (EMT) that results in reduced expression of epithelial markers, which means that, using anti-EpCAM antibodies as single recognition ligands might result in false negative findings [53, 54]. Thus, there is a need for broad-spectrum enrichment and accurate discrimination of CTCs based on specific cocktails of epithelial, mesenchymal, tumor (such as HER2 and EGFR)- and/or tissue (such as PSA and mammaglobin)-specific markers [38]. SERS provides tremendous spectral multiplexing capacity for simultaneous detection of different targets due to its narrow peak width (~1-2 nm, about 10-100 times narrower than fluorescence emission bands) [20, 22], and therefore is becoming a highly desirable technique for CTCs discrimination and subtyping.
(A) Schematic of SERS-active NPs with various shapes for CTCs detection. (B) TEM images of Au nanospheres-based SERS tags, Au nanorods-based SERS tags, Au nanostars-based SERS tags, and their sensitivity for CTCs detection. Adapted with permission from [51], copyright 2016 American Chemical Society.
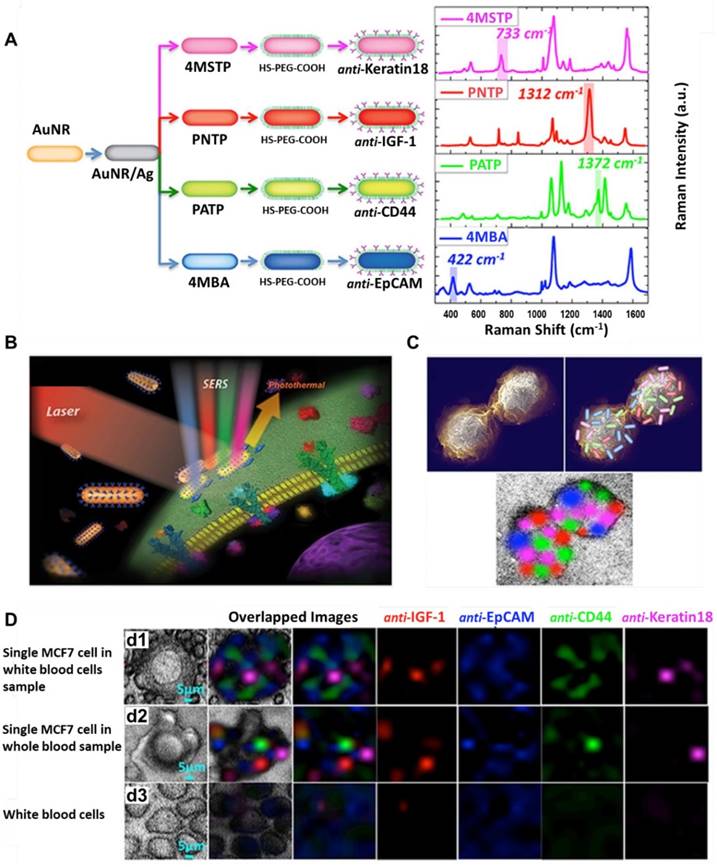
Nima et al. fabricated a series of SERS probes by functionalizing Ag-Au nanorods with four different Raman-active molecules ((4-mercaptobenzoic acid [4MBA], p-aminothiophenol [PATP], p-nitrothiophenol [PNTP], and 4-(methylsulfanyl) thiophenol [4MSTP]), and conjugating them with four kinds of antibodies specific to breast cancer markers (anti-EpCAM, anti-CD44, anti-Keratin 18 and anti-Insulin-like growth factor antigen), respectively (Figure 9). The Raman signals of the reporter molecules on the Ag-Au nanorods were more than two orders of magnitude higher than those on conventional Au nanorods. Using this highly sensitive and specific SERS-labeled antibody cocktail, they could detect single breast cancer cells in unprocessed human blood [55]. In another study performed by Wang et al., SERS-fluorescence joint spectral-encoded magnetic nanoprobes were fabricated and used for multiplexed cancer cell separation [56]. The nanoprobes have four main components: silica-coated magnetic nanobeads as the inner core, Au@Ag nanorods as the SERS generator, CdTe quantum dots as the fluorescent agent, and antibodies modified on the outside layer as recognition groups. Two Raman reporter molecules (5,5′-dithiobis (2-nitrobenzoic acid) [DTNB] and 4MBA) and two quantum dots (CdTe 614 and CdTe 512) were combined to encode the probes. Four antibodies/proteins (anti-HER2, transferrin, anti-CD3 and anti-prostate-specific membrane antigen (PSMA)) were conjugated onto the probes to target SKBR3, Hela, Jurkat T and LNCaP cells, respectively. By utilizing the above four human cancer cell lines and one normal cell line MRC-5 as model cells, the authors proved that the nanoprobes can specifically separate cancer cells from normal ones and potentially could be used for high throughput analysis and cancer diagnosis. However, the authors pointed out that the limitation of this system is that the nanoprobes precipitated and produced signals no matter if a target cell was captured or not, and they suggested to separate the magnetic cores from the probes in future work [56].
Detection of multiple surface markers on CTCs using SERS. (A) Schematic and Raman spectra of four antibody-modified SERS nanotags. (B) Schematic of breast cancer cell surface targeting by four SERS tags and a SERS/photothermal detection technique. (C) Schematic of 2D multi-color SERS data correlation with SERS tag' distribution on the cell surface. (D) Multicolor SERS analysis of a single MCF-7 cell among WBCs (d1), a single MCF-7 cell in whole blood (d2), and WBCs only (d3). Adapted with permission from [55], copyright 2014 Springer Nature.
(A) Experimental workflow for the use of four melanoma surface marker antibodies (MCSP, MCAM, ErbB3, and LNGFR)-modified SERS tags to monitor CTC surface marker expression. (B) CTC populations in response to treatment: the frequency distribution of each marker can signal how diverse the cell populations are in terms of surface marker expression levels. (C) CTC signature in response to treatment: the relationship between the average Raman intensities of each surface marker represents the CTC signature. Adapted with permission from [59], copyright 2018 Springer Nature.
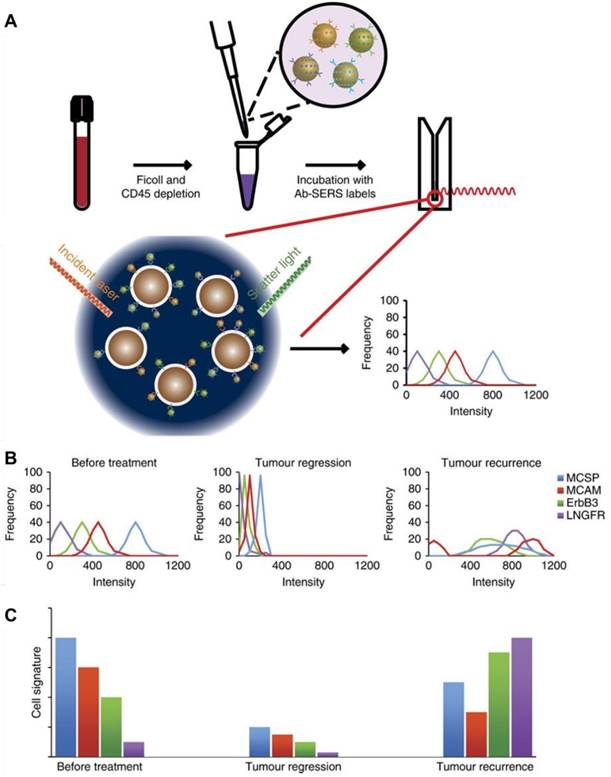
Recent studies have indicated that the molecular phenotype of CTCs evolves dynamically with the progression of disease and during treatment [38]. For example, ER- CTCs have been detected in patients with breast cancer who had ER+ primary carcinomas [57], while HER2- breast cancer was found to acquire a HER2+ subpopulation after multiple courses of therapy [58]. Therefore, real-time monitoring of phenotypic evolution of CTCs is crucial for obtaining vital tumor biological information for treatment guidance. Recently, Tsao and coworkers developed a SERS-based technique to characterize the phenotype changes of melanoma cell lines as well as CTCs from clinical melanoma patients during immunological or molecular targeted therapies [59]. Four Raman reporter molecule-surface marker pairings were decorated on the surface of Au NPs (Figure 10): 4MBA with melanoma-chondroitin sulphate proteoglycan (MCSP); 2,3,5,6-tetrafluoro-4-mercaptobenzoic acid (TFMBA) with melanoma cell adhesion molecule (MCAM); 4-mercapto-3-nitro benzoic acid (MNBA) with erythroblastic leukaemia viral oncogene homologue 3 (ErbB3); and 4-mercaptopyridine (MPY) with low-affinity nerve growth factor receptor (LNGFR). After testing the assay specificity and sensitivity using cell lines with known expression of surface markers—SK-MEL-28, MCF-7, SKBR3, BM-MSC, etc. —the SERS probes were applied to monitor cellular phenotypic changes of melanoma cell lines harboring BRAF mutations in response to BRAF inhibitor (PLX4720), showing that all three tested cell lines formed distinct subpopulations after drug treatment. Then the authors collected blood samples from ten stage-IV melanoma patients serially during the course of treatment and monitored the CTC signature change. Based on the multiplexed SERS detection and analysis, they found that CTC populations shifted after treatment with dabrafenib and trametinib for 40 days and discriminated drug-resistant clones that show different CTC phenotypes [59]. Compared with current CTC detection technologies such as CellSearch system and CTC Chip, the proposed SERS-based technique is extremely sensitive (10 cells in 10 mL of blood), highly multiplexed (simultaneous monitoring of several surface protein expression profiles) and simple (does not need initial enrichment of CTCs), and therefore holds great potential to be translated into clinical use for disease and treatment monitoring.
Capture, detection and release of CTCs
Besides enumeration and subtype characterization of CTCs, downstream analyses such as single cell genomics/ transcriptomics, or ex vivo culture of CTCs for drug sensitivity tests may provide more comprehensive information for personalized cancer therapy [8]. In a SERS-coding microsphere suspension chip designed by Li et al., folic acid as a recognition molecule was immobilized on magnetic composite microspheres through a disulfide bond and used to capture CTCs. After that, 90% of the CTCs were eluted within 20 min by incubation with glutathione which breaks disulfide bonds, and SERS labels on the cells assisted in conveniently recognizing the captured/ recovered cells [60]. More recently, Ruan et al. established a supersensitive CTC analysis system using folic acid-conjugated triangular silver nanoprisms and folic acid-conjugated superparamagnetic iron oxide nanoparticles (SPION) as SERS probes and CTC capturing agents, respectively (Figure 11A). A LOD as low as 1 cell/mL was achieved, and after adding excess folic acid the captured CTCs could be released for further cell expansion and phenotype identification [61].
Capture, detection and release of CTCs. (A) Schematic of the preparation of a supersensitive CTC analysis system based on Ag nanoprisms and SPION (a), and its application to the capture, enrichment, detection, and release of CTCs (b). Adapted with permission from [61], copyright 2018 American Chemical Society. (B) Schematic of the selective detection and analysis of CTCs and circulating cancer stem cells for monitoring tumorigenesis and metastasis.
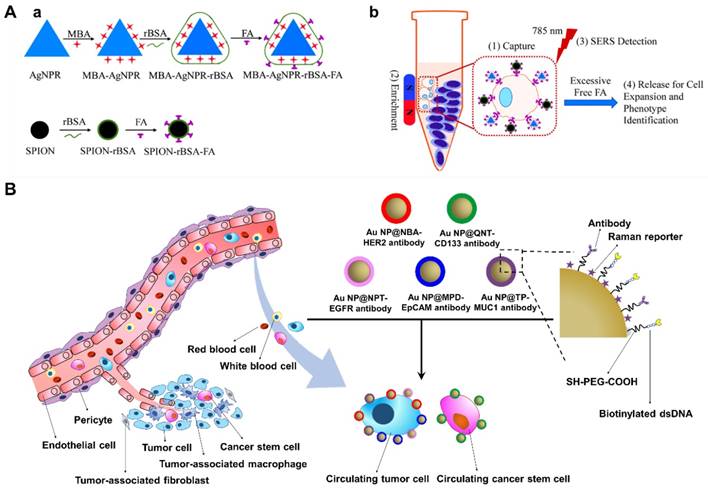
Circulating cancer stem cells (CCSCs) as a rare type of CTCs, have arisen as a useful resource for monitoring and characterizing both cancers and their metastatic derivatives. Based on SERS detection combined with a microfluidic chip, Cho and coworkers developed a new technique for selective isolation and non-invasive analysis of CCSCs in complete blood samples. Au NPs were first labeled with 5 different Raman reporter molecules: thiophenol (TP), Nile blue A (NBA), 1-naphthalenethiol (NPT), 4-mercaptopyridine (MPD) and 2-quinolinethiol (QNT). The obtained SERS labels were then coated with a PEG layer and individually conjugated with antibodies against 5 different surface markers (anti-CD133, anti-EpCAM, anti-EGFR, anti-HER2 and anti-MUC1) to distinguish CCSCs and several major breast cancer CTC subtypes (Figure 11B). A biotinylated dsDNA was also conjugated on the surface of the SERS probes for later recognition of streptavidin on the CCSC-chip, thus when the SERS probes-labeled cell suspensions flow through the microfluidic channel on the chip, the labeled CCSCs can be captured through biotin-streptavidin reaction and simultaneously detected by SERS (93% accuracy), followed by restriction enzyme digestion of dsDNA to release the cells. The authors then utilized their new method to predict tumor metastasis by screening blood samples from xenograft models and, upon CCSC detection, they found that all the tested subjects exhibited liver metastasis [62].
Exosomes
Exosomes are small extracellular vesicles (~30-150 nm in diameter) that originate from multivesicular bodies and are released into the extracellular environment by fusion of the compartments with the plasma membrane [10]. Exosomes were first isolated by Johnstone et al. in the 1970s and in the following decades were considered to be only involved in the removal of unnecessary substances [63]. After their function as intercellular messengers were demonstrated in several studies recently, exosomes have gained enormous attention both as disease markers for diagnosis and as delivery vehicles for therapy [11, 64-66]. The surface molecules on exosomes mediate their recognition of recipient cells [67]. Once attached to a target cell, exosomes can induce signaling via receptor-ligand interaction or be internalized into the target cells and deliver their content (DNA, RNA, protein and lipids) into the cytosol for cell-cell communication [10]. Exosomes can be released from almost all cell types; among them, tumor-derived exosomes are of particular interest due to their important role in cancer development, metastasis, regulation of immune responses, and induction of angiogenesis [68-70].
Current techniques for exosomes isolation and detection
Exosomes are present in most body fluids such as blood, urine, saliva and ascites, and hold great potential to be used as promising biomarkers for liquid biopsy-based cancer diagnosis. In recent years, a large variety of techniques have been developed for exosome detection, most of which require an initial isolation step to purify exosomes from the complex biological milieu using separation methods such as differential ultracentrifugation, ultrafiltration, precipitation using water-excluding polymers, immunoaffinity capture, and microfluidics-based techniques [71]. In 2006, Thery et al. set up the ultracentrifugation protocol for purifying exosomes from cell culture supernatants and biological fluids that is currently considered the gold standard and represents one of the most commonly used and reported techniques in exosome isolation [72]. Additionally, based on the small size of exosomes, ultrafiltration using nanomembranes has also been introduced for exosome isolation [73]. Compared with ultracentrifugation, ultrafiltration is relatively faster and does not rely on specialized equipment; however, the shear stress might cause deterioration and the exosomes might be lost due to trapping in the pores of the filters. Exosome precipitation using polymers such as PEG is another method that is easy to perform and has been proved to be more efficient than ultracentrifugation and nanomembrane concentration [73]. Several isolation kits based on precipitation are now comercially available; nevertheless, their applications are limited due to the coisolation of non-exosome materials such as proteins, which influence subsequent analysis. Immunological separation based on selective recognition of proteins on the membrane of exosomes (such as tetraspanins CD81, CD9, CD63 and cancer-related proteins HER2, EGFR) using corresponding antibodies-modified magnetic beads enables specific and fast purification of exosomes; however, it remains difficult to be applied to large-volume samples. Recently, rapid advances in microfabrication technology have offered the opportunity to fabricate complicated microfluidics-based devices and integrate them with the above-mentioned size/immunoaffinity-based techniques as well as acoustic/electrokinetic sorting methods, allowing fast separation of exosomes from a large number of samples [74, 75]. After purification, the exosomes are usually characterized using techniques such as ELISA, flow cytometry, nanoparticle tracking analysis (NTA), microfluidic or electrochemistry-based approaches to evaluate the number of exosomes or their expression levels of disease-related proteins. Progress in the aforementioned exosome isolation and detection techniques has been summarized in several comprehensive reviews [18, 71, 76].
In recent years, plasmonic nanomaterials have been intensively employed for quantitative and qualitative analysis of exosomes. For example, Liang et al. described a rapid, ultrasensitive and inexpensive nanoplasmon-enhanced scattering (nPES) assay that directly quantifies tumor-derived extracellular vesicles from as little as 1 μL of plasma. In this assay, extracellular vesicles are first captured by antibodies modified on a sensor chip and when antibody-conjugated gold nanospheres and nanorods bind with the vesicles concurrently, the formed Au nanosphere-vesicle-Au nanorod complexes produce a local plasmon effect that shifts the spectra of scattered light while increasing the scattering intensity [77]. Besides the nPES assay, plasmonic nanomaterials have been designed as SERS tags for simultaneous detection of multiple cancerous exosomes, and have also been applied as enhancement substrates for exosomes classification using label-free SERS analysis methods [78]. In the following sections, we will summarize recent studies using SERS-based techniques for exosome detection and analysis.
Schematic of the preparation of aptamer-modified magnetic beads, SERS tags and the work flow of SERS-based detection of exosomes. Adapted with permission from [80], copyright 2018 The Royal Society of Chemistry.
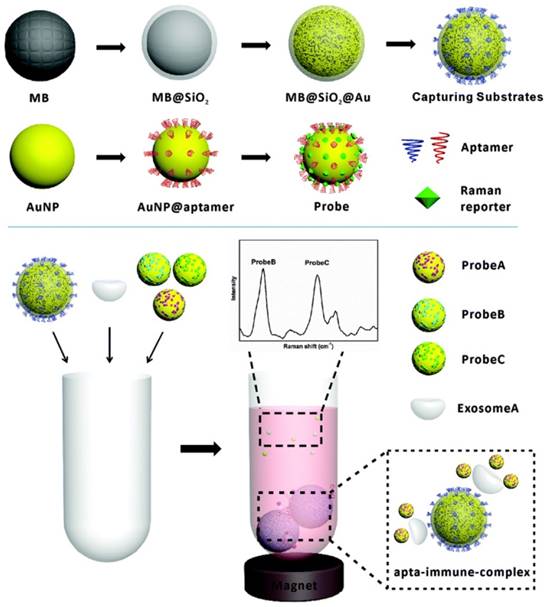
Detection of exosomes by SERS tags
Although SERS tags have been largely applied for the detection of CTCs and disease-related biomolecules in the past decade, their first application to the detection of tumor-derived exosomes appeared only two years ago. In 2016, Cui's group fabricated anit-CD63 antibody-modified magnetic NPs and anti-HER2 antibody-modified Au@Ag nanorods, which can bind various surface proteins on the membrane of tumor-derived exosomes. In the presence of the target exosomes, a sandwich-type immunocomplex is formed and, after precipitation using a magnet, the SERS signals are measured. With this assay, exosomes from a human breast cancer cell line SKBR3 and a human fetal lung fibroblast cell line MRC5 were studied, proving that tumor-derived exosomes can be qualitatively and quantitatively detected, with a LOD of 1200 exosomes in duration of 2 h [79]. To simultaneously detect multiple kinds of exosomes, the same group improved their method by mixing exosome samples with CD63 aptamer-modified magnetic nanobeads and three kinds of SERS nanoprobes that were labelled with different Raman reporter molecules and modified with aptamers against different tumor markers (HER2, CEA and PSMA). When one kind of target exosome is present, only the specific SERS probes will recognize the target exosomes and form a sandwich-type complex with the magnetic nanobeads. After the complexes are precipitated by a magnet, decreased signals of the corresponding SERS probes in the supernatant will be detected while the signals of the other non-relevant SERS probes remain unchanged. When three kinds of exosomes are present simultaneously, the SERS intensities of the three types of the probes will all decrease (Figure 12). Using this method, LOD values of 32, 73, and 203 exosomes per microliter were achieved for exosomes secreted by SKBR3, Tb4 and LNCaP cells, respectively. In addition, blood samples from breast cancer, colorectal cancer and prostate cancer patients were tested, demonstrating the great potential of this method to be applied to early stage screening of cancers [80].
(A) Experimental workflow of the fabrication of an antibody array and the detection of exosomes by the antibody array and SERS tags. (B) Average SERS spectra of exosomes at different concentrations captured with CD63 antibodies, average SERS spectra from exosomes using different capture antibodies, and a colorimetric comparison of protein expressions on cancer and normal cells based on SERS detection results. Adapted with permission from [81], copyright 2018 Ivyspring.
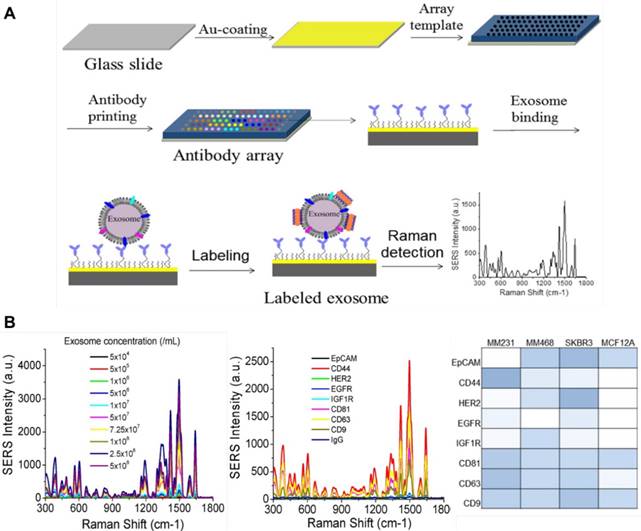
In another study performed by Kwizera et al., an antibody array was printed on a standard gold-coated glass slide to fabricate a miniaturized device for exosomes capture (Figure 13). By using gold nanorods labelled with QSY21 as SERS tags to detect the exosomes, a LOD of 2000 exosomes per microliter was obtained and more than 80 purified samples could be analyzed on a single device within 2 h. The expression levels of serval surface protein markers (EpCAM, CD44, HER2, EGFR, IGF1R, CD81, CD63 and CD9) were analyzed, showing that exosomes derived from MDA-MB-231, MDA-MB-468, and SKBR3 breast cancer cells give distinct protein profiles compared to exosomes derived from MCF12A normal breast cells. The authors also found that exosomes in the plasma from HER2-positive breast cancer patients exhibit significantly higher levels of HER2 and EpCAM than those from healthy donors, suggesting the diagnostic potential of these markers for breast cancer diagnostics [81]. More recently, Li et al. developed an ultrasensitive SERS immunoassay for exosome-based diagnosis and classification of pancreatic cancer. In this assay, polydopamine (PDA) was self-polymerized on the surface of both glass slides and SERS tags (Au@Ag-PATP) and antibodies against different target proteins on exosomes (migration inhibitor factor (MIF), glypican 1 (GPC1), CD63, EGFR) were individually encapsulated into the PDA layer. Then, exosomes derived from pancreatic cancer or healthy control patient samples were captured on the chip surface followed by recognition of the SERS tags to form a “chip-exosome-SERS tag” sandwich structure. Based on the specific recognition and strong signal of the SERS tags, a single exosome in 2 μL sample could be detected. In addition, by analyzing 2 μL of clinical serum sample, the MIF antibody-based immunoassay could not only discriminate pancreatic cancer patients from healthy individuals, but also distinguish metastasized tumors from metastasis-free tumors and different stages of tumor node metastasis [82].
Label-free SERS detection of exosomes
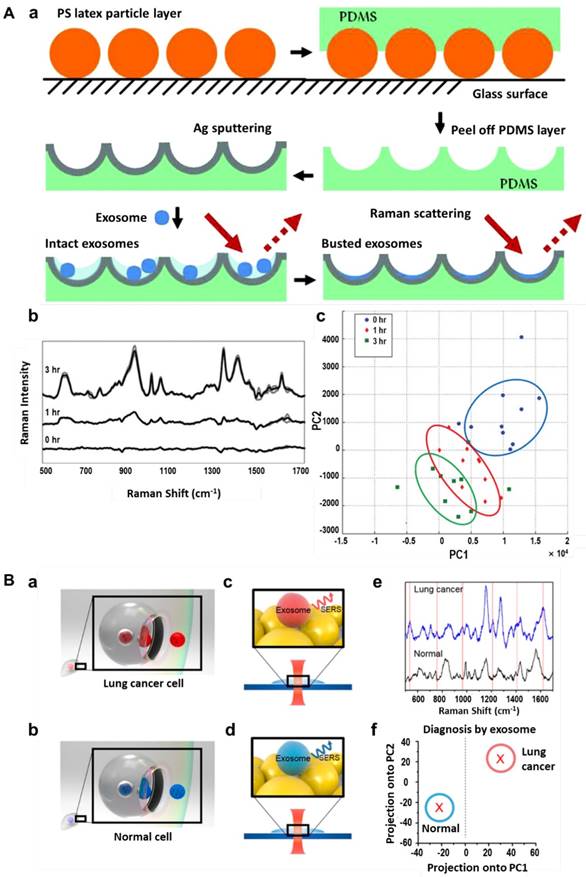
Among the various techniques for exosome characterization, ELISA, flow cytometry, SERS tags and other labelling tools provide information of only targeted biological components in the exosomes, while other techniques like NTA and TEM provide only physical information such as size distribution of the exosomes. Genomic, proteomic and lipidomic approaches may provide comprehensive molecular information of the exosomal content, but these methods usually require complicated and time-consuming protocols and are cost inefficient since a large amount of sample is needed for a single measurement [83]. Raman spectroscopy provides the intrinsic molecular information of biological samples and, as in SERS, these signals can be greatly enhanced when the analytes are located in close proximity to plasmonic nanomaterials [32]. To obtain enhanced Raman signals of exosomes, Tirinato et al. decorated nano-geometry-based phonics structures on a super-hydrophobic surface, with which exosomes were conveniently concentrated and conveyed onto the SERS-active area. SERS spectra analysis indicated that exosomes derived from healthy cells (CCD841-CoN) exhibited a higher lipid content while the ones derived from tumor cells (HCT116) exhibited a richer RNA content [84]. In a later study performed by Lee et al., a silver film-coated nanobowl platform was fabricated to capture exosomes secreted by SKOV3 cells and served as a substrate to enhance the Raman signals (Figure 14A). SERS spectra of exosomes purified by two separation methods (total exosome isolation reagent (TEIR) and ultracentrifugation) were recorded and statistically analyzed with principal component analysis (PCA). They found that the TIER kit might purify a certain kind of exosomes more selectively than ultracentrifugation but produced strong background SERS peaks which needed to be subtracted. New SERS peaks were found to appear during the drying process, indicating that the initially intact exosomes ruptured over time and released their molecular contents. This time-dependent evolution of SERS peaks enabled analysis of exosomal membrane compositions as well as the contents inside the exosomes [83]. Other metallic substrates such as a bimetallic nanoplasmonic gap-mode substrate fabricated by assembling Ag nanocubes on Au nanorods array [85], Au nanospheres-deposited cover glass (Figure 14B) [86], silver-coated recordable compact disk [87], positively-charged small Au NPs [88] and integrin-specific peptide ligand-modified Ag NPs [89] have also been applied for analysis of intrinsic Raman spectra of exosomes. In most of the cases, patterned metallic surfaces instead of colloidal NPs were applied for SERS detection of exosomes, probably due to the similar size range of SERS NPs and exosomes and the difficulty in linking exosomes in close proximity to the NP surface. In a recent study performed by Stremersch and coworkers, 10 nm positively-charged Au NPs were prepared by coating a 4-dimethylaminopyridine layer on the NPs. These NPs can be electrostatically adsorbed onto the surface of exosomes to form a plasmonic shell, thereby enabling generation of enhanced Raman signals of individual vesicles in suspension. By performing partial least squares discriminant analysis (PLS-DA) on the obtained spectra, vesicles from different origins (B16F10 melanoma cells and red blood cells) were distinguished [88].
Different from SERS tags-based detections, label-free SERS detection usually obtains complicated spectra due to the complexity of biological samples and, therefore, requires arduous spectral analysis to obtain biomedically relevant information. In most cases, the SERS spectra of exosomes from cancer patients with reference to healthy people are recorded and compared, and the peaks in the difference spectra are tentatively assigned to certain components such as lipid, phospholipid, nucleic acid, polysaccharide, amino acids and proteins according to previous literatures. However, SERS signals greatly depend on interactions among sample, metal substrate and laser. Even for exosomes derived from the same cell lines, the intensity and shape of SERS spectra might be varied when different parts of exosomes are located in the plasmonically active area. To statistically classify the origin of exosomes, PCA analysis is usually performed to compact the information contained in the spectra into a limited set of principal components, and the scores for individual measurements may provide a quantitative metric to detect differences between complex spectra. Alternatively, by training with Raman spectra of pure samples, a PLS-DA model can be built and used to discriminate different types of vesicles.
Circulating tumor DNA
ctDNA is a class of cell-free DNA (cfDNA) that is derived from apoptotic and necrotic neoplastic cells in the tumor microenvironment or is released from lysed CTCs. They are relatively small molecules with a short length of ~100-200 nucleic acid base pairs corresponding to 30-60 nm [19, 90]. In 1977, Leon et al. found that the cfDNA levels in cancer patients were significantly higher than those in healthy individuals [91], but this phenomenon gained little attention until 1994 when mutated K-ras sequences were detected in the blood of pancreatic cancer patients [92]. After that, a panel of abnormal DNAs such as mutation of oncogene and antioncogene, loss of heterozygosity of chromosome, and DNA methylation, have been discovered in cancer patients. Recent studies have demonstrated that these ctDNAs present in body fluids are closely related to tumor burden, relapse, therapy response, and resistance, which provides new molecular markers for cancer diagnosis, therapeutic effect monitoring and prognosis prediction [14]. Moreover, the half-life of ctDNAs is shorter than 2 h, and therefore enables dynamic cancer genotyping and real-time monitoring of treatment response [93, 94].
Label-free SERS detection of exosomes. (A) Schematic of a silver film-coated nanobowl substrate preparation and its use in SERS analysis of intact and ruptured exosomes (a). Time-dependent SERS spectra of exosomes derived from the SKOV3 cell line (b) and principal component analysis of the SERS spectra (c). Adapted with permission from [83], copyright 2015 The Royal Society of Chemistry. (B) Schematic of exosomes released from lung cancer cells (a) and normal cells (b), and SERS detection of the two types of exosomes (c, d). e, SERS spectra of exosomes released from lung cancer cells (blue) and normal cells (black). f, Exosome classification by PCA of SERS spectra. Adapted with permission from [86], copyright 2017 American Chemical Society.
Isolation of cfDNA for detection of tumor-specific alterations is relatively easy compared with purification of CTCs and exosomes. Typically, 5-10 mL of blood sample is extracted in an anticoagulant tube and centrifuged to obtain plasma containing cfDNAs. Although the amount of cfDNAs in plasma may be 2-4 times lower than that in serum [95], it is recommended to extract cfDNAs from plasma due to the lower interference from lysed cellular DNA. It is important that preparation of cfDNAs should be completed promptly after blood draw because they are not stable as a result of the existence of DNase in the blood [96]. Over the past decades, continuous efforts have been devoted to the development of valid methods for analyzing ctDNA, which is challenging due to the small fraction of tumor-specific DNA among the enormous total cfDNAs (<1.0% in many cases). Current technologies for the analysis of ctDNAs can be broadly categorized as targeted and untargeted approaches. Targeted approaches utilize PCR-based, digital PCR-based or sequencing-based techniques to detect specific known somatic mutations/epigenetic alterations that have been discovered in a primary tumor, such as BRAF, KRAS, TP53, PIK3CA and methylated CpGs. On the other hand, untargeted approaches employing genome-wide methods such as whole genome sequencing, personalized analysis of rearranged ends (PARE), and digital karyotyping, are important for the discovery of novel disease markers. Progress in these techniques has been summarized in several recent reviews [93, 97]. Here, we will introduce two SERS-based strategies for the detection of targeted ctDNAs: combining enzymatic amplification with SERS tags, and “click” SERS reaction.
ctDNAs detection by enzymatic amplification combined with SERS tags
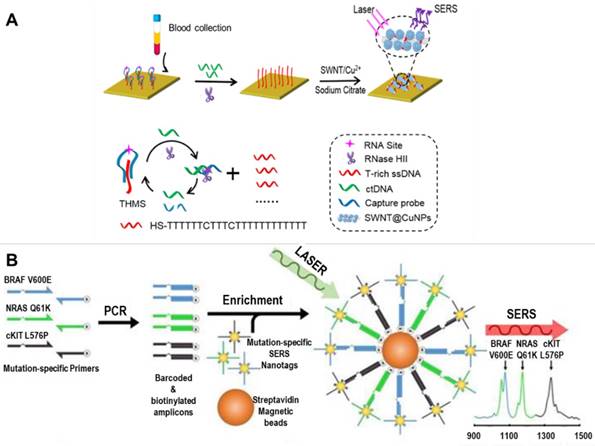
Zhou and coworkers developed a novel strategy to detect single-stranded ctDNA by combining a molecular recognition unit (triple-helix molecular switch, THMS) and, a signal amplification unit (RNase HII enzyme) with T-rich ssDNA-mediated SERS enhancement of single-walled carbon nanotubes (SWNTs). As shown in Figure 15A, the presence of target ctDNA leads to release of T-rich ssDNA from THMS. With the aid of HII enzyme-assisted amplification, a large number of T-rich ssDNAs could be adsorbed onto the surface of SWNTs by π-π stacking interactions and serve as a template for in situ growth of copper NPs. The Cu NPs decorations greatly enhance the G-band peak of the SWNTs due to the enhanced electromagnetic field between Cu NPs and SWNTs. This method enables detection of DNA point mutations (KRAS G12DM) with a sensitivity of 0.3 fM, and needs a sample volume of only 5 μL [98]. In another study performed by Wee et al., a multiplexed PCR/SERS detection method was designed for simultaneous analysis of 3 DNA point mutations in melanoma (BRAF V600E, c-Kit L576P and NRAS Q61K). In this assay, Au NPs modified with Raman reporter molecules and thiolated DNA oligonucleotides were used as SERS nanoprobes to recognize the barcoded-amplicons, and then were enriched using magnetic beads for SERS detection (Figure 15B). This assay enables highly specific and sensitive (10 mutant alleles from a background of 10,000 wild type sequences) detection of multiple ctDNAs with the convenience of standard PCR, and may potentially be translated into clinical applications [99].
SERS-based detection of circulating tumor DNA. (A) Schematic of SWNT-based SERS assay coupling with RNase HII-assisted amplification for highly sensitive detection of ctDNA in human blood. Adapted with permission from [98], copyright 2016 American Chemical Society. (B) Schematic of a multiplexed PCR/SERS assay: multiplex mutation-specific primers were used to amplify tumor DNA, the amplicons were then tagged with mutation-specific SERS nanotags and enriched using magnetic beads. After that, Raman detection was performed for evaluation of the mutation based on the corresponding unique spectral peaks. Adapted with permission from [99], copyright 2016 Ivyspring.
(A) Schematic of the concept of “Click SERS” analogous to click chemistry. (B) a, Schematic of a 15-well plate for 10-plex DNA detection and top view of the detection wells with full spectra when 10 targets are added. b, TEM images of dimer formation. c, Acquired “Click” spectra presented in the wells during sample detection. Adapted with permission from [100], copyright 2018 American Chemical Society.
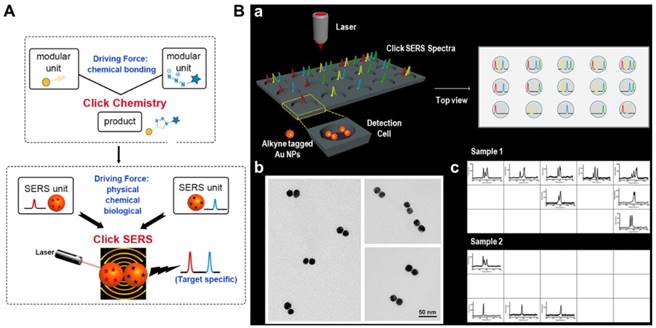
ctDNAs detection by “Click” SERS
Most recently, Zeng et al. reported a new readout technique for DNA detection, the so-called “Click” SERS, which is based on Raman scattered light splice derived from SERS tag assemblies [100]. Analogous to the reaction of small molecule units in click chemistry, two SERS tags form a dimer upon simultaneous recognition of a target molecule, leading to combinatorial output of signals from individual NPs (Figure 16A). In order to simplify the spectral analysis and avoid interference from biological milieu, triple bond-containing reporters with single and narrow emission in the biological Raman silent region (~1800-2600 cm-1) were synthesized and employed to label the SERS tags. In contrast to the conventional “sole code related to sole target” readout method, “Click” SERS relies on the number and positions rather than the intensity of combinatorial emissions, and thus the multiplexing capacity can be continuously increased as long as more distinguishable emissions are employed (Figure 16B). In a proof of concept study, the authors successfully detected 10-plex DNAs synchronously using this strategy. Apart from DNA molecules, other substances such as thrombin, ATP, Hg2+ in solution and receptor proteins on cell membranes could also be detected/ imaged by target-simulated assembly of SERS tags [100].
Circulating microRNAs
MicroRNAs are small non-coding RNAs (18-25 nucleotides) that regulate gene expression by paring to the 3' untranslated region (UTR) of a set of mRNAs [101]. Since discovery of the first miRNA in Caenorhabditis elegans, over 30,000 mature miRNAs have been identified in 206 species [102, 103]. miRNAs are involved in regulating a wide range of biological and pathological processes including development, metabolism, signal transduction, and tumor progress. Correlations between miRNAs and different types of cancer have been well-documented in several recent reviews, showing that certain species of miRNAs play key roles in tumorigenesis, progression and metastasis [15, 16, 103-105]. For example, miR-25 and miR-223 were proved to be relevant in the tumorigenesis of NSCLC [106], while miR-155 was found to be significantly elevated in breast cancer patients [107]. Due to their broad existence and stability in many body fluids including blood, urine, saliva, tear, and cerebrospinal fluid, circulating miRNAs are emerging as a class of novel biomarkers for cancer detection and prognosis [108].
Circulating miRNAs are usually extracted from body fluids using acid guanidinium thiocyanate-phenol-chloroform and then isolated by either precipitation or column-based purification before detection. Current approaches for circulating miRNA analysis include qRT-PCR and miRNA arrays, which may quantitatively characterize target miRNAs, and next-generation sequencing, which may help to identify novel tumor-related miRNAs [109]. However, due to the low abundance of miRNAs in total RNA samples, these traditional methods are limited in detection sensitivity and specificity. New techniques that can simultaneously detect multiple target miRNAs with improved sensitivity and specificity are highly desired for clinical applications of circulating miRNAs.
Label-free SERS detection of circulating miRNA
SERS as a sensitive spectroscopic technique was introduced into the field of miRNA detection in 2008 when Driskell et al. utilized silver nanorod arrays as a metal substrate for label-free detection and classification of miRNAs. In this work, 5 unrelated human miRNAs and 8 members of the Let-7 miRNA family were synthesized and directly adsorbed onto the silver nanorod arrays. The SERS spectra of related and unrelated miRNAs were detected in near-real time, and miRNA patterns were classified using PLS-DA analysis with high accuracy [110]. By generation of PLS models for two-, three-, and five-component mixtures from massive spectra covering a wide concentration range of each miRNA, the same group could quantitatively detect individual miRNA in multicomponent mixtures [111]. In a later study, Abell et al. employed an array-patterned Ag nanorods chip to obtain reproducible SERS signals and used least squares analysis for quantitative determination of the relative ratios of the four nucleotide components A, C, G, and T/U in miRNA sequences. Using this method, subtle changes in the SERS spectra of a clinically relevant miRNA before and after hybridization were captured [112].
Detection of circulating miRNA by SERS tags
Compared with label-free analysis, SERS tags have been more frequently employed for ultrasensitive and multiplexed detection of circulating miRNAs, mainly through the following three strategies: formation of a sandwich structure, signal turn “on/off”, and hybridization chain reaction (HCR)-induced signal amplification.
A sandwich structure is usually composed of a detection probe, a target molecule and a capture unit. In a study performed by Guven et al., miR-21 was either directly immobilized onto a gold slide or using a capture probe for hybridization, and then SERS tags modified with detection probes were added and further hybridized with the targets. The detection limits of the direct and sandwich assays were both around 1 nM; however, the sandwich assay provided better selectivity [113]. Later, Su and coworkers synthesized DNA-mediated Au-Ag nanomushrooms with interior nanogaps as ultrasensitive detection probes. The DNA on the NPs act as both gap DNA for SERS signal enhancement and probe DNA for hybridization with target sequences. Raman reporter molecules were adsorbed onto the NPs either by using a Raman reporter-labeled alkanethiol probe DNA or co-assembling thiol-containing Raman reporter molecules with the probe DNA, and capture DNA was modified on the surface of magnetic NP through biotin-streptavidin reaction (Figure 17A). In the presence of target sequences (miR21, miR31, miR141), sandwich structures (Au-Ag nanomushroom-target RNA-magnetic NP) are formed and SERS signals can be detected at a miRNA concentration as low as 1 pM [114]. Similarly, Zhou et al. fabricated nanogap-based SERS tags encoded with Raman dyes and synthesized hollow silver microspheres using bacteria as templates. The SERS tags and Ag microspheres were modified with detection DNA probes and capture DNA probes, respectively. Simultaneous detection of multiple liver cancer-related miRNAs (miR223, miR21, miR122) was performed with high sensitivity (detection limit of 10 fM) and specificity, demonstrating the potential of this assay in clinical diagnosis [115].
The second strategy, turning “on” or “off” the signal in response to the presence of target sequences, is usually achieved using molecular beacons (MBs). Typical MBs are single-stranded DNA molecules that consist of a stem-and-loop structure doubly labeled with a fluorophore and a quencher group on each end. In the absence of targets, MBs are in the “off” position due to the close proximity of the fluorophore with the quencher group; Upon binding with the target, the hairpin is opened and the fluorescence is turned “on”. Based on their simple operation and, high sensitivity and selectivity, fluorescent MBs have been widely used in biosensing [116]. Song et al. combined MBs with SERS for multiple detection of 3 lung cancer-related miRNAs, i.e., miR-21, miR-486 and miR-375. Three MBs complementary to the target miRNAs were prepared and functionalized with a thiol group at their 3' end and Raman reporter molecules at their 5' end, and then adsorbed onto an Ag nanorod array substrate. In the presence of target miRNAs, the molecular beacons opened and the SERS signals changed from “on” to “off” (Figure 17B). By monitoring the change in SERS signal, the concentrations of miR-21, miR-486 and miR-375 in human serum were simultaneously detected at LODs of 393 aM, 176 aM and 144 aM, respectively [117]. In another assay, Wang et al. developed an “off” to “on” signal switch, which they called “inverse Molecular Sentinel (iMS)” nanoprobes to distinguish from the previous “on” to “off” switch. In this assay, a single-stranded DNA served as a “placeholder” strand to hybridize with the stem-loop probe and keep the Raman reporter molecules away from the metal surface. Upon exposure to a target miRNA, the placeholder DNA left the MB probe, allowing the stem-loop structure to close and Raman reporter molecules to move onto the metal surface, and thereby, SERS signals were yielded [118]. In a recent work performed by He et al., the “off” to “on” SERS nanoprobes were further combined with padlock probe-based exponential rolling circle amplification (P-ERCA) for ultrasensitive detection of miR-155. This assay exhibited a wide linear range of 100 aM to 100 pM with a LOD of 70.2 aM, indicating its potential application for clinical diagnostics [119].
SERS-based detection of circulating microRNA. (A) Schematic of the synthesis of mushroom-like Au-Ag SERS probes by either using a Raman reporter-labeled alkanethiol probe DNA (1), or co-assembling thiol-containing Raman reporter molecules with the probe DNA (2), and formation of the sandwich complexes by hybridization of target DNA/RNA with capture beads and SERS probes. Adapted with permission from [114], copyright 2017 American Chemical Society. (B) Schematic of the preparation and application of the molecular beacon-functionalized SERS sensor (signal turn “on/off”) for simultaneously measuring multiple miRNAs. Adapted with permission from [117], copyright 2016 The Royal Society of Chemistry. (C) Design scheme of enzyme-free quadratic SERS signal amplification for circulating microRNA detection in human serum via miRNA-triggered hybridization chain reaction and Ag+-mediated cascade amplification. Adapted with permission from [121], copyright 2015 The Royal Society of Chemistry.
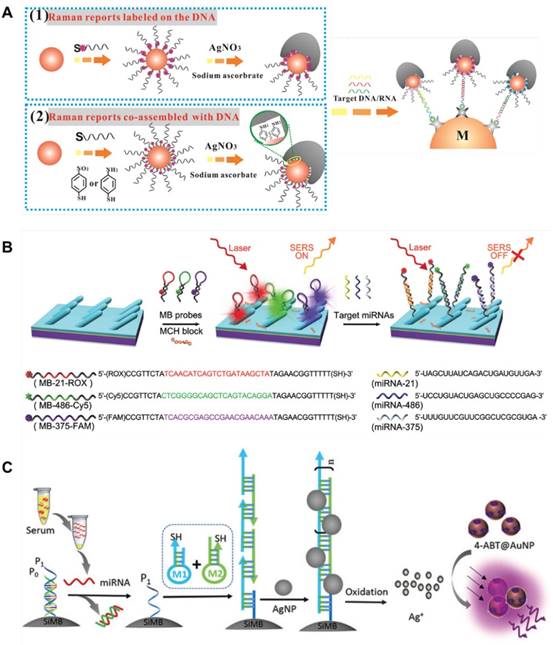
Hybridization chain reaction is an enzyme-free, room temperature linear amplification approach that was first introduced by Robert et al. in 2004 [120]. In this approach, stable species of DNA hairpins coexist in solution until the introduction of initiator strands triggers a cascade of hybridization events that yields nicked double helices analogous to alternating copolymers. Compared with enzyme-based amplification strategies, HCR is simple in operation and cost-effective, only using the DNA single strand for in situ adjustment of the length of double-stranded DNA. Zheng and coworkers combined miRNA-triggered HCR reaction with Ag+-mediated cascade amplification to detect circulating miRNAs in serum. As shown in Figure 17C, the target miRNA triggered fabrication of long self-assembled DNA polymer via HCR on the surface of silica microbeads, inducing multiple Ag NPs conjugation, which worked as a primary amplification element. Then, the Ag NPs were dissolved into silver ions, which can control the gaps between neighboring Raman reporter-encoded Au NPs to form ''hot-spots'' and produce enhanced SERS signals. The quadratic amplification allowed a detection limit of miRNA as low as 0.3 fM. Using this assay, miR-21 in sera of chronic lymphocytic leukemia (CCL) patients and healthy donors was detected, indicating increased expression of miR-21 in CCL patients [121]. In a later study performed by Liu et al., locked nucleic acid (LNA) capture sequences were bolted on a chip surface to recognize miR-21. Upon addition of target molecules, high-efficiency cascade HCR was triggered and hairpin structure-modified, 4,4′-biphenyldithiol (DBDT)-coded Ag NP dimers were assembled into a large aggregate. As a result, extremely bright SERS signals were generated, allowing detection of a single miR-21 at the single-cell level [122]. Similarly, Li et al. designed a target-triggered strand displacement-hybridization chain reaction (TSD-HCR). In this reaction, miR-141, an epithelial-associated miRNA expressed in a wide range of common human cancers including breast, lung, and prostate cancer, was used as a model to optimize the experimental conditions. This assay enabled monitoring of miR-141 from human breast cancer cells down to 0.17 fM, with a wide linear range from 10-15 to 10-7 M [123].
Cancer-related protein
For decades, panels of blood protein markers including carcinoembryonary antigen (CEA), alpha-fetoprotein (AFP), PSA, CA 19-9, CA15-3, CA 125, EGFR, etc. have been applied for prognostic tests, monitoring of cancer recurrence, and for predicting therapeutic response [17, 124-127]. The expressions of these markers in the blood are frequently elevated several months before imaging abnormalities, and therefore could effectively guide triage of patients into optimal treatment strategies and facilitate personalized therapy [126]. Compared with other circulating tumor markers, proteins are easily recovered from blood plasma and serum, but the high abundance of blood proteins such as albumin and the broad size/concentration ranges of various protein molecules complicate the detection of low-abundant protein biomarkers. Identification and quantification of protein markers is normally performed using immunological techniques such as Western blot (WB) or ELISA. The target proteins are captured by corresponding antibodies or aptamers to generate a colorimetric or fluorescence signal proportional to the quantity of protein in the samples. Although robust, these methods are tedious, expensive and, in the case of WB, semi-quantitative and with low sensitivity. Compared to enzyme-labeled antibodies broadly used in traditional ELISA, SERS tags-labeled antibodies/aptamers usually provide a higher detection sensitivity and enable simultaneous detection of multiple protein markers, which helps to increase the accuracy of disease diagnosis. During the past decade, a variety of SERS-based platforms have been established for ultrasensitive and multiplexed detection of cancer-related proteins, employing different mechanisms such as SERS signal generation upon formation of a sandwich structure, SERS signal turn “off” upon dissociation of core-satellite assemblies, enzyme-induced generation/amplification of SERS signal, target molecule binding-induced Raman frequency shift, etc. A couple of examples will be presented for each strategy in the following section.
SERS signal generation upon formation of a sandwich structure
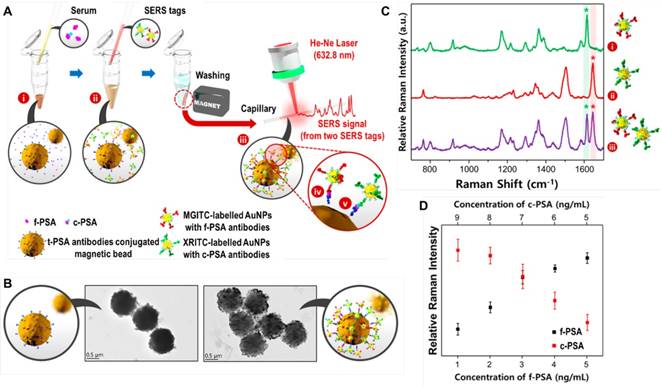
In SERS-based detection assays of target proteins, a sandwich structure is usually formed from a SERS tag, target protein, and a magnetic bead or a nanostructured metal surface. Choo's group has utilized a SERS-based immunoassay for simultaneous detection of two lung cancer markers, CEA and AFP, directly in the sera of patients. In this assay, hollow gold nanospheres were labeled with malachite green isothiocyanate (MGITC) and X-rhodamine-5-(and-6)-isothiocyanate (XRITC), and conjugated with anti-CEA and anti-AFP antibodies, respectively. The SERS labels and antibodies-modified magnetic beads formed sandwich immuno-complexes in the presence of CEA and AFP, and the Raman signals were then measured [128]. In a recent study performed by the same group, a similar immunoassay was developed for the determination of free to total (f/t) PSA ratio to improve the diagnostic performance of prostate cancer. PSA screening has been applied in the diagnosis of prostate cancer for more than 20 years [129]; however, it has been found that the PSA blood test is not specific for prostate cancer because other factors including benign prostatic hyperplasia or prostatitis can also cause an increase in total PSA (t-PSA) [130]. To solve this problem, the f/t PSA ratio can be additionally used to discriminate prostate cancer from benign prostatic diseases. In the study of SERS-based detection of the f/t PSA ratio, Au NPs were labeled with MGITC and XRITC, and conjugated to anti-free PSA(f-PSA) antibodies and anti-complexed PSA (c-PSA) antibodies, respectively, while magnetic beads were modified with anti-t-PSA antibodies (Figure 18). The SERS-based immunoassay provided a LOD of 0.012 ng/mL for f-PSA and 0.15 ng/mL for c-PSA. In the detection of 30 clinical samples, the SERS-based assay showed comparable results with parallel electrochemiluminescence (ECL) detection, with a small sample volume (<10 μL), a short assay time (<1 h), and a smaller standard derivation, therefore holding strong potential for accurate diagnosis of prostate cancer in the clinic [131]. To avoid the washing step and simplify the detection process, Gao et al. combined a SERS-based immunoassay with a droplet-based microfluidic system embedded with a rectangular magnetic bar. In the presence of PSA targets, immunocomplexes formed in the droplets. Then, the system segregated free SERS tags and magnetic immunocomplexes by splitting the droplets into two smaller parts: the supernatant part containing free SERS tags and the other part containing magnetic immunocomplexes. By detecting the SERS signals in the supernatant droplets, PSA in the serum was quantitatively and automatically detected with a LOD below 0.1 ng/mL, without any washing steps [132].
In most sandwich immuno-complexes, the SERS tags are smaller than the magnetic beads. Instead, Song et al. synthesized novel gold mesoflowers with an average diameter of ~770 nm and a highly rough surface as SERS substrates for detection of human IgG. The gold mesoflowers exhibited strong SERS effects and could detect human IgG in a wide range between 1 ng/mL and 1 fg/mL, with a LOD of 1 fg/mL [133]. Other than magnetic beads, glass or metal surfaces have also been frequently used to capture target proteins. For example, Domenici et al. encapsulated glass slides with the bacterial blue-copper protein azurin, which binds P53 proteins. SERS tags linked to P53 were then captured by azurin on the slide and selectively detected wild- type/mutated P53 at concentrations as low as 500 fM in human serum [134]. A three-dimensional hierarchical plasmonic nanostructure endows much higher sensitivity than a planar surface. Li et al. compared the performance of SERS immunosensors with different configurations, i.e., Au nanospheres with Au film, Au nanospheres with Au triangle nanoarray, and Au nanostars with Au triangle nanoarray. The last one exhibited the highest sensitivity with a LOD of 7 fg/mL toward human IgG and was successfully applied for detection of VEGF in plasma from clinical breast cancer patients [135]. In a recent study, Song et al. fabricated Au-coated butterfly wings with natural 3D hierarchical sub-micrometer structures as SERS substrates. The Au layer was ~40-70 nm in thickness, formed a much higher hotspot density than the plain surface, and enabled reliable and sensitive detection of tumor marker CEA in clinical biofluid samples [136].
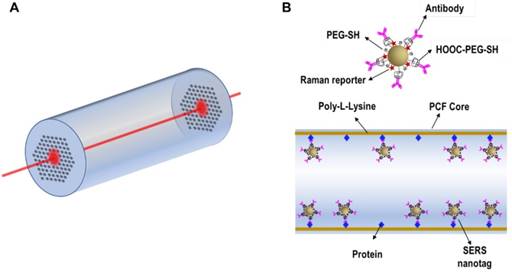
A special material that has been exploited for SERS detection of target proteins in a low sample volume is hollow core photonic crystal fiber (HCPCF). Conventional optical fibers have been combined with SERS for in vivo sensing/imaging, showing desired flexibility but usually lacking detection sensitivity. As shown in Figure 19A, HCPCF enables the incorporation of liquid analytes and SERS tags into air holes and provides a long laser light-analyte interaction length, thus allowing highly sensitive detection of biomolecules from extremely low sample volumes. Olivo et al. developed a highly sensitive protein sensing methodology by combining SERS with HCPCF, using EGFR as a model analyte. The inner wall of the core of HCPCF was first modified with poly-L-Lysine, then EGFR was immobilized on the wall. After dipping into the suspension of anti-EGFR antibody-conjugated SERS tags, the tags specifically bound with the target proteins (Figure 19B). This sensing method enables detection of ∼100 pg protein in a sample volume of ∼10 nL [137]. In a following work by the same research group, two potential hepatocellular carcinoma markers, AFP and alpha-1-antitrypsin (A1AT), were simultaneously detected using Cy5-labeled SERS tags and MGITC-labeled SERS tags in combination with HCPCF in a sample volume of only 20 nL. However, the authors pointed out that two challenges were faced in this platform: non-specific binding of the SERS tags onto the inner wall of the fiber, and incorporation of the samples into the cladding holes along with the hollow core, which lead to alteration of the bandgap and hence affected light guidance inside the core. A potential solution is selectively collapsing the cladding walls to realize liquid-core PCF that allows the liquid analytes to fill only the central hollow core, and better controls should also be provided to avoid non-specific signals [138].
SERS-based detection of PSA. (A) Schematic of a SERS-based assay for the simultaneous detection of f-PSA and c-PSA: (i) mixing of f-PSA, c-PSA, and t-PSA antibody-conjugated magnetic beads; (ii) addition of SERS nanotags to form sandwich immunocomplexes; (iii) separation of magnetic immunocomplexes using a magnetic bar; simultaneous detection of (iv) f-PSA and (v) c-PSA. (B) TEM images of magnetic beads before and after the formation of magnetic immunocomplexes at a 5:5 molar ratio of f-PSA and c-PSA. (C) Raman spectra of (i) f-PSA antibody/MGITC-labeled AuNPs, (ii) c-PSA antibody/XRITC-labeled AuNPs and (iii) their 1:1 (V/V) mixture. (D) Raman intensity variations for different molar ratios of f-PSA and c-PSA (9:1, 8:2, 7:3, 6:4, and 5:5). Adapted with permission from [131], copyright 2017 American Chemical Society.
(A) Schematic of HCPCF as a SERS platform. (B) Schematic of the binding of anti-EGFR antibody-conjugated SERS tags to the target proteins immobilized on the inner wall of the core of HCPCF.
(A) Scheme of a SERS sensor for the detection of mucin-1 based on Au NRs-Ag NPs core-satellite assemblies. (B) TEM images of Au NRs-Ag NPs core-satellite assemblies with different concentrations of mucin-1. (C) SERS spectra and standard curve of mucin-1 detection. Adapted with permission from [140], copyright 2015 The Royal Society of Chemistry.
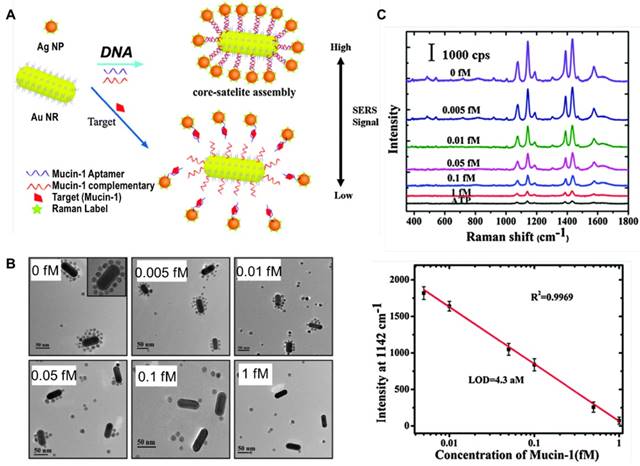
SERS signal turn “off” upon dissociation of core-satellite assemblies
A signal turn “off” strategy has been designed for the detection of target protein based on competitive binding of an aptamer with target protein and the partial complementary sequence. Ma et al. fabricated a core-satellite structure comprised of a 37 nm Au@Ag NP as a core and 10 nm Au NPs as satellites, using an aptamer specific to PSA and its partial complementary DNA as linker molecules. The assemblies had intense electromagnetic hot-spots and emitted strong SERS signals. Following addition of PSA, the aptamer bound with PSA, leading to release of satellite NPs from the core NP and decrease in SERS signals. By detecting the decline in SERS signals, a LOD of 4.8 aM against PSA was achieved [139]. In a following study by the same group, they used Au nanorod as the core, Ag NPs as satellites, and an aptamer against mucin-1 and its partial complementary sequence as linkers to fabricate core-satellite SERS sensors (Figure 20). These assemblies enabled a LOD of 4.3 aM and a wide linear range of 0.005-1 fM for mucin-1 detection [140].
(A) Schematic of a Ag NPs aggregation-based SERS signal generation system. Adapted with permission from [142], copyright 2015 American Chemical Society. (B) a, Schematic of SERS tags binding on BMFON after MMP-2/MMP-7 enzyme cleavage of peptides on BMFON and AuNPs. b, SEM images of (i) clean BMFON substrate (ii) peptide-shielded avidin-conjugated BMFON and (iii) SERS tags bound to BMFON after enzyme cleavage. Adapted with permission from [144], copyright 2015 Optical Society of America.
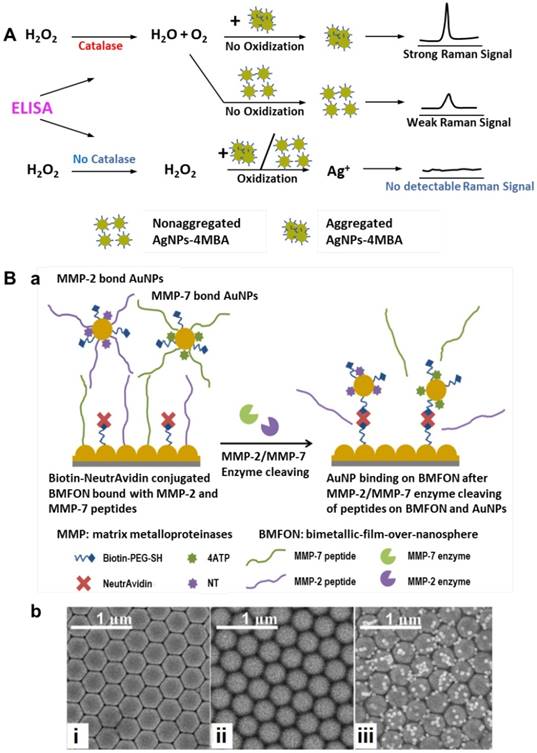
Enzyme-induced generation/amplification of SERS signal
In conventional ELISA detection, a signal is generated by the conversion of the enzyme substrate into a colored molecule, the absorbance of which is then measured with a plate reader. By combining enzyme labels in ELISA with hydrogen peroxide-mediated controlled growth of gold NPs, Stevens et al. developed an ultrasensitive assay for detection of biomarkers with the naked eye, which was termed plasmonic ELISA [141]. Similarly, Liang and coworkers demonstrated a new method that use oxidation-induced dissolution of aggregated SERS tags for ultrasensitive analyte detection. The mechanism is shown in Figure 21 A: proper aggregation of SERS tags produces “hot spots” and generates bright SERS signals, which can be reduced by hydrogen peroxide through oxidization of Ag NPs to Ag+. In the presence of target molecules, catalase-labeled antibodies bind to the plate and consume hydrogen peroxide in solution; therefore, the Raman signal is indirectly proportional to the amount of analyte. Using this assay, PSA and the adrenal stimulant ractopamine were detected in whole serum and urine at ultralow concentrations of 10-9 and 10-6 ng/mL, respectively [142]. In another enzyme-based SERS detection strategy proposed by Olivo and co-workers, formation of hemoglobin-haptoglobin (Hb-Hp) complex was utilized to catalyze the reaction of 3,3′,5,5′-tetramethylbenzidine (TMB) substrate to generate the final product of strongly SERS-active TMB2+. Haptoglobin is an acute phase plasma glycoprotein that is widely gaining application as a prognostic ovarian cancer biomarker. In this assay, a linear increase in the SERS signal of TMB2+ was observed with increasing concentrations of Hb-Hp complex from 50 nM to 34 µM, and Hp in clinical samples was quantified, showing higher sensitivity compared with conventional ELISA method [143].
Matrix metalloproteinases (MMP) are a family of zinc-dependent endopeptidases, among which, over expression of MMP-2 and MMP-7 has been reported to correlate closely with carcinomas such as breast cancer, lung cancer, pancreatic cancer, gastric cancer and prognosis of colorectal carcinoma. Gong et al. designed a platform for the detection of MMP-2 and MMP-7 based on corresponding enzymatic reactions. In this platform, avidin-modified bimetallic-film-over-nanosphere (BMFON) substrate and biotin-modified SERS tags are shielding by MMP peptides; in the presence of corresponding enzymes, the peptides are cleaved, leading to the binding of BMFON with SERS tags and thus strong SERS signals are generated (Figure 21B). This method enabled detection of individual enzymes ranging from 1 ng/mL to 40 µg/mL, and multiplexed detection of MMP-2 and MMP-7 was realized through simultaneous incubation with 4-aminothiophenol (4ATP)- and 2-naphthalenethiol (NT)-labeled peptide-modified SERS tags [144].
Target molecules binding-induced Raman frequency shift
In most of the afore-mentioned SERS tags-based detection techniques, the amount of target molecule is quantitatively analyzed according to the intensity of SERS signal. In an entirely different way, Olivo et al. observed that the Raman frequencies of an antibody-conjugated SERS-active molecule can be affected when binding to its target antigen. The frequency shifts were attributed to structural deformations of the reporter molecule as a result of the binding event, which means that, a single antibody-conjugated SERS reporter molecule could behave as a nanomechanical biosensor (Figure 22A). Based on this phenomenon, the SERS response of anti-influenza-H1-conjugated 4ATP to mechanical stresses induced upon binding to influenza-H1 (H1) and the SERS response of anti-p53-conjugated 6-MP upon binding to P53 antigen were investigated. A detection limit of around 2.5 nM was obtained, a level comparable with conventional ELISA [145]. In a following study by the same group, the LOD of H1 was further improved to ~10 pM by fabricating a random silver film as a highly sensitive SERS substrate [146]. Later, Tang et al. developed a wet chemical silver NP film fabrication for SERS substrates and employed microcontact printing to define ordered domains of chemisorbed Raman reporters on the substrate (Figure 22B); simultaneous detection of AFP and GPC-3 was realized with a LOD down to subpicomolar concentrations (10-13 M) [147]. Instead of using antibodies for target molecules recognition, Pazos et al. designed a SERS-active peptide-conjugate (MB-H1) by attaching a 4-mercaptobenzoyl (MB) unit to the side chain amino group of a lysine residue on the H1 peptide. The MB provided a large SERS cross-section and bound to the metal NPs through silver-thiol interaction, while the H1 peptide selectively linked with c-MYC protein. The peptide /c-MYC recognition was translated into measurable alterations of SERS spectra (ratio between the out-of-plane CCH deformation at 756 cm-1 and the in-plane ring breathing at 1075 cm-1), and enabled fast and reliable detection of c-MYC in blood [148].
Apart from conjugating recognition groups with Raman reporter molecules, Balzerova et al. developed a method directly using antibody-modified Fe3O4@ Ag nanocomposites for label-free determination of human IgG in blood samples obtained by finger prick (Figure 22C). The antibody-modified Fe3O4@Ag nanocomposites generated Raman peaks at 1650, 1539, and 1350 cm-1, which were induced by amide I, II and III vibrations, respectively. Upon binding of human IgG onto the nanocomposites, the intensities and ratios of the spectral bands changed in correlation to the amount of IgG, enabling detection of IgG at concentrations above 600 fg/mL [149].
Diagnosis based on label-free SERS detection of body fluids
CTCs, exosomes, circulating nucleic acids and cancer-related proteins are promising biomarkers present in body fluids. As described above, plenty of SERS-based strategies have been proposed in combination with specific recognition/ separation/ amplification techniques for qualitative/ quantitative analysis of individual targets (Table 1). On the other hand, vibrational spectra of bulk body fluids which can be directly characterized using label-free SERS analysis, may also provide valuable information for cancer diagnosis and have been intensively studied by several research groups in the last decade [31, 32]. Recent investigations on cancer diagnostics based on label-free SERS analysis of body fluids are listed in Table 2.
Circulating cancer biomarkers detected by SERS-based techniques.
| Circulating cancer biomarkers | Detection targets | Detection method | Detection results | Reference |
|---|---|---|---|---|
| CTC | SKBR3 cells | Magnetic bead-EpCAM antibody in combination with NanoplexTM biotag-HER2 antibody | LOD: 10 cells/mL in buffer; LOD: 50 cells/mL in blood | [43] |
| HeLa cells | Magnetic NP-folic acid in combination with SERS NP(S440/S220)-folic acid, magnetic trapping | LOD: 300 cells/mL | [44] | |
| SKBR3 cells | Au-coated iron oxide NPs-QSY21-EpCAM antibody and Au-coated iron oxide NPs-QSY21-HER2 antibody, magnetic trapping | LOD: 1-2 cells/mL in blood | [45] | |
| DLD-1 cells | Magnetic bead-KDED2a-3 aptamer in combination with Au-MBA-KDED2a-3 aptamer | Capture efficiency in buffer: 73%; capture efficiency in blood: 55% | [46] | |
| HepG2 cells; blood sample from patient with liver cancer | Fe3O4@Ag NP-ASGPR antibody in combination with AuAg nanorod-DTNB-GPC3 antibody | LOD: 1 cell/mL in blood; linear relationship from 1 to 100 cells/ mL | [47] | |
| NSCLC cells (NCI-H1650) | Nitrocellulose membrane-EpCAM antibody in combination with Au NP-MBA-EpCAM antibody | LOD: 20 cells/mL in buffer | [48] | |
| Tu212cells; H292 cells; MDA-MB-231 cells; blood samples from SCCHN patients | Au NP-QSY-PEG-EGF peptide | LOD: 5 cells/mL in blood; identified CTCs in the peripheral blood of 19 SCCHN patients with a range of 1-720 CTCs/mL in whole blood | [49] | |
| HeLa cells; MCF-7 cells | Au NP-MBA-folic acid | LOD: 5 cells/mL in blood; linear range of 5-500 cells/mL | [50] | |
| HeLa cells | Au NP-MBA-folic acid; Au nanorod-MBA-folic acid; Au nanostar-MBA-folic acid | LOD: 1 cell/mL in blood | [51] | |
| MCF-7 cells | AuNR@Ag-4MSTP-keratin18 antibody; AuNR@Ag-PNTP-IGF-1 antibody; AuNR@Ag-PATP-CD44 antibody; AuNR@Ag-4MBA-EpCAM antibody | Extremely high specificity of a single cancer cell within 7 million blood cells without any enrichment or separation | [55] | |
| SKBR3 cells; Hela cells; Jurkat T cells; LNCaP cells | Magnetic bead@AuAg NR-DTNB@CdTe614-HER2 antibody; magnetic bead@AuAg NR-4MBA@CdTe614-Transferrin; magnetic bead@AuAg NR-4MBA@CdTe512-CD3 antibody; magnetic bead@AuAg NR-DTNB@CdTe512-PSMA antibody | Simultaneous multiple cancer cell separation from a large population of normal cells | [56] | |
| SK-MEL-28 cells; MCF-7 cells; SKBR3 cells; BM-MSC cells; etc; blood samples from melanoma patients | Au NP-MBA-MCSP antibody; Au NP-TFMBA-MCAM antibody; Au NP-MNBA-ErbB3 antibody; Au NP-MPY-LNGFR antibody | LOD: 10 cells in 10 mL blood; monitored phenotypic changes of melanoma cell lines during molecular targeted treatment and CTCs signature changes of 10 stage-IV melanoma patients receiving immunological or molecular targeted therapies | [59] | |
| Hela cells | Magnetic NP@PMAA-SS-folic acid in combination with SERS probe-folic acid | LOD: 10-20 cells/mL in blood; recycle 90% cells within 20 min eluted by glutathione solution | [60] | |
| HeLa cells; MCF-7 cells | SPION-folic acid in combination with Ag nanoprism-MBA-folic acid | LOD: 1 cell/mL in blood; CTCs can be further released via adding excess free folic acid | [61] | |
| MCF-7 cells; MDA-MB-231 cells; SKBR3 cells; Human breast CCSC cells | Streptavidin-modified CCSC chip in combination with biotinylated dsDNA-modified SERS probes: Au NP-QNT-CD133 antibody; Au NP-NBA-HER2 antibody; Au NP-NPT-EGFR antibody; Au NP-MPD-EpCAM antibody; Au NP-TP-MUC1 antibody | CCSCs can be captured through biotin-streptavidin reaction and simultaneously subtyped by SERS (93% accuracy), followed by restriction enzyme digestion of dsDNA to release the cells | [62] | |
| Exosomes | Exosomes derived from SKBR3 cells and MRC5 cells | Magnetic bead-CD63 antibody in combination with AuAg NR-DTNB-HER2 antibody | LOD: 1200 exosomes | [79] |
| Exosomes derived from SKBR3 cells, Tb4 cells and LNCaP cells | Magnetic bead-CD63 antibody with Au NP-DTNB-HER2 aptamer, Au NP-MMC-CEA aptamer, Au NP-2NAT-PSMA aptamer | LOD: 32 exosomes/μL SKBR3, 73 exosomes/μL Tb4, 203 exosomes/μL LNCaP | [80] | |
| Exosomes derived from MDA-MB-231 cells, MDA-MB-468 cells, SKBR3 cells and MCF12A cells | Au array-EpCAM, CD44, HER2, EGFR, IGF1R, CD81, CD63 and CD9 antibodies, in combination with Au NR-QSY21 | LOD: 2000 exosomes/μL | [81] | |
| Exosomes derived from PANC-01 and HPDE6-C7 cells; exosomes derived from pancreatic ductal adenocarcinoma patients and from healthy volunteers | Glass slides coated with PDA and conjugated with MIF/GPC1/CD63/EGFR antibodies; Au@Ag-PATP-PDA-MIF/GPC1/CD63/EGFR antibodies | LOD: 1 exosome in 2 μL sample | [82] | |
| Exosomes derived from CCD841-CoN cells and HCT116 cells | Silicon micropillars tailored with randomly distributed silver nanograins assemblies, label-free SERS detection | Exosomes from CCD841-CoN (healthy colon cells) show a high presence of lipid signals whereas exosomes from HCT116 (tumor colon cells) exhibit a high presence of RNA | [84] | |
| Exosomes secreted by SKOV3 cells | Silver film-coated nanobowl platform, label-free SERS detection and PCA analysis | Compared SERS spectra from UC-purified exosomes and TIER-purified exosomes, and monitored changes of SERS spectra during drying process | [83] | |
| Exosomes derived from lung normal (NL-20, BEAS-2B, and L929) and cancer (PC-9, H1975, and HCC827) cell lines | Ag nanocubes on an Au nanorod array substrate, label-free SERS detection | Lung normal exosomes showed strong SERS signals of protein, nucleic acid, and lipids, whereas lung cancer exosomes exhibited strong SERS signals of protein | [85] | |
| Exosomes from lung cancer cells (H1299 and H522) and normal (alveolar) cells | Au NPs on cover glass, label-free SERS detection and PCA analysis | Lung cancer cell-derived exosomes were clearly distinguished from normal cell-derived exosomes by 95.3% sensitivity and 97.3% specificity | [86] | |
| Exosomes from A549 cells | Silver coated CD-R and DVD-R recordable disks, label-free SERS detection | SERS signals were obtained with all the structures, especially intense for the DVD-R substrates | [87] | |
| Exosomes from RBC and B16F10 melanoma cancer cells | Negatively-charged exosomes functionalized with positively-charged 10 nm Au NPs on their surface; label-free SERS detection and PLS-DA analysis | Exosomes from different origin can be distinguished, even when present in the same mixture | [88] | |
| Exosomes from SKOV-3 cells and Jurkat cells | Ag NPs-LXY30 peptide for capturing α3ß1 integrin over-expressed exosomes and label-free SERS detection | Raman peaks specific to SKOV‐3 exosomes were detected, while exosomes from Jurkat cells showed no specific SERS signals | [89] | |
| ctDNA | KRAS G12DM | SWNT-based SERS assay coupling with RNase HII-assisted amplification | LOD: 0.3 fM | [98] |
| BRAF V600E, c-Kit L576P and NRAS Q61K | PCR with magnetic bead-streptavidin, Au NP-MBA-BRAF V600E probe, Au-MMC-NRAS Q61K probe, Au-MNBA-c-Kit L576P robe | 10 mutant alleles from a background of 10,000 wild-type sequences | [99] | |
| Model target DNA molecules | “Click SERS”: two SERS tags form a dimer upon simultaneous recognition of a target molecule, leading to combinatorial output of signals from individual NPs | 10-plex biomarkers detection synchronously | [100] | |
| microRNA | miR-21 | Au slide-miR21 probe (direct assay or sandwich assay); Au NR-DTNB-miR21 probe | LOD: 0.36 nM direct assay, 0.85 nM sandwich assay | [113] |
| miR-21, miR-31, miR-141 | Magnetic bead-capture DNAs, Au Ag nanomushrooms-NBT/ROX/4-ABT/Cy3-probe DNAs | LOD: 1 pM | [114] | |
| miR-21, miR-122, miR-223 | Ag microsphere-capture DNAs, Au RNNP-44DP/4ATP/DTNB-probe DNAs | LOD: 10 fM | [115] | |
| miR-21, miR-486, miR-375 | Au NR arrays, molecular beacon-21-ROX, molecular beacon-486-Cy5, molecular beacon-375-FAM | LOD: 393 aM miR-21 in serum, 176 aM miR-486 in serum, 144 aM miR-375 in serum | [117] | |
| miR-21, miR-34a | AuNS@Ag-inverse molecular sentinel nanoprobes -Cy5/Cy5.5 | Linear range of 10-500 ng total small RNA sample | [118] | |
| miR-155 | Co@C/PEI/Ag-Hairpin DNA-Cy5 in combination with padlock probe-base exponential rolling circle amplification | LOD: 70.2 aM | [119] | |
| miR-21 | Ag+-mediated cascade amplification of Au NP-4-ABT in combination with miRNA-triggered hybridization chain reaction | LOD: 0.3 fM | [121] | |
| miR-21 | Ag NP dimer-DBDT-hairpin in combination with miRNA-triggered hybridization chain reaction | LOD: single miR-21 at the single-cell level | [122] | |
| miR-141 | Au NP-Rox DNA in combination with target triggered stand displacement-hybridization chain reaction | LOD: 0.17 fM | [123] | |
| Protein | CEA and AFP | Magnetic bead-CEA/AFP antibody; hollow Au nanosphere-MGITC-CEA antibody, hollow Au nanosphere-XRITC-AFP antibody | LOD:1.67 ng/mL CEA, 1.56 ng/mL AFP | [128] |
| f-PSA and c-PSA | Magnetic bead-t-PSA antibody; Au NP-MGITC-f-PSA antibody, Au NP-XRITC-c-PSA | LOD: 0.012 ng/mL f-PSA, 0.15 ng/mL c-PSA | [131] | |
| PSA | Magnetic bead-PSA antibody and AuNP-MGITC-PSA antibody, in combination with droplet microfluidics | LOD: 0.1 ng/mL | [132] | |
| Human IgG | Au mesoflower-4MBA-antibody, Au-coated-magnetic NP-antibody | LOD: 1 fg/mL | [133] | |
| P53 | Glass slide-azurin, Au NP-4ATP | LOD: 500 fM | [134] | |
| Human IgG and VEGF | Au film-antibody or Au triangle array-antibody in combination with Au nanosphere-MGITC-antibody or Au nanostar-MGITC-antibody | LOD: 7 fg/mL Human IgG | [135] | |
| CEA | Au-coated butterfly wing-antibody in combination with RhodG-tagged CEA aptamer | Linear relationship in the range of 10-104 ng/mL | [136] | |
| EGFR | Au NP-MGITC-EGFR antibody in combination with HCPCF | LOD: 100 pg in a sample volume of ∼10 nL | [137] | |
| AFP and A1AT | Au NP-Cy5-AFP antibody and Au NP-MGITC-A1AT antibody, in combination with HCPCF | Multiplex detection in a sample volume of ∼20 nL | [138] | |
| PSA | 37 nm Au@Ag NP-PSA aptamer as core, 10 nm Au NP-complementary DNA as satellite, 4-NTP as Raman reporter molecules | LOD: 4.8 aM; linear range of 0.01-5 fM | [139] | |
| Mucin-1 | Au NR-complementary DNA as core, Au NP-4ATP-mucin-1 aptamer as satellite | LOD: 4.3 aM; Linear range of 0.005-1 fM | [140] | |
| PSA and adrenal stimulant ractopamine | Aggregated Ag NPs-4MBA in combination with ELISA | LOD: 10-9 ng/mL PSA in whole serum; 10-6 ng/mL adrenal stimulant ractopamine in urine | [142] | |
| Haptoglobin | Catalysis of TMB to SERS-active TMB2+ by formation of hemoglobin-haptoglobin complex | Linear range of 50 nM-34 µM | [143] | |
| MMP-2 and MMP-7 | Avidin-modified BMFON substrate and biotin-modified SERS tags shielded by MMP peptides | Linear range of 1 ng/mL-40 µg/mL | [144] | |
| Influenza-H1 and p53 | Influenza-H1 antibody-conjugated 4ATP and p53 antibody-conjugated 6-MP adsorbed on a bimetallic Au/Ag surface | LOD: 2.2 nM influenza-H1, 2.5 nM p53 | [145] | |
| Influenza-H1 | Influenza-H1 antibody-conjugated 4ATP adsorbed on a random silver film | LOD: 10 pM | [146] | |
| AFP and GPC-3 | AFP antibody-DSNB and GPC-3 antibody-MBA adsorbed in ordered domains on a silver NP film | LOD: 10-13 M | [147] | |
| c-MYC | SiO2@Ag bead-MB-H1, capture of the c-MYC target by MB-H1 results in alterations of the SERS spectra | Analogous detection limit with ELISA | [148] | |
| Human IgG | Fe3O4@Ag-IgG antibody, label-free analysis | Direct determination of IgG at concentrations from 600 fg/mL | [149] |
Cancer diagnosis based on label-free SERS detection of biofluids.
| Cancer types | Biofluids | Samples | SERS substrates | Laser (nm) | Data analysis | Detection results | Reference |
|---|---|---|---|---|---|---|---|
| Oral cancer | Saliva | Oral cancer patients (n=5); healthy volunteers (n=5) | A closely-packed gold particle film | 632.8 | Abnormal SERS peaks | Sensitivity: 70% | [150] |
| Oral squamous cell carcinoma | Serum | Oral squamous cell carcinoma patients (n=135); patients with old maxillofacial fracture and healthy volunteers (n=145) | Gold colloids (55 nm) | 633 | PCA-LDA | Sensitivity: 80.7%, specificity: 84.1% | [152] |
| Serum | Oral squamous cell carcinoma patients (n=135) with different tumor stages and histologic grades | Gold colloids (55 nm) | 633 | OPLS-DA | Classification of T stages accuracy: ~80%; classification of N stages accuracy: 85.9%; classification of different histological grades accuracy: ~90% | [153] | |
| Nasopharyngeal cancer | Plasma | Patients with pathologically confirmed nasopharyngeal carcinomas (n=43); healthy volunteers (n=33) | Silver colloids (34±5 nm) | 785 | PCA-LDA | Sensitivity: 90.7%, specificity: 100% | [151] |
| Plasma | Nasopharyngeal cancer patients with T1 stage (n=25); nasopharyngeal cancer patients with T2-T4 stage (n=75); healthy volunteers (n=60) | Gold colloids (43±5 nm) | 785 | PCA-LDA | Classification between T1 stage cancer and normal sensitivity: 84%, specificity: 83.3%; classification between T2-T4 stage cancer and normal sensitivity: 92%, specificity: 95% | [162] | |
| Gastric cancer | Plasma | Gastric cancer patients (n=32); healthy volunteers (n=33) | Silver colloids (34±5 nm) | 785 | PCA-LDA | Non-polarized laser sensitivity: 71.9%, specificity: 72.7%; linear-polarized laser sensitivity: 75%, specificity: 87.9%; right-handed circularly polarized laser sensitivity: 81.3%, specificity: 78.8%; left-handed circularly polarized laser sensitivity: 100%, specificity: 97% | [163] |
| Colorectal cancer | Serum | Colorectal cancer patients (n=38); healthy volunteers (n=45) | Gold colloids (43±6 nm) | 785 | PCA-LDA | Sensitivity: 90.7%, specificity: 100% | [154] |
| Lung cancer | Saliva | Lung cancer patients (n=21); healthy volunteers (n=20) | Silver colloids (34±5 nm) | 632.8 | PCA-LDA | Sensitivity: 78%, specificity: 83% | [168] |
| Cervical cancer | Plasma | Cervical cancer patients (n=60); healthy volunteers (n=50) | Silver colloids (34±5 nm) | 785 | PCA-LDA | Sensitivity: 96.7%, specificity: 92% | [164] |
| Esophageal cancer | Serum | Esophageal cancer patients (n= 30); healthy volunteers (n=31) | Silver colloids | 785 | PCA; SVM; PCA-SVM | PCA-SVM sensitivity: 83.3%, specificity: 86.7% | [155] |
| Urine | Esophagus cancer patients (n = 56); healthy volunteers (n = 36) | Silver colloids (42±5 nm) | 785 | PCA-LDA | Sensitivity: 89.3%, specificity: 83.3% | [166] | |
| Plasma | Esophageal cancer patients (n=36); healthy volunteers (n=50) | Silver colloids (34±5 nm) | 785 | PCA-LDA; SVM | Sensitivity: 94.4%, specificity: 100% | [165] | |
| Prostate cancer | Serum | Prostate cancer patients (n=93); healthy volunteers (n=68) | Silver colloids (50 nm) | 785 | PCA-LDA; SVM | Sensitivity: 97.8%, specificity: 100% | [156] |
| Urine | Prostate cancer patients (n=9); healthy volunteers (n=9) | Gold colloids (50 nm) | 785 | PCA-LDA | Sensitivity: 100%, specificity: 89% | [167] | |
| Expressed prostatic secretion and serum | Prostate cancer patients (n=20); patients with benign prostatic hyperplasia (n=20) | Silver colloids | 633 | PCA-LDA | Expressed prostatic secretion sensitivity: 75%, specificity: 75%; serum sensitivity: 60%, specificity: 76.5% | [157] | |
| Breast cancer | Serum | Breast cancer patients with lymph node involvement (pTxN+, n=20); breast cancer patients without lymph node involvement (pT1N0, n=20); healthy volunteers (n=20) | Silver colloids (23 nm) | 785 | PCA-LDA | Classification of healthy and cancer patients sensitivity: 92 %, specificity: 85%; classification of cancer stage sensitivity: ≥80%, specificity: ≥80% | [158] |
| Gastrointestinal cancer | Serum | Gastric cancer patients (n=12); colorectal cancer patients (n=12); patients with benign diseases (n=12) | Silver nanoscale hexagonal columns on the surface of a phosphor bronze chip | 632.8 | Peak heights of the SERS spectra | The peak heights of SERS spectra from patients with benign diseases were significantly lower than those from patients with gastric/colorectal cancer | [159] |
| Cancer screening | Serum | Patients with liver cancer, colonic cancer, esophageal cancer, nasopharyngeal cancer, gastric cancer (n=130); healthy volunteers (n=113) | Silver colloids | 785 | PCA-LDA; SVM; PCA-SVM | Sensitivity: 93.1%, specificity: 99.1% | [160] |
| Liver disease analysis | Serum | Patients with hepatopathy (n=333); patients with esophageal cancer (n=99); healthy volunteers (n=304) | Silver colloids (71.2±7.7 nm) | 785 | PCA-LDA | Accuracy: 95.33% | [161] |
In 2007, Kah et al. reported that the SERS spectra of saliva from closely-packed Au NP films was differentiable between those acquired from normal individuals and oral cancer patients, showing the promise of SERS-based biofluid assays for early diagnosis of oral cancer [150]. In 2010, Feng et al. found that SERS spectra from the plasma of nasopharyngeal cancer patients and healthy subjects could be separated into two distinct clusters using PCA, and linear discriminate analysis (LDA) based on the PCA-generated features could differentiate the nasopharyngeal cancer SERS spectra from normal SERS spectra with a high sensitivity (90.7%) and specificity (100%) [151]. Since these studies, SERS analyses of varied body fluids have been largely performed for diagnosis of different cancer types. Serum [152-161] and plasma [151, 162-165], which contain abundant proteins, nucleic acids, lipids and metabolites, are the two most investigated biofluids, while urine [166, 167] and saliva [150, 168] have also been analyzed in several studies.
The SERS signals of body fluids are generated by the bio-substances adsorbed onto the surface of metallic nanostructures, typically at a distance less than 10 nm. A near-infrared excitation at 632.8 or 785 nm is usually adopted to avoid interference from autofluorescence and strong resonance Raman signals from hemoglobin and carotenoids, which largely exist in blood. In a general approach, body fluids from cancer patients and healthy controls are collected; the SERS spectra of each sample are recorded and then compared. Tentative assignments of the major vibrational bands in the spectra are performed according to existing literatures, and discrimination between the spectra from different groups is preponderantly done by multivariate statistics. Silver or gold colloids are utilized as SERS substrates more often than nanostructured surfaces, possibly because of the facile synthesis of colloidal NPs and the simplicity of adsorbing biomolecules on metal surface by mixing. Statistical analysis methods such as PCA, PCA-linear discriminate analysis (PCA-LDA), Support Vector Machine (SVM), and SVM in combination with PCA (PCA-SVM) have been employed to develop effective diagnostic algorithms for classification of SERS spectra between normal and cancer patients [164-166] and for classification of patients with different tumor stages [153, 162].
Conclusion
Summary
SERS has become a powerful vibrational spectroscopic and imaging technique over the last decades, and its applications in chemical, material and, in particular, biomedical fields are rapidly increasing. In this review, we have summarized recently reported SERS-based strategies for supersensitive and facile detection of liquid biopsy (i.e., CTCs, exosomes, ctDNAs, circulating miRNAs, tumor-related proteins, and bulk biofluids) in the last years. As shown in Table 1, the first study using SERS for detection of CTCs was published in 2008 by Sha et al. [43], with a LOD of 50 cells per milliliter of blood; after 10 years, a single cell in 1 mL of peripheral blood sample can be rapidly identified [47, 61] and phenotypic evolution of CTCs can be sensitively monitored in real-time during treatment [59]. The detection sensitivity of exosomes and other circulating cancer markers has also been prominently improved with the rapid development of nanomaterial fabrication techniques, capturing reagents, as well as advances in Raman microspectroscopic instrumentation. Cancer diagnostic studies based on label-free SERS analysis of bulk body fluid began in 2007, with a sensitivity of 70% by looking for abnormal peaks in SERS spectra obtained from saliva [150], while in later years a couple diagnostic algorithms with much higher accuracy in spectral discrimination have been developed (Table 2).
(A) Schematic of mechanical deformation in an antibody-conjugated 4ATP sensor. Adapted with permission from [145], copyright 2012 American Chemical Society. (B) Schematic of microcontact printing to define ordered domains of chemisorbed Raman reporters on the substrate (left) and target molecules-induced Raman frequency shift (right). Adapted with permission from [147], copyright 2016 American Chemical Society. (C) Scheme showing label-free determination of human IgG in blood samples obtained by finger prick. Adapted with permission from [149], copyright 2014 American Chemical Society.
Ultrasensitive and rapid detection of multiple biomarkers at a low concentration and a low sample volume is highly desired for early diagnosis. A couple of immunoassays based on colorimetric or fluorescence methods have been developed and are extensively used to detect biomarkers in the clinic. ELISA is one of the most common immunoassay methods. It offers a detection limit of ng-μg of protein in a sample volume of ~100 μL, with a duration time of several hours. In contrast, SERS enables detection of multiple biomarkers with a LOD about two orders of magnitude lower [147], an assay time of less than one hour, at a sample volume of ~10 μL [131]. And, when combined with photonic crystal fiber, ∼100 pg of protein in an ∼10 nL sample could be detected [137]. In addition, SERS enables simultaneous detection of multiple targets [100], and avoids autofluorescence from bio-samples and photobleaching that often limit the application of fluorescence-based assays. CellSearch system is currently the only FDA-approved system for clinical CTC detection, which is based on selection of putative CTCs using EpCAM antibodies-modified magnetic beads and enumeration according to immunostaining (cytokeratin+/DAPI+/CD45-). However, the enrichment process based on a single marker might induce false positive/negative results due to the heterogenous properties of CTCs [52-54]. Instead, by using SERS probes encoded with different colors and modified with antibodies corresponding to multiple surface markers, the phenotype evolution of CTCs during treatment could be monitored in nearly real time without prior separation [59]. Beside the SERS tags-based detection platforms, label-free SERS analysis is another powerful tool to be utilized potentially for cancer diagnosis. By using plasmonic nanostructures to enhance the intrinsic vibrational spectra of biomolecules, fingerprint information can be obtained from either purified circulating markers such as exosomes [86] and nucleic acids [112], or directly from bulk body fluids [32]. Differentiation of patients and healthy individuals could be achieved through spectral discrimination using multivariate statistics. Although these molecular changes can be monitored also by other techniques such as WB, high-performance liquid chromatography (HPLC) and mass spectrometry (MS), the detection is usually labor-intensive, cost ineffective and time consuming. In the case of MS, ion-suppression effect and non-universal ionization efficiency would further complicate analysis of such complex samples [150]. In summary, as an ultrasensitive, versatile and facile detection tool, SERS offers the possibility to simultaneously detect multiple biomarkers in body fluids and obtain intrinsic fingerprint information of biomolecules. Therefore, it holds great potential to be broadly applied in clinical liquid biopsy analysis.
Challenges and outlooks
In spite of the promising outcomes demonstrated in recent research works, SERS has not been successfully implemented into the clinic yet. To further accelerate translation of SERS-based techniques into biomedical applications, several issues regarding the manufacturing of SERS probes, obtaining reliable results during detection and data analysis processing, as well as instrument development need to be addressed.
The first challenge is large-scale production of sensitive SERS probes with high reproducibility and long-term storage stability. Spherical Au and Ag NPs are relatively easy to prepare and have been widely used as SERS substrates in the past. However, previous studies have indicated that NP monomers are not SERS active [36, 169]; the ensemble SERS signals are mainly dominated by NP aggregates, which is hard to control. Anisotropic NPs such as nanorods, nanocubes, nanostars, and rationally designed NP assemblies (such as dimers and core-satellite structures) have attracted more attention, because extremely high electric fields can be obtained at the sharp edges or in particle conjunctions (the so-called “hotspots”) [30]. On the other hand, NPs tend to agglomerate during long-term storage and Raman reporter molecules might disassociate from the NPs. Therefore, proper surface modification is necessary to keep the probes stable. Silica encapsulation and PEG coating are two general ways frequently used for the protection of SERS tags. Silica-encapsulated Raman reporter molecules-encoded Au spherical NPs are commercially available from Cabot Security Materials [170]. However, uniform encapsulation of anisotropic NPs or assemblies is more difficult than for spherical NPs. For label-free detection, it has been found that an ultrathin coating layer of silica/aluminum oxide (shell-isolated nanoparticle-enhanced Raman spectroscopy, SHINERS) [171] or a monolayer of iodide [172] on the metal nanoparticles can dramatically improve the reproducibility of SERS signals. Nevertheless, large-scale production of homogeneous SERS NPs with ultrasensitivity, high stability and acceptable costs has not been realized yet.
Second, to evaluate the concentrations of varied circulating biomarkers in liquid biopsy, reliable quantitative detection results must be obtained. The SERS signals can be influenced by a couple of factors during the detection process, such as aggregation, non-specific binding of the NP probes, and improper detection conditions. PEGylation is a commonly used method to protect NPs from aggregation and minimize nonspecific binding. Still, fluctuations in the SERS signals have been frequently observed due to the complex compositions of body fluids, especially when multiple targets are simultaneously detected. To improve the reliability of SERS signals, NP probes with embedded internal standards have recently been introduced to correct the fluctuating samples and measuring conditions [173]. Meanwhile, Wang et al. reported that a ratiometric quantification assay using an equimolar mixture of targeted NPs and non-targeted NPs together for immunostaining allows unambiguous assessment of molecular expression on tissue specimens [174]. This assay could be extended to liquid biopsy analysis for minimizing the effect of non-specific binding in future studies. It should also be noted that high laser power density may elevate the local temperature or produce high-energy hot carriers, leading to desorption, photodecomposition, or photobleaching of the biomolecules [37, 175]. Therefore, one should be very cautious to control the laser power density to obtain reliable quantitative results.
Third, assay reproducibility and reliability in large clinical sample pools needs to be investigated. In two of the most recent studies, samples from 10 melanoma patients and 71 pancreatic cancer patients were investigated for CTC phenotyping [59] and exosomes classification [82], respectively, indicating the great potential of these two SERS tags-based liquid biopsy methods for clinical applications. For label-free SERS detection-based liver disease diagnosis, Shao et al. collected serum from 333 patients with hepatopathy, 99 patients with esophageal cancer, and 304 normal individuals, among which the classification accuracy reached 93% by statistical analysis. However, in most of the “proof of concept” studies, only simulated biofluids or a limited number of clinical samples were tested. The actual performance of the proposed SERS-based assays needs to be verified in larger trials of clinical specimens. In addition, as SERS-based assays provide a higher sensitivity than other clinically used detection techniques and enable simultaneous detection of multiple targets, the “cutoff” concentrations of some potential biomarkers or biomarker cocktails, above which it is considered malignant, need to be rigorously ascertained by extensive SERS analysis of large clinical cohorts.
Fourth, developing highly sensitive, easy to handle and cost-effective Raman spectrophotometers is a critical step for the translation of SERS techniques into practical applications. Most of the in-laboratory SERS studies were conducted using complicated confocal Raman microscopes or custom-built instruments, which are not widely equipped in healthcare institutions. Fortunately, researchers have realized this aspect and some miniaturized SERS-based medical devices are being developed. For example, Zavaleta et al. built a SERS-based endoscope that fits into the instrument channels of conventional white light endoscopes, which is ideally suited for detection of tumor-targeting SERS probes during routine endoscopy [170]. To promote clinical application of SERS probes in phenotype monitoring of CTCs, Tsao et al. has attempted to obtain SERS spectra of patient samples using a hand-held Raman spectrometer [59]. Further efforts to integrate SERS probes with portable Raman instruments will to a large extent facilitate clinical applications of SERS-based liquid biopsy techniques, especially for point-of-care tests.
In 2015, Renishaw Diagnostics released a SERS-based platform (the RenDx Fungiplex assay) for the detection of 12 different fungal pathogens ex vivo [23]. We believe that with the continuous joint efforts contributed by multi-disciplinary researchers as well as clinicians, the SERS-based detection techniques will be further improved and steadily moved towards broad clinical applications.
Abbreviations
A1AT: alpha-1-antitrypsin; AFP: alpha-fetoprotein; ASGPR: anti-asialoglyco protein receptor; 4ATP: 4-aminothiophenol; CCL: chronic lymphocytic leukemia; CCSCs: circulating cancer stem cells; CEA: carcino-embryonic antigen; cfDNA: cell-free DNA; CTCs: circulating tumor cells; ctDNAs: circulating tumor DNAs; DBDT: 4,4′-biphenyldithiol; DTNB: 5,5′-dithiobis (2-nitrobenzoic acid); ECL: electrochemiluminescence; EGF: epidermal growth factor; EMT: epithelial-to-mesenchymal transition; EpCAM: epithelial cell adhesion molecule; ErbB3: erythroblastic leukaemia viral oncogene homologue 3; EVs: extracellular vesicles; GPC1: glypican 1; GPC3: glypican 3; HCC: hepatocellular carcinoma; HCPCF: hollow core photonic crystal fiber; HCR: hybridization chain reaction; HER2: human epidermal growth factor receptor-2; HPLC: high-performance liquid chromatography; iMS: inverse molecular sentinel; ISET: isolation by size of epithelial tumor cells; LDA: linear discriminant analysis; LNA: locked nucleic acid; LNGFR: low-affinity nerve growth factor receptor; LOD: limit of detection; MB: 4-mercaptobenzoyl; MBs: molecular beacons; 4MBA: 4-mercaptobenzoic acid; MCAM: melanoma cell adhesion molecule; MCSP: melanoma-chondroitin sulphate proteoglycan; MGITC: malachite green isothiocyanate; MIF: migration inhibitor factor; MNBA: 4-mercapto-3-nitro benzoic acid; MPD: 4-mercaptopyridine; MPY: 4-mercaptopyridine; MS: Mass spectrometry; 4MSTP: 4-(methylsulfanyl) thiophenol; NBA: nile blue A; nPES: nanoplasmon-enhanced scattering; NPT: 1-naphthalenethiol; NSCLC: nonsmall-cell lung cancer; NT: 2-naphthalenethiol; NTA: nanoparticle tracking analysis; OPLS-DA: orthogonal partial least squares discriminant analysis; PARE: personalized analysis of rearranged ends; PATP: p-aminothiophenol; PCA: principal component analysis; PDA: polydopamine; P-ERCA: padlock probe-based exponential rolling circle amplification; PLS-DA: partial least squares discriminant analysis; PNTP: p-nitrothiophenol; PSA: prostate specific antigens; PSMA: prostate-specific membrane antigen; QNT: 2-quinolinethiol; rBSA: reductive bovine serum albumin; SCCHN: squamous cell carcinoma of the head and neck; SERS: surface-enhanced Raman spectroscopy; SHINERS: shell-isolated nanoparticle-enhanced Raman spectroscopy; SPION: superparamagnetic iron oxide nanoparticles; SVM: support vector machine; SWNTs: single-walled carbon nanotubes; TEIR: total exosome isolation reagent; TFMBA: 2,3,5,6-tetrafluoro -4-mercaptobenzoic acid; THMS: triple-helix molecular switch; TMB: 3,3′,5,5′-tetramethylbenzidine; TSD-HCR: triggered strand displacement-hybridization chain reaction; TP: thiophenol; UTR: untranslated region; WB: Western blot; XRITC: X-rhodamine-5-( and-6)-isothiocyanate.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant Nos. 81871487; 81502297).
Competing Interests
The authors have declared that no competing interest exists.
References
1. McGranahan N, Swanton C. Clonal heterogeneity and tumor evolution: past, present, and the future. Cell. 2017;168:613-28
2. Perakis S, Speicher MR. Emerging concepts in liquid biopsies. BMC Med. 2017;15:75
3. Alix-Panabieres C, Pantel K. Clinical applications of circulating tumor cells and circulating tumor DNA as liquid biopsy. Cancer Discov. 2016;6:479-91
4. Hofman P. Liquid biopsy for early detection of lung cancer. Curr Opin Oncol. 2017;29:73-8
5. Aro K, Wei F, Wong DT, Tu M. Saliva liquid biopsy for point-of-care applications. Front Public Health. 2017;5:77
6. Siravegna G, Marsoni S, Siena S, Bardelli A. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol. 2017;14:531-48
7. Di Meo A, Bartlett J, Cheng Y, Pasic MD, Yousef GM. Liquid biopsy: a step forward towards precision medicine in urologic malignancies. Mol Cancer. 2017;16:80
8. Vicki Plaks, Charlotte D. Koopman, Werb Z. Circulating tumor cells. Science. 2013;341:1186-8
9. Cabel L, Proudhon C, Gortais H, Loirat D, Coussy F, Pierga JY. et al. Circulating tumor cells: clinical validity and utility. Int J Clin Oncol. 2017;22:421-30
10. Tkach M, Thery C. Communication by extracellular vesicles: where we are and where we need to go. Cell. 2016;164:1226-32
11. Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654-9
12. Li W, Li C, Zhou T, Liu X, Liu X, Li X. et al. Role of exosomal proteins in cancer diagnosis. Mol Cancer. 2017;16:145
13. Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, Kaye J. et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523:177-82
14. Heitzer E, Ulz P, Geigl JB. Circulating tumor DNA as a liquid biopsy for cancer. Clin Chem. 2015;61:112-23
15. Hamam R, Hamam D, Alsaleh KA, Kassem M, Zaher W, Alfayez M. et al. Circulating microRNAs in breast cancer: novel diagnostic and prognostic biomarkers. Cell Death Dis. 2017;8:e3045
16. Santangelo A, Tamanini A, Cabrini G, Dechecchi MC. Circulating microRNAs as emerging non-invasive biomarkers for gliomas. Ann Transl Med. 2017;5:277
17. Duffy MJ, McDermott EW, Crown J. Blood-based biomarkers in breast cancer: from proteins to circulating tumor cells to circulating tumor DNA. Tumour Biol. 2018;40:1010428318776169
18. Hyun KA, Kim J, Gwak H, Jung HI. Isolation and enrichment of circulating biomarkers for cancer screening, detection, and diagnostics. Analyst. 2016;141:382-92
19. Ferhan AR, Jackman JA, Park JH, Cho N-J, Kim D-H. Nanoplasmonic sensors for detecting circulating cancer biomarkers. Adv Drug Deliv Rev. 2018;125:48-77
20. Lane LA, Qian X, Nie S. SERS nanoparticles in medicine: from label-free detection to spectroscopic tagging. Chem Rev. 2015;115:10489-529
21. Cialla-May D, Zheng XS, Weber K, Popp J. Recent progress in surface-enhanced Raman spectroscopy for biological and biomedical applications: from cells to clinics. Chem Soc Rev. 2017;46:3945-61
22. Schlücker S. SERS microscopy: nanoparticle probes and biomedical applications. Chemphyschem. 2009;10:1344-54
23. Jamieson LE, Asiala SM, Gracie K, Faulds K, Graham D. Bioanalytical measurements enabled by surface-enhanced Raman scattering (SERS) probes. Annu Rev Anal Chem (Palo Alto Calif). 2017;10:415-37
24. Fleischmann M, Hendra PJ, McQuillan AJ. Raman spectra of pyridine adsorbed at a silver electrode. Chem Phys Lett. 1974;26:163-6
25. Jeanmaire DL, Duyne RPV. Surface raman spectroelectrochemistry: part I. heterocyclic, aromatic, and aliphatic amines adsorbed on the anodized silver electrode J Electroanal Chem Interfacial Electrochem. 1977;84:1-20
26. Nie S, Emory SR. Probing single molecules and single nanoparticles by surface-enhanced Raman scattering. Science. 1997;275:1102-6
27. Kneipp K, Wang Y, Kneipp H, Perelman LT, Itzkan I, Dasari RR. et al. Single molecule detection using surface-enhanced Raman scattering (SERS). Phys Rev Lett. 1997;78:1667
28. Schlücker S. Surface-enhanced Raman spectroscopy: concepts and chemical applications. Angew Chem Int Ed Engl. 2014;53:4756-95
29. Xie W, Schlücker S. Medical applications of surface-enhanced Raman scattering. Phys Chem Chem Phys. 2013;15:5329-44
30. Wang Y, Yan B, Chen L. SERS tags: novel optical nanoprobes for bioanalysis. Chem Rev. 2013;113:1391-428
31. Zheng XS, Jahn IJ, Weber K, Cialla-May D, Popp J. Label-free SERS in biological and biomedical applications: Recent progress, current challenges and opportunities. Spectrochim Acta A Mol Biomol Spectrosc. 2018;197:56-77
32. Bonifacio A, Cervo S, Sergo V. Label-free surface-enhanced Raman spectroscopy of biofluids: fundamental aspects and diagnostic applications. Anal Bioanal Chem. 2015;407:8265-77
33. Qian X, Peng XH, Ansari DO, Yin-Goen Q, Chen GZ, Shin DM. et al. In vivo tumor targeting and spectroscopic detection with surface-enhanced Raman nanoparticle tags. Nat Biotechnol. 2008;26:83-90
34. Zavaleta CL, Smith BR, Walton I, Doering W, Davis G, Shojaei B. et al. Multiplexed imaging of surface enhanced Raman scattering nanotags in living mice using noninvasive Raman spectroscopy. Proc Natl Acad Sci U S A. 2009;106:13511-6
35. Maiti KK, Dinish US, Samanta A, Vendrell M, Soh K-S, Park S-J. et al. Multiplex targeted in vivo cancer detection using sensitive near-infrared SERS nanotags. Nano Today. 2012;7:85-93
36. Wang Y, Schlücker S. Rational design and synthesis of SERS labels. Analyst. 2013;138:2224-38
37. Zong C, Xu M, Xu LJ, Wei T, Ma X, Zheng XS. et al. Surface-enhanced Raman spectroscopy for bioanalysis: reliability and challenges. Chem Rev. 2018;118:4946-80
38. Alix-Panabieres C, Pantel K. Challenges in circulating tumour cell research. Nat Rev Cancer. 2014;14:623-31
39. Shen Z, Wu A, Chen X. Current detection technologies for circulating tumor cells. Chem Soc Rev. 2017;46:2038-56
40. Bhana S, Wang Y, Huang X. Nanotechnology for enrichment and detection of circulating tumor cells. Nanomedicine (Lond). 2015;10:1973-90
41. Hosseini H, Obradovic MM, Hoffmann M, Harper KL, Sosa MS, Werner-Klein M. et al. Early dissemination seeds metastasis in breast cancer. Nature. 2016;540:552-28
42. Rawal S, Yang Y-P, Cote R, Agarwal A. Identification and quantitation of circulating tumor cells. Annu Rev Anal Chem (Palo Alto Calif). 2017;10:321-43
43. Sha MY, Xu H, Natan MJ, Cromer R. Surface-enhanced Raman scattering tags for rapid and homogeneous detection of circulating tumor cells in the presence of human whole blood. J Am Chem Soc. 2008;130:17214-5
44. Shi W, Paproski RJ, Moore R, Zemp R. Detection of circulating tumor cells using targeted surface-enhanced Raman scattering nanoparticles and magnetic enrichment. J Biomed Opt. 2014;19:056014
45. Bhana S, Chaffin E, Wang Y, Mishra SR, Huang X. Capture and detection of cancer cells in whole blood with magnetic-optical nanoovals. Nanomedicine (Lond). 2014;9:593-606
46. Sun C, Zhang R, Gao M, Zhang X. A rapid and simple method for efficient capture and accurate discrimination of circulating tumor cells using aptamer conjugated magnetic beads and surface-enhanced Raman scattering imaging. Anal Bioanal Chem. 2015;407:8883-92
47. Pang YF, Wang CW, Xiao R, Sun ZW. Dual-selective and dual-enhanced SERS nanoprobes strategy for circulating hepatocellular carcinoma cells detection. Chemistry. 2018;24:7060-7
48. Zhang P, Zhang R, Gao M, Zhang X. Novel nitrocellulose membrane substrate for efficient analysis of circulating tumor cells coupled with surface-enhanced Raman scattering imaging. ACS Appl Mater Interfaces. 2014;6:370-6
49. Wang X, Qian X, Beitler JJ, Chen ZG, Khuri FR, Lewis MM. et al. Detection of circulating tumor cells in human peripheral blood using surface-enhanced Raman scattering nanoparticles. Cancer Res. 2011;71:1526-32
50. Wu X, Luo L, Yang S, Ma X, Li Y, Dong C. et al. Improved SERS nanoparticles for direct detection of circulating tumor cells in the blood. ACS Appl Mater Interfaces. 2015;7:9965-71
51. Wu X, Xia Y, Huang Y, Li J, Ruan H, Chen T. et al. Improved SERS-active nanoparticles with various shapes for CTC detection without enrichment process with supersensitivity and high specificity. ACS Appl Mater Interfaces. 2016;8:19928-38
52. Pantel K, Denève E, Nocca D, Coffy A, Vendrell JP, Maudelonde T. et al. Circulating epithelial cells in patients with benign colon diseases. Clin Chem. 2012;58:936-40
53. Hong Y, Zhang Q. Phenotype of circulating tumor cell: face-off between epithelial and mesenchymal masks. Tumour Biol. 2016;37:5663-74
54. Yu M, Bardia A, Wittner BS, Stott SL, Smas ME, Ting DT. et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science. 2013;339:580-4
55. Nima ZA, Mahmood M, Xu Y, Mustafa T, Watanabe F, Nedosekin DA. et al. Circulating tumor cell identification by functionalized silver-gold nanorods with multicolor, super-enhanced SERS and photothermal resonances. Sci Rep. 2014;4:4752
56. Wang Z, Zong S, Chen H, Wang C, Xu S, Cui Y. SERS-fluorescence joint spectral encoded magnetic nanoprobes for multiplex cancer cell separation. Adv Healthc Mater. 2014;3:1889-97
57. Babayan A, Hannemann J, Spotter J, Muller V, Pantel K, Joosse SA. Heterogeneity of estrogen receptor expression in circulating tumor cells from metastatic breast cancer patients. PLoS One. 2013;8:e75038
58. Jordan NV, Bardia A, Wittner BS, Benes C, Ligorio M, Zheng Y. et al. HER2 expression identifies dynamic functional states within circulating breast cancer cells. Nature. 2016;537:102-6
59. Tsao SC, Wang J, Wang Y, Behren A, Cebon J, Trau M. Characterising the phenotypic evolution of circulating tumour cells during treatment. Nat Commun. 2018;9:1482
60. Li D, Zhang Y, Li R, Guo J, Wang C, Tang C. Selective capture and quick detection of targeting cells with SERS-coding microsphere suspension chip. Small. 2015;11:2200-8
61. Ruan H, Wu X, Yang C, Li Z, Xia Y, Xue T. et al. A supersensitive CTC analysis system based on triangular silver nanoprisms and SPION with function of capture, enrichment, detection, and release. ACS Biomater Sci Eng. 2018;4:1073-82
62. Cho HY, Hossain MK, Lee JH, Han J, Lee HJ, Kim KJ. et al. Selective isolation and noninvasive analysis of circulating cancer stem cells through Raman imaging. Biosens Bioelectron. 2018;102:372-82
63. Johnstone RM. Revisiting the road to the discovery of exosomes. Blood Cells Mol Dis. 2005;34:214-9
64. Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M. et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470-6
65. Van Niel G, D'Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol. 2018;19:213-28
66. EL Andaloussi S, Mäger I, Breakefield XO. et al. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat Rev Drug Discov. 2013;12:347-57
67. Hoshino A, Costa-Silva B, Shen TL, Rodrigues G, Hashimoto A, Tesic Mark M. et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527:329-35
68. Azmi AS, Bao B, Sarkar FH. Exosomes in cancer development, metastasis, and drug resistance: a comprehensive review. Cancer Metastasis Rev. 2013;32:623-42
69. Robbins PD, Morelli AE. Regulation of immune responses by extracellular vesicles. Nat Rev Immunol. 2014;14:195-208
70. Ludwig N, Whiteside TL. Potential roles of tumor-derived exosomes in angiogenesis. Expert Opin Ther Targets. 2018;22:409-17
71. Li P, Kaslan M, Lee SH, Yao J, Gao Z. Progress in Exosome Isolation Techniques. Theranostics. 2017;7:789-804
72. Thery C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006 Chapter 3: Unit 3 22
73. Rood IM, Deegens JK, Merchant ML, Tamboer WP, Wilkey DW, Wetzels JF. et al. Comparison of three methods for isolation of urinary microvesicles to identify biomarkers of nephrotic syndrome. Kidney Int. 2010;78:810-6
74. Lee K, Shao H, Weissleder R, Lee H. Acoustic purification of extracellular microvesicles. ACS Nano. 2015;9:2321-7
75. Ibsen SD, Wright J, Lewis JM, Kim S, Ko SY, Ong J. et al. Rapid isolation and detection of exosomes and associated biomarkers from plasma. ACS Nano. 2017;11:6641-51
76. Liu C, Yang Y, Wu Y. Recent advances in exosomal protein detection via liquid biopsy biosensors for cancer screening, diagnosis, and prognosis. AAPS J. 2018;20:41
77. Liang K, Liu F, Fan J, Sun D, Liu C, Lyon CJ. et al. Nanoplasmonic quantification of tumor-derived extracellular vesicles in plasma microsamples for diagnosis and treatment monitoring. Nat Biomed Eng. 2017;1:pii 0021
78. Huang X, O'Connor R, Kwizera EA. Gold Nanoparticle Based Platforms for Circulating Cancer Marker Detection. Nanotheranostics. 2017;1:80-102
79. Zong S, Wang L, Chen C, Lu J, Zhu D, Zhang Y. et al. Facile detection of tumor-derived exosomes using magnetic nanobeads and SERS nanoprobes. Anal Methods. 2016;8:5001-8
80. Wang Z, Zong S, Wang Y, Li N, Li L, Lu J. et al. Screening and multiple detection of cancerous exosomes using a SERS-based method. Nanoscale. 2018;10:9053-62
81. Kwizera EA, O'Connor R, Vinduska V, Williams M, Butch ER, Snyder SE. et al. Molecular detection and analysis of exosomes using surface-enhanced Raman scattering gold nanorods and a miniaturized device. Theranostics. 2018;8:2722-38
82. Li TD, Zhang R, Chen H, Huang ZP, Ye X, Wang H. et al. An ultrasensitive polydopamine bi-functionalized SERS immunoassay for exosome-based diagnosis and classification of pancreatic cancer. Chem Sci. 2018;9:5372-82
83. Lee C, Carney RP, Hazari S, Smith ZJ, Knudson A, Robertson CS. et al. 3D plasmonic nanobowl platform for the study of exosomes in solution. Nanoscale. 2015;7:9290-7
84. Tirinato L, Gentile F, Di Mascolo D, Coluccio ML, Das G, Liberale C. et al. SERS analysis on exosomes using super-hydrophobic surfaces. Microelectron Eng. 2012;97:337-40
85. Sivashanmugan K, Huang W, Lin C, Liao J, Lin C, Su W. et al. Bimetallic nanoplasmonic gap-mode SERS substrate for lung normal and cancer-derived exosomes detection. J Taiwan Inst Chem Eng. 2017;80:149-55
86. Park J, Hwang M, Choi B, Jeong H, Jung JH, Kim HK. et al. Exosome classification by pattern analysis of surface-enhanced Raman spectroscopy data for lung cancer diagnosis. Anal Chem. 2017;89:6695-701
87. Avella-Oliver M, Puchades R, Wachsmann-Hogiu S, Maquieira A. Label-free SERS analysis of proteins and exosomes with large-scale substrates from recordable compact disks. Sens Actuators B Chem. 2017;252:657-62
88. Stremersch S, Marro M, Pinchasik BE, Baatsen P, Hendrix A, De Smedt SC. et al. Identification of individual exosome-like vesicles by surface enhanced Raman spectroscopy. Small. 2016;12:3292-301
89. Lee C, Carney R, Lam K, Chan JW. SERS analysis of selectively captured exosomes using an integrin-specific peptide ligand. J Raman Spectrosc. 2017;48:1771-6
90. Underhill HR, Kitzman JO, Hellwig S, Welker NC, Daza R, Baker DN. et al. Fragment length of circulating tumor DNA. PLoS Genet. 2016;12:e1006162
91. Leon SA, Shapiro B, Sklaroff DM, MJ Y. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977;37:646-50
92. Sorenson GD, Pribish DM, Valone FH, Memoli VA, Bzik DJ, SL Y. Soluble normal and mutated DNA sequences from single-copy genes in human blood. Cancer Epidemiol Biomarkers Prev. 1994;3:67-71
93. Zou Z, Qi P, Qing Z, Zheng J, Yang S, Chen W. et al. Technologies for analysis of circulating tumor DNA: progress and promise. Trends Analyt Chem. 2017;97:36-49
94. Heidary M, Auer M, Ulz P, Heitzer E, Petru E, Gasch C. et al. The dynamic range of circulating tumor DNA in metastatic breast cancer. Breast Cancer Res. 2014;16:421
95. Jung M, Klotzek S, Lewandowski M, Fleischhacker M, Jung K. Changes in concentration of DNA in serum and plasma during storage of blood samples. Clin Chem. 2003;49:1028-9
96. Qin Z, Ljubimov VA, Zhou C, Tong Y, Liang J. Cell-free circulating tumor DNA in cancer. Chin J Cancer. 2016;35:36
97. Gorgannezhad L, Umer M, Islam MN, Nguyen NT, Shiddiky MJA. Circulating tumor DNA and liquid biopsy: opportunities, challenges, and recent advances in detection technologies. Lab Chip. 2018;18:1174-96
98. Zhou Q, Zheng J, Qing Z, Zheng M, Yang J, Yang S. et al. Detection of circulating tumor DNA in human blood via DNA-mediated surface-enhanced Raman spectroscopy of single-walled carbon nanotubes. Anal Chem. 2016;88:4759-65
99. Wee EJ, Wang Y, Tsao SC, Trau M. Simple, sensitive and accurate multiplex detection of clinically important melanoma DNA mutations in circulating tumour DNA with SERS nanotags. Theranostics. 2016;6:1506-13
100. Zeng Y, Ren JQ, Shen AG, Hu JM. Splicing nanoparticles-based "click" SERS could aid multiplex liquid biopsy and accurate cellular imaging. J Am Chem Soc. 2018;140:10649-52
101. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281-97
102. Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843-54
103. Cheng G. Circulating miRNAs: roles in cancer diagnosis, prognosis and therapy. Adv Drug Deliv Rev. 2015;81:75-93
104. Valentino A, Reclusa P, Sirera R, Giallombardo M, Camps C, Pauwels P. et al. Exosomal microRNAs in liquid biopsies: future biomarkers for prostate cancer. Clin Transl Oncol. 2017;19:651-7
105. Khoury S, Tran N. Circulating microRNAs potential biomarkers for common malignancies. Biomark Med. 2015;9:131-51
106. Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K. et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997-1006
107. Roth C, Rack B, Muller V, Janni W, Pantel K, Schwarzenbach H. Circulating microRNAs as blood-based markers for patients with primary and metastatic breast cancer. Breast Cancer Res. 2010;12:R90
108. Schwarzenbach H, Nishida N, Calin GA, Pantel K. Clinical relevance of circulating cell-free microRNAs in cancer. Nat Rev Clin Oncol. 2014;11:145-56
109. Armand-Labit V, Pradines A. Circulating cell-free microRNAs as clinical cancer biomarkers. Biomol Concepts. 2017;8:61-81
110. Driskell JD, Seto AG, Jones LP, Jokela S, Dluhy RA, Zhao YP. et al. Rapid microRNA (miRNA) detection and classification via surface-enhanced Raman spectroscopy (SERS). Biosens Bioelectron. 2008;24:923-8
111. Driskell JD, Primera-Pedrozo OM, Dluhy RA, Zhao Y, Tripp RA. Quantitative surface-enhanced Raman spectroscopy based analysis of microRNA mixtures. Appl Spectrosc. 2009;63:1107-14
112. Abell JL, Garren JM, Driskell JD, Tripp RA, Zhao Y. Label-free detection of micro-RNA hybridization using surface-enhanced Raman spectroscopy and least-squares analysis. J Am Chem Soc. 2012;134:12889-92
113. Guven B, Dudak FC, Boyaci IH, Tamer U, Ozsoz M. SERS-based direct and sandwich assay methods for mir-21 detection. Analyst. 2014;139:1141-7
114. Su J, Wang D, Norbel L, Shen J, Zhao Z, Dou Y. et al. Multicolor gold-silver nano-mushrooms as ready-to-use SERS probes for ultrasensitive and multiplex DNA/miRNA detection. Anal Chem. 2017;89:2531-8
115. Zhou W, Tian YF, Yin BC, Ye BC. Simultaneous surface-enhanced Raman spectroscopy detection of multiplexed microRNA biomarkers. Anal Chem. 2017;89:6120-8
116. Zheng J, Yang R, Shi M, Wu C, Fang X, Li Y. et al. Rationally designed molecular beacons for bioanalytical and biomedical applications. Chem Soc Rev. 2015;44:3036-55
117. Song CY, Yang YJ, Yang BY, Sun YZ, Zhao YP, Wang LH. An ultrasensitive SERS sensor for simultaneous detection of multiple cancer-related miRNAs. Nanoscale. 2016;8:17365-73
118. Wang H-N, Crawford BM, Fales AM, Bowie ML, Seewaldt VL, Vo-Dinh T. Multiplexed detection of microRNA biomarkers using SERS-based inverse molecular sentinel (iMS) nanoprobes. J Phys Chem C Nanomater Interfaces. 2016;120:21047-55
119. He Y, Yang X, Yuan R, Chai Y. "Off" to "On" surface-enhanced Raman spectroscopy platform with padlock probe-based exponential rolling circle amplification for ultrasensitive detection of microRNA 155. Anal Chem. 2017;89:2866-72
120. Dirks RM, Pierce NA. Triggered amplification by hybridization chain reaction. Proc Natl Acad Sci U S A. 2004;101:15275-8
121. Zheng J, Ma D, Shi M, Bai J, Li Y, Yang J. et al. A new enzyme-free quadratic SERS signal amplification approach for circulating microRNA detection in human serum. Chem Commun (Camb). 2015;51:16271-4
122. Liu H, Li Q, Li M, Ma S, Liu D. In Situ Hot-Spot Assembly as a General Strategy for Probing Single Biomolecules. Anal Chem. 2017;89:4776-80
123. Li X, Ye S, Luo X. Sensitive SERS detection of miRNA via enzyme-free DNA machine signal amplification. Chem Commun (Camb). 2016;52:10269-72
124. Stoss O, Henkel T. Biomedical marker molecules for cancer - current status and perspectives. Drug Discov Today. 2004;3:228-37
125. Lim SY, Lee JH, Diefenbach RJ, Kefford RF, Rizos H. Liquid biomarkers in melanoma: detection and discovery. Mol Cancer. 2018;17:8
126. Shimada H, Noie T, Ohashi M, Oba K, Takahashi Y. Clinical significance of serum tumor markers for gastric cancer: a systematic review of literature by the Task Force of the Japanese Gastric Cancer Association. Gastric Cancer. 2014;17:26-33
127. Norman Zamcheck, Pusztaszeri G. CEA, AFP and other potential tumor markers. CA Cancer J Clin. 1975;25:204-14
128. Chon H, Lee S, Yoon SY, Chang SI, Lim DW, Choo J. Simultaneous immunoassay for the detection of two lung cancer markers using functionalized SERS nanoprobes. Chem Commun (Camb). 2011;47:12515-7
129. Pron G. Prostate-Specific Antigen (PSA)-based population screening for prostate cancer: an evidence-based analysis. Ont Health Technol Assess Ser. 2015;15:1-64
130. Kim EH, Andriole GL. Prostate-specific antigen-based screening: controversy and guidelines. BMC Med. 2015;13:61
131. Cheng Z, Choi N, Wang R, Lee S, Moon KC, Yoon SY. et al. Simultaneous detection of dual prostate specific antigens using surface-enhanced Raman scattering-based immunoassay for accurate diagnosis of prostate cancer. ACS Nano. 2017;11:4926-33
132. Gao R, Cheng Z, deMello AJ, Choo J. Wash-free magnetic immunoassay of the PSA cancer marker using SERS and droplet microfluidics. Lab Chip. 2016;16:1022-9
133. Song C, Min L, Zhou N, Yang Y, Su S, Huang W. et al. Synthesis of novel gold mesoflowers as SERS tags for immunoassay with improved sensitivity. ACS Appl Mater Interfaces. 2014;6:21842-50
134. Domenici F, Bizzarri AR, Cannistraro S. Surface-enhanced Raman scattering detection of wild-type and mutant p53 proteins at very low concentration in human serum. Anal Biochem. 2012;421:9-15
135. Li M, Cushing SK, Zhang J, Suri S, Evans R, Petros WP. et al. Three-dimensional hierarchical plasmonic nano-architecture enhanced surface-enhanced Raman scattering immunosensor for cancer biomarker detection in blood plasma. ACS Nano. 2013;7:4967-76
136. Song G, Zhou H, Gu J, Liu Q, Zhang W, Su H. et al. Tumor marker detection using surface enhanced Raman spectroscopy on 3D Au butterfly wings. J Mater Chem B. 2017;5:1594-600
137. U SD, Fu CY, Soh KS, Ramaswamy B, Kumar A, Olivo M. Highly sensitive SERS detection of cancer proteins in low sample volume using hollow core photonic crystal fiber. Biosens Bioelectron. 2012;33:293-8
138. Dinish US, Balasundaram G, Chang YT, Olivo M. Sensitive multiplex detection of serological liver cancer biomarkers using SERS-active photonic crystal fiber probe. J Biophotonics. 2014;7:956-65
139. Ma W, Yin H, Xu L, Wu X, Kuang H, Wang L. et al. Ultrasensitive aptamer-based SERS detection of PSAs by heterogeneous satellite nanoassemblies. Chem Commun (Camb). 2014;50:9737-40
140. Feng J, Wu X, Ma W, Kuang H, Xu L, Xu C. A SERS active bimetallic core-satellite nanostructure for the ultrasensitive detection of Mucin-1. Chem Commun (Camb). 2015;51:14761-3
141. De la Rica R, Stevens MM. Plasmonic ELISA for the ultrasensitive detection of disease biomarkers with the naked eye. Nat Nanotechnol. 2012;7:821-4
142. Liang J, Liu H, Huang C, Yao C, Fu Q, Li X. et al. Aggregated silver nanoparticles based surface-enhanced Raman scattering enzyme-linked immunosorbent assay for ultrasensitive detection of protein biomarkers and small molecules. Anal Chem. 2015;87:5790-6
143. Perumal J, Balasundaram G, Mahyuddin AP, Choolani M, Olivo M. SERS-based quantitative detection of ovarian cancer prognostic factor haptoglobin. Int J Nanomedicine. 2015;10:1831-40
144. Gong T, Kong KV, Goh D, Olivo M, Yong KT. Sensitive surface enhanced Raman scattering multiplexed detection of matrix metalloproteinase 2 and 7 cancer markers. Biomed Opt Express. 2015;6:2076-87
145. Kho KW, Dinish US, Olivo M. Frequency shifts in SERS for biosensing. ACS Nano. 2012;6:4892-902
146. Perumal J, Kong KV, Dinish US, Bakker RM, Olivo M. Design and fabrication of random silver films as substrate for SERS based nano-stress sensing of proteins. RSC Adv. 2014;4:12995-3000
147. Tang B, Wang J, Hutchison JA, Ma L, Zhang N, Guo H. et al. Ultrasensitive, multiplex Raman frequency shift immunoassay of liver cancer biomarkers in physiological media. ACS Nano. 2016;10:871-9
148. Pazos E, Garcia-Algar M, Penas C, Nazarenus M, Torruella A, Pazos-Perez N. et al. Surface-enhanced Raman scattering surface selection rules for the proteomic liquid biopsy in real samples: efficient detection of the oncoprotein c-MYC. J Am Chem Soc. 2016;138:14206-9
149. Balzerova A, Fargasova A, Markova Z, Ranc V, Zboril R. Magnetically-assisted surface enhanced raman spectroscopy (MA-SERS) for label-free determination of human immunoglobulin G (IgG) in blood using Fe3O4@Ag nanocomposite. Anal Chem. 2014;86:11107-14
150. Kah JCY, Kho KW, Lee CGL, Sheppard CJR, Shen ZX, Soo KC. et al. Early diagnosis of oral cancer based on the surface plasmon resonance of gold nanoparticles. Int J Nanomedicine. 2007;2:785-98
151. Feng S, Chen R, Lin J, Pan J, Chen G, Li Y. et al. Nasopharyngeal cancer detection based on blood plasma surface-enhanced Raman spectroscopy and multivariate analysis. Biosens Bioelectron. 2010;25:2414-9
152. Tan Y, Yan B, Xue L, Li Y, Luo X, Ji P. Surface-enhanced Raman spectroscopy of blood serum based on gold nanoparticles for the diagnosis of the oral squamous cell carcinoma. Lipids Health Dis. 2017;16:73
153. Xue L, Yan B, Li Y, Tan Y, Luo X, Wang M. Surface-enhanced Raman spectroscopy of blood serum based on gold nanoparticles for tumor stages detection and histologic grades classification of oral squamous cell carcinoma. Int J Nanomedicine. 2018;13:4977-86
154. Lin D, Feng S, Pan J, Chen Y, Lin J, Chen G. et al. Colorectal cancer detection by gold nanoparticle based surface-enhanced Raman spectroscopy of blood serum and statistical analysis. Opt Express. 2011;19:13565-77
155. Li S, Zeng Q, Li L, Zhang Y, Wan M, Liu Z. et al. Study of support vector machine and serum surface-enhanced Raman spectroscopy for noninvasive esophageal cancer detection. J Biomed Opt. 2013;18:027008
156. Li S, Zhang Y, Xu J, Li L, Zeng Q, Lin L. et al. Noninvasive prostate cancer screening based on serum surface-enhanced Raman spectroscopy and support vector machine. Appl Phys Lett. 2014;105:091104
157. Shao X, Pan J, Wang Y, Zhu Y, Xu F, Shangguan X. et al. Evaluation of expressed prostatic secretion and serum using surface-enhanced Raman spectroscopy for the noninvasive detection of prostate cancer, a preliminary study. Nanomedicine. 2017;13:1051-9
158. Cervo S, Mansutti E, Del Mistro G, Spizzo R, Colombatti A, Steffan A. et al. SERS analysis of serum for detection of early and locally advanced breast cancer. Anal Bioanal Chem. 2015;407:7503-9
159. Ito H, Inoue H, Hasegawa K, Hasegawa Y, Shimizu T, Kimura S. et al. Use of surface-enhanced Raman scattering for detection of cancer-related serum-constituents in gastrointestinal cancer patients. Nanomedicine. 2014;10:599-608
160. Li SX, Zhang YJ, Zeng QY, Li LF, Guo ZY, Liu ZM. et al. Potential of cancer screening with serum surface-enhanced Raman spectroscopy and a support vector machine. Laser Phys Lett. 2014;11:065603
161. Shao L, Zhang A, Rong Z, Wang C, Jia X, Zhang K. et al. Fast and non-invasive serum detection technology based on surface-enhanced Raman spectroscopy and multivariate statistical analysis for liver disease. Nanomedicine. 2018;14:451-9
162. Lin D, Pan J, Huang H, Chen G, Qiu S, Shi H. et al. Label-free blood plasma test based on surface-enhanced Raman scattering for tumor stages detection in nasopharyngeal cancer. Sci Rep. 2014;4:4751
163. Feng S, Chen R, Lin J, Pan J, Wu Y, Li Y. et al. Gastric cancer detection based on blood plasma surface-enhanced Raman spectroscopy excited by polarized laser light. Biosens Bioelectron. 2011;26:3167-74
164. Feng S, Lin D, Lin J, Li B, Huang Z, Chen G. et al. Blood plasma surface-enhanced Raman spectroscopy for non-invasive optical detection of cervical cancer. Analyst. 2013;138:3967-74
165. Li D, Feng S, Huang H, Chen W, Shi H, Liu N. et al. Label-free detection of blood plasma using silver nanoparticle based surface-enhanced Raman spectroscopy for esophageal cancer screening. J Biomed Nanotechnol. 2014;10:478-84
166. Huang S, Wang L, Chen W, Feng S, Lin J, Huang Z. et al. Potential of non-invasive esophagus cancer detection based on urine surface-enhanced Raman spectroscopy. Laser Phys Lett. 2014;11:115604
167. Del Mistro G, Cervo S, Mansutti E, Spizzo R, Colombatti A, Belmonte P. et al. Surface-enhanced Raman spectroscopy of urine for prostate cancer detection: a preliminary study. Anal Bioanal Chem. 2015;407:3271-5
168. Li X, Yang T, Lin J. Spectral analysis of human saliva for detection of lung cancer using surface-enhanced Raman spectroscopy. J Biomed Opt. 2012;17:037003
169. Zhang Y, Walkenfort B, Yoon JH, Schlücker S, Xie W. Gold and silver nanoparticle monomers are non-SERS-active: a negative experimental study with silica-encapsulated Raman-reporter-coated metal colloids. Phys Chem Chem Phys. 2015;17:21120-6
170. Zavaleta CL, Garai E, Liu JT, Sensarn S, Mandella MJ, Van de Sompel D. et al. A Raman-based endoscopic strategy for multiplexed molecular imaging. Proc Natl Acad Sci U S A. 2013;110:E2288-97
171. Li JF, Huang YF, Ding Y, Yang ZL, Li SB, Zhou XS. et al. Shell-isolated nanoparticle-enhanced Raman spectroscopy. Nature. 2010;464:392-5
172. Xu LJ, Zong C, Zheng XS, Hu P, Feng JM, Ren B. Label-free detection of native proteins by surface-enhanced Raman spectroscopy using iodide-modified nanoparticles. Anal Chem. 2014;86:2238-45
173. Shen W, Lin X, Jiang C, Li C, Lin H, Huang J. et al. Reliable quantitative SERS analysis facilitated by core-shell nanoparticles with embedded internal standards. Angew Chem Int Ed Engl. 2015;54:7308-12
174. Wang YW, Khan A, Som M, Wang D, Chen Y, Leigh SY. et al. Rapid ratiometric biomarker detection with topically applied SERS nanoparticles. Technology (Singap World Sci). 2014;2:118-32
175. Wang X-P, Walkenfort B, König L, Kasimir-Bauer S, Schlücker S. Fast and reproducible iSERS microscopy of single HER2-positive breast cancer cells using gold nanostars as SERS nanotags. Faraday Discuss. 2017;205:377-86
Author contact
![]() Corresponding author: Dr. Yuying Zhang, Medical School of Nankai University, 94 Weijin Road, 300071 Tianjin, China. Email: yuyingzhangedu.cn
Corresponding author: Dr. Yuying Zhang, Medical School of Nankai University, 94 Weijin Road, 300071 Tianjin, China. Email: yuyingzhangedu.cn
 Global reach, higher impact
Global reach, higher impact