13.3
Impact Factor
Theranostics 2018; 8(22):6263-6273. doi:10.7150/thno.29025 This issue Cite
Research Paper
An enzyme-free biosensor for sensitive detection of Salmonella using curcumin as signal reporter and click chemistry for signal amplification
1. Key Laboratory of Agricultural Information Acquisition Technology, Ministry of Agriculture, China Agricultural University, Beijing, China
2. Key Laboratory on Modern Precision Agriculture System Integration Research, Ministry of Education, China Agricultural University, Beijing, China
3. Department of Biological and Agricultural Engineering, University of Arkansas, Fayetteville, AR, US
4. College of Veterinary, South China Agricultural University, Guangzhou, China
Received 2018-8-6; Accepted 2018-10-17; Published 2018-11-29
Abstract
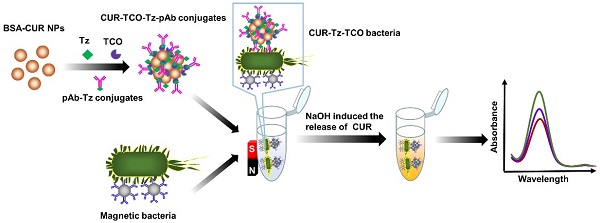
In this study, an enzyme-free biosensor was developed for sensitive and specific detection of Salmonella typhimurium (S. typhimurium) using curcumin (CUR) as signal reporter and 1,2,4,5-tetrazine (Tz)-trans-cyclooctene (TCO) click chemistry for signal amplification.
Methods: Nanoparticles composed of CUR and bovine serum albumin (BSA) were formulated and reacted with Tz and TCO to form Tz-TCO-CUR conjugates through Tz-TCO click chemistry. Then, the Tz-TCO-CUR conjugates were functionalized with polyclonal antibodies (pAbs) against S. typhimurium to form CUR-TCO-Tz-pAb conjugates. Magnetic nanoparticles (MNPs) conjugated with monoclonal antibodies (mAbs) against S. typhimurium through streptavidin-biotin binding were used to specifically and efficiently separate S. typhimurium from the background by magnetic separation. CUR-TCO-Tz-pAb conjugates were reacted with the magnetic bacteria to form CUR-Tz-TCO bacteria. Finally, CUR was released quickly from the CUR-Tz-TCO bacteria in the presence of NaOH, and the color change was measured at the characteristic wavelength of 468 nm for bacteria quantification.
Results: A linear relationship between absorbance at 468 nm and concentration of S. typhimurium from 102 to 106 CFU/mL was found. The lower detection limit was calculated to be as low as 50 CFU/mL and the mean recovery was 107.47% for S. typhimurium in spiked chicken samples.
Conclusion: This biosensor has the potential for practical applications in the detection of foodborne pathogens.
Keywords: enzyme-free biosensor, curcumin, Tz/TCO click reaction, signal amplification, Salmonella typhimurium
Introduction
Over decades, foodborne pathogens have caused numerous illnesses and led to serious public health issues and huge economic losses [1]. Salmonella is a major foodborne pathogenic bacteria that often causes more serious clinical manifestations than other known pathogenic bacteria, such as Escherichia coli, Listeria, Vibrio and Yersinia etc. [2]. Among more than 2400 serotypes of Salmonella, Salmonella typhimurium (S. typhimurium) can cause acute gastroenteritis in human, swine, cattle, poultry, etc., and has received most attention [3, 4]. Existing methods for detecting S. typhimurium, such as culture, polymerase chain reaction (PCR) and enzyme-linked immunosorbent assay (ELISA), either are time-consuming and lab-intensive, require complex DNA extraction procedures, or lack sensitivity and accuracy. Thus, it is of great importance to develop new, rapid, specific and sensitive methods for the detection of S. typhimurium in food samples to ensure food safety.
In the past decades, tremendous efforts have been made to develop various biosensors for detection of foodborne pathogenic bacteria, such as electrochemical biosensors [5-7], quartz crystal microbalance (QCM) biosensors [8, 9], surface plasmon resonance (SPR) biosensors [10, 11], optical biosensors [12, 13], etc. These biosensors have shown the advantages of high sensitivity, good specificity, short detection time, simple operation and low cost. Different enzymes, such as glucose oxidase (GOD) [14], urease [15, 16], horseradish peroxidase (HRP) [17] and invertase [18], have been used for signal amplification to improve the sensitivity of these biosensors in the detection of foodborne pathogens. However, the high cost of these enzymes and the complicated procedures for these enzyme-based assays had limited practical applications for screening of foodborne pathogens to some degree. Additionally, enzymatic catalysis often requires a certain time and appropriate temperature, resulting in longer detection times and a more complicated environment. More importantly, the enzymes have often been conjugated with nanomaterials or modified on nanostructures, resulting in partial loss of their activity. Thus, it is of great significance to find alternatives to enzymes in the development of biosensors for rapid, sensitive and low-cost detection of foodborne pathogens.
Signal amplification is often the most important challenge for biosensors to achieve a desirable sensitivity in the detection of foodborne pathogens. Recently, layer-by-layer bio-orthogonal reactions have been reported for bio-conjugation as a signal amplifier [19, 20]. Among these bio-orthogonal reactions, the click reaction between 1,2,4,5-tetrazine (Tz) and trans-cyclooctene (TCO) has gained increasing attention due to its high reaction rate and good biological compatibility [21, 22]. The click molecules (TCO and Tz) can provide functional groups to react with amino groups of such proteins as BSA or enzymes to form Tz or TCO functionalized proteins, and they can further react with each other through high cycloaddition reaction between Tz and TCO, leading to the aggregation of the monodisperse small protein particles into large protein assemblies which can greatly amplify the signals to enhance the sensitivity [23].
Curcumin (CUR), a natural yellow polyphenol pigment extracted from ginger family plants[24], is a sensitive signal reporter with an intrinsic allochroic effect induced by NaOH, resulting in obvious color change[25]. CUR also has a better binding ability with BSA molecules than resveratrol and genistein [26]. Besides, CUR has relatively stable structure under alkaline conditions[27]. To the best of our knowledge, only one paper using CUR as a signal reporter has been published, which used CUR for the detection of cardiac troponin biomarkers by a dual-readout ELISA method [25]. Thus, the combination of the click reaction between Tz and TCO and CUR might be promising for the development of novel biosensors for the sensitive detection of foodborne pathogens.
In this study, a novel enzyme-free biosensor using Tz-TCO click reaction for straightforward signal amplification and CUR as the signal reporter was developed for sensitive and rapid detection of S. typhimurium. As shown in Figure 1, streptavidin-modified magnetic nanoparticles (MNPs) were mixed with biotinylated monoclonal antibodies (mAbs) against Salmonella to form immune MNPs for specifically separating and efficiently concentrating the target S. typhimurium cells from the background to form magnetic bacteria. Separately, bovine serum albumin (BSA)-complexed CUR nanoparticles (BSA-CUR NPs) were prepared. The click molecules (TCO or Tz) with N-hydroxysuccinimide-activated functional groups were reacted with the amino groups of the BSA-CUR NPs to form Tz-BSA-CUR NPs and TCO-BSA-CUR NPs, respectively. Then, the Tz-BSA-CUR NPs and TCO-BSA-CUR NPs were assembled layer-by-layer using Tz-TCO click reaction to form Tz-TCO-CUR conjugates. Next, Tz-functionalized polyclonal antibodies (pAbs) against Salmonella were modified onto the surface of the Tz-TCO-CUR conjugates to form CUR-TCO-Tz-pAb conjugates. The CUR-TCO-Tz-pAb conjugates were then incubated with the magnetic bacteria to form CUR-Tz-TCO bacteria and the supernatant was removed by magnetic separation. Sodium hydroxide (NaOH) was used to resuspend the CUR-Tz-TCO bacteria and release the CUR on the CUR-Tz-TCO bacteria. The CUR color change was measured using an optical detector to determine the concentration of S. typhimurium in the sample.
Methods
Materials
Phosphate buffered saline (PBS, 10 mM, pH 7.4) from Sigma-Aldrich (St. Louis, MO, USA) was used as buffer solution. CUR from Aladdin (Shanghai, China) was used as signal reporter. Sodium hydroxide from Sinopharm Chemical Reagent (Shanghai, China) was prepared with a concentration of 0.1 M to release the CUR. BSA from Coolaber (Beijing, China) was used for blocking and also for preparing the BSA-CUR NPs (20 mg/mL). Ethanol (99%, v/v) from Deen Reagent (Tianjin, China) was used for dissolving the CUR. TCO-PEG4-NHS ester (TCO) and tetrazine-sulfo-NHS ester (Tz) from Click Chemistry Tools Bioconjugate Technology (Scottsdale, AZ, USA) were used for the click reaction. Streptavidin-modified MNPs with a diameter of 150 nm from Ocean NanoTech (Springdale, AR, USA) were used for separating S. typhimurium from the samples. mAbs against Salmonella from Meridian (Memphis, TN, USA) were used to specifically capture the target bacteria. The biotin labeling kit from Elabscience (Wuhan, China) was used for biotinylating the mAbs. pAbs against Salmonella from Abcam (Cambridge, MA, USA) were used for specific reaction with the target bacteria. All the solvents and chemicals were from Sinopharm Chemical (Shanghai, China) and were of analytical grade. Deionized water produced by Advantage A10 from Millipore (18.2 MΩ∙cm, Billerica, MA, USA) was used to prepare all the solutions.
Schematic of the proposed biosensor for rapid and sensitive detection of S. typhimurium.
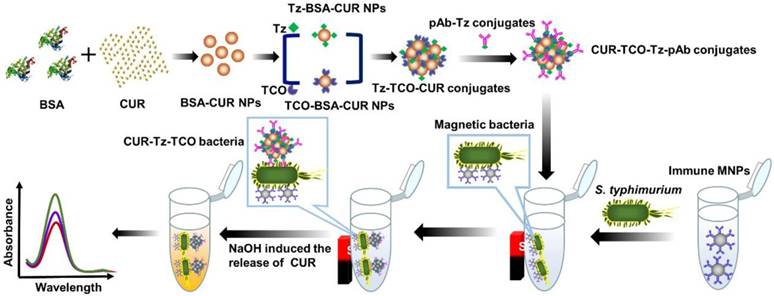
Preparation of the bacterial cultures
In this study, S. typhimurium was used as the target bacteria while Escherichia coli O157:H7 (E. coli O157:H7, ATCC43888), Listeria monocytogenes (L. monocytogene, ATCC13932) from American Type Culture Collection (ATCC), Staphylococcus aureus (S. aureus, CICC10001), Bacillus cereus (B. cereus, CICC10041) and Vibrio cholera (V. cholera, CICC23795) from China Center of Industrial Culture Collection (CICC) were used as the non-target bacteria. They were all stored at -20 °C with glycerol and first revived in Luria-Bertani (LB) medium (Aoboxing Biotech, Beijing, China) at 37 °C with shaking at 180 rpm. After incubation for 16-18 h, the bacteria cultures were serially 10-fold diluted with sterile PBS to obtain bacteria at concentrations of 102-106 CFU/mL.
Preparation of immune MNPs
mAbs were first modified with biotin and then conjugated to streptavidin-modified MNPs to form the immune MNPs for separation and concentration of S. typhimurium cells from the sample. First, NH2-reactive biotin was dissolved in dimethyl formamide (DMF) to a concentration of 10 mM. 100 μg of the mAbs was mixed with 1.33 μL of the NH2-reactive biotin and shaken for 30 min. Then, the mixture was filtered (10 kDa) and centrifuged at 12,000 ×g for 10 min at 4 °C, followed by washing with the labeling buffer to remove excess biotin to obtain biotinylated mAbs at a concentration of 0.5 mg/mL. Finally, 4 μg of the biotinylated mAbs were conjugated with 20 μg of the streptavidin-modified MNPs at 15 rpm for 45 min, and magnetically separated for 2 min to remove excess mAbs to obtain the immune MNPs. The immune MNPs were stored at 4 °C for further use.
Preparation of BSA-CUR NPs
BSA-CUR NPs were synthesized based on the desolvation method reported by Sadeghi et al. [28] with some modifications. Briefly, 10 mL of BSA (20 mg/mL, pH 9.0) prepared with deionized water was first added into 4 mg of CUR dissolved in 1 mL of ethanol, which was used as the desolvating agent, and then mixed by magnetic stirring for 4 h. Finally, the BSA-CUR NPs were collected by centrifugation at 16,000 × g for 15 min, washed 3 times with deionized water and stored at 4 °C for further use.
Preparation of pAb-Tz conjugates, Tz-BSA-CUR NPs and TCO-BSA-CUR NPs
For the purpose of signal amplification, Tz and TCO click reaction was used to layer-by-layer assemble the BSA-CUR NPs. Prior to synthesizing these NPs, DMF was used to prepare Tz and TCO reagents at a concentration of 10 mM. The proteins (BSA-CUR NPs or pAbs) and the labeling reagents (Tz or TCO) were first mixed at a molar ratio of 1:40 followed by incubation for 45 min. Next, 20 μL of Tris-HCl (500 mM, pH 8.0) was added for 5 min to terminate the reaction and then the mixture was then centrifuged at 16,000 × g and 4 °C for 15 min, and washed 3 times with deionized water to remove excess labeling reagents. Finally, the Tz-BSA-CUR NPs, TCO-BSA-CUR NPs and pAb-Tz conjugates were resuspended in PBS and stored at 4 °C for further use.
Preparation of CUR-TCO-Tz-pAb conjugates
CUR-TCO-Tz-pAb conjugates were layer-by-layer assembled based on the Tz-TCO click reaction between the Tz-BSA-CUR NPs, TCO-BSA-CUR NPs and pAb-Tz conjugates for signal amplification. Briefly, 1 mL of TCO-BSA-CUR NPs and 200 μL of Tz-BSA-CUR NPs were mixed and incubated for 45 min, and then the mixture was centrifuged at 16,000 × g and 4 °C for 15 min, followed by washing 3 times with deionized water. The Tz-TCO-CUR conjugates were re-suspended in 1 mL of PBS. Next, 40 μg of pAb-Tz conjugates was added to the Tz-TCO-CUR conjugate mixture and incubated for 45 min. The CUR-TCO-Tz-pAb conjugates were formed after centrifugation at 16,000 × g and 4 °C for 15 min and washing 3 times with deionized water. Finally, the CUR-TCO-Tz-pAb conjugates were resuspended in 1 mL of PBS and stored at 4 °C for further use.
Separation and concentration of the target bacteria
Separation and concentration of the target bacteria from the sample were conducted using the immune MNPs. Prior to testing, 20 μg of the immune MNPs (1 mg/mL) were washed with PBS using a magnetic separator (Aibit Biotech, MS0206, Jiangyin, China) at its maximum magnetic strength of ~1.1 T and re-suspended in 50 μL of PBS. Then, the immune MNPs were mixed with 500 μL of the bacterial samples at different concentrations (102-106 CFU/mL) and stirred at 15 rpm for 45 min to form the magnetic bacteria through antigen-antibody binding. Finally, the magnetic bacteria were washed 3 times with PBS to remove the sample background, and re-suspended in 200 μL of PBS.
Detection of the target bacteria
Detection of the target bacteria was based on the formation of CUR-Tz-TCO bacteria, release of the CUR from the CUR-Tz-TCO bacteria, and optical measurement of the color change. First, 200 μL of the magnetic bacteria was mixed with 100 μL of the CUR-TCO-Tz-pAb conjugates and incubated for 45 min to form CUR-Tz-TCO bacteria through antigen-antibody binding, followed by washing 3 times with PBS to remove excess CUR-TCO-Tz-pAb conjugates. Next, 400 μL of NaOH (0.1 M, pH 12.0) was added and the CUR in the CUR-Tz-TCO bacteria was rapidly released, resulting in the color of the solution changing from colorless to light yellow. Finally, the absorbance was measured at the characteristic wavelength of 468 nm using an optical detector (USB 4000, Ocean Optics, Dunedin, FL, USA) to determine the concentration of the target bacteria.
To evaluate the feasibility of this proposed biosensor, chicken meat purchased from the local supermarket were used as real sample model. First, 25 g of chicken meat was added into 225 mL of sterile PBS and homogenized (BagMixer, Interscience, Mourjou, France) for 3 min to prepare the chicken sample. Then, S. typhimurium cells were added into the supernatant of the chicken sample to obtain spiked chicken samples with bacterial concentrations ranging from 102 to 106 CFU/mL. Finally, the spiked chicken samples were separated and detected using the proposed biosensor.
Results and discussion
Measurement of the absorbance of CUR in the presence of NaOH
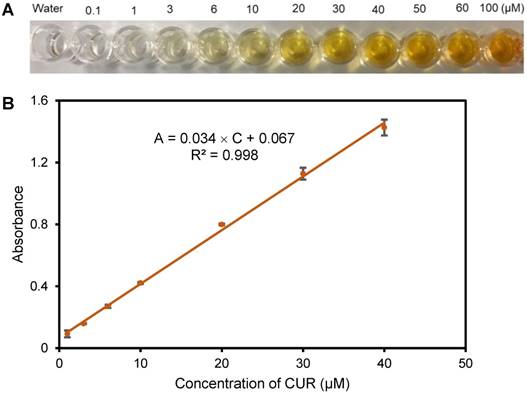
The proposed biosensor for target bacteria is based on measurement of the solution color change due to release of CUR from CUR-Tz-TCO bacteria, thus it was essential to verify that CUR absorbance is concentration dependent. Therefore, solutions of different concentrations of CUR, from 0.1 μM to 100 μM, were prepared in the presence of NaOH (0.1 M, pH 12.0). As shown in Figure 2A, it was obvious that the color of the CUR solutions became darker and darker when the concentration of CUR increased. The solution color was further measured using a USB4000 optical detector at the characteristic wavelength of 468 nm. As shown in Figure 2B, the absorbance (A) at 468 nm had a good linear relationship with concentration (C) in the range of 1 μM to 40 μM, which could be expressed as A = 0.034 × C + 0.067 (R2=0.998).
Characterization of BSA-CUR NPs and Tz-TCO-CUR conjugates
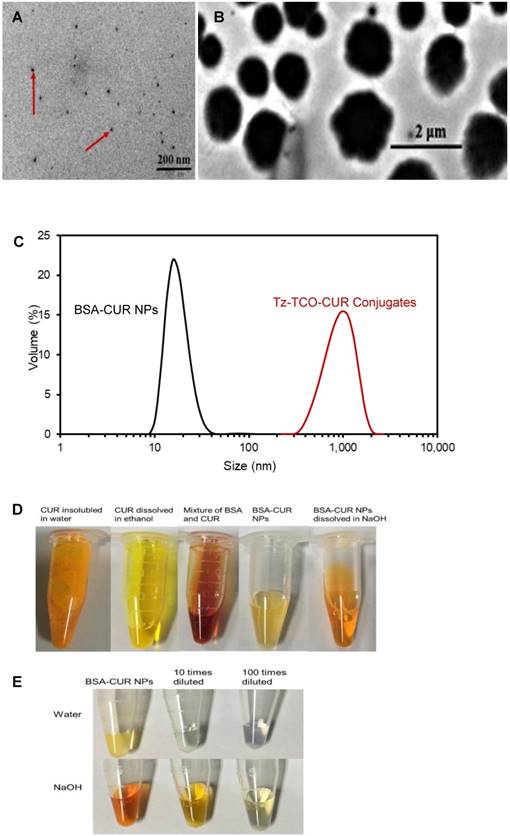
The BSA-CUR NPs and Tz-TCO-CUR conjugates are the key materials for the proposed biosensor. Since BSA with 583 amino acids has lots of groups (such as amino groups) to react with Tz/TCO [29, 30], it was used in this study to prepare BSA-CUR NPs and then form the Tz-TCO-CUR conjugates through Tz-TCO click reaction. The BSA-CUR NPs and Tz-TCO-CUR conjugates were characterized using both transmission electron microscopy (TEM) and dynamic light scattering (DLS). TEM images of the BSA-CUR NPs and Tz-TCO-CUR conjugates are shown in Figure 3A and Figure 3B, respectively, and the DLS results are shown in Figure 3C. The results indicate that the BSA-CUR NPs were spherical with the size around 20 nm (Z-Average: 52 ± 9 nm; PDI: 0.558 ± 0.043) and the Tz-TCO-CUR conjugates were much larger with the size around 1 μm (Z-Average: 1114 ± 59 nm; PDI: 0.629 ± 0.178). This proved the successful click reaction between Tz and TCO. Additionally, as shown in Figure 3D, CUR has poor solubility in water but good solubility in ethanol and NaOH. When BSA and CUR were mixed, the color of the mixture was originally red and turned into light yellow when the BSA-CUR NPs were completely synthesized. Moreover, the synthesized BSA-CUR NPs were diluted 10 times and 100 times and triggered by NaOH. As shown in Figure 3E, obvious color changes were observed after NaOH was added, indicating the successful loading of CUR on the BSA-CUR NPs.
(A) Photographs of solutions of CUR in the presence of NaOH. (B) The linear relationship between absorbance at 468 nm and concentration of CUR (N=3).
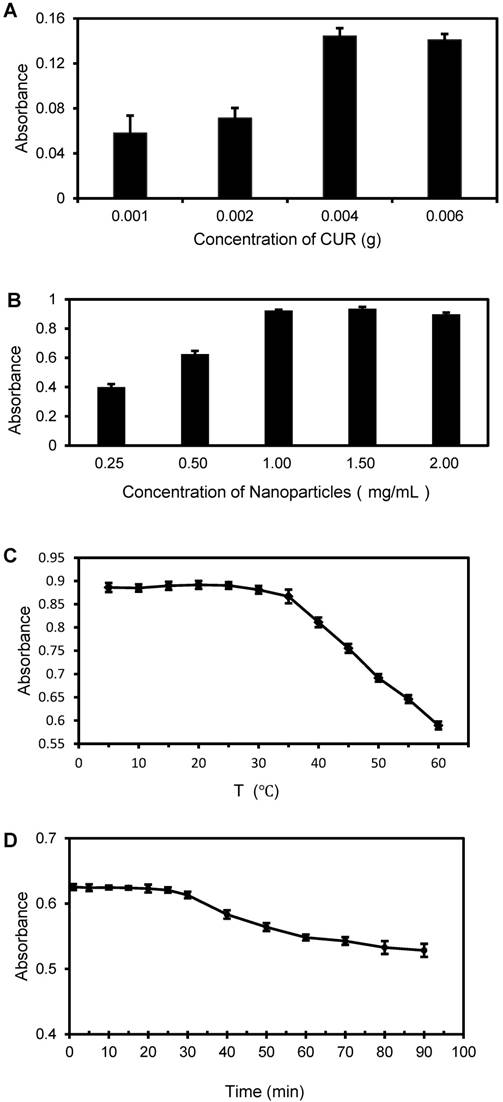
Optimization of the proposed biosensor
The complexation reaction between BSA and CUR takes place in the vicinity of Trp-212 and Trp-134, which are located in the hydrophobic binding pocket and on the surface of BSA molecules, respectively, indicating that CUR can be both encapsulated inside BSA molecules and conjugated on the surface of BSA molecules[26]. Thus, the complexation efficiency of CUR in the BSA-CUR NPs has a great impact on the sensitivity of this proposed biosensor. Thus, different amounts of CUR ranging from 1 mg to 6 mg in 1 mL of ethanol were reacted with 10 mL of BSA (20 mg/mL, pH 9.0) to form the nanoparticles. Then, the BSA-CUR NPs were centrifuged at 16,000 × g and 4 °C for 15 min and the supernatant was collected. Finally, the supernatant was diluted 100 times with NaOH and its absorbance was measured at the wavelength of 468 nm to calculate the concentration of unbound CUR. The difference between the absorbance of the mixture of CUR and BSA and that of the unbound CUR in the supernatant was used to calculate the encapsulation efficiency of CUR. As shown in Figure 4A, when the amount of CUR increased from 1 mg to 4 mg, the complexation efficiency increased obviously and did not increase when the initial amount increased to 6 mg, indicating that 4 mg of CUR saturated the BSA molecules. Therefore, the optimal amount of 4 mg CUR was used in this study to prepare the BSA-CUR NPs. At this CUR amount, the absorbance of the mixture of CUR and BSA was 0.648, while that of the unbound CUR in the supernatant was 0.504, indicating that the absorbance of CUR on the nanoparticles was 0.144, and thus the complexation efficiency of CUR was about 22%.
It is also important to optimize the amount of CUR-TCO-Tz-pAb conjugates because it can greatly influence signal amplification of this proposed biosensor. Thus, different amounts of CUR-TCO-Tz-pAb conjugates (CUR content: from 0.25 mg/mL to 2 mg/mL, 100 μL) were used to react with 200 μL of the magnetic bacteria (1.0 ×106 CFU/mL) and the absorbance of CUR released from the CUR-Tz-TCO bacteria was measured at the characteristic wavelength of 468 nm. As shown in Figure 4B, the absorbance increased from 0.4 to 0.9 when the amount of CUR-TCO-Tz-pAb conjugates was increased from 0.25 mg/mL to 1 mg/mL, and it slightly decreased when the volume was increased to 2 mg/mL. This slight decrease in absorbance might be due to the increase in steric hindrance resulting from excessive labeling. Therefore, the optimal concentration of 1 mg/mL for CUR-TCO-Tz-pAb conjugates was used in this study.
Furthermore, the impact of temperature on the signal response of CUR was also investigated. As shown in Figure 4C, CUR in the presence of NaOH shows a wide tolerance range of 0-35 °C with stable absorbance, and the absorbance decreased slowly when the temperature increased from 35 °C to 60 °C. This reduction in absorbance might be due to structural decomposition of CUR caused by high temperature. In a previous report by Miao et al, the impact of pH on the signal response of CUR was investigated and the authors found that the pH value of 12.0 was the best for releasing CUR [25]. Therefore, room temperature and NaOH with a pH of 12.0 were used in this study.
Additionally, for diagnostic purposes it is important to see how the absorbance of CUR changes over time when it is released from the CUR-Tz-TCO bacteria. As shown in Figure 4D, when the time was increased to 25 min, the absorbance of the released CUR remained steady, and began to decrease slowly when the time was increased further. This small decrease in absorbance (~0.1 for 1.5 h) might be attributed to the prolonged presence of the strong alkaline and exposure to light. Therefore, a release time of 5 min was selected in this study.
TEM images of (A) BSA-CUR NPs and (B) Tz-TCO-CUR conjugates. (C) DLS results for the size distribution of BSA-CUR NPs and Tz-TCO-CUR conjugates. (D) The color change during the preparation of BSA-CUR NPs. (E) The color of BSA-CUR NPs in water and NaOH.
(A) Optimization of the encapsulation of CUR (N=3). (B) Optimization of the volume of CUR-TCO-Tz-pAb conjugates (N=3). (C) The impact of temperature on the signal response of CUR. (D) The absorbance of CUR changed over time when it was released.
Comparison of this proposed biosensor with some recently reported enzymatic biosensors and real-time PCR.
| Methods | Enzymes | Linear Range (CFU/mL) | Detection Limit (CFU/mL) | Ref |
|---|---|---|---|---|
| Electrochemical | Urease | 1.6×102 to 1.6×104 | 1.6×102 | [32] |
| Optical | Urease | 4.3×102 to 4.3×106 | 1.0×102 | [16] |
| Glucose meter | Invertase | 1.0×102 to 4.3×106 | 7.9×101 | [18] |
| ELISA | Peroxidase | 1.0×104 to 2.8×106 | 9.0×102 | [33] |
| Chemiluminescence | Glucose oxidase | 4.3×103 to 4.3×105 | 1.2×103 | [34] |
| Real-time PCR | - | 2.5×103 to 2.5×106 | 2.5×103 | [35] |
| This biosensor | - | 1.0×102 to 1.0×106 | 5.0×101 |
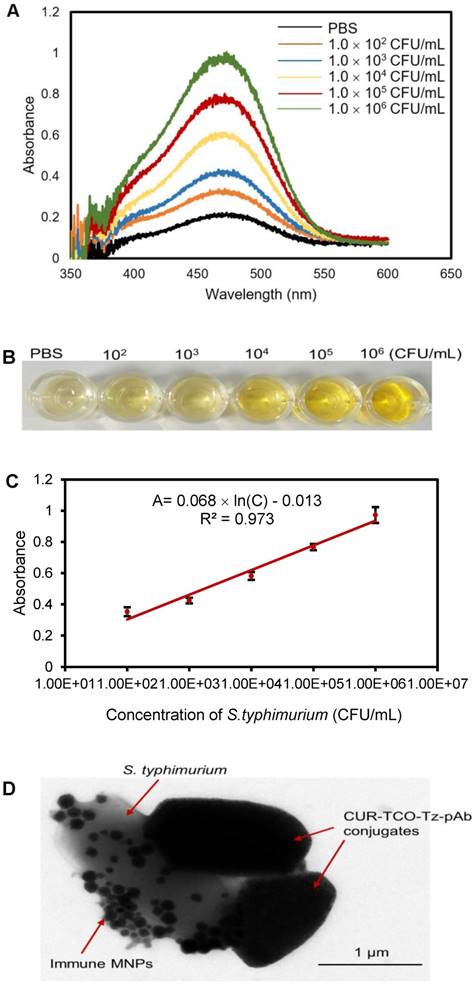
Standard curve for detection of S. typhimurium
A standard curve was measured for the proposed biosensor to determine the concentration of S. typhimurium in unknown samples. Different concentrations of the target S. typhimurium cells ranging from 102 CFU/mL to 106 CFU/mL were detected using the proposed biosensor under optimal conditions. The absorbance and the color images are shown in Figure 5A and Figure 5B, respectively. It could be seen that the bacteria samples with higher concentrations had greater color changes and stronger absorbances at the characteristic wavelength of 468 nm. As shown in Figure 5C, a linear relationship between absorbance (A) at 468 nm and concentration (C) of the target bacteria was found and could be expressed as A = 0.068 × ln(C) - 0.013 (R2=0.973).
Additionally, PBS was used as a negative control to determine the limit of detection (LOD) of the biosensor. Five negative controls were first tested using this biosensor and a mean absorbance of 0.220 with a standard deviation of 0.013 was obtained. An absorbance of 0.259 for the negative control was calculated by adding three standard deviations to the PBS mean absorbance. Finally, the absorbance of the negative control was plugged into the linear model to calculate the LOD of this biosensor, which was determined to be 50 CFU/mL. This LOD is much lower than that of some recently reported biosensors using different enzymes as signal reporters for detection of foodborne pathogenic bacteria (Table 1). The higher sensitivity of this proposed biosensor might be attributed to the following aspects: first, CUR is a sensitive signal reporter with an intrinsic allochroic effect induced by NaOH, resulting in obvious color change [25]; secondly, CUR was complexed with the nanoparticles and could be easily released from the CUR-Tz-TCO bacteria without signal loss [31]; thirdly, Tz/TCO click reaction was used to form the Tz-TCO-CUR conjugates and greatly improved the loading of CUR, resulting in more released CUR and stronger detection signal [21].
To further confirm the formation of CUR-Tz-TCO bacteria, TEM was also conducted. As shown in Figure 5D, CUR-Tz-TCO bacteria were successfully formed.
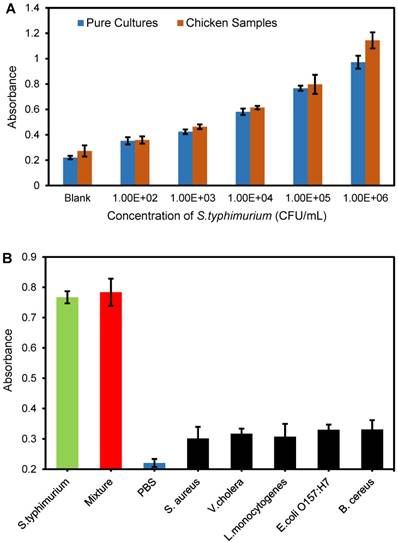
Detection of S. typhimurium in spiked chicken samples
To further evaluate the applicability of this proposed biosensor for detection of S. typhimurium, chicken meat was purchased from the local supermarket and used as a real sample model. Prior to testing, the chicken samples were prepared according to China's food safety national standards, i.e., 25 g of chicken meat were added into 225 mL of PBS and homogenized to obtain the supernatant. Different concentrations of target S. typhimurium cells were added into the supernatant to prepare spiked chicken samples with bacterial concentrations ranging from 102 to 106 CFU/mL. Three parallel tests at each concentration were conducted using this proposed biosensor. The absorbance at the wavelength of 468 nm was collected and shown in Figure 6A. The absorbances of the bacteria-spiked chicken samples were slightly higher than those of the pure bacterial cultures at the same bacteria concentration. This might be because the large number of proteins, fats, and other molecules in the chicken samples possibly increased non-specific adsorption of the CUR-TCO-Tz-pAb conjugates onto the target bacteria, resulting in increased release of CUR.
Recovery (R) of the target bacteria was calculated as the ratio of the concentration of target bacteria in the spiked chicken sample (Ns) to that of the bacteria in the pure culture (Nc): R = Ns / Nc × 100%. The recoveries for different concentrations from 102 CFU/mL to 106 CFU/mL of the target bacteria are shown in Table 2 with a mean recovery of 107.47%. This verified the applicability of this proposed biosensor for detection of S. typhimurium in real samples.
The recovery of S. typhimurium in spiked chicken samples (N=3)
| S. typhimurium concentration (CFU/mL) | Absorbance of chicken samples | Absorbance of pure culture | Recovery (%) |
|---|---|---|---|
| 1.0×102 | 0.359 | 0.353 | 101.64 |
| 1.0×103 | 0.463 | 0.424 | 108.92 |
| 1.0×104 | 0.614 | 0.582 | 105.38 |
| 1.0×105 | 0.798 | 0.767 | 103.97 |
| 1.0×106 | 1.144 | 0.972 | 117.46 |
(A) Absorbance spectra and (B) photographs of solutions of S. typhimurium incubated with the biosensor. (C) Calibration curve of the proposed biosensor (N=3). (D) TEM image of CUR-Tz-TCO bacteria.
The specificity of this proposed biosensor was evaluated using E. coli O157:H7, L. monocytogenes, S. aureus, B. cereus and V. cholerae as non-target bacteria. Additionally, a mixture of target bacteria and non-target bacteria was prepared as well. Three parallel tests on E. coli O157:H7, L. monocytogenes, S. aureus, B. cereus, V. cholerae, and S. typhimurium at the same concentration of 105 CFU/mL and a mixture of all these bacteria were conducted using this biosensor. The results are shown in Figure 6B. It is obvious that the absorbances of the non-target bacteria are much lower than those of the target bacteria, and slightly higher than that of the negative control (PBS), indicating that this proposed biosensor has good specificity. Non-specific nanoparticle adsorption to bacteria cells possibly resulted in the higher absorbance of the non-target bacteria than the negative control. In addition, the absorbance of the mixture was slightly higher than that for S. typhimurium, indicating that the non-specific adsorption to the non-target bacteria had little interference on the detection of S. typhimurium.
(A) Detection of chicken samples spiked with S. typhimurium (N=3). (B) Detection of L. monocytogenes, E. coli O157:H7, S. aureus, B. cereus, V. cholera, S. typhimurium and their mixture at the same concentration of 105 CFU/mL, and the negative control (N=3).
Conclusion
In this study, a novel enzyme-free biosensor using CUR as the signal reporter and Tz/TCO click chemistry for signal amplification has been successfully developed for sensitive, specific and rapid detection of S. typhimurium. The CUR released from the CUR-Tz-TCO bacteria was demonstrated to be a great signal reporter for this biosensor that could dramatically induce an obvious color change. Tz/TCO click reaction was used to form CUR-Tz-TCO-pAb conjugates with more CUR, and this could effectively improve the sensitivity of the biosensor. More importantly, the proposed biosensor simultaneously retained the merits of CUR as signal reporter for sensitive optical analysis and the high specific surface area of Tz-TCO-CUR conjugates for signal amplification without loss of the optical activity of CUR. This proposed biosensor has the potential to be extended for detection of other foodborne pathogens or biological targets by changing the biological recognition elements.
Abbreviations
CUR: curcumin; DMF: dimethyl formamide; ELISA: enzyme-linked immunosorbent assay; GOD: glucose oxidase; HRP: horseradish peroxidase; MNPs: magnetic nanoparticles; mAbs: monoclonal antibodies against Salmonella; PCR: polymerase chain reaction; pAbs: polyclonal antibodies against Salmonella; Tz: 1,2,4,5-tetrazine; TCO: trans-cyclooctene.
Acknowledgements
This study was supported in part by the National Key Research and Development of China (2016YFD0500706), the Walmart Foundation (SA1703161) and the Walmart Food Safety Collaboration Center.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Scharff RL. Economic burden from health losses due to foodborne illness in the United States. J Food Protect. 2012;75:123-31
2. Dar MA, Ahmad SM, Bhat SA, Ahmed R, Urwat U, Mumtaz PT. et al. Salmonella typhimurium in poultry: a review. World Poultry Sci J. 2017;73:345-54
3. Antunes P, Mourao J, Campos J, Peixe L. Salmonellosis: the role of poultry meat. Clin Microbiol Infect. 2016;22:110-21
4. Thung TY, Mahyudin NA, Basri DF, Wan Mohamed Radzi CW, Nakaguchi Y, Nishibuchi M. et al. Prevalence and antibiotic resistance of Salmonella Enteritidis and Salmonella Typhimurium in raw chicken meat at retail markets in Malaysia. Poultry Sci. 2016;95:1888-93
5. Sheikhzadeh E, Chamsaz M, Turner APF, Jager EWH, Beni V. Label-free impedimetric biosensor for Salmonella Typhimurium detection based on poly [pyrrole-co-3-carboxyl-pyrrole] copolymer supported aptamer. Biosens Bioelectron. 2016;80:194-200
6. Mutreja R, Jariyal M, Pathania P, Sharma A, Sahoo DK, Suri CR. Novel surface antigen based impedimetric immunosensor for detection of Salmonella typhimurium in water and juice samples. Biosens Bioelectron. 2016;85:707-13
7. Xu M, Wang R, Li Y. Rapid detection of Escherichia coli O157:H7 and Salmonella Typhimurium in foods using an electrochemical immunosensor based on screen-printed interdigitated microelectrode and immunomagnetic separation. Talanta. 2016;148:200-8
8. Wang L, Wang R, Chen F, Jiang T, Wang H, Slavik M. et al. QCM-based aptamer selection and detection of Salmonella typhimurium. Food Chem. 2017;221:776-82
9. Yu X, Chen F, Wang R, Li Y. Whole-bacterium SELEX of DNA aptamers for rapid detection of E.coli O157:H7 using a QCM sensor. J Biotechnol. 2018;266:39-49
10. Nguyen HH, Yi SY, Woubit A, Kim M. A portable surface plasmon resonance biosensor for rapid detection of Salmonella typhimurium. Appl Sci ConverTechnol. 2016;25:61-5
11. Nagai H, Tomioka K, Okumura S. Optimal conditions for the asymmetric polymerase chain reaction for detecting food pathogenic bacteria using a personal SPR sensor. Appl Biochem Biotech. 2018;3:1-15
12. Duan N, Chang B, Zhang H, Wang Z, Wu S. Salmonella typhimurium detection using a surface-enhanced Raman scattering-based aptasensor. Int J Food Microbiol. 2016;218:38-43
13. Kim SU, Batule BS, Mun H, Shim W-B, Kim M-G. Ultrasensitive colorimetric detection of Salmonella enterica Typhimurium on lettuce leaves by HRPzyme-integrated polymerase chain reaction. Food Control. 2018;84:522-8
14. Kwon SJ, Kim D, Lee I, Nam J, Kim J, Dordick JS. Sensitive multiplex detection of whole bacteria using self-assembled cell binding domain complexes. Anal Chimica Acta. 2018;1030:156-65
15. Wang L, Huang F, Cai G, Yao L, Zhang H, Lin J. An electrochemical aptasensor using coaxial capillary with magnetic nanoparticle, urease catalysis and PCB electrode for rapid and sensitive detection of Escherichia coli O157:H7. Nanotheranostics. 2017;1:403-14
16. Chen Q, Huang F, Cai G, Wang M, Lin J. An optical biosensor using immunomagnetic separation, urease catalysis and pH indication for rapid and sensitive detection of Listeria monocytogenes. Sensors Actuat B -Chemical. 2018;258:447-53
17. Qiao Z, Lei C, Fu Y, Li Y. An antimicrobial peptide-based colorimetric bioassay for rapid and sensitive detection of E. coli O157:H7. RSC Advances. 2017;7:15769-75
18. Huang F, Zhang H, Wang L, Lai W, Lin J. A sensitive biosensor using double-layer capillary based immunomagnetic separation and invertase-nanocluster based signal amplification for rapid detection of foodborne pathogen. Biosens Bioelectron. 2018;100:583-90
19. Haun JB, Devaraj NK, Hilderbrand SA, Lee H, Weissleder R. Bioorthogonal chemistry amplifies nanoparticle binding and enhances the sensitivity of cell detection. Nat Nanotechnol. 2010;5:660-5
20. Chan J, Dodani SC, Chang CJ. Reaction-based small-molecule fluorescent probes for chemoselective bioimaging. Nat Chem. 2012;4:973-84
21. Liu Z, Xianyu Y, Zheng W, Zhang J, Luo Y, Chen Y. et al. T1-mediated nanosensor for immunoassay based on an activatable MnO2 nanoassembly. Anal Chem. 2018;90:2765-71
22. Ran B, Xianyu Y, Dong M, Chen Y, Qian Z, Jiang X. Bioorthogonal reaction-mediated ELISA using peroxide test strip as signal readout for point-of-care testing. Anal Chem. 2017;89:6113-9
23. Xianyu Y, Wu J, Chen Y, Zheng W, Xie M, Jiang X. Controllable assembly of enzymes for multiplexed lab-on-a-chip bioassays with a tunable detection range. Angew Chem. 2018;57:7503-7
24. Naksuriya O, Okonogi S, Schiffelers RM, Hennink WE. Curcumin nanoformulations: a review of pharmaceutical properties and preclinical studies and clinical data related to cancer treatment. Biomaterials. 2014;35:3365-83
25. Miao L, Zhu C, Jiao L, Li H, Du D, Lin Y. et al. Smart drug delivery system-inspired enzyme-linked immunosorbent assay based on fluorescence resonance energy transfer and allochroic effect induced dual-modal colorimetric and fluorescent detection. Anal Chem. 2018;90:1976-82
26. Bourassa P, Kanakis CD, Tarantilis P, Pollissiou MG, Tajmirriahi HA. Resveratrol, genistein, and curcumin bind bovine serum albumin. J Phys Chem B. 2010;114:3348-54
27. Anand P, Kunnumakkara AB, Newman RA, Aggarwal BB. Bioavailability of curcumin: Problems and Promises. Mol Pharm. 2007;4:807-18
28. Sadeghi R, Moosavi-Movahedi AA, Emam-jomeh Z, Kalbasi A, Razavi SH, Karimi M. et al. The effect of different desolvating agents on BSA nanoparticle properties and encapsulation of curcumin. J Nanopart Res. 2014;16:901-14
29. Nosrati H, Salehiabar M, Manjili HK, Danafar H, Davaran S. Preparation of magnetic albumin nanoparticles via a simple and one-pot desolvation and co-precipitation method for medical and pharmaceutical applications. Int J Biol Macromol. 2018;108:909-15
30. Bourassa P, Kanakis CD, Tarantilis P, Pollissiou MG, Tajmir-Riahi HA. Resveratrol, genistein, and curcumin bind bovine serum albumin. J Phys Chemi B. 2010;114:3348-54
31. Ye R, Zhu C, Song Y, Song J, Fu S, Lu Q. et al. One-pot bioinspired synthesis of all-inclusive protein-protein nanoflowers for point-of-care bioassay: detection of E. coli O157:H7 from milk. Nanoscale. 2016;8:18980-6
32. Chen Q, Wang D, Cai G, Xiong Y, Li Y, Wang M. et al. Fast and sensitive detection of foodborne pathogen using electrochemical impedance analysis, urease catalysis and microfluidics. Biosens Bioelectron. 2016;86:770-6
33. Zhu L, He J, Cao X, Huang K, Luo Y, Xu W. Development of a double-antibody sandwich ELISA for rapid detection of Bacillus Cereus in food. Sci Rep. 2016;6:16092
34. Zhang Y, Tan C, Fei R, Liu X, Zhou Y, Chen J. et al. Sensitive chemiluminescence immunoassay for E. coli O157:H7 detection with signal dual-amplification using glucose oxidase and laccase. Anal Chem. 2014;86:1115-22
35. Wang M, Yang J, Gai Z, Huo S, Zhu J, Li J. et al. Comparison between digital PCR and real-time PCR in detection of Salmonella typhimurium in milk. Int J Food Microbiol. 2018;266:251-6
Author contact
![]() Corresponding author: Dr. Jianhan Lin, Phone/Fax: +86-10-62737599; Email: jianhanedu.cn
Corresponding author: Dr. Jianhan Lin, Phone/Fax: +86-10-62737599; Email: jianhanedu.cn
 Global reach, higher impact
Global reach, higher impact