13.3
Impact Factor
Theranostics 2018; 8(22):6248-6262. doi:10.7150/thno.28721 This issue Cite
Research Paper
CD44ICD promotes breast cancer stemness via PFKFB4-mediated glucose metabolism
1. School of Medicine, Nankai University, Tianjin 300071, China;
2. The First Affiliated Hospital of Chongqing Medical University, Chongqing 400016, China;
3. Department of Surgery, Chinese PLA General Hospital, Beijing 100071, China;
4. Department of Pathology and Institute of Precision Medicine, Jining Medical University, Jining 272067, China;
5. Department of Medicine, Vanderbilt University Medical Center, Nashville, Tennessee 37212, USA;
6. Department of Reproductive Medicine, School of Medicine and Moores Cancer Center, University of California, San Diego, La Jolla, California 39216, USA;
7. Tianjin Key Laboratory of Tumor Microenvironment and Neurovascular Regulation, Tianjin 300071, China;
8. Project Collaborative Innovation Center for Biotherapy of Ministry of Education 2011, Tianjin 300071, China.
Received 2018-7-24; Accepted 2018-10-20; Published 2018-11-29
Abstract
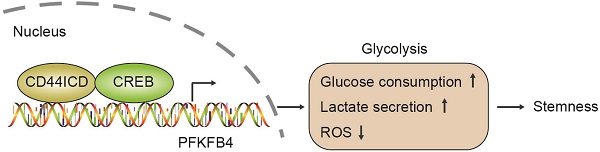
CD44 is a single-pass cell surface glycoprotein that is distinguished as the first molecule used to identify cancer stem cells in solid tumors based on its expression. In this regard, the CD44high cell population demonstrates not only the ability to regenerate a heterogeneous tumor, but also the ability to self-regenerate when transplanted into immune-deficient mice. However, the exact role of CD44 in cancer stem cells remains unclear in part because CD44 exists in various isoforms due to alternative splicing.
Methods: Gain- and loss-of-function methods in different models were used to investigate the effects of CD44 on breast cancer stemness. Cancer stemness was analyzed by detecting SOX2, OCT4 and NANOG expression, ALDH activity, side population (SP) and sphere formation. Glucose consumption, lactate secretion and reactive oxygen species (ROS) levels were detected to assess glycolysis. Western blot, immunohistochemical staining, ELISA and TCGA dataset analysis were performed to determine the association of CD44ICD and PFKFB4 with clinical cases. A PFKFB4 inhibitor, 5MPN, was used in a xenograft model to inhibit breast cancer development.
Results: In this report, we found that the shortest CD44 isoform (CD44s) inhibits breast cancer stemness, whereas the cleaved product of CD44 (CD44ICD) promotes breast cancer stemness. Furthermore, CD44ICD interacts with CREB and binds to the promoter region of PFKFB4, thereby regulating PFKFB4 transcription and expression. The resultant PFKFB4 expression facilitates the glycolysis pathway (vis-à-vis oxidative phosphorylation) and promotes stemness of breast cancer. In addition, we found that CD44ICD and PFKFB4 expressions are generally up-regulated in the tumor portion of breast cancer patient samples. Most importantly, we found that 5MPN (a selective inhibitor of PFKFB4) suppresses CD44ICD-induced tumor development.
Conclusion: CD44ICD promotes breast cancer stemness via PFKFB4-mediated glycolysis, and therapies that target PFKFB4 (e.g., 5MPN therapy) may lead to improved outcomes for cancer patients.
Keywords: PFKFB4, CD44ICD, stemness, glucose metabolism
Introduction
CD44 is a type I transmembrane glycoprotein that plays a role in cell proliferation, differentiation, adhesion, migration, and survival [1-3]. More importantly, CD44 acts as a cell surface marker to identify, isolate, and enrich cancer stem cells in many different types of cancer including breast, colon, liver, ovarian, and pancreatic cancers [4]. In addition, CD44 undergoes a two-step proteolytic cleavage in the ecto-domain and intramembrane domain that releases an intracellular domain (CD44ICD) [5]. Subsequently, CD44ICD then interacts with various stemness factors, which include SOX2, NANOG, and OCT4 in breast cancer [6].
Cancer stem cells encompass a small distinct population of cancer cells that possess properties of self-renewal and differentiation into multiple cell types [7]. The presence of cancer stem cells serves as the primary driver for cancer recurrence or relapse [8]. A number of reports have revealed that cancer stem cells preferentially utilize the glycolysis pathway rather than oxidative phosphorylation to maintain homeostasis even in the face of oxygen abundance [9-12].
The 6-phosphofructo-2-kinase/fructose-2, 6-biphosphatase 4 (PFKFB4) enzyme belongs to a family of bi-functional enzymes that adjusts the balance between 6-phosphofructose (F6P) and fructose-2, 6-biphosphate (F26P). PFKFB4 not only catalyzes the synthesis of F26P from F6P and ATP, but also catalyzes the hydrolysis of F26P to F6P and orthophosphate. Consequently, PFKFB4 dynamically regulates the levels of F26P, which is a powerful allosteric activator of phosphofructokinase1 (PFK1), a critical enzyme in the glycolysis pathway [13].
In this study, we investigated the role of CD44 on breast cancer stemness from a glucose metabolism point of view. We found the following: 1) CD44s inhibits breast cancer stemness, but CD44ICD promotes breast cancer stemness; 2) CD44ICD promotes breast cancer stemness via PFKFB4-mediated glycolysis; 3) CD44ICD interacts with the promoter region of PFKFB4, which depends on CREB; 4) CD44ICD expression and PFKFB4 expression increase in the tumor portion of breast cancer patient samples; 5) 5MPN (a specific inhibitor of PFKFB4) suppresses breast cancer development. Our findings suggest a potential strategy whereby 5MPN either as a single agent or in combination with currently available treatments may lead to an improved therapeutic outcome for cancer patients.
Methods
Cell culture
Human breast cancer cell lines MDA-MB-231, MCF7, SUM159 and MDA-MB-468 were cultured in high glucose DMEM (Biological Industries, Kibbutz Beit-Haemek, Israel) containing 10% FBS (Biological Industries) and 100 U/mL penicillin-streptomycin (Hyclone, Logan, UT, USA). Human breast cancer cell line ZR-75-1 cells were cultured in RPMI 1640 (Biological Industries) containing 10% FBS, 2.5 g/L glucose (Sigma-Aldrich, St. Louis, MO, USA), 1.5 g/L sodium bicarbonate (Thermo Fisher Scientific, Waltham, MA, USA), 0.1 g/L sodium pyruvate (Thermo Fisher Scientific) and 100 U/mL penicillin-streptomycin. Human breast cancer cell line SKBR3 cells were cultured in DMEM containing 10% FBS, 1.5 g/L sodium bicarbonate and 100 U/mL penicillin-streptomycin. Murine breast cancer cell lines EMT6 and EO771 cells were cultured in RPMI 1640 containing 10% FBS and 100 U/mL penicillin-streptomycin. Cells were maintained at 37 °C in a humidified atmosphere with 5% CO2.
Tumor and blood specimens
Human breast cancer samples and blood were obtained from the Chinese PLA General Hospital. All tumor samples were invasive ductal carcinoma of the breast that were confirmed histologically. This study was approved by the institutional ethics committees of PLA General Hospital. All patients signed a consent form.
Vector construction and stable cell lines establishment
For gene overexpression, DNA sequences encoding human CD44s and PFKFB4 were PCR-amplified from MDA-MB-231 cDNA and cloned into the pLV-EF1α-MCS-IRES-Bsd plasmid (Biosettia, San Diego, CA, USA). DNA sequences encoding CD44s-Flag, CD44∆67, CD44ICD and CD44ICD-Flag were PCR-amplified from CD44s overexpression plasmid and cloned into the pLV-EF1α-MCS-IRES-Bsd plasmid. To generate kinase dead K173A mutant of PFKFB4, PFKFB4-mut primers were used. For gene silencing, short hairpin RNAs (shRNAs) targeting human CD44, PFKFB4, CREB and murine Cd44 were cloned into the pLV-H1-EF1α-puro plasmid (Biosettia). The lentiviruses carrying the overexpression vectors, gene silencing vectors or empty vectors were produced according to manufacturer's instruction. Lentivirus-containing medium was applied to cells in the presence of 8 μg/mL polybrene for 48 h, prior to selection with 10 μg/mL blasticidin or 1 μg/mL puromycin for a week to establish stable cell lines. The primers and shRNAs are listed in Table S1, and the CD44s cDNA sequence is in Supplementary Material.
CRISPR/Cas9 KO system
20 nucleotides guide sequences were designed using the https://chopchop.rc.fas.harvard.edu/ webpage. Single-guide RNAs (sgRNAs) were cloned into LentiCRISPR vector (#51761, Addgene, Cambridge, MA, USA). Lentivirus was prepared using 293T cells, and lentiviral-rich conditioned media was applied to EO771 cells in the presence of polybrene for 48 h, prior to selection with 1 μg/mL puromycin for 3 days. Then, cells were seeded into 96-well plates at an average density of 0.4 cells/well. Single clones were expanded and screened by immunoblotting and genomic sequencing. The murine Cd44 sgRNAs were: oligo1:5'-caccGCAGGTTACATTCAAATCTG-3' and oligo2: 5'-aaacCAGATTTGAATGTAACCTGC-3'.
Cell viability
Cell viability was determined using cell counting kit-8 (CCK-8; Dojindo Molecular Technologies, Rockville, MD, USA). Briefly, cells were seeded into 96-well plates at a density of 3×103 cells/well. CCK-8 reagent was added into the wells at indicated time points and incubated at 37 °C for 2 h. The absorbance was measured at 450 nm in a microplate reader (Promega, Madison, WI, USA).
Apoptosis assay
About 1×106 cells were seeded into each well of a 6-well plate and treated with 2 μg/mL cisplatin (Sigma-Aldrich) for 48 h. Cell apoptosis was quantified using FITC Annexin V Apoptosis Detection Kit (BD Biosciences, San Jose, CA, USA) according to the manufacturer's instructions.
Aldefluor assay
ALDEFLUOR kit (STEMCELL Technologies, Vancouver, Canada) was used to measure ALDH enzymatic activity. Briefly, 2.5×105 cells were suspended in ALDRFLUOR assay buffer containing ALDH1 substrate and incubated at 37 °C for 45 min. Stained cells were analyzed on FACS Calibur flow cytometer (BD Biosciences), and data analysis was performed using Flowjo software (Tree Star, Inc., Ashland, OR, USA). DEAB, a specific ALDH inhibitor, served as a negative control.
Side population assay
Cells were suspended at a density of 1×106 cells/mL and then incubated with 5 μg/mL Hoechst 33342 (Sigma-Aldrich) at 37 °C for 60 min. Samples were analyzed on LSRFortessa flow cytometer (BD Biosciences), and data analysis was performed using Flowjo software (Tree Star, Inc.). Verapamil (Sigma-Aldrich) served as a negative control.
Sphere formation assay
About 1×103 cells were seeded onto ultra-low attachment 48-well plates (Corning, NY, USA) in the presence of serum-free DMEM or RPMI 1640 supplemented with 1× B27 (Invitrogen, Carlsbad, CA, USA), 20 ng/mL human epidermal growth factor (Invitrogen), and 20 ng/mL basic fibroblast growth factor (Invitrogen) for 7 days. Then, the spheres were counted.
Measurement of glucose consumption and lactate secretion
ZR-75-1 and SKBR3 cells were seeded in 96-well plates at a density of 1×104 cells/well and incubated in low glucose RPMI 1640 or DMEM, respectively. Twenty-four hours later, glucose concentrations in the media were determined using a glucose colorimetric assay kit (BioVision, Milpitas, CA, USA) following the manufacturer's instructions. For assessment of lactate secretion, cells were cultured in RPMI 1640 or DMEM, respectively. Thirty-six hours later, lactate secretion was assessed using a lactate colorimetric assay kit (BioVision) according to the manufacturer's protocol.
Measurement of reactive oxygen species
Reactive oxygen species (ROS) were detected by staining the cells with the ROS-sensitive probe CM-H2DCFDA (Thermo Fisher Scientific) according to the manufacturer's instructions. Briefly, cells were incubated with 5 μM CM-H2DCFDA/ PBS at 37 °C for 30 min and washed twice with PBS. The mean fluorescence intensity of CM-H2DCFDA was determined by FACS Calibur flow cytometer (BD Biosciences) as an indicator of ROS production.
Quantitative RT-PCR
Total RNA was extracted using TRIZOL (Invitrogen), and reverse transcription was performed using the TransScript First-Strand cDNA Synthesis SuperMix Kit (TransGen Biotech, Beijing, China) according to the manufacturer's recommendations. qPCR was performed using a CFXTM real-time thermal cycler (Bio-Rad, Hercules, CA, USA) using a TransStart Top Green qPCR SupperMix kit (TransGen Biotech). The primers used are listed in Table S2.
Western blot
Detection of protein expression by western blot was carried out according to the established protocols described previously [14]. Western blot was performed using the following antibodies: SOX2 (sc-20088) and β-actin (sc-47778) antibodies were purchased from Santa Cruz Biotechnology (Dallas, Texas, USA). OCT4 (ab19857), NANOG (ab80892) and PFKFB4 (ab137785) antibodies were purchased from Abcam (Cambridge, UK). CD44 (3570), Flag (8146s) and CREB (9197s) antibodies were purchased from Cell Signaling Technology (Danvers, MA, USA).
Enzyme-linked immunosorbent assay (ELISA)
Soluble CD44 (sCD44) in human serum was quantified using the CD44std (Human) ELISA kit (Abnova, Walnut, CA, USA) according to the manufacturer's protocol. Briefly, the serum was added in duplicate to wells of microtiter plates coated with horseradish peroxidase-conjugated CD44s. Then, the absorbance was measured at 450 nm using a microplate reader (Promega).
Chromatin immunoprecipitation (ChIP)
ChIP assay was performed using an EZ-ChIP kit (Millipore Corp, Billerica, MA, USA) following the manufacturer's instructions. In brief, cells grown in 10 cm dishes were cross-linked with 1% formaldehyde for 10 min at room temperature and the reaction was stopped with glycine. After sonication and centrifugation, the supernatant was collected for anti-Flag immunoprecipitation. Anti-RNA polymerase and anti-rabbit IgG were also used as a positive or negative control, respectively. Semi-quantitative RT-PCR was performed to detect DNA fragments of the PFKFB4 promoter region. The primers used are listed in Table S3.
Co-immunoprecipitation
Total proteins were extracted from ZR-75-1 cells expressing CD44s-Flag or CD44ICD-Flag and protein concentrations were determined by BCA protein assay. Briefly, 500 μg of cell lysates were incubated with 5 μg of anti-Flag antibody (Cell Signaling Technology) overnight at 4 °C with rotation. The protein-antibody mixture was then incubated with pre-washed protein G-agarose beads (CWBIO, Beijing, China) for an additional 4 h. The beads were washed twice with lysis buffer and resuspended in 2× Laemmli buffer. After protein denature, 5 μL of the beads supernatant was separated on SDS-PAGE gel for western blot to detect FLAG, SOX2, Oct4, Nanog, and CREB. 10 μg of protein lysate was subjected to western blot as an input.
Dual-luciferase reporter assay
ZR-75-1 cells were co-transfected with human PFKFB4 promoter firefly luciferase and CD44ICD expression plasmids using Lipofectamine Reagent (Invitrogen). Thirty-six hours later, luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer's protocol. Luciferase activity was normalized to Renilla luciferase activity.
Animal experiments
ZR-75-1 cells (2×106) expressing either CD44s, CD44ICD, or control vector were injected into the second mammary fat pad of 8-week-old female NOD/ SCID mice (n=5 per group). Also, 5×105 EO771 cells expressing CD44KO, CD44KO+CD44s, CD44KO+CD44ICD or WT, respectively, were injected into the second mammary fat pad of 8-week-old female C57BL/6 mice. Tumor volume was measured twice a week and calculated using the standard formula: length × width2 / 2. To examine the lung metastases foci, lung tissues were fixed with 4% PFA for 24 h and stained with hematoxylin and eosin. Metastasis was evaluated by counting the metastatic foci in three randomly selected fields. Mice were purchased from Vital River Laboratory Animal Technology Co. Ltd (Beijing, China) and maintained in a specific pathogen-free facility. Animal use complied with Nankai University Animal Welfare Guidelines. All of the animal experiments were approved by the Nankai University Animal Care and Use Committee.
Animal experiment with 5MPN
CD44 knockout EO771 cells infected with lentivirus encoding CD44ICD (CD44KO+CD44ICD) or control virus (CD44KO+MCS) were injected into the fourth fat pad of 8-week-old female C57BL/6 mice (5×105 cells/mice; n=5 per group). When the tumor size reached about 100 mm3, mice were treated with 120 mg/kg 5MPN (a specific inhibitor of PFKFB4) every two days by oral gavage for 10 days. Tumor size, tumor weight and lung metastases were evaluated using the methods mentioned above.
Statistical analysis
Kaplan-Meier survival curves were created using the log-rank test for TCGA data to compare the PFKFB4 high group with the PFKFB4 low group. All data were analyzed using GraphPad Prism5 software (GraphPad Software, San Diego, CA, USA). Results are expressed as mean ± SD with the exception of human sample and animal model data, which are expressed as mean ± SEM. P values were calculated using a two-tailed Student's t-test (two groups) or one-way ANOVA (more than 2 groups) unless otherwise noted. The results were considered statistically significant when *P<0.05, **P<0.01, ***P<0.001.
Results
CD44s inhibits breast cancer stemness, whereas CD44ICD promotes breast cancer stemness
CD44 serves as a cell surface marker for cancer stem cells in many types of cancer, including breast, ovarian, lung, and pancreatic cancers. However, the exact role of CD44 on cancer cell stemness remains unclear. Besides the standard isoform of CD44 (CD44s), CD44 also expresses as other isoforms due to inclusion of variant exons or alternative splicing. In this study, we clarified the role of CD44s, CD44Δ67, and CD44ICD isoforms on breast cancer stemness. The CD44Δ67 isoform has a truncated cytoplasmic domain that includes exon19 instead of exon20 and occurs naturally [15].
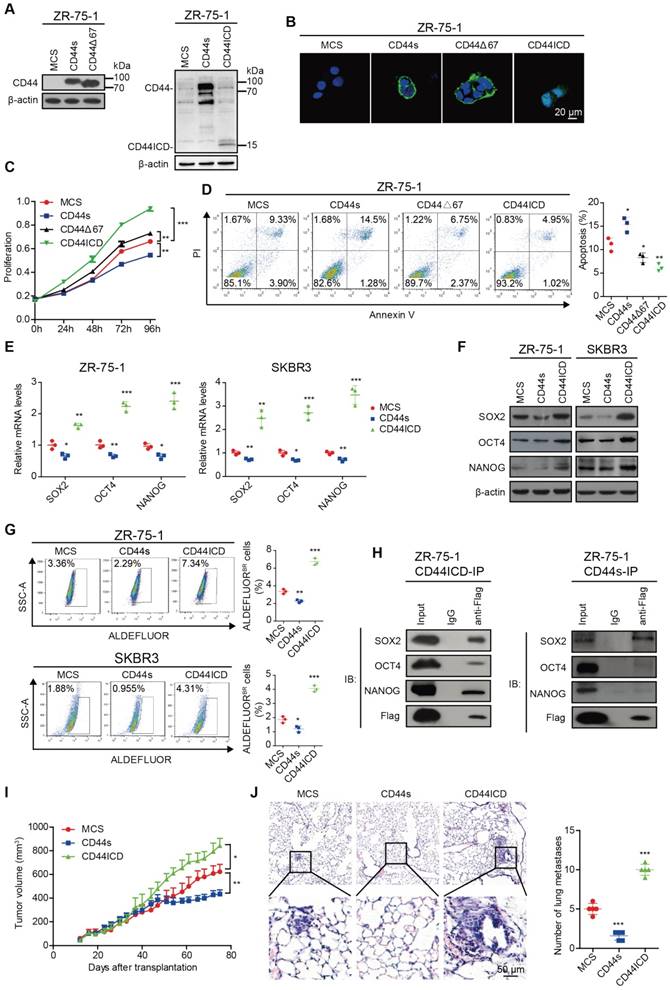
We first ectopically expressed CD44s, CD44Δ67, and CD44ICD in CD44-negative ZR-75-1 human breast cancer cells (Figure 1A and Figure S1A) and then analyzed the effect of their ectopic expression on localization, cell proliferation, and apoptosis. Our results indicated that reconstituted CD44s and CD44Δ67 localize on the cell membrane, whereas reconstituted CD44ICD localizes in the nucleus (Figure 1B). A similar localization pattern also occurs in CD44-negative SKBR3 human breast cancer cells (Figure S1B-C). In addition, our results indicated that CD44s ectopic expression inhibits cell proliferation in ZR-75-1 breast cancer cells. However, CD44ICD and CD44Δ67 ectopic expression stimulates cell proliferation in ZR-75-1 breast cancer cells. The pro-proliferation effect of CD44Δ67 is not as dramatic when compared to CD44ICD ectopic expression (Figure 1C and Figure S1D). Our results also indicated that CD44s ectopic expression advances apoptosis in ZR-75-1 breast cancer cells. However, CD44ICD and CD44Δ67 ectopic expression suppresses apoptosis in ZR-75-1 breast cancer cells. The anti-apoptosis effect of CD44Δ67 is not as dramatic when compared to CD44ICD ectopic expression (Figure 1D). Therefore, we focused our further investigation on the role of CD44s and CD44ICD on breast cancer stemness.
We next explored the role of CD44s and CD44ICD on various stem cell markers. On one hand, our results showed that CD44s ectopic expression decreases SOX2, OCT4, and NANOG expression at both mRNA and protein levels in ZR-75-1 and SKBR3 breast cancer cells (Figure 1E-F and Figure S1E). In addition, CD44s ectopic expression attenuates the percentage of ALDEFLUOR+ cells in ZR-75-1 and SKBR3 breast cancer cells (Figure 1G). CD44s ectopic expression also decreases the percent side population and sphere formation ability (Figure S1F-G). On the other hand, our results showed that CD44ICD ectopic expression increases SOX2, OCT4, and NANOG expression at both mRNA and protein levels in ZR-75-1 and SKBR3 breast cancer cells (Figure 1E-F and Figure S1H). In addition, CD44ICD ectopic expression enhances the percentage of ALDEFLUOR+ cells in ZR-75-1 and SKBR3 breast cancer cells (Figure 1G). CD44ICD ectopic expression also increases the percent side population and sphere formation ability (Figure S1I-J). We observed a similar effect of CD44s versus CD44ICD on the percent side population and sphere formation ability of MDA-MB-231 cells (Figure S1K-L).
Since previous reports have indicated that CD44ICD associates and activates stem cell markers (i.e., NANOG, OCT4, SOX2) [6, 16], we then examined interaction of stem cell markers with CD44s versus CD44ICD. The co-immunoprecipitation results showed that CD44s interacts mainly with SOX2 (not OCT4 or NANOG) and that CD44ICD interacts with SOX2, OCT4, and NANOG in ZR-75-1 breast cancer cells (Figure 1H). The CD44ICD interaction with SOX2, OCT4, and NANOG may partially explain why CD44ICD plays a more impactful role on stemness than CD44s.
The ZR-75-1 xenograft mouse model results showed that CD44s ectopic expression decreases tumor volume and metastatic foci in the lung (Figure 1I-J). However, CD44ICD ectopic expression increases tumor volume and metastatic foci in the lung (Figure 1I-J). These in vivo xenograft mouse model results confirm our in vitro results that CD44s plays an inhibitory role, whereas CD44ICD plays a stimulatory role, in tumorigenesis of breast cancer.
Our studies also showed that CD44 knockdown increases Sox2, Oct4, and Nanog expression at both mRNA and protein levels in MDA-MB-231 and EMT6 breast cancer cells (Figure S2A-B). CD44 knockdown also increases the percent side population and sphere formation ability (Figure S2C-D).
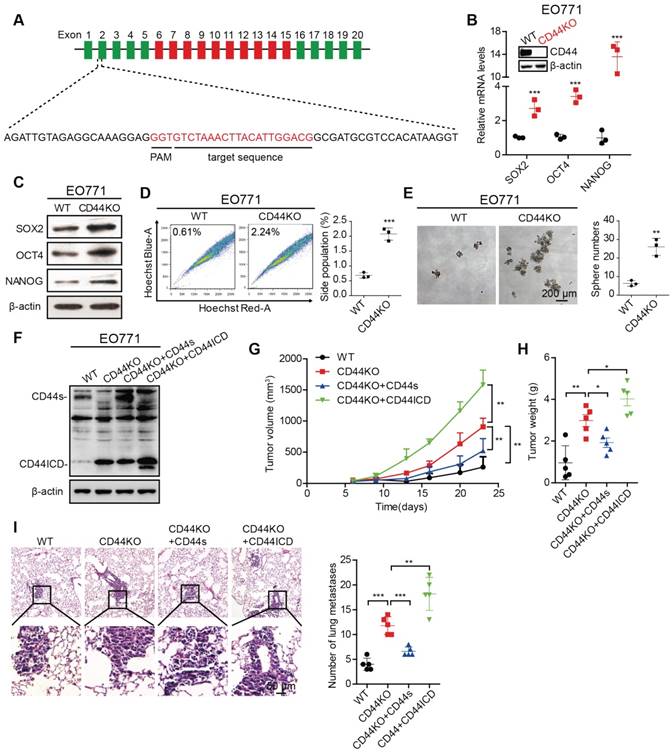
To further confirm the above findings, we generated a CD44 knockout (CD44KO) murine mammary carcinoma EO771 cell line using CRISPR/Cas9 technique (Figure 2A). We found that CD44KO increases Sox2, Oct4, and Nanog expression at both mRNA and protein levels, which confirmed our previously mentioned findings (Figure 2B-C). CD44KO also increases the percent side population and sphere formation ability (Figure 2D-E). In addition, CD44KO increases tumor volume, tumor weight, and metastatic foci in the lung (Figure 2F-I). We also found that reconstituted CD44s in CD44KO-EO771 cells decreases tumor volume, tumor weight, and metastatic foci in the lung (Figure 2F-I), whereas, reconstituted CD44ICD in CD44KO-EO771 cells increases tumor volume, tumor weight, and metastatic foci in the lung (Figure 2F-I). We observed that re-constitution of CD44ICD in CD44KO-EO771 cells leads to an increase in stem cell marker (i.e., Sox2, Oct4, Nanog) mRNA and protein expression (Figure S2E-F) and an increase in sphere formation ability (Figure S2G). These results using CD44KO-EO771 cells further confirm our earlier results that CD44s plays an inhibitory role, whereas CD44ICD plays a stimulatory role for tumorigenesis in breast cancer.
CD44ICD promotes stem cell-like characteristics of breast cancer cells. (A) Western blot analysis of CD44s, CD44∆67 and CD44ICD expression in ZR-75-1 cells. β-actin serves as a loading control. (B) Immunofluorescence staining of CD44s, CD44∆67 and CD44ICD in ZR-75-1 cells. (C) CCK-8 assay of ZR-75-1 cells reconstituted with CD44s, CD44∆67, and CD44ICD. (D) PI-Annexin V double staining assay of ZR-75-1 cells reconstituted with CD44s, CD44∆67, or CD44ICD. (E) Quantitative PCR (qPCR) analysis of SOX2, OCT4 and NANOG expression in ZR-75-1 and SKBR3 cells. (F) Western blot analysis of SOX2, OCT4 and NANOG expression in ZR-75-1 and SKBR3 cells. (G) Flow cytometric analysis of ALDH activity in ZR-75-1 and SKBR3 cells reconstituted with CD44s or CD44ICD. (H) Co-immunoprecipitation of CD44ICD or CD44s ZR-75-1 cells. The western blot in the left panel qualitatively shows that CD44ICD (anti-Flag) co-immunoprecipitated with SOX2, OCT4 and NANOG in CD44ICD ZR-75-1 cells. The western blot in the right panel qualitatively shows that CD44s (anti-Flag) co-immunoprecipitated with SOX2, but not OCT4 or NANOG in CD44s ZR-75-1 cells. Mouse IgG antibody serves as a negative control. (I) Tumor volume of ZR-75-1 xenograft mouse model. (J) Representative images of lung metastasis in ZR-75-1 xenograft mouse model (left panel). Quantification of total tumor metastasis to the lung based on incidence of dissemination from primary tumors (right panel). One-way ANOVA was used for statistical analysis and data are shown as mean ± SD except for (I-J) (mean ± SEM). Data are representative of at least three independent experiments.
CD44 knockout enhances stem cell-like characteristics of breast cancer cells. (A) Illustration of the sgRNA genomically targeted sequence in mouse Cd44 locus. Constant exons are depicted as green bars, variant exons are depicted as red bars, and introns are depicted as black lines. PAM: protospacer adjacent motif. (B) qPCR analysis of Sox2, Oct4 and Nanog expression in CD44 knockout (CD44KO) EO771 cells versus wild type controls (WT). Establishment of CD44KO EO771 stable cell line was confirmed by western blot (inset). (C) Western blot analysis of SOX2, OCT4, and NANOG expression in CD44KO EO771 cells versus WT. β-actin serves as a loading control. (D) Flow cytometric analysis of side population (SP) in CD44KO EO771 cells versus WT. (E) Sphere formation ability of EO771 cells in CD44KO EO771 cells versus WT. (F) Western blot analysis of CD44s or CD44ICD re-expression in CD44KO EO771 cells. (G) Tumor volume of EO771-derived CD44 cell lines injected into C57BL/6 mice. (H) Tumor weight of EO771-derived CD44 cell lines injected into C57BL/6 mice. (I) Representative images of lung metastasis (left panel). Quantification of total tumor metastasis to the lung based on incidence of dissemination from primary tumors (right panel). For (D-E), Student's t-test was used for statistical analysis and data are shown as mean ± SD. For (G-I), one-way ANOVA was used for statistical analysis and data are shown as mean ± SEM. Data are representative of at least three independent experiments.
CD44ICD promotes glycolysis and up-regulates PFKFB4 expression by binding to its promoter region
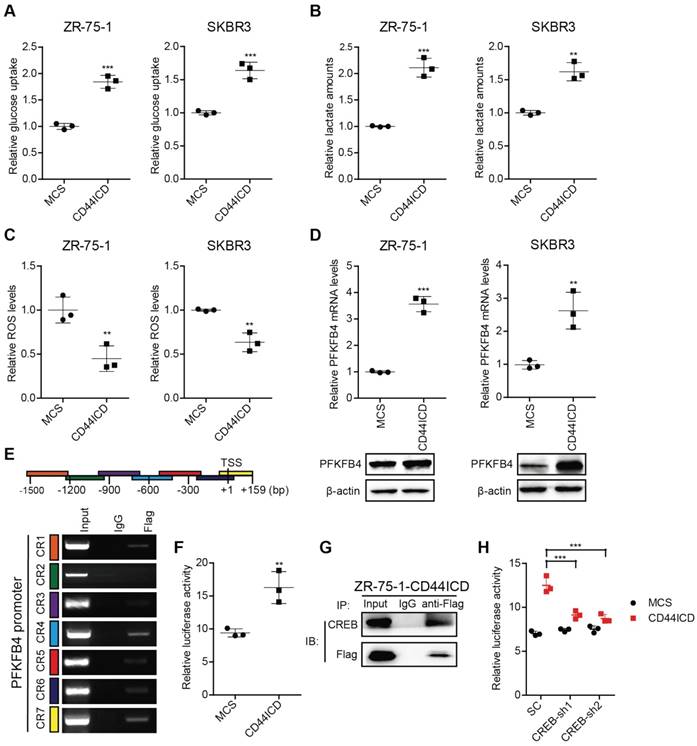
A distinctive property associated with cancer stem cells (CSCs) is that CSCs show high glycolytic activity but low oxidative phosphorylation activity. The characteristics of glycolysis include not only increased glucose consumption and lactate secretion, but also a decreased production of reactive oxygen species (ROS) [17]. To verify the impact of CD44ICD on breast cancer stemness, we examined glucose consumption, lactate secretion, and ROS production. Our results indicate that CD44ICD ectopic expression not only increases glucose consumption and lactate secretion, but also decreases ROS production (Figure 3A-C). This finding strongly suggests that CD44ICD promotes breast cancer stemness from a metabolic perspective.
In this regard, we found that CD44ICD ectopic expression specifically up-regulates PFKFB4 (a key enzyme in the glycolysis pathway) mRNA levels, but not ENO2 and ALDOC mRNA levels (Figure S3A). In addition, PFKFB4 mRNA levels increase in five different human breast cancer cell lines when cultured in spheres compared to monolayer (Figure S3B). Thus, we investigated whether PFKFB4 is involved in CD44ICD-mediated breast cancer stemness. Our results showed that CD44ICD ectopic expression significantly up-regulates PFKFB4 expression at both mRNA and proteins levels in ZR-75-1 and SKBR3 breast cancer cells (Figure 3D). Moreover, our results showed that CD44ICD interacts with the PFKFB4 promoter region using the ChIP assay (Figure 3E) and that CD44ICD induces PFKFB4 transcription using a luciferase reporter system (Figure 3F). Previous studies from other laboratories have indicated that CD44ICD interacts with the CREB transcription factor in thyroid cancer cells [18]. Our study confirmed that CD44ICD interacts with the CREB transcription factor in ZR-75-1 breast cancer cells (Figure 3G). In addition, we found that CREB knockdown decreases CD44ICD-induced PFKFB4 transcription (Figure 3H). This suggests that CD44ICD may function as a co-transcription factor and interact with CREB to regulate PFKFB4 transcription.
CD44ICD promotes glycolysis and up-regulates PFKFB4 expression. (A) Analysis of glucose uptake in CD44ICD ZR-75-1 and SKBR3 cells. (B) Analysis of lactate secretion in CD44ICD ZR-75-1 and SKBR3 cells. (C) Analysis of ROS levels in CD44ICD ZR-75-1 and SKBR3 cells. (D) Analysis of PFKFB4 expression by qPCR and western blot in CD44ICD ZR-75-1 and SKBR3 cells. (E) ChIP-PCR analysis of Flag marks (CD44ICD) at the PFKFB4 promoter region in ZR-75-1 cells. Schematic diagram of PFKFB4 promoter regions (top). Mouse IgG serves as a negative control. (F) Relative firefly-luciferase activity of PFKFB4 in CD44ICD ZR-75-1 cells. Firefly luciferase activity was normalized to Renilla luciferase activity for all samples to yield relative luciferase activity. (G) Co-immunoprecipitation in CD44ICD ZR-75-1 cells. This western blot qualitatively shows that CD44ICD (anti-Flag) co-immunoprecipitated with CREB in CD44ICD ZR-75-1 cells. Mouse IgG serves as a negative control. (H) Relative firefly-luciferase activity of PFKFB4 in CREB knockdown ZR-75-1 cells. This graph shows that the relative firefly-luciferase activity decreases in CREB knockdown (CREB-sh1 and CREB-sh2) ZR-75-1 cells versus control cells (SC). For (A-D, F), Student's t-test was used for statistical analysis, for (H), one-way ANOVA was used for statistical analysis and data are shown as mean ± SD. Data are representative of at least three independent experiments.
PFKFB4 enhances stem cell-like characteristics of breast cancer cells via up-regulation of glycolysis. (A) Analysis of glucose uptake in ZR-75-1 and SKBR3 cells. The left graph shows that glucose uptake decreases in PFKFB4 knockdown (PFKFB4-sh1 and PFKFB4-sh2) ZR-75-1 cells versus vector control (SC). The right graph shows that glucose uptake increases in PFKFB4 ectopic expression (PFKFB4-WT) SKBR3 cells versus vector control (MCS), while glucose uptake does not increase in PFKFB4 kinase dead mutant ectopic expression (PFKFB4-mut) SKBR3 cells versus MCS. (B) Analysis of lactate secretion in ZR-75-1 and SKBR3 cells. (C) Analysis of ROS levels in ZR-75-1 and SKBR3 cells. (D) Western blot analysis of SOX2, OCT4, NANOG and PFKFB4 expression in ZR-75-1 and SKBR3 cells. (E) Flow cytometric analysis of ALDH activity in ZR-75-1 and SKBR3 cells. (F) Sphere formation ability of ZR-75-1 and SKBR3 cells. (G) Western blot analysis of SOX2, OCT4 and NANOG expression in PFKFB4 ectopic expression (PFKFB4) SKBR3 cells. (H) Flow cytometric analysis of ALDH activity in PFKFB4 SKBR3 cells. (I) Sphere formation ability of PFKFB4 SKBR3 cells. One-way ANOVA was used for statistical analysis and data are shown as mean ± SD. Data are representative of at least three independent experiments.
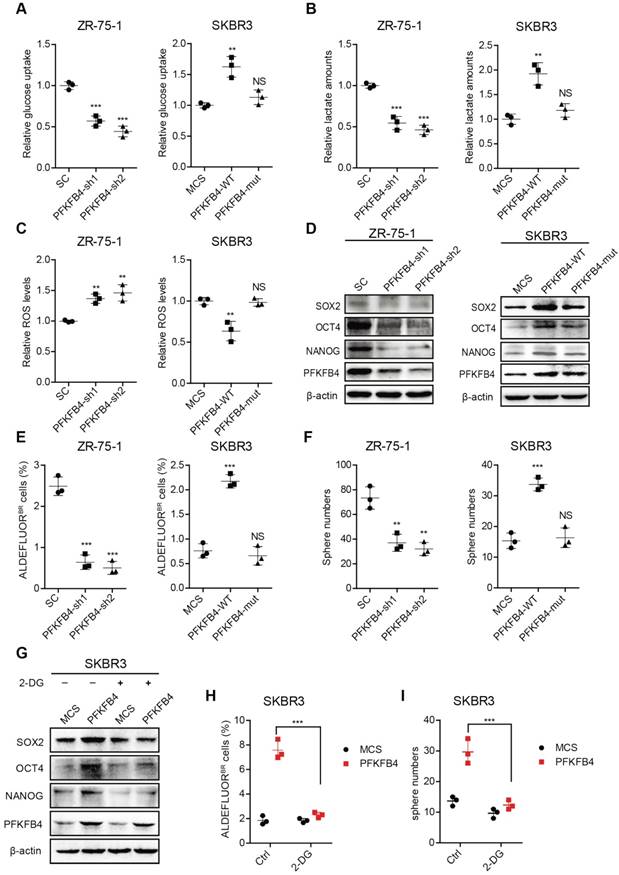
PFKFB4 promotes breast cancer stemness via enhancing glycolysis and the PFKFB4K173A mutation is important for PFKFB4 function
To further explore the impact of PFKFB4 on breast cancer stemness, we performed PFKFB4 knockdown experiments. We found that PFKFB4 knockdown in ZR-75-1 breast cancer cells produces the following effects: 1) decreases glucose consumption and lactate secretion, but enhances ROS production (Figure 4A-C); 2) decreases SOX2, OCT4, and NANOG expression (Figure 4D); 3) decreases the percentage of ALDEFLUOR+ cells (Figure 4E and Figure S3C); and, 4) decreases sphere formation ability (Figure 4F and Figure S3D). However, PFKFB4 ectopic expression in SKBR3 breast cancer cells produces the following opposite effects: 1) increases glucose consumption and lactate secretion, but reduces ROS production (Figure 4A-C); 2) increases SOX2, OCT4, and NANOG expression (Figure 4D); 3) increases the percentage of ALDEFLUOR+ cells (Figure 4E and Figure S3C); and, 4) increases sphere formation ability (Figure 4F and Figure S3D). These findings indicate that PFKFB4 regulates breast cancer stemness via its metabolic function in glycolysis. Previous reports indicate that the mutation of Lys173 to Ala in PFKFB4 results in a reduction of both ATP binding and F6P binding [19]. In this regard, we found that PFKFB4K173A ectopic expression in SKBR3 breast cancer cells produces the following effects: 1) does not increase glucose consumption and lactate secretion, or reduce ROS production (Figure 4A-C); 2) does not increase SOX2, OCT4, and NANOG expression (Figure 4D); 3) does not increase the percentage of ALDEFLUOR+ cells (Figure 4E and Figure S3C); and, 4) does not increase sphere formation ability (Figure 4F and Figure S3D). These findings suggest that the PFKFB4 kinase activity is important for both glycolysis and maintenance of breast cancer stemness.
To further confirm that PFKFB4 regulates breast cancer stemness via glycolysis, we utilized 2-deoxy-D-glucose (2-DG), a competitive inhibitor for the production of glucose-6-phosphate from glucose. Our results showed that 2-DG treatment impedes the PFKFB4-mediated increase in SOX2, OCT4, and NANOG expression (Figure 4G), the percentage of ALDEFLUOR+ cells (Figure 4H and Figure S3E), and sphere formation ability in SKBR3 breast cancer cells (Figure 4I and Figure S3E).
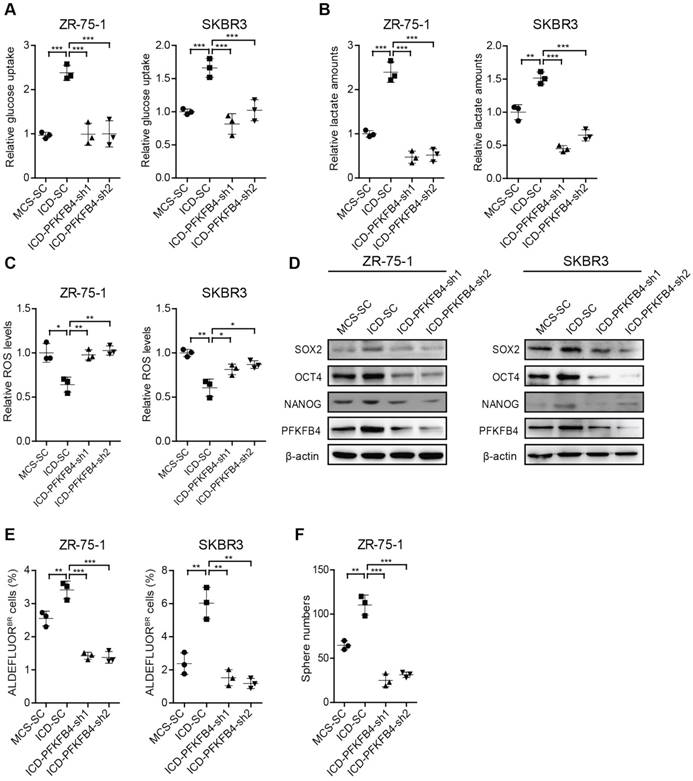
PFKFB4 is required for CD44ICD-mediated up-regulation of glycolysis and stemness
To further substantiate that CD44ICD promotes breast cancer stemness via PFKFB4, we knocked down PFKFB4 in CD44ICD-overexpressed ZR-75-1 and SKBR3 breast cancer cells. Our results showed that a reduction of PFKFB4 in CD44ICD-overexpressed ZR-75-1 or SKBR3 cells results in the following effects: 1) reduces glucose uptake and lactate secretion, and induces ROS production (Figure 5A-C); 2) decreases SOX2, OCT4, and NANOG expression (Figure 5D); 3) decreases the percentage of ALDEFLUOR+ cells (Figure 5E and Figure S4A); and 4) decreases sphere formation ability (Figure 5F and Figure S4B). In summary, our above-mentioned results indicate that CD44ICD promotes breast cancer stemness via PFKFB4 (a direct downstream molecule of CD44) and its metabolic involvement in glycolysis.
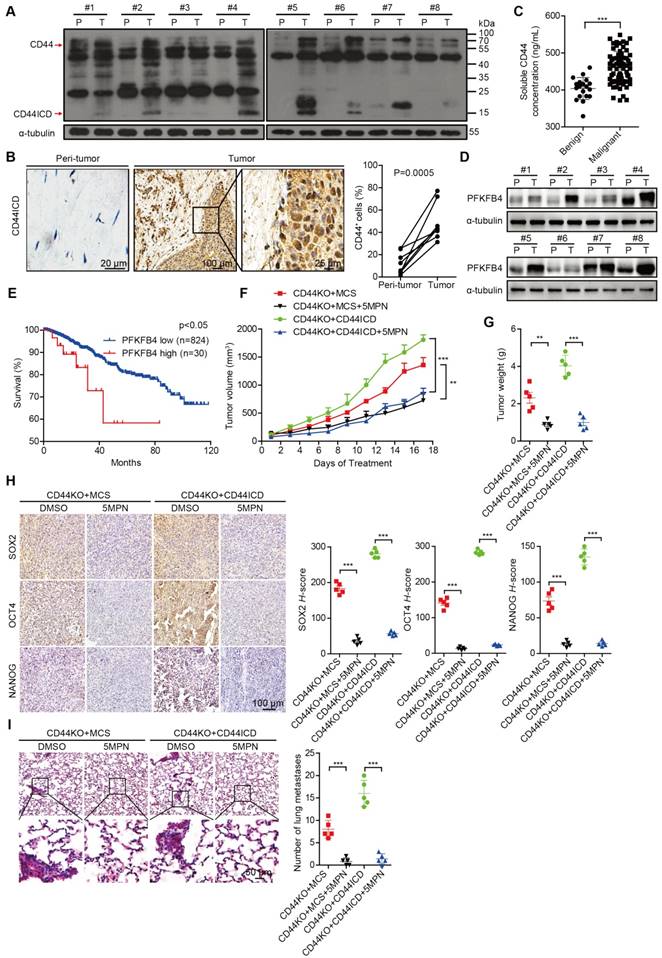
CD44ICD and PFKFB4 expression levels in human breast cancer samples are elevated and 5MPN inhibits breast cancer development
To substantiate the relevance of our laboratory findings to clinical cases, we collected 8 pairs of human breast cancer samples that included tumor tissue and peri-tumor tissue. We detected elevated CD44ICD expression in tumor tissue compared to peri-tumor tissue in matched pairs (Figure 6A-B). We also found elevated levels of soluble CD44 in periphery blood drawn from malignant breast cancer patients compared to benign patients, which suggests that more CD44 cleavage occurs in malignant patients (Figure 6C). Moreover, we detected elevated PFKFB4 expression levels in tumor tissue compared to peri-tumor tissue in matched pairs (Figure 6D). Interestingly, elevated PFKFB4 expression levels correlate with poor survival outcome based on the TCGA dataset (Figure 6E). Most importantly, the administration of 5-(n-(8- methoxy-4-quinolyl) amino) pentyl nitrate (5MPN; a selective inhibitor of PFKFB4) significantly suppressed tumor growth and tumor weight in CD44ICD-overexpressed EO771 tumors (Figure 6F-G). 5MPN administration also significantly attenuated SOX2, OCT4, and NANOG expression in CD44ICD-overexpressed EO771 tumors (Figure 6H). In addition, administration of 5MPN significantly decreased metastatic foci in the lung (Figure 6I). These results suggest that 5MPN functions as a selective inhibitor of PFKFB4 and inhibits CD44ICD-promoted breast cancer development.
Discussion
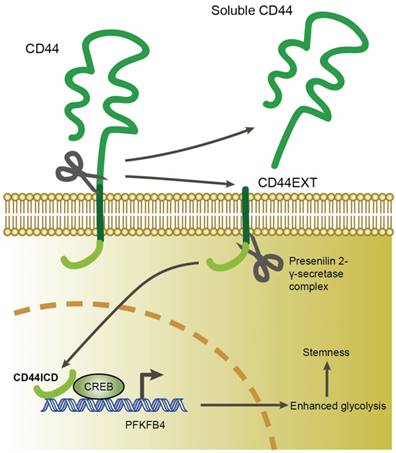
In this study, we found that CD44ICD (the cleaved product of CD44) interacts with the CREB transcription factor and binds to the promoter region of PFKFB4, which thereby regulates PFKFB4 transcription and expression. The resultant PFKFB4 expression facilitates the glycolysis pathway (vis-à-vis oxidative phosphorylation) and promotes stemness features of breast cancer (Figure 7).
CD44 is a popular marker used to identify and enrich stem cells in many different types of cancer including breast, colon, liver, ovarian, pancreatic cancers [4, 20-23]. However, the exact role of CD44 in cancer stem cells remains unclear in part because CD44 exists in various isoforms as a result of alternative splicing [24]. CD44v isoform expression (especially CD44v6) increases in colon, prostate, pancreatic, non-small cell lung, and gastric cancers [25-29]. In addition, CD44v isoform expression serves as a diagnostic or prognostic marker for tumor development [30-34]. CD44s is the shortest CD44 isoform without insertion of any variant exons and is widely expressed in both cancer and normal tissues [1]. However, the exact role of CD44s in cancer remains unclear since CD44s expression variably correlates with an increased likelihood of tumor promotion. In addition, CD44s and CD44v6 play opposing roles in tumor promotion as shown by immunostaining of human breast cancer patient samples [35]. And, CD44v isoforms (but not the CD44s isoform) promote adenoma initiation in Apc(Min/+) mice as indicated by knockin expression of either CD44v4-10 isoforms or the CD44s isoform [36]. In our studies, we showed that CD44s inhibits breast cancer stemness using gain-of-function and loss-of-function methods in different models. This suggests that CD44v may outperform the effects of CD44s and result in tumor development. Moreover, Shuo et al. has reported that CD44 knockout in the re-programmed liver cancer cell C3A model increases CSC stemness and promotes differentiation, which confirms our findings via a loss-of-function method [37].
PFKFB4 is required for CD44ICD-mediated up-regulation of glycolysis and stemness. (A) Analysis of glucose uptake in ZR-75-1 and SKBR3 cells. This graph shows that glucose uptake increases in CD44ICD ectopic expression (ICD-SC) ZR-75-1 and SKBR3 cells versus vector controls (MCS-SC), while glucose uptake decreases in PFKFB4 knockdown ICD (ICD-PFKFB4-sh1 and ICD-PFKFB4-sh2) ZR-75-1 and SKBR3 cells versus SC (ICD-SC). (B) Analysis of lactate secretion in ZR-75-1 and SKBR3 cells. (C) Analysis of ROS levels in ZR-75-1 and SKBR3 cells. (D) Western blot analysis of SOX2, OCT4 and NANOG expression in ZR-75-1 and SKBR3 cells. (E) Flow cytometric analysis of ALDH activity in ZR-75-1 and SKBR3 cells. (F) Sphere formation ability of ZR-75-1 cells. One-way ANOVA was used for statistical analysis and data are shown as mean ± SD. Data are representative of at least three independent experiments.
CD44ICD and PFKFB4 levels correlate with clinical malignancy of breast cancer patients and 5MPN inhibits tumor development. (A) Western blot analysis of CD44ICD expression in 8 matched pairs of human breast cancer samples including tumor tissues and peri-tumor tissues. (B) Representative images of CD44ICD immunohistochemical staining in 8 matched pairs of human breast cancer samples including tumor tissues and peri-tumor tissues. The graph on the right shows the statistical results. (C) Concentration of soluble CD44 in the serum of patients with benign (n=19) or malignant (n=70) breast tumors. (D) Western blot analysis of PFKFB4 expression in 8 matched pairs of human breast cancer samples including tumor tissues and peri-tumor tissues. (E) Kaplan-Meier analysis for overall survival of breast cancer patients according to PFKFB4 expression in the TCGA dataset. Low- and high-PFKFB4 expression groups were separated according to the mean expression value of all samples studied. The P-value was calculated using the Mantel-Haenszel test. (F) Tumor volume of ICD EO771 cells (in a CD44KO genetic background) injected into the 4th fat pad of C57BL/6 mice treated with 5MPN or vehicle control. (G) Tumor weight of ICD EO771 cells (in a CD44KO genetic background) injected into the 4th fat pad of C57BL/6 mice treated with 5MPN or vehicle control. (H) Representative images of SOX2, OCT4 and NANOG immunohistochemical staining in indicated EO771 tumors from C57BL/6 mice treated with 5MPN or vehicle control (left panel). Quantification of SOX2, OCT4, and NANOG staining based on H-score (right panel). (I) Representative images of lung metastasis of ICD EO771 cells (in a CD44KO genetic background) injected into the 4th pair fat pad of C57BL/6 mice treated with 5MPN or vehicle control (left panel). Quantification of total tumor metastasis to the lung based on incidence of dissemination from primary tumors (right panel). For (B-C), Student's t-test was used for statistical analysis and data are shown as mean ± SD. For (F-I), one-way ANOVA was used for statistical analysis and data are shown as mean ± SEM. Data are representative of at least three independent experiments.
A proposed model for CD44ICD-mediated regulation of stemness in breast cancer cells. CD44ICD is produced upon sequential proteolytic cleavages of CD44. The released CD44ICD translocates from the cytoplasm to the nucleus and then interacts with CREB to bind to the PFKFB4 promoter region, which causes PFKFB4 expression. The resultant PFKFB4 promotes breast cancer cell stemness via up-regulation of glycolysis.
We also found that CD44ICD promotes tumor development via up-regulation of cancer stemness, which confirms the findings of Bourguignon et al. [38] and Cho et al. [6] that CD44ICD physically associates with and activates stem cell markers (i.e., NANOG, OCT4, SOX2). CD44ICD translocates into the nucleus and regulates the transcription of a number of genes in a manner similar to Notch-ICD. Other reports demonstrate that the nuclear localization of CD44ICD is important for the transcriptional activation of various stemness factors (e.g., Nanog, Sox2, Oct4, c-Myc) [6, 37]. CD44ICD sustains cell proliferation in thyroid cancer and promotes neoplastic transformation of rat fibroblasts [18, 39]. More importantly, the early responsive genes of CD44ICD encode for critical enzymes (e.g., PDK1, PFKFB4) in the glycolysis metabolic pathway, which indicates that CD44ICD acts as the gatekeeper of the Warburg effect [40]. In this regard, we showed that CD44ICD functions as a cotranscription factor, interacts with CREB binding to the promoter region of PFKFB4, and thereby regulates PFKFB4 expression. This accumulating evidence suggests that PFKFB4 is an early responsive gene of CD44ICD and that PFKFB4 mediates not only CD44-induced glycolysis but also breast cancer stemness.
The PFKFB4 gene plays the most prominent role in the metabolic survival of brain CSCs, as shown by RNA interference screening establishing the complete human kinome and phosphatome that identify genes and pathways relevant for the survival of brain CSCs [41]. PFKFB4 deficiency induces AMPK activation, which thereby inhibits the mTOR pathway and promotes tumor cell apoptosis [36]. In this regard, PFKFB4 not only maintains the survival of brain CSCs but also associates with a shorter survival time in primary glioblastoma patients. PFKFB4 also activates the oncogenic steroid receptor co-activator-3 (SRC-3) to promote aggressive metastatic breast cancer [42]. Moreover, PFKFB4 expression levels are higher in prostatic small cell neuroendocrine carcinoma PC-3 cells that exhibit glycolytic features compared to adenocarcinoma LNCaP cells, which suggests PFKFB4 involvement in glycolysis [17]. We showed that PFKFB4 expression alters glucose consumption, lactate secretion, and ROS production of breast cancer cells, thereby indicating an alteration of the glycolysis pathway. In addition, PFKFB4 expression assists in the maintenance of breast cancer stemness by impacting the glycolysis metabolic pathway using 2-DG (a competitive inhibitor for the production of glucose-6-phosphate from glucose). Most important, we found that Lys173 of PFKFB4 kinase activity plays an essential role in the glycolysis metabolic pathway and the maintenance of breast cancer stemness.
Hypoxia is a fundamental pathophysiological condition in the microenvironment of solid tumors and occurs when the oxygen supply from the bloodstream does not meet the cellular demand [43-45]. Hypoxia levels correlate with PFKFB4 expression in human lung adenocarcinoma xenografts [46]. In addition, hypoxia levels correlate with increased PFKFB4 mRNA and protein expression, which are required for hypoxia-induced F2,6P production, glucose uptake, and glycolysis. Hypoxia-inducible factor 1 (HIF1) is a master transcription factor that responds to hypoxia and prevents cell differentiation [47]. HIF1α trans-activates hypoxia-responsive elements (HRE)-D of the PFKFB4 promoter region under hypoxia conditions in human bladder cancer cells [48]. In addition, HIF1α expression associates with PFKFB4 expression in human bladder cancer specimens [48]. The Gene Ontology analysis of CD44ICD-targeted “early response” genes showed that many of the genes activated by HIF1α were also activated by CD44ICD [40]. So, although hypoxia elicits downstream events that lead to the maintenance of CSC stemness, CD44ICD also elicits downstream events that lead to the maintenance of CSC stemness but under both normal oxygen and hypoxia conditions.
We showed that 5MPN administration significantly suppresses tumor growth, tumor weight, and lung metastasis in CD44ICD-overexpressed EO771 cells using an EO771 murine breast carcinoma model. 5MPN was screened as a selective inhibitor of PFKFB4 by structure-based virtual computational screening and was shown to possess high oral bioactivity. 5MPN suppresses glycolysis and proliferation in multiple human cancer cell lines, but not in non-transformed epithelial cells in vitro. Furthermore, 5MPN suppresses glucose uptake and tumor growth in mice injected with Lewis lung carcinoma cells [49]. PFKFB4 is not only a key participant in the glycolysis metabolic pathway, but also shows high expression levels in many different cancers [50-53]. Consequently, this allows us to speculate that a clinical trial using 5MPN either as a single agent or in combination with currently available treatment may lead to an improved therapeutic outcome for cancer patients.
Abbreviations
CD44ICD: CD44 intracytoplasmic domain; CD44s: CD44 standard form; CREB: cAMP response element-binding protein; CSCs: cancer stem cells; DMEM: Dulbecco's Modified Eagle's medium; ELISA: enzyme-linked immunosorbent assay; FBS: fetal bovine serum; F26P: fructose-2, 6-biphosphate; F6P: 6-phosphofructose; MCS: multiple cloning site; 5MPN: 5-(n-(8-methoxy-4-quinolyl)amino)pentyl nitrate; OCT4: octamer-binding transcription factor 4; PFKFB4: 6-phosphofructo-2-kinase/fructose-2, 6-biphosphatase 4; ROS: reactive oxygen species; RT-PCR: reverse transcription polymerase chain reaction; SC: scrambled control; SD: standard deviation; SEM: standard error of mean; shRNA: short hairpin RNA; SOX2: SRY (sex determining region Y)-box 2; TCGA: the cancer genome atlas; WT: wild type.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
This work was supported by National Natural Science Foundation of China 81301856 (NL) and the S&T plan project of Tianjin (12JCYBJC30900).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Zoller M. CD44: can a cancer-initiating cell profit from an abundantly expressed molecule? Nat Rev Cancer. 2011;11:254-67
2. Zoller M. CD44, hyaluronan, the hematopoietic stem cell, and leukemia-initiating cells. Front Immunol. 2015;6:235
3. Chanmee T, Ontong P, Kimata K, Itano N. Key roles of hyaluronan and its CD44 receptor in the stemness and survival of cancer stem cells. Front Oncol. 2015;5:180
4. Medema JP. Cancer stem cells: the challenges ahead. Nat Cell Biol. 2013;15:338-44
5. Okamoto I, Kawano Y, Murakami D, Sasayama T, Araki N, Miki T. et al. Proteolytic release of CD44 intracellular domain and its role in the CD44 signaling pathway. J Cell Biol. 2001;155:755-62
6. Cho Y, Lee HW, Kang HG, Kim HY, Kim SJ, Chun KH. Cleaved CD44 intracellular domain supports activation of stemness factors and promotes tumorigenesis of breast cancer. Oncotarget. 2015;6:8709-21
7. Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105-11
8. Visvader JE, Lindeman GJ. Cancer stem cells: current status and evolving complexities. Cell Stem Cell. 2012;10:717-28
9. Alvero AB, Montagna MK, Sumi NJ, Joo WD, Graham E, Mor G. Multiple blocks in the engagement of oxidative phosphorylation in putative ovarian cancer stem cells: implication for maintenance therapy with glycolysis inhibitors. Oncotarget. 2014;5:8703-15
10. Ciavardelli D, Rossi C, Barcaroli D, Volpe S, Consalvo A, Zucchelli M. et al. Breast cancer stem cells rely on fermentative glycolysis and are sensitive to 2-deoxyglucose treatment. Cell Death Dis. 2014;5:e1336
11. Feng W, Gentles A, Nair RV, Huang M, Lin Y, Lee CY. et al. Targeting unique metabolic properties of breast tumor initiating cells. Stem Cells. 2014;32:1734-45
12. Xie H, Hanai J, Ren JG, Kats L, Burgess K, Bhargava P. et al. Targeting lactate dehydrogenase-a inhibits tumorigenesis and tumor progression in mouse models of lung cancer and impacts tumor-initiating cells. Cell Metab. 2014;19:795-809
13. Yalcin A, Telang S, Clem B, Chesney J. Regulation of glucose metabolism by 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatases in cancer. Exp Mol Pathol. 2009;86:174-9
14. Li J, Yin J, Shen W, Gao R, Liu Y, Chen Y. et al. TLR4 promotes breast cancer metastasis via Akt/GSK3beta/beta-Catenin pathway upon LPS stimulation. Anat Rec (Hoboken). 2017;300:1219-29
15. Jiang H, Peterson RS, Wang W, Bartnik E, Knudson CB, Knudson W. A requirement for the CD44 cytoplasmic domain for hyaluronan binding, pericellular matrix assembly, and receptor-mediated endocytosis in COS-7 cells. J Biol Chem. 2002;277:10531-8
16. Bourguignon LYW, Wong G, Earle C, Chen L. Hyaluronan-CD44v3 interaction with Oct4-Sox2-Nanog promotes miR-302 expression leading to self-renewal, clonal formation, and cisplatin resistance in cancer stem cells from head and neck squamous cell carcinoma. J Biol Chem. 2012;287:32800-24
17. Li W, Cohen A, Sun Y, Squires J, Braas D, Graeber TG. et al. The role of CD44 in glucose metabolism in prostatic small cell neuroendocrine carcinoma. Mol Cancer Res. 2016;14:344-53
18. De Falco V, Tamburrino A, Ventre S, Castellone MD, Malek M, Manie SN. et al. CD44 proteolysis increases CREB phosphorylation and sustains proliferation of thyroid cancer cells. Cancer Res. 2012;72:1449-58
19. Yuen MH, Wang XL, Mizuguchi H, Uyeda K, Hasemann CA. A switch in the kinase domain of rat testis 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase. Biochemistry. 1999;38:12333-42
20. Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983-8
21. Dalerba P, Dylla SJ, Park IK, Liu R, Wang X, Cho RW. et al. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci U S A. 2007;104:10158-63
22. Yang ZF, Ho DW, Ng MN, Lau CK, Yu WC, Ngai P. et al. Significance of CD90+ cancer stem cells in human liver cancer. Cancer Cell. 2008;13:153-66
23. Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V. et al. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030-7
24. Ponta H, Sherman L, Herrlich PA. CD44: from adhesion molecules to signalling regulators. Nat Rev Mol Cell Biol. 2003;4:33-45
25. Wielenga VJ, Heider KH, Offerhaus GJ, Adolf GR, van den Berg FM, Ponta H. et al. Expression of CD44 variant proteins in human colorectal cancer is related to tumor progression. Cancer Res. 1993;53:4754-6
26. Ghatak S, Hascall VC, Markwald RR, Misra S. Stromal hyaluronan interaction with epithelial CD44 variants promotes prostate cancer invasiveness by augmenting expression and function of hepatocyte growth factor and androgen receptor. J Biol Chem. 2010;285:19821-32
27. Gotoda T, Matsumura Y, Kondo H, Saitoh D, Shimada Y, Kosuge T. et al. Expression of CD44 variants and its association with survival in pancreatic cancer. Jpn J Cancer Res. 1998;89:1033-40
28. Miyoshi T, Kondo K, Hino N, Uyama T, Monden Y. The expression of the CD44 variant exon 6 is associated with lymph node metastasis in non-small cell lung cancer. Clin Cancer Res. 1997;3:1289-97
29. Zavros Y. Initiation and Maintenance of Gastric Cancer: A focus on CD44 variant isoforms and cancer stem cells. Cell Mol Gastroenterol Hepatol. 2017;4:55-63
30. Gotoda T, Matsumura Y, Kondo H, Ono H, Kanamoto A, Kato H. et al. Expression of CD44 variants and prognosis in oesophageal squamous cell carcinoma. Gut. 2000;46:14-9
31. Xin Y, Grace A, Gallagher MM, Curran BT, Leader MB, Kay EW. CD44V6 in gastric carcinoma: a marker of tumor progression. Appl Immunohistochem Mol Morphol. 2001;9:138-42
32. Hoshimoto K, Yamauchi N, Takazawa Y, Onda T, Taketani Y, Fukayama M. CD44 variant 6 in endometrioid carcinoma of the uterus: its expression in the adenocarcinoma component is an independent prognostic marker. Pathol Res Pract. 2003;199:71-7
33. Lee SC, Harn HJ, Lin TS, Yeh KT, Liu YC, Tsai CS. et al. Prognostic significance of CD44v5 expression in human thymic epithelial neoplasms. Ann Thorac Surg. 2003;76:213-8 discussion 8
34. Hagiwara M, Kikuchi E, Kosaka T, Mikami S, Saya H, Oya M. Variant isoforms of CD44 expression in upper tract urothelial cancer as a predictive marker for recurrence and mortality. Urol Oncol. 2016;34:337.e19-26
35. Wu XJ, Li XD, Zhang H, Zhang X, Ning ZH, Yin YM. et al. Clinical significance of CD44s, CD44v3 and CD44v6 in breast cancer. J Int Med Res. 2015;43:173-9
36. Zeilstra J, Joosten SP, van Andel H, Tolg C, Berns A, Snoek M. et al. Stem cell CD44v isoforms promote intestinal cancer formation in Apc(min) mice downstream of Wnt signaling. Oncogene. 2014;33:665-70
37. Han S, Guo J, Liu Y, Zhang Z, He Q, Li P. et al. Knock out CD44 in reprogrammed liver cancer cell C3A increases CSCs stemness and promotes differentiation. Oncotarget. 2015;6:44452-65
38. Bourguignon LY, Wong G, Earle C, Chen L. Hyaluronan-CD44v3 interaction with Oct4-Sox2-Nanog promotes miR-302 expression leading to self-renewal, clonal formation, and cisplatin resistance in cancer stem cells from head and neck squamous cell carcinoma. J Biol Chem. 2012;287:32800-24
39. Pelletier L, Guillaumot P, Freche B, Luquain C, Christiansen D, Brugiere S. et al. Gamma-secretase-dependent proteolysis of CD44 promotes neoplastic transformation of rat fibroblastic cells. Cancer Res. 2006;66:3681-7
40. Miletti-Gonzalez KE, Murphy K, Kumaran MN, Ravindranath AK, Wernyj RP, Kaur S. et al. Identification of function for CD44 intracytoplasmic domain (CD44-ICD): modulation of matrix metalloproteinase 9 (MMP-9) transcription via novel promoter response element. J Biol Chem. 2012;287:18995-9007
41. Goidts V, Bageritz J, Puccio L, Nakata S, Zapatka M, Barbus S. et al. RNAi screening in glioma stem-like cells identifies PFKFB4 as a key molecule important for cancer cell survival. Oncogene. 2012;31:3235-43
42. Dasgupta S, Rajapakshe K, Zhu B, Nikolai BC, Yi P, Putluri N. et al. Metabolic enzyme PFKFB4 activates transcriptional coactivator SRC-3 to drive breast cancer. Nature. 2018;556:249-54
43. Keith B, Simon MC. Hypoxia-inducible factors, stem cells, and cancer. Cell. 2007;129:465-72
44. Brown JM, Wilson WR. Exploiting tumour hypoxia in cancer treatment. Nat Rev Cancer. 2004;4:437-47
45. Harris AL. Hypoxia-a key regulatory factor in tumour growth. Nat Rev Cancer. 2002;2:38-47
46. Chesney J, Clark J, Klarer AC, Imbert-Fernandez Y, Lane AN, Telang S. Fructose-2,6-bisphosphate synthesis by 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 4 (PFKFB4) is required for the glycolytic response to hypoxia and tumor growth. Oncotarget. 2014;5:6670-86
47. Nurwidya F, Takahashi F, Minakata K, Murakami A, Takahashi K. From tumor hypoxia to cancer progression: the implications of hypoxia-inducible factor-1 expression in cancers. Anat Cell Biol. 2012;45:73-8
48. Zhang H, Lu C, Fang M, Yan W, Chen M, Ji Y. et al. HIF-1alpha activates hypoxia-induced PFKFB4 expression in human bladder cancer cells. Biochem Biophys Res Commun. 2016;476:146-52
49. Chesney J, Clark J, Lanceta L, Trent JO, Telang S. Targeting the sugar metabolism of tumors with a first-in-class 6-phosphofructo-2-kinase (PFKFB4) inhibitor. Oncotarget. 2015;6:18001-11
50. Minchenko OH, Opentanova IL, Ogura T, Minchenko DO, Komisarenko SV, Caro J. et al. Expression and hypoxia-responsiveness of 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 4 in mammary gland malignant cell lines. Acta Biochim Pol. 2005;52:881-8
51. Minchenko OH, Ochiai A, Opentanova IL, Ogura T, Minchenko DO, Caro J. et al. Overexpression of 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase-4 in the human breast and colon malignant tumors. Biochimie. 2005;87:1005-10
52. Minchenko OH, Ogura T, Opentanova IL, Minchenko DO, Ochiai A, Caro J. et al. 6-Phosphofructo-2-kinase/fructose-2,6-bisphosphatase gene family overexpression in human lung tumor. Ukr Biokhim Zh. 2005;77:46-50
53. Houddane A, Bultot L, Novellasdemunt L, Johanns M, Gueuning MA, Vertommen D. et al. Role of Akt/PKB and PFKFB isoenzymes in the control of glycolysis, cell proliferation and protein synthesis in mitogen-stimulated thymocytes. Cell Signal. 2017;34:23-37
Author contact
![]() Corresponding authors: Dr. Na Luo, School of Medicine, Nankai University, 94 Weijin Road, Tianjin 300071, China. E-mail: luon11edu.cn Tel. (086) 022-23509482 or Dr. Dwayne G. Stupack, Department of Reproductive Medicine, School of Medicine and Moores Cancer Center, University of California, San Diego, La Jolla, California 39216, USA. E-mail: dstupackedu
Corresponding authors: Dr. Na Luo, School of Medicine, Nankai University, 94 Weijin Road, Tianjin 300071, China. E-mail: luon11edu.cn Tel. (086) 022-23509482 or Dr. Dwayne G. Stupack, Department of Reproductive Medicine, School of Medicine and Moores Cancer Center, University of California, San Diego, La Jolla, California 39216, USA. E-mail: dstupackedu
 Global reach, higher impact
Global reach, higher impact