13.3
Impact Factor
Theranostics 2018; 8(21):5915-5928. doi:10.7150/thno.28018 This issue Cite
Research Paper
Radiomic signature of 18F fluorodeoxyglucose PET/CT for prediction of gastric cancer survival and chemotherapeutic benefits
1. Department of General Surgery, Nanfang Hospital, Southern Medical University, 1838 North Guangzhou Avenue, Guangzhou, China;
2. Nanfang PET Center, Nanfang Hospital, Southern Medical University, Guangzhou, Guangdong, 510515, China;
3. School of Biomedical Engineering and Guangdong Provincal Key Laboratory of Medical Image Processing, Southern Medical University, Guangzhou, Guangdong, 510515, China;
4. Guangdong Key Laboratory of Liver Disease Research, the 3rd Affiliated Hospital of Sun Yat-sen University, Guangzhou, 510630, China;
5. Department of Infectious Disease, the 3rd Affiliated Hospital of Sun Yat-sen University, Guangzhou, China.
*These authors contributed equally to this work.
Received 2018-6-20; Accepted 2018-10-22; Published 2018-11-12
Abstract
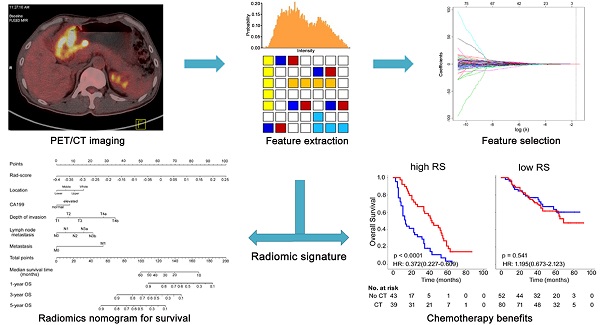
We aimed to evaluate whether radiomic feature-based fluorine 18 (18F) fluorodeoxyglucose (FDG) positron emission tomography (PET) imaging signatures allow prediction of gastric cancer (GC) survival and chemotherapy benefits.
Methods: A total of 214 GC patients (training (n = 132) or validation (n = 82) cohort) were subjected to radiomic feature extraction (80 features). Radiomic features of patients in the training cohort were subjected to a LASSO cox analysis to predict disease-free survival (DFS) and overall survival (OS) and were validated in the validation cohort. A radiomics nomogram with the radiomic signature incorporated was constructed to demonstrate the incremental value of the radiomic signature to the TNM staging system for individualized survival estimation, which was then assessed with respect to calibration, discrimination, and clinical usefulness. The performance was assessed with concordance index (C-index) and integrated Brier scores.
Results: Significant differences were found between the high- and low-radiomic score (Rad-score) patients in 5-year DFS and OS in training and validation cohorts. Multivariate analysis revealed that the Rad-score was an independent prognostic factor. Incorporating the Rad-score into the radiomics-based nomogram resulted in better performance (C-index: DFS, 0.800; OS, 0.786; in the training cohort) than TNM staging system and clinicopathologic nomogram. Further analysis revealed that patients with higher Rad-scores were prone to benefit from chemotherapy.
Conclusion: The newly developed radiomic signature was a powerful predictor of OS and DFS. Moreover, the radiomic signature could predict which patients could benefit from chemotherapy.
Keywords: gastric cancer, prognosis, chemotherapy, predictive signature, PET/CT
Introduction
Gastric cancer (GC) is the fifth most common cancer and the third leading cause of cancer death worldwide [1]. Surgical resection remains the main curative method for GC, but the high rate of relapse in patients with advanced gastric cancer makes it important to consider adjuvant treatments [2-4]. Recent studies showed that GC patients who received chemotherapy have improved survival [2, 4, 5]. However, the survival rates for many patients were still low despite initial high response rates [4, 6]. Therefore, there is an urgent need for a precise classification of GC that can be used to better predict patient outcomes and chemotherapy responses. The current prognostic model used by clinicians for risk stratification and treatment management of GC relies mainly on the tumor node metastasis (TNM) staging system. However, large variations in clinical outcomes have been reported among patients even with the same stage and similar treatment because of tumor heterogeneity, suggesting the current prognostic model could not provide full prognostic information.
Imaging with fluorine 18 (18F) fluorodeoxyglucose (FDG) positron emission tomography (PET) is a well-established method for staging GC, because it allows detection of distant metastases and lymph nodes involved [7]. Conventional PET imaging metrics such as maximum standardized uptake value (SUVmax) and total lesion glycolysis (TLG) have been reported to be valuable prognostic factors in patients with GC [8, 9]. Recent studies showed that metabolically active tumor volume (MATV) is a prognostic factor in patients with GC [10]. However, despite intensive investigation of these and other imaging metrics, the predictive value of these metrics to allow accurate discrimination of different risk groups appears to be limited. More sophisticated tools that improve on existing imaging metrics are needed.
Radiomics is an emerging approach that converts imaging data into a high-dimensional mineable feature space using a large number of automatically applied data-characterization algorithms [11, 12]. By extracting a large number of putative imaging features, radiomics enables the noninvasive profiling of tumor heterogeneity, which may ultimately correlate with clinical outcomes [11-16]. This approach has provided insights for personalized medicine in oncological practices associated with tumor detection, prognosis, subtype classification, lymph node metastasis, distant metastasis, and therapeutic response evaluation in many types of cancer with widely available imaging data such as CT or 18F-FDG PET [11, 12, 15, 17-20].
We hypothesize that automated analysis of quantitative imaging features coupled with appropriate statistical modeling may lead to improved prognostic value compared with that of conventional imaging metrics. Here, we adopt a quantitative radiomic approach to extract imaging features from pretreatment 18F-FDG PET scans. The purpose of this study was to identify quantitative imaging biomarkers from 18F-FDG PET for predicting survival and chemotherapy benefit in patients with GC.
Methods
Patient population
Under approval from the institutional review board, we retrospectively reviewed records and images of patients with gastric cancer (n = 214) who underwent total or partial radical gastrectomy at Nanfang Hospital of Southern Medical University (Guangzhou, China) between January 2010 and December 2014. Tumor staging was performed on the basis of the American Joint Committee on Cancer TNM Staging Manual, 8th Edition. Inclusion criteria were biopsy-confirmed primary GC, PET/CT performed fewer than 15 days before surgery, availability of follow-up data and clinicopathologic characteristics, no history of cancer treatment, and appropriate patient informed consent. We excluded patients if they had received previous treatment with any anticancer therapy. The end points of this study were DFS and OS. DFS was defined as the time from the time of surgery until either the date of disease progression, which refers to tumor relapse, distal metastasis, or death, or until the date that the patient was last known to be free of progression. OS was defined as the time to death from any cause. The minimum follow-up period was 42 months after surgery, while the maximum follow-up time was 98 months. Patients were postoperatively followed up with abdomen CT every 6-12 months for the first 2 years and then annually, according to the follow-up protocol of our institution. Baseline information for each patient with GC, including age, sex, TNM stage, tumor size, location, differentiation, lauren type, CEA, CA199, postsurgical chemotherapy and follow-up data (follow-up duration and survival), and time of baseline PET/CT imaging and surgery, were obtained from medical records (Table 1). Ethical approval was obtained for this retrospective analysis, and the need to obtain informed consent was waived.
PET/CT imaging
GC patients who underwent PET/CT were imaged after a 6-8 h fast and checked for a blood glucose level in the range of 3.6-10 mM. The blood glucose level was monitored by finger stick immediately before the injection of 18F FDG. 18F FDG with a radiochemical purity greater than 95% was manufactured automatically using the tracer synthesis system of a Tracerlab FXF-N (GE Healthcare). Patients were injected with 161-361 MBq (4.35-9.76 mCi, 150 μCi/kg), and imaging was performed 60 min (59 ± 3 min, range 53-62 min) later with a PET/CT scanner (GE Discovery LS PET/CT scanner (GE Healthcare, Waukesha, Wisconsin, USA) or Biograph mCT scanner (Siemens, Germany)). CT images were collected in the helical acquisition mode. In the same scanning locations and generally in 6-8 bed positions, PET data were acquired with 3-5 min of acquisition time per bed position. The complete PET/CT examination required approximately 1.5 h, including patient setup, tracer uptake, and image acquisition. We defined 132 patients imaged with the GE Discovery LS PET/CT scanner as the training cohort and 82 patients imaged with the Biograph mCT PET/CT scanner as the validation cohort.
PET images were reconstructed using standard ordered-subset expectation maximization (OSEM). The reconstruction thicknesses of the CT images were 4.25 mm (GE Healthcare) and 3.0 mm (Siemens). The fields of views (FOV) were 500 mm (Siemens) and 700 mm (GE Healthcare), and matrix sizes were 512×512 for Siemens and GE systems. The OSEM algorithm (3 iterations and 21 subsets) was used for PET image reconstruction, resulting in voxel sizes of 4.07×4.07×2 mm3 (Siemens) and 4.3×4.3×4.25 mm3 (GE Healthcare). The CT image voxel sizes were 0.97×0.97×3 mm3 (Siemens) and 1.95×1.95×5 mm3 (GE Healthcare). Images were corrected for attenuation with a CT-based attenuation correction method. The PET and CT images were individually transferred to Xeleris (GE Healthcare) or Syngo MMWP (Siemens) workstations, respectively, to display frame-on-frame fusion images. Tumor manual segmentation was then performed based on consensus reached by two expert physicians, and checked by authors Q.Y. and Q.W. on the PET/CT images with ITK-SNAP software (www.itksnap.org) [21, 22]. To verify whether the features extracted from these two machines on the same patients were not significant different, we selected 30 patients' images from the two machines firstly. The inter-scanner agreement of feature extraction was evaluated by using an intraclass correlation coefficient (ICC) [23]. The strength of agreement was evaluated as follows: an ICC value of less than 0.20 indicated poor agreement; 0.21-0.40, fair agreement; 0.41-0.60, moderate agreement; 0.61-0.80, good agreement; and 0.81-1.0, excellent agreement [24].
Image feature extraction
We calculated a total of 80 quantitative features from each volume of interest (VOI) of each patient's PET image to characterize intratumor heterogeneity and complexity. The feature pool included 14 first-order intensity features, 9 shape features, and 57 second- and higher-order textural features, which are summarized in Table S1. In this work, we investigated four types of texture features on the basis of gray-level co-occurrence matrices (GLCM), gray-level run length matrix (GLRLM), gray-level size zone matrix (GLSZM), and neighborhood gray-tone difference matrix wavelet decompositions (NGTDM). 26, 13, 13 and 5 features were extracted from GLCM, GLRLM, GLSZM and NGTDM, respectively. The detailed mathematical definitions of all imaging features listed in Supplementary Materials. All radiomic features were extracted in Matlab R2012a (The MathWorks Inc.) using an available radiomic analysis package (https://github.com/mvallieres/radiomics/).
The SUV image was discretized by 0.1 SUV unit bin width according to the following equation [25]: SUVDis(x) = | SUV(x)/0.1| - min(|SUV(x)/0.1|) + 1, where SUV(x) is the SUV of voxel x and SUVDis(x) is the discretized value of voxel x. The discretization step is necessary to generate matrices whose size (defined by the maximum SUVDis(x)) highly impacts computation, and is used to reduce image noise and generate a constant intensity resolution so that textural features from different patients are comparable.
Feature selection
We applied the least absolute shrinkage and selection operator (LASSO) algorithm jointly with the Cox survival model to implement a nested feature selection scheme based on the association between every feature and patients' DFS in the training cohort, and then a multiple-feature-based radiomic signature, the radiomic score (Rad-score), was constructed for predicting patients' DFS in the training cohort [3, 26, 27]. The “glmnet” package was used to perform the LASSO Cox regression model analysis [28, 29]. Complete details are provided in Supplementary Materials.
Validation of radiomic signature
The potential association of the radiomic signature with DFS and OS was first assessed in the training cohort and then validated in the validation cohorts by using Kaplan-Meier survival analysis. The optimum cutoff score of the Rad-score was selected on the basis of the association with the patients' DFS using X-tile software (version 3.6.1) in the training cohort [3, 30], and patients were classified into high- or low-Rad-score groups for DFS and OS analyses. Then, the same threshold values were applied to the validation cohorts. Stratified analyses were performed to explore the potential association of the radiomic signature with DFS and OS using subgroups within TNM stage from all patients. Evaluation of the radiomic signature as an independent biomarker was performed by integrating clinicopathologic risk factors into the multivariable Cox proportional hazards model using a backward stepwise approach.
Assessment of incremental value of radiomic signature in individual DFS and OS estimation
To demonstrate the incremental value of the radiomic signature to the TNM staging system and other clinicopathologic risk factors for individualized assessment of DFS and OS, both a radiomics nomogram and a clinicopathologic nomogram were developed in the training cohort. The radiomics nomogram incorporated the radiomic signature and the prognostic clinicopathologic risk factors. The clinicopathologic nomogram incorporated only the prognostic clinicopathologic risk factors. The incremental value of the radiomic signature to the TNM staging system and other clinicopathologic risk factors was assessed with respect to calibration, discrimination, reclassification, and clinical usefulness. The performance of the radiomics nomogram was compared with that of both the TNM staging system and the clinicopathologic nomogram.
To compare the predicted survival with the actual survival, calibration curves were generated. To quantify the discrimination performance, Harrell's concordance index (C-index) was measured [31]. The Akaike information criterion (AIC) was calculated to assess the risk of overfitting. Finally, a decision curve analysis determined the clinical usefulness of the radiomics nomogram by quantifying the net benefits at different threshold probabilities [32].
The overall model performance of the radiomic model was also assessed by calculating prediction errors over time from all patients by using the integrated Brier score (IBS) (prediction error curves function of the “pec” package in R software used with the “Boot- 632plus” split method with 1000 iterations) [33, 34], which represents a weighted average of the squared distances between the observed survival status and the predicted survival probability of a model and can range from 0, for a perfect model, to 0.25, for a noninformative model with a 50% incidence of the outcome. The IBS of the radiomic model was compared with the IBS of the clinicopathologic model and TNM stage.
Demographics and clinicopathologic characteristics of patients with gastric cancer.
| Variables | Training cohort (n = 132) | Validation cohort (n = 82) | ||
|---|---|---|---|---|
| N | % | N | % | |
| Gender | ||||
| Male | 88 | 66.7% | 61 | 74.4% |
| Female | 44 | 33.3% | 21 | 25.6% |
| Age (years) | ||||
| <60 | 65 | 49.2% | 47 | 57.3% |
| ≧60 | 67 | 50.8% | 35 | 42.7% |
| Tumor size (cm) | ||||
| <4 | 47 | 35.6% | 27 | 32.9% |
| ≧4 | 85 | 64.4% | 55 | 67.1% |
| Tumor location | ||||
| Upper | 47 | 35.6% | 29 | 35.4% |
| Middle | 25 | 18.9% | 9 | 11.0% |
| Lower | 46 | 34.8% | 29 | 35.4% |
| Whole | 14 | 10.6% | 15 | 18.3% |
| Differentiation status | ||||
| Well | 17 | 12.9% | 18 | 22.0% |
| Moderate | 25 | 18.9% | 10 | 12.2% |
| Poor and undifferentiated | 90 | 68.2% | 54 | 65.9% |
| Lauren type | ||||
| Intestinal type | 60 | 45.5% | 33 | 40.2% |
| Diffuse or mixed type | 72 | 54.5% | 49 | 59.8% |
| CEA | ||||
| Elevated | 19 | 14.4% | 16 | 19.5% |
| Normal | 113 | 85.6% | 66 | 80.5% |
| CA199 | ||||
| Elevated | 25 | 18.9% | 23 | 28.0% |
| Normal | 107 | 81.1% | 59 | 72.0% |
| Depth of invasion | ||||
| T1 | 29 | 22.0% | 8 | 9.8% |
| T2 | 4 | 3.0% | 10 | 12.2% |
| T3 | 10 | 7.6% | 5 | 6.1% |
| T4a | 71 | 53.8% | 49 | 59.8% |
| T4b | 18 | 13.6% | 10 | 12.2% |
| Lymph node metastasis | ||||
| N0 | 45 | 34.1% | 27 | 32.9% |
| N1 | 16 | 12.1% | 9 | 11.0% |
| N2 | 20 | 15.2% | 15 | 18.3% |
| N3a | 35 | 26.5% | 22 | 26.8% |
| N3b | 16 | 12.1% | 9 | 11.0% |
| Distant metastasis | ||||
| M0 | 116 | 87.9% | 75 | 91.5% |
| M1 | 16 | 12.1% | 7 | 8.5% |
| TNM stage | ||||
| I | 29 | 22.0% | 12 | 14.6% |
| II | 19 | 14.4% | 18 | 22.0% |
| III | 68 | 51.5% | 45 | 54.9% |
| IV | 16 | 12.1% | 7 | 8.5% |
| Chemotherapy | ||||
| No | 60 | 45.5% | 35 | 42.7% |
| Yes | 72 | 54.5% | 47 | 57.3% |
Statistical analysis
We compared two groups using the t-test for continuous variables and chi-square test for categorical variables. All radiomic features were normalized by transforming the data into new scores with a mean of 0 and a SD of 1 (z-score transformation). The Kaplan-Meier method and log-rank test were used to estimate DFS and OS. Multivariate analyses were performed using the Cox proportional hazards model. Calibration plots were generated to explore the performance characteristics of the nomograms. The decision curve analysis (DCA) was used to evaluate the clinical usefulness of the nomograms. Nomograms and calibration plots were done with the rms package of R software [35]. Statistical analysis was conducted with R software (version 3.1.0) and SPSS software (version 19.0). A two-sided P value < 0.05 was considered significant.
Results
Patient characteristics and radiomic signature construction
The clinicopathologic characteristics of patients are listed in Table 1. Of the 214 patients included in the study, 149 (69.6%) were men, and the median (interquartile range, IQR) age of all patients was 58.0 (52.5-68.0) years. Patients in the training (132 of 214 (61.7%)) and validation (82 of 214 (38.3%)) cohorts were balanced for survival, with a median (IQR) DFS and OS of 36.0 (12.0-51.25) months and 38.0 (18.75-60.0) months for the training cohort and 37.5 months (14.75-65.25) and 42.0 months (21.0-66.5) for the validation cohort (log-rank P = 0.316 and 0.716, respectively), and for baseline clinicopathologic characteristics (Table 1). The inter-scanner agreements of all metrics calculated on the basis of the reader's two measurements were good to excellent, with ICCs ranging from 0.72 to 0.98.
We used the LASSO Cox regression model to build a prognostic classifier, which selected 3 potential predictors from the 80 features identified in the training cohort (Figures S1-3). The radiomic signature was constructed, including a Rad-score calculation formula: Rad-score = -0.10385119 × Hist_Var - 0.00885129 × Hist_Entropy - 0.01904336 × LGRE_GLRLM.
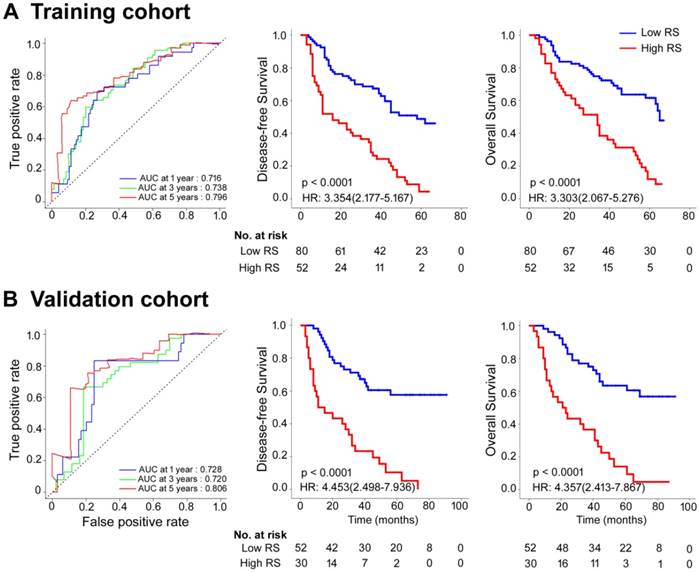
The optimum cutoff generated by the X-tile plot was 0.06 (Figure S4). Accordingly, patients were classified into a low-Rad-score group (Rad-score < 0.06) and a high-Rad-score group (Rad-score ≥ 0.06). We assessed the prognostic accuracy of the Rad-score in the training cohort using time-dependent receiver operator characteristics (ROC) analysis at different follow-up times (Figure 1A). The 5-year DFS and OS were 28.8% and 37.5%, respectively, for the low-Rad-score group and 3.8% and 9.6%, respectively, for the high-Rad-score group (hazard ratios (HRs) 3.354 (2.177-5.167) and 3.303 (2.067-5.276), all P<0.0001; Figure 1A). We then performed the same analyses in the validation cohort and similar results were observed (HRs 4.453 (2.498-7.936) and 4.357 (2.413-7.867), all P<0.0001; Figure 1B). Meanwhile, we also performed time-dependent ROC analysis and survival analysis for SUVmax, MATV, TLG, and TNM stage (Figures S5-8). Table S2 lists the relationships between the Rad-score and clinicopathological characteristics (Table S2).
Multivariate Cox regression analyses for disease-free survival and overall survival in the training, validation, and combined cohorts of patients with gastric cancer.
| Variables | Disease-free survival | Overall survival | ||
|---|---|---|---|---|
| HR (95%CI) | p | HR (95%CI) | p | |
| Training cohort | ||||
| TNM stage | 1.483 (1.247-1.763) | <0.0001 | 1.570 (1.288-1.915) | <0.0001 |
| Rad-score | 3.995 (2.560-6.234) | <0.0001 | 4.087 (2.502-6.676) | <0.0001 |
| CA199 | 1.912 (1.135-3.219) | 0.015 | 1.932 (1.108-3.369) | 0.020 |
| Validation cohort | ||||
| TNM stage | 1.739 (1.253-2.413) | 0.001 | 1.650 (1.184-2.299) | 0.003 |
| Rad-score | 5.197 (2.646-10.207) | <0.0001 | 4.942 (2.459-9.930) | <0.0001 |
| Location | 0.002 | 0.002 | ||
| Upper | Reference | Reference | ||
| Middle | 2.550 (0.841-7.735) | 0.098 | 1.757 (0.528-5.845) | 0.358 |
| Lower | 0.929 (0.420-2.055) | 0.855 | 1.171 (0.519-2.640) | 0.704 |
| Whole | 4.497 (1.956-10.341) | <0.0001 | 5.029 (2.164-11.690) | <0.0001 |
| CA199 | 2.753 (1.454-5.210) | 0.002 | 2.435 (1.289-4.601) | 0.006 |
| Training + validation cohort | ||||
| TNM stage | 1.544 (1.323-1.801) | <0.0001 | 1.553 (1.309-1.842) | <0.0001 |
| Rad-score | 4.169 (2.868-6.062) | <0.0001 | 3.954 (2.655-5.889) | <0.0001 |
| Chemotherapy | 0.638 (0.451-0.901) | 0.011 | 0.665 (0.461-0.961) | 0.030 |
| CA199 | 2.169 (1.463-3.215) | <0.0001 | 2.182 (1.446-3.294) | <0.0001 |
| Location | 0.036 | 0.040 | ||
| Upper | Reference | Reference | ||
| Middle | 1.530 (0.880-2.661) | 0.132 | 1.265 (0.690-2.320) | 0.448 |
| Lower | 1.128 (0.735-1.730) | 0.581 | 1.143 (0.730-1.792) | 0.559 |
| Whole | 2.021 (1.240-3.294) | 0.005 | 2.158 (1.297-3.591) | 0.003 |
Rad-score measured by time-dependent ROC curves and Kaplan-Meier survival in the training and validation cohorts. (A) Training cohort. (B) Validation cohort. We used AUCs at 1, 3, and 5 years to assess prognostic accuracy in the training and validation cohorts. We calculated P-values using the log-rank test. Data are AUC or P-value. AUC: area under the curve; HR: hazard ratio; ROC: receiver operator characteristic; RS: radiomic score.
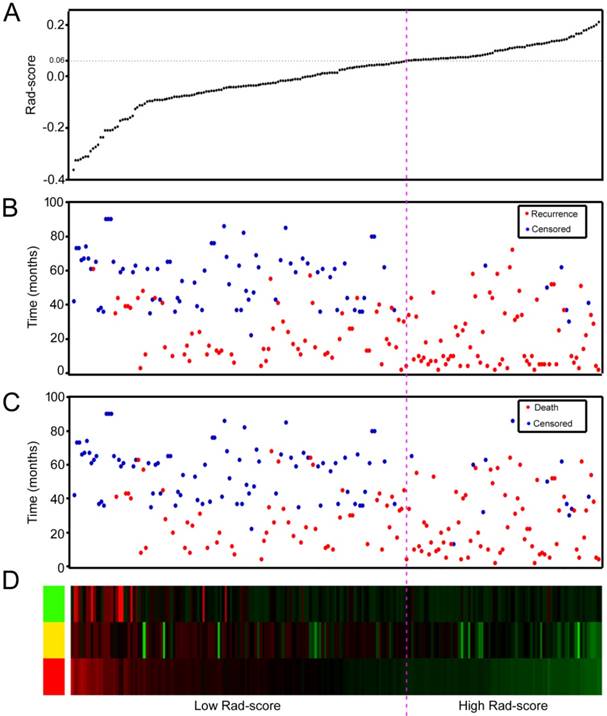
We also assessed the distribution of Rad-score, recurrence and survival statuses, and the expression of the 3 radiomic features in the training and validation cohorts (Figure 2 and Figures S9-10). Patients with higher Rad-scores were more likely to have recurrence and death. Multivariate Cox regression analysis after adjustment for clinicopathological variables revealed that the Rad-score remained a powerful and independent prognostic factor for DFS and OS in the training and validation cohorts (Table 2 and Table S3). In addition, the selected 3 features were significantly associated with DFS and OS (Figure S11); however, the AUC (area under the curves) values of the radiomic signature were higher than any single feature in the training and validation cohorts (Figure S12).
To further determine whether Rad-score could stratify patients by TNM stage, we evaluated the prognostic value of Rad-score in GC patients with TNM stage I + II and stage III + IV disease (Figure S13). High-Rad-score patients with stage I + II or stage III + IV disease had a significantly shorter DFS and OS than patients with low Rad-scores.
Assessment of incremental value of radiomic signature in individual DFS and OS performance
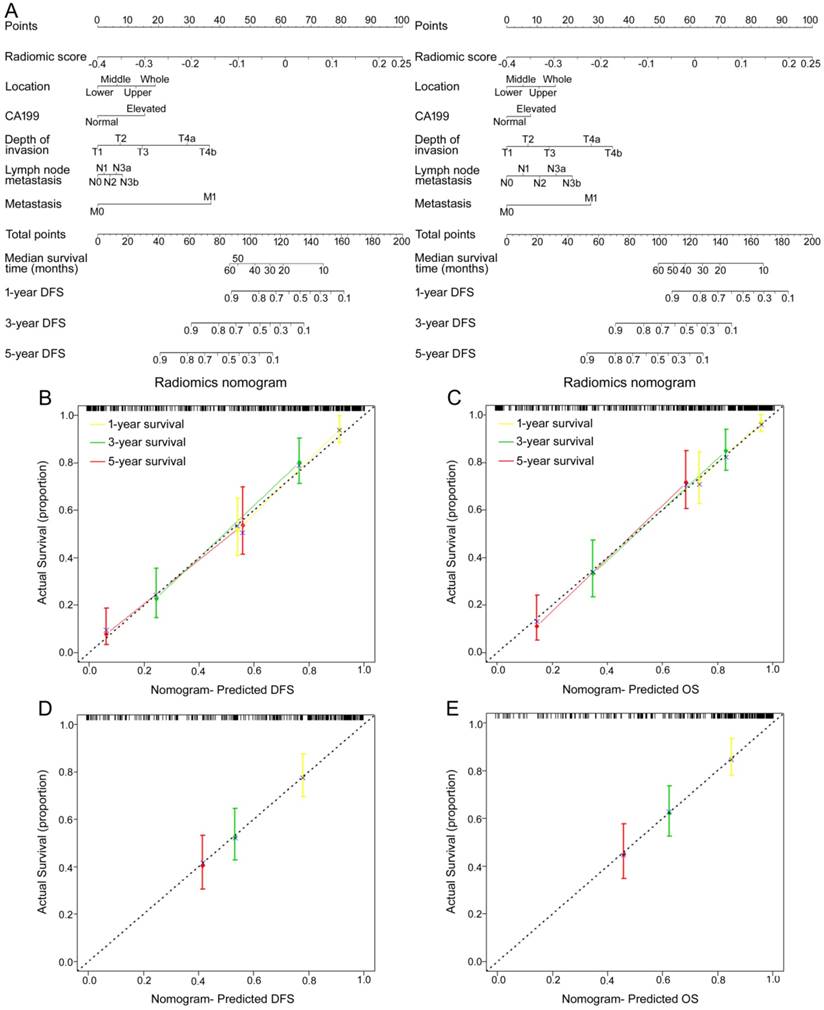
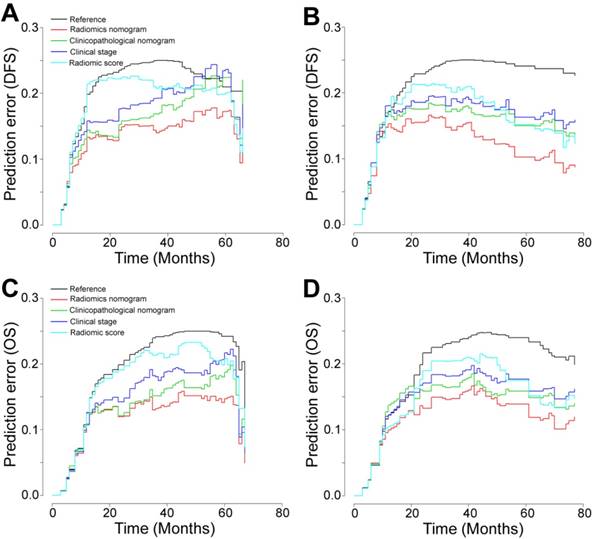
To provide the clinician with a quantitative method to predict patients' probability of 1-, 3- and 5-year DFS and OS, and to demonstrate the incremental value of the radiomic signature to the TNM staging system for individualized assessment of DFS and OS, both a radiomics nomogram and a clinicopathologic nomogram were built in the training cohort (Figure 3A and Figure S14A). We selected 5 clinicopathologic risk factors in the models, including tumor location, CA199, depth of invasion, lymph nodes metastasis, and distal metastasis, which were significantly associated with survival. The radiomics nomograms for DFS and OS are presented in Figure 3A. The calibration curves of the nomograms at 1, 3, or 5 years showed good agreement between the estimations with the radiomics nomogram and actual observations in the training and validation cohorts (Figure 3B-E). We also constructed two clinicopathologic nomograms for DFS and OS only using these clinicopathologic risk factors (Figure S14A). C-index, IBS and AIC estimates for the different models are listed in Table S4. Compared with either the TNM staging system or the clinicopathologic nomogram, the radiomics nomogram showed a better discrimination capability in the training and validation cohorts (Table S4). The radiomics nomogram, as assessed according to both the IBS (lower values indicating better model performance), the C-index (higher values indicating better discriminative ability) and AIC (lower values indicating better model performance) performed better (DFS: IBS 0.132, C-index 0.800, AIC 665.15; OS: IBS 0.117, C-index 0.786, AIC 576.61; in the training cohort) than the TNM staging system (DFS: IBS 0.169, C-index 0.717, AIC 704.44; OS: IBS 0.144, C-index 0.710, AIC 605.59) or the clinicopathologic nomogram (DFS: IBS 0.152, C-index 0.762, AIC 686.87; OS: IBS 0.128, C-index 0.761, AIC 593.37; Table S4). The corresponding prediction error curves of all Cox models are shown in Figure 4. The C-indexes of the radiomic signature to predict DFS and OS were significantly higher than those of SUVmax and MATV in the training and validation cohorts (all P<0.001 for DFS and OS in the training and validation cohorts). The C-indexes of the radiomic signature were not higher than those of TNM stage, but the C-indexes of the radiomic nomogram were significantly higher than those of TNM stage (all P<0.001 for DFS and OS in the training and validation cohorts).
Rad-score analysis of 214 GC patients in the combined training and validation cohorts (n = 214). (A) Rad-score distribution. (B) Recurrence status of GC patients. (C) Survival status of GC patients. (D) Color-gram of the expression profiles of 3 radiomic features in GC patients. Rows represent 3 radiomic features and columns represent patients. Magenta dotted line represents the Rad-score cutoff dividing the patients into high- and low-Rad-score groups.
Use of the constructed radiomics nomogram to estimate DFS and OS for GC, along with the assessment of the model calibration. (A) Radiomics nomogram to estimate DFS (left) and OS (right). To determine how many points toward the probability of DFS and OS the patient receives for his or her Rad-score, locate the patient's Rad-score on the Rad-score axis, draw a line straight upward to the point axis, repeat this process for each variable, sum the points achieved for each of the risk factors, locate the final sum on the Total Point axis, and draw a line straight down to find the patient's probability of DFS and OS. Calibration curves for the radiomics nomograms of DFS (left, (B, D)) and OS (right, (C, E)) show the calibration of each model in terms of the agreement between the estimated and the observed 1-, 3-, and 5-year outcomes. Nomogram-estimated DFS or OS is plotted on the x-axis; the observed DFS or OS is plotted on the y-axis. The diagonal dotted line is a perfect estimation by an ideal model, in which the estimated outcome perfectly corresponds to the actual outcome. The solid line is the performance of the nomogram: a closer alignment with the diagonal dotted line represents a better estimation.
Prediction error curves for each model in the study for stratifying DFS and OS in the training and validation cohorts. Prediction error curves for the (A, C) training cohort and the (B, D) validation cohort (lower prediction errors indicate higher model accuracy).
In addition, we also developed nomograms combining PET conventional metrics (SUVmax/MATV/TLG) and Rad-score with clinical features, and only combining PET conventional metrics (SUVmax/MATV/TLG) with clinical features, for DFS and OS respectively (Figures S15-16). We also found that the combination of PET conventional metrics, Rad-score, and clinical features (C-index, training cohort: DFS 0.803 (0.760-0.846), OS 0.791 (0.739-0.842); validation cohort: DFS 0.792 (0.728-0.857), OS 0.801 (0.740-0.863)) did not improve the classification results even more compared with the radiomics nomogram (training cohort: DFS P=0.460, OS P=0.461; validation cohort: DFS P=0.396, OS P=0.488). However, the C-indexes of the radiomics nomogram were higher than the C-indexes of nomograms only combining PET conventional metrics with clinical features (C-index, training cohort: DFS 0.778 (0.732-0.824), OS 0.773 (0.720-0.826); validation cohort: DFS 0.761 (0.695-0.827), OS 0.754 (0.688-0.821)).
Clinical use
The decision curve analysis showed that the radiomics nomogram had a faintly higher net benefit than the clinicopathologic nomogram and the TNM staging system across the majority range of reasonable threshold probabilities, especially for threshold probabilities 0.1-0.3 and >0.7 (Figure S17).
Radiomic signature and chemotherapy
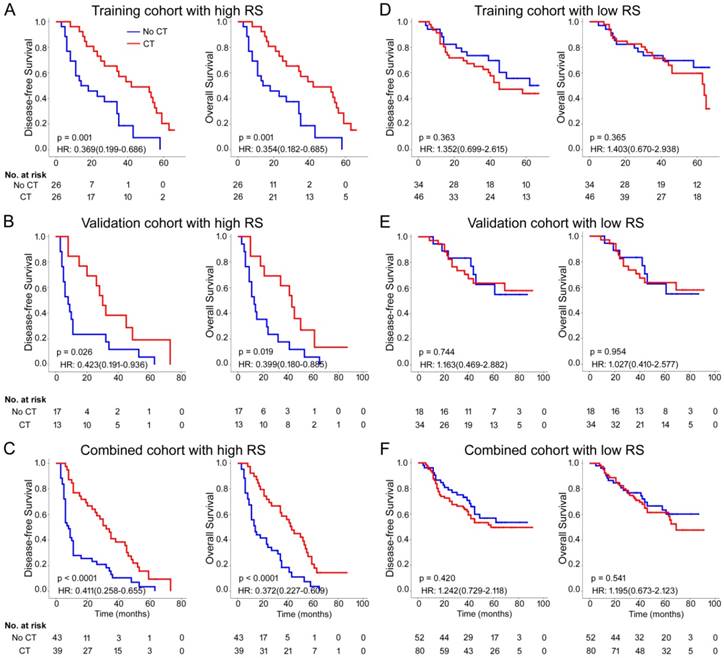
Previous data suggest that image features are associated with chemotherapy efficacy; thus, we evaluated the benefit of chemotherapy according to the level of Rad-score in patients who received postsurgical chemotherapy. For patients who did or did not receive chemotherapy, Rad-score was associated with DFS and OS in training and validation cohorts (Figure S18). High Rad-score seemingly had a greater association with the prognosis of patients who received chemotherapy than patients who did not receive chemotherapy (Figure S18). Therefore, we performed a subset analysis according to Rad-score. A test for an interaction between Rad-score and chemotherapy indicated that, both in the training and validation cohorts, the benefit from chemotherapy was greater among patients with high Rad-scores (training cohort: DFS 0.369 (0.199-0.686) HR 0.001, OS 0.354 (0.182-0.685) HR 0.001; validation cohort: DFS 0.423 (0.191-0.936) HR 0.026, OS 0.399 (0.180-0.885) HR 0.019; combined cohort: DFS 0.411 (0.258-0.655) HR <0.001, OS 0.372 (0.227-0.609) HR <0.001; all P < 0.0001 for interaction; Table 3) than among those with low Rad-scores. The corresponding Kaplan-Meier survival curves for patients with GC, which comprehensively compared low with high Rad-scores by treatment, are shown in Figure 5. The results from the subset analysis using Rad-score classifier revealed that chemotherapy significantly increased OS and DFS in the high-Rad-score group (training cohort: P=0.001 and P=0.001; validation cohort: P=0.026 and P=0.019; combined cohort: P<0.0001 and P<0.0001), but had no significant effect in the low-Rad-score group (Figure 5). Consequently, these results suggested that GC patients with high Rad-scores could benefit from chemotherapy.
Treatment interaction with Rad-score for disease-free survival and overall survival in patients with gastric cancer.
| Rad-score | CT | No CT | Disease-free survival | Overall survival | ||||
|---|---|---|---|---|---|---|---|---|
| CT vs. No CT,HR (95% CI) | P | P value for interaction | CT vs. No CT,HR (95% CI) | P | P value for interaction | |||
| Training cohort (n = 132) | ||||||||
| High Rad-score | 26 | 26 | 0.369 (0.199-0.686) | 0.002 | <0.0001 | 0.354 (0.182-0.685) | 0.002 | <0.0001 |
| Low Rad-score | 46 | 34 | 1.352 (0.699-2.615) | 0.371 | 1.403 (0.670-2.938) | 0.369 | ||
| Validation cohort (n = 82) | ||||||||
| High Rad-score | 13 | 17 | 0.423 (0.191-0.936) | 0.034 | <0.0001 | 0.399 (0.180-0.885) | 0.024 | <0.0001 |
| Low Rad-score | 34 | 18 | 1.163 (0.469-2.882) | 0.745 | 1.027 (0.410-2.577) | 0.954 | ||
| Combined cohort (n = 214) | ||||||||
| High Rad-score | 39 | 43 | 0.411 (0.258-0.655) | 0.0002 | <0.0001 | 0.372 (0.227-0.609) | 0.0001 | <0.0001 |
| Low Rad-score | 80 | 52 | 1.242 (0.729-2.118) | 0.425 | 1.195 (0.673-2.123) | 0.554 | ||
CT: chemotherapy.
Chemotherapy benefits in gastric cancer compared using disease-free survival (DFS) and overall survival (OS). Kaplan-Meier survival curves for patients with gastric cancer in different Rad-score subgroups, which were stratified by the receipt of chemotherapy. (A) Training cohort (n=132), (B) validation cohort (n=82), (C) combined cohort (n=214). CT: chemotherapy; RS: radiomic score.
Furthermore, we analyzed the association of SUVmax, MATV and TNM stage with survival benefits from chemotherapy (Figures S19-20). However, the survival benefits of chemotherapy were not obviously different for low and high SUVmax or MATV. Additionally, the signature based on SUVmax and MATV didn't significantly improve the predictive value and was not associated with survival benefits of chemotherapy (Figures S21-22).
Discussion
Accurate prognostic assessment is essential for the selection of appropriate treatment. Because GC is a clinically heterogeneous disease with large variations in clinical outcomes even among patients with the same stage [6, 36, 37], we sought to improve the prediction of GC prognosis by developing a novel radiomic signature to categorize patients into low- and high-Rad-score groups with large differences in 5-year OS and DFS. Cox regression analysis showed the radiomic signature was an independent prognostic factor for OS and DFS, even after adjustment for TNM stage and clinicopathologic characteristics. Although TNM staging is crucial to assess prognosis and establish a treatment strategy, the staging is performed mainly on the basis of anatomical information [3, 4]. In contrast, Radiomics refers to the comprehensive quantification of tumor phenotypes by applying a large number of quantitative image features, which may reflect changes of human tissues at the cellular and genetic levels and provide different information from TNM staging [11, 12, 38-41]. In stratified analyses with TNM stage, radiomic signature can distinguish patients into low- and high-risk groups with significant differences in OS and DFS, supporting the prognostic value of the signature and allowing clinicians to potentially identify candidates for systemic approaches with greater effectiveness to improve treatment outcomes. Moreover, incorporation of radiomic signature into TNM staging system could add some prognostic information to better identify patients with different outcomes, and the radiomic nomogram is a good witness. These results suggested that the radiomic signature reinforced the prognostic ability of TNM stage, thereby adding prognostic value to TNM staging. Ultimately, patients with the same stage might be classified into different risk groups on the basis of the radiomic models, and thus be treated with different approaches to improve their survival outcomes.
Current guidelines recommend advanced GC patients receive chemotherapy as a standard component, whereas respectable studies have reported that a subset of patients can't obtain benefit from current chemotherapy [2-4]. Therefore, accurately identifying candidates for chemotherapy would improve the survival rate and lead to more personalized therapy. Several studies have evaluated the potential of texture features for treatment response assessment [18, 20, 42-45]. Ahn et al. showed that CT texture analysis is useful for prediction of therapeutic response after cytotoxic chemotherapy in patients with liver metastasis from colorectal cancer [18]. Also, heterogeneous texture features on CT images were associated with better survival in HER2-positive patients who received trastuzumab-based treatment [42]. Kickingereder et al. developed a radiomic signature of MRI images for the identification of patients who may derive the most benefit from antiangiogenic therapy [43]. Recently, Jiang et al. developed a radiomic signature consisted of 19 features from CT images, which could predict which patients with stage II and III GC benefit from chemotherapy [20]. Furthermore, a multiparametric approach using fully integrated 18F-FDG PET/MRI was feasible for patients with unresectable gastric cancers [44]. In addition, K (trans) and iAUC values can be used as early predictive markers for chemotherapy response [44]. Pinker et al. deemed multiparametric 18F-FDG PET/MRI enables insights into tumor biology on multiple levels and provides information on tumor heterogeneity, which has the potential to improve planning of chemo-radiation therapy [45]. In the present study, we showed that chemotherapy provided a better survival benefit to patients classified as high Rad-score, which was developed based on multiparametric 18F-FDG PET; thus, further use of Rad-score enabled more accurate identification of patients who might benefit from chemotherapy. For patients with low Rad-scores, more effective systemic approaches to improve treatment outcomes need to be identified. Assignment of treatment based in part on tumor molecular signatures is an increasingly promising approach [3, 46, 47]. Previous studies have shown that tumor infiltrated immune cells, microRNA and gene signatures were associated with chemotherapeutic response in GC [3, 48, 49]. However, these signatures have not been widely introduced into clinical practice as initially expected due to the difficulty of pre-operative use and repeated detection, the variability of measurements in microarray assays etc., and the requirement for analytical expertise [3, 48, 49]. The radiomic signature is a reliable non-invasive approach, which could be repeatedly detected at different stages of the disease. For these reasons, non-invasive methods, such as molecular imaging, for predicting chemotherapy sensitivity have great clinical relevance; they have the potential to evaluate the effect of neoadjuvant chemotherapy. The mechanism of the relationship between radiomic features and chemotherapy has not been researched thoroughly, but it may be associated with the strong correlation of intratumor heterogeneous radiomic features with tumor cell cycling pathways and infiltration of immune cells [12, 50], and further radio-genomics studies may provide additional information and strategies for treatment [51].
Intratumor heterogeneity has been suggested to correlate with worse patient outcome and to manifest on multiple spatial scales (e.g., at molecular or genetic, cellular, and tissue levels) [6, 19, 36, 52]. In this study, the extraction of advanced PET imaging features allowed us to assess intratumor heterogeneity quantitatively on a macroscopic tissue scale. For predicting survival, the radiomic signature allowed quantification of regional heterogeneity of the PET image. Whether these radiomic imaging features have clear underlying biologic relevance must be investigated further.
Previous studies have explored the relations between SUVmax and GC outcomes. Celli et al. showed that patients with a high SUVmax demonstrated a worse overall survival, and metabolic signature of 18F-FDG PET/CT is a better predictor of biologic tumor aggressiveness than histologic signature [8]. Also, SUVmax was significantly higher in the HER2-negative group than in the HER2-positive group [53]. Furthermore, nodal SUVmax measured by preoperative 18F-FDG PET/CT is an independent prognostic factor for OS and combining nodal SUVmax with pT/pN staging can improve survival prediction precision in GC patients [9]. Additionally, a radiomic signature for either CT, PET, or PET/CT images has been identified and validated for the prediction of disease-free survival in patients with non-small cell lung cancer treated by surgery [54]. However, the prognostic value of SUVmax, SUVmean, MATV and TLG for patients with GC remains controversial [55, 56]. Both SUVmax and SUVmean can reflect the SUV values of the tumor lesions [55, 57, 58]. Although SUVmax and SUVmean are highly correlated, there are still some differences between their prognostic values [55, 57, 58]. A meta-analysis including eight studies with 1080 patients showed that there was a significant relationship between high SUVmax and poor prognosis, however, the relationship was not significant between SUVmean and poor prognosis [55]. This may be explained by that when using the SUVmean, it is more likely for us to neglect the larger SUVs among the regions of interest [55, 59]. In this study, SUVmean was not associated with survival in patients of training cohort, validation and total cohorts, respectively. Although SUVmax was also not significantly associated with survival in patients of either training cohort or validation cohort, respectively, SUVmax was significantly associated with survival in all patients (n=214) of total cohort including training and validation cohorts (DFS, P=0.036; OS, P=0.01; Table S3). MATV and TLG of tumor were also not significantly associated with DFS and OS in this study. Additionally, we found that the combination of Rad-score, PET conventional metrics (SUVmax/MATV/TLG), and clinical features did not improve the classification results even more compared with the radiomics nomogram combining Rad-score and clinical features. In addition, to our knowledge, the association between radiomic features of PET images and survival or chemotherapy benefit of GC patients has not been evaluated. In this study, we developed a radiomic feature of PET images that could successfully stratify GC patients into high- and low-Rad-score groups with significant differences in DFS and OS. The radiomics nomograms performed better than the traditional staging system and clinicopathologic nomograms, demonstrating well the incremental value of the radiomic signature for individualized DFS and OS estimation.
The limitations of this study are the multistep postprocessing workflow and statistical processing. With the use of customized high-performance and parallel computing, postprocessing time is expected to be shortened substantially in the future. Although the LASSO Cox analysis appears promising, further improvement and validation of tailored feature selection methods is required for the novel type of data that arise in radiomic analyses. The limitations of the study also include the relatively small sample size and the retrospective nature of the data collection. As the relatively small number of patients in the training cohort, we may not be able to screen out the most valuable and stable features, and the developed radiomic signature may not be the best and most effective. Clearly, we should include more patients to adjust and validate the model in prospective studies of multicenter clinical trials. Other predictive features may be included to improve performance of this model. Additionally, as more specific patients and tumor information becomes routinely collected in the future, such as genetic information and other molecular tumor markers, use of these types of predictive models will become increasingly important [3, 37, 47, 60]. Besides, in subsequent studies, we also will focus on a subgroup of patients without significant clinicopathologic predictors for survival and collect more patients to reduce overfitting, and then validate our findings from similar patients from other medical centers. Still, the radiomic signature was a powerful predictor of OS and DFS, and patients with higher Rad-scores were prone to benefit from chemotherapy, both in the training and validation cohorts. To justify the clinical usefulness, we also assessed whether the radiomic models-assisted decisions would improve patient outcomes, and decision curve analysis was applied in this study. In the future, prospective multicenter validation of the risk model proposed here will be required to confirm its value for survival stratification of patients with newly diagnosed GC. Furthermore, relationships among imaging signatures and genomic sequencing should be assessed in the future and are currently not available on the same scale as are imaging and clinical data.
In conclusion, we identified a radiomic signature that can effectively predict survival and add prognostic value to the traditional staging system. Moreover, the radiomic signature might be a useful predictive tool to predict patient benefit from chemotherapy. Additionally, the radiomics nomogram may serve as a potential tool to guide individual care.
Abbreviations
AIC: the Akaike information criterion; AUC: area under the curve; CA199: carbohydrate antigen 199; CEA: carcinoembryonic antigen; C-index: concordance index; DCA: decision curve analysis; DFS: disease-free survival; 18F: fluorine 18; FDG: fluorodeoxyglucose; GC: gastric cancer; GLCM: gray-level co-occurrence matrices; GLRLM: gray-level run length matrix; GLSZM: gray-level size zone matrix; HR: hazard ratio; IBS: integrated Brier score; ICC: intraclass correlation coefficient; IQR: interquartile range; LASSO: the least absolute shrinkage and selection operator; MATV: metabolically active tumor volume; NGTDM: neighborhood gray-tone difference matrix wavelet decompositions; OS: overall survival; OSEM: ordered-subset expectation maximization; PET: positron emission tomography; PLD: partial likelihood deviance; Rad-score: radiomic score; ROC: receiver operator characteristics; SE: standard error; SUVmax: maximum standardized uptake value; TNM: tumor node metastasis; TLG: total lesion glycolysis; VOI: volume of interest.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
This work was supported by grants from: National Natural Science Foundation of China 81672446, 81600510, 81370575, 81570593. The National Key Research and Development Program of China 2017YFC0108300. Natural Science Foundation of Guangdong Province, 2014A030313131. Science and Technology Planning Project of Guangzhou, 2014B020228003, 2014B030301041, 2015A030312013. Key Clinical Specialty Discipline Construction Program. Director's Foundation of Nanfang Hospital, 2016B010.
Authors' Contributions
G.L. and Q.W. were responsible for the study design and participated in evaluation of results. Y.J., Q.Y., Y.H., J.Y., T.L., W.Liu and H.C. participated in collection of study materials or patients. Y.J., Q.Y., W.Lv, S.X., W.H., Z.S., L.Z., L.L. and J.M. participated in collection and assembly of data. Y.J., Q.Y., W.Lv and S.X. did the data analysis and interpretation. Y.J., Q.Y., S.X. and T.L. drafted the manuscript. G.L., Q.W., J.Y., T.L., W.Liu, L.L., J.M. and L.Z. proofread the manuscript for important intellectual content. All authors contributed to manuscript preparation. All authors reviewed the report and approved the final version.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87-108
2. Noh SH, Park SR, Yang HK, Chung HC, Chung IJ, Kim SW. et al. Adjuvant capecitabine plus oxaliplatin for gastric cancer after D2 gastrectomy (CLASSIC): 5-year follow-up of an open-label, randomised phase 3 trial. Lancet Oncol. 2014;15:1389-1396
3. Jiang Y, Zhang Q, Hu Y, Li T, Yu J, Zhao L. et al. ImmunoScore signature: A prognostic and predictive tool in gastric cancer. Ann Surg. 2018;267:504-513
4. Jiang Y, Li T, Liang X, Hu Y, Huang L, Liao Z. et al. Association of adjuvant chemotherapy with survival in patients with stage II or III gastric cancer. JAMA Surg. 2017;152:e171087
5. Group G, Paoletti X, Oba K, Burzykowski T, Michiels S, Ohashi Y. et al. Benefit of adjuvant chemotherapy for resectable gastric cancer: a meta-analysis. JAMA. 2010;303:1729-1737
6. Razzak M. Genetics: new molecular classification of gastric adenocarcinoma proposed by The Cancer Genome Atlas. Nat Rev Clin Oncol. 2014;11:499
7. De Raffele E, Mirarchi M, Cuicchi D, Lecce F, Cola B. Evolving role of FDG-PET/CT in prognostic evaluation of resectable gastric cancer. World J Gastroenterol. 2017;23:6923-6926
8. Celli R, Colunga M, Patel N, Djekidel M, Jain D. Metabolic signature on 18F-FDG PET/CT, HER2 status, and survival in gastric adenocarcinomas. J Nucl Med Technol. 2016;44:234-238
9. Song BI, Kim HW, Won KS, Ryu SW, Sohn SS, Kang YN. Preoperative standardized uptake value of metastatic lymph nodes measured by 18F-FDG PET/CT improves the prediction of prognosis in gastric cancer. Medicine (Baltimore). 2015;94:e1037
10. Na SJ, o JH, Park JM, Lee HH, Lee SH, Song KY. et al. Prognostic value of metabolic parameters on preoperative 18F-fluorodeoxyglucose positron emission tomography/ computed tomography in patients with stage III gastric cancer. Oncotarget. 2016;7:63968-63980
11. Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images are more than pictures, They Are Data. Radiology. 2016;278:563-577
12. Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Carvalho S. et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5:4006
13. O'Connor JP, Aboagye EO, Adams JE, Aerts HJ, Barrington SF, Beer AJ. et al. Imaging biomarker roadmap for cancer studies. Nat Rev Clin Oncol. 2017;14:169-186
14. Kumar V, Gu YH, Basu S, Berglund A, Eschrich SA, Schabath MB. et al. Radiomics: the process and the challenges. Magnetic Resonance Imaging. 2012;30:1234-1248
15. Coroller TP, Grossmann P, Hou Y, Rios Velazquez E, Leijenaar RT, Hermann G. et al. CT-based radiomic signature predicts distant metastasis in lung adenocarcinoma. Radiother Oncol. 2015;114:345-350
16. Banerjee S, Wang DS, Kim HJ, Sirlin CB, Chan MG, Korn RL. et al. A computed tomography radiogenomic biomarker predicts microvascular invasion and clinical outcomes in hepatocellular carcinoma. Hepatology. 2015;62:792-800
17. Altazi B, Fernandez D, Zhang G, Biagioli M, Moros E. Prediction of cervical cancer treatment response using radiomics features based on F18-FDG uptake in PET images. Medical Physics. 2015;42:3326-3326
18. Ahn SJ, Kim JH, Park SJ, Han JK. Prediction of the therapeutic response after FOLFOX and FOLFIRI treatment for patients with liver metastasis from colorectal cancer using computerized CT texture analysis. Eur J Radiol. 2016;85:1867-1874
19. Huang Y, Liu Z, He L, Chen X, Pan D, Ma Z. et al. Radiomics signature: A potential biomarker for the prediction of disease-free survival in early-stage (I or II) non-small cell lung cancer. Radiology. 2016;281:947-957
20. Jiang Y, Chen C, Xie J, Wang W, Zha X, Lv W. et al. Radiomics signature of computed tomography imaging for prediction of survival and chemotherapeutic benefits in gastric cancer. EBioMedicine. 2018;36:171-182
21. Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC. et al. User-guided 3D active contour segmentation of anatomical structures: Significantly improved efficiency and reliability. Neuroimage. 2006;31:1116-1128
22. Lu L, Lv W, Jiang J, Ma J, Feng Q, Rahmim A. et al. Robustness of radiomic features in [(11)C]choline and [(18)F]FDG PET/CT imaging of nasopharyngeal carcinoma: impact of segmentation and discretization. Mol Imaging Biol. 2016;18:935-945
23. McAlinden C, Khadka J, Pesudovs K. Statistical methods for conducting agreement (comparison of clinical tests) and precision (repeatability or reproducibility) studies in optometry and ophthalmology. Ophthalmic and Physiological Optics. 2011;31:330-338
24. Zou X, Luo Y, Li Z, Hu Y, Li H, Tang H. et al. Volumetric apparent diffusion coefficient histogram analysis in differentiating intrahepatic mass-forming cholangiocarcinoma from hepatocellular carcinoma. J Magn Reson Imaging. 2018 [Epub ahead of print]
25. Leijenaar RT, Nalbantov G, Carvalho S, van Elmpt WJ, Troost EG, Boellaard R. et al. The effect of SUV discretization in quantitative FDG-PET Radiomics: the need for standardized methodology in tumor texture analysis. Sci Rep. 2015;5:11075
26. Zhang JX, Song W, Chen ZH, Wei JH, Liao YJ, Lei J. et al. Prognostic and predictive value of a microRNA signature in stage II colon cancer: a microRNA expression analysis. Lancet Oncol. 2013;14:1295-1306
27. Tibshirani R. Regression shrinkage and selection via the lasso: a retrospective. Journal of the Royal Statistical Society Series B-Statistical Methodology. 2011;73:273-282
28. Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33:1-22
29. Simon N, Friedman J, Hastie T, Tibshirani R. Regularization paths for cox's proportional hazards model via coordinate descent. J Stat Softw. 2011;39:1-13
30. Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004;10:7252-7259
31. Gonen M, Heller G. Concordance probability and discriminatory power in proportional hazards regression. Biometrika. 2005;92:965-970
32. Vickers AJ, Cronin AM, Elkin EB, Gonen M. Extensions to decision curve analysis, a novel method for evaluating diagnostic tests, prediction models and molecular markers. BMC Med Inform Decis Mak. 2008;8:53
33. Gerds TA, Schumacher M. Efron-type measures of prediction error for survival analysis. Biometrics. 2007;63:1283-1287
34. Mogensen UB, Ishwaran H, Gerds TA. Evaluating random forests for survival analysis using prediction error curves. J Stat Softw. 2012;50:1-23
35. Harrell FE Jr, Lee KL, Califf RM, Pryor DB, Rosati RA. Regression modelling strategies for improved prognostic prediction. Stat Med. 1984;3:143-152
36. McLean MH, El-Omar EM. Genetics of gastric cancer. Nat Rev Gastroenterol Hepatol. 2014;11:664-674
37. Jiang Y, Xie J, Han Z, Liu W, Xi S, Huang L. et al. Immunomarker support vector machine classifier for prediction of gastric cancer survival and adjuvant chemotherapeutic benefit. Clin Cancer Res. 2018 [Epub ahead of print]
38. Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RG, Granton P. et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48:441-446
39. Li H, Zhu Y, Burnside ES, Huang E, Drukker K, Hoadley KA. et al. Quantitative MRI radiomics in the prediction of molecular classifications of breast cancer subtypes in the TCGA/TCIA data set. NPJ Breast Cancer. 2016;2:16012
40. Yamamoto S, Huang D, Du L, Korn RL, Jamshidi N, Burnette BL. et al. Radiogenomic analysis demonstrates associations between (18)F-fluoro-2-deoxyglucose PET, prognosis, and epithelial-mesenchymal transition in non-small cell lung cancer. Radiology. 2016;280:261-270
41. Sun R, Limkin EJ, Vakalopoulou M, Dercle L, Champiat S, Han SR. et al. A radiomics approach to assess tumour-infiltrating CD8 cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: an imaging biomarker, retrospective multicohort study. Lancet Oncol. 2018;19:1180-1191
42. Yoon SH, Kim YH, Lee YJ, Park J, Kim JW, Lee HS. et al. Tumor heterogeneity in human epidermal growth factor receptor 2 (HER2)-positive advanced gastric cancer assessed by CT texture analysis: Association with survival after Trastuzumab treatment. PLoS One. 2016;11:e0161278
43. Kickingereder P, Gotz M, Muschelli J, Wick A, Neuberger U, Shinohara RT. et al. Large-scale radiomic profiling of recurrent glioblastoma identifies an imaging predictor for stratifying anti-angiogenic treatment response. Clin Cancer Res. 2016;22:5765-5771
44. Lee DH, Kim SH, Im SA, Oh DY, Kim TY, Han JK. Multiparametric fully-integrated 18-FDG PET/MRI of advanced gastric cancer for prediction of chemotherapy response: a preliminary study. Eur Radiol. 2016;26:2771-2778
45. Pinker K, Andrzejewski P, Baltzer P, Polanec SH, Sturdza A, Georg D. et al. Multiparametric [18F]fluorodeoxyglucose/ [18F]fluoromisonidazole positron emission tomography/ magnetic resonance imaging of locally advanced cervical cancer for the non-invasive detection of tumor heterogeneity: a pilot study. PLoS One. 2016;11:e0155333
46. Zhu CQ, Ding K, Strumpf D, Weir BA, Meyerson M, Pennell N. et al. Prognostic and predictive gene signature for adjuvant chemotherapy in resected non-small-cell lung cancer. J Clin Oncol. 2010;28:4417-4424
47. Jiang Y, Liu W, Li T, Hu Y, Chen S, Xi S. et al. Prognostic and predictive value of p21-activated kinase 6 associated support vector machine classifier in gastric cancer treated by 5-fluorouracil/oxaliplatin chemotherapy. EBioMedicine. 2017;22:78-88
48. Cheong JH, Yang HK, Kim H, Kim WH, Kim YW, Kook MC. et al. Predictive test for chemotherapy response in resectable gastric cancer: a multi-cohort, retrospective analysis. Lancet Oncol. 2018;19:629-638
49. Chia NY, Tan P. Molecular classification of gastric cancer. Ann Oncol. 2016;27:763-769
50. Thawani R, McLane M, Beig N, Ghose S, Prasanna P, Velcheti V. et al. Radiomics and radiogenomics in lung cancer: a review for the clinician. Lung Cancer. 2018;115:34-41
51. Mankoff DA, Farwell MD, Clark AS, Pryma DA. How imaging can impact clinical trial design molecular imaging as a biomarker for targeted cancer therapy. Cancer Journal. 2015;21:218-224
52. Giganti F, Antunes S, Salerno A, Ambrosi A, Marra P, Nicoletti R. et al. Gastric cancer: texture analysis from multidetector computed tomography as a potential preoperative prognostic biomarker. Eur Radiol. 2017;27:1831-1839
53. Chen RH, Zhou X, Liu JJ, Huang G. Relationship between F-18-FDG PET/CT findings and HER2 expression in gastric cancer. Journal of Nuclear Medicine. 2016;57:1040-1044
54. Kirienko M, Cozzi L, Antunovic L, Lozza L, Fogliata A, Voulaz E. et al. Prediction of disease-free survival by the PET/CT radiomic signature in non-small cell lung cancer patients undergoing surgery. Eur J Nucl Med Mol Imaging. 2018;45:207-217
55. Wu Z, Zhao J, Gao P, Song Y, Sun J, Chen X. et al. Prognostic value of pretreatment standardized uptake value of F-18-fluorodeoxyglucose PET in patients with gastric cancer: a meta-analysis. BMC Cancer. 2017;17:275
56. Lee JW, Lee SM, Lee MS, Shin HC. Role of 18F-FDG PET/CT in the prediction of gastric cancer recurrence after curative surgical resection. Eur J Nucl Med Mol Imaging. 2012;39:1425-1434
57. Choi J, Kim JW, Jeon TJ, Lee IJ. The 18F-FDG PET/CT response to radiotherapy for patients with spinal metastasis correlated with the clinical outcomes. PLoS One. 2018;13:e0204918
58. Liao S, Penney BC, Wroblewski K, Zhang H, Simon CA, Kampalath R. et al. Prognostic value of metabolic tumor burden on 18F-FDG PET in nonsurgical patients with non-small cell lung cancer. Eur J Nucl Med Mol Imaging. 2012;39:27-38
59. Ozturk K, Gencturk M, Caicedo-Granados E, Li F, Cayci Z. Utility of FDG PET/CT in the characterization of sinonasal neoplasms: Analysis of standardized uptake value parameters. AJR Am J Roentgenol. 2018:1-7 [Epub ahead of print]
60. Li TJ, Jiang YM, Hu YF, Huang L, Yu J, Zhao LY. et al. Interleukin-17-producing neutrophils link inflammatory stimuli to disease progression by promoting angiogenesis in gastric cancer. Clin Cancer Res. 2017;23:1575-1585
Author contact
![]() Corresponding authors: Guoxin Li, Department of General Surgery, Nanfang Hospital, Southern Medical University, 1838 North Guangzhou Avenue, Guangzhou, 510515, China; Tel and Fax: +86-20-6278 7170; E-mail: gzliguoxincom or jiangymbestcom; Quanshi Wang, Nanfang PET Center, Nanfang Hospital, Southern Medical University, 1838 Guangzhou Avenue North, Guangzhou, Guangdong Province 510515, China Tel/fax: +86-20-6164 2127; e-mail: wqslphnet; Jiang Yu, Department of General Surgery, Nanfang Hospital, Southern Medical University, 1838 North Guangzhou Avenue, Guangzhou, 510515, China; Tel and Fax: +86-20-6278 7170; E-mail: balbccom; Tuanjie Li, Department of General Surgery, Nanfang Hospital, Southern Medical University, 1838 North Guangzhou Avenue, Guangzhou, 510515, China; Tel and Fax: +86-20-6278 7170; E-mail: ehbhltjcom.
Corresponding authors: Guoxin Li, Department of General Surgery, Nanfang Hospital, Southern Medical University, 1838 North Guangzhou Avenue, Guangzhou, 510515, China; Tel and Fax: +86-20-6278 7170; E-mail: gzliguoxincom or jiangymbestcom; Quanshi Wang, Nanfang PET Center, Nanfang Hospital, Southern Medical University, 1838 Guangzhou Avenue North, Guangzhou, Guangdong Province 510515, China Tel/fax: +86-20-6164 2127; e-mail: wqslphnet; Jiang Yu, Department of General Surgery, Nanfang Hospital, Southern Medical University, 1838 North Guangzhou Avenue, Guangzhou, 510515, China; Tel and Fax: +86-20-6278 7170; E-mail: balbccom; Tuanjie Li, Department of General Surgery, Nanfang Hospital, Southern Medical University, 1838 North Guangzhou Avenue, Guangzhou, 510515, China; Tel and Fax: +86-20-6278 7170; E-mail: ehbhltjcom.
 Global reach, higher impact
Global reach, higher impact