13.3
Impact Factor
Theranostics 2018; 8(21):5855-5869. doi:10.7150/thno.27285 This issue Cite
Research Paper
A circular transcript of ncx1 gene mediates ischemic myocardial injury by targeting miR-133a-3p
1. Institute for Translational Medicine, Qingdao University, Qingdao 266021, China;
2. Department of comprehensive internal medicine, Affiliated Hospital, Qingdao University, Qingdao 266000, China;
3. School of Basic Medical Sciences, Qingdao University, Qingdao 266071, China.
Received 2018-5-16; Accepted 2018-10-6; Published 2018-11-12
Abstract
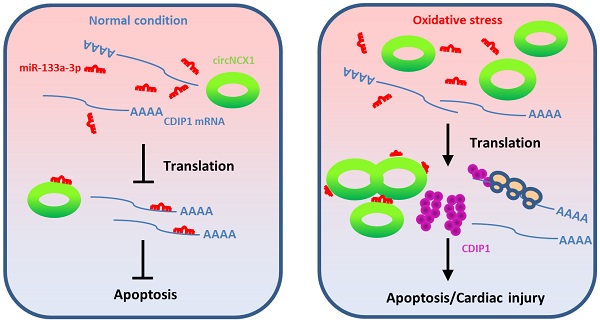
Non-coding RNAs (ncRNAs) are considered major players in physiological and pathological processes based on their versatile regulatory roles in different diseases including cardiovascular disease. Circular RNAs (circRNAs), a newly discovered class of RNAs, constitute a substantial fraction of the mammalian transcriptome and are abundantly expressed in the cardiovascular system. However, the regulatory functions of these circRNAs in ischemic cardiac disease remain largely unknown. Here, we investigated the role of a circRNA transcribed from the sodium/calcium exchanger 1 (ncx1) gene, named circNCX1, in oxidative stress-induced cardiomyocyte apoptosis during ischemic myocardial injury.
Methods: Divergent polymerase chain reaction (PCR) was conducted to amplify the circRNA. The circular structure of circNCX1 was verified by Sanger sequencing and RNase R digestion. The subcellular localization of circNCX1 was detected by fluorescence in situ hybridization (FISH). To test the expression pattern and function of circNCX1 during oxidative stress, H9c2 cells and neonatal rat cardiomyocytes were treated with H2O2 or hypoxia-reoxygenation (H/R). Mechanistically, the interaction of circNCX1 with miRNA was examined by AGO2-IP and RNA pull-down assays. The regulatory role of circNCX1 in target gene expression was tested by western blot and luciferase reporter assays. At the animal level, we constructed a myocardial ischemia-reperfusion (I/R) mouse model to analyze the effect of circNCX1 on heart function, cardiomyocyte apoptosis and cardiac remodeling.
Results: circNCX1 was increased in response to reactive oxygen species (ROS) and promotes cardiomyocyte apoptosis by acting as an endogenous miR-133a-3p sponge. Due to competitive binding of circNCX1 to miR-133a-3p, the suppressive activity of pro-apoptotic gene cell death-inducing protein (CDIP1) by miR-133a-3p was reduced. Knockdown of circNCX1 in murine cardiomyocytes and heart tissues reduced the levels of CDIP1 and attenuated the apoptosis and I/R injury.
Conclusions: Our findings reveal a novel regulatory pathway that comprises circNCX1, miR-133a-3p and CDIP1, that is involved in cardiomyocyte apoptosis. This pathway may serve as a potential therapeutic avenue for ischemic heart diseases.
Keywords: circular RNA, miR-133a, CDIP1, cardiomyocyte apoptosis, ischemic cardiomyopathy
Introduction
Cardiovascular disease is one of the leading causes of death in humans [1]. Mammalian cardiomyocytes constitute most of the heart by mass and possess negligible proliferation ability; hence, they are classified as terminally differentiated cells [1]. Once cardiomyocyte damage is inflicted by ischemia, reactive oxygen species (ROS), toxins or any other factors, dead cardiomyocytes are replaced by fibroblasts, eventually leading to remodeling, dysfunction of the ventricle, and even death [1]. Apoptosis or programmed cell death has been reported to be a pivotal form of cell death in ischemia and reperfusion injury and is considered a key modulator in the failing heart [2]. Therefore, a deeper understanding of the mechanisms of cardiomyocyte apoptosis is critical to prevent heart injury and treat heart disease. To date, multiple apoptotic signaling cascades in ischemia and reperfusion injury have been identified [1, 2]. Recently, non-coding RNAs such as microRNAs (miRNAs) and long non-coding RNAs (lncRNAs) have been reported to be involved in myocardial apoptosis [3-6]. For instance, miR-499-5p protects cardiomyocytes from apoptosis by targeting calcineurin [5], and cardiac lncRNA CARL interacts with miR-539 to regulate apoptosis [6]. However, considerably large efforts are needed to fully understand the molecular mechanism of myocardial apoptosis regarding RNA levels. Additionally, a newly discovered class of RNAs, called circRNAs, is emerging as another important modulator in the physiology and pathology of different human organs, including the heart, and remains to be explored.
circRNAs are single-stranded and covalently closed molecules [7]. They have been recently identified by RNA sequencing (RNA-Seq) technology in many species, including humans, mice, rats, zebrafish, worms, fruit flies, plants, and yeasts [8, 9]. Exonic circRNAs (ecircRNAs) and exon-intron circRNAs (eicircRNAs) are generated by the back-splicing of precursor mRNA (pre-mRNA) with 3'-5' links throughout the molecule. However, intronic circRNAs (ciRNAs) are distinct species with an independent mode of generation and the presence of a 2'-5' carbon linkage at the splicing branch point [7]. The expressions of circRNAs, which are affected by the cis-elements in the flanking introns and trans-factors of their host genes, showed a tissue- and developmental stage-specific pattern [10].
circRNAs have been reported to play critical roles in biological processes. For instance, accumulated evidence has revealed the participation of circRNAs in developmental and pathological processes such as cancer, Alzheimer's disease and neural development [11-14]. circRNAs function by serving as miRNA sponges, binding proteins and translational regulators. For example, the interaction of a circRNA, CDR1as, with miR-7 has been reported to inhibit the activity of miR-7 during brain development [9, 15]. A circular transcript of the muscleblind (mbl) gene that binds MBL has been found to suppress its function [16]. Moreover, eicircRNAs EIciEIF3 and EIciPAIP2 have been shown to associate with U1 snRNP to promote the transcription of host genes [17]. Furthermore, circRNAs possess protein-coding potential [18-21].
RNA-Seq data published recently showed that numerous circRNAs were abundant in the heart, and some circRNAs such as circHIPK3 even exhibit higher expression than the linear transcripts of their host gene [22], suggesting their roles in cardiac physiological and pathological processes [23]. It was further shown by data that among these circRNAs, some were differentially expressed during heart development or disease, implying the relationship between circRNAs and heart development and disease [24, 25]. To date, the importance of circRNAs for the heart has been gradually discovered. For instance, increased expression levels of circASXL1, circCAMSAP1 and circHIPK3 are related to fetal heart development [26]. Disorder of Ttn circRNAs generation is closely associated with dilated cardiomyopathy (DCM) [27]. Down-regulation of heart-related circRNA (HRCR) induces cardiac hypertrophy [28]. However, the function and molecular mechanisms of cardiac circRNAs remain unclear. Therefore, the regulatory functions of circRNAs in heart disease in general and ischemic cardiac disease in particular, need to be further explored.
Here, we investigated the role of circNCX1 (a circRNA transcribed from the 2nd exon of the sodium/calcium exchanger 1 (ncx1) gene) in oxidative stress-induced cardiomyocyte apoptosis during ischemic myocardial injury. It is abundantly expressed in the heart and shows differential expression levels during cardiomyopathy. Our study showed that circNCX1 is induced by apoptotic stress and promotes the expression of pro-apoptosis factor cell death-inducing protein (cdip1) by targeting miR-133a-3p, eventually leading to the death of cardiomyocytes both in vitro and in vivo. Our findings reveal a novel role for circRNA in regulating cardiomyocyte apoptosis and the potency of the circNCX1/miR-133a-3p/CDIP1 axis as a therapeutic target for the treatment of cardiovascular diseases.
Results
circNCX1 is abundant in cardiomyocytes
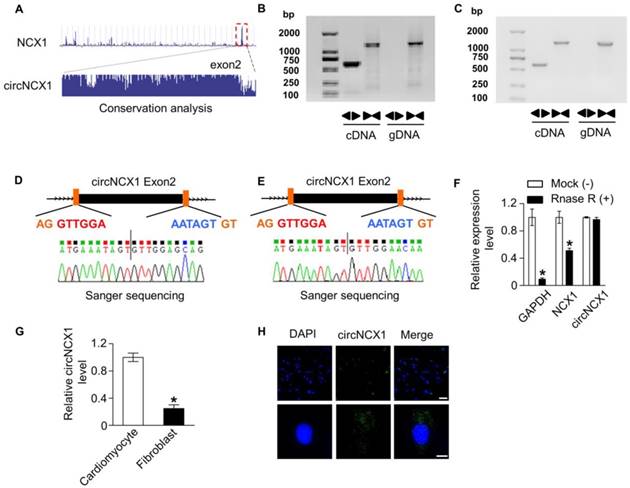
First, we screened circRNAs that were conserved and highly expressed in the heart from an online database and published data [24, 29, 30]. The heart can generate excessive ROS under pathological conditions. Hydrogen peroxide (H2O2) has been used to generate ROS to mimic ischemic cardiomyocytes death in vitro [31]. To investigate the potential involvement of these circRNAs in ROS-induced cardiomyocyte apoptosis, we analyzed their expression in H9c2 cells treated with 100 μM H2O2. Six circRNAs were detected by qRT-PCR; among them, one circRNA transcribed from the 2nd exon of the ncx1 gene was dramatically elevated upon H2O2 treatment (Figure S1A). This circRNA was highly conserved and was renamed circNCX1 (Figure 1A). We detected the expression of this transcript as circRNAs by amplifying it from the cDNA of mouse and rat myocardial tissue using the divergent primer strategy (Figure 1B-C). Furthermore, the amplified product of circNCX1 was sequenced using Sanger-sequencing to validate the circularized junction of the circRNAs (Figure 1D-E). Because circRNAs are resistant to RNase R treatment, we performed RNase R digestion assay. Both linear RNA transcripts (NCX1 and GAPDH mRNA) were significantly degraded compared with circNCX1, further validating the circular nature of the circNCX1 transcript (Figure 1F). Next, we compared the circNCX1 level in neonatal rat cardiomyocytes and cardiac fibroblasts by qRT-PCR. circNCX1 was found to be enriched in cardiomyocytes (Figure 1G). We further detected the subcellular localization of circNCX1 in H9c2 cells. circNCX1 was predominantly localized in the cytoplasm as shown by fluorescence in situ hybridization (FISH) against circNCX1 (Figure 1H).
Verification of circNCX1. (A) circNCX1 was generated from the 2nd exon of the ncx1 gene. Sequence analysis of PhyloP showed that circNCX1 is conserved. circNCX1 exists in mouse (B) and rat (C) myocardial tissue. Clear single bands were amplified from the cDNA of mouse and rat myocardial tissue by divergent primers, while they could not be amplified from gDNA. Sanger-Seq validated the head-to-tail junction of mouse (D) and rat (E) circNCX1. (F) RNAs from mouse cardiac tissue were incubated with RNase R or buffer only (Mock). After digestion, the RNAs were purified. The levels of circNCX1, NCX1 mRNA and GAPDH mRNA were analyzed by qRT-PCR. *P < 0.05 versus Mock. n=3. (G) RNAs were obtained from neonatal rat cardiomyocytes and cardiac fibroblasts. The levels of circNCX1 in cardiomyocytes and fibroblasts were analyzed by qRT-PCR. *P < 0.05 versus cardiomyocytes. n=3. (H) circNCX1 identified by specific probes are shown in green. DAPI-stained nuclei are shown in blue. Scale bars: upper, 30 μm; lower, 10 μm.
circNCX1 promotes apoptosis in cardiomyocytes
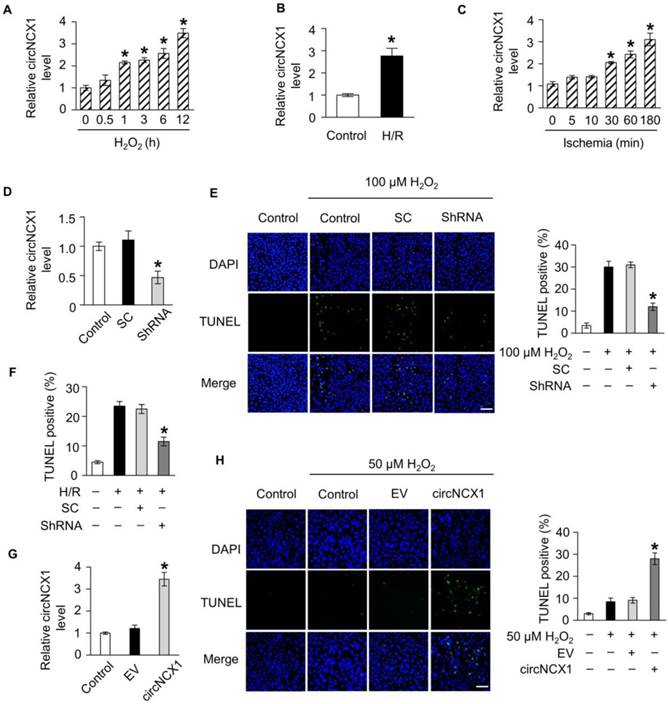
To understand the role of circNCX1 in the heart, we further detected its expression level under oxidative stress. A time-dependent increase in circNCX1 level was observed in H9c2 cells upon treatment with 100 μM H2O2 (Figure 2A). Hypoxia/reoxygenation (H/R) treatment also increased the circNCX1 level in H9c2 cells (Figure 2B). We also confirmed this finding in neonatal cardiomyocytes, and the same results were observed (Figure S2A-B). To further investigate the expression pattern of circNCX1 in the pathogenesis of cardiac infarction, we used a mouse model of myocardial infarction (MI). circNCX1 was also upregulated during myocardial ischemia in vivo (Figure 2C). Thus, circNCX1 is upregulated under pathological conditions. The increase in circNCX1 level led us to consider whether it was related to cardiomyocyte apoptosis. To verify our hypothesis, we performed a series of validation assays using loss-of-function and gain-of-function experiments. ShRNAs that specifically target the junction region of circNCX1 were utilized to knockdown circNCX1 expression in H9c2 cells (Figure 2D). Silencing of circNCX1 efficiently attenuated the apoptosis of H9c2 cells induced by H2O2 (Figure 2E) and H/R (Figure 2F). Next, we over-expressed circNCX1 in H9c2 cells successfully, as shown in Figure 2G. We found that treatment of H9c2 cells with a low concentration of H2O2 did not show any significant induction in expression levels of circNCX1 (Figure S3A) and only slightly induced apoptosis of H9c2 cells, while over-expression of circNCX1 significantly sensitized the H9c2 cells to H2O2 treatment (Figure 2H). As expected, silencing or over-expression of circNCX1 in neonatal cardiomyocytes had the same effects (Figure S3B-D). We further found that circNCX1 showed no effect on the mRNA levels of its host gene (Figure S4A-B). Taken together, these results showed that circNCX1 promotes apoptosis in cardiomyocytes.
circNCX1 promotes cardiomyocyte apoptosis. (A) H9c2 cells were treated with 100 μΜ H2O2. Total RNA was isolated and reverse-transcribed. The circNCX1 level was analyzed by qRT-PCR. *P < 0.05 versus 0 h. n=3. (B) H9c2 cells were treated with H/R. The circNCX1 level was analyzed by qRT-PCR. *P < 0.05 versus control. n=3. (C) Total RNA of mouse ischemic cardiac tissue was isolated and reverse-transcribed. The circNCX1 level was analyzed by qRT-PCR. *P < 0.05 versus 0 min. n=6. (D) H9c2 cells were transfected with the shRNA vector. The expression level of circNCX1 was analyzed by qRT-PCR. SC: scramble control; ShRNA: shRNA targeting the junction parts of circNCX1. *P < 0.05 versus SC. n=3. (E) H9c2 cells were transfected with the shRNA vector and were treated with 100 μΜ H2O2 for 12 h. Cell apoptosis was analyzed by the TUNEL assay. Representative images are shown on the left. The apoptosis rate calculated from three independent experiments is shown on the right. Green, TUNEL-positive nuclei; blue, DAPI-stained nuclei. Scale bars, 50 μm. *P < 0.05 versus SC. n=3. (F) H9c2 cells were transfected with the shRNA vector and treated with H/R. Cell apoptosis was analyzed by the TUNEL assay. *P < 0.05 versus SC. n=3. (G) H9c2 cells were transfected with the circNCX1 expression vector. The expression level of circNCX1 was analyzed by qRT-PCR. EV: empty vector. *P < 0.05 versus EV. n=3. (H) H9c2 cells were transfected with the circNCX1 overexpression vector and were treated with 50 μΜ H2O2 for 12 h. Cell apoptosis was detected by the TUNEL assay. Scale bars, 50 μm. *P < 0.05 versus EV. n=3.
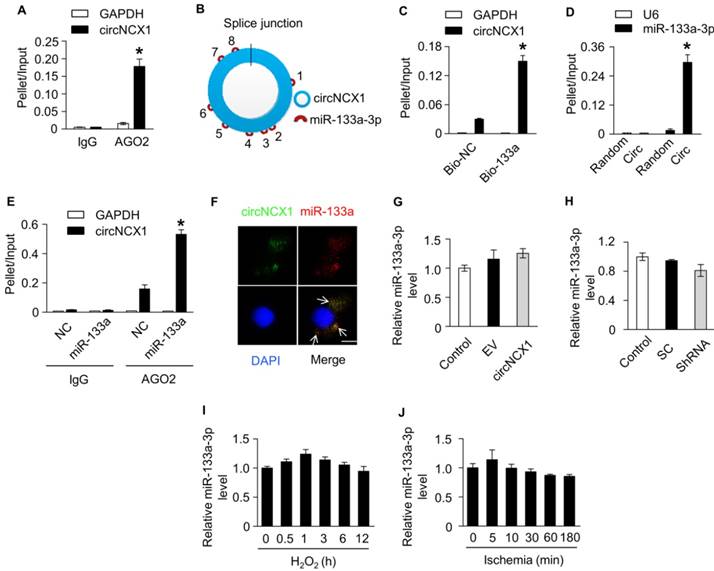
circNCX1 interacts with miR-133a-3p. (A) Immunoprecipitation of AGO2 (control, mouse IgG) in H9c2 cells was performed. The associated RNAs were purified. The levels of circNCX1 and GAPDH mRNA were analyzed by qRT-PCR. The relative pellet/ input ratios were calculated. *P < 0.05 versus GAPDH. n=3. (B) circNCX1 contains 8 potential binding sites of miR-133a-3p. (C) Biotin-labeled miR-133a-3p mimics or control mimics were transfected into H9c2 cells. The level of total and streptavidin-captured GAPDH mRNA and circNCX1 were respectively analyzed by qRT-PCR. The relative pellet/input ratios were calculated. Bio-NC: biotin-labeled control mimics. *P < 0.05 versus bio-NC. n=3. (D) The levels of total and pulled-down miR-133a-3p and U6 by circNCX1 probe or control probe were analyzed by qRT-PCR. The relative pellet/input ratios were calculated. Random: scrambled control probes; Circ: circNCX1 probes. *P < 0.05 versus Random. n=3. (E) Immunoprecipitation of AGO2 (control, mouse IgG) in miR-133a-3p or control mimics (NC)-transfected H9c2 cells was performed. The associated RNAs were purified. The levels of circNCX1 and GAPDH mRNA were analyzed by qRT-PCR. The relative pellet/input ratios were calculated. *P < 0.05 versus NC. n=3. (F) circNCX1 identified by specific probes is shown in green. miR-133a-3p is shown in red. DAPI-stained nuclei are shown in blue. Scale bar, 10 μm. (G) H9c2 cells were transfected with the circNCX1 expression vector. The expression level of miR-133a-3p was analyzed by qRT-PCR. n=3. (H) H9c2 cells were transfected with the shRNA vector. The expression level of miR-133a-3p was analyzed by qRT-PCR. n=3. (I) H9c2 cells were treated with 100 μΜ H2O2. The expression level of miR-133a-3p was analyzed by qRT-PCR. n=3. (J) The expression level of miR-133a-3p in mouse ischemic cardiac tissue was analyzed by qRT-PCR. n=6.
circNCX1 acts as a sponge for miR-133a-3p
It was reported that circRNAs containing introns largely accumulate in the nucleus and regulate gene transcription, while exonic circRNAs can act as miRNAs or protein sponges to inhibit their activities in the cytoplasm [10]. Our data showed that circNCX1 was predominantly localized in the cytoplasm. To assess whether circNCX1 acts as a sponge for miRNAs, we performed AGO2 immunoprecipitation using the lysates of H9c2 cells. Compared with IgG, the AGO2 antibody specifically enriched circNCX1, suggesting that circNCX1 might have interactions with miRNAs (Figure 3A). Bioinformatic software RNAhybrid [32] was implemented to identify hypothetical miRNA binding sites present within the sequence of circNCX1. We found that circNCX1 contains eight different predicted binding sites for miR-133a-3p, suggesting the possibility of interactions between circNCX1 and miR-133a-3p (Figure 3B and Figure S5). miR-133a-3p was found to be involved in regulating cardiac hypertrophy and heart development [33]. To validate the interactions between circNCX1 and miR-133a-3p, we carried out RNA pull-down assays and observed higher enrichment of circNCX1 in the miR-133a-3p-captured fraction compared with the negative control (Figure 3C). Moreover, we employed an inverse pull-down assay to test whether circNCX1 could bind miR-133a-3p. DNA probes specific to the junction part of circNCX1 pulled down a significantly higher amount of circNCX1 than the scrambled probes, verifying the efficiency of the probes (Figure S6). Additionally, more miR-133a-3p was detected in the circNCX1 probe-captured fraction than that from the scrambled probes (Figure 3D). The results of RNA pull-down assays demonstrated that circNCX1 and miR-133a-3p could interact with each other. AGO2-IP also demonstrated the association of miR-133a-3p with circNCX1 (Figure 3E). We also found co-localization between circNCX1 and miR-133a-3p in the cytoplasm by FISH analysis (Figure 3F). We further measured the expression levels of miR-133a-3p in H9c2 cells with overexpression (Figure 3G) or knockdown of circNCX1 (Figure 3H), but no significant change was observed in the expression levels of miR-133a-3p. Our results also showed that miR-133a-3p did not affect the expression levels of circNCX1 (Figure S7A-B). Additionally, we did not observe significant differential expression of miR-133a-3p during apoptosis induced upon treating H9c2 cells with H2O2 (Figure 3I) or in myocardial tissues during ischemia (Figure 3J). Taken together, these results showed that circNCX1 acts as a sponge for miR-133a-3p without regulating the expression levels of miR-133a-3p.
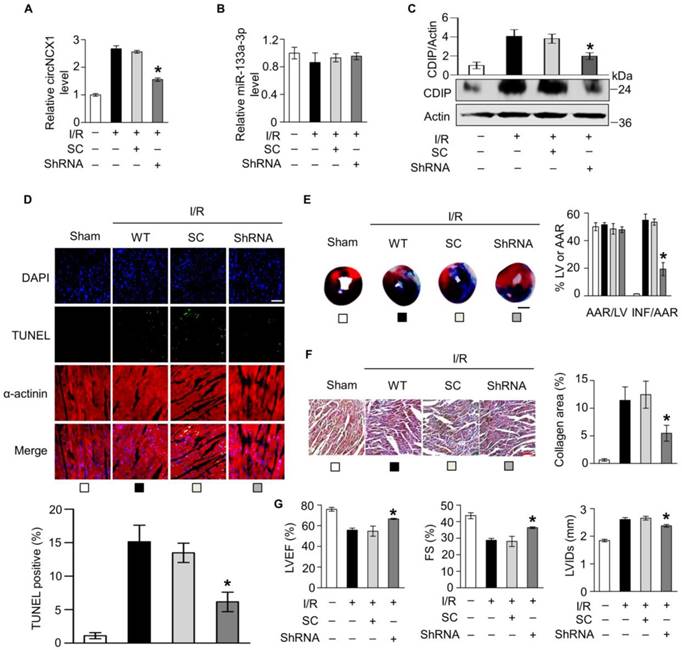
Next, we employed a miR-133a-3p inhibitor to test whether endogenous miR-133a-3p is involved in the apoptotic program of H2O2. Knockdown of miR-133a-3p sensitized cells to undergo apoptosis (Figure 4A-B). This finding indicated that miR-133a-3p can protect cardiomyocytes from apoptosis and led us to suspect that circNCX1 regulates apoptosis in cardiomyocytes by interacting with miR-133a-3p. To test this suspicion, we overexpressed circNCX1 in cells and observed that it sensitized cells to H2O2 treatment, while miR-133a-3p showed a strong inhibitory effect on apoptosis in the presence of circNCX1 (Figure 4C). Conversely, silencing of circNCX1 inhibited cell apoptosis, but this effect was significantly abolished by knockdown of miR-133a-3p (Figure 4D). These data demonstrate that increased expression levels of circNCX1 promote cardiomyocyte apoptosis by interacting with miR-133a-3p and inhibiting the activity of miR-133a-3p.
circNCX1 increases cardiomyocyte apoptosis by inhibiting miR-133a-3p. (A) H9c2 cells were transfected with the miR-133a-3p inhibitor (Inhibitor-133a) or control inhibitor (Inhibitor-nc) and were treated with 50 μM H2O2 for 12h. The levels of cleaved (Cl) and full-length (Fl) caspase-3 were analyzed by western blotting. β-Actin was selected as a reference. A representative image is shown at the bottom. The relative protein level calculated by ImageJ from three independent experiments is shown at the top. *P < 0.05 versus inhibitor-nc. (B) H9c2 cells were transfected with miR-133-3p inhibitors and were treated with 50 μM H2O2 for 12 h. Cell apoptosis was detected by the TUNEL assay. *P < 0.05 versus Inhibitor-nc. n=3. (C) circNCX1 expression vector and miR-133a-3p mimics (or control mimics) were co-transfected into H9c2 cells. Cells were treated with 50 μM H2O2 for 12 h. Cell apoptosis was detected by TUNEL assay. *P < 0.05 versus Mimic-nc. n=3. (D) circNCX1 shRNA vector and miR-133a-3p inhibitors (or control inhibitors) were co-transfected into H9c2 cells. Cells were treated with 100 μM H2O2 for 12 h. Cell apoptosis was detected by the TUNEL assay. *P < 0.05 versus inhibitor-nc. n=3.
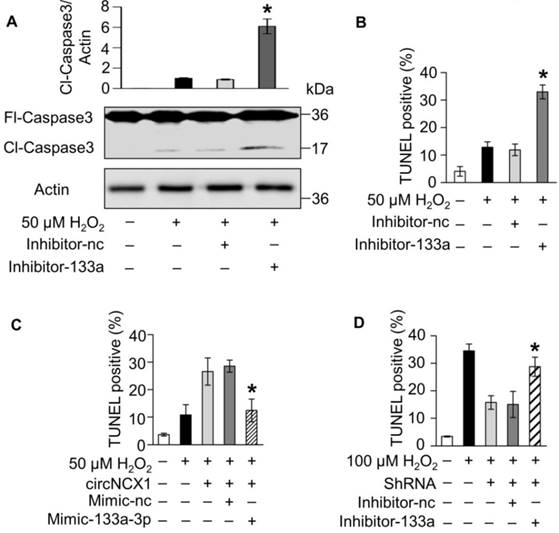
miR-133a-3p suppresses apoptosis by targeting CDIP1. (A) A conserved binding site of miR-133a-3p exists in the 3' UTR of cdip1. The expression of CDIP1 in 100 μM H2O2-treated H9c2 cells (n=3, *P < 0.05 versus 0 h) (B) and ischemic cardiac tissue (n=6, *P < 0.05 versus 0 min) (C) was analyzed by western blotting. (D) H9c2 cells were transfected with miR-133a-3p mimics and were treated with 100 μM H2O2 for 12 h. The expression of CDIP1 was analyzed by western blotting. *P < 0.05 versus Mimic-nc. n=3. (E) HEK293 cells were transfected with the pGL3 vector containing the wild type CDIP1 3' UTR or mutant 3' UTR as well as miR-133a-3p mimics or control mimics. The cells were harvested, and luciferase activity was measured. *P < 0.05 versus Mimic-nc. n=3. (F) CDIP1 siRNA or control oligo were transfected into H9c2 cells. Cells were treated with 100 μM H2O2 for 12 h. Cell apoptosis was detected by the TUNEL assay. CDIP1-sc: control oligo. *P < 0.05 versus CDIP1-sc. (G) miR-133a-3p inhibitor as well as CDIP1 siRNA or control oligo were co-transfected into H9c2 cells. Cells were treated with 50 μM H2O2 for 12 h. Cell apoptosis was detected by the TUNEL assay. *P < 0.05 versus CDIP1-sc.
miR-133a-3p protects cardiomyocytes from apoptosis by targeting CDIP1
To elucidate the molecular mechanism by which miR-133a-3p regulates apoptosis, we analyzed its potential targets using TargetScan [34] and RNAhybrid [32]. The 3' UTR of the cell death-inducing protein (CDIP1) gene harbors the conserved 8-mer seed region of miR-133a-3p (Figure 5A). CDIP1 was identified previously as an apoptotic inducer [35-37]; however, whether CDIP1 is involved in cardiomyocyte apoptosis remains to be explored. We found that the protein levels of CDIP1 were induced in H9c2 cells exposed to H2O2 treatment (Figure 5B) and mouse cardiac tissue under myocardial infarction in a time-dependent manner (Figure 5C), while the increasing level of CDIP1 protein was remarkably reduced by miR-133a-3p (Figure 5D). However, we observed no change in the mRNA expression levels of CDIP1 in miR-133a-3p mimic-transfected H9c2 cells compared with the negative control (Figure S8). These findings suggested that miR-133a-3p plays an inhibitory role in the expression of CDIP1 at the translational level during apoptosis. Next, we constructed a luciferase assay system by inserting the wild-type or miR-133a-3p binding site mutant 3'-UTR of rat cdip1 (Figure S9) between the ORF of the luciferase gene and poly-A signal and performed a luciferase assay. A significant reduction in luciferase activity was observed in the miR-133a-3p mimic-transfected groups compared with the control group, whereas this reduction was significantly abolished by mutation of the 3'-UTR (Figure 5E). These results demonstrated that miR-133a-3p could suppress the translation of CDIP1 by interacting with the 3' UTR of the cdip1 gene. Furthermore, we employed siRNAs of CDIP1 to investigate the effect of CDIP1 in the apoptotic program of H2O2 (Figure S10). Our results showed that knockdown of CDIP1 attenuates apoptosis induced by treating H9c2 cells with H2O2, indicating that CDIP1 promotes cardiomyocyte apoptosis (Figure 5F). Therefore, we hypothesized that miR-133a-3p may protects cardiomyocytes from apoptosis by inhibiting the apoptotic activity of CDIP1. As expected, it was shown that inhibition of miR-133a-3p sensitizes H9c2 cells to H2O2 exposure, while simultaneous knockdown of CDIP1 and miR-133a-3p significantly rescues the cells from apoptosis (Figure 5G). Taken together, these data revealed that miR-133a-3p protects cardiomyocytes from apoptosis by targeting pro-apoptotic CDIP1.
circNCX1 promotes cardiomyocyte apoptosis via the miR-133a-3p/CDIP1 axis
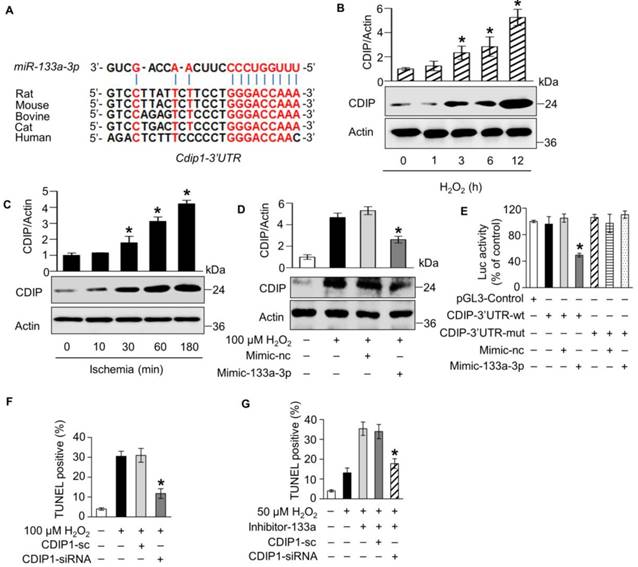
Our results showed that circNCX1 promotes cardiomyocyte apoptosis by interacting with miR-133a-3p, and miR-133a-3p showed anti-apoptotic activity in cardiomyocytes by targeting the pro-apoptotic gene cdip1. Therefore, we hypothesized that the interactions of circNCX1 with miR-133a-3p may rescue the inhibition of CDIP1 by miR-133a-3p and eventually lead to cardiomyocyte apoptosis. First, we employed the luciferase reporter assay to detect the effects of circNCX1 on the expression levels of CDIP1. The luciferase assay showed that circNCX1 could increase the luciferase activity inhibited by miR-133a-3p (Figure 6A). Moreover, over-expression of circNCX1 in H9c2 cells caused up-regulation of CDIP1 (Figure 6B); conversely, silencing of circNCX1 down-regulated CDIP1 as shown by immunoblotting assay (Figure 6C). It was further shown that the induced expression of CDIP1 by circNCX1 was attenuated by miR-133a-3p mimics (Figure 6D). Conversely, silencing of miR-133a-3p rescued the inhibition of CDIP1 levels caused by circNCX1 knockdown (Figure 6E). These results demonstrated that circNCX1 induces CDIP1 expression by inhibiting the activity of miR-133a-3p. We further assessed the relationship of circNCX1 and CDIP1 in cardiomyocyte apoptosis. Our results showed that knockdown of CDIP1 significantly reduced the apoptosis of H9c2 cells that was induced by circNCX1 (Figure 6F). These data demonstrated that circNCX1 acts as a pro-apoptotic factor by indirectly targeting CDIP1 via the sponge activity of miR-133a-3p in cardiomyocytes.
circNCX1 promotes cardiomyocyte apoptosis by increasing the CDIP1 level. (A) HEK293 cells were co-transfected with the pGL3 vector containing the 3' UTR of CDIP1, miR-133a-3p mimics and circNCX1 expression vector (or empty vector). The cells were harvested and luciferase activity was measured. *P < 0.05 versus EV. n=3. (B) H9c2 cells were transfected with the circNCX1 expression vector and were treated with 50 μM H2O2 for 12 h. The expression of CDIP1 was analyzed by western blotting. *P < 0.05 versus EV. n=3. (C) H9c2 cells were transfected with the shRNA expression vector and treated with 100 μM H2O2 for 12 h. The expression of CDIP1 was analyzed by western blotting. *P < 0.05 versus circNCX1-sc. n=3. (D) H9c2 cells were co-transfected with the circNCX1 over-expression vector and miR-133a-3p mimics. Cells were treated with 50 μM H2O2 for 12 h. The expression of CDIP1 was analyzed by western blotting. *P < 0.05 versus Mimic-nc. n=3. (E) H9c2 cells were co-transfected with the shRNA vector and miR-133a-3p inhibitors. Cells were treated with 100 μM H2O2 for 12 h. The expression of CDIP1 was analyzed by western blotting. *P < 0.05 versus Inhibitor-nc. n=3. (F) H9c2 cells were co-transfected with the circNCX1 over-expression vector and CDIP1 siRNAs. Cells were treated with 50 μM H2O2 for 12 h. Cell apoptosis was detected by the TUNEL assay. *P < 0.05 versus CDIP1-sc. n=3.
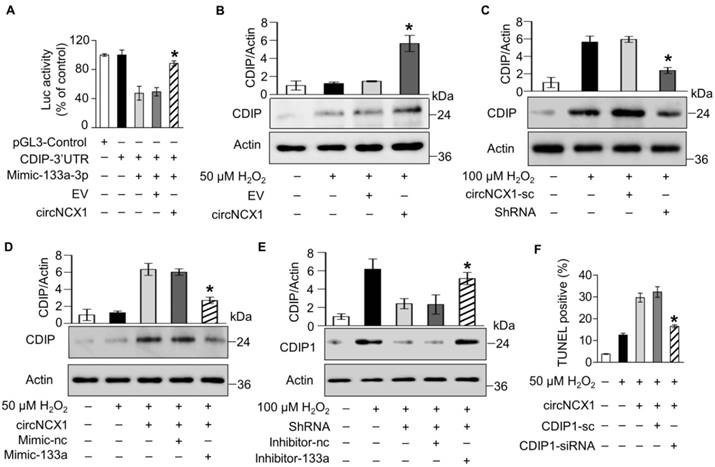
Silencing of circNCX1 in the heart attenuates cardiac ischemia-reperfusion injury. (A) Mouse hearts were infected by shRNA adenovirus. The infected mice were subjected to sham operation or I/R. The expression level of circNCX1 in mouse hearts was analyzed by qRT-PCR. *P < 0.05 versus SC. n=6. (B) The expression level of miR-133a-3p in mouse hearts was analyzed by qRT-PCR. n=6. (C) The expression level of CDIP1 in mouse hearts was analyzed by western blotting. *P < 0.05 versus SC. n=6. (D) Representative images of ventricular myocardium sections from mice exposed to sham operation or I/R. Green, TUNEL-positive nuclei; blue, DAPI-stained nuclei; red, cardiomyocytes labeled with antibody to α-actinin; scale bar, 50 μm. Quantitative analysis of apoptosis is shown at the bottom. *P < 0.05 versus SC. n=6. (E) Infarct sizes (right) and representative images of midventricular myocardial slices (left) from mice exposed to I/R or sham operation. AAR: area at risk; LV: left ventricular area; INF: infarct area. Scale bars, 2 mm. *P < 0.05 versus SC. n = 6. (F) Collagen areas of mice subjected to I/R or sham operation. Scale bars, 20 μm. *P < 0.05 versus SC. n = 6. (G) Echocardiographic analysis of left ventricular dimensions and cardiac function in mice exposed to I/R. LVIDs: systolic left ventricular internal diameters; FS: fractional shortening of the left ventricular diameter; LVEF: left ventricular ejection fraction. *P < 0.05 versus SC. n = 6.
Inhibition of circNCX1 protects the heart from ischemia-reperfusion damage
Ischemia-reperfusion (I/R) was documented to induce ROS production and apoptosis in the heart [5]. Because circNCX1 promotes apoptosis in cardiomyocytes, we next validated whether inhibition of circNCX1 could protect cardiac tissue from I/R damage in a mouse model. We knocked down the expression of circNCX1 in the mouse heart by adenoviral infection (Figure S11A). Our results showed no significant change in the expression levels of miR-133a-3p in the mouse heart of the circNCX1 knocked-down group compared with the control group (Figure S11B). Next, the mice were subjected to sham or I/R operation. Adenovirus harboring circNCX1 shRNA significantly reduced I/R-induced expression levels of circNCX1 (Figure 7A) without altering the miR-133a-3p level (Figure 7B). Our results showed that the induced expression of CDIP1 due to the up-regulation of circNCX1 during I/R damage was reversed in the circNCX1 shRNA-bearing mouse group compared with the negative control group (Figure 7C). Moreover, the loss of circNCX1 in circNCX1 shRNA-bearing mice attenuated I/R-induced cardiomyocyte apoptosis (Figure 7D) and myocardial infarction (Figure 7E). Furthermore, our results showed that silencing of circNCX1 significantly ameliorated myocardial fibrosis, as revealed by increased collagen deposition (Figure 7F) and improved cardiac function in circNCX1 shRNA-bearing mice (Figure 7G). Taken together, these results demonstrated that circNCX1 promotes myocardial I/R injury and its silencing ameliorates myocardial I/R damage in mice.
Discussion
Here, we investigated a circRNA, circNCX1, that is transcribed from the ncx1 gene and showed that it promotes cardiomyocyte apoptosis by regulating the miR-133a-3p-CDIP1 axis in vitro. Conversely, silencing of circNCX1 attenuates cardiomyocyte apoptosis in both H2O2-exposed neonatal cardiomyocytes and H9c2 cells. We further revealed that silencing of circNCX1 in mouse heart significantly ameliorates myocardial ischemia-reperfusion damage.
circRNAs usually act as miRNA sponges and are involved in miRNA-mediated post-transcriptional regulation. It was reported that the circRNA CDR1as/ciRS-7 harbors 63 conserved miR-7 binding sites and regulates brain development by actively binding miR-7 [9, 15]. Sex-determining region Y (Sry) was reported to possess 16 putative miR-138 target sites that serve as miR-138 sponge [15]. Although most circRNAs do not possess as many miRNA-binding sites as reported in CDR1as or Sry, accumulated evidence has shown that circRNAs act as miRNA sponges and regulate their function. For instance, the circRNA encoded by the hipk3 transcript harbors only two binding sites for miR-124 but can significantly inhibit miR-124 and regulate its function, suggesting that the circRNA sponge activity for microRNA binding and its regulation may not need numerous miRNA binding sites [22]. Furthermore, it remains controversial whether circRNAs are involved in post-transcriptional regulation of miRNAs. It was proven that miRNAs play regulatory roles by forming RNA-induced silencing complexes (RISC) with other ribonucleoproteins [38]. They guide Argonaute 2 (AGO2), the core subunit of RISC, to identify and silence their complementary targets [39]. The stable AGO2-miRNA complexes result in a long half-life of miRNAs [40]. Therefore, circRNAs that possess lower sequence complementarity with target miRNAs may not destabilize miRNA complexes and may not have a direct effect on the miRNA decay machinery, but they still can act as a miRNA-sponge and compete for miRNA binding with miRNA target sites to suppress miRNA activity [41]. For instance, it was reported that circMTO1 suppresses hepatocellular carcinoma progression by acting as a sponge for miR-9 in hepatocellular carcinoma tissues but without affecting the expression levels of miR-9 [12]. However, if miRNAs are highly complementary to the target transcripts, they still have a likely chance to be released from the complex [40, 42] and be degraded quickly by some AGO2-associated enzymes such as exonuclease DIS3L2 [43]. Some circRNAs have been reported to destabilize miRNA complexes due to their high complementarity for miRNAs. For instance, circRNA CDR1as have been reported to significantly down-regulate miR-671 [44]. Our study demonstrated that circNCX1 binds with miR-133a-3p to inhibit its activity without affecting the expression levels of miR-133a-3p. All eight different binding sites of miR-133a-3p in the circNCX1 sequence are partially complementary to miR-133a-3p, suggesting that circNCX1 may act as a sponge for miR-133a-3p and may not be involved in miR-133a-3p decaying activity.
Expression of miR-133a was reported previously in both heart and skeletal muscles [45, 46]. Moreover, miR-133a plays an important role in heart and muscle development, as well as in diseases including hypertrophic cardiomyopathy, DCM and heart failure [47]. However, the miR-133a upstream regulatory factors involved in cardiomyocyte apoptosis are not known yet. Our data showed no significant differential expression of miR-133a-3p during cardiomyocyte apoptosis in response to both H2O2 treatment in vitro and ischemia injury in vivo, suggesting that miR-133a might not be regulated at the transcriptional level during apoptosis. In this study, we discovered that circNCX1 interacts with miR-133a-3p to inhibit its anti-apoptotic activity, indicating a novel upstream regulatory factor of miR-133a-3p. We also suggest that circNCX1-miR-133a may play a regulatory role in cardiac development and DCM.
A study reported that miR-133a-3p inhibits cardiomyocyte apoptosis by targeting caspase-9 [48]. By contrast, caspase-9 was found to be not regulated by miR-133a at either the transcriptional or protein levels in miR-133a-deficient hearts [49]. These contradictory results added a piece to the puzzle concerning the apoptosis regulatory mechanism by miR-133a [47]. We identified CDIP1 as a new target of miR-133a-3p during apoptosis. A conserved binding region for miR-133a-3p is located in the 3' UTR of the CDIP1 gene. Our results showed that miR-133a-3p reduces apoptosis by binding to the 3' UTR of CDIP1 and suppressing the expression of CDIP1 at the translational level, acting as the downstream regulatory pathway of miR-133a-3p during apoptosis. CDIP1 is a pro-apoptotic signal transducer that links intrinsic and death receptor-mediated extrinsic apoptotic signaling [35, 36]. It up-regulates TNF-α and directs apoptosis upon cell exposure to stress. CDIP1 has also been reported to be a key signal transducer between mitochondrial apoptosis and ER stress-mediated apoptosis [37]. Moreover, CDIP1 showed abundant expression levels in the heart and brain [50]. As an important pro-apoptotic protein, CDIP1 may significantly contribute to the development of heart pathologies. However, the involvement of CDIP1 in cardiomyocyte apoptosis and myocardial injury has not been explored yet. Here, we demonstrated for the first time that CDIP1 promotes cardiomyocyte apoptosis. The knockdown of CDIP1 attenuated cardiomyocyte apoptosis. Furthermore, we revealed the novel upstream circNCX1-miR-133a-3p regulatory axis for the posttranslational regulation of CDIP1 during apoptosis in cardiomyocytes exposed to H2O2 treatment or myocardial ischemia injury.
Intriguingly, it was reported that one circRNA that acts as a miRNA sponge might be involved in other biological or pathological processes by interacting with proteins. For instance, a circRNA transcribed from the foxo3 gene sponges a handful of miRNAs to modulate tumor growth and angiogenesis [51], but it also functions by binding proteins. Additionally, due to the effect of the cellular environment on the circRNA dynamic tertiary structure, the proteins interacting with circRNA might be different in various cells, tissues, and developmental or pathological stages [52]: circFoxo3 promotes cardiac senescence by interacting with Id-1, E2F1, HIF1α, and FAK [53], while it retards cell cycle progression via forming ternary complexes with p21 and CDK2 [54]. Therefore, we suppose that circNCX1 might also play other roles. The binding partners of circNCX1 in different cells and tissues need to be further explored in a future study.
RNA binding proteins and splicing factors such as QKI [55], MBL [16], ADAR [56], DHX9 [57], HnRNP family and SR proteins [58], can regulate the biogenesis of circRNAs. In heart disease, these factors might affect the pathological process by regulating circRNA biogenesis. For example, reduction of QKI by doxorubicin could inhibit the expression of anti-apoptotic circTtn, leading to cardiomyocyte death [59]. RBM20 regulates the generation of several DCM-related Ttn circRNAs. Loss of RBM20 causes DCM [27]. We noticed that the 2nd exon of the ncx1 gene is flanked by long introns (approximately 80 kb and 100 kb respectively), implicating the broad targeting region for regulatory factors. Therefore, we suppose that the induction of circNCX1 during cardiomyocyte apoptosis is regulated by splicing factors or RNA binding proteins. The upstream regulatory roles of circNCX1 will be our future research direction.
Conclusions
In summary, this study for the first time discovered the function of circNCX1 and revealed a novel regulatory pathway that comprises circNCX1, miR-133a-3p, and CDIP1, which is involved in cardiomyocyte apoptosis. Our results demonstrated the mechanism of the circNCX1/miR-133a-3p/CDIP1 regulatory pathway, in which circNCX1 acts as a miR-133a-3p sponge by inhibiting the regulation of miR-133a-3p on CDIP1. Our results also showed that the pro-apoptotic protein CDIP1 exacerbates ischemic myocardial injury due to the loss of the translational inhibitory effect of miR-133a-3p on CDIP1 because of its competitive binding to circNCX1 in cardiomyocytes. This sheds new light on the understanding of the complex molecular mechanism of apoptotic cell death. In conclusion, the circNCX1/miR-133a-3p/CDIP1 regulatory pathway serves as a potential therapeutic avenue for ischemic heart disease.
Methods
Cell culture and treatment
Fetal cardiomyocyte-derived H9c2 cell line (American Type Culture Collection) was cultured in Dulbecco's modified Eagle's medium (GIBCO, Grand Island, NY, USA) supplemented with 10% fetal bovine serum, 100 U/mL penicillin, 100 μg/mL streptomycin and 110 mg/mL sodium pyruvate in a humidified atmosphere containing 5% CO2 at 37 °C. Cells were treated with 50 μM or 100 μM H2O2 for 12 h except as otherwise indicated. Neonatal rat cardiomyocytes and cardiac fibroblast were isolated from 2-day-old Wistar rats and prepared as previously described [5]. H/R was performed to induce apoptosis. Briefly, cells were placed in an anoxic chamber with a water-saturated atmosphere composed of 5% CO2 and 95% N2. After 6 h hypoxia, the cells were subjected to reoxygenation for 12 h.
Apoptosis assays
Apoptosis was determined by TUNEL using the TUNEL Apoptosis Detection kit (Yisheng, Shanghai, China). The detection procedures were in accordance with the kit instructions. Samples were mounted with mounting medium containing 4', 6'-diamidino-2-phenylindole (DAPI; Vector Laboratories) to stain nuclei and examined with a Zeiss LSM510 META microscope. The percentage of apoptotic nuclei was calculated by counting the total number of TUNEL-stained nuclei divided by total DAPI-positive nuclei.
Vector construction
The circNCX1 vector was synthesized as previous studies described [15]. We inserted the circNCX1 sequence along with the endogenous flanking sequence (1 kilobase (kb) upstream) into pcDNA3.1. Then, we copied part of the upstream flanking sequence and inserted it in an inverted orientation downstream.
Adenoviral constructions
The sequence of short hairpin siRNA (shRNA) for mouse circNCX1 was 5′-AATAGTGTTGGAGCAGTTGGA -3′ and that for rat circNCX1 was 5′-AATAGTGTTGGAACAATTGGA-3′. A scramble form was used as a control: 5′-ACTACCGTTGTTATAGGTG -3′. The adenoviruses harboring shRNA or their scrambled forms were constructed using the pSilencer Adeno 1.0-CMV System (Ambion, GrandIsland, NY, USA) according to the instructions provided in the kit. All constructs were amplified in HEK293 cells.
Animal experiments
8-week-old male adult C57BL/6 mice were used to perform experiments in vivo. All experiments were performed according to protocols approved by the Institute Animal Care Committee. Five days before the operation, the mice were infected with adenoviruses. circNCX1-shRNA adenoviruses (2×1010 MOI) were injected with a catheter from the apex of the LV into the aortic root, while the aorta and pulmonary arteries were cross-clamped. The clamp was maintained for 20 s while the heart pumped against a closed system. I/R operation, cardiac function, and Evan's blue dye analysis were performed as previously described [5].
RNA interference (RNAi)
Small interfering RNA (siRNA) oligonucleotides specific for CDIP1 were designed using Ambion's siRNA design tool, and purchased from GenePharma Co. Ltd (Shanghai, China). The siRNA sequences used were: CDIP1 siRNA, 5'-CCUACAUGCCUGCAGGUUUdTdT-3'; scramble CDIP1 siRNA (CDIP1-sc), 5'-GGAUUUCAAUCGCGUUGAAdTdT-3'. The specificity of the oligonucleotides was confirmed through comparing with all other sequences in Genbank using Nucleotide BLAST. Transfection of siRNAs was performed using Lipofectamine 2000 (Thermo Fisher, Waltham, MA, USA) according to the manufacturer's instructions.
Cell transfection with miRNA duplexes or inhibitors
miR-133a-3p duplexes were synthesized by GenePharma Co. Ltd. The miR-133a-3p mimic sequence was 5'-UUUGGUCCCCUUCAACCAGCUG-3'. The mimic control sequence was 5'-UUUUCCGAACGUUCACGUTT-3'. Antisense oligonucleotides (inhibitors) were used to inhibit endogenous miR-133a-3p expression. The inhibitor sequence was 5'-CAGCUGGUUGAAGGGGACCAAA-3'. The inhibitor control sequence was 5'-CAGUACUUUUGUGUAGUACAA-3'. Cells were transfected with miRNA duplexes (100 nM) or inhibitor (100 nM) using Lipofectamine 2000 (Thermo Fisher) according to the manufacturer's instructions.
Fluorescence in situ hybridization
Cardiomyocytes were grown to about 70% confluence at the time of fixation. After prehybridization (1× PBS/0.5% Triton X-100), cells were hybridized as described [22]. The sequence of the circNCX1 probe for FISH was 5′-FAM-AATTGTTCCAACACTATTTCATCATTCTGGAA-3′. The sequence of the miR-133a-3p probe was 5′-ROX-CAGCTGGTTGAAGGGGACCAAA-3′.
Divergent PCR
The head-to-tail junction part of circNCX1 was validated by PCR with divergent primers. Convergent primers were used as control. The specificity of the PCR amplification was confirmed by agarose gel electrophoresis. The sequences of mouse circNCX1 divergent primers were: forward, 5′-GGTGGAGGGGAAGACTTTG-3′; reverse, 5′-GCAGCACTTCCCACGATG-3′. The sequences of mouse circNCX1 convergent primers were: forward, 5′-CATCGTGGGAAGTGCTGC-3′; reverse, 5′-CAAAGTCTTCCCCTCCACC-3′. The sequences of rat circNCX1 divergent primers were: forward, 5′-GAGAGCATTGGCATCATGGAGGTG-3′; reverse, 5′-CATCAAGGTCAGGTTGGACACAGTC-3′. The sequences of rat circNCX1 convergent primers were: forward, 5′- GACTGTGTCCAACCTGACCTTGATG-3′; reverse, 5′- CACCTCCATGATGCCAATGCTCTC-3′.
Quantitative real-time PCR (qRT-PCR)
Stem-loop qRT-PCR for mature miRNAs was performed as previously described [28] on a CFX96 Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). Total RNA was extracted using Trizol reagent. After DNAse I (Takara, Tokyo, Japan) treatment, RNA was reverse transcribed with reverse transcriptase (Takara, Tokyo, Japan). The expression of miR-133a-3p was normalized to that of U6. The sequence of the miR-133a-3p-specific reverse-transcription primer was 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCAGCTGGT-3′. The sequences of miR-133a-3p primers were: forward, 5′-ACACTCCAGCTGGGTTTGTCCCCTTCAAC-3′; reverse, 5′-TGGTGTCGTGGAGTCG-3′. The sequences of U6 primers were: forward, 5′-CTCGCTTCGGCAGCACA-3′; reverse, 5′-AACGCTTCACGAATTTGCGT-3′. Divergent primers were designed for circNCX1. The sequences of circNCX1 primers were: forward, 5′-ATTGAAGGCACAGCCCGAGGTG-3′; reverse, 5′-CCCATTGAAACATTGGGTGGGAGAC-3′. The sequences of NCX1 mRNA primers were: forward, 5′-CCCGTCTGGTGGAGATGAGTGAGAA-3′; reverse, 5′-TTGCTGGTCAGTGGCTGCTTGTC-3′. The sequences of CDIP1 mRNA primers were: forward, 5′-GGCCATTACCACCAAGATCTCCTAC-3′; reverse, 5′-GTGCGTCACATCCTTGAAGTCATTG-3′. The sequences of GAPDH primers were: forward, 5′-GTCGTGGAGTCTACTGGCGTCTTCA-3′; reverse, 5′-TCGTGGTTCACACCCATCACAAACA-3′. The expressions of circNCX1, NCX1 and CDIP1 mRNA were normalized to that of GAPDH. The specificity of the PCR amplification was confirmed by agarose gel electrophoresis.
Reporter constructs and luciferase assay
The fragment of CDIP1 3' UTR cDNA containing the miR-133a-3p binding site was amplified by PCR. The cloning primers were: forward, 5'-CGCTTGTGCTAACAGAGCCAGACTT-3'; reverse, 5'-TGATGACACTCCTTGCCCACTTCCT-3'. The amplified products were cloned into PGL3 vector (Promega, Madison, WI, USA) immediately downstream of the stop codon of the luciferase gene. The mutant of CDIP1 was constructed using Takara MutanBEST kit (Takara, Tokyo, Japan). The primers for generating mutations were: forward, 5'-AAATGGTCAAGCACCCAGAGGCATCAACA-3'; reverse, 5'-AGGAAGAATAAGGACCAGCCTGA-3'. For luciferase assay performed in HEK-293T, cells in 24-well plates were co-transfected with 200 ng/well luciferase reporter constructs, 400 ng/well miR-133a-3p mimic or mimic control using Lipofectamine 2000. 5 ng/well SV-Renilla luciferase plasmids served as the internal control. Cells were harvested at 24 h after transfection and the luciferase activity was detected using the Dual Luciferase Reporter Assay kit (Promega) according to the manufacturer's instructions. 30 μL protein samples were analyzed in a luminometer. Firefly luciferase activities were normalized to Renilla luciferase activity.
Immunoblotting
Cells were lysed for 30 min on ice in RIPA lysis buffer (Solarbio, Beijing, China) containing 0.1 mM PMSF and a protease inhibitor (Roche). Samples were subjected to 12% SDS-PAGE and transferred to PVDF membranes. Blots were probed using the primary antibodies including anti-Caspase-3, anti-CDIP (Cell Signaling, Danvers, USA) and anti-β-actin (Santa Cruz Biotechnology). After four washes with PBS-Tween 20, horseradish peroxidase-conjugated secondary antibodies were added. The signals were detected with Pierce® ECL western blotting substrate (Pierce, Rockford, IL, USA) according to the directions of the manufacturer.
Pull-down assay with biotinylated miRNA
Pull-down assay was performed as we previously described [28]. Briefly, we synthesized miR-133a-3p and scrambled single-strand RNAs, which were 5'-biotin labeled. H9c2 cells were transfected with biotinylated miRNAs and harvested 24 h after transfection. The cells were washed with PBS followed by brief vortexing, and incubated in a lysis buffer (20 mM Tris-HCL, pH 7.5, 200 mM NaCl, 2.5 mM MgCl2, 60 U/mL RNase inhibitor, 1 mM DTT, protease inhibitor) on ice for 30 min. The lysates were precleared by centrifugation, and 50 μL of the samples was aliquoted for input. The remaining lysates were incubated with streptavidin agarose beads (Invitrogen). To prevent non-specific binding of RNA and protein complexes, the beads were coated with 1% RNase-free BSA (Sigma) and 0.5 mg/mL yeast tRNA (Sigma). The beads were incubated at 4 °C for 3 h, washed twice with ice-cold lysis buffer, three times with low-salt buffer (0.1% SDS, 1% Trition X-100, 2 mM EDTA, 20 mM Tris-HCl pH 8.0, 150 mM NaCl), and once with high-salt buffer (0.1% SDS, 1% Trition X-100, 2 mM EDTA, 20 mM Tris-HCl pH 8.0, 500 mM NaCl). The bound RNAs were purified using Trizol for the analysis. The levels of circNCX1 and GAPDH in the RNA samples were detected by qRT-PCR. GAPDH was selected as a negative control. The primer sequences for qRT-PCR detection are described above.
Pull-down assay with biotinylated circNCX1 probe
The biotinylated DNA probe complementary to the junction part of circNCX1 was synthesized and dissolved in 500 μL of wash/binding buffer (0.5M NaCl, 20 mM Tris-HCl, pH 7.5, and 1 mM EDTA). The sequence of circNCX1 probe was: 5'-bio-GAGACTTCCAATTGTTCCAACACTATTTCATCATTC-3'. The scrambled control probe was: 5'-bio-TGATGTCTAGCGCTTGGGCTTTG-3'. The probes were incubated with streptavidin-coated agarose beads at 25 °C for 2 h to generate probe-coated beads. Lysates of H9c2 cells were incubated with probe-coated beads, and after washing with the wash/binding buffer, the RNA complexes bound to the beads were eluted and extracted. miR-133a-3p and circNCX1 were analyzed by qRT-PCR using the purified RNAs as the templates. U6 and GAPDH were selected as negative controls.
AGO2 immunoprecipitation
miR-133a-3p mimics and control mimics were transfected into H9c2 cells. AGO2 immunoprecipitation was performed using an AGO2-specific antibody (Cell signaling technology), and an IgG antibody was selected as negative control. In brief, cells were lysed in 150 mM KCl, 25 mM Tris-HCl, pH 7.4, 5 mM EDTA, 0.5% Triton X-100, and 5 mM DTT supplemented with RNase inhibitor (Takara, Japan) and proteinase inhibitor cocktail (Roche Applied Science). The lysate was mixed with antibody-coupled Sepharose beads and left under rotation for 4 h at 4 °C. Beads were subsequently washed six times in lysis buffer and the RNA was extracted using Trizol reagent (Invitrogen).
Statistical analyses
Data are expressed as mean ± S.E.M. of at least three independent experiments. One-way analysis of variance was used for multiple comparisons. A value of P < 0.05 was considered significant.
Abbreviations
ADAR: double-stranded RNA-specific adenosine deaminase; AGO2: Argonaute 2; CDIP1: cell death-inducing protein 1; CDK2: cyclin-dependent kinase 2; circRNA: circular RNA; CiRNA: intronic circular RNA; DCM: dilated cardiomyopathy; DHX9: DEAH box protein 9; DIS3L2: DIS3-like exonuclease 2; DTT: dithiothreitol; E2F1: transcription factor E2F1; EcircRNA: exonic circular RNA; ECL: enhanced chemiluminescence; EDTA: ethylenediaminetetraacetic acid; EicircRNA: exon-intron circular RNA; FAK: focal adhesion kinase 1; FISH: fluorescence in situ hybridization; GAPDH: glyceraldehyde-3-phosphate dehydrogenase; HIF1α: hypoxia-inducible factor 1-alpha; HnRNP: heterogeneous nuclear ribonucleoprotein; HRCR: heart-related circular RNA; H/R: hypoxia-reoxygenation; Id-1: inhibitor of DNA binding 1; IP: immunoprecipitation; I/R: ischemia-reperfusion; LncRNA: long non-coding RNA; MBL: muscleblind; miRNA: microRNA; NcRNA: non-coding RNA; NCX1: sodium/calcium exchanger 1; p21: cyclin-dependent kinase inhibitor 1; PCR: polymerase chain reaction; Pre-mRNA: precursor mRNA; QKI: quaking; qRT-PCR: quantitative real-time polymerase chain reaction; RBM20: RNA-binding protein 20; RISC: RNA-induced silencing complexes; RNA-Seq: RNA sequencing; ROS: reactive oxygen species; SR: serine/arginine-rich-splicing factor; Sry: sex-determining region Y; TNF-α: tumor necrosis factor alpha; tRNA: transfer RNA; Ttn: titin; TUNEL: terminal deoxynucleotidyl transferase dUTP nick end labeling; UTR: untranslated region.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
This work was supported by the national natural science foundation of China (81622005), Shandong Provincial Natural Science Foundation (JQ201815), China Postdoctoral Science Foundation (2017M622140), Qingdao Postdoctoral Foundation (2016075) and Entrepreneurship and Innovation Leading Talent Program of Qingdao (13-CX-3).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Dorn GW. Apoptotic and non-apoptotic programmed cardiomyocyte death in ventricular remodeling. Cardiovasc Res. 2009;81(3):465-73
2. van Empel VP, Bertrand AT, Hofstra L, Crijns HJ, Doevendans PA, De Windt LJ. Myocyte apoptosis in heart failure. Cardiovasc Res. 2005;67(1):21-29
3. Li PF. MicroRNAs in cardiac apoptosis. J Cardiovasc Transl Res. 2010;3(3):219-224
4. Archer K, Broskova Z, Bayoumi AS, Teoh JP, Davila A, Tang YL. et al. Long Non-Coding RNAs as Master Regulators in Cardiovascular Diseases. Int J of Mol Sci. 2015;16(10):23651-67
5. Wang JX, Jiao JQ, Li Q, Long B, Wang K, Liu JP. et al. miR-499 regulates mitochondrial dynamics by targeting calcineurin and dynamin-related protein-1. Nat Med. 2011;17(1):71-78
6. Wang K, Long B, Zhou LY, Liu F, Zhou QY, Liu CY. et al. CARL lncRNA inhibits anoxia-induced mitochondrial fission and apoptosis in cardiomyocytes by impairing miR-539-dependent PHB2 downregulation. Nat Commun. 2014;5:3596
7. Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32:453-461
8. Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J. et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141-157
9. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A. et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333-338
10. Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17(4):205-211
11. Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73:5609-5612
12. Han D, Li JX, Wang HM, Su XP, Hou J, Gu Y. et al. Circular RNA mto1 acts as the sponge of mir-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66(4):1151-1164
13. Lukiw WJ. Circular RNA (circRNA) in Alzheimer's disease (AD). Front Genet. 2013;4:307
14. Venø MT, Hansen TB, Venø ST, Clausen BH, Grebing M, Finsen B. et al. Spatio-temporal regulation of circular RNA expression during porcine embryonic brain development. Genome Biol. 2015;16:245
15. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK. et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384-388
16. Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M. et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55-66
17. Li Z, Huang C, Bao C, Chen L, Lin M, Wang X. et al. Exonintron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256-264
18. Legnini I, Timoteo GD, Rossi F, Morlando M, Briganti F, Sthandier O. et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell. 2017;66:22-37
19. Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L. et al. Translation of circRNAs. Mol Cell. 2017;66:9-21
20. Yang Y, Fan XJ, Mao MW, Song XW, Wu P, Zhang Y. et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017;27:626-641
21. Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao F. et al. Novel Role of FBXW7 Circular RNA in Repressing Glioma Tumorigenesis. J Natl Cancer Inst. 2018;110(3):304-315
22. Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B. et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215
23. Li M, Ding W, Sun T, Tariq MA, Xu T, Wang JX. Biogenesis of circular RNAs and their roles in cardiovascular development and pathology. FEBS J. 2017;285(2):220-232
24. Werfel S, Nothjunge S, Schwarzmayr T, Strom TM, Meitinger T, Engelhardt S. Characterization of circular RNAs in human, mouse and rat hearts. J Mol Cell Cardiol. 2016;98:103-107
25. Devaux Y, Creemers EE, Boon RA, Werfel S, Thum T, Engelhardt S. et al. Circular RNAs in heart failure. Eur J Heart Fail. 2017;19(6):701-709
26. Szabo L, Morey R, Palpant NJ, Wang PL, Afari N, Jiang C. et al. Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol. 2015;16:126
27. Khan MA, Reckman YJ, Aufiero S, van den Hoogenhof MM, van der Made I, Beqqali A. et al. RBM20 Regulates Circular RNA Production From the Titin Gene. Circ Res. 2016;119(9):996-1003
28. Wang K, Long B, Liu F, Wang JX, Liu CY, Zhao B. et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J. 2016;37(33):2602-2611
29. Glažar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA. 2014;20(11):1666-1670
30. Tan WL, Lim BT, Anene-Nzelu CG, Ackers-Johnson M, Dashi A, See K. et al. A landscape of circular RNA expression in the human heart. Cardiovasc Res. 2017;113:298-309
31. Wang JX, Zhang XJ, Li Q, Wang K, Wang Y, Jiao JQ. et al. MicroRNA-103/107 Regulate Programmed Necrosis and Myocardial Ischemia/Reperfusion Injury Through Targeting FADD. Circ Res. 2015;117(4):352-363
32. Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10(10):1507-1517
33. Carè A, Catalucci D, Felicetti F, Bonci D, Addario A, Gallo P. et al. MicroRNA-133 controls cardiac hypertrophy. Nat Med. 2007;13(5):613-618
34. Agarwal V, Bell G W, Nam J W, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4:e05005
35. Brown L, Ongusaha PP, Kim HG, Nuti S, Mandinova A, Lee JW. et al. CDIP, a novel pro-apoptotic gene, regulates TNFalpha-mediated apoptosis in a p53-dependent manner. EMBO J. 2007;26(14):3410-3422
36. Brown-Endres L, Schoenfeld D, Tian F, Kim HG, Namba T, Muñoz-Fontela C. et al. Expression of the p53 target CDIP correlates with sensitivity to TNFα-induced apoptosis in cancer cells. Cancer Res. 2012;72(9):2373-2382
37. Namba T, Tian F, Chu K, Hwang SY, Yoon KW, Byun S. et al. CDIP1-BAP31 complex transduces apoptotic signals from endoplasmic reticulum to mitochondria under endoplasmic reticulum stress. Cell Rep. 2013;5(2):331-339
38. Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates posttranscriptional gene silencing in Drosophila cells. Nature. 2000;404:293-296
39. Rivas FV, Tolia NH, Song JJ, Aragon JP, Liu J, Hannon GJ. et al. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat Struct Mol Biol. 2005;12:340-349
40. De N, Young L, Lau PW, Meisner NC, Morrissey DV, MacRae IJ. Highly Complementary Target RNAs Promote Release of Guide RNAs from Human Argonaute2. Mol Cell. 2013;50(3):344-355
41. Denzler R, McGeary SE, Title AC, Agarwal V, Bartel DP, Stoffel M. Impact of MicroRNA Levels, Target-Site Complementarity, and Cooperativity on Competing Endogenous RNA-Regulated Gene Expression. Mol Cell. 2016;64(3):565-579
42. de la Mata M, Gaidatzis D, Vitanescu M, Stadler MB, Wentzel C, Scheiffele P. et al. Potent degradation of neuronal miRNAs induced by highly complementary targets. EMBO Rep. 2015;16(4):500-511
43. Haas G, Cetin S, Messmer M, Chane-Woon-Ming B, Terenzi O, Chicher J. et al. Identification of factors involved in target RNA-directed microRNA degradation. Nucleic Acids Res. 2016;44(6):2873-2887
44. Piwecka M, Glažar P, Hernandezmiranda LR, Memczak S, Wolf SA, Rybak-Wolf A. et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science. 2017;357(6357):eaam8526
45. Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214-220
46. Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM. et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228-233
47. Meder B, Katus HA, Rottbauer W. Right into the heart of microRNA-133a. Genes Dev. 2008;22(23):3227-3231
48. He B, Xiao J, Ren AJ, Zhang YF, Zhang H, Chen M. et al. Role of miR-1 and miR-133a in myocardial ischemic postconditioning. J Biomed Sci. 2011;18(1):22
49. Liu N, Bezprozvannaya S, Williams AH, Qi X, Richardson JA, Bassel-Duby R. et al. microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev. 2008;22(23):3242-3254
50. Bhalla K, Eyre HJ, Whitmore SA, Sutherland GR, Callen DF. C16orf5, a novel proline-rich gene at 16p13.3, is highly expressed in the brain. J Hum Genet. 1999;44(6):383-387
51. Yang W, Du WW, Li X, Yee AJ, Yang BB. Foxo3 activity promoted by non-coding effects of circular RNA and Foxo3 pseudogene in the inhibition of tumor growth and angiogenesis. Oncogene. 2016;35(30):3919-3931
52. Du WW, Zhang C, Yang W, Yong T, Awan FM, Yang BB. Identifying and Characterizing circRNA-Protein Interaction. Theranostics. 2017;7(17):4183-4191
53. Du WW, Yang W, Chen Y, Wu ZK, Foster FS, Yang Z. et al. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur Heart J. 2016;38(18):1402-1412
54. Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK. Nucleic Acids Res. 2016;44(6):2846-2858
55. Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA. et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125-1134
56. Ivanov A, Memczak S, Wyler E, Torti F, Porath HT, Orejuela MR. et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015;10:170-177
57. Aktas T, Ilık IA, Maticzka D, Bhardwaj V, Rodrigues CP, Mittler G. et al. Dhx9 suppresses rna processing defects originating from the alu invasion of the human genome. Nature. 2017;544:115-119
58. Kramer MC, Liang D, Tatomer DC, Gold B, March ZM, Cherry S. et al. Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev. 2015;29:2168-2182
59. Gupta SK, Garg A, Bär C, Chatterjee S, Foinquinos A, Milting H. et al. Quaking Inhibits Doxorubicin-Mediated Cardiotoxicity Through Regulation of Cardiac Circular RNA Expression. Circ Res. 2018;122(2):246-254
Author contact
![]() Corresponding author: Prof. Jianxun Wang, center for regenerative medicine, Institute for Translational Medicine, Qingdao University, Qingdao, 266021, China. Tel: +86-532-82991791, Fax: +86053282991791 Email: wangjxedu.cn
Corresponding author: Prof. Jianxun Wang, center for regenerative medicine, Institute for Translational Medicine, Qingdao University, Qingdao, 266021, China. Tel: +86-532-82991791, Fax: +86053282991791 Email: wangjxedu.cn
 Global reach, higher impact
Global reach, higher impact