13.3
Impact Factor
Theranostics 2018; 8(20):5772-5783. doi:10.7150/thno.28949 This issue Cite
Research Paper
An Aptamer-Based Probe for Molecular Subtyping of Breast Cancer
1. State Key Laboratory of Bioelectronics, National Demonstration Center for Experimental Biomedical Engineering Education (Southeast University), School of Biological Science and Medical Engineering, Southeast University, Nanjing 210096, P. R. China
2. School of Chemistry and Chemical Engineering, Southeast University, Nanjing 211189, P. R. China
3. Department of Pathology, Third Xiangya Hospital, Central South University, Changsha 410013, P. R. China
4. The Medical School, Ningbo University, Ningbo 315211, P. R. China
5. Economical Forest Cultivation and Utilization of 2011 Collaborative Innovation Center in Hunan Province, Hunan Key Laboratory of Biomedical Nanomaterials and Devices, Hunan University of Technology, Zhuzhou 412007, P. R. China
6. Department of Medical Imaging, Jinling Hospital, School of Medicine, Nanjing University, Nanjing 210002, P.R. China
7. State Key Laboratory of Analytical Chemistry for Life Science, School of Chemistry and Chemical Engineering, Nanjing University, Nanjing 210093, P.R. China
*These three authors contributed equally to this work.
Received 2018-8-2; Accepted 2018-10-12; Published 2018-11-10
Abstract
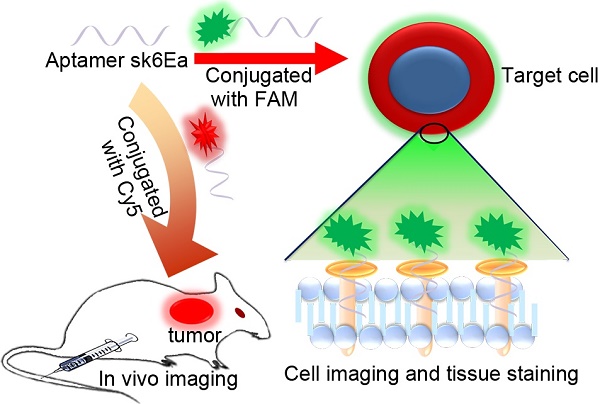
Molecular subtyping of breast cancer is of considerable interest owing to its potential for personalized therapy and prognosis. However, current methodologies cannot be used for precise subtyping, thereby posing a challenge in clinical practice. The aim of the present study is to develop a cell-specific single-stranded DNA (ssDNA) aptamer-based fluorescence probe for molecular subtyping of breast cancer.
Methods: Cell-SELEX method was utilized to select DNA aptamers. Flow cytometry and confocal microscopy were used to study the specificity, binding affinity, temperature effect on the binding ability and target type analysis of the aptamers. In vitro and in vivo fluorescence imaging were used to distinguish the molecular subtypes of breast cancer cells, tissue sections and tumor-bearing mice.
Results: Six SK-BR-3 breast cancer cell-specific ssDNA aptamers were evolved after successive in vitro selection over 21 rounds by Cell-SELEX. The Kd values of the selected aptamers were all in the low-nanomolar range, among which aptamer sk6 showed the lowest Kd of 0.61 ± 0.14 nM. Then, a truncated aptamer-based probe, sk6Ea, with only 53 nt and high specificity and binding affinity to the target cells was obtained. This aptamer-based probe was able to 1) differentiate SK-BR-3, MDA-MB-231, and MCF-7 breast cancer cells, as well as distinguish breast cancer cells from MCF-10A normal human mammary epithelial cells; 2) distinguish HER2-enriched breast cancer tissues from Luminal A, Luminal B, triple-negative breast cancer tissues, and adjacent normal breast tissues (ANBTs) in vitro; and 3) distinguish xenografts of SK-BR-3 tumor-bearing mice from those of MDA-MB-231 and MCF-7 tumor-bearing mice within 30 min in vivo.
Conclusion: The results suggest that the aptamer-based probe is a powerful tool for fast and highly sensitive subtyping of breast cancer both in vitro and in vivo and is also very promising for the identification, diagnosis, and targeted therapy of breast cancer molecular subtypes.
Keywords: aptamer, cell-SELEX, molecular subtype, breast cancer, imaging
Introduction
Breast cancer is a highly heterogeneous tumor conventionally classified into four molecular subtypes, i.e., Luminal A, Luminal B, HER2-enriched, and triple-negative breast cancer, the latter of which has become the most frequently diagnosed life-threatening cancer among women [1-5]. In clinical practice, precise molecular subtyping of breast cancer is a powerful approach for preventing its progression and plays a vital role in the delivery of personalized medicine. Currently, the most commonly used method for the clinical classification of breast cancer molecular subtypes is based on immunohistochemistry (IHC). Although IHC is the gold standard for pathological diagnosis [6], the antibodies used in IHC are highly heterogeneous, costly, and unstable, thus limiting their applications [7]. Molecular imaging can contribute to breast cancer subtyping due to its high sensitivity, ease of operation, and noninvasive nature [8-10]. However, specific probes for molecular imaging are extremely scarce, thus limiting the utility of this method for precise molecular subtyping of breast cancer. Therefore, the development of novel and highly specific molecular probes for precisely differentiating breast cancer molecular subtypes is needed.
Aptamers, often termed “chemical antibodies”, are single-stranded DNA or RNA oligonucleotides evolved from a wide range of targets, such as proteins, small molecules, ions, cells, and even tissues [11-16]. Aptamers have been widely applied in various biomedical fields, such as biosensors [17, 18], molecular imaging [19, 20], disease detection and diagnosis [21-23], drug delivery [24, 25], targeted therapy [26-28], and tissue engineering [29]. Compared with antibodies, aptamers have several advantages, such as high specificity, high binding affinity, low molecular weight, low toxicity, rapid tissue penetration, low immunogenicity, simple and reproducible synthesis, good stability, and ease of modification [30-32]. Although aptamers evolved from protein targets perform well in in vitro applications, they do not recognize native protein targets effectively in vivo owing to the fact that the protein targets used for aptamer selection are mostly purified proteins or epitope peptides [33, 34], which differ from native proteins. Cell-based systematic evolution of ligands by exponential enrichment (Cell-SELEX) is a relatively new method for developing aptamers that specifically bind to whole living cells. Although aptamers have been evolved through Cell-SELEX to recognize various kinds of cancer cells [35-42] and other cell types have been widely investigated, aptamer evolution for the molecular subtyping of human breast cancer has not been realized. Moreover, as reported aptamers were all evolved from one cancer molecular subtype through positive selection without negative counter selection or with only a single negative control cell for negative counter selection, they can only distinguish cancer molecular subtypes from control cells and cannot differentiate among cancer cells of various molecular subtypes due to the lack of specificity. For example, Gijs et al. [43] developed two novel DNA aptamers targeting the HER2 receptor using an adherent whole-cell SELEX approach with five rounds of positive selection. Although both of the aptamers were able to bind to HER2-overexpressing cells (SK-BR-3 and SKOV3 cells) and HER2-positive tumor tissue samples, they could not distinguish Luminal B and HER2-enriched breast cancer among the four breast cancer molecular subtypes very well. Moreover, protein heterogeneity can also affect aptamer specificity, thus further limiting the clinical value of aptamers. To overcome such problems, multiple negative counter selection using several cancer cells is essential to develop aptamers with excellent specificity and high binding ability to their targets for breast cancer molecular subtyping applications.
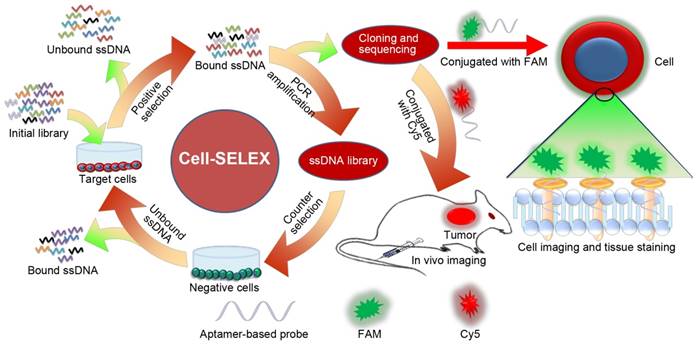
Herein, we developed an excellent cell-specific single-stranded DNA (ssDNA) aptamer-based fluorescence probe for precise molecular subtyping of breast cancer via an improved Cell-SELEX method. As shown schematically in Figure 1, SK-BR-3 breast cancer cells were chosen as the target cell, while MCF-7 and MDA-MB-231 breast cancer cells and MCF-10A human normal mammary epithelial cells were utilized as negative control cells. After 21 successive rounds of selection, six ssDNA aptamer probes capable of specifically binding to SK-BR-3 breast cancer cells were identified. The specificity and binding affinity of these aptamers were systematically investigated, demonstrating that aptamer sk6 exhibited both the best specificity and the highest binding affinity among the six aptamer candidates. Due to the fact that the recognition domain of an aptamer is usually composed of only a few nucleotides [44], aptamer sk6 was subsequently truncated and optimized. Consequently, a new aptamer probe, sk6Ea, composed of only 53 nt and exhibiting similar recognition ability to that of sk6, was obtained. The specificity, binding affinity, effects of temperature, target-type, and ability of sk6Ea to differentiate breast cancer molecular subtypes were also systematically investigated. The results indicated that the aptamer sk6Ea had higher specificity against SK-BR-3 breast cancer cells and could not only distinguish breast cancer molecular subtypes both in vitro and in vivo but also differentiate SK-BR-3 breast cancer cells from other cancer cells and normal cells when compared with other aptamers correlated with breast cancer. To the best of our knowledge, sk6Ea is the first aptamer-based probe for the molecular subtyping of breast cancer and is a promising candidate for the identification, diagnosis, and targeted therapy of breast cancer molecular subtypes.
Schematic illustration of selection of aptamers for molecular subtyping of breast cancer using Cell-SELEX method. Cell-SELEX procedures: First, the initial ssDNA pool was incubated with SK-BR-3 cells for positive selection. Then, the binding sequences were eluted and PCR-amplified for secondary pool preparation, followed by incubation with MCF-7, MDA-MB-231, and MCF-10A cells for counter selection to remove nonspecific binding sequences. Finally, sequences specifically binding to SK-BR-3 cells were eluted and PCR-amplified for DNA cloning and sequencing to obtain aptamer candidates.
Methods
Materials and reagents
SK-BR-3, MCF-7, and MDA-MB-231 human breast cancer cell lines as well as the MCF-10A human normal mammary epithelial cell line were obtained from the American Type Culture Collection. All cell lines were cultured at 37 °C in a humid atmosphere with 5% CO2. MCF-7 and SK-BR-3 cells were cultured in RPMI-1640 (Gibco, USA) supplemented with 10% fetal bovine serum (Gibco, USA). MDA-MB-231 cells were cultured in Dulbecco's modified Eagle's medium (Gibco, USA) supplemented with 10% fetal bovine serum. MCF-10A cells were cultured in MEGM (Lonza, Switzerland) supplemented with 100 ng/mL cholera toxin (Sigma, USA). The Cell-SELEX library and primers were purchased from Sangon Biotech Co., Ltd. (Shanghai, China). The library contained a randomized region of 40 nt flanked by two constant regions of 23 nt on both sides for primer annealing and PCR amplification (5′-AGCAGAGTTCACGACCCGATAAG-N40-GAGTTACATACCAATCGTCGCAG-3′). For PCR amplification, the forward primer was labeled with FAM at the 5′ end (5′-FAM-AGCAGAGTTCACGACCCGATAAG-3′) to monitor the enrichment of the library pool by flow cytometry (ACEA NovoCyteTM Series, ACEA, USA), and the reverse primer was labeled with biotin at the 5′ end (5′-biotin-CTGCGACGATTGGTATGTAACTC-3′) to isolate the sense strands using streptavidin-coated magnetic nanoparticles (SA@MNPs) for subsequent selection rounds. The SA@MNPs were synthesized according to our previous report [45]. The agarose used for electrophoretic analysis was purchased from Sangon Biotech Co., Ltd. (Shanghai, China). The binding buffer utilized in the Cell-SELEX process was prepared with phosphate-buffered saline (PBS, pH 7.4) supplemented with 4.5 g/L glucose, 5 mM MgCl2, 0.1 mg/mL yeast tRNA, and 1 mg/mL bovine serum albumin. The washing buffer was prepared with PBS supplemented with 4.5 g/L glucose and 5 mM MgCl2.
Cell-SELEX procedures
In this Cell-SELEX process, SK-BR-3 breast cancer cells were used as target cells. The initial DNA library (10 nmols) was thoroughly dissolved in 500 µL of pure water, followed by denaturation at 95 °C for 5 min and immediate cooling on ice for 10 min. Then, the initial library was uniformly mixed with 2× binding buffer before incubation with about 9×106 SK-BR-3 cells (cell viability was measured at about 92% using flow cytometry) cultured in a 100-mm cell culture dish on a rotary shaker at 4 °C for 1 h. After incubation, the cells were washed thrice with 3 mL of washing buffer. Afterwards, 500 µL of TE buffer (Sangon Biotech Co., Ltd., Shanghai China) was added, and SK-BR-3 cells were harvested with a cell scraper and transferred to a 1.5-mL centrifuge tube. Then, the target cell-binding ssDNA sequences were extracted by heating the solution in a boiling water bath for 10 min and subsequently centrifuging at 15,000 rcf for 5 min. The collected ssDNA sequences were amplified by PCR with FAM-labeled forward primer and biotin-labeled reverse primer (14-22 cycles of 5 min at 95 °C, 30 s at 95 °C, 30 s at 64 °C, and 30 s at 72 °C, followed by 5 min at 72 °C) to prepare the evolved aptamer pool. Then, electrophoretic analysis of the PCR products was performed after each Cell-SELEX round to guarantee the successful enrichment of the aptamer pool. Thereafter, the double-stranded DNA products were purified by incubation with SA@MNPs for 30 min and denatured with 0.2 M NaOH, followed by acid neutralization and desalting. The separated FAM-labeled ssDNA sequences were used as a new aptamer pool for the next round of selection.
After four rounds of successive selection with the target cells, counter selection was performed to remove nonspecific binding sequences after the fifth selection round. The aptamer pool (about 265.1 pmol) was first successively incubated with about 9×106 negative cells (MCF-7, MDA-MB-231, and MCF-10A), followed by subsequent incubation with the target SK-BR-3 cells (about 9×106) using the unbound ssDNA sequences in the supernatant solution for positive selection. To improve the specificity and binding affinity of aptamers, the positive incubation time was decreased from 60 min to 15 min, while the negative incubation time was increased from 30 min to 90 min (for incubation with all negative cells); the washing time was increased gradually from 5 min to 30 min as the number of selection rounds increased. Meanwhile, the amount of the aptamer pool used in the selection rounds was also gradually decreased from 10 nmol to 30 pmol.
Flow cytometry was used to monitor the enrichment of the selected pools. For each selected pool, 250 nM of FAM-labeled ssDNA pools were incubated with 1×105 target or negative cells in 200 µL of binding buffer at 4 °C for 15 min. After incubation, the cells were washed twice with 1 mL of washing buffer, followed by resuspension in 500 µL of washing buffer. Finally, 1×104 target cells were counted with a FACScan cytometer (ACEA NovoCyteTM Series, ACEA, USA) in triplicate to measure fluorescence intensity on the cell surface. Meanwhile, target cells incubated with the FAM-labeled initial library pool and target cells only were used as the negative control and blank control, respectively.
When the fluorescence signal of the target cells incubated with the last two pools was dramatically stronger than that of the target cells incubated with the initial pool with no further increase, the enrichment process of the library was completed. The last pool was then PCR-amplified for cloning and sequencing. In total, 30 random positive clones were selected and sequenced (Sangon Biotech Co., Ltd., Shanghai China). A multiple sequence alignment and secondary structures of the resulting 30 sequences were analyzed by DNAMAN software [12]. Finally, the aptamers were identified and chemically labeled with FAM or Cy5 for further analysis.
Selectivity analysis of the selected aptamers
The target SK-BR-3, MCF-7, and MDA-MB-231 breast cancer cells; MCF-10A human normal breast epithelial cells; HepG2 hepatocellular carcinoma cells; L02 normal hepatic cells; A549 lung cancer cells; HeLa cells; and A375 melanoma cells were cultured and incubated with the selected aptamers to investigate their specificity using a FACScan cytometer, as described above. In detail, a total of 1×105 cells were washed with cold washing buffer, followed by incubation with FAM-labeled aptamers (250 nM) in 200 µL of binding buffer at 4 °C for 15 min. Then, the cell/aptamer complexes were washed twice with 1 mL of washing buffer, followed by resuspension in 500 µL of washing buffer. Finally, 1×104 cells were counted with a FACScan cytometer (ACEA NovoCyteTM Series) to measure fluorescence intensity on the cell surface.
Binding affinity analysis of the selected aptamers
To measure the binding affinity of the selected aptamers, as determined by their dissociation constant (Kd) values, 1×105 SK-BR-3 cells were washed with cold washing buffer, followed by incubation with FAM-labeled aptamers at various concentrations in 200 µL of binding buffer at 4 °C for 15 min. After incubation, the cell/aptamer complexes were washed twice with 1 mL of washing buffer, followed by resuspension in 500 µL of washing buffer. Finally, 1×104 SK-BR-3 cells were counted with a FACScan cytometer (ACEA NovoCyteTM Series) to measure fluorescence intensity on the cell surface. All binding assays were performed in triplicate. Relative fluorescence intensity was obtained by subtracting the background fluorescence from SK-BR-3 cells alone, followed by mathematical fitting of the dependence of fluorescence intensity of SK-BR-3 cells on probe concentration according to the following equation:
Y = Bmax × X / (Kd + X),
using Origin software (OriginLab) [36].
Confocal microscopy
SK-BR-3, MCF-7, and MDA-MB-231 breast cancer cells and MCF-10A human normal breast epithelial cells were seeded in 20-mm glass-bottom cell culture dishes at 1×105 cells and cultured at 37 °C in a humid atmosphere with 5% CO2 for 24 h. After washing with cold washing buffer, the cells were incubated with FAM-labeled aptamers or library (250 nM) in 200 µL of binding buffer at 4 °C for 15 min. Then, the cells were washed thrice with 1 mL of washing buffer, followed by imaging using a Leica TCS SP8 confocal microscope (Leica, Germany).
Effect of temperature on binding ability
Since aptamer sk6Ea was selected at 4 °C, it was important to evaluate its recognition ability at room temperature and physiological temperature. Therefore, the binding affinity of aptamer sk6Ea with SK-BR-3 target cells was investigated at reaction temperatures of 4 °C, 25 °C, and 37 °C.
Target-type analysis
Although aptamer sk6Ea can specifically bind to target cells, the specific target type was unknown. Therefore, either 0.25% trypsin or 0.1 mg/mL proteinase K was used to dissociate target SK-BR-3 cells for 5 min, followed by incubation with 250 nM of aptamer sk6Ea in 200 µL of binding buffer at 4 °C for 15 min. After incubation, the cell/aptamer complexes were thoroughly washed with washing buffer and analyzed by flow cytometry.
Fluorescence staining of breast cancer tissue sections
We then investigated whether aptamer sk6Ea could identify breast cancer tissue sections of different subtypes. Formalin-fixed paraffin-embedded breast cancer tissue sections, including Luminal A, Luminal B, triple-negative/basal-like, HER2-enriched, and adjacent normal breast tissues (ANBTs), were supplied by the Third Xiangya Hospital (Changsha, China); signed informed consent was obtained from either the patient or from the next of kin. The embedded tissue sections were pretreated as described previously [46, 47]. Briefly, tissue sections were deparaffinized in xylene (3 × 10 min) and then rehydrated using a graded ethanol series (100%, 100%, 95%, 85%, and distilled water for 5 min each). After washing with PBS (pH 8.0), the hydrated tissue sections were heated in boiling EDTA buffer (0.1 M, pH 8.0) at high pressure for 2 min, followed by cooling to room temperature to retrieve antigens. Then, the prepared tissue sections were blocked with a binding buffer containing 0.05% tRNA for 60 min, followed by incubation with 250 nM FAM-labeled aptamer sk6Ea or library in 100 μL of binding buffer at 4 °C for 15 min. Afterwards, the sections were washed thrice with PBS, dehydrated, stained with DAPI, and sealed. Finally, fluorescence microscopy (Olympus BX51, Japan) was used to analyze the sections.
In vivo and ex vivo organ imaging
To evaluate the in vivo applications of aptamer sk6Ea, in vivo and ex vivo imaging of different breast cancer xenograft-bearing mice was carried out according to the animal management guidelines of the Ministry of Health of the PRC and the guidelines for the care and use of animals of the Southeast University Laboratory Animal Center. Five-week-old female athymic BALB/c (BALB/c-nude) mice were purchased from Shanghai Lingchang Biotechnology Co., Ltd (China). The nude mice were subcutaneously injected with 5×106 SK-BR-3, MCF-7, or MDA-MB-231 cells in the side of the neck. Afterwards, the tumors were allowed to grow for about 20 to 30 days until they reached 0.5-1.5 cm in diameter.
For in vivo fluorescence imaging, tumor-bearing BALB/c nude mice were anesthetized with both a tranquilizer and an anesthetic, followed by intravenous injection with 200 μL of physiological saline containing 2 nmol of Cy5-labeled aptamer sk6Ea or library via the tail vein. Then, an IVIS Lumina XRMS Series III (PerkinElmer, USA) was used to obtain fluorescence images of live mice at different time points. For ex vivo fluorescence imaging, tumor-bearing mice were intravenously injected with 2 nmol of Cy5-labeled aptamer sk6Ea or library, after which the mice were sacrificed by cervical dislocation under narcosis at 30 min post-injection. Following dissection, fluorescence images of the dissected tumor tissues were collected with the IVIS Lumina XRMS Series III in vivo imaging system, as described above.
Results
Selection of aptamers against SK-BR-3 breast cancer cells
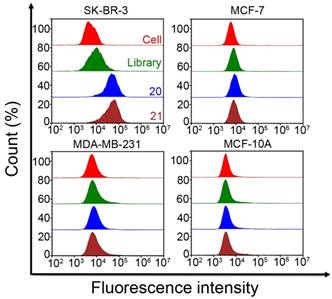
For the Cell-SELEX procedure, the initial ssDNA pool was incubated with SK-BR-3 cells alone for positive selection to ensure the binding of ssDNA sequences to the target cells. From the fifth round of selection onward, negative control cells were introduced for counter selection to remove nonspecific binding sequences, after which the unbound sequences were incubated with the target cells for positive selection. Finally, the ssDNA sequences bound to the target cells were eluted and amplified using PCR for the next round of selection. In total, 21 rounds of successive selection were conducted before the Cell-SELEX process was finally completed. The PCR result of each selection round was monitored by electrophoresis, and the electrophoretogram demonstrated that the length of the target band was about 86 bp, equal to that of the initial library (Figure S1). Meanwhile, flow cytometry was used to monitor the enrichment of the ssDNA pool. As shown in Figure 2, the fluorescence signal of the target cells incubated with FAM-labeled ssDNA pools from the 21st round was significantly stronger than that of cells incubated with the initial library and cells alone. However, the fluorescence signal was approximately constant between the 20th and 21st round of selection and did not increase thereafter. Compared with the negative control cells incubated with the FAM-labeled initial ssDNA pool, no significant increase in fluorescence intensity was observed among the negative control cells (MCF-7, MDA-MB-231, and MCF-10A) after incubation with FAM-labeled ssDNA pools from the 20th and 21st rounds. Moreover, confocal microscopy of the cells incubated with the FAM-labeled ssDNA pool from the 21st round also showed that this ssDNA pool was capable of binding to SK-BR-3 target cells with high binding affinity and specificity (Figure S2). These results demonstrated that the enrichment of the target cell-binding ssDNA pool had terminated by the 21st round of selection and that counter selection had successfully removed unbound sequences. Finally, the ssDNA pool of the 21st round was PCR-amplified, cloned, and sequenced.
In total, 30 random positive clones were selected for sequencing. A total of six ssDNA aptamer probes with a length of 86 nt were identified using multiple sequence alignment analysis by DNAMAN software. Considering the important role that secondary structures play in the recognition capacity of aptamers [48, 49], the secondary structures of the six selected aptamer probes were also analyzed and found to be composed of various stem-loop and hairpin structures (Figure S3). Probes sk1, sk2, sk3, sk4, sk5, and sk6 accounted for 50%, 3.3%, 13.3%, 16.7%, 13.3%, and 3.3% of all positive clones, respectively (Table 1). All six probes were then synthesized and labeled with FAM for further study and analysis.
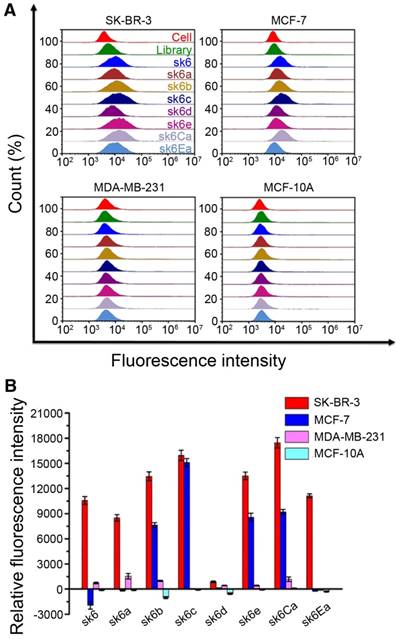
Selectivity of the selected aptamer candidates
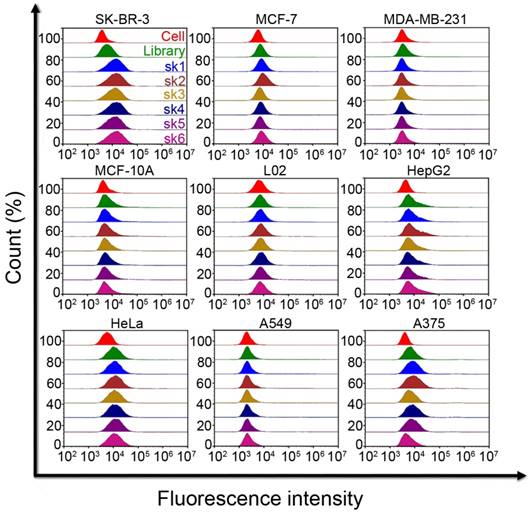
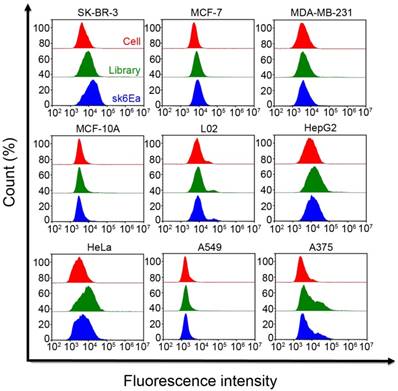
The selectivity of each of the six probes was determined by flow cytometry and confocal microscopy. First, the specificities of the probes to breast cancer cells were investigated. All six FAM-labeled probes were incubated with SK-BR-3, MCF-7, MDA-MB-231, and MCF-10A breast cancer cells individually, followed by monitoring of the fluorescence signal using flow cytometry and confocal microscopy. The FAM-labeled initial library was utilized as a negative control under the same conditions. When incubated with SK-BR-3 cells, all six FAM-labeled probes exhibited markedly higher fluorescence intensities than that of the FAM-labeled initial library. By contrast, when incubated with MCF-7, MDA-MB-231, and MCF-10A cells, all six FAM-labeled aptamers showed fluorescence intensities similar to that of the initial library, indicating significantly weaker binding compared to that of SK-BR-3 cells (Figure 3 and Figure S4-S7). These results indicate the high specificity of the selected aptamer probes for SK-BR-3 target cells, while the probes bound only weakly or not at all to the negative control MCF-7, MDA-MB-231, and MCF-10A cells. To further investigate the specificity of the six probes in the presence of other cancer cells, five other tumor cells, including HepG2 hepatocellular carcinoma cells, L02 normal hepatic cells, A549 lung cancer cells, HeLa cells, and A375 melanoma cells, were incubated with all FAM-labeled aptamers individually. Again, the fluorescence intensities of the cell-aptamer complexes were monitored by flow cytometry. Probes sk1, sk2, sk4, and sk5 all showed some nonspecific binding to A375 cancer cells compared with the initial library, while probes sk3 and sk6 showed almost no binding to all eight types of negative control cells (Figure 3). These results indicated that probes sk3 and sk6 exhibited higher specificity for SK-BR-3 target cells than for other aptamer candidates, indicating their potential for discriminating SK-BR-3 breast cancer cells from other breast cancer cell subtypes and other cancer cells.
Sequences of identified aptamers.
| Aptamer | Sequence (5'-3') | Rate |
|---|---|---|
| sk1 | AGCAGAGTTCACGACCCGATAAGGGGAAAACACATTTCGGATCTCCGTTGTCGCCCTGATACCGAGTTACATACCAATCGTCGCAG | 50% |
| sk2 | AGCAGAGTTCACGACCCGATAAGGGCGATGCCGATCCCTGTGGCCGTAGGGACAGTCCCGCTAGAGTTACATACCAATCGTCGCAG | 3.3% |
| sk3 | AGCAGAGTTCACGACCCGATAAGGGGAAAACACATTTCGGATCTCCGTTGTCGCCCTGGTACCGAGTTACATACCAATCGTCGCAG | 13.3% |
| sk4 | AGCAGAGTTCACGACCCGATAAGGGGAAAATACATTTCGGATCTCCGTTGTCGCCCTGATACCGAGTTACATACCAATCGTCGCAG | 16.7% |
| sk5 | AGCAGAGTTCACGACCCGATAAGGGGAAAATACATTTCGGATCTCCGTTGTCGCCCTGGTACCGAGTTACATACCAATCGTCGCAG | 13.3% |
| sk6 | AGCAGAGTTCACGACCCGATAAGGGCGATGCCGATCCCTGTGGCCGTAGGGGCAGTCCCGCTAGAGTTACATACCAATCGTCGCAG | 3.3% |
Binding ability of enriched pools to different cells. Fluorescence intensities of SK-BR-3 and negative control cells incubated with FAM-labeled ssDNA pools from the initial library and the 20th and 21st rounds, as well as that of non-incubated cells. The final concentration of both enriched pools and initial library was 250 nM.
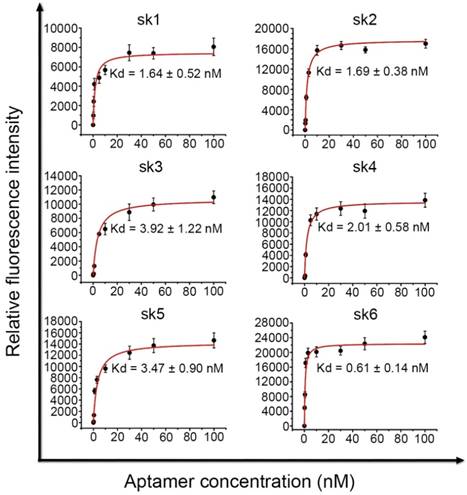
Binding affinity of the selected aptamer candidates
To determine the binding affinity of each of the selected probes, SK-BR-3 cells were incubated with all six FAM-labeled probes in different concentrations, followed by measurement of their relative fluorescence intensity using flow cytometry. The Kd values of the six probes were all below 4 nM. However, sk6 showed better binding affinity to SK-BR-3 cells than the other five probes, with a Kd of 0.61 ± 0.14 nM (Figure 4). The above results indicated that the binding affinity and specificity of probe sk6 for SK-BR-3 cells were stronger than those of the other five probes. Therefore, probe sk6 was chosen for further analysis.
Selectivity of six aptamer candidates. Fluorescence intensities of SK-BR-3 and negative control cells incubated with six FAM-labeled aptamers or initial library. The final concentration of both aptamers and initial library was 250 nM.
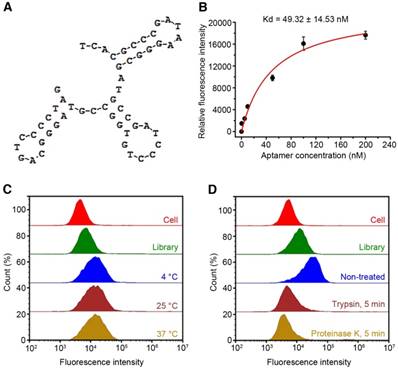
Sequence truncation of the aptamer and its identification
To make the probe sequence shorter, a total of seven truncated aptamer probes were obtained from probe sk6 by removing nucleotides not involved in the formation of the stem-loop or hairpin structures (Table S1). SK-BR-3, MCF-7, and MDA-MB-231 breast cancer cells and MCF-10A human normal breast epithelial cells were incubated with all FAM-labeled truncated probes, followed by flow cytometry analysis. Except for probe sk6d, all six of the other truncated probes were able to bind to SK-BR-3 cells (Figure 5A-B). However, probe sk6Ea, with a length of 53 nt and three typical stem-loop structures (Figure 6A), showed the best selectivity for SK-BR-3 cells among the seven truncated probes, as it bound only to SK-BR-3 cells, with much weaker binding to MCF-7, MDA-MB-231, and MCF-10A cells (Figure 5A-B). Other cancer cells, including HepG2 hepatocellular carcinoma cells, L02 normal hepatic cells, A549 lung cancer cells, HeLa cells, and A375 melanoma cells, were also incubated with FAM-labeled probe sk6Ea to investigate its selectivity. However, the fluorescence signal in the presence of these cell types was significantly weaker than that in the presence of SK-BR-3 target cells, indicating the high selectivity of probe sk6Ea (Figure 7). Next, the binding ability of probe sk6Ea was subsequently assessed using flow cytometry after incubating the target cells with FAM-labeled probe sk6Ea at different concentrations. The Kd of probe sk6Ea was 49.32 ± 14.53 nM (Figure 6B), which was somewhat higher than that of full-length probe sk6. Nonetheless, probe sk6Ea still showed good specificity and favorable binding ability to the target cells, making it ideal for the recognition of SK-BR-3 breast cancer cells.
Influence of temperature on the binding ability of aptamer sk6Ea
In another experiment, due to the fact that these full-length aptamer probes were selected at 4 °C while clinical detection and therapeutic applications are usually carried out at room temperature or physiological temperature, the influence of incubation temperature on the recognition ability of the probe was investigated by incubating FAM-labeled probe sk6Ea with SK-BR-3 cells at temperatures of 4 °C, 25 °C, and 37 °C, followed by analysis using flow cytometry. The fluorescence signals of sk6Ea-cell complexes at the three temperatures were very similar, and all fluorescence signals were higher than those of the initial library and cells alone (Figure 6C). These results indicated that probe sk6Ea was able to effectively recognize SK-BR-3 cells at 4 °C, 25 °C, and 37 °C, further suggesting its potential for future applications involving in vitro detection and in vivo diagnostics and targeted therapy.
Target-type determination of aptamer sk6Ea
Moreover, the specific target of probe sk6Ea was investigated. To do this, FAM-labeled probe sk6Ea was incubated with trypsin- or proteinase K-treated SK-BR-3 cells and then analyzed using flow cytometry. Both treatments resulted in a dramatic decrease in the probe's ability to recognize SK-BR-3 breast cancer cells compared with that of non-treated SK-BR-3 cells (Figure 6D), suggesting that the binding target of probe sk6Ea is likely to be a cytomembrane protein.
Dissociation constants (Kd) of the selected aptamers.
Molecular subtyping of breast cancer cells
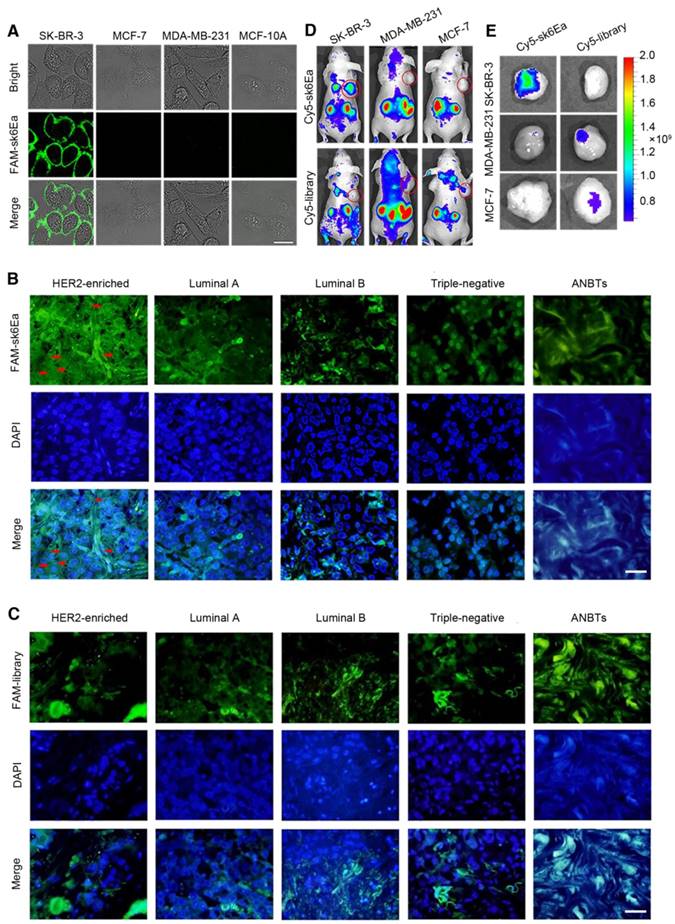
The utility of aptamer-based probe sk6Ea for molecular subtyping of breast cancer was systematically studied. Firstly, SK-BR-3, MCF-7, and MDA-MB-231 breast cancer cells and MCF-10A human normal breast epithelial cells were incubated with FAM-labeled sk6Ea or FAM-labeled library and assessed using confocal microscopy. The fluorescence signal in the presence of SK-BR-3 cells was significantly greater than that emitted by probes in the presence of MCF-7, MDA-MB-231, and MCF-10A cells (Figure 8A). As a negative control, all cells incubated with FAM-labeled library showed no, or negligible, fluorescence signal under the same conditions (Figure S8). These results indicate that probe sk6Ea is able to distinguish SK-BR-3 breast cancer cells from MDA-MB-231 and MCF-7 breast cancer cells, as well as MCF-10A human normal mammary epithelial cells.
Molecular subtyping of breast cancer tissues
Next, to assess the ability of probe sk6Ea to distinguish breast cancer tissues of different molecular subtypes, fluorescence microscopy was applied to analyze FAM-labeled sk6Ea- or FAM-labeled library-stained breast cancer tissues, while DAPI was used to visualize the cell nucleus. In total, 66 cases of clinical human breast cancer tissue sections, including 19 cases of HER2-enriched, 17 cases of triple-negative, 10 cases of Luminal A, and 10 cases of Luminal B breast cancer tissues, as well as 10 cases of ANBTs, were systematically tested and statistically analyzed (Table 2). The fluorescence signal on the cytomembrane of HER2-enriched breast cancer tissue sections (arrow) was significantly stronger than that on Luminal A, Luminal B, and triple-negative breast cancer tissue sections and ANBTs (Figure 8B). As a negative control, FAM-labeled library staining was performed on the four subtypes of breast cancer tissue sections and ANBTs, and all exhibited negligible fluorescence signal on the cytomembrane (Figure 8C). Statistical analysis of these results showed that the positivity rate for HER2-enriched tissues was 78.95%, which was significantly higher than that of the initial library (10.53%). Meanwhile, the positivity rates for FAM-labeled sk6Ea-stained Luminal A, Luminal B, and triple-negative breast cancer tissues and ANBTs were no higher than 23.53% and were thus lower than that of FAM-labeled library-stained tissues (30%) (Table 2). Therefore, the positive results in sk6Ea-stained Luminal A, Luminal B, and triple-negative breast cancer tissues and ANBTs might represent nonspecific binding. These results indicate that probe sk6Ea is able to distinguish HER2-enriched breast tissues from Luminal A, Luminal B, and triple-negative breast cancer tissues and ANBTs to some extent. However, when performing breast cancer tissue staining with aptamers, there is considerable nonspecific binding, which can be difficult to deal with despite the existence of various blocking measures. We speculate that the nonspecific binding of aptamers to formalin-fixed paraffin-embedded breast cancer tissue sections may result from a change in cytomembrane permeability, which limits the selectivity of the probe. Therefore, to promote tissue staining applications using aptamers, new effective blocking methods are essential.
Recognition ability of truncated aptamers. The concentration of both aptamers and initial library was 250 nM.
Secondary structure (A), Kd (B), effects of temperature (C), and target-type analysis (D) of aptamer sk6Ea.
Selectivity of aptamer sk6Ea for different cells. The final concentration of both aptamers and initial library was 250 nM.
Molecular subtyping of breast cancer tissues using aptamer sk6Ea.
| ssDNA | Tissue section | Number of samples | Positivity rate |
|---|---|---|---|
| FAM-labeled sk6Ea | HER2-enriched | 19 | 78.95% |
| Luminal A | 10 | 10% | |
| Luminal B | 10 | 20% | |
| Triple-negative | 17 | 23.53% | |
| ANBTs | 10 | 20% | |
| FAM-labeled library | HER2-enriched | 19 | 10.53% |
| Luminal A | 10 | 30% | |
| Luminal B | 10 | 20% | |
| Triple-negative | 17 | 17.65% | |
| ANBTs | 10 | 30% |
In vivo and ex vivo molecular subtyping of breast cancer xenografts
Furthermore, to determine whether probe sk6Ea could recognize breast cancer molecular subtypes in vivo, Cy5-labeled sk6Ea was injected into nude mice xenografts of SK-BR-3, MDA-MB-231, and MCF-7 cells through the tail vein. Fluorescence images were taken to detect the fluorescence intensity at the tumor sites at different time points post-injection. As shown in Figure S9-S11, at 30 min after injection with Cy5-labeled sk6Ea, the fluorescence signal at the tumor sites in SK-BR-3 tumor-bearing mice increased significantly compared to that in MDA-MB-231 and MCF-7 tumor-bearing mice post-injection. Fluorescence in SK-BR-3 tumor-bearing mice continued to increase until 1 h post-injection and significantly decreased at 3 h post-injection. As a negative control, injection of Cy5-labeled library resulted in almost no significant fluorescence signal at the tumor sites in SK-BR-3 tumor-bearing mice and only very weak fluorescence signal at the tumor sites in MDA-MB-231 and MCF-7 tumor-bearing mice at 30 min post-injection (Figure 8D-E). Collectively, these results demonstrate that probe sk6Ea specifically targeted the xenografts of SK-BR-3 tumor-bearing mice but not the xenografts of MDA-MB-231 and MCF-7 tumor-bearing mice in vivo within 30 min post-injection, suggesting that probe sk6Ea has significant potential for applications involving the molecular subtyping, diagnosis, and targeted therapy of breast cancer.
Application of aptamer sk6Ea for molecular subtyping of breast cancer. (A) Molecular subtyping of breast cancer cells. Scale bar = 20 µm. (B) Molecular subtyping of breast cancer tissues. (C) FAM-labeled library-stained breast cancer tissues of different subtypes. All pictures were taken under a fluorescence microscope at ×400 magnification. Scale bar = 50 µm. (D) In vivo molecular subtyping of breast cancer cells xenografted into mice. (E) Ex vivo imaging of xenograft tumors from mice bearing different breast cancer cells.
Discussion
Although the aptamer sk6Ea has potential for the molecular subtyping of breast cancer, there are still some challenges to be addressed before the development of aptamer sk6Ea-based theranostic tools for breast cancer. First, while aptamer sk6Ea can distinguish HER2-enriched breast cancer from three other breast cancer subtypes and ANBTs, it lacked the ability to further differentiate among Luminal A, Luminal B, and triple-negative breast cancer. To achieve this purpose, additional breast cancer molecular subtype-specific aptamers should be developed. The use of a combination of several aptamers for detection, namely aptamer-based multicolor imaging [50, 51], could enable simple identification of breast cancer molecular subtypes. In addition, new biomarkers should be developed for breast cancer, especially for triple-negative breast cancer, which currently has no suitable biomarkers and is increasing in incidence among patients; this could also be realized using the Cell-SELEX method. The second challenge is that the target of aptamer sk6Ea still needs to be validated and is still being systematically studied. Although the above results indicate that the target of sk6Ea may be the HER2 protein, no competition was observed between sk6Ea and anti-HER2 polyclonal antibodies (Poly-anti-HER2) (Figure S12). In consideration of the heterogeneity of proteins, additional evidence and tests are still required. Third, while aptamer sk6Ea shows strong potential for breast cancer diagnostics and therapy, it exhibits instability in vivo and is excreted 4-5 h post-injection. This is likely because: 1) aptamers are highly sensitive to nucleases in vivo and can be degraded very easily [52]; and 2) the small size and molecular mass of aptamers allow them to be eliminated rapidly via the kidneys [41]. To address these problems, chemical modifications of aptamers with fluoro-, amino-, O-methyl, and other groups, are usually applied [28, 53-55]. Moreover, spiegelmers (L-aptamers), which are the enantiomers of wild-type RNA aptamers (D-aptamers), have been reported to be intrinsically insensitive to nucleases and thus to possess better physiological stability in vivo [56-58]. Finally, aptameric structural modulation was recently reported to improve the physiological stability of aptamers for in vivo applications. For instance, Tan et al. [59, 60] developed a circular bivalent aptamer for cancer cell recognition and in vivo tumor imaging. Thereafter, they further used a supramolecular approach to modulate the interaction of the circular bivalent aptamer with molecular therapeutics for enhanced delivery of intracellular proteins for targeted cancer therapy. The above methods designed to improve the physiological stability of aptamers may provide effective strategies for accelerating the clinical application of aptamer sk6Ea-based theranostics.
In summary, we have developed a new aptamer-based probe for molecular subtyping of breast cancer, both in vitro and in vivo. The probe is able to distinguish SK-BR-3 breast cancer cells from MDA-MB-231 and MCF-7 breast cancer cells, as well as MCF-10A human normal mammary epithelial cells. It is also able to distinguish HER2-enriched breast cancer tissues from Luminal A, Luminal B, and triple-negative breast cancer tissues and ANBTs, as well as xenografts of SK-BR-3 tumor-bearing mice from those of MDA-MB-231 and MCF-7 tumor-bearing mice in vivo. These results indicate that probe sk6Ea may be an ideal candidate for molecular subtyping, diagnosis, and targeted therapy of human breast cancer in clinical practice.
Abbreviations
ANBTs: adjacent normal breast tissues; Cell-SELEX: cell-based systematic evolution of ligands by exponential enrichment; D-aptamers: wild-type RNA aptamers; IHC: immunohistochemistry; L-aptamers: spiegelmers; Poly-anti-HER2: anti-HER2 polyclonal antibodies; ssDNA: single stranded DNA.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
This work was supported by the State key Basic Research Program of the PRC (2014CB744501), the National Key Research and Development Program of China (2017YFA0205301), the NSF of China (61527806, 61471168 and 61871180) and open Funding of State Key Laboratory of Oral Diseases (SKLOD2018OF02).
Competing Interests
The authors have declared that no competing interest exists.
References
1. DeSantis CE, Fedewa SA, Goding Sauer A, Kramer JL, Smith RA, Jemal A. Breast cancer statistics, 2015: convergence of incidence rates between black and white women. CA Cancer J Clin. 2016;66:31-42
2. Ward EM, DeSantis CE, Lin CC, Kramer JL, Jemal A, Kohler B. et al. Cancer statistics: breast cancer in situ. CA Cancer J Clin. 2015;65:481-95
3. Toss A, Cristofanilli M. Molecular characterization and targeted therapeutic approaches in breast cancer. Breast Cancer Res. 2015;17:60
4. Harbeck N, Gnant M. Breast cancer. Lancet. 2017;389:1134-50
5. Liu M, Li Z, Yang J, Jiang Y, Chen Z, Ali Z. et al. Cell-specific biomarkers and targeted biopharmaceuticals for breast cancer treatment. Cell Prolif. 2016;49:409-20
6. Wolff AC, Hammond MEH, Hicks DG, Dowsett M, McShane LM, Allison KH. et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. Arch Pathol Lab Med. 2014;138:241-56
7. Groff K, Brown J, Clippinger AJ. Modern affinity reagents: recombinant antibodies and aptamers. Biotechnol Adv. 2015;33:1787-98
8. Li X, Kim J, Yoon J, Chen XY. Cancer-associated, stimuli-driven, turn on theranostics for multimodality imaging and therapy. Adv Mater. 2017;29:1606857
9. Rao L, Bu LL, Cai B, Xu JH, Li A, Zhang WF. et al. Cancer cell membrane-coated upconversion nanoprobes for highly specific tumor imaging. Adv Mater. 2016;28:3460-3466
10. Gebhart G, Lamberts LE, Wimana Z, Garcia C, Emonts P, Ameye L. et al. Molecular imaging as a tool to investigate heterogeneity of advanced HER2-positive breast cancer and to predict patient outcome under trastuzumab emtansine (T-DM1): the ZEPHIR trial. Ann Oncol. 2016;27:619-24
11. Fang X, Tan W. Aptamers generated from cell-SELEX for molecular medicine: a chemical biology approach. Accounts Chem Res. 2010;43:48-57
12. Xi Z, Huang R, Li Z, He N, Wang T, Su E. et al. Selection of HBsAg-specific DNA aptamers based on carboxylated magnetic nanoparticles and their application in the rapid and simple detection of Hepatitis B Virus infection. ACS Appl Mater Interfaces. 2015;7:11215-23
13. Li S, Xu H, Ding H, Huang Y, Cao X, Yang G. et al. Identification of an aptamer targeting hnRNP A1 by tissue slide-based SELEX. J Pathol. 2009;218:327-36
14. Lyu Y, Chen G, Shangguan D, Zhang L, Wan S, Wu Y. et al. Generating cell targeting aptamers for nanotheranostics using Cell-SELEX. Theranostics. 2016;6:1440-52
15. Wei Q, Nagi R, Sadeghi K, Feng S, Yan E, Ki SJ. et al. Detection and spatial mapping of mercury contamination in water samples using a smart-phone. ACS Nano. 2014;8:1121-9
16. Dunn MR, Jimenez RM, Chaput JC. Analysis of aptamer discovery and technology. Nat Rev Chem. 2017;1:0076
17. Huo Y, Qi L, Lv XJ, Lai T, Zhang J, Zhang ZQ. A sensitive aptasensor for colorimetric detection of adenosine triphosphate based on the protective effect of ATP-aptamer complexes on unmodified gold nanoparticles. Biosens Bioelectron. 2016;78:315-20
18. Feng L, Chen Y, Ren J, Qu X. A graphene functionalized electrochemical aptasensor for selective label-free detection of cancer cells. Biomaterials. 2011;32:2930-7
19. Zhao Z, Fan H, Zhou G, Bai H, Liang H, Wang R. et al. Activatable fluorescence/MRI bimodal platform for tumor cell imaging via MnO2 nanosheet-aptamer nanoprobe. J Am Chem Soc. 2014;136:11220-3
20. Zhang J, Smaga LP, Satyavolu NSR, Chan J, Lu Y. DNA aptamer-based activatable probes for photoacoustic imaging in living mice. J Am Chem Soc. 2017;139:17225-8
21. Shastri A, McGregor L, Liu Y, Harris V, Nan HQ, Mujica M. et al. An aptamer-functionalized chemomechanically modulated biomolecule catch-and-release system. Nat Chem. 2015;7:447-54
22. Zheng FY, Cheng Y, Wang J, Lu J, Zhang B, Zhao YJ. et al. Aptamer-functionalized barcode particles for the capture and detection of multiple types of circulating tumor cells. Adv Mater. 2014;26:7333-8
23. Wu Y, Sefah K, Liu H, Wang R, Tan W. DNA aptamer-micelle as an efficient detection/delivery vehicle toward cancer cells. Proc Natl Acad Sci USA. 2010;107:5-10
24. Meng H, Liu H, Kuai H, Peng RZ, Mo LT, Zhang XB. Aptamer-integrated DNA nanostructures for biosensing, bioimaging and cancer therapy. Chem Soc Rev. 2016;45:2583-602
25. Zhu GZ, Zheng J, Song EQ, Donovan M, Zhang KJ, Liu C. et al. Self-assembled, aptamer-tethered DNA nanotrains for targeted transport of molecular drugs in cancer theranostics. Proc Natl Acad Sci USA. 2013;110:7998-8003
26. McNamara JO, Andrechek ER, Wang Y, Viles KD, Rempel RE, Gilboa E. et al. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat Biotechnol. 2006;24:1005-15
27. Mor-Vaknin N, Saha A, Legendre M, Carmona-Rivera C, Amin MA, Rabquer BJ. et al. DEK-targeting DNA aptamers as therapeutics for inflammatory arthritis. Nat Commun. 2017;8:14252
28. Zhou J, Rossi J. Aptamers as targeted therapeutics: current potential and challenges. Nat Rev Drug Discov. 2017;16:181-202
29. Hu XX, Wang YL, Tan YN, Wang J, Liu HY, Wang YQ. et al. A difunctional regeneration scaffold for knee repair based on aptamer-directed cell recruitment. Adv Mater. 2017;29:1605235
30. Xi Z, Huang R, Deng Y, He N. Progress in selection and biomedical applications of aptamers. J Biomed Nanotechnol. 2014;10:3043-62
31. Wang R, Zhu G, Mei L, Xie Y, Ma H, Ye M. et al. Automated modular synthesis of aptamer-drug conjugates for targeted drug delivery. J Am Chem Soc. 2014;136:2731-4
32. Toh SY, Citartan M, Gopinath SC, Tang TH. Aptamers as a replacement for antibodies in enzyme-linked immunosorbent assay. Biosens Bioelectron. 2015;64:392-403
33. Mahlknecht G, Maron R, Mancini M, Schechter B, Sela M, Yarden Y. Aptamer to ErbB-2/HER2 enhances degradation of the target and inhibits tumorigenic growth. Proc Natl Acad Sci USA. 2013;110:8170-75
34. Liu Z, Duan JH, Song YM, Ma J, Wang FD, Lu X. et al. Novel HER2 aptamer selectively delivers cytotoxic drug to HER2-positive breast cancer cells in vitro. J Trans Med. 2012;10:148
35. Zhao Z, Xu L, Shi X, Tan W, Fang X, Shangguan D. Recognition of subtype non-small cell lung cancer by DNA aptamers selected from living cells. Analyst. 2009;134:1808-14
36. Sefah K, Shangguan D, Xiong X, O'Donoghue MB, Tan W. Development of DNA aptamers using Cell-SELEX. Nat Protoc. 2010;5:1169-85
37. Li WM, Bing T, Wei JY, Chen ZZ, Shangguan DH, Fang J. Cell-SELEX-based selection of aptamers that recognize distinct targets on metastatic colorectal cancer cells. Biomaterials. 2014;35:6998-7007
38. Hung LY, Wang CH, Hsu KF, Chou CY, Lee GB. An on-chip Cell-SELEX process for automatic selection of high-affinity aptamers specific to different histologically classified ovarian cancer cells. Lab Chip. 2014;14:4017-28
39. Meng H-M, Fu T, Zhang X-B, Tan W. Cell-SELEX-based aptamer-conjugated nanomaterials for cancer diagnosis and therapy. Natl Sci Rev. 2015;2:71-84
40. Daniels DA, Chen H, Hicke BJ, Swiderek KM, Gold L. A tenascin-C aptamer identified by tumor cell SELEX: systematic evolution of ligands by exponential enrichment. Proc Natl Acad Sci USA. 2003;100:15416-21
41. Liu M, Yu X, Chen Z, Yang T, Yang D, Liu Q. et al. Aptamer selection and applications for breast cancer diagnostics and therapy. J Nanobiotechnol. 2017;15:81
42. Wu X, Zhao Z, Bai H, Fu T, Yang C, Hu X. et al. DNA aptamer selected against pancreatic ductal adenocarcinoma for in vivo imaging and clinical tissue recognition. Theranostics. 2015;5:985-94
43. Gijs M, Penner G, Blackler GB, Impens NREN, Baatout S, Luxen A. et al. Improved aptamers for the diagnosis and potential treatment of HER2-positive cancer. Pharmaceuticals. 2016;9:29
44. He X, Guo L, He J, Xu H, Xie J. Stepping library-based post-SELEX strategy approaching to the minimized aptamer in SPR. Anal Chem. 2017;89:6559-66
45. Mou X, Li T, Wang J, Ali Z, Zhang Y, Chen Z. et al. Genetic variation of BCL2 (rs2279115), NEIL2 (rs804270), LTA (rs909253), PSCA (rs2294008) and PLCE1 (rs3765524, rs10509670) genes and their correlation to gastric cancer risk based on universal tagged arrays and Fe3O4 magnetic nanoparticles. J Biomed Nanotechnol. 2015;11:2057-66
46. Pu Y, Liu Z, Lu Y, Yuan P, Liu J, Yu B. et al. Using DNA aptamer probe for immunostaining of cancer frozen tissues. Anal Chem. 2015;87:1919-24
47. Zeng Z, Zhang P, Zhao N, Sheehan AM, Tung CH, Chang CC. et al. Using oligonucleotide aptamer probes for immunostaining of formalin-fixed and paraffin-embedded tissues. Mod Pathol. 2010;23:1553-8
48. Hermann T, Patel D. Adaptive recognition by nucleic acid aptamers. Science. 2000;287:820-5
49. Ren X, Gelinas AD, von Carlowitz I, Janjic N, Pyle AM. Structural basis for IL-1alpha recognition by a modified DNA aptamer that specifically inhibits IL-1alpha signaling. Nat Commun. 2017;8:810
50. Wang S, Kong H, Gong XY, Zhang SC, Zhang XR. Multicolor imaging of cancer cells with fluorophore-tagged aptamers for single cell typing. Anal Chem. 2014;86:8261-6
51. Zhao BJ, Wu P, Zhang H, Cai CX. Designing activatable aptamer probes for simultaneous detection of multiple tumor-related proteins in living cancer cells. Biosens Bioelectron. 2015;68:763-70
52. Healy JM, Lewis SD, Kurz M, Boomer RM, Thompson KM, Wilson C. et al. Pharmacokinetics and biodistribution of novel aptamer compositions. Pharm Res. 2004;21:2234-46
53. Siddiqui MAA, Keating GM. Pegaptanib: in exudative age-related macular degeneration. Drugs. 2005;65:1571-7
54. Burmeister PE, Lewis SD, Silva RF, Preiss JR, Horwitz LR, Pendergrast PS. et al. Direct in vitro selection of a 2 '-O-methyl aptamer to VEGF. Chem Biol. 2005;12:25-33
55. Nimjee SM, White RR, Becker RC, Sullenger BA. Aptamers as therapeutics. Annu Rev Pharmacol. 2017;57:61-79
56. Vater A, Klussmann S. Toward third-generation aptamers: spiegelmers and their therapeutic prospects. Curr Opin Drug Discov Devel. 2003;6:253-61
57. Sczepanski JT, Joyce GF. Specific inhibition of microrna processing using L-RNA aptamers. J Am Chem Soc. 2015;137:16032-37
58. Olea C, Weidmann J, Dawson PE, Joyce GF. An L-RNA aptamer that binds and inhibits RNase. Chem Biol. 2015;22:1437-41
59. Kuai H, Zhao Z, Mo L, Liu H, Hu X, Fu T. et al. Circular bivalent aptamers enable in vivo stability and recognition. J Am Chem Soc. 2017;139:9128-31
60. Jiang Y, Pan XS, Chang J, Niu WJ, Hou WJ, Kuai HL. et al. Supramolecularly engineered circular bivalent aptamer for enhanced functional protein delivery. J Am Chem Soc. 2018;140:6780-84
Author contact
![]() Corresponding authors: hndengyancom (Yan Deng); cjr.luguangming163.com (Guangming Lu); nyhe1958com (Nongyue He).
Corresponding authors: hndengyancom (Yan Deng); cjr.luguangming163.com (Guangming Lu); nyhe1958com (Nongyue He).
 Global reach, higher impact
Global reach, higher impact