13.3
Impact Factor
Theranostics 2018; 8(20):5634-5644. doi:10.7150/thno.27776 This issue Cite
Research Paper
Engineered RNase P Ribozymes Effectively Inhibit the Infection of Murine Cytomegalovirus in Animals
1. Department of Biotechnology, College of Life Science and Technology, Jinan University, Guangzhou, Guangdong 510632, China
2. School of Public Health, University of California, Berkeley, CA 94720
3. School of Medicine, St. George's University, Grenada, West Indies
4. Taizhou Institute of Virology, Taizhou, Jiangsu 225300, China
5. Jiangsu Affynigen Biotechnolgies Inc, Taizhou, Jiangsu 225300, China
6. Guangzhou Qinheli Biotechnolgies Inc, Guangzhou, Guangdong 510600, China
7. School of Pharmacy, Shandong University of Traditional Chinese Medicine, Jinan, Shandong 250355, China
*These authors contribute equally to this study.
Received 2018-6-10; Accepted 2018-10-3; Published 2018-11-9
Abstract
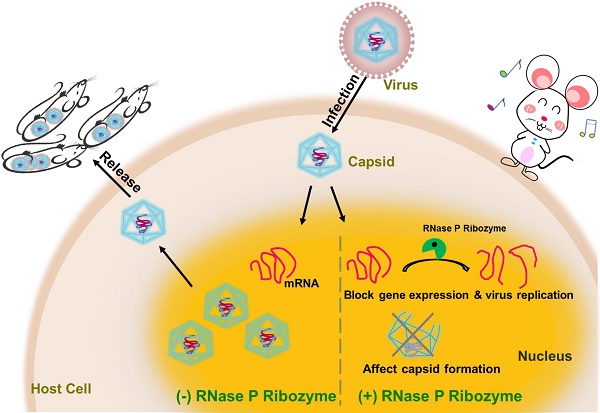
Rationales: Gene-targeting ribozymes represent promising nucleic acid-based gene interference agents for therapeutic application. We previously used an in vitro selection procedure to engineer novel RNase P-based ribozyme variants with enhanced targeting activity. However, it has not been reported whether these ribozyme variants also exhibit improved activity in blocking gene expression in animals.
Methods and Results: In this report, R388-AS, a new engineered ribozyme variant, was designed to target the mRNA of assemblin (AS) of murine cytomegalovirus (MCMV), which is essential for viral progeny production. Variant R338-AS cleaved AS mRNA sequence in vitro at least 200 times more efficiently than ribozyme M1-AS, which originated from the wild type RNase P catalytic RNA sequence. In cultured MCMV-infected cells, R338-AS exhibited better antiviral activity than M1-AS and decreased viral AS expression by 98-99% and virus production by 15,000 fold. In MCMV-infected mice, R388-AS was more active in inhibiting AS expression, blocking viral replication, and improving animal survival than M1-AS.
Conclusions: Our results provide the first direct evidence that novel engineered RNase P ribozyme variants with more active catalytic activity in vitro are also more effective in inhibiting viral gene expression in animals. Moreover, our studies imply the potential of engineering novel RNase P ribozyme variants with unique mutations to improve ribozyme activity for therapeutic application.
Keywords: antisense RNA, ribozyme, antiviral, gene targeting, gene therapy
Introduction
Human cytomegalovirus (CMV), called HCMV, is an important and common human virus causing severe infections in individuals with compromised immune system such as AIDS patients and transplant recipients [1]. However, no vaccines and few anti-HCMV drugs are currently available in clinics, which dictates an urgent need for developing novel therapeutics. Murine cytomegalovirus (MCMV) infection of mice provides a useful animal model for studying innovative antiviral therapeutics in vivo [1-3]. For example, the CB17 severe combined immunodeficient (SCID) mice are highly susceptible to MCMV infection due to the absence of functional T and B lymphocytes [4, 5]. This animal model can be used for screening novel anti-CMV agents in vivo.
Nucleic acid-based gene silencing technologies targeting mRNA sequences of choice represent promising gene-targeting strategies [6, 7]. For example, ribozymes were used to target HIV-1 mRNA sequences and reduce HIV-1 growth in cultured cells [8]. Small interfering RNAs (siRNAs) were also used to reduce virus growth by targeting viral gene sequences [6, 9, 10]. Therefore, nucleic acid-based, sequence-specific gene targeting strategies can be employed as new antiviral therapeutics.
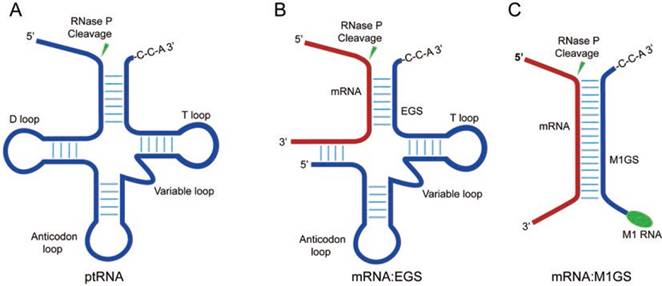
RNase P of Escherichia coli is a ribonucleoprotein complex consisting of a catalytically active RNA subunit (M1 RNA) and a protein subunit (C5 protein) [11, 12]. Moreover, M1 RNA can be engineered to target tRNA substrates and specific mRNA sequences of choice (Figure 1) [13-16]. A sequence-specific ribozyme, M1GS RNA, which can be designed by attaching an external guide sequence (EGS) to the 3'
3'  terminus of M1 RNA (Figure 1C), is active in specifically cleaving the target gene sequences in cultured cells [17, 18]. Compared with other antisense technologies [6, 7], the M1 ribozyme-based tool has its own unique advantages because M1 RNA is catalytically efficient [11, 12]. Engineered M1GS ribozymes were used to specifically target viral mRNAs and reduce viral growth in cultured cells infected with human viruses such as influenza virus, HCMV, and herpes simplex virus 1 (HSV-1) [18-20]. Furthermore, RNase P ribozymes were also studied for blocking viral gene expression and replication in mice [21].
terminus of M1 RNA (Figure 1C), is active in specifically cleaving the target gene sequences in cultured cells [17, 18]. Compared with other antisense technologies [6, 7], the M1 ribozyme-based tool has its own unique advantages because M1 RNA is catalytically efficient [11, 12]. Engineered M1GS ribozymes were used to specifically target viral mRNAs and reduce viral growth in cultured cells infected with human viruses such as influenza virus, HCMV, and herpes simplex virus 1 (HSV-1) [18-20]. Furthermore, RNase P ribozymes were also studied for blocking viral gene expression and replication in mice [21].
Improving ribozyme cleaving activity is essential for its utilization as a useful gene-targeting approach. New ribozyme variants, selected by using an in vitro selection procedure [22-24], were more active than the ribozyme derived from the wildtype M1 RNA sequence [19]. However, whether these selected ribozyme variants also perform better than the wildtype M1 RNA-derived ribozyme in animals has not been reported. In this study, using MCMV-infected SCID mice as the animal model, we showed that a selected ribozyme variant, R388-AS, was more efficient in knocking down viral gene expression and inhibiting viral infection in cultured cells and in mice than ribozyme M1-AS, which was derived from the wildtype M1 RNA. Ribozyme variant R388-AS was generated to cleave the mRNA of MCMV assemblin (AS) [25, 26], which encodes a protease essential for viral capsid maturation and growth [27, 28]. In vitro studies showed that R388-AS effectively cleaved the target mRNA sequence, as well as knocking down AS expression and viral replication in cultured cells. When plasmids expressing ribozymes were injected into SCID mice by a modified “hydrodynamic transfection” procedure [29-31], AS expression and viral replication in these mice were also inhibited and mice lived longer. Our study indicates that ribozyme variants constructed in this study can be used as a class of novel and innovative anti-HCMV therapeutics.
cleaving activity is essential for its utilization as a useful gene-targeting approach. New ribozyme variants, selected by using an in vitro selection procedure [22-24], were more active than the ribozyme derived from the wildtype M1 RNA sequence [19]. However, whether these selected ribozyme variants also perform better than the wildtype M1 RNA-derived ribozyme in animals has not been reported. In this study, using MCMV-infected SCID mice as the animal model, we showed that a selected ribozyme variant, R388-AS, was more efficient in knocking down viral gene expression and inhibiting viral infection in cultured cells and in mice than ribozyme M1-AS, which was derived from the wildtype M1 RNA. Ribozyme variant R388-AS was generated to cleave the mRNA of MCMV assemblin (AS) [25, 26], which encodes a protease essential for viral capsid maturation and growth [27, 28]. In vitro studies showed that R388-AS effectively cleaved the target mRNA sequence, as well as knocking down AS expression and viral replication in cultured cells. When plasmids expressing ribozymes were injected into SCID mice by a modified “hydrodynamic transfection” procedure [29-31], AS expression and viral replication in these mice were also inhibited and mice lived longer. Our study indicates that ribozyme variants constructed in this study can be used as a class of novel and innovative anti-HCMV therapeutics.
Materials and Methods
Ethics Statement. Animal studies were either approved by the Animal Care and Use Committee of the University of California-Berkeley (Protocol #R240) or by the Animal Care and Use Committee of the College of Life Sciences and Technology, Jinan University (Guangzhou, China).
RNase P and its ribozyme substrates. (A) a precursor tRNA (ptRNA); (B) A complex of a target mRNA with an EGS; (C) a target mRNA hybridized with an M1GS ribozyme.
In vitro studies of M1GS ribozymes. The dimethyl sulphate (DMS)-accessible regions [18, 32, 33] of AS mRNA were mapped following previously described procedures [34, 35]. NIH3T3 cells were infected with MCMV Smith strain (MOI = 2). At 18 hours post infection, cells were treated with 1% DMS-containing culture media for 10 mins, washed with PBS, and lysed with buffer A (10 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1.5 mM MgCl2, 0.2% NP40). We isolated total RNAs and performed primer extension assays to identify the DMS modification sites, as explained earlier [34, 35].
We performed PCR to construct the DNA template of substrate as-39 with 5' primer as-39-5-AF25 (5'-GGAATTCTAATACGACTCACTATAG-3') and 3' primer as-39-3 (5'-CGGGATCCT GGCGGGACCGCGGGCGACGAGTTTGGATTCTATAGTGAGTCGTATTA-3'). PCR was used to generate the R388-AS, M1-AS, C-R388-AS, and C-MI-AS ribozyme sequences from plasmid pFL117, p388, and pC102, which include the M1 RNA, variant R388, and mutant C102 ribozyme sequence respectively [19, 36, 37]. The 5' and 3' primers for the PCR reactions were Rb-AS-AF25 (5'-GGAATTCTAATACGACTCACTATAG-3') and Rb-AS3 (5'-CCCGCTCGAGAAAAAATGGTGTCGCCCGCGGTCCCGCCTGTGGAATTGTG-3') respectively. Ribozymes and as-39 were made using in vitro transcription procedures [19, 36, 37].
We performed single-turnover kinetic analyses of in vitro cleavage reactions in buffer A (50 mM Tris, pH 7.5; 100 mM NH4Cl, 100 mM MgCl2), as explained earlier [19, 36, 37]. We generated plots of observed cleavage rate (kobs) vs. ribozyme concentration using a Kaleidagraph program (Synergy Software, Reading, PA) and determined the overall cleavage rate (kcat/Km)s [19, 36, 37].
We determined the equilibrium dissociation constants (Kd) of complexes of the ribozymes and substrate as-39 in buffer B (50 mM Tris, pH 7.5; 100 mM NH4Cl, 100 mM CaCl2, 3% glycerol, 0.1% xylene cyanol, 0.1% bromophenol blue), as explained earlier [38]. The samples were allowed to bind and then loaded on a non-denaturing gel for electrophoresis. We generated plots of percent of product bound versus M1GS RNA concentration to calculate Kd value. Experiments were performed in triplicate and repeated three times [19, 36, 37].
Antibodies, viruses, and cells. MCMV (Smith strain) and mouse NIH3T3 cells were obtained from American Type Culture Collection (ATCC) (Manassas, VA). The anti-MCMV and anti-actin antibodies have been described previously [4, 21]. Growth of MCMV and NIH3T3 cells were performed, following procedures as explained earlier [4, 21].
Cells expressing EGSs. DNAs of retroviral vector LXSN-M1GS were transfected into NIH3T3 cells. The M1GS-expressing cells were selected using neomycin (500 µg/ml) and cloned. We performed Northern blot experiments to assay EGS and mouse RNase P RNA expression [19, 36, 37]. The cytotoxic effect of M1GS expression was studied using 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT; Sigma), following the procedures explained earlier [39].
Assaying viral mRNA and protein expression. The levels of viral mRNAs were investigated using Northern blot experiments with [32P]-labelled DNA probes containing the MCMV DNA sequence or the DNA sequence coding for mouse RNase P RNA [19, 36, 37]. The levels of viral proteins and actin were investigated using Western blot experiments, with anti-MCMV and anti-actin antibodies and a chemiluminescent substrate (GE Healthcare, Shanghai, China and Sunnyvale, CA). The expression levels were analyzed using a STORM840 Phosphorimager [19, 36, 37]. The assays were performed in triplicate and repeated three times.
Studies in animals. Eight week-old CB17 SCID mice (at least five animals per group) (Jackson Laboratory, Bay Harbor, Maine) were infected intraperitoneally with MCMV (1x104 PFU/animal), and hydrodynamically transfected with different LXSN-EGS DNAs (20 µg) at 48 hours postinfection [29-31]. We performed hydrodynamic transfection every 3 days following the first injection. To assess the transfection efficiency, the expressions of M1GS RNA and green fluorescent protein (GFP) from tissues were investigated using Northern blot analyses and fluorescent microscopy.
Animal survivability was assessed by examining the mice 2 times a day for mortality for at least 100 days [4, 21]. Different organs (i.e. spleens, livers, lungs, and salivary glands) were collected from mice at 1, 3, 7, 10, 14, and 21 days postinfection. Viral gene expression and growth in these organs were assayed by Northern/Western blots analyses and plaque assays, respectively, as explained previously [19, 36, 37].
Analysis of MCMV growth with plaque assays. Tissues and cells were collected and sonicated to generate virus stocks as explained earlier [4, 40]. Virus titers, as plaque-forming unit (PFU) per milliliter of homogenated cell lysates and tissues, were determined using plaque assays in NIH3T3 cells [4, 21]. Each plaque assay was performed in triplicate and repeated three times. The detection limit of homogenated cells and tissues was 10 PFU/ml and samples negative at a 10-1 dilution were designated a titer of 10 (101) PFU/ml [4, 21].
Results
MCMV AS mRNA sequence cleaved efficiently by ribozymes in vitro. MCMV assemblin (AS) encodes a protease required for formation of mature viral genomic DNA-containing capsid and therefore, is essential for the production of viral progeny [25, 26]. Ribozymes were engineered to target the MCMV AS mRNA sequence. An in vivo mapping approach developed previously was used to choose an accessible binding site for the ribozymes with high catalytic activity [18, 33]. A position, 306 nucleotides downstream from the AS translational initiation codon [41], which was accessible for modification by dimethyl sulphate (DMS) in MCMV-infected cells (data not shown), was designed as the ribozyme targeting site.
By using an in vitro selection procedure, ribozyme variants being more active in targeting the HSV-1 thymidine kinase (TK) mRNA than the wild-type M1 RNA-derived ribozyme were isolated [19]. Mutations were introduced into the active domains (such as nucleotides 30-61, 110-140, and 290-332) of M1 RNA [42, 43] and ribozymes with highly efficient cleavage activities were isolated [19]. A ribozyme variant, R388-AS, was generated and was most efficient in slicing the assemblin mRNA sequence in vitro (see below, Table 1). Compared to the wild-type M1 RNA, R388 comprises of three point mutations: G33 -> C33, C158 -> G158, and C292 -> A292 [19]. Here we examined the catalytic activity of R338-AS in vitro and investigated its ability to block assemblin expression in MCMV-infected cells and mice.
Kinetic analysis (values of (kcat/Km)s and Kd) in cleavage of substrate as-39 by M1GS ribozymes.
| Enzyme | kcat/Km (µM-1·min-1) | Kd (nM) |
|---|---|---|
| M1-AS | 0.13±0.06 | 0.41±0.06 |
| C-M1-AS | <5x10-6 | 0.43±0.06 |
| R388-AS | 27.3.±0.06 | 0.02±0.01 |
| C-R338-AS | <5x10-6 | 0.02±0.01 |
| M1-TK | <5x10-6 | ND |
Experiments were performed in triplicate and repeated three times as described in Methods and Materials. “ND”: not determined.
Ribozyme R388-AS and M1-AS were constructed by covalently joining an 18 nt long guide sequence to the 3' end of R388 RNA and M1 RNA, respectively. Control ribozymes C-R388-AS and C-M1-AS were generated in a similar way but had no catalytic activity due to the introduction of point mutations (A347C348→C347U348, C353C354C355G356→G353G354A355U356) in the P4 catalytic domain of these ribozymes [21, 36]. These four ribozymes (R388-AS, M1-AS, C-R388-AS and C-M1-AS) had the same guide sequence targeting the assemblin mRNA sequence.
RNA substrate as-39, which contained a 39 nt long assemblin mRNA target region, was cleaved by ribozyme R388-AS and M1-AS (Table 1). To the contrary, no cleavage of substrate as-39 by C-R388-AS and C-M1-AS was detected (Table 1). The catalytic activity (kcat/Km)s results indicated that R388-AS had about 200 times higher activity than M1-AS (Table 1). To study whether different catalytic activities of these four ribozymes were due to the difference in binding affinities to substrate as-39, the binding affinities, measured as the dissociation constant (Kd), were studied by gel shift assays. Ribozymes C-R388-AS and C-M1-AS had similar binding affinities as R388-AS and M1-AS, respectively (Table 1). Furthermore, R338-AS and C-R388-AS bound to as-39 about 20 times better than M1-AS and C-M1-AS, suggesting that the point mutations of R388 (G33 -> C33, C158 -> G158, and C292 -> A292) may enhance substrate binding. Thus, C-R388-AS and C-M1-AS could be used as antisense effect controls for R388-AS and M1-AS, respectively, since they had similar binding affinity as R388-mAP and M1-mAP but without catalytic activity.
Ribozymes expressed in cultured cells. The R388-AS, M1-AS, C-R388-AS, and C-M1-AS coding sequences were cloned into retroviral vector LXSN, which had expressed M1GS RNA efficiently previously [18, 44, 45]. The constructed LXSN-M1GS DNAs were transfected into PA317 cells to produce retroviral vectors which were then transduced into NIH3T3 cells [45], and cell lines expressing the ribozymes were cloned.
Ribozyme expression in NIH3T3 cells. RNA fractions (30 µg) from cells expressing different M1GS RNAs were analyzed in northern blot experiments using probes containing M1GS RNA (lanes 1-4) and mouse RNase P RNA (mP1 RNA)(internal control) (lanes 5-8) sequences.
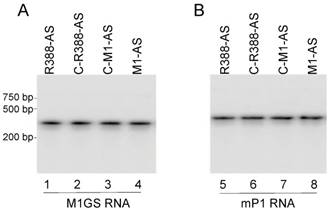
Another cell line expressing ribozyme M1-TK, which was constructed to target HSV-1 TK mRNA sequence, was used as a control with mismatched guide sequence [18]. No catalytic activity of ribozyme M1-TK to cleave substrate as-39 was detected in vitro (data not shown). Northern analysis was performed to examine M1GS RNA expression in each cell line with mouse RNase P RNA (mP1 RNA) as the internal control (Figure 2) [46, 47]. Only cell lines expressing ribozymes with similar levels were used for subsequent studies. Using the MTT assays, we observed that all ribozyme-expressing cell lines and cell lines only transfected with blank LXSN vector as control were similar in cellular growth for 90 days (data not shown), suggesting ribozymes were not toxic to the cells.
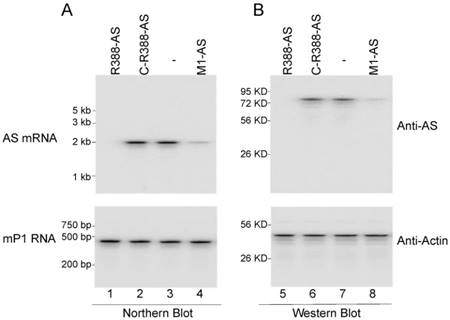
Ribozyme-mediated reduction of MCMV gene expression and growth in cultured cells. Northern analyses were carried out to examine MCMV assemblin expression in cells expressing different ribozymes, with mouse RNase P RNA (mP1 RNA) as the internal control (Figure 3A). A decline of 99% and 81% in assemblin mRNA expression was detected in cells expressing R388-AS and M1-AS, respectively (Figure 3, lanes 1 and 4, Table 2). In contrast, a reduction of less than 10% was found in cells expressing C-R388-AS, C-M1-AS, or M1-TK, which was probably caused by antisense effect (Figure 3, lane 2, Table 2) (data not shown). These results suggest that the substantial reduction of AS mRNA expression in cells expressing R388-AS and M1-AS was due to ribozyme-mediated cleavage. Using Northern analyses and rapid amplification cDNA ends (RACE) PCR assays, we detected no specific fragments of viral mRNAs cleaved by ribozymes in these cells, possibly because these cleavage products, which were RNAs lacking of either a 5' cap structure or a 3' poly (A) sequence, were extremely unstable and quickly hydrolyzed intracellularly.
Western analyses were performed to study the AS protein expression in cells expressing different ribozymes, with actin as the internal control (Figure 3). A decline of about 98% and 79% in the level of AS protein was found in cells with R388-AS and M1-AS, respectively (Figure 3, lanes 5 and 8, Table 2). To the contrary, only 10% reduction was found in cells with C-R388-AS, C-M1-AS, or M1-TK, which was probably due to the antisense effect (Figure 3, lane 6, Table 2) (data not shown).
Ribozyme effect on viral mRNA and protein expression in NIH3T3 cells. The parental cells (-) and cells expressing different ribozymes (R388-AS, C-R388-AS, and M1-AS) were infected with MCMV (MOI=1). (A) RNA fractions (45 µg) were prepared from cells at 18 hours postinfection and were assayed in northern blot experiments for the expression of AS mRNA and mP1 RNA expression (internal control) (lanes 1-4). (B) Protein fractions (50 µg) were prepared from cells at 24 hours postinfection and assayed in Western blot experiments for the expression of AS protein and mouse actin (internal control) (lanes 5-8).
Levels of inhibition of viral genes in cells expressing ribozymes, as compared to the levels of inhibition in cells that did not express a ribozyme (NIH3T3).
| Viral gene | Gene class | NIH3T3 | C-M1-AS | C-R388-AS | M1-AS | R388-AS | M1-TK |
|---|---|---|---|---|---|---|---|
| M36 mRNA | IE | 0% | 0% | 0% | 0% | 0% | 0% |
| MCP mRNA | Early | 0% | 0% | 0% | 0% | 0% | 0% |
| AS mRNA | Late | 0% | 7% | 6% | 81±6% | 99±7% | 0% |
| gB mRNA | Late | 0% | 0% | 0% | 0% | 0% | 0% |
| ie1 protein | IE | 0% | 0% | 0% | 0% | 0% | 0% |
| M112 protein | Early/Late | 0% | 0% | 0% | 0% | 0% | 0% |
| AS protein | Late | 0% | 6% | 7% | 79±7% | 98±7% | 0% |
| M99 protein | Late | 0% | 0% | 0% | 0% | 0% | 0% |
Experiments were performed in triplicate and repeated three times as described in Methods and Materials. The values of standard deviation for these results are less than 5%.
Plaque assays were carried out to examine MCMV replication in cells expressing different ribozymes. Virus stocks were prepared 1-5 days post-infection and virus titers were examined. At 4 days post-infection, a repression of at least 15,000 and 1,200-fold in viral yield was found in cells expressing R388-AS and M1-AS, respectively (Figure 4). No significant knockdown was detected in cells expressing C-R388-AS, C-M1-AS, or M1-TK (Figure 4). Hence, R388-AS and M1-AS appeared to target viral AS mRNA and reduce MCMV replication.
Ribozyme effect on MCMV growth in NIH3T3 cells. Experiments were performed in triplicate and repeated three times and the details were described in Methods and Materials.
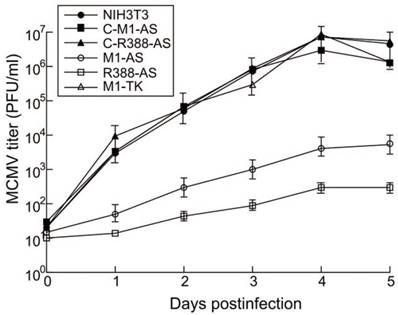
It is known that AS plays no role in regulating the expression of other viral genes, including immediate-early (IE), early, and late genes [1], so their expression was not expected to change as a result of M1GS-mediated inhibition of AS expression. To further study the effects of the constructed ribozymes on viral gene expression, we examined the mRNA expression of MCMV M36 (an IE gene), major capsid protein (MCP) (an early gene), and gB (a late gene) with northern blot analysis, and the protein expression of viral ie1 (an IE gene), M112 (an early/late gene), and M99 (a late gene) with western blot analysis. We noticed no appreciable difference in their expression among the M1GS-expressing cells and the parental NIH3T3 cells (Table 2). Hence, R388-AS and M1-AS expression appeared to suppress AS expression but not the expression of other MCMV genes. These results further suggest that the reduced virus production is probably due to the AS expression reduction induced by M1GS ribozymes but not from the changes of the expression of other viral genes.
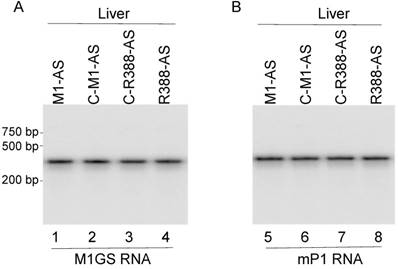
Ribozyme-mediated reduction of MCMV gene expression and growth in animals. SCID mice are good animal models for MCMV infection studies as reported previously [1, 4, 5]. To study the inhibition of MCMV growth in vivo by the constructed ribozymes, hydrodynamic transfection of the constructed LXSN-M1GS plasmids along with a GFP expression reporter construct was given to SCID mice that were intraperitoneally infected 24 hours earlier with MCMV, and repeated every 72 hours [29-31]. Ribozyme expression was assayed by northern blot analyses, using mouse RNase P RNA as the internal control (Figure 5). The transfection procedure seemed to be successful as ribozyme and GFP were detected in different tissues of mice (e.g. livers and spleens) (Figure 5) (data not shown), consistent with previous studies showing the gene-transfer effect of the hydrodynamic transfection method [29-31].
Ribozyme expression in the livers of SCID mice. MCMV-infected mice were hydrodynamically transfected with PBS containing M1GS constructs as described in Materials and Methods. RNA (45 μg) were isolated from the livers of the mice at 14 days postinfection and were analyzed in northern blot experiments using probes containing M1GS RNA (lanes 1-4) and mouse RNase P RNA (mP1 RNA) (internal control) (lanes 5-8) sequences.
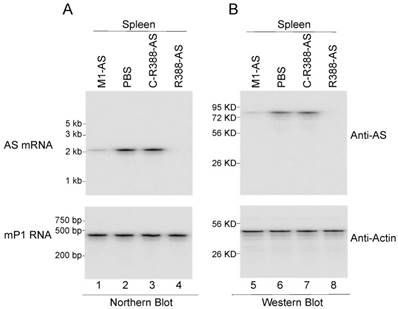
Three different sets of experiments were performed to study the ribozyme effect on MCMV growth in vivo. The first set of experiments was to examine whether viral mRNA and protein expression were inhibited in mice treated with different ribozymes. At 14 days post-infection, viral assemblin mRNA and protein expression was observable in the spleens of mice receiving control ribozymes (i.e. C-R388-AS, C-M1-AS, and M1-TK) (data not shown), while reduced AS expression was observed in mice receiving R388-AS and M1-AS (Figure 6). Furthermore, we observed no difference in AS expression between mice receiving no ribozymes and the control ribozymes (i.e. C-R388-AS, C-M1-AS, or M1-TK) (Figure 6) (data not shown). These results indicated that the viral assemblin mRNA and protein expression were knocked down in mice receiving functional ribozymes R388-AS and M1-AS. Moreover, R388-AS exhibited higher efficiency than M1-AS in blocking AS expression in mice.
Ribozyme effect on viral mRNA and protein expression in SCID mice. MCMV-infected mice were hydrodynamically transfected with PBS only (PBS) and PBS containing different M1GS constructs as described in Materials and Methods. (A) RNA fractions (45 µg) were prepared from the spleens at 14 days postinfection and were assayed in northern blot experiments for the expression of AS mRNA and mP1 RNA expression (internal control) (lanes 1-4). (B) Protein fractions (70 µg) were prepared from the spleens at 14 days postinfection and assayed in Western blot experiments for the expression of AS protein and mouse actin (internal control) (lanes 5-8).
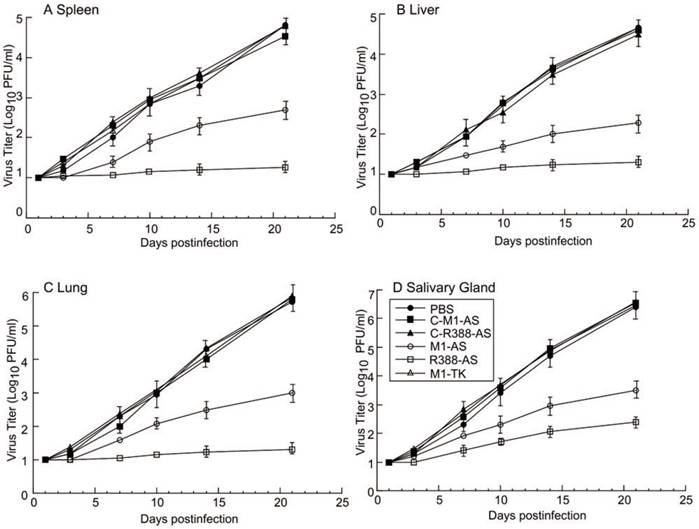
The second set of experiments was to study viral growth in mice treated with different ribozymes. Because of our observations of the inhibition of viral AS expression in mice treated with ribozymes R388-AS and M1-AS, it was reasonable to suggest that MCMV replication would be inhibited in these mice. To determine whether this was the case, viral load in different organs was examined at different time points post-infection. The viral titers in mice receiving the control ribozymes (i.e. C-R388-AS, C-M1-AS, or M1-TK) were similar to those in mice receiving phosphate-buffered saline (PBS) only (Figure 7). On the other hand, the viral titers in mice receiving R388-AS and M1-AS were consistently lower at every time point. At 21 days post-infection, the viral titers in the spleens, livers, lungs, and salivary glands of the mice receiving R388-AS were lower than those in mice receiving no ribozyme by 3,000-, 2,000-, 4,000-, and 15,000-fold, respectively, while those in the spleens, livers, lungs, and salivary glands of the mice receiving M1-AS were lower than mice receiving no ribozyme by 100-, 200-, 800-, and 1,000-fold, respectively (Figure 7). These findings suggested that the expression of R388-AS and M1-AS inhibited viral infection in vivo. Moreover, R388-AS inhibited viral infection more effectively than M1-AS in mice.
Ribozyme effect on MCMV growth in SCID mice. MCMV-infected mice were hydrodynamically transfected with PBS only (PBS) and PBS containing different M1GS constructs as described in Materials and Methods. Spleens (A), livers (B), lungs (C), and salivary glands (D) were harvested at 1, 3, 7, 10, 14, and 21 days postinfection, and viral titers were determined with plaque assays. Experiments were performed in triplicate and repeated three times.
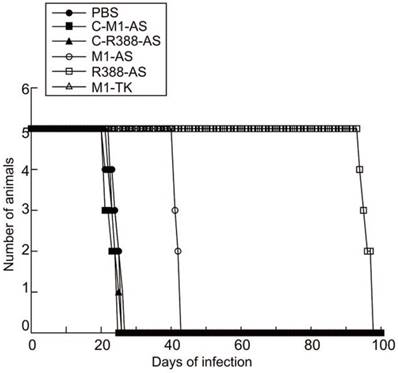
The third set of experiments was to examine the ribozyme effect on the survival of the MCMV-infected mice. It is conceivable that the survival of the mice would increase if viral replication was blocked in mice treated with ribozymes R388-AS and M1-AS. To determine if this was the case, the survival rate of the animals receiving ribozymes R388-AS and M1-AS was studied and compared to animals receiving PBS only or the control ribozymes C-R388-AS, C-M1-AS, or M1-TK. All non-infected animals given LXSN-M1GS survived and showed no signs of distress for at least 100 days (data not shown). Control ribozymes C-R388-AS, C-M1-AS, or M1-TK had no effect on improving survivability of MCMV-infected mice compared with those treated with PBS only, which died within 26 to 27 days post-infection (Figure 8). On the contrary, the life span of MCMV-infected mice receiving R388-AS and M1-AS improved substantially as no animals died until 95 and 43 days post-infection, respectively (Figure 8). These results suggested that MCMV gene expression and viral replication were inhibited in animals receiving R388-AS, and M1-AS, leading to overall improved survival of the mice.
Ribozyme effect on survivability of MCMV-infected SCID mice. MCMV-infected mice were hydrodynamically transfected with PBS only (PBS) and PBS containing different M1GS constructs as described in Materials and Methods. Animal survival was monitored for at least 100 days postinfection.
Discussion
The RNase P ribozyme-based approach demonstrates an innovative strategy for gene silencing for its highly efficient knockdown of the target RNA sequences of choice [11, 12]. The simple design of the guide sequence makes M1GS ribozyme a novel and useful tool for antiviral applications. However, the knowledge is limited regarding the rate-limiting step of M1GS ribozyme-based cleavage in cultured cells and in vivo and how to improve catalytic activity of ribozyme-mediated cleavage. In this report, a ribozyme, R388-AS, was designed to target the MCMV AS mRNA sequence. We hypothesize that the effectiveness of the ribozyme-mediated cleavage in cultured cells and in vivo is mediated by its catalytic efficiency in vitro (i.e. kcat/Km). For this hypothesis to be true, an increase in the catalytic efficiency of ribozymes should lead to better cleavage effect on targeted mRNA in cultured cells and mice.
Our study provides direct evidence that a ribozyme variant (i.e. R388-AS) selected in vitro is about 200-fold more active [kcat/Km] than the ribozyme derived from wild type M1 RNA (i.e. M1-AS) in targeting the AS mRNA sequence in vitro. Also, R388-AS exhibited better efficacy than M1-AS in knocking down assemblin expression and viral replication in cells and in mice. In contrast, we observed minimal knockdown (<10%) in AS expression and viral replication in cells and mice expressing control ribozymes C-R388-AS, C-M1-AS, or M1-TK (Figures 4 and 7, Table 2). Control ribozymes C-R388-AS and C-M1-AS had comparable binding affinities to substrate as-39 as R388-AS and M1-AS, respectively (Table 1). However, they exhibited little activity to cleave the substrate because of the mutations at their P4 catalytic sequences that abolished catalytic activity (Table 1). These findings indicated that the detected inhibition of assemblin expression and MCMV replication in cells and mice expressing R388-AS and M1-AS could be credited to ribozyme activity for cleaving the target AS mRNA but not to the antisense effect or additional nonspecific effects from the ribozymes. Likewise, the experiments indicated that R388-AS, which had better catalytic activity [i.e. kcat(apparent)/Km(apparent)] than M1-AS in vitro, was also more active than M1-AS in repressing MCMV gene expression and replication in cell and in mice. These findings support the hypothesis that improving the catalytic activity of ribozymes in vitro may enhance ribozyme mediated cleaving effects in cells and in animals.
Our studies also demonstrated that variant R388-AS was successfully expressed and functional in inhibiting MCMV AS expression and replication in cells and in animals. First, we could detect noticeable levels of R388-AS RNA in cultured cells and different organs of mice such as livers and spleens. Second, we found no difference in the MTT experiments with regards to the growth and viability of the cell lines with ribozymes compared to the parental cells for a three-month period. Mice receiving LXSN-M1GS plasmids grew with no adverse symptoms and were similar to mice receiving PBS only for a three-month period (data not shown). These findings suggest that expressing these ribozymes may be non-toxic to cells and animals. Third, R388-AS specifically repressed assemblin expression, but did not alter the expression of other MCMV genes such as M36, mie1, MCP, M112, and M99 (Table 2). Furthermore, R388-AS seemed to be effective in cleaving AS mRNA in mice. Inhibited AS gene expression, repressed viral titers, and improved survivability were detected in mice receiving R388-AS and M1-AS but not from mice receiving PBS only or control ribozymes C-R388-AS, C-M1-AS, or M1-TK (Figures 6-8) (data not shown). These findings implied that R388-AS cleaved its target AS mRNA in vivo, leading to inhibition of AS expression, decreased MCMV replication, and enhanced survivability of the infected animals.
Hydrodynamic transfection via tail vein injection is one of the best ways to deliver plasmid DNAs to the livers and spleens with great efficiency [29-31]. Although it is not suitable in clinical settings, the hydrodynamic transfection technique is an excellent method to deliver novel agents into animals for study in vivo [29-31]. The ribozyme-based technology is unique and attractive as this method employs M1 RNA, a catalytic RNA found in nature [11, 12], to cleave the target mRNAs. The activity of M1 RNA as well as M1GS RNAs has been shown to be enhanced by C5 protein from E. coli and several human proteins [12]. Furthermore, previous studies as well as this study showed that M1GSs effectively repress human and viral gene expression, and that they may be as effective in knocking down gene expression as small interfering RNAs (siRNAs) and other ribozymes [15, 20, 48-52]. Thus, M1GSs represent a novel and attractive class of nucleic acid-based gene interfering agents for therapeutic application. Further studies on the effectiveness of the M1GS-based approach and other nucleic acid-based gene interfering approaches should reveal the advantages and shortcomings of these gene-targeting strategies.
One of the most important findings in our current study was the isolation of ribozyme variant R388-AS. The biochemistry of RNase P ribozymes was studied [12, 53] and the three-dimensional structures of several RNase P catalytic RNAs were reported [42, 43, 54]. Several conserved regions including nucleotides 1-61 and 270-332 constitute the active site. Other conserved regions such as nucleotides 73-100 and 110-200 consist of the binding domain for a tRNA. However, how M1GS ribozymes interact with a target mRNA is not well-known. R388 had three point mutations (i.e. (G33 -> C33, C158 -> G158, and G292 -> A292) that have previously not been reported. It is currently not known how these point mutations increase the activity of R388-AS to cleave its target mRNA substrate. Our previous studies of M1GS ribozyme variants suggest that point mutations can independently enhance substrate binding and the rate of chemical cleavage [55]. Our results showed that R388-AS exhibited higher overall cleavage rate (kcat/Km)s and better binding affinity (Kd) (Table 1), suggesting the possible roles of the point mutations of this variants in modulating step of cleavage and substrate binding. Further characterization of R388 and other variants will elucidate the mRNA-cleaving mechanism of M1GS and advance the use of these ribozymes for various applications.
Because CMV is a DNA virus, genome editing approaches such as the transcription activator-like effector nuclease (TALEN) and CRISPR/Cas9 systems may be advantageous by targeting the viral genomic DNA [56, 57]. However, the efficacy of these genome editing approaches in reducing viral genomic DNA levels in animals has not yet been determined [58], while knocking down viral essential mRNA expression by numerous methods such as RNAi and ribozymes has yielded impressive inhibition of viral infection and replication [6, 12, 17, 59]. Further studies should be carried out to evaluate the effectiveness of both genome editing approaches and mRNA-targeting strategies. These studies, as well as further investigations on increasing ribozymes cleavage activity and specificity, will lead to develop more effective ribozymes for better gene-silencing applications.
Acknowledgements
We are grateful to Marco Paliza-Corre, Hao Gong, and Ting Wang for critical comments, reagents, and technical assistance.
This research has been supported by grants from Guangdong Innovative and Entepreneurial Research Team Program (No. 2014ZT05S136), the National Mega Project on Major Infectious Disease Prevention (2012ZX10002006-003 and 2012ZX10004-207), National Mega Project on Major Drug Development (2009ZX09103-678, 2012ZX09102301-004, 2012ZX09103301-20, 2013ZX09102-031, 2014ZX09509001-001), National Natural Science Foundation of China (No. 21708014), Antiviral Cooperative Innovation Center of Traditional Chinese Medicine at Shandong Province (XTCX2014B01-09), Natural Science Foundation of Guangdong Province, China (No. 2015A030310515), Project for Construction of Guangzhou Key Laboratory of Virology (No. 201705030003), National Small Business Innovation and Research (SBIR) Program of China, the Technology R & D Program of Jiangsu Province, China (BG2007035 and BG2008662), Open Research Fund Program of the State Key Laboratory of Virology of China (2015KF011 and 2018IOV006), a grant-in aid program for cultivate and innovation from Jinan University (No. 21616302), and NIH (RO1-AI041927, RO1-AI091536, RO1-DE023935, and RO1-DE025462).
Author Contributions Statement: W.L, Y.L, Y.W, R.L, P.T, W.T., Z.Y, Y.W, X.S, X.X, S.L, and F.L conceived and designed the experiments. W.L, Y.L, Y.W, R.L, P.T, W.T., Z.Y, Y.W, X.S, performed the experiments. W.L, Y.L, Y.W, R.L, P.T, W.T., Z.Y, Y.W, X.S, X.X, S.L, and F.L analyzed the data: W.L, Y.L, Y.W, R.L, P.T, W.T., Z.Y, Y.W, and X.S contributed reagents/materials/analysis tools. W.L, Y.L, X.X, S.L, and F.L wrote the paper. All authors reviewed the manuscript.
Conflict of Interests
The authors declare that three authors, Z.Y, Y.W, and X.S, are employed by commercial companies, Jiangsu Affynigen Biotechnologies, Inc., and Guangzhou Qinheli Biotechnologies, Inc. This does not alter the authors' adherence to Theranostics policies. All other authors declare no competing financial interests.
References
1. Mocarski ES, Shenk T, Griffiths PD, Pass RF. Cytomegaloviruses. In: Knipe DM, Howley PM, Cohen JI, al. e, editors. Fields Virology. Philadelphia, Pa.: Lippincott-William & Wilkins. 2013;2:1960-2014
2. Collins TM, Quirk MR, Jordan MC. Biphasic viremia and viral gene expression in leukocytes during acute cytomegalovirus infection of mice. J Virol. 1994;68:6305-11
3. Katzenstein DA, Yu GS, Jordan MC. Lethal infection with murine cytomegalovirus after early viral replication in the spleen. J Infect Dis. 1983;148:406-11
4. Abenes G, Chan K, Lee M, Haghjoo E, Zhu J, Zhou T. et al. Murine cytomegalovirus with a transposon insertional mutation at open reading frame m155 is deficient in growth and virulence in mice. J Virol. 2004;78:6891-9
5. Pollock JL, Virgin HWt. Latency, without persistence, of murine cytomegalovirus in the spleen and kidney. J Virol. 1995;69:1762-8
6. Scherer LJ, Rossi JJ. Approaches for the sequence-specific knockdown of mRNA. Nat Biotechnol. 2003;21:1457-65
7. Stein CA, Cheng YC. Antisense oligonucleotides as therapeutic agents-is the bullet really magical? Science. 1993;261:1004-12
8. Sarver N, Cantin EM, Chang PS, Zaia JA, Ladne PA, Stephens DA. et al. Ribozymes as potential anti-HIV-1 therapeutic agents. Science. 1990;247:1222-5
9. Wiebusch L, Truss M, Hagemeier C. Inhibition of human cytomegalovirus replication by small interfering RNAs. J Gen Virol. 2004;85:179-84
10. Jacque JM, Triques K, Stevenson M. Modulation of HIV-1 replication by RNA interference. Nature. 2002;418:435-8
11. Evans D, Marquez SM, Pace NR. RNase P: interface of the RNA and protein worlds. Trends Biochem Sci. 2006;31:333-41
12. Gopalan V, Altman S. RNase P:structure and catalysis. In: (ed.) Gesteland R, Cech T, Atkins J. The RNA World. Cold Spring Harbor: Cold Spring Harbor Laboratory Press. 2006 Chapter 6.1(online only at http://rna.cshl.edu)
13. Altman S, Kirsebom LA. Ribonuclease P. In: Gesteland RF, Cech TR, Atkins JF, editors. The RNA World. Cold Spring Harbor: Cold Spring Harbor Press. 1999;5:351-80
14. Frank DN, Pace NR. Ribonuclease P: unity and diversity in a tRNA processing ribozyme. Annu Rev Biochem. 1998;67:153-80
15. Guerrier-Takada C, Li Y, Altman S. Artificial regulation of gene expression in Escherichia coli by RNase P. Proc Natl Acad Sci U S A. 1995;92:11115-9
16. Forster AC, Altman S. External guide sequences for an RNA enzyme. Science. 1990;249:783-6
17. Kim K, Liu F. Inhibition of gene expression in human cells using RNase P-derived ribozymes and external guide sequences. Biochim Biophys Acta. 2007;1769:603-12
18. Liu F, Altman S. Inhibition of viral gene expression by the catalytic RNA subunit of RNase P from Escherichia coli. Genes Dev. 1995;9:471-80
19. Kilani AF, Trang P, Jo S, Hsu A, Kim J, Nepomuceno E. et al. RNase P ribozymes selected in vitro to cleave a viral mRNA effectively inhibit its expressionin cell culture. J Biol Chem. 2000;275:10611-22
20. Plehn-Dujowich D, Altman S. Effective inhibition of influenza virus production in cultured cells by external guide sequences and ribonuclease P. Proc Natl Acad Sci U S A. 1998;95:7327-32
21. Bai Y, Trang P, Li H, Kim K, Zhou T, Liu F. Effective inhibition in animals of viral pathogenesis by a ribozyme derived from RNase P catalytic RNA. Proc Natl Acad Sci U S A. 2008;105:10919-24
22. Gold L, Polisky B, Uhlenbeck O, Yarus M. Diversity of oligonucleotide functions. Annu Rev Biochem. 1995;64:763-97
23. Joyce GF. Directed molecular evolution. Sci Am. 1992;267:90-7
24. Szostak JW. In vitro genetics. Trends Biochem Sci. 1992;17:89-93
25. Sloan JH, Loutsch JM, Boyce SY, Holwerda BC. Expression and characterization of recombinant murine cytomegalovirus protease. J Virol. 1997;71:7114-8
26. Loutsch JM, Galvin NJ, Bryant ML, Holwerda BC. Cloning and sequence analysis of murine cytomegalovirus protease and capsid assembly protein genes. Biochem Biophys Res Commun. 1994;203:472-8
27. Dunn W, Chou C, Li H, Hai R, Patterson D, Stolc V. et al. Functional profiling of human cytomegalovirus genome. Proc Natl Acad Sci U S A. 2003;100:14223-8
28. Welch AR, Woods AS, McNally LM, Cotter RJ, Gibson W. A herpesvirus maturational proteinase, assemblin: identification of its gene, putative active site domain, and cleavage site. Proc Natl Acad Sci U S A. 1991;88:10792-6
29. Zhang G, Budker V, Wolff JA. High levels of foreign gene expression in hepatocytes after tail vein injections of naked plasmid DNA. Hum Gene Ther. 1999;10:1735-7
30. Song E, Lee SK, Wang J, Ince N, Ouyang N, Min J. et al. RNA interference targeting Fas protects mice from fulminant hepatitis. Nat Med. 2003;9:347-51
31. Liu F, Song Y, Liu D. Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Ther. 1999;6:1258-66
32. Ares M Jr, Igel AH. Lethal and temperature-sensitive mutations and their suppressors identify an essential structural element in U2 small nuclear RNA. Genes Dev. 1990;4:2132-45
33. Zaug AJ, Cech TR. Analysis of the structure of Tetrahymena nuclear RNAs in vivo: telomerase RNA, the self-splicing rRNA intron, and U2 snRNA. RNA. 1995;1:363-74
34. Bai Y, Li H, Vu G, Gong H, Umamoto S, Zhou T. et al. Salmonella-mediated delivery of RNase P ribozymes for inhibition of viral gene expression and replication in human cells. Proc Natl Acad Sci U S A. 2010;107:7269-74
35. Trang P, Lee M, Nepomuceno E, Kim J, Zhu H, Liu F. Effective inhibition of human cytomegalovirus gene expression and replication by a ribozyme derived from the catalytic RNA subunit of RNase P from Escherichia coli. Proc Natl Acad Sci U S A. 2000;97:5812-7
36. Kim JJ, Kilani AF, Zhan X, Altman S, Liu F. The protein cofactor allows the sequence of an RNase P ribozyme to diversify by maintaining the catalytically active structure of the enzyme. RNA. 1997;3:613-23
37. Trang P, Kim K, Zhu J, Liu F. Expression of an RNase P ribozyme against the mRNA encoding human cytomegalovirus protease inhibits viral capsid protein processing and growth. J Mol Biol. 2003;328:1123-35
38. Pyle AM, McSwiggen JA, Cech TR. Direct measurement of oligonucleotide substrate binding to wild-type and mutant ribozymes from Tetrahymena. Proc Natl Acad Sci U S A. 1990;87:8187-91
39. Jiang X, Chen YC, Gong H, Trang P, Lu S, Liu F. Ribonuclease P-mediated inhibition of human cytomegalovirus gene expression and replication induced by engineered external guide sequences. RNA biology. 2012;9:1186-95
40. Lee M, Xiao J, Haghjoo E, Zhan X, Abenes G, Tuong T. et al. Murine cytomegalovirus containing a mutation at open reading frame M37 is severely attenuated in growth and virulence in vivo. J Virol. 2000;74:11099-107
41. Rawlinson WD, Farrell HE, Barrell BG. Analysis of the complete DNA sequence of murine cytomegalovirus. J Virol. 1996;70:8833-49
42. Reiter NJ, Osterman A, Torres-Larios A, Swinger KK, Pan T, Mondragon A. Structure of a bacterial ribonuclease P holoenzyme in complex with tRNA. Nature. 2010;468:784-9
43. Torres-Larios A, Swinger KK, Krasilnikov AS, Pan T, Mondragon A. Crystal structure of the RNA component of bacterial ribonuclease P. Nature. 2005;437:584-7
44. Bertrand E, Castanotto D, Zhou C, Carbonnelle C, Lee NS, Good P. et al. The expression cassette determines the functional activity of ribozymes in mammalian cells by controlling their intracellular localization. RNA. 1997;3:75-88
45. Miller AD, Rosman GJ. Improved retroviral vectors for gene transfer and expression. Biotechniques. 1989;7:980-90
46. Li K, Williams RS. Cloning and characterization of three new murine genes encoding short homologues of RNase P RNA. J Biol Chem. 1995;270:25281-5
47. Altman S, Wesolowski D, Puranam RS. Nucleotide sequences of the RNA subunit of RNase P from several mammals. Genomics. 1993;18:418-22
48. Yuan Y, Hwang ES, Altman S. Targeted cleavage of mRNA by human RNase P. Proc Natl Acad Sci U S A. 1992;89:8006-10
49. Zhu J, Trang P, Kim K, Zhou T, Deng H, Liu F. Effective inhibition of Rta expression and lytic replication of Kaposi's sarcoma-associated herpesvirus by human RNase P. Proc Natl Acad Sci U S A. 2004;101:9073-8
50. Ma M, Benimetskaya L, Lebedeva I, Dignam J, Takle G, Stein CA. Intracellular mRNA cleavage induced through activation of RNase P by nuclease-resistant external guide sequences. Nat Biotechnol. 2000;18:58-61
51. Hnatyszyn H, Spruill G, Young A, Seivright R, Kraus G. Long-term RNase P-mediated inhibition of HIV-1 replication and pathogenesis. Gene Ther. 2001;8:1863-71
52. Xia C, Chen YC, Gong H, Zeng W, Vu GP, Trang P. et al. Inhibition of hepatitis B virus gene expression and replication by ribonuclease P. Mol Ther. 2013;21:995-1003
53. Kazantsev AV, Pace NR. Bacterial RNase P: a new view of an ancient enzyme. Nat Rev Microbiol. 2006;4:729-40
54. Mondragon A. Structural studies of RNase P. Annu Rev Biophys. 2013;42:537-57
55. Zou H, Lee J, Umamoto S, Kilani AF, Kim J, Trang P. et al. Engineered RNase P ribozymes are efficient in cleaving a human cytomegalovirus mRNA in vitro and are effective in inhibiting viral gene expression and growth in human cells. J Biol Chem. 2003;278:37265-74
56. Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096
57. Stone D, Niyonzima N, Jerome KR. Genome editing and the next generation of antiviral therapy. Human Genetics. 2016;135:1071-82
58. van Diemen FR, Lebbink RJ. CRISPR/Cas9, a powerful tool to target human herpesviruses. Cellular Microbiology. 2017:19
59. Santoro SW, Joyce GF. A general purpose RNA-cleaving DNA enzyme. Proc Natl Acad Sci U S A. 1997;94:4262-6
Author contact
![]() Corresponding author: Dr. Fenyong Liu, School of Public Health, University of California, Berkeley, CA 94720, Tel: +1-510-643-2436, Fax: +1-510-643-9955, Email: liu_fyedu Dr. Xiwen Xing, Department of Biotechnology, Jinan University, Guangzhou, Guangdong 510632, China, Tel: +86-20-85220219, Fax: +86-20-85222616, Email: xingxiwenedu.cn Dr. Sangwei Lu, School of Public Health, University of California, Berkeley, CA 94720, Tel: +1-510-643-4986, Fax: +1-510-643-0896, Email: sangweiedu
Corresponding author: Dr. Fenyong Liu, School of Public Health, University of California, Berkeley, CA 94720, Tel: +1-510-643-2436, Fax: +1-510-643-9955, Email: liu_fyedu Dr. Xiwen Xing, Department of Biotechnology, Jinan University, Guangzhou, Guangdong 510632, China, Tel: +86-20-85220219, Fax: +86-20-85222616, Email: xingxiwenedu.cn Dr. Sangwei Lu, School of Public Health, University of California, Berkeley, CA 94720, Tel: +1-510-643-4986, Fax: +1-510-643-0896, Email: sangweiedu
Short Title: Antiviral RNase P ribozyme
 Global reach, higher impact
Global reach, higher impact