13.3
Impact Factor
Theranostics 2018; 8(19):5213-5230. doi:10.7150/thno.28001 This issue Cite
Research Paper
PIWI-interacting RNA-54265 is oncogenic and a potential therapeutic target in colorectal adenocarcinoma
1. State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Sun Yat-sen University Cancer Center, Guangzhou 510060, P. R. China
2. Department of Colorectal Surgery, Sun Yat-sen University Cancer Center, Guangzhou 510060, P. R. China;
3. Department of Pathology, Sun Yat-sen University Cancer Center, Guangzhou 510060, P. R. China;
4. Department of Cancer Genetics, Medical College of Soochow University, Suzhou 215123, P. R. China;
5. Department of Etiology and Carcinogenesis, National Cancer Center/National Clinical Research Center/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing 100021, P. R. China;
6. Department of Colorectal Surgery, National Cancer Center/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing 100021, P. R. China;
7. Department of Radiation Oncology, National Cancer Center/National Clinical Research Center/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing 100021, P. R. China;
8. Department of Medical Oncology, National Cancer Center/National Clinical Research Center/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing 100021, P. R. China;
9. Department of Pathology, National Cancer Center/ National Clinical Research Center/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing 100021, P. R. China;
10. Department of Medical Oncology, Sun Yat-sen University Cancer Center, Guangzhou 510060, P. R. China.
#Dongmei Mai, Peirong Ding and Liping Tan contributed equally to this work.
Received 2018-6-19; Accepted 2018-8-30; Published 2018-10-6
Abstract
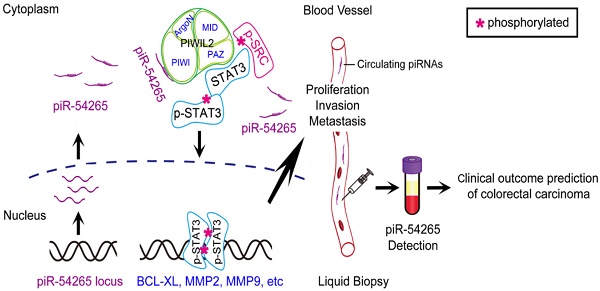
Although PIWI-interacting RNAs (piRNAs) have recently been linked to human diseases, their roles and functions in malignancies remain unclear. This study aimed to investigate the significance of some piRNAs in colorectal cancer (CRC).
Methods: We first analyzed the expression profile of piRNAs in CRC using the TCGA and GEO databases. The top 20 highly expressed piRNAs were selected and tested in our CRC tumor and non-tumor tissue samples. We then examined the relevance of the significantly differentially expressed piRNA to the CRC outcomes in 218 patients receiving postoperative chemotherapy and 317 patients receiving neoadjuvant chemotherapy. A series of biochemical and molecular biological assays were conducted to elucidate the functional mechanism of a piRNA of interest in CRC. Furthermore, experiments with mice xenografts were performed to evaluate the therapeutic effect of an inhibitor specific to the piRNA.
Results: We found that among the examined 20 piRNAs, only piRNA-54265 was overexpressed in CRC compared with non-tumor tissues and higher levels in tumor or in serum were significantly associated with poor survival in patients. Functional assays demonstrated that piRNA-54265 binds PIWIL2 protein and this is necessary for the formation of PIWIL2/STAT3/phosphorylated-SRC (p-SRC) complex, which activates STAT3 signaling and promotes proliferation, metastasis and chemoresistance of CRC cells. Treatment with a piRNA-54265 inhibitor significantly suppressed the growth and metastasis of implanted tumors in mice.
Conclusion: These results indicate that piRNA-54265 is an oncogenic RNA in CRC and thus might be a therapeutic target.
Keywords: piRNA, biomarker, STAT3, therapeutic target, colorectal cancer
Introduction
Colorectal cancer (CRC) takes the third place in men and the second place in women across all types of cancer in terms of incidence and is also one of the leading causes of cancer-related death in the world [1, 2]. To date, early detection or diagnosis of sporadic CRC is still difficult because there are no effective markers and measures. Therefore, many patients in clinics have late-stage disease, and their prognosis is dismal due to the lack of effective therapy [3]. The molecular mechanism for CRC progression is largely unknown, impeding the development of more effective novel therapies for CRC. Presently, neoadjuvant chemotherapy using 5-fluorouracil (5-FU) and/or oxaliplatin combined with surgical resection is routinely recommended for advanced CRC, which may improve survival of patients [4]. However, the curative effects and clinical outcomes in patients treated with the regimen vary greatly and many patients have resistance to the drugs [5]. The mechanism underlying resistance to chemotherapeutic drugs has not been fully elucidated and thus few applicable biomarkers can be used in the clinic to predict the curative effect of chemotherapy [6]. These contexts highlight a need to elucidate the mechanism underlying CRC progression and to identify therapeutic targets or predictive biomarkers for personalized treatment of CRC.
Small non-coding RNAs (ncRNAs) have attracted special attention because of their roles as key regulators of gene expression and can be easily detected in blood samples [7-9]. PIWI-interacting RNAs (piRNAs) have been discovered as a new class of small ncRNAs [10, 11], and to date, there are more than 20,000 piRNAs identified [12, 13]. Although piRNAs were originally perceived as germline-specific [10], they are also expressed in somatic tissues [14]. The multifaceted somatic functions of piRNAs have been demonstrated [15-17], and piRNA-dependent epigenetic regulation has enlightened compelling studies on the roles of piRNAs in human diseases including cancer [18, 19]. Although little is known about the functions of piRNAs in human cancer, emerging studies have suggested that piRNAs may play an important role in cancer development and progression and may serve as diagnostic and prognostic biomarkers [19-22].
Here, we identify piRNA 54265 (piR-54265) as an oncogenic piRNA that plays an important role in CRC development and progression. Our biochemical assays demonstrate that piR-54265 specifically binds PIWIL2, facilitating the formation of PIWIL2/STAT3/phosphorylated-SRC complex [23] and phosphorylation of STAT3, which consequently promote proliferation and metastasis of CRC cells. We show that overexpressed piR-54265 prevents CRC cells from undergoing apoptosis and is correlated with resistance to anticancer agents and poor prognosis in CRC patients. Treatment targeting piR-54265 shows efficacy in inhibiting xenograft tumor growth and metastasis in mice. Serum piR-54265 levels are up-regulated in a stage-dependent manner, and high piR-54265 level is associated with poor response to chemotherapy and poor survival in CRC patients. All these findings suggest that this oncogenic piRNA might be a therapeutic target and could be used as a circulating tumor RNA for predicting clinical outcomes of CRC.
Methods
Clinical specimens
This study consisted of four independent cohorts (Figure S1) from Sun Yat-sen University Cancer Center (SYSUCC, Guangzhou), the Affiliated Hospitals of Soochow University (AHSU, Suzhou) and Cancer Hospital, Chinese Academy of Medical Sciences (CHCAMS, Beijing). A total of 218 paired cancer and normal tissue (at least 5 cm away from tumor) and matched serum specimens from CRC patients who underwent proctocolectomy and postoperative chemotherapy at the SYSUCC (SYSUCC1, N=110) and AHSU (N=108) between 2008 and 2012. All CRC were histopathologically diagnosed and tumor stage was classified according to the 7th edition of AJCC Cancer Staging System [24]. All patients received standard FOLFOX or XELOX chemotherapy for 6 months after surgery. The tissue and blood samples from each individual were collected during the operation. Tissue samples were immediately frozen and stored in liquid nitrogen until use. Serum was isolated from blood samples and stored at -80 °C. The basic demography characteristics and clinical information of these patients were obtained from medical records (Table S1). Overall survival (OS) time of individuals with CRC was measured as the date of cancer diagnosis to the date of last follow-up or death while progression-free survival (PFS) time was defined as the date of treatment start to the date of tumor progression. The follow-up information was acquired from inpatient and outpatient records and follow-up telephone calls. The date of last follow-up at SYSUCC was 2016/5/10 with a median follow-up time of 65.8 months, while the date of last follow-up at AHSU was 2017/4/23 with a median follow-up time of 56.6 months.
Another 317 serum specimens from CRC patients who suffered from advanced CRC treated with neoadjuvant chemoradiotherapy were recruited at SYSUCC (SYSUCC2, N=215, Table S2) and CHCAMS (N=102, Table S3) between 2008 and 2013. All patients recruited at CHCAMS suffered from locally advanced rectal carcinoma without distant metastasis. They received a total radiation dose of 50 Gy applied in 25 fractions over 5 weeks concurrent with standard XELOX regimen prior to surgery. The response to the chemotherapy was evaluated by Tumor Regression Grade (TRG) [25, 26] using surgically removed tissue samples. TRG scores were: TRG 1, no residual cancer; TRG 2, rare residual cancer cells; TRG 3, fibrosis outgrowing residual cancer; TRG 4, residual cancer outgrowing fibrosis and TRG5, absence of regression changes (Figure S2). Patients recruited at SYSUCC suffered from either colon (N=10) or rectal cancer (N=205) and received standard FOLFOX or XELOX regimen concurrent with radiotherapy of total 50 Gy applied in 25 fractions over 5 weeks. All patients having measurable tumor lesions and CT/MRI scan obtained before treatment received a second CT/MRI scan around one month after chemotherapy. Response to therapy was evaluated by measuring the rate of change of tumor size based on CT/MRI scanning and classified into 4 groups according to the revised RECIST guideline (version 1.1) [27]: complete response (CR, disappearance of all tumor), partial response (PR, at least a 30% decrease in the sum of the largest diameter of tumor), stable disease (SD, neither sufficient shrinkage to qualify for PR nor sufficient increase to qualify for progressive disease), and progressive disease (PD, at least a 20% increase in the sum of the largest diameter of tumor). Vein blood sample (5 mL) from each patient was taken prior to the radio-chemotherapy and serum sample was isolated and stored at -80 °C for analysis.
Written informed consent was obtained from each subject and this study was approved by the Internal Review Boards of SYSUCC, AHSU and CHCAMS.
Public data mining and piRNA selection
Small RNA sequencing data of 616 colon and rectal adenocarcinoma were downloaded from the Cancer Genomics Hub (https://www.cghub.ucsc.edu/) with the permission of the Data Access Committee. Independent colon and rectal adenocarcinoma data containing 50 samples were acquired from the GEO (https://www.ncbi.nlm.nih.gov/geo, accession number GSE63119). To process the TCGA data, we extracted raw and unaligned read sequences and quality scores (FASTQ format) from BAM files using the “bamToFastq” command, a subcommand of the genome arithmetic toolset “BEDtools2” (v2.25.0). To process GEO data, raw reads were transformed into FASTQ files with the SRA Toolkit (https://www.ncbi.nlm.nih.gov/sra/docs/toolkitsoft/). Subsequently, the above two resultant files were trimmed based on the criterions of “Phred quality score ≥20” and “reads length ≥21 nucleotides” to get high-quality reads corresponding to piRNAs via FASTX-Toolkit (v0.0.13) (https://hannonlab.cshl.edu/fastx_toolkit/). Custom piRNA reference transcriptome information was obtained from the functional RNA database (v3.4) according to the genomic coordinates of piRNA sequences [28, 29]. Both TCGA and GEO reads were then realigned or remapped by the STAR software (v020201) with “hg19” as a reference genome and one mismatch allowed [30]. Counts per million mapped reads (CPM) was calculated to measure the abundance of piRNAs using HTseq-count (v0.9.1) [31] and R package edger (v3.18.1) [32] and only the piRNAs with CPM ≥1 and expression in more than 20% of tumor samples were considered to be adequately expressed and adopted for further analyses. There were 210 and 85 piRNAs that were respectively defined as “expressed” in the TCGA and GEO sets; intersecting resulted in 81 co-expressed piRNAs. These 81 piRNAs were then ranked according to their expression levels (mean value of CPM in all samples) in the more reliable TCGA set, the top 20 of which are shown in Figure S3A.
Copy number alteration and methylation analysis
The gene expression, somatic copy number alterations (SCNA) and DNA methylation data for CRC (colorectal adenocarcinoma, COADREAD) were obtained from TCGA database (PMID: 22810696). DNA copy number alterations including deletion, neutral and amplification for piR-54265 were determined by the copy number of segments covering the gene. Since the two DNA methylation probes of cg07941567 (chr20: 2637452) and cg08216131 (chr20: 2637503) were close to the locus of piR-54265 (chr20: 2637585-2637614) and right in its promoter region, the methylation status was employed for differential methylation analysis.
Cell lines and cell culture
Human CRC cell lines, HCT116 and LoVo, and human embryo kidney cell line 293T were purchased from the Cell Bank of Type Culture Collection of Chinese Academy of Sciences, Shanghai Institute of Biochemistry and Cell Biology. All cell lines were authenticated by DNA finger printing analysis. All cells were free of mycoplasma infection. HCT116, LoVo and HEK293T cells were respectively maintained in McCoy's 5A, F-12K and DMEM medium supplemented with 10% fetal bovine serum in an atmosphere of 5% CO2 at 37 °C.
Cytoplasmic and nuclear fractionation
Subcellular fractionation of cells was performed as described previously [33, 34]. Briefly, cytoplasmic fractions were extracted in ice-cold NP40 buffer containing 24% sucrose. Nuclear fractions were lysed with RIPA buffer supplemented with protease and phosphatase inhibitor cocktail (Roche).
Northern blot analysis
Approximately 15 µg of total RNA isolated from CRC cell lines or serum samples was subjected to formaldehyde gel electrophoresis and transferred to an Amersham Hybond-N+ membrane (GE Healthcare). After pre-hybridization for 30 min, the membrane was hybridized overnight at 68 °C in buffer containing the denatured digoxigenin-labeled RNA probes synthesized by Bersinbio (Table S4). After washing, signal on the membrane was detected using an Odyssey infrared scanner (Li-Cor, Lincoln).
Measurement of absolute piR-54265 copy number per cell
The accurate piR-54265 copy number per cell was measured by using an absolute RT-qPCR method. We used 8 different dilutions of synthetic piR-54265 to formulate a standard curve, in which each Ct value represents an exact concentration of piR-54265. The exact piR-54265 copy number per cell was then calculated on the basis of molecular weight and cell count.
RNA extraction and RT-qPCR analysis
Total RNA from cell lines and clinical tissues was extracted with Trizol reagent (Invitrogen), while total RNA from human serum samples was extracted with the MagMAX™ mirVana™ Total RNA Isolation Kit (Applied Biosystems). Reverse transcription reactions were conducted with oligo (dT) or specific miRNA/piRNA stem-loop RT primers using the Revert Aid First Strand cDNA Synthesis Kit (Thermo). Relative RNA levels determined by RT-qPCR were measured on the Light Cycler 480 II using the SYBR Green method. The primer sequences used are shown in Table S5. For quantification of piRNAs, U6 small nuclear RNA was employed as an internal control in both cell lines and tissue samples. Serum samples were spiked in synthesized cel-miR-39 and cel-miR-54 (RuiBiotech) as exogenous controls. All experiments were performed in three biological replicates. The relative expression of RNAs was calculated as the power value (2-△△Ct).
Plasmids, lentivirus production and transduction
To establish CRC cells overexpressing piR-54265, HCT116 and LoVo cells were infected with recombinant lentiviruses designated piR-54265, which express pi-sno57 because of the insertion of SNORD57 precursor downstream of CMV promoter in pLent-Puro-GFP (Vigene Biosciences), a lentiviral vector or an empty pLent-Puro-GFP vector, which severs as control. A short hairpin RNA (shRNA) specifically targeting piR-54265 was synthesized and inserted into the pSIH1-Puro-GFP lentiviral shRNA vector (Obio Technology) to produce lentivirus in HEK293T cells. A scrambled shRNA was used as control. The overexpression or knockdown of piR-54265 in cells was verified by qRT-PCR.
Expression vector construction
To construct expression vectors for FLAG-tagged PIWIL2, cDNAs encoding PIWIL2 or various truncated PIWIL2 were synthesized and subcloned into the NheI and BamHI sites of the pcDNA3.1(+)-3FLAG vector (Obio Technology). This approach was also used for the construction of HIS-tagged STAT3.
Cell viability and colony formation assays
Cell viability was measured using the CCK-8 kit (Dojindo Labs). Colony formation ability was determined by counting the number of cells in 6-well cell-culture clusters with complete growth media after fixing with methanol and staining with crystal violet.
Cell cycle and apoptosis analysis
Flow cytometry (ACEA NovoCyte) was used to determine cell cycle and apoptosis. Briefly, for cell cycle analysis, cells were collected and fixed in 70% ethanol overnight at 4 °C. Single-cell suspensions were labeled with propidium iodide (PI, 50 μg/mL; Keygen Biotech) and analyzed by flow cytometry. For detection of apoptosis, cells were stained with Annexin V and PI using the Annexin V-FITC Apoptosis Detection kit (Invitrogen), and the percentage of apoptotic cells was determined by flow cytometry.
In vitro invasion and migration assays
Invasion assay was done in a 24-well Millicell chamber. The 8-μm pore inserts were coated with 30 μg of Matrigel (BD Biosciences). Cells (LoVo, 2×104; HCT116, 8×104) were added to coated filters in 200 μL of serum-free medium in triplicate wells. 500 μL medium containing 20% fetal bovine serum was added to the lower chamber as chemo-attractant. After 20 h at 37 °C in a 5% CO2 incubator, the Matrigel coating on the upper surface of the filter was removed. Cells that migrated through the filters were fixed with methanol, stained with 0.5% crystal violet, and photographed. Cell number on three random fields was counted. The migration assay was conducted in a similar fashion without coating with Matrigel.
RNA immunoprecipitation (RIP) assay
RIP experiments were performed using the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore). Antibodies from Abcam against PIWIL1 (ab12337) or STAT3 (ab119352), from Santa Cruz against PIWIL2 (sc-67502), PIWIL3 (sc-398779) or PIWIL4 (sc-68932) and from Cell Signaling Technology against p-SRC (p-Tyr416, D4964) were used. Total RNA (input control) and isotype control (IgG) for each antibody were assayed simultaneously. The co-precipitated RNAs were detected by RT-qPCR.
RNA pull-down assay
RNA pulldown assays were performed according to the manufacturer's instructions of Pierce™ GST Protein Interaction Pull-Down Kit (Thermo Fisher). Briefly, commercially synthesized, biotinylated piR-54265 (Genepharma) or its antisense was incubated with cellular protein extracted from ordinary CRC cells or CRC cells transfected with whole-length PIWIL2 or truncated PIWIL2. Streptavidin beads were then added and recovered total proteins associated with piR-54265 were subjected to Western blot analysis.
Cell lysis and immunoprecipitation
Cells transfected with STAT3-6×HIS and PIWIL2-3×FLAG (whole length or truncated constructs) were lysed with 1× RIPA buffer supplemented with Protease/Phosphatase Inhibitor Cocktail (Pierce). Lysates were cleared by centrifugation and the supernatants were prepared for immunoblotting or immunoprecipitation with antibodies described below. Immunoblot signal was detected using Immobilon Western Chemiluminescent HRP Substrate (Millipore).
Western blot analysis
Protein extracts from cells or immunoprecipitation samples were prepared using detergent-containing lysis buffer. Total protein (50 µg) was subjected to SDS-PAGE and transferred to PVDF membrane (Millipore). Antibodies against STAT3 (ab119352), p-STAT3 (p-Y705, ab76315), SRC (ab109381), BCL-XL (ab32370), active CASPASE-3 (ab32351), MMP2 (ab86607) or MMP9 (ab58803) were from Abcam. Antibodies against cleaved CASPASE-7 (p20, D198; orb159339) or cleaved CASPASE-9 (p35, D315; orb159343) were from Biorbyt. Antibodies against PIWIL2 (sc-67502) or p-SRC (p-Tyr416, D4964) were from Santa Cruz or Cell Signaling Technology, respectively. Antibody against FLAG tag (F1804) or 6×HIS tag (SAB2702218) was from Sigma, while antibody against β-ACTIN (60008-1-Ig) was from Proteintech. Membranes were incubated overnight at 4 °C with primary antibody and visualized with Immobilon Western Chemiluminescent HRP Substrate (Millipore).
In vitro chemosensitivity assay
CRC cells were seeded in 96-well plates (5,000 cells/well) and about 14 h later right after cell adherence, oxaliplatin (Eloxatin) or 5-FU (Tianjin) were added to cells in a concentration gradient as indicated in the figure legends. After culturing for an additional 72 h, cell viability was measured by CCK8 assay. The results were expressed as percentage of cell survival on the basis of the difference between the optical density at the start and end of drug exposure. Drug concentration corresponding to a reduction in cell survival by 50% compared with that of control cells (IC50) was calculated from plots of drug concentration versus proportion of cells that survived. All analyses were performed in three experiments and each had three replicates.
Animal experiments
Animal experiments were carried out in compliance with approved protocols and guidelines from the Institutional Animal Care and Use Committee of SYSUCC. To examine the effects of piR-54265 on subcutaneous xenograft growth, BALB/c nude mice (5 mice/group; Beijing Vital River Laboratory Animal Technology) were subcutaneously injected with 0.1 mL of cell suspension containing 2×106 cells in the rear flank. When a tumor was palpable, it was measured every other day and its volume was calculated according to the formula volume = length × width2 × 0.5. Groups of animals with subcutaneous xenograft were also treated with chemotherapeutic agents to test the effects of piR-54265 on chemo-sensitivity of CRC cells. Oxaliplatin (6 mg/kg), 5-FU (100 mg/kg) or a combination of these two agents was intraperitoneally injected to mice once a week for 4 weeks since the day when the xenograft reached 250 mm3 (designated day 0). Tumor growth suppression was quantified every three days by volume measurement.
We also treated subcutaneous xenografts with antagopiR54265, a synthesized piR-54265 inhibitor (Ribobio; Table S4) by intra-tumor injection. Briefly, when tumors implanted in both left and right rear mouse flanks reached a certain volume, 50 μg of AntagopiR54265 in 50 μL phosphate buffered saline was directly injected into the tumor on one flank. The tumor on the other flank was injected with AntagopiR NC (Ribobio; Table S4) as control. Treatment was delivered every three days for 22 days and tumor volume was measured before each injection. Mice were sacrificed at the end of the experiment and the xenografts were stripped and photographed. Haematoxylin and eosin (H&E) staining was then performed for histological examination of CRC tumor.
To generate a metastasis model, luciferase-labeled HCT116 or LoVo cells (2×106) with piR-54265 overexpression or knockdown were injected into the tail vein of mice. The metastases were detected using a Living Image® system (Perkin Elmer) and the quantitative data were expressed as photon flux. At the end of the experiment, systemic organs were isolated for histopathological examination of CRC tumor. We also treated metastatic tumors with antagopiR54265. When tumors reached 1×106 radiance (p/s/cm2/sr), antagopiR54265 (80 mg/kg body weight) only or antagopiR54265 plus 5-FU (100 mg/kg body weight) was intravenously given to mice one time per day in the first three days, and after that one time per week for 3 weeks. Control mice were treated with AntagopiR NC in the same way. Tumor growth and spread was quantified once every week and the body weight and survival time of animals in each group were recorded.
Immunohistochemical analysis
Mice xenograft tumors were selected for examination of CRC cancer by haematoxylin and eosin (H&E) staining and for immunohistochemical staining (IHC) of p-STAT3, BCL-XL, active CASPASE 3, cleaved CASPASE 7, cleaved CASPASE 9, MMP2 and MMP9. All antibodies were the same as those used for Western blotting analysis except for MMP2 (ab97779) and MMP9 (ab38898), which were from Abcam. Briefly, for IHC, the sections were incubated with primary antibody at 4 °C overnight and then detected with the ABC Kit (Pierce).
Statistical analysis
Cox proportional hazard models were used to identify independent variables. HRs and 95% CIs were calculated with age, sex, tumor location, tumor stage and treatment as covariates. Kaplan-Meier estimate with log-rank test was used to compare patient survival by different piR-54265 expression levels. Linear regression was performed to examine the correlation of piR-54265 expression levels in tumor and serum and the correlation was considered significant when P<0.05 and r>0.30. Chi-square test was used to examine the relationships between piR-54265 expression levels and chemotherapy efficacy in CRC patients. Receiver operating characteristic curve was constructed to quantify the performance of serum piR-54265 levels as a biomarker by assessing its sensitivity, specificity and respective areas under the curve with 95% CI in a binary classifier system. We investigated the optimum cutoff value for diagnosis with the maximal Youden index (J). For functional analysis, results are presented as mean ± SEM of three or more experiments. The comparison of mean between two groups or more than two groups was conducted by using t-test or one-way ANOVA. Data in abnormal distributions were analyzed by nonparametric tests. All statistical analyses were performed using SPSS 20.0 (IBM, US) and P<0.05 was considered significant.
Results
piR-54265 is overexpressed in CRC and associated with clinical outcomes of patients
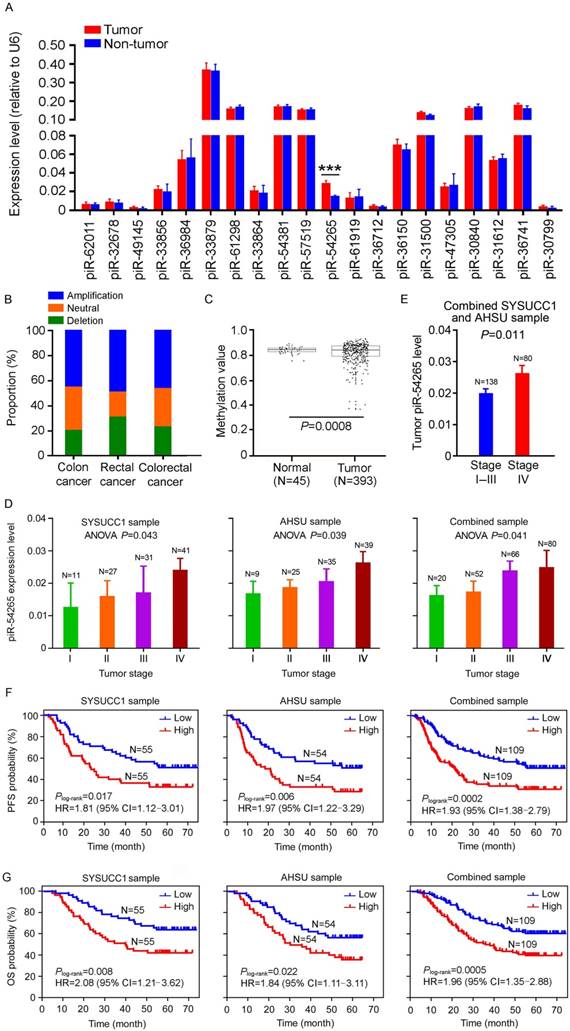
We firstly examined the expression patterns of piRNAs in CRC samples from TCGA and GEO databases and then performed RT-qPCR to assess the levels of the top 20 highly expressed piRNAs (Figure S3A) in a set of 110 paired CRC and non-tumor tissues collected at SYSUCC (SYSUCC1, Guangzhou). We found that, among these top 20 piRNAs, only piR-54265 was significantly overexpressed in tumor tissues compared with their non-tumor counterparts (Figure 1A) and this result was verified in another set of 108 samples consisting of paired CRC and non-tumor tissues from the AHSU (Suzhou; Figure S3B). Additional analysis of piR-54265 gene in TCGA data showed significant amplification and lower methylation in CRC specimens (Figure 1B-C), supporting our results. We then performed a series of experiments to characterize piR-54265 in two CRC cell lines. The existence of piR-54265 29 nucleotides in size was confirmed by RNA blot analysis (Figure S3C) and the absolute copy number of this piRNA was 800 to 1,000 copies per cell (Figure S3D) and the majority of piR-54265 was in the cytosol with a small amount being in the nucleus (Figure S3E).
We also examined the effect of piR-54265 level on clinical outcomes in our CRC patient sets (Table S1) and found that overexpression of this piRNA was significantly more elevated in advanced CRC (Figure 1D) and metastatic CRC (Figure 1E). Kaplan-Meier estimate showed that CRC patients with high piR-54265 level had significantly shorter progression-free survival (PFS) time and overall survival (OS) time than patients with low piR-54265 level in both SYSUCC1 and AHSU patient sets and combined samples (Figure 1F-G). Cox regression analysis with sex, age, smoking status, drinking status and family history as covariates further demonstrated piR-54265 level was an independent risk factor for PFS (HR=1.96, 95% CI=1.37-2.80) and OS (HR=1.99, 95% CI=1.35-2.93) in combined samples (Table 1 and Table 2). We further performed the analysis stratified by tumor location and TNM stage and found that higher level of piR-54265 was associated with shorter survival time in patients with either colon or rectal cancer (Figure S4A). However, with respect to TNM stage, higher level of piR-54265 was only associated with shorter survival time in patients with advanced cancer (Figure S4B). We did not find any correlations between piR-54265 level in CRC and clinical parameters other than TNM stage (Table 3) and survival time.
Univariate and multivariate Cox regression analyses for progression-free survival in CRC patients recruited at SYSUCC and AHSU who received 5-FU and oxaliplatin-based postoperative chemotherapy (N=218)
| Variable | Univariate | Multivariate | |||||
|---|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | ||
| Age at diagnosis (≥60 y vs. <60 y) | 1.11 | 0.78-1.58 | 0.564 | - | - | - | |
| Gender (male vs. female) | 1.00 | 0.67-1.49 | 0.991 | - | - | - | |
| Family history (yes vs. no) | 1.33 | 0.82-2.13 | 0.246 | - | - | - | |
| Smoking (ever vs. never) | 1.34 | 0.25-2.20 | 0.251 | - | - | - | |
| Drinking (ever vs. never) | 0.90 | 0.52-1.56 | 0.714 | - | - | - | |
| TNM stage (III/IV vs. I/II) | 1.94 | 1.56-2.41 | <0.001 | 1.97 | 1.59-2.44 | <0.001 | |
| Tumor location (colon vs. rectum) | 1.15 | 0.92-1.43 | 0.221 | - | - | - | |
| piR-54265 level (high vs. low) | 1.99 | 1.38-2.86 | <0.001 | 1.96 | 1.37-2.80 | <0.001 | |
AHSU: the Affiliated Hospitals of Soochow University (Suzhou, China); CI: confidence interval; HR: hazard ratio; SYSUCC: Sun Yat-sen University Cancer Center (Guangzhou, China).
Univariate and multivariate Cox regression analyses of overall survival in CRC patients recruited at SYSUCC and AHSU who received 5-FU and oxaliplatin-based postoperative chemotherapy (N=218)
| Variable | Univariate | Multivariate | |||||
|---|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | ||
| Age at diagnosis (≥60 y vs. <60 y) | 1.27 | 0.87-1.87 | 0.216 | - | - | - | |
| Gender (male vs. female) | 1.17 | 0.75-1.82 | 0.489 | - | - | - | |
| Family history (yes vs. no) | 1.18 | 0.69-2.01 | 0.551 | - | - | - | |
| Smoking (ever vs. never) | 1.61 | 0.93-2.79 | 0.086 | - | - | - | |
| Drinking (ever vs. never) | 0.81 | 0.45-1.48 | 0.496 | - | - | - | |
| TNM stage (III/IV vs. I/II) | 2.06 | 1.61-2.62 | <0.001 | 2.06 | 1.63-2.62 | <0.001 | |
| Tumor location (colon vs. rectum) | 1.41 | 1.10-1.82 | 0.007 | 1.39 | 1.09-1.79 | 0.009 | |
| piR-54265 level (high vs. low) | 2.02 | 1.36-3.01 | 0.001 | 1.99 | 1.35-2.93 | <0.001 | |
AHSU: the Affiliated Hospitals of Soochow University (Suzhou, China); CI: confidence interval; HR: hazard ratio; SYSUCC: Sun Yat-sen University Cancer Center (Guangzhou, China).
Associations between piR-54265 levels in tumor tissues and clinicopathological characteristics in CRC patients recruited at SYSUCC and AHSU who received 5-FU and oxaliplatin-based postoperative chemotherapy
| Variable | SYSUCC sample | AHSU sample | Combined sample | ||||||
|---|---|---|---|---|---|---|---|---|---|
| High (N=55) | Low (N=55) | P | High (N=54) | Low (N=54) | P | High (N=109) | Low (N=109) | P | |
| Age, mean (S.D.) | 56.3(1.76) | 58.9(1.66) | 0.268 | 59.5(1.26) | 55.2(1.35) | 0.106 | 57.5(1.26) | 57.6(1.24) | 0.671 |
| Age at diagnosis, N (%) | 0.341 | 0.083 | 0.588 | ||||||
| <60 | 30(54.5) | 25(45.5) | 23(42.6) | 32(59.3) | 53(48.6) | 57(52.3) | |||
| ≥60 | 25(45.5) | 30(54.5) | 31(57.4) | 22(40.7) | 56(51.4) | 52(47.7) | |||
| Sex, N (%) | 0.699 | 0.117 | 0.409 | ||||||
| M | 33(60.0) | 31(56.4) | 28(51.9) | 36(66.7) | 61(55.9) | 67(61.5) | |||
| F | 22(40.0) | 24(43.6) | 26(48.1) | 18(33.3) | 48(44.1) | 42(38.5) | |||
| Family history, N (%) | 0.161 | 0.302 | 0.837 | ||||||
| Yes | 2(3.6) | 7(12.7) | 11(20.4) | 7(12.9) | 13(11.9) | 14(12.8) | |||
| No | 53(96.4) | 48(87.3) | 43(79.6) | 47(87.1) | 96(88.1) | 95(87.2) | |||
| Smoking, N (%) | 0.381 | 0.837 | 0.455 | ||||||
| Yes | 16(29.1) | 12(21.8) | 18(33.3) | 17(31.5) | 34(31.2) | 29(26.6) | |||
| No | 39(70.9) | 43(78.2) | 36(66.7) | 37(68.5) | 75(68.8) | 80(73.4) | |||
| Drinking, N (%) | 0.178 | 0.661 | 0.209 | ||||||
| Yes | 16(29.1) | 10(18.2) | 15(27.8) | 13(24.1) | 31(28.4) | 23(21.1) | |||
| No | 39(70.9) | 45(81.8) | 39(72.2) | 41(75.9) | 78(71.6) | 86(78.9) | |||
| TNM stage, N (%) | 0.016 | 0.013 | 0.001 | ||||||
| I/II | 13(27.3) | 25(41.8) | 11(20.4) | 23(38.9) | 24(22.1) | 48(40.4) | |||
| III/IV | 42(72.7) | 30(58.2) | 43(79.6) | 31(61.1) | 85(77.9) | 61(59.6) | |||
| Tumor location, N (%) | 0.127 | 0.701 | 0.416 | ||||||
| Colon | 33(60.0) | 25(45.5) | 27(50.0) | 29(53.7) | 60(55.1) | 54(49.5) | |||
| Rectum | 22(40.0) | 30(54.5) | 27(50.0) | 25(46.3) | 49(44.9) | 55(50.5) | |||
AHSU: the Affiliated Hospitals of Soochow University (Suzhou, China); SYSUCC: Sun Yat-sen University Cancer Center (Guangzhou, China).
piR-54265 is overexpressed in CRC and associated with clinical outcomes. (A) Expression levels of the top 20 highly expressed piRNAs in CRC and paired non-tumor tissue samples from 110 individuals recruited at SYSUCC. Only piR-54265 shows significantly higher levels in CRC than in normal tissues (mean ± SEM; ***, P<0.001). (B) DNA copy number alterations of piR-54265 gene locus in CRC from TCGA database. (C) Lower methylation status of piR-54265 gene locus in CRC than in normal tissues based on TCGA database. (D) piR-54265 expression levels in CRC with different tumor stages (mean ± SEM). P-values by one-way ANOVA show significant difference among four groups. (E) Tumor piR-54265 expression levels of CRC in stage I-III and stage IV by RT-qPCR (mean ± SEM). P-values by Student's t-test. (F-G) Kaplan-Meier estimates of progression-free survival (PFS) time (F) and overall survival (OS) time (G) in CRC patients with different piR-54265 levels in tumor. HR: hazard ratio; CI: confidence interval.
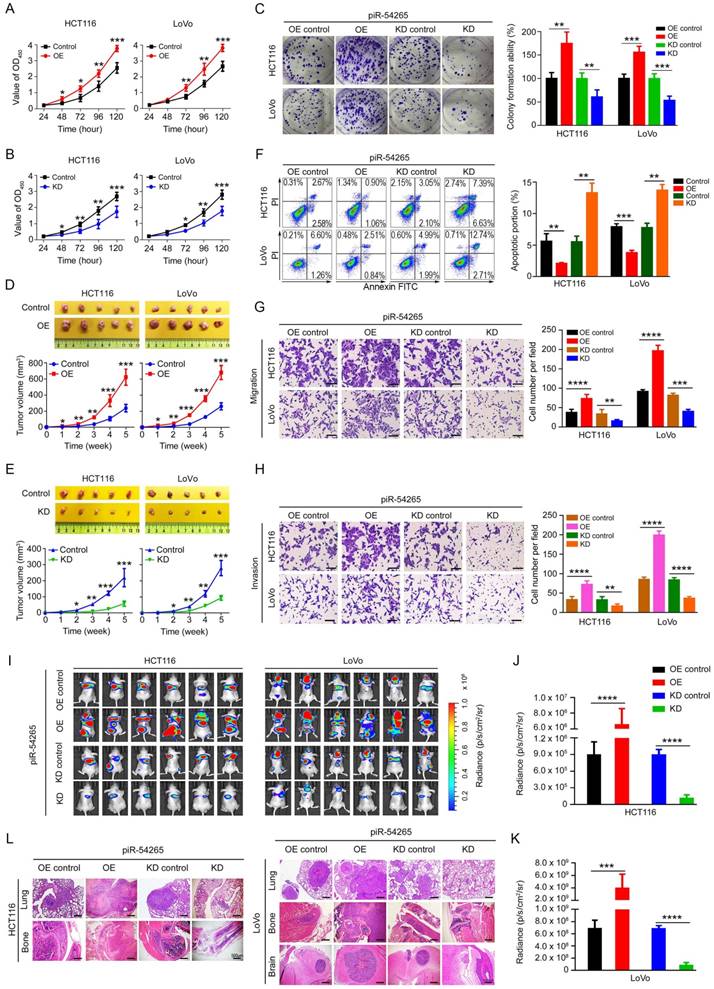
piR-54265 promotes CRC cell proliferation and invasiveness. (A-B) Overexpression of piR-54265 (A) promoted but knockdown of piR-54265 (B) suppressed CRC cell proliferation (mean ± SEM, N=6; *, P<0.05; **, P<0.01 and ***, P<0.001). (C) Effects of piR-54265 expression on the colony formation ability of CRC cells. Representative images (left panel) and quantification (right panel) of colony formation ability (mean ± SEM). (D-E) Effects of piR-54265 expression on subcutaneous xenograft growth in mice. Images of xenograft tumors (upper panels) and tumor growth curves as a function of time (lower panels). (F) Effects of piR-54265 on CRC cell apoptosis. Flow cytometry images (left panel) and quantitative statistics (mean ± SEM, N=3; **, P<0.01; ***, P<0.001) (right panel). (G-H) Effects of piR-54265 on the abilities of migration (G) and invasion (H) of CRC cells in vitro. Representative images of transwell assays (left panels) and quantitative statistics (mean ± SEM, N=3; **, P< 0.01; ***, P<0.001; ****, P<0.0001) (right panels). Scale bars, 100 μm. (I-L) Effects of piR-54265 expression on metastasis of CRC cells in mice. Luminescence imaging of metastases (I) and quantification (J-K) of radiance intensity (mean ± SEM, N=6 in each group). (L) H&E staining images of pathological sections of metastatic cancers in the lung, brain and bone in mice (scale bars, 500 μm). *, P<0.05; **, P<0.01; ***, P<0.001 and ****, P<0.0001 by Student's t-test. OE: overexpression; KD: knockdown.
piR-54265 promotes CRC cell proliferation and invasiveness
We next examined the effects of piR-54265 on malignant phenotypes of CRC cells by ectopic expression of piR-54265 or its antisense (Figure S5A-B). We observed that overexpression of piR-54265 in CRC cells substantially increased the rate of cell proliferation (Figure 2A), whereas knockdown of piR-54265 had the reverse results (Figure 2B). The colony formation ability in both CRC cells was markedly enhanced by piR-54265 overexpression but substantially suppressed by piR-54265 knockdown (Figure 2C). We injected CRC cells into the rear flank of nude mice and found that the growth rate of xenograft overexpressing piR-54265 was significantly higher than that of controls (Figure 2D). In contrast, the growth rate of xenograft with piR-54265 knockdown was far slower than that of controls (Figure 2E). Flow cytometry analysis showed significantly decreased apoptosis in cells overexpressing piR-54265 compared with control cells; conversely, cells with piR-54265 knockdown had a significant increase in apoptosis (Figure 2F). Neither overexpression nor knockdown of piR-54265 had a significant effect on the cell cycle progression (Figure S5C), suggesting that the promoting effects of piR-54265 on CRC cell proliferation may be through inhibiting apoptosis.
We then investigated the effects of piR-54265 on the capability of CRC cell migration and invasion in vitro and in vivo. We found that overexpression of piR-54265 significantly enhanced but knockdown of piR-54265 significantly suppressed the migration and invasion abilities of CRC cells in vitro (Figure 2G-H). Furthermore, we found that CRC cells with stable piR-54265 overexpression had remarkably increased lung metastasis compared with control cells as measured by luminescence imaging. In contrast, cells with stable knockdown of piR-54265 expression had significantly reduced lung metastasis compared with control cells (Figure 2I-K and Figure S6). In addition to lung metastasis, overexpression of piR-54265 promoted CRC cells localized in other organs and tissues such as brain and bone (Figure 2L). Taken together, these results indicate that piR-54265 is an oncogenic piRNA capable of promoting proliferation and metastasis of CRC cells.
piR-54265 binds PIWIL2 and facilitates PIWIL2/STAT3/p-SRC complex formation
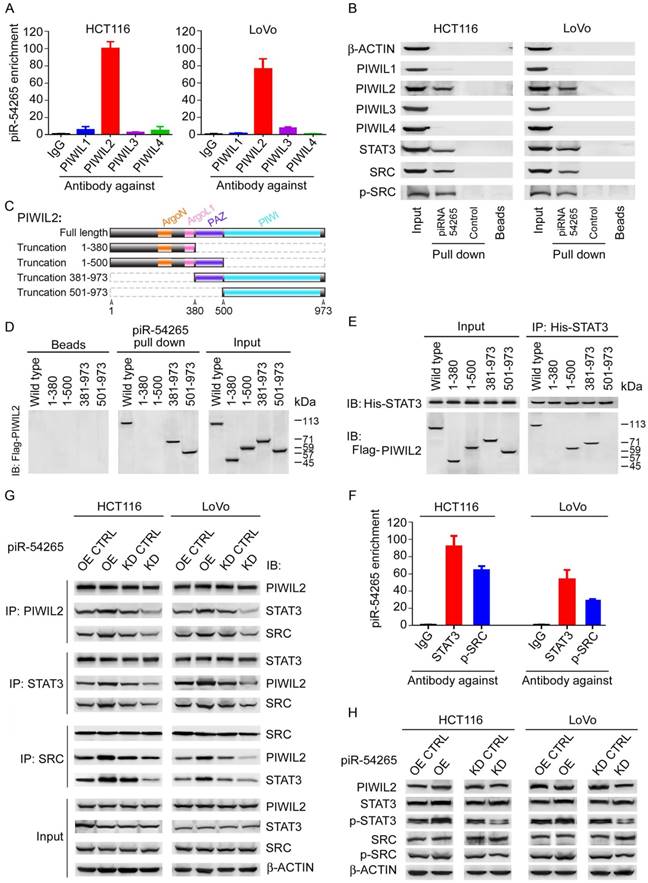
Having observed that piR-54265 promotes the malignant phenotypes of CRC cells, we next wanted to seek the molecular mechanism for this action. Since ncRNAs may be involved in the cis regulation of target genes located at the nearby genomic locus [35], we first examined the mRNA levels of 9 genes within about 2 mega-bases centering the gene generating piR-54265 (Figure S7A) and found no significant difference when piR-54265 expression was knocked down as compared with controls (Figure S7B), suggesting that piR-54265 might not function in this way. Another possible function we conjectured might be that piR-54265 interacts with PIWI proteins to perform the function as its name defines. Indeed, by RIP-qPCR and pulldown assays, we demonstrated that piR-54265 specifically binds PIWIL2, a member of the PIWI family (Figure 3A-B). Further pulldown assays using truncated PIWIL2 fragments and biotin-labeled piR-54265 showed that the piRNA binds the PIWI domain of PIWIL2 protein (Figure 3C-D).
We then investigated the molecular consequences of the interaction between piR-54265 and PIWIL2. Previous studies have shown that PIWIL2 can directly recruit STAT3 to form PIWIL2/STAT3/p-SRC complex [23]. Co-immunoprecipitation assays confirmed that the PAZ domain of PIWIL2 is required for its interaction with STAT3 (Figure 3E). By using RNA pulldown and RIP assays, we further verified the association of piR-54265 with both STAT3 and p-SRC (Figure 3B-F). However, what is important is that we found that the PIWIL2/STAT3/p-SRC interaction in CRC cells varied when the piR-54265 expression was perturbed, although the levels of both PIWIL2, total STAT3 and SRC proteins were roughly constant (Figure 3G-H); these results indicated that piR-54265 may facilitate the formation of PIWIL2/STAT3/p-SRC complex.
piR-54265 promotes oncogenic STAT3 signaling
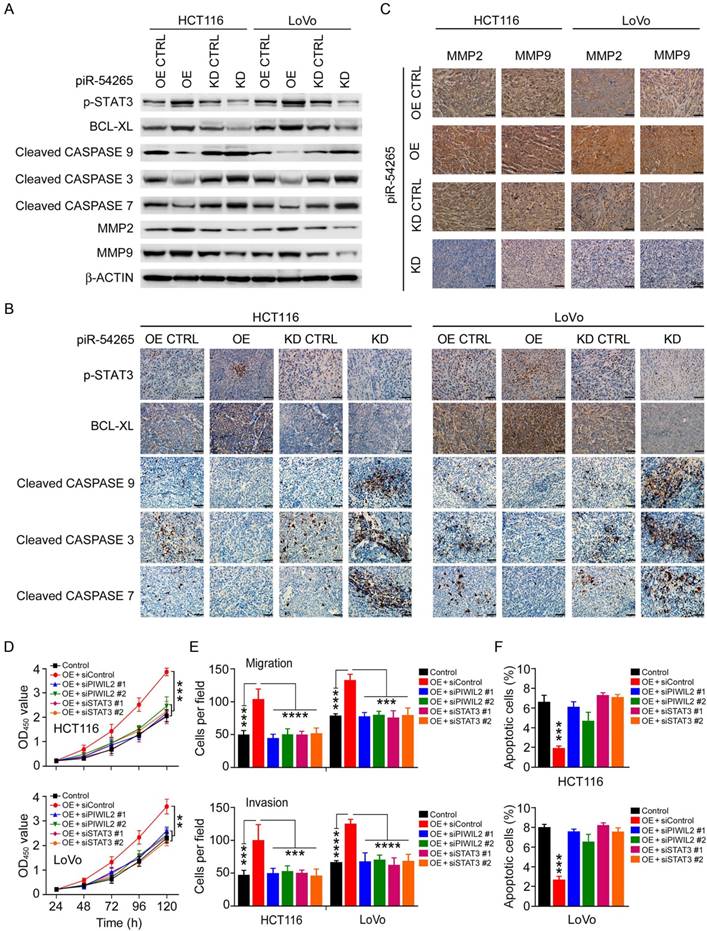
Because piR-54265 overexpression or knockdown in CRC cells appeared to substantially affect only the level of phosphorylated STAT3 (p-STAT3) but no other proteins (Figure 3H), we postulated that piR-54265 may function via activating STAT3 signaling pathway. We therefore examined the impact of piR-54265 on the expression or activation of STAT3-dependent molecules, focusing on those related to apoptosis and metastasis in CRC cells and xenograft tumors in mice. We found that CRC cells overexpressing piR-54265 had substantial up-regulation of p-STAT3 and BCL-XL but down-regulation of cleaved-CASP9, -CASP3 and -CASP7 compared with control cells; the reverse results were obtained in cells with piR-54265 knockdown (Figure 4A). Immunohistochemical staining of these proteins in subcutaneous xenograft tumors formed by the same CRC cells also provided consistent results (Figure 4B). Furthermore, we observed a substantial increase in the expression levels of metastasis-causing molecules, matrix metalloproteinase-2 (MMP2) and -9 (MMP9), which have been shown to be regulated by STAT3 [36, 37], in xenograft tumors where piR-54265 was overexpressed (Figure 4C). The reverse results were also seen in xenograft tumors when piR-54265 was knocked down (Figure 4B-C).
piR-54265 interacts with PIWIL2, facilitating PIWIL2/STAT3/p-SRC formation. (A) RNA immunoprecipitation assays showed specific bind of piR-54265 to PIWIL2. Results are mean ± SEM of piR-54265 enrichment relative to input from three independent experiments. (B) Biotin-labeled piR-54265 RNA pulldown coupled with western blot analysis revealed interaction of piR-54265 with PIWIL2, STAT3 and p-SRC. (C) Schematic of the domain structure of PIWIL2 protein. (D) Biotin-labeled piR-54265 RNA pulldown from lysates containing FLAG-tagged full-length or truncated PIWIL2 protein coupled with Western blot analysis revealed the interaction of piR-54265 with PIWIL2 via the PIWI domain in PIWIL2. (E) Co-immunoprecipitation assays revealed the interaction of STAT3 with PIWIL2 via the PAZ domain in PIWIL2. (F) RNA immunoprecipitation assays showed specific association of piR-54265 with STAT3 and p-SRC in CRC cells. Results are mean ± SEM of piR-54265 enrichment relative to input from three independent experiments. (G) Reciprocal immunoprecipitation assays showed that the interaction among PIWIL2, STAT3 and SRC in CRC cells was affected by piR-54265 expression. (H) Effects of overexpression or knockdown of piR-54265 on the expression levels of STAT3 and SRC and their phosphorylated forms detected by western blot.
We then performed rescue assays to verify that the effects of piR-54265 on CRC cell are via PIWIL2 and STAT3. The results showed that knockdown of PIWIL2 or STAT3 expression (Figure S8A) significantly inhibited proliferation, migration and invasion of CRC cells overexpressing piR-54265 (Figure 4D-F and Figure S8B-C), suggesting important roles of PIWIL2 and STAT3 in piR-54265-induced malignant phenotypes of CRC cells.
piR-54265 enhances oncogenic STAT3 signaling. (A) Effects of piR-54265 on expression and/or activation of proliferation and metastasis-related STAT3 downstream modules in CRC cells detected by western blot. (B-C) Immunohistochemical (IHC) staining of proliferation- and metastasis-related STAT3 downstream modules in mouse xenograft tumors of CRC cells with overexpression or knockdown of piR-54265. IHC staining of proliferation-related molecules (B) and metastasis-related molecules (C). 200×; scale bars, 50 μm. (D) Knockdown of PIWIL2 or STAT3 affects piR-54265-induced CRC cell proliferation (each point in the curve represents mean ± SEM; N=6; **, P<0.01; ***, P<0.001). (E) Knockdown of PIWIL2 or STAT3 affects the piR-54265-induced migration (upper panel) and invasion (lower panel) abilities of CRC cells (mean ± SEM; N=6. ***, P<0.001; ****, P<0.0001). See also Figure S8B. (F) Knockdown of PIWIL2 or STAT3 affects piR-54265-induced CRC cell apoptosis (mean ± SEM; N=3; ***, P<0.001). See also Figure S8C. OE: overexpression; KD: knockdown.
piR-54265 is a therapeutic target in mouse xenograft tumors. (A-B) Effect of piR-54265 overexpression (OE) or knockdown (KD) on chemosensitivity of CRC cells to 5-FU (A) or oxaliplatin (B) as presented in statistic histograms of IC50 values. (mean ± SEM, N=6; ***, P<0.001; ****, P<0.0001). (C-D) Effects of piR-54265 overexpression (OE) or knockdown (KD) on apoptosis of CRC cells induced by 5-FU (C) or oxaliplatin (D). Results are mean ± SEM (N=3). **, P<0.01; ***, P<0.001. See also Figure S8A. (E-F) Effects of piR-54265 on sensitivity of mouse subcutaneous xenograft tumors to chemotherapeutic agents. Overexpression of piR-54265 (OE) conferred xenograft tumor tolerance toward oxaliplatin (oxal) and 5-FU (E). Knockdown of piR-54265 (KD) conferred xenograft tumor sensitivity to oxaliplatin and 5-FU (F). Arrows indicate the time of drug treatment. All P<0.05 for repeated measurement of a general linear model. (G) Timeline schematic for intra-tumor treatment of mouse subcutaneous xenografts with antagopiR54265. Colored arrows indicate the times when different events occurred. (H-I) Injection of antagopiR54265 into xenograft tumors significantly suppressed tumor growth. Images of xenograft tumors with or without injection of antagopiR54265 (H) and the growth curves of xenografts (I). Each point in the curves presents mean ± SEM (N=5). *, P<0.05; **, P<0.01 compared with each control. (J) Timeline schematic for treatment of mouse metastatic tumors with antagopiR54265 alone or combination with 5-FU. Colored arrows indicate the times when different events occurred. (K-L) Injection of antagopiR54265 via tail vein significantly suppressed tumor spread and metastasis. Radiant images of metastatic tumors in mice (K) and quantitation of tumor metastasis indicated by radiance intensity (L). Each point in the curve is mean ± SEM of radiance (N=5). *, P<0.05; **, P<0.01 and ***, P<0.001. (M) Survival times of mice with metastatic tumors treated with or without antagopiR54265 or antagopiR54265 plus 5-FU. All log-rank P<0.05 compared with control group.
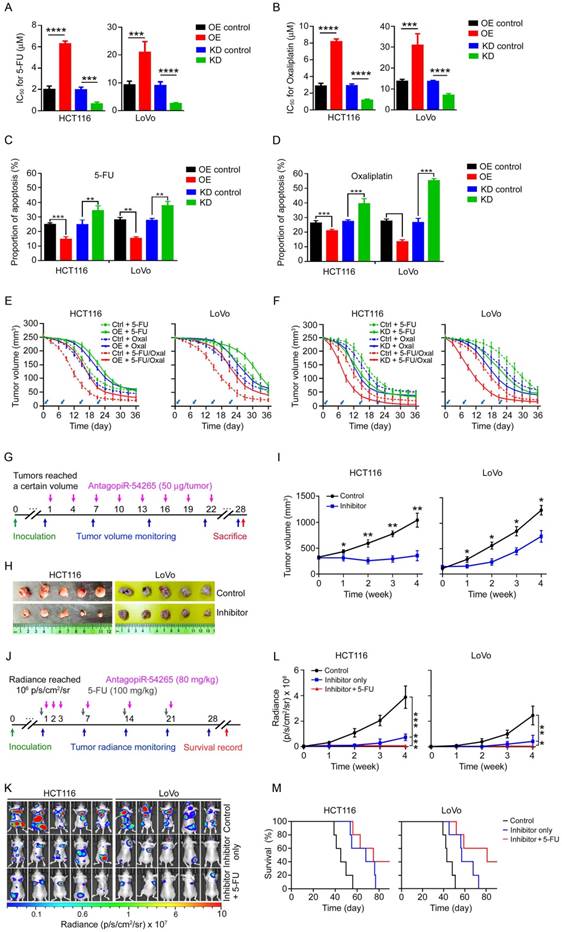
piR-54265 is a therapeutic target in mouse xenograft tumors
Since overexpression of piR-54265 prevents CRC cells from undergoing apoptosis and was associated with prognosis of CRC patients who received postoperative chemotherapy, we thus investigated whether this piRNA is involved in chemo-sensitivity. We observed that CRC cells overexpressing piR-54265 had significantly higher half maximal inhibitory concentrations (IC50) toward 5-FU and oxaliplatin, while cells with piR-54265 knockdown had significantly lower IC50 compared with their controls (Figure 5A-B). Flow cytometry analysis demonstrated that the insensitivity to the chemotherapeutic agents of CRC cells overexpressing piR-54265 was due to a decrease in the apoptosis induced by the agents (Figure 5C-D and Figure S9A). These in vitro results encouraged us to perform experimental chemotherapy using 5-FU, oxaliplatin or combined regimen in a mouse xenograft tumor model. We found that subcutaneous tumors formed from CRC cells overexpressing piR-54265 were more resistant to the chemotherapy than tumors derived from the cells without piR-54265 overexpression (Figure 5E), while subcutaneous tumors with knockdown of piR-54265 had a better response to these agents (Figure 5F). These results indicate that overexpression of piR-54265 plays an important role in conferring chemotherapy insensitivity to CRC cells, and targeting piR-54265 might provide a new approach for CRC treatment.
To test whether targeting piR-54265 might suppress tumor growth and metastasis, we conducted experimental therapy in mouse subcutaneous xenograft and metastatic tumor models using antagopiR54265, a specific chemically modified piR-54265 inhibitor with suitable pharmacokinetic properties for in vivo study. We injected antagopiR54265 or scramble sequence directly into subcutaneous tumors (Figure 5G) and observed that after 5-8 injections, subcutaneous tumors treated with antagopiR54265 had significantly reduced tumor volume compared with those treated with scramble controls (Figure 5H-I). We also injected this antagopiR54265 or its control through the tail vein into blood to treat metastatic tumors when tumor sizes reached a certain threshold (Figure 5J) and similar results were obtained showing that mice treated with antagopiR54265 had significantly reduced tumor burdens and longer survival time compared with controls (Figure 5K-M). Furthermore, combined treatment of antagopiR54265 with 5-FU had greater efficacy than treatment with each agent alone (Figure 5K-M). In addition, treatment of mice with antagopiR54265 showed no apparent side-effects because no significant differences in animal body-weight gain were seen over the course of intra-tumor (Figure S9B) or intravenous injection (Figure S9C), implying a low toxicity of this piRNA inhibitor.
piR-54265 occurs in serum of CRC patients and predicts clinical outcomes
Since small ncRNAs can be found in blood samples and might serve as feasible biomarkers, we examined whether piR-54265 can be detected in serum of CRC patients. By Northern blotting assays of randomly selected serum samples, we verified the existence of piR-54265 (Figure 6A). We then established a RT-qPCR approach to determine the serum piR-54265 level and found that piR-54265 was relatively stable in serum by analyzing samples stored for different times and at room temperature (Figure 6B). We found that the serum piR-54265 levels were up-regulated in a CRC stage-dependent manner and the levels were highest in patients with metastatic CRC (Figure 6C). The piR-54265 levels in patients' serum were significantly correlated with that in tumor tissues (Figure 6D). We also investigated correlations between serum piR-54265 levels and PFS or OS in our patient sets and the results showed that, like the piR-54265 levels in tumors, serum piR-54265 levels were significantly correlated with survival times in both sets and combined samples (Figure 6E). Because piR-54265 is involved in sensitivity to chemotherapeutic agents, we further examined the correlation between serum piR-54265 levels and curative efficacy of preoperative neoadjuvant chemotherapy of 5-FU and oxaliplatin in CRC patients. We found that patients with low serum piR-54265 level had significantly better response to chemotherapy than those with high serum piR-54265 level in patients recruited at SYSUCC (SYSUCC2, N=215, P=0.010; Table S2 and Figure 6F) and at Cancer Hospital, Chinese Academy of Medical Sciences (CHCAMS, N=102, P=0.019; Table S3 and Figure 6G) and combined samples (N=317, P<0.0001; Figure 6H). Next, we evaluated the diagnostic performance of serum piR-54265 level in predicting curative effect of treatment of 5-FU and oxaliplatin in patients. The receiver operating characteristic (ROC) curves built with 215 cases at SYSUCC2 as the model set, 102 cases at CHCAMS as the validation set and combined samples of both sets yielded area under the ROC curves (AUCs) of 0.819 (P<0.0001, 95% CI=0.761-0.868), 0.808 (P<0.0001, 95% CI=0.718-0.879) and 0.811 (P<0.0001, 95% CI=0.764-0.853), respectively, indicating a highly significant association between serum piR-54265 levels and disease progression or control rate of chemotherapy in CRC patients (Figure 6I). Based on the selection principle of maximum value of Youden's J statistic, the optimal cutoff power of serum piR-54265, 0.0119, was achieved from the combined sample corresponding to a sensitivity of 66.7% and specificity of 88.5%, which effectively or accurately discriminates the disease progressors from non-progressors including clinical beneficiaries with CR, PR or SD after neoadjuvant chemotherapy. Different cutoff values employed different sensitivities and specificities, which could be selected according to the clinical applications (Table 4).
Discussion
In the present study, we have identified for the first time that piR-54265 is an oncogenic piRNA in human CRC. The overexpressed piR-54265 may function as an essential mediator to activate STAT3 signaling, consequently enhancing proliferation and promoting invasion and metastasis of CRC cells. We have demonstrated in mice xenograft models that treatment with piR-54265 inhibitor is effective in suppressing tumor growth and metastasis. Remarkably, we have further found that the levels of piR-54265 in CRC tissues is correlated with the levels in serum of patients, both of which are highly associated with clinical outcomes such as resistance to chemotherapy and poor survival in CRC patients. Together, these findings shed new light on the important role of some piRNAs in the development and progression of human cancer and highlight the potential use of certain piRNAs as treatment targets and valuable diagnostic and prognostic biomarkers for cancer.
Although piRNAs account for the major proportion of small ncRNAs in cells and have been proved to be involved in somatic functions [15, 16], little has been known about their roles in human cancer. Emerging reports have shown aberrant expressions of certain piRNAs in some types of human cancer, indicating that this class of ncRNAs may be important in cancer development and progression [38-40]. In this study, we selected the top 20 highly expressed piRNAs in CRC tissues from TCGA and GEO databases and examined their expression levels in our sample sets. Among the 20 piRNAs, only piR-54265 was significantly overexpressed in CRC tissues compared with normal tissues, suggesting that piR-54265 may play an oncogenic role in a CRC tissue-specific manner. We have also investigated the biological functions of piR-54265 in CRC and demonstrated that piR-54265 is able to specifically interact with the PIWI domain of PIWIL2 protein and mediates the formation of PIWIL2/STAT3/p-SRC complex, in which STAT3 is phosphorylatively activated by p-SRC [23]. This newly identified piR-54265 function in CRC cells might serve as an example for studies on other piRNAs and other types of human cancer. Many studies have shown STAT3 as an oncogenic signaling that causes anti-apoptotic and pro-metastatic effects [41, 42]. Indeed, we confirmed the overexpression and/or aberrant activation of STAT3 downstream modules such as the anti-apoptotic BCL-XL and pro-metastatic MMP2 and MMP9 pathways, which are substantially enhanced by piR-54265. These findings extend our current knowledge about the biological functions of piRNAs and their important roles in human cancer.
Sensitivity and specificity for curative effect of 5-FU and oxaliplatin-based chemotherapy of CRC by cutoff points of serum piR-54265 level
| Serum piR-54265 level (Power value) | SYSUCC sample CR/PR/SD (N=200) versus PD (N=15) | CHCAMS sample TRG1/2/3 (N=87) versus TRG4/5 (N=15) | Combined sample* CR/PR/SD (N=287) versus PD (N=30) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Sensitivity | Specificity | Youden's index# | Sensitivity | Specificity | Youden's index# | Sensitivity | Specificity | Youden's index# | |
| ≥0.00018 | 100.0 | 0.0 | 0.000 | 100.0 | 0.0 | 0.000 | 100.0 | 0.0 | 0.000 |
| >0.00765 | 73.3 | 51.0 | 0.243 | 80.0 | 63.1 | 0.431 | 80.0 | 53.3 | 0.333 |
| >0.01072 | 73.3 | 81.5 | 0.482 | 66.7 | 85.1 | 0.518 | 66.7 | 82.6 | 0.493 |
| >0.01181 | 66.7 | 87.0 | 0.537 | 66.7 | 90.8 | 0.575** | 66.7 | 86.3 | 0.531 |
| >0.01186 | 66.7 | 87.5 | 0.542 | 65.8 | 90.8 | 0.566 | 66.7 | 88.5 | 0.552** |
| >0.01227 | 66.7 | 90.0 | 0.567** | 53.3 | 93.1 | 0.464 | 60.0 | 90.6 | 0.506 |
| >0.01455 | 40.0 | 95.0 | 0.351 | 53.3 | 100 | 0.533 | 46.7 | 96.5 | 0.432 |
*In combined samples, TRG1/2/3 was combined with CR/PR/SD and TRG4/5 was combined with PD for analysis.
**The highest J in respective ROC curves.
#Youden index (J) = Sensitivity + Specificity - 1. Higher J indicates higher probability of an informed decision in multiclass case statistically.
CHCAMS: Cancer Hospital, Chinese Academy of Medical Sciences (Beijing, China); SYSUCC: Sun Yat-sen University Cancer Center (Guangzhou, China).
Serum piR-54265 predicts clinical outcomes of CRC patients. (A) Northern blot of piR-54265 in randomly selected serum samples from individuals with CRC confirmed its existence and expected molecular size of 29 nucleotides. (B) Stability of serum piR-54265 under different storage conditions. piR-54265 levels in sera of 8 randomly selected individuals with CRC were detected: 1, at day 1 (2017/04/16); 2, after storage at -80 °C for about 7 months (2017/11/21); and 3, after storage at room temperature for 80 h (mean ± SEM, N=3; all differences are not significant). (C) Serum piR-54265 levels in tumors of different stages (mean ± SEM). P-value by one-way ANOVA shows difference among four groups, Stage I, II, III and IV (left panel). P-value by Student's t-test shows the significant difference between Stage I-III and IV (right panel). (D) The levels of piR-54265 in serum were positively correlated with the levels of piR-54265 in tumors from patients recruited at SYSUCC, AHSU and combined samples. (E) Kaplan-Meier estimates of progression-free survival time (upper panel) and overall survival time (lower panel) in CRC patients with different serum piR-54265 levels. HR: hazard ratio; CI: confidence interval. (F-H) Outcomes of 5-FU and oxaliplatin-based neoadjuvant chemotherapy in patients with CRC as a function of serum piR-54265 level. Shown are the results of SYSUCC2 sample (F), CHCAMS sample (G) and combined samples (H). P-value by Chi-square test. (I) Diagnostic performance of serum piR-54265 levels in predicting curative effect of 5-FU and oxaliplatin-based chemotherapy by receiver operating characteristic (ROC) curves analyses in the two patient sets and combined samples. AUC: area under ROC curves. See also Table 4.
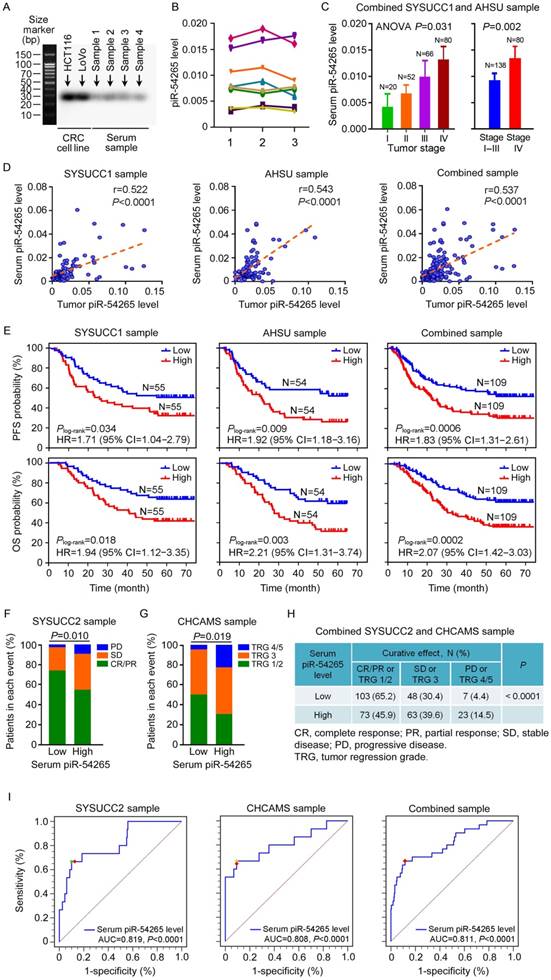
Another interesting finding is that piR-54265 is involved in drug-sensitivity of CRC cells. We found that CRC cells with piR-54265 overexpression were resistant while those with piR-54265 knockdown were sensitive to 5-FU and oxaliplatin, two first-line drugs routinely used for chemotherapy of advanced CRC [4]. The underlying mechanism seems to be the anti-apoptotic role of piR-54265. Serum piR-54265 levels are positively correlated with tumor piR-54265 levels in CRC patients. We observed a better response to the chemotherapeutic regimen in individuals with low serum piR-54265 level compared with those with high serum piR-54265 level. These findings would be important and promising in terms of developing a simple molecular biomarker for predicting the efficacy of neoadjuvant chemotherapy based on 5-FU and oxaliplatin for advanced CRC, since the curative effects of this therapy vary greatly in individuals [5] and there are currently no effective measures to predict who will benefit from the therapy.
Because piR-54265 shows oncogenic effect and upregulates in a stage-dependent manner, we assumed that it may be a valuable therapeutic target for CRC. By carrying out experimental treatment in xenograft tumor models with a synthetic inhibitor of piR-54265, we found that administration of antagopiR54265 directly into the tumor or vein substantially reduced the growth and metastasis of xenografts in mice. Furthermore, combined administration of antagopiR54265 with 5-FU showed an increased anticancer effect. Interestingly, we observed that administration of antagopiR54265 to mice did not show apparent side-effects, which is reflected by the similar bodyweight gain compared with control mice treated with a scrambled sequence. Although further investigations should be performed to systematically evaluate the potential toxicity caused by the inhibition of piR-54265, the results in the current study suggest that piR-54265 may be a druggable target for effective treatment of CRC.
It is critical to find out an approach to better capture the variability that defines the clinical behavior of CRC. In this context, our identification of piR-54265 in CRC has revealed a new level of biological complexity having the potential to define a subgroup that has a differential response to neoadjuvant chemotherapy and survival time. Therefore, it is rational to infer that piR-54265 could be built into molecular diagnostic strategies for risk stratification of CRC. It is fascinating that piR-54265 can readily and easily be detected in the serum of individuals with CRC and the level reflects well the level in CRC tissues and can also predict the differential clinical outcomes. Thus, serum piR-54265 is a promising surrogate that may serve as a noninvasive biomarker for predicting the efficacy of neoadjuvant chemotherapy and clinical outcomes in CRC patients.
In conclusion, we have identified piR-54265 as an oncogenic piRNA in CRC and elucidated its underlying molecular mechanism for driving CRC cell malignant phenotypes. Suppression of CRC cell growth and metastasis in mice by treatment with piR-54265 inhibitor highlights that piR-54265 may be a novel therapeutic target for CRC. Furthermore, serum piR-54265 may serve as a prediction marker for response to chemotherapy and prognosis in CRC patients. However, although this study was designed with three independent patient sets for validation, studies with prospective design and multicenter replication are needed to validate our findings. In addition, it would be interesting to explore whether piR-54265 also plays the same role in other types of human cancer.
Abbreviations
AUC: area under the ROC curve; CPM: counts per million mapped reads; CRC: colorectal cancer; 5-FU: 5-fluorouracil; IC50: half maximal inhibitory concentration; OS: overall survival; Oxal: oxaliplatin; PFS: progression-free survival; piRNA: PIWI-interacting RNA; p-SRC: phosphorylated SRC; p-STAT3: phosphorylated STAT3; RECIST: response evaluation criteria in solid tumors; ROC: receiver operating characteristic curve; SCNA: somatic copy number alteration; TRG: tumor regression grade.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
This work was supported by grants from National Key Research and Development Program of China (2016YFC1302700 to W. J.), National Key Basic Research and Development Program (2014CB542004 to W. T.), Sun Yat-sen University Intramural Funds (to D. L. and to J. Zheng), National Young Top-notch Talent Support Program (to J. Zheng), Young Elite Scientists Sponsorship Program by CAST (2017QNRC001 to J. Zheng), Guangdong Province Universities and Colleges Pearl River Scholar Funded Scheme (2017, to J. Zheng), and Science Foundation for Distinguished Young Scholars in Jiangsu (BK20160008, to Y. Zhou).
Author Contributions
J. Zheng, R. X. and D. L. conceptualized and supervised the research. D. M., P. D. and L. T. designed and performed most experiments. Z. P., C. L., W. J., R. X., L. P., Y. Zheng, Y. Zhou, W. L., S. W., W. T., Z. Zhou, A. Z., Y. L. and C. W. were responsible for patient recruitment, biospecimen sampling, clinical data collection and analysis at SYSUCC, AHSU or CHCAMS, respectively. J. Zhang, R. B. and X. L. provided technique support. M. L. and S. Z. contributed to histopathological and immune-histochemical analyses. Z. L., Z. Zuo, Q. Z. and Y. Y. were engaged in statistical and bioinformatics analyses. D. M. and J. Zheng prepared the manuscript. D. L. reviewed and approved the manuscript.
Availability of data
The key raw data in this study have been uploaded onto the Research Data Deposit of Sun Yat-sen University Cancer Center (Approval number RDDB2018000409, http://www.researchdata.org.cn).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7-30
2. Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64:104-17
3. Carethers JM, Jung BH. Genetics and Genetic Biomarkers in Sporadic Colorectal Cancer. Gastroenterology. 2015;149:1177-90 e3
4. Cunningham D, Atkin W, Lenz HJ, Lynch HT, Minsky B, Nordlinger B. et al. Colorectal cancer. Lancet. 2010;375:1030-47
5. Zhang Y, Chen Z, Li J. The current status of treatment for colorectal cancer in China: A systematic review. Medicine. 2017;96:e8242
6. Kulendran M, Stebbing JF, Marks CG, Rockall TA. Predictive and prognostic factors in colorectal cancer: a personalized approach. Cancers. 2011;3:1622-38
7. Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857-66
8. Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K. et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997-1006
9. Hur K, Toiyama Y, Okugawa Y, Ide S, Imaoka H, Boland CR. et al. Circulating microRNA-203 predicts prognosis and metastasis in human colorectal cancer. Gut. 2017;66:654-65
10. Grivna ST, Beyret E, Wang Z, Lin H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 2006;20:1709-14
11. Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199-202
12. Sai Lakshmi S, Agrawal S. piRNABank: a web resource on classified and clustered Piwi-interacting RNAs. Nucleic Acids Res. 2008;36:D173-7
13. Zhang P, Si X, Skogerbo G, Wang J, Cui D, Li Y. et al. piRBase: a web resource assisting piRNA functional study. Database (Oxford). 2014;2014:bau110
14. Yan Z, Hu HY, Jiang X, Maierhofer V, Neb E, He L. et al. Widespread expression of piRNA-like molecules in somatic tissues. Nucleic Acids Res. 2011;39:6596-607
15. Ross RJ, Weiner MM, Lin H. PIWI proteins and PIWI-interacting RNAs in the soma. Nature. 2014;505:353-9
16. Iwasaki YW, Siomi MC, Siomi H. PIWI-interacting RNA: its biogenesis and functions. Annu Rev Biochem. 2015;84:405-33
17. Peng JC, Lin H. Beyond transposons: the epigenetic and somatic functions of the Piwi-piRNA mechanism. Curr Opin Cell Biol. 2013;25:190-4
18. Siddiqi S, Matushansky I. Piwis and piwi-interacting RNAs in the epigenetics of cancer. J Cell Biochem. 2012;113:373-80
19. Ng KW, Anderson C, Marshall EA, Minatel BC, Enfield KS, Saprunoff HL. et al. Piwi-interacting RNAs in cancer: emerging functions and clinical utility. Mol Cancer. 2016;15:5
20. Mei Y, Wang Y, Kumari P, Shetty AC, Clark D, Gable T. et al. A piRNA-like small RNA interacts with and modulates p-ERM proteins in human somatic cells. Nat Commun. 2015;6:7316
21. Zhong F, Zhou N, Wu K, Guo Y, Tan W, Zhang H. et al. A SnoRNA-derived piRNA interacts with human interleukin-4 pre-mRNA and induces its decay in nuclear exosomes. Nucleic Acids Res. 2015;43:10474-91
22. Assumpcao CB, Calcagno DQ, Araujo TM, Santos SE, Santos AK, Riggins GJ. et al. The role of piRNA and its potential clinical implications in cancer. Epigenomics. 2015;7:975-84
23. Lu Y, Zhang K, Li C, Yao Y, Tao D, Liu Y. et al. Piwil2 suppresses p53 by inducing phosphorylation of signal transducer and activator of transcription 3 in tumor cells. PLoS One. 2012;7:e30999
24. Sobin LH, Compton CC. TNM seventh edition: what's new, what's changed: communication from the International Union Against Cancer and the American Joint Committee on Cancer. Cancer. 2010;116:5336-9
25. Suarez J, Vera R, Balen E, Gomez M, Arias F, Lera JM. et al. Pathologic response assessed by Mandard grade is a better prognostic factor than down staging for disease-free survival after preoperative radiochemotherapy for advanced rectal cancer. Colorectal Dis. 2008;10:563-8
26. Mandard AM, Dalibard F, Mandard JC, Marnay J, Henry-Amar M, Petiot JF. et al. Pathologic assessment of tumor regression after preoperative chemoradiotherapy of esophageal carcinoma. Clinicopathologic correlations. Cancer. 1994;73:2680-6
27. Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R. et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer. 2009;45:228-47
28. Kin T, Yamada K, Terai G, Okida H, Yoshinari Y, Ono Y. et al. fRNAdb: a platform for mining/annotating functional RNA candidates from non-coding RNA sequences. Nucleic Acids Res. 2007;35:D145-8
29. Mituyama T, Yamada K, Hattori E, Okida H, Ono Y, Terai G. et al. The Functional RNA Database 3.0: databases to support mining and annotation of functional RNAs. Nucleic Acids Res. 2009;37:D89-92
30. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15-21
31. Anders S, Pyl PT, Huber W. HTSeq - a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166-9
32. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139-40
33. Zheng J, Huang X, Tan W, Yu D, Du Z, Chang J. et al. Pancreatic cancer risk variant in LINC00673 creates a miR-1231 binding site and interferes with PTPN11 degradation. Nat Genet. 2016;48:747-57
34. Li W, Zheng J, Deng J, You Y, Wu H, Li N. et al. Increased levels of the long intergenic non-protein coding RNA POU3F3 promote DNA methylation in esophageal squamous cell carcinoma cells. Gastroenterology. 2014;146:1714-26 e5
35. Guil S, Esteller M. Cis-acting noncoding RNAs: friends and foes. Nat Struct Mol Biol. 2012;19:1068-75
36. Xie TX, Wei D, Liu M, Gao AC, Ali-Osman F, Sawaya R. et al. Stat3 activation regulates the expression of matrix metalloproteinase-2 and tumor invasion and metastasis. Oncogene. 2004;23:3550-60
37. Song Y, Qian L, Song S, Chen L, Zhang Y, Yuan G. et al. Fra-1 and Stat3 synergistically regulate activation of human MMP-9 gene. Mol Immunol. 2008;45:137-43
38. Yan H, Wu QL, Sun CY, Ai LS, Deng J, Zhang L. et al. piRNA-823 contributes to tumorigenesis by regulating de novo DNA methylation and angiogenesis in multiple myeloma. Leukemia. 2015;29:196-206
39. He X, Chen X, Zhang X, Duan X, Pan T, Hu Q. et al. An LncRNA (GAS5)/ SnoRNA-derived piRNA induces activation of TRAIL gene by site-specifically recruiting MLL/COMPASS-like complexes. Nucleic Acids Res. 2015;43:3712-25
40. Fu A, Jacobs DI, Hoffman AE, Zheng T, Zhu Y. PIWI-interacting RNA 021285 is involved in breast tumorigenesis possibly by remodeling the cancer epigenome. Carcinogenesis. 2015;36:1094-102
41. Al Zaid Siddiquee K, Turkson J. STAT3 as a target for inducing apoptosis in solid and hematological tumors. Cell Res. 2008;18:254-67
42. Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer. 2009;9:798-809
Author contact
![]() Corresponding authors: Jian Zheng (zhengjianorg.cn) or Ruihua Xu (xurhorg.cn)
Corresponding authors: Jian Zheng (zhengjianorg.cn) or Ruihua Xu (xurhorg.cn)
 Global reach, higher impact
Global reach, higher impact