13.3
Impact Factor
Theranostics 2018; 8(18):5178-5199. doi:10.7150/thno.27798 This issue Cite
Research Paper
Targeted imaging and inhibition of triple-negative breast cancer metastases by a PDGFRβ aptamer
1. Istituto per l'Endocrinologia e l'Oncologia Sperimentale "G. Salvatore" (IEOS), CNR, Naples, Italy.
2. Istituto di Biostrutture e Bioimmagini, CNR, Naples, Italy.
3. Pathology Unit, Istituto Nazionale Tumori-IRCCS-Fondazione G. Pascale, Naples, Italy.
4. Functional Genomics, Istituto Nazionale Tumori-IRCCS-Fondazione G. Pascale, Naples, Italy.
Received 2018-6-11; Accepted 2018-9-3; Published 2018-10-6
Abstract
While the overall mortality for breast cancer has recently declined, management of triple-negative breast cancer (TNBC) is still challenging because of its aggressive clinical behavior and the lack of targeted therapies. Genomic profiling studies highlighted the high level of heterogeneity of this cancer, which comprises different subtypes with unique phenotypes and response to treatment. Platelet-derived growth factor receptor β (PDGFRβ) is an established mesenchymal/stem cell-specific marker in human glioblastoma and, as recently suggested, it may uniquely mark breast cancer cells with stem-like characteristics and/or that have undergone epithelial-mesenchymal transition.
Methods: Immunohistochemical analysis for PDGFRβ expression was performed on a human TNBC tissue microarray. Functional assays were conducted on mesenchymal-like TNBC cells to investigate the effect of a previously validated PDGFRβ aptamer on invasive cell growth in three-dimensional culture conditions, migration, invasion and tube formation. The aptamer was labeled with a near-infrared (NIR) dye and its binding specificity to PDGFRβ was assessed both in vitro (confocal microscopy and flow cytometry analyses) and in vivo (fluorescence molecular tomography in mice bearing TNBC xenografts). A mouse model of TNBC lung metastases formation was established and NIR-labeled PDGFRβ aptamer was used to detect lung metastases in mice untreated or intravenously injected with unlabeled aptamer.
Results: Here, we present novel data showing that tumor cell expression of PDGFRβ identifies a subgroup of mesenchymal tumors with invasive and stem-like phenotype, and propose a previously unappreciated role for PDGFRβ in driving TNBC cell invasiveness and metastases formation. We show that the PDGFRβ aptamer blocked invasive growth and migration/invasion of mesenchymal TNBC cell lines and prevented TNBC lung metastases formation. Further, upon NIR-labeling, the aptamer specifically bound to TNBC xenografts and detected lung metastases.
Conclusions: We propose PDGFRβ as a reliable biomarker of a subgroup of mesenchymal TNBCs with invasive and stem-like phenotype as well as the use of the PDGFRβ aptamer as a high efficacious tool for imaging and suppression of TNBC lung metastases. This study will allow for the significant expansion of the current repertoire of strategies for managing patients with more aggressive TNBC.
Keywords: aptamer, PDGFRβ, invasiveness, mesenchymal TNBC subtype, metastases.
Introduction
Triple-negative breast cancer (TNBC), which comprises ~15% of all breast cancers, lacks expression of the estrogen receptor, progesterone receptor and human epidermal growth factor receptor 2 (HER2), thus excluding targeted therapy against these proteins [1]. Still, no single best target has been so far identified, and cytotoxic chemotherapy remains the only available treatment for patients with TNBC in both the early and advanced stages [2]. Unfortunately, after an initial response, recurrent tumors, which are resistant to therapy and highly metastatic, commonly develop, leading to shorter overall survival than other breast cancer subtypes [3].
The major obstacle in TNBC management is the high heterogeneity of this disease consisting of several subtypes with a unique biological pathway and distinct clinical behavior and response to current therapies [4-6]. Thus, discovering biomarkers for patient stratification is a prioritized issue to establish specific intervention for TNBC.
Platelet-derived growth factor receptor β (PDGFRβ) is a transmembrane receptor tyrosine kinase required for the mesenchymal lineage commitment during embryogenesis [7]. Its expression in normal adults is restricted to fibroblasts, vascular smooth muscle cells, endothelial and perivascular cells, where it plays important roles in wound healing and tissue repair, inflammation and angiogenesis [8]. It has been ascertained that overexpression of PDGFRβ in endothelial cells and tumor-associated stromal cells occurs in different human cancers, where PDGFRβ-dependent signaling contributes to angiogenesis and tumor progression [9-12]. More recently, PDGFRβ expression in cancer cells, mainly in populations characterized by a mesenchymal/stem and poorly differentiated phenotype, has been correlated with aggressiveness, resistance to therapy and/or recurrence in heterogeneous human cancers, including glioblastoma (GBM) [13, 14], colorectal cancer [15], tongue squamous cell carcinoma [16], medulloblastoma [17] and pancreatic neuroendocrine tumors [18]. Still, PDGFRβ has been shown to be a potent mediator of Foxq1-promoted stemness traits and chemoresistance in mammary epithelial cells [19] and its expression is induced during epithelial-to-mesenchymal transition (EMT) in breast cancer cells [20]. These experimental evidences suggest that the expression of PDGFRβ in breast cancer may be a unique feature of cancer cells that possess stem cell characteristics and/or that have undergone EMT, two features associated with chemoresistance and aggressiveness, thus encouraging therapeutic options in targeting this receptor. More recently, PDGFRβ has been shown to mediate endothelial cell differentiation of mesenchymal TNBC cells and vasculogenic mimicry (VM) [21].
To date, several tyrosine kinase inhibitors against PDGFRβ have been tested in TNBC treatment, but they failed to produce the expected results [22, 23]. Thus, developing new strategies to specifically target PDGFRβ-positive TNBC is crucial to improve patient's chances of survival.
An emerging class of compounds for anti-cancer therapy are single-stranded oligonucleotide aptamers, which interact with high affinity with their specific protein targets [24]. We previously generated a nuclease-resistant RNA aptamer, named Gint4.T, targeting PDGFRβ with high affinity with a dissociation constant (Kd) value of 9.6 nM for the extracellular domain of PDGFRβ in a soluble form (EC-PDGFRβ). We validated it as an inhibitor of GBM-derived tumor growth [25] and a delivery agent for drug-loaded nanoparticles to intracranial GBM in mice [26]. This aptamer resulted in highly efficacious targeting and inhibition of PDGFRβ expressed on bone marrow-derived mesenchymal stem cells (BM-MSCs), thus blocking their homing to the tumor and pro-metastatic action in TNBC [12].
Here, by analyzing a large cohort of human TNBC samples for PDGFRβ expression in tumor cells, we found that PDGFRβ expression identified a specific patient subgroup, characterized by mesenchymal tumors with invasive phenotype. Importantly, the treatment of mesenchymal-like (ML) TNBC cell lines with PDGFRβ aptamer blocks their invasive growth in three-dimensional (3D) culture conditions, cell migration and invasion, and shows strong anti-metastatic activity in a mouse model of TNBC lung metastases. Still, a near-infrared (NIR) fluorescent version of the aptamer is able to specifically bind to TNBC xenografts and detect lung metastases. To the best of our knowledge, this is the first report of a highly efficacious agent for molecular targeted noninvasive imaging and suppression of metastatic TNBC in the lungs.
Together, our findings indicate a crucial role for PDGFRβ in the metastatic behavior of ML TNBC and provide critical endpoints for the clinical translation of a cell-targeted RNA smart drug to manage PDGFRβ-positive TNBC.
Methods
Patients and specimens, tissue microarray (TMA) building and immunohistochemistry analysis
From 2003 to 2013, 200 patients who underwent a tumorectomy, quadrantectomy or metastectomy at the National Cancer Institute ″Fondazione Giovanni Pascale″ of Naples, Italy, were enrolled into this study. All cases of TNBC and non-TNBC samples were reviewed according to WHO classification criteria, as previously reported [27]. The cohort included patients with an average age of 57 years, ranging from 24 to 93. The histological samples included 178 (89%) grade III and 22 (11%) grade II. Tumor size was lower than 2 cm in 44.7% (85/190) of the samples, between 2 and 5 cm in 46.3% (88/190) and larger than 5 cm in 8.9% (17/190). For 10 cases, we were not able to retrieve that information. Metastatic lymph nodes (LNM) were found in 42.3% of patients (83/196) at surgery. This information for 4 patients was lost. Distant metastases were found in 27.5% of patients (38/138) and were not present in 72.5% of cases (100/138). For 62 cases we were unable to recover this information. The expression of proliferation factor Ki-67 was high (>20%) in 82.3% (158/192), and low (≤20%) in 17.7% of cases (34/192). This information for 8 patients was lost.
TMA was built as previously reported [27] using the most representative areas from each single case with one replicate.
Immunohistochemical staining was done on TMA slides (4 μm) from formalin-fixed, paraffin-embedded tissues, as previously reported [27], by using primary antibodies against PDGFRβ on a total of 200 samples (dilution 1:50, rabbit monoclonal antibody, Cell Signaling Technology Inc., Danvers, MA) and vimentin on a total of 178 samples (dilution 1:700, mouse monoclonal antibody, Leica Biosystem, Wetzlar, Germany) for 1 h. The remaining 22 samples were not evaluable for vimentin staining for technical reasons.
The PDGFRβ-positive cases were also stained for matrix metallopeptidase 9 (MMP-9, dilution 1:150, overnight incubation, goat polyclonal antibody, R&D System Europe, Ltd, Abingdon, UK) and aldehyde dehydrogenase 1‑A1 (ALDH1A1, dilution 1:100, 1 h incubation, rabbit monoclonal antibody, Abcam, Cambridge, MA), on whole sections.
Results were interpreted using a light microscope. Ten fields on each of two cores and at least >500 cells were analyzed for each sample. Two pathologists independently evaluated the intensity, extent and subcellular distribution of the immunostaining. PDGFRβ expression was interpreted as positive when membranous and/or cytoplasmic staining was observed. Staining was scored as follows: negative (absence of staining) and positive (≥1%) in tumor cells.
For PDGFRβ mRNA expression analysis and correlation with molecular subtypes and specific gene signatures, the Genomics Analysis and Visualization platform [28] was used as previously described [29]. The data set used (GSE76124) included 198 TNBC tumors from MD Anderson Cancer Center (Houston, Texas) [6].
Cell culture
Growth conditions for human TNBC MDA-MB-231 and BT-549 cells, and HER2-positive breast cancer BT-474 cells (American Type Culture Collection, Manassas, VA) were previously reported [30]. MDA-MB-231 cell line expressing green fluorescent protein (GFP), herein indicated as MDA-MB-231/GFP, was produced as previously described [12].
Western blot
Cell lysates preparation and Western blot analyses were performed as previously reported [31]. Filters were probed with the indicated primary antibodies: anti-phospho-PDGFRβ (Tyr771, indicated as pPDGFRβ), anti-PDGFRβ, anti-phospho-Akt (Ser473, indicated as pAkt) (Cell Signaling Technology Inc.) and anti-vinculin (N-19) (Santa Cruz Biotechnology, Santa Cruz, CA).
RNA extraction and reverse transcription quantitative polymerase chain reaction (RT-qPCR)
RT-qPCR was performed as previously described [31]. Primers used were: PDGFRβ, Fwd 5′-AGGACACGCAGGAGGTCAT-3′, Rev 5′-TTCTGCCAAAGCATGATGAG-3′; MMP-9, Fwd 5′-GACTTGGCAGTGGAGACTGCGGGCA-3′, Rev 5′-GACCCCACCCC TCCTTGACAGGCAA-3′; Snail, Fwd 5′-CCTCCCTGTCAGATGAGGAC-3′, Rev 5′-CCAGGCTGAGGTATTCCTTG-3′; Caldesmon, Fwd 5′-GAGCGTCGCAGAGAACTTAGA-3′, Rev 5′-TCCTCTGGTAGGCGATTCTTT-3′. The following primers were used for internal control: β-actin, Fwd 5′-CAAGAGATGGCCACGGCTGCT-3′, Rev 5′-TCCTTCTGCATCCTGTCGGCA-3′; Glyceraldehyde-3-phosphate dehydrogenase, Fwd 5′-AAACAGAAGGCAGCTTTACGATG-3′, Rev 5′-AAATGTTCTGATCCAGTAGCG-3′.
Aptamers and NIR labeling
2′ F-Pyrimidines (2′F-Py) RNA anti-PDGFRβ Gint4.T aptamer, the scrambled (Scr) sequence used as a negative control, and the corresponding versions containing a C6-NH2 5′ terminal modification were synthesized by TriLink Biotechnologies (San Diego, CA). The sequences of the aptamers and handling protocols prior to each treatment were previously reported [12].
For imaging studies, 5′-(C6-NH2)-Gint4.T and 5′-(C6-NH2)-Scr were labeled with VivoTag-S 680 NIR-dye (MW: 1439 g/mol) (PerkinElmer, Waltham, MA). RNAs were incubated with VivoTag-S 680 (RNA/dye ratio, 1/20) in 50 mM carbonate-bicarbonate buffer (pH 8.5) for 2 h at 25 °C under slow mixing conditions. Non-reacted free dye was removed by using centrifugal filter devices (Amicon Ultra-0.5 mL 10,000 MW-cutoff centrifugal filter, Millipore, Billerica, MA). Aptamer labeling with NIR-dye was confirmed by ultraviolet-visible (UV-Vis) spectroscopy and by electron spray ionization (ESI) mass spectrometry (MS) using an Agilent 6230 time-of-flight (TOF) mass spectrometer (Agilent Technologies, Cernusco Sul Naviglio, Italy). Direct infusion analysis was performed on a 30 µM solution of the conjugate derivative in 50 mM carbonate-bicarbonate buffer (pH 8.5).
RNA stability in human serum
Human serum stability of NIR-Gint4.T and NIR-Scr was evaluated by incubating 10 μM NIR-2′F-Py-RNAs in Dulbecco's phosphate-buffered saline (DPBS) containing 80% human serum at 37 °C up to 72 h. RNA from all time points was mixed with formamide RNA loading dye and stored at -80 °C. All samples were resolved by polyacrylamide gel electrophoresis (PAGE) under denaturing condition (15% polyacrylamide with 7 M urea). The gel was stained with ethidium bromide and UV exposed to visualize the RNA bands.
Cell growth in 3D cultures
MDA-MB-231 and BT-549 cells (7×104 cells/well) in RPMI-1640 containing 2% fetal bovine serum (FBS) (Invitrogen, Carlsbad, CA) and 2% growth factor-reduced Matrigel (BD Biosciences, Franklin Lakes, NJ) were seeded into 6-well plates pre-coated with Matrigel (500 µL per well) with or without aptamer. After 3 and 7 days, cells were analyzed under a phase-contrast microscope. Colonies having two or more invasive protrusions were counted as a measure of invasive behavior in at least 10 fields per condition. Following 7 days, cell/Matrigel mixture was recovered from the plates using cold DPBS-EDTA (5 mM) and a polyethylene cell lifter and transferred to a 15 mL conical tube. The tubes were gently shaken in a cold room for 1 h to dissolve the Matrigel and then centrifuged for 5 min at 200 ×g at 4 °C. RNA was extracted from cell pellets by TRIzol and then processed for RT-qPCR, as described above.
Tube formation assay
Tube formation assay and immunofluorescence analysis of vascular endothelial (VE)-cadherin (Cell Signaling Technology Inc.) were performed on MDA-MB-231 and BT-549 cells, as previously reported [30].
Cell migration and invasion
For transwell migration assay, MDA-MB-231, BT-549 and BT-474 cells were serum starved overnight in the presence of Gint4.T or Scr. After starvation, cells (5×104 in 100 μL serum-free medium per well) were seeded into the upper chamber of a 24-well transwell (Transwell filters 8 μm pore size; Corning Incorporate, Corning, NY) in the presence of Gint4.T or Scr and exposed to medium containing 10% FBS (lower chamber), as inducer of migration.
The transwell invasion assay was performed as the migration assay except that cells (1×105 in 100 μL serum-free medium per well) were plated on the Matrigel-coated (diluted 1:3 in serum-free medium) filters of a transwell chamber. After incubation at 37 °C in humidified 5% CO2 for the indicated times, cells were visualized by staining with 0.1% crystal violet in 25% methanol and photographed. Stained cells were lysed in 1% sodium dodecyl sulfate and absorbance at 595 nm was measured on a microplate reader.
Cell viability and proliferation
Viability of MDA-MB-231 and BT-474 cells (4.0×103 cells/well, 96-well plates) was assessed by CellTiter 96 AQueous One Solution Cell Proliferation Assay (Promega BioSciences Inc., San Luis Obispo, CA) according to the manufacturer's instructions.
For growth curves experiments, MDA-MB-231 cells (5×103 cells/3.5-cm plate) were either mock-treated or treated with Gint4.T or Scr and then counted through the Bürker chamber at the indicated time points.
Cell targeting with NIR-aptamer
Binding of NIR-Gint4.T to the cells was assessed by confocal microscopy and flow cytometry.
For confocal microscopy, MDA-MB-231 and BT-474 cells (105 cells/well in 24-well), previously seeded on a coverslip for 24 h, were incubated with NIR-Gint4.T or NIR-Scr (500 nM-final concentration) in the presence of 100 μg/mL polyinosine (Sigma-Aldrich, Milan, Italy) as nonspecific competitor. After 5 min at room temperature (RT), cells were fixed with 4% paraformaldehyde in DPBS for 20 min. In co-localization experiments, non-permeabilized cells were subjected to blocking in 10% FBS/DPBS for 20 min at RT. Cells were incubated with anti-PDGFRβ (R&D system) antibody, washed three times in DPBS and incubated with Alexa Fluor 488 Anti-Goat (Invitrogen). Finally, after three further washes in DPBS, cells were incubated with 1.5 μM 4′,6-Diamidino-2-phenylindole (DAPI, D9542, Sigma-Aldrich) and mounted with glycerol/DPBS. Samples were visualized by Zeiss LSM 700 META confocal microscopy equipped with a Plan-Apochromat 63x/1.4 Oil DIC objective.
For flow cytometry, MDA-MB-231 and BT-474 cells were trypsinized and washed twice with 500 μL DPBS. MDA-MB-231 cells (3.5×105) were mock treated or incubated with increasing concentrations of NIR-Gint4.T or NIR-Scr (5 nM, 10 nM, 25 nM, 50 nM, 100 nM, 200 nM and 500 nM) in the presence of 100 μg/mL polyinosine (Sigma-Aldrich) as nonspecific competitor. After 15 min at RT, cells were washed twice with 500 μL DPBS, suspended in 500 μL DPBS and analyzed by flow cytometry (BD Accuri™ C6). The mean fluorescence intensity (MFI) of the cells labeled by NIR-Gint4.T was used to calculate specific binding by subtracting the MFI of nonspecific binding from NIR-Scr. The apparent Kd of PDGFRβ aptamer-cell interaction was obtained by fitting the dependence of fluorescence intensity of specific binding on the aptamer concentration to the equation Y = Bmax × X / (Kd + X), using GraphPad Prism version 6.00. Binding of NIR-Gint4.T or NIR-Scr to negative control BT-474 cells (3.5×105) was analysed as above by using 500 nM aptamer concentration.
Tumor targeting with NIR-aptamer
All experimental procedures complied with the European Communities Council directives (2010/63/EU) and national regulations (D.L. 116/92) and were performed in accordance with National Institutes of Health (NIH) recommendations. The present study was approved by the Italian Ministry of Health (authorization number 38/2015-01-28). All efforts were made to minimize animal suffering and the number of animals necessary to produce reliable results. All experimental procedures described were performed under general anesthesia with 2% isoflurane in 100% oxygen at 0.8 L/min. All mice were maintained on a diet with a purified, alfalfa-free rodent chow for 15 days before fluorescence imaging to minimize fluorescence in the gut.
Subcutaneous TNBC xenograft models were realized as previously described [32, 33]. Briefly, 10×106 MDA-MB-231 target cells or BT-474 non-target cells were re-suspended in 0.1 mL of 1:1 mix of physiological saline and Matrigel and subcutaneously (s.c.) injected into the right (MDA-MB-231 cells) or left flank (BT-474 cells) of five-week-old female Balb/c nude mice that weighed around 18-20 g (Charles River, Milan, Italy). Once tumors became palpable (established), approximately 50 mm3 [volume = 0.5 × long diameter × (short diameter)2], mice were randomized into groups (five animals per group). NIR-Gint4.T or NIR-Scr (1 nmol/70 µL) were injected into the lateral tail vein of mice maintained under isoflurane anesthesia using a catheter to improve the likelihood of a successful injection. The body temperature of the animals was held constant during probe administration with a heating pad. At the specified time (15 min, 2 h, 4 h and 24 h) post NIR-Gint4.T or NIR-Scr injection, isoflurane-anesthetized mice were subjected to fluorescence molecular tomography (FMT) scanning using an FMT 4000 imaging system (PerkinElmer) as previously reported [12]. Briefly, mice were placed in a biplanar imaging cassette supplied with the instrument and trans-illuminated with laser light. A single fiber-coupled laser diode scans the surface of the animal at a user-adjustable pitch and span in X and Y. The laser power and exposure time at each scan point are automatically adjusted by the system to provide high signal-to-noise while avoiding saturation. Resulting transmission and fluorescence patterns were captured with a thermoelectrically cooled CCD camera (excitation, Ex: 670; emission, Em: 690). The total scan times were on the order of 2-4 min (depending on the region-of-interest). FMT 4000 has 4 distinct laser diodes provided at 635 nm, 670 nm, 750 nm and 785 nm and it has a laser output power of 80 mW for each wavelength. The entire laser spectral energy is condensed within <3 nm FWHM to ensure sufficient spectral power density for deep tissue penetration and subsequent quantification. The position and intensity of fluorescence sources were reconstructed in two-dimensions (2D, epifluorescence) and/or 3D (tomography) using the TrueQuant software package (PerkinElmer), supplied with the FMT4000. Three-dimensional volumes-of-interest (VOIs) were drawn around tumor and adjoining background non-tumor area, and the total amount of fluorescence (pmol) was automatically calculated relative to internal standards generated with known concentrations of the appropriate probe (NIR-Gint4.T, NIR-Scr or VivoTag-S 680). A threshold was applied to all animals equal to 30% of the maximum value of fluorescence in the adjoining non-tumor area. As a control, mice injected with 1 nmol VivoTag-S 680 were analyzed under the same experimental conditions.
To confirm the specificity of in vivo uptake of NIR-Gint4.T, blocking studies with a large excess of unlabeled Gint4.T were done in nude mice bearing MDA-MB-231 tumors. Briefly, animals underwent a first FMT scan following the injection of NIR-Gint4.T (100 pmol). After 48 h, when the tumor-specific fluorescence signal became undetectable, they were pretreated for 20 min with unlabeled Gint4.T (5000 pmol) and then subjected to a second FMT scan using the same NIR-Gint4.T (100 pmol). In all imaging studies, the scan area was focused in close proximity to the tumor area to improve the spatial resolution of acquired images and a VOI was drawn around the tumor. Statistical significance was determined using the two-tailed unpaired Student's t-test. Differences were considered significant at P < 0.05.
Imaging and inhibition of TNBC lung metastases with PDGFRβ aptamer
To assess the ability of NIR-Gint4.T to detect lung metastases, a TNBC model of lung metastases was established by injecting MDA-MB-231/GFP cells (6×105 in 100 μL DPBS) into the tail vein of female Balb/c nude mice. After 42 days, mice (three per group) were injected with 1 nmol NIR-Gint4.T or NIR-Scr and imaged at the specified time by FMT, as described above. In addition, 1 nmol VivoTag-S 680 or ProSense 680 (PerkinElmer) were intravenously (i.v.) injected in mice and used as negative and positive controls, respectively, for lung metastases detection.
To assess the effect of Gint4.T on the inhibition of lung metastases, female Balb/c nude mice were randomized into Gint4.T and Scr treatment groups (n=4) and injected in the tail vein with MDA-MB-231/GFP cells (6×105 in 100 μL DPBS) prior to being incubated with 400 pmol Gint4.T or Scr for 30 min at 37 °C. Systemic i.v. administration of the aptamers (1400 pmol) was continued as schematized in Figure 8. After 42 days from cell injection, mice were i.v. injected with 1 nmol NIR-Gint4.T and imaged at 2 h by FMT, as described above.
Ex vivo analysis
After imaging studies, mice were euthanized and tumors, liver, kidney, spleen, muscle and heart (tumor targeting study) or lungs (metastases imaging/inhibition study) were excised and imaged by 2D FMT analysis.
For the metastases imaging/inhibition analyses, one lung from each animal was stored in 10% neutral buffered formalin for paraffin embedding, sectioning (4 μm) and staining for hematoxylin and eosin (H&E), PDGFRβ, Ki-67, MMP-9 and vimentin, at the same experimental conditions as for human tissues. The other lung was embedded in O.C.T. (optimal cutting temperature) compound for sectioning, and 10 μm frozen sections were incubated with DAPI to stain nuclei. Subsequently, sections were prepared for GFP detection by a Zeiss LSM 700 META confocal microscope equipped with a N-Achroplan 10×/0.25 objective. Liver and axillary lymph nodes were stained for H&E, as described above.
In vivo pharmacokinetic (PK) of NIR-Gint4.T
NIR-Gint4.T (1 nmol/70 µL) was administered intravenously through the tail vein of three Balb/c nude mice. Blood was collected from the orbital sinus immediately before administration (0 h) and at 0.083, 0.25, 0.5, 1, 3, 5 and 24 h after administration. Samples were centrifuged for 10 min at 1000 ×g and plasma was stored at -80 °C until the assay. For the detection of aptamer, 5 μL plasma was directly reverse transcribed and PCR amplified with iQ SYBR Green Supermix (Bio-Rad, Hercules, CA) in the presence of Gint4.T 5′ and 3′ primers, as previously described [26]. Reactions were all done in triplicate. The quantity of the amplified product was extrapolated from a standard amplification curve obtained with NIR-Gint4.T.
Statistical analysis
Student's t-test and Pearson's χ2 test were used to determine whether a relationship exists between the categorical variables included in the TMA-related study by Statistical Package for Social Science v. 20 software (SPSS Inc., Chicago, IL, USA). All correlations within the public dataset were assessed by Pearson's χ2 test or one-way analysis of variance (ANOVA) through the R2 platform [28].
All other statistical values were defined using GraphPad Prism version 6.00 for Windows by t-test. P value < 0.05 was considered significant for all analyses.
Results
PDGFRβ expression in tumor cells identifies a subgroup of human mesenchymal TNBC
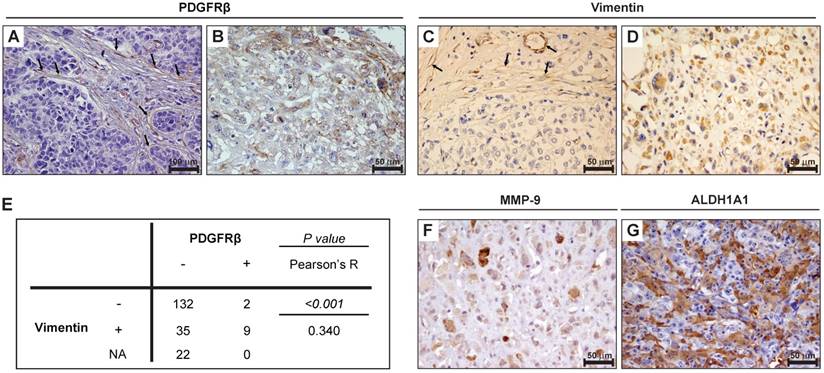
To analyze PDGFRβ expression in TNBC and correlate it with clinicopathological and molecular features, we performed immunohistochemical analyses on a human TNBC tissue microarray comprising 200 TNBC samples [27]. As expected, in all TNBC samples, PDGFRβ expression was detected in mesenchymal stromal cells and vascular endothelial cells (see the arrows in Figure 1A), which was used as internal positive control for staining. Importantly, we found that tumor cells of 11 TNBC samples (5.5%) expressed PDGFRβ either at the cell membrane or cytoplasmic level (Figure 1B and Table 1), whereas in all the other samples there was no expression of PDGFRβ in tumor cells (Figure 1A). Despite the low number of PDGFRβ-positive cases, a significant correlation was found between PDGFRβ expression and the pre-menopausal status, and a linear-to-linear association was found between PDGFRβ expression and the age and grading (Table 1). We also stained the TMA for the mesenchymal marker vimentin and considered its expression on tumor cells (Figure 1D), using stromal/endothelial cells, stained positively (Figure 1C, see the arrows), as internal positive control. A significant positive correlation between vimentin and PDGFRβ expression on tumor cells (Figure 1E) was found, suggesting the association of PDGFRβ expression in TNBC tumor cells with the mesenchymal TNBC subtype. The 11 PDGFRβ-positive cases were also stained for either matrix metallopeptidase MMP-9 (Figure 1F) or aldehyde dehydrogenase ALDH1A1 (Figure 1G): all were positive for at least one of them (Table 2), thus indicating a high invasive and stem-like phenotype for this subgroup of TNBC.
PDGFRβ expression in TNBC and correlation with clinicopathological features.
| PDGFRβ-positive expression | |||
|---|---|---|---|
| TNBC cases | n. 200 | 11 | |
| Variables | pa | ||
| Age | |||
| <40 | 23 | 3 | 0.043 |
| 40-60 | 93 | 6 | |
| >60 | 84 | 2 | |
| Menopausal status | |||
| Pre-menopause | 74 | 8 | 0.015 |
| Post-menopause | 126 | 3 | |
| Histotype | |||
| CDI | 168 | 7 | 0.126 |
| CLI | 10 | 2 | |
| Medullary | 8 | 0 | |
| Metaplastic | 7 | 1 | |
| CDLI | 7 | 1 | |
| Grading | |||
| 1 | 1 | 1 | 0.001 |
| 2 | 22 | 3 | |
| 3 | 177 | 7 | |
| LN metastases | |||
| Negative | 115 | 5 | 0.278 |
| Positive | 82 | 6 | |
| NA | 3 | ||
| Distant metastases | |||
| Absent | 68 | 4 | 0.350 |
| Present | 37 | 4 | |
| NA | 95 | ||
| Tumor size | |||
| 1 | 85 | 4 | 0.698 |
| 2-3 | 100 | 7 | |
| NA | 15 | ||
a Fisher exact test value is indicated for groups of two variables, while linear-to-linear association value is indicated for groups with more than two variables. NA: not available
Summary of the immunohistochemical results for MMP-9 and ALDH1A1 staining in the 11 PDGFRβ-positive cases (T1-T11).
| MMP-9 | ALDH1A1 | |
|---|---|---|
| T1 | Neg | Pos |
| T2 | Pos | Pos |
| T3 | Pos | Pos |
| T4 | Pos | Neg |
| T5 | Pos | Neg |
| T6 | Neg | Pos |
| T7 | Pos | Neg |
| T8 | Pos | Pos |
| T9 | Pos | Pos |
| T10 | Pos | Pos |
| T11 | Pos | Neg |
ALDH1A1: aldehyde dehydrogenase 1‑A1; MMP-9: matrix metallopeptidase-9.
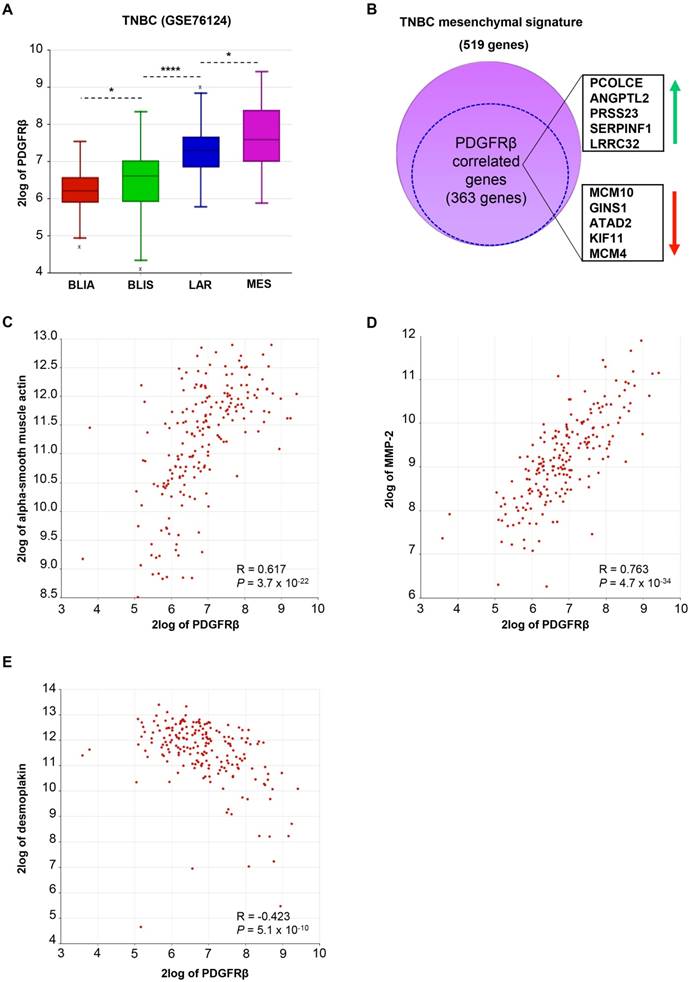
To further study the correlation of PDGFRβ with a specific TNBC subtype, we analyzed PDGFRβ gene expression in a public dataset of 198 TNBC samples molecularly characterized and subdivided in four subtypes, including luminal-androgen receptor (LAR), mesenchymal (MES), basal-like immune-suppressed (BLIS), and basal-like immune-activated (BLIA) [6]. As shown in Figure 2A, PDGFRβ gene expression significantly correlates with the TNBC molecular subtype and, as expected, is highest in the mesenchymal subtype. Consistently, 70% of genes belonging to the signature of the mesenchymal TNBC subtype are significantly correlated to PDGFRβ expression (Figure 2B and Table S1). Besides these, we found that PDGFRβ positively correlates with mesenchymal markers, such as vimentin (Figure S1A) and alpha-smooth muscle actin (Figure 2C), as well as with genes associated with an invasive phenotype, such as those encoding for MMP-2 (Figure 2D) and MMP-9 (Figure S1B), while negatively correlating with epithelial markers, such as desmoplakin, E-cadherin, CD24, occludin (Figure 2E, Table S1 and Figure S1C) and many others that are downregulated in the mesenchymal TNBC subtype (Table S1). Further, by restricting our analyses to the mesenchymal TNBC subtype, we found many highly significant associations between PDGFRβ expression and mesenchymal stem cell-specific markers, including Endoglin (ENG), Thy-1 cell surface antigen (THY), and others (Table 3).
Taking together protein and gene expression data, PDGFRβ appears to be a specific biomarker of a TNBC subgroup with mesenchymal/stem phenotype.
Mesenchymal/stem cell-specific genes [4, 34] correlated with PDGFRβ in the mesenchymal subtype.
| R | P | |
|---|---|---|
| ENG | 0.709 | 2.5e-08 |
| THY1 | 0.707 | 2.7e-08 |
| MCAM | 0.539 | 9.1e-05 |
| NT5E | 0.533 | 1.2e-04 |
| ITGAV | 0.528 | 1.4e-04 |
| KDR | 0.422 | 3.2e-03 |
Correlations were analyzed by Pearson's χ2 test through the R2 platform [28]. Significance of correlation was assessed by r-value (R) and P value (P).
ENG: endoglin; ITGAV: integrin alpha V; KDR: kinase insert domain receptor; MCM4: minichromosome maintenance 4; NT5E: 5'-nucleotidase ecto; THY1: Thy-1 cell surface antigen.
Effect of PDGFRβ aptamer on invasive cell growth in 3D culture conditions
Due to the propensity of tumor cells with a mesenchymal phenotype to migrate and invade, and the under-explored role of PDGFRβ in TNBC progression, we asked whether inhibition of PDGFRβ activity by a specific PDGFRβ aptamer would result in inhibition of TNBC cell invasiveness and metastases formation. To this aim, we took advantage of MDA-MB-231 and BT-549 cell lines, which are representative of the mesenchymal subtype [4]. In agreement with their poorly differentiated phenotype, these cells share high expression levels of PDGFRβ and vimentin, and undetectable E-cadherin ([30] and Figure S2).
Tumor cells expression of PDGFRβ is associated with a mesenchymal/stem phenotype in TNBC patients. Images of representative human TNBC cases stained for PDGFRβ (A-B) and vimentin (C-D). In (A) and (C), the arrows identify stromal/endothelial cells with PDGFRβ and vimentin expression, respectively. (E) Statistical association between PDGFRβ and vimentin expression on tumor cells. Data were analyzed by Pearson's χ2 test and the resultant P value and R coefficient are reported. (F-G) Images of representative PDGFRβ-positive TNBCs stained for MMP-9 (F) and ALDH1A1 (G). (A) Magnification 20 ×, scale bar = 100 μm. (B-D, F-G) Magnification 40 ×, scale bar = 50 μm. ALDH1A1: aldehyde dehydrogenase 1‑A1; MMP-9: matrix metallopeptidase-9; NA: not analyzed; PDGFRβ: platelet-derived growth factor receptor β.
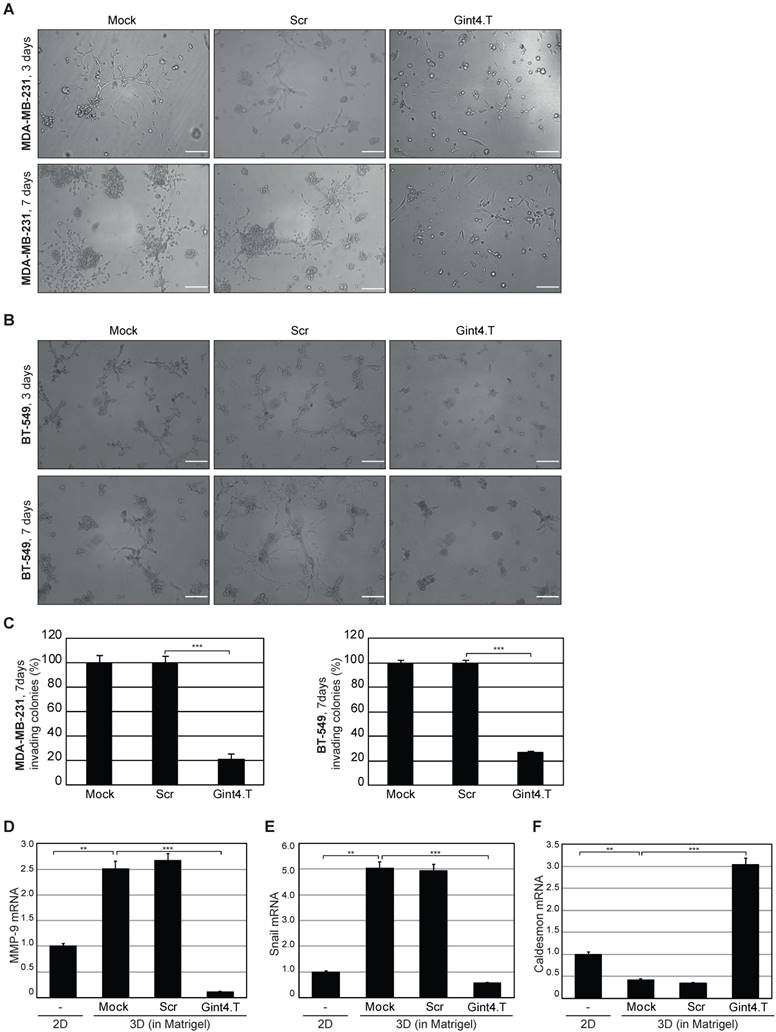
Gint4.T aptamer was previously validated as a specific inhibitor of PDGFRβ on human BM-MSCs [12] and GBM cells [25, 26, 31]. First, we verified, in MDA-MB-231 and BT-549 cells, the ability of the aptamer to inhibit platelet-derived growth factor-BB (PDGF-BB)-dependent activation of PDGFRβ and downstream PI3K-Akt pathway (Figure S3), reported to be involved in metastases formation from breast cancers [35, 36]. Next, we determined whether the inhibition of PDGFRβ activation results in impairment of invasive growth of MDA-MB-231 and BT-549 cells embedded in Matrigel (the standard 3D culture environment). When cultured in 3D, these cell lines acquire a typical invasive phenotype, which strictly stems from their remarkable degree of plasticity, and have a different response to various signaling inhibitors and chemotherapeutics compared to when grown as monolayers on tissue culture plastic [37]. Thus, the two cell lines were grown in Matrigel in the absence or in the presence of 200 nM Gint4.T or a scrambled sequence (Scr) used as negative control, and their invasive growth was assessed up to 7 days. As shown, starting from 3 days, mock- and Scr-treated cells displayed invasive growth and developed spider-like protrusions extending into the Matrigel (Figure 3A-B, upper left and middle panels). The invasive morphological phenotype, for both cell lines, with stellate projections that bridge multiple cell colonies became prominent after 7 days in culture (Figure 3A-B, lower left and middle panels). Interestingly, Gint4.T (Figure 3A-B, right panels) inhibited the formation of cell projections at both 3 and 7 days, impairing the development of colonies that were drastically fewer, smaller in size and with no protrusions as compared with well-developed branching colonies observed in control cells (Mock and Scr). In the presence of Gint4.T, we observed more than 70% reduction of colonies having two or more invasive protrusions compared to the controls (Figure 3C). Previous studies have shown that MDA-MB-231 cells express MMP-9, which is up-regulated by cell contact to Matrigel during cell invasiveness [38, 39]. Accordingly, we found that MMP-9 expression was favored in Matrigel-based 3D with respect to 2D cell cultures, as assessed by RT-qPCR analyses on RNA extracted from cells harvested from Matrigel following 7 days of culture (Figure 3D). Importantly, inhibiting PDGFRβ by Gint4.T aptamer caused a strong reduction of MMP-9 mRNA, while no decrease was obtained in the presence of Scr treatment. Further, we got similar results when we analyzed mRNA expression of Snail (Figure 3E), a transcription factor critical for EMT, which has been found to be associated with the transition from a non-invasive to an invasive phenotype and regulation of MMPs in different human cancers [40]. Conversely, we found opposite results for the expression of Caldesmon (Figure 3F), in perfect agreement with its role in inhibiting invasion and MMPs release [41].
In silico meta-analysis of PDGFRβ gene expression in human TNBC tissues. (A) Box-plot comparing PDGFRβ expression in four TNBC subtypes according to Burstein MD et al. [6] and statistical analysis of the differences among them. The data were analyzed by one-way analysis of variance (ANOVA) through the R2 web platform. (B) Schematic representation of the overlap between the MES signature and genes correlated to PDGFRβ in the MES subtype (the dotted circle). The top ten shared genes (5 upregulated and 5 downregulated) are framed on the side. For the complete list of the shared genes see Table S1. (C-E) XY-plots correlating PDGFRβ and alpha-smooth muscle actin (C), MMP-2 (D) and desmoplakin (E) in the MES subtype. Correlations were analyzed by Pearson's χ2 test through the R2 platform [28]. Significance of correlation was assessed by r-value (R) and P value (P). ANGPTL2: angiopoietin-like protein 2; ATAD2: ATPase family, AAA domain containing 2; BLIA: basal-like immune-activated; BLIS: basal-like immune-suppressed; GINS1: GINS Complex Subunit 1; KIF11: kinesin family member 11; LAR: luminal-androgen receptor; LRRC32: leucine rich repeat containing 32; MCAM: melanoma cell adhesion molecule; MCM10: minichromosome maintenance 10; MES: mesenchymal; MMP-2: matrix metallopeptidase-2; PCOLCE: procollagen c-endopeptidase enhancer; PDGFRβ: platelet-derived growth factor receptor β; PRSS23: protease serine 23; SERPINF1: serpin family F member 1; TNBC: triple-negative breast cancer.
PDGFRβ aptamer inhibits invasive growth of mesenchymal TNBC cells in 3D cultures. Representative phase-contrast images of MDA-MB-231 (A) and BT-549 (B) cells grown in Matrigel in the absence (Mock) or in the presence of 200 nM Gint4.T or Scr for the indicated times. The aptamers treatment was renewed every 48 h. At least three independent experiments were performed. Magnification 10 ×, scale bar = 200 μm. (C) Invasive ability of MDA-MB-231 and BT-549 cells is expressed as percentage of invading colonies at 7 days considering the mock-treated control cells as 100%. (D-F) RNA extracted from MDA-MB-231 cells grown in 2D (left untreated) or in Matrigel (mock-treated or treated as in (A)) for 7 days was analyzed by RT-qPCR for the indicated genes and mRNA relative expression was reported. (C-F) Bars depict mean ± SD of three independent experiments. ***P < 0.001; **P < 0.01. MMP-9: matrix metallopeptidase-9; 3D: three-dimensional; 2D: two-dimensional.
It has been shown that the plasticity of TNBC with a mesenchymal phenotype is essential for VM, a perfusion mechanism distinct from classical angiogenesis and associated with metastatic progression [42]. In agreement with their poorly differentiated phenotype, MDA-MB-231 and BT-549 cells have a high tendency to transdifferentiate into endothelial cells, thus forming VM [30], and PDGFRβ is involved in mediating their endothelial differentiation capability [21, 43]. Accordingly, we found that PDGFRβ aptamer strongly prevented the ability of these cells, grown on Matrigel monolayer, to form VE-positive channel structures, a hallmark of VM (Figure S4).
Effect of PDGFRβ aptamer on TNBC cell migration and invasion
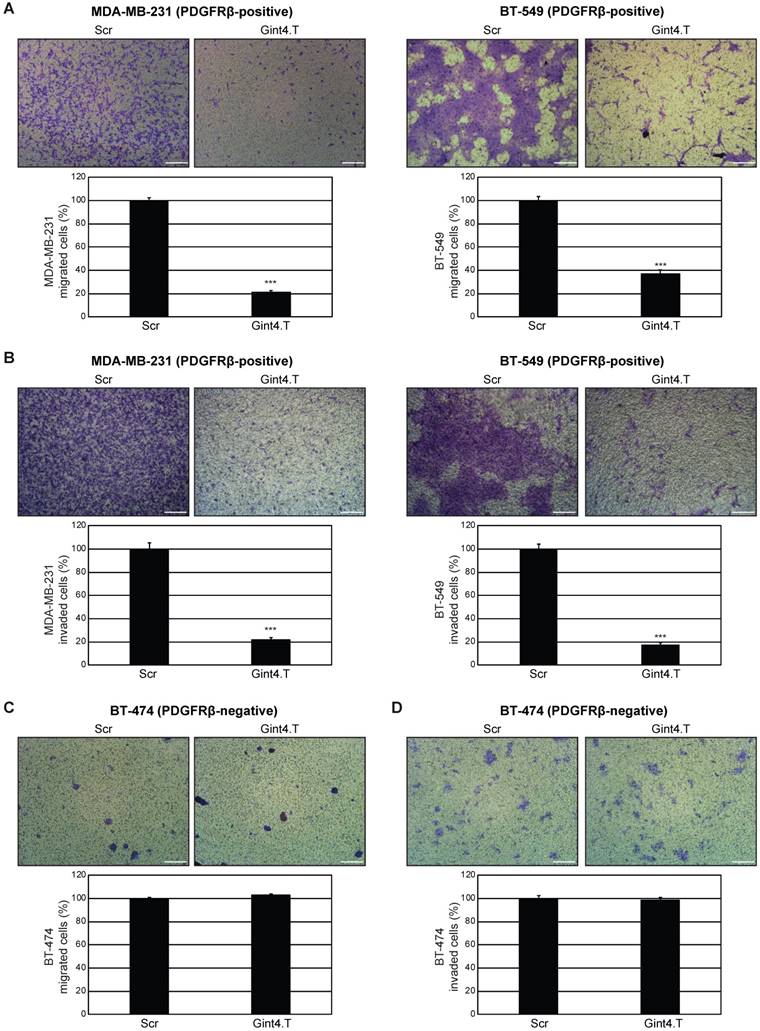
As a next step, we investigated whether PDGFRβ aptamer affects the migration and invasion of TNBC cells. Migration was analyzed by a transwell migration assay, which assesses the chemotactic capacity of cells. In the presence of Gint4.T, cell migration stimulated by serum was reduced by approximately 80% in MDA-MB-231 and 65% in BT-549 cells at 24 h, as compared to Scr-treated cells (Figure 4A). Then, we examined the effect of Gint4.T on the ability of the cells to invade through a Matrigel-coated membrane using a transwell invasion assay, which mimics the whole process of cell invasion of basement membranes. Using this assay, we observed that the MDA-MB-231 and BT-549 cell invasion rate in the presence of Gint4.T was decreased by 80% at 72 h, compared to Scr-treated cells (Figure 4B). Consistent with previous results [44], a human epithelial HER2-positive breast cancer cell line, BT-474, lacking detectable levels of endogenous PDGFRβ (Figure S2 and [30]) displayed very poor migration (Figure 4C) and invasion (Figure 4D) capability, which was not affected by Gint4.T treatment (Figure 4C-D). We checked that the aptamer-mediated decrease in migration and invasion of TNBC cells occurred independently of cell proliferation. Indeed, as shown in Figure S5A, no effect on viability of MDA-MB-231 cells was observed up to 72 h in the presence of Gint4.T treatment, whereas about 20% inhibition occurred at 96 h. Conversely, no effect was observed up to 96 h with the scrambled aptamer. Importantly, Gint4.T did not affect viability of BT-474 control cells (Figure S5A). Furthermore, as assessed by growth curve experiments, the prolonged treatment of MDA-MB-231 cells with Gint4.T caused about 60% inhibition of cell proliferation at day 9 with respect to mock- and Scr-treated cells (Figure S5B).
Altogether, these results show that Gint4.T aptamer drastically hampers the migration and invasion capability of mesenchymal PDGFRβ-positive MDA-MB-231 and BT-549 cells, and has an effect on MDA-MB-231 cell proliferation only after a prolonged treatment.
Determination of the tumor-targeting selectivity of the NIR-labeled PDGFRβ aptamer
In order to evaluate the ability of Gint4.T to target PDGFRβ in TNBC animal models, we labeled the aptamer, which contains an amino group at its 5´ extremity, with an amine-reactive fluorescent NIR dye (VivoTag-S 680), which allows for minimal scattering, low autofluorescence and deep tissue penetration. We checked the labeling efficiency of NIR-Gint4.T and NIR-Scr negative control prior to performing our studies (Figure S6A-B). First, we verified the capacity of NIR-Gint4.T to specifically bind to PDGFRβ-expressing TNBC cells. Analysis of binding by flow cytometry of MDA-MB-231 cells incubated with different concentrations of NIR-Gint4.T gave an apparent Kd of 57.48 ± 7.8 nM (Figure 5A, left panel), thus confirming that the aptamer, upon conjugation to NIR dye, maintains high-affinity targeting to PDGFRβ-positive TNBC cells. Further, we observed that the binding of NIR-Gint4.T to MDA-MB-231 cells was competed by EC-PDGFRβ (Figure 5A, right panel), thus supporting the ability of the aptamer to specifically recognize cancer cells through binding to the extracellular domain of PDGFRβ. Accordingly, no binding of NIR-Gint4.T to non-mesenchymal PDGFRβ-negative BT-474 cells was assessed at concentrations as high as 500 nM (Figure 5B). The specific binding of NIR-Gint4.T to MDA-MB-231 cells was further confirmed by confocal imaging. As shown in Figure 5C and Figure S7A-B, NIR-Gint4.T aptamer specifically localized at the membrane level with PDGFRβ on MDA-MB-231 cells after 5 min incubation. As expected, very little to no NIR-Gint4.T-associated signal was observed on PDGFRβ-negative BT-474 cells (Figure 5D).
To definitively confirm that the labeling of the aptamer does not affect its target recognition, we also verified its inhibiting effect on ligand-dependent phosphorylation of the receptor and downstream Akt in both MDA-MB-231 and BT-549 cells (Figure S7C).
PDGFRβ aptamer inhibits mesenchymal TNBC cell migration and invasion. (A) MDA-MB-231 and BT-549 (PDGFRβ-positive) cell migration toward 10% FBS was analyzed by transwell migration assay in the presence of 200 nM Gint4.T or Scr for 24 h. (B) Invasion of MDA-MB-231 and BT-549 cells toward 10% FBS through Matrigel was carried out in the presence of 200 nM Gint4.T or Scr for 72 h. (C) Migration and (D) invasion of control BT-474 (PDGFRβ-negative) cells were analyzed as in (A) and (B), respectively. (A-D) Photographs of a representative experiment are shown. Magnification 10 ×, scale bar = 200 μm. Data are presented as percentage of migrated or invaded cells in the presence of Gint4.T compared with Scr control. Bars depict mean ± SD of three independent experiments. ***P < 0.001.
NIR-Gint4.T aptamer specifically binds to human PDGFRβ-positive TNBC cells. (A) Left panel: Binding curve of NIR-Gint4.T on MDA-MB-231 cells for calculation of the apparent Kd of aptamer-cell interaction. Binding was analyzed using flow cytometry. The nonspecific binding value for NIR-Scr sequence was subtracted from every data point. Bars depict mean ± SD of three independent experiments. Right panel: MDA-MB-231 cells were mock-treated or incubated with 25 nM NIR-Gint4.T prior to incubation with 50 nM EC-PDGFRβ for 30 min at RT, and binding was analyzed by flow cytometry. (B) Control BT-474 (PDGFRβ-negative) cells were mock-treated or incubated with 500 nM NIR-Gint4.T or NIR-Scr and binding was analyzed by flow cytometry. (C) Following 5 min incubation with 500 nM NIR-Gint4.T or NIR-Scr, MDA-MB-231 cells were fixed and labeled with anti-PDGFRβ antibody without permeabilization, visualized by confocal microscopy and photographed. Representative 2D and 3D images constructed using Z-scan are shown. NIR-aptamer, PDGFRβ and nuclei are visualized in red, green and blue, respectively. Co-localization results appear yellow in the merged images. (D) Following 5 min incubation with NIR-Gint4.T or NIR-Scr, BT-474 cells were visualized by confocal microscopy and photographed. (A-D) At least three independent experiments were performed. All digital images were captured at the same setting to allow direct comparison of staining patterns. Magnification 63 ×, scale bar = 20 μm. DAPI: 4′,6-Diamidino-2-phenylindole; Kd: dissociation constant; MFI: mean fluorescence intensity; NIR: near-infrared; PDGFRβ: platelet-derived growth factor receptor β; 3D: three-dimensional; 2D: two-dimensional.

Selective imaging of PDGFRβ-positive tumors by NIR-Gint4.T. (A) Nude mice bearing MDA-MB-231 xenografts were i.v. injected with 1 nmol of either NIR-Gint4.T or NIR-Scr and analyzed with FMT at the indicated times. Top: Representative volume renderings taken at the same color gating for NIR-Gint4.T and NIR-Scr (n = 5)-injected mice are shown. Bottom, left: The amount of fluorescence (pmol) was quantified in specific VOIs encompassing the tumor in the animal. Bars depict mean ± SD. ***P < 0.001; **P < 0.01; *P < 0.05 (n = 5). Bottom, right: Shown are representative images of the tumors excised from mice 24 h post NIR-Gint4.T and NIR-Scr injection. (B) Mice bearing PDGFRβ-positive (left flank) or PDGFRβ-negative (right flank) tumors were i.v. injected with 1 nmol of either NIR-Gint4.T (MDA-MB-231 and BT-474 tumors) or NIR-Scr (MDA-MB-231 tumors) and analyzed with FMT at 2 h following injection. (C) Representative images of FMT obtained 2 h after injection of 100 pmol NIR-Gint4.T in the absence (left) or in the presence (right) of pretreatment with a large excess of unlabeled Gint4.T. Notably, the ten-fold reduced amount of NIR-Gint4.T is efficacious to detect the tumor. (D) Analyses of the in vitro serum stability of the NIR-RNAs. Denaturing PAGE of NIR-Gint4.T and NIR-Scr following incubation with 80% human serum at the indicated times. Depicted results represent one of three typical experiments performed. Gint4.T and Scr contain 2´F-Py in all the sequence to increase nuclease resistance. (E) Plasma pharmacokinetic profile of NIR-Gint4.T aptamer. Concentration of aptamer is shown as a function of time following a single i.v. injection in Balb/c mice. Data are presented as the mean ± SD (n = 3 for each administration). Note that we obtained a pharmacokinetic profile of unlabeled Gint4.T aptamer superimposable to that of NIR-Gint4.T. Em: emission; Ex: excitation; NIR: near-infrared; PDGFRβ: platelet-derived growth factor receptor β.
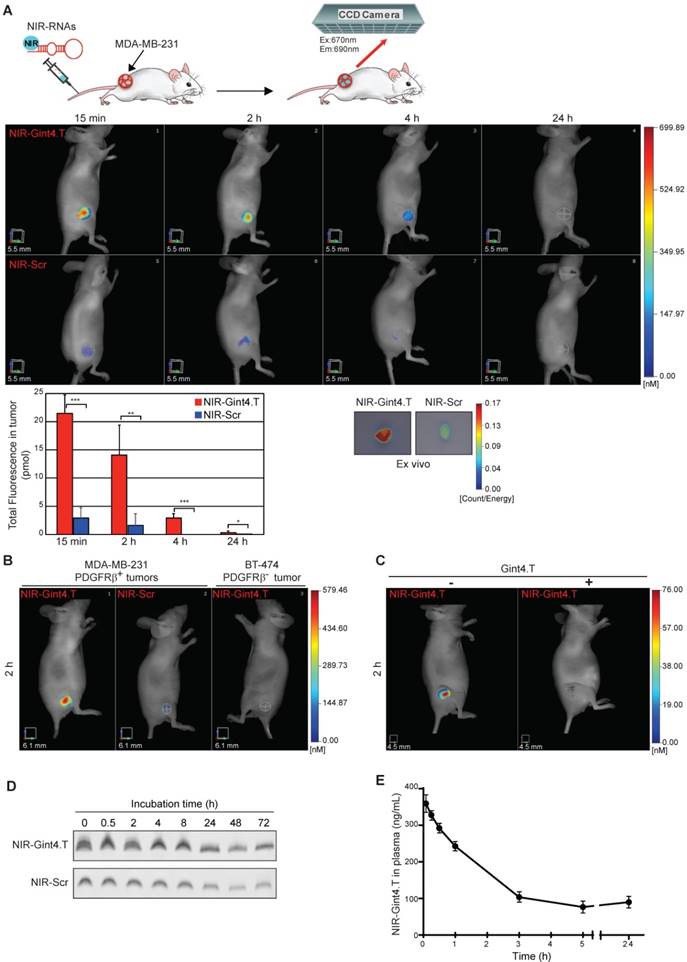
Next, to determine whether NIR-Gint4.T retains its recognition ability in vivo, NIR-Gint4.T and NIR-Scr (1 nmol or approximately 0.59 mg aptamer/kg mean body-weight) were i.v. injected into mice bearing subcutaneous MDA-MB-231 tumors and 3D non-invasive imaging was performed over 24 h by FMT (Figure 6A). This technique allows quantification of the total amount of fluorescence in the tumor VOI. As shown, NIR-Gint4.T-associated fluorescence signal was strongly detected in the tumors at 15 min after administration, maintained high up to 2 h and significantly decreased starting from 4 h up to 24 h post-injection, but remained still higher compared to NIR-Scr control. Indeed, NIR-Scr-associated signal, at the same experimental points, was very low or undetectable in the tumors, thus revealing that NIR-Gint4.T, but not the control sequence, efficiently targeted tumor sites in vivo. Fifteen minutes after injection, the amount of NIR-Gint4.T in the tumor (approximately 2.15% of the injected dose) was around 7.4-fold higher compared to NIR-Scr (21.44 ± 3.29 vs. 2.90 ± 1.9 pmol) (Figure 6A). Accordingly, ex vivo 2D imaging of the tumors, harvested 24 h after NIR-labeled RNAs injection, revealed higher fluorescence intensity from NIR-Gint4.T than from NIR-Scr (Figure 6A). Furthermore, the biodistribution of NIR-Gint4.T in major organs other than the tumor was also examined at 24 h post-injection by ex vivo imaging. As shown in Figure S8, mice injected with NIR-Gint4.T exhibited more accumulation in tumor than heart, muscle, liver or spleen. A strong fluorescence signal was observed in the kidney due to non-specific accumulation caused by the small size of the aptamers that makes them susceptible to renal filtration.
We verified that VivoTag-S 680 dye alone failed to visualize tumors (Figure S9A). Further, when mice bearing s.c. tumors derived from PDGFRβ-negative BT-474 cells were injected with NIR-Gint4.T, no fluorescence signal was detected (Figure 6B), thus indicating the tumor-targeting selectivity of the probe. To further confirm the specificity of NIR-Gint4.T in visualizing PDGFRβ-expressing cells in vivo, we performed competitive studies using the unlabeled Gint4.T aptamer. As shown (Figure 6C), the tumor accumulation of NIR-Gint4.T was prevented by pre-treatment of mice with a large excess (1/50 molar ratio of NIR labeled-to-unlabeled Gint4.T) of the corresponding unlabeled aptamer. In the blocking studies, 3D non-invasive imaging at 24 h revealed that NIR-Gint4.T was predominantly cleared from the body via the kidneys (data not shown).
Because of the presence of 2'F-Py in the entire sequence of the RNAs, NIR-Gint4.T and NIR-Scr are highly resistant to nucleases. As shown (Figure 6D), they were stable at least up to 8 h in human serum and then slowly degraded with a similar stability profile. Further, the plasma pharmacokinetic profile of NIR-Gint4.T (Figure 6E) following a single i.v. injection in Balb/c mice was compatible with the absence of bulky moieties appended to the aptamer sequence [45, 46].
Overall, these data indicate that, upon labeling with a NIR probe, the Gint4.T aptamer is able to specifically image TNBC xenografts provided they express PDGFRβ.
NIR-labeled PDGFRβ aptamer detects TNBC metastases
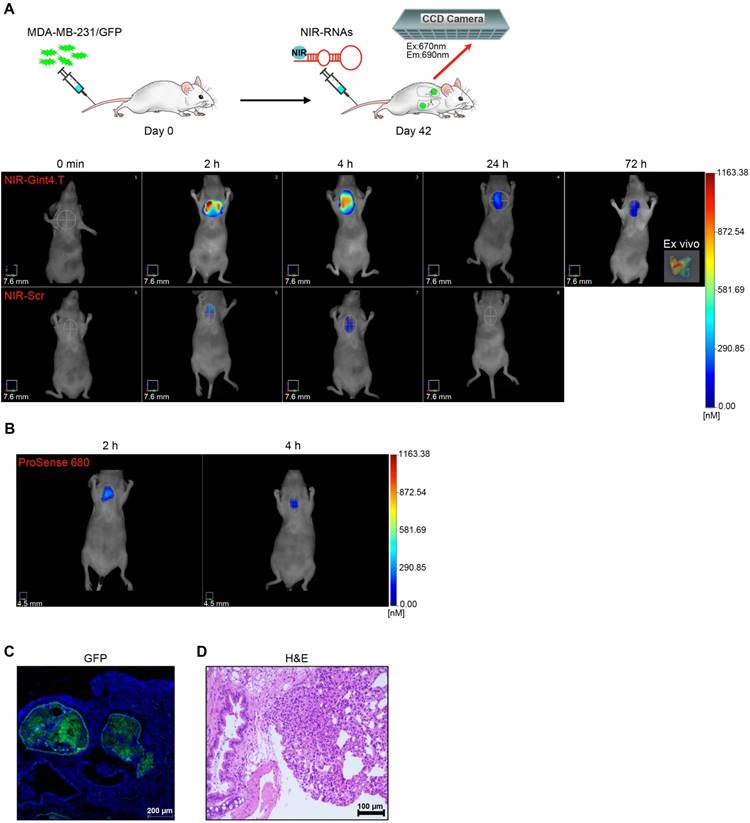
With the promising results obtained in the xenograft model, we extended the use of NIR-Gint4.T to the detection of TNBC metastases by using a lung metastases model established by tail vein injection of GFP-labeled MDA-MB-231 cells into athymic nude mice [47]. Forty-two days after cell injection, mice were imaged for metastases formation by FMT at different time points (starting from 2 h up to 72 h) after injection of NIR-Gint4.T or NIR-Scr. As shown in Figure 7A, there was high uptake of NIR-Gint4.T in metastatic lungs, as assessed by the strong fluorescence signal detected at 2 h, which then slowly declined but was still detectable at 72 h. Indeed, ex vivo imaging analysis showed a persistent signal in lungs dissected at 72 h (inset). As expected, no signal was revealed in metastatic lungs of mice injected with VivoTag-S 680 alone (Figure S9B) and, in addition, NIR-Gint4.T did not detect lungs of healthy mice (Figure S9C).
The presence of metastases was confirmed by the lung uptake of ProSense 680 (Figure 7B), a cathepsin-activatable fluorescent imaging agent commonly used to detect the metastatic process [48]. In addition, the metastases originating from GFP-labeled TNBC cells in the lungs of NIR-Gint4.T imaged mice were verified by confocal microscopy analysis (Figure 7C) and H&E staining (Figure 7D) of lung sections.
Effect of PDGFRβ aptamer on the inhibition of TNBC metastases
To assess whether Gint4.T aptamer was able not only to detect lung metastases upon NIR conjugation but also to inhibit metastases formation, GFP-labeled MDA-MB-231 cells were pre-treated with Gint4.T or Scr (400 pmol) prior to being injected into nude mice. Then, systemic i.v administration of the aptamers (1400 pmol) was continued for 28 days as schematized in Figure 8A. Two weeks after the last treatment (at day 42), metastases formation was analyzed by FMT using NIR-Gint4.T as the probe. Importantly, a significant reduction of NIR-Gint4.T amount (about 80%) was detected in the lungs of mice that received Gint4.T treatment compared to those treated with Scr (Figure 8A). This result indicates the ability of PDGFRβ aptamer to drastically inhibit metastases formation. In order to confirm the anti-metastatic function of Gint4.T, soon after imaging mice were sacrificed and lungs were explanted and analyzed for fluorescence and histological examination. Accordingly, we detected a strong reduction of both GFP-labeled (Figure 8B) and PDGFRβ-positive (Figure 8C) metastatic TNBC cells in the lung sections from Gint4.T-treated mice (bottom row) compared to the Scr group (top row).
Selective imaging of TNBC lung metastases by NIR-Gint4.T. (A) Forty-two days following i.v. injection of MDA-MB-231/GFP cells, nude mice were i.v. injected with 1 nmol of either NIR-Gint4.T or NIR-Scr and analyzed with FMT at the indicated times. Representative volume renderings taken at the same color gating from NIR-Gint4.T and NIR-Scr (n = 3)-injected mice are shown. Inset: Representative image of lung excised at 72 h post-injection of NIR-Gint4.T. (B) In vivo imaging of ProSense 680 in lungs. Representative sections of lungs excised at 72 h post-injection of NIR-Gint4.T were analyzed by (C) confocal microscopy (GFP) and (D) H&E staining as indicated. In (C), Nuclei were stained with DAPI. (C) Magnification 10 ×, scale bar = 200 μm. (D) Magnification 20 ×, scale bar = 100 μm. Em: emission; Ex: excitation; GFP: green fluorescent protein; H&E: hematoxylin and eosin; NIR: near-infrared.
PDGFRβ aptamer inhibits TNBC lung metastases. (A) Mice were injected with MDA-MB-231/GFP cells prior to being subjected to a short incubation with 400 pmol of either Gint4.T or Scr, and then i.v. injected (black arrows) with 1400 pmol aptamers every 24 h until day 3, after which, administration was reduced to three times for the second week and once for the following two weeks. At day 42, mice were analyzed with FMT at 2 h post-injection of 1 nmol NIR-Gint4.T. Right: Representative images of Scr (n = 4) and Gint4.T (n = 4)-treated mice are shown. Left: Quantification of the amount of in vivo NIR-Gint4.T-associated fluorescence (pmol) in the lungs. Bars depict mean ± SD. **P < 0.01 (n = 4). Representative sections of lungs from Gint4.T and Scr-treated mice after NIR-Gint4.T imaging analyzed by (B) confocal microscopy (GFP), (C) immunostaining with PDGFRβ antibodies and (D) H&E staining. (E) Metastatic foci in H&E lung sections (≥5 random fields per section, two sections per animal) were counted under magnification 10 ×. **P < 0.01. In (B), Nuclei were stained with DAPI. In (B) and (D), bottom row, the arrows identify cancer cells dispersed in the lung tissue. Representative sections of lungs from Scr-treated mice after NIR-Gint4.T imaging analyzed by immunostaining with (F) Ki-67, (G) MMP-9 and (H) vimentin antibodies. (B-D, F-H) Magnification 10 ×, scale bar = 200 μm. Inset: magnification 40 ×, scale bar = 50 μm. Em: emission; Ex: excitation; GFP: green fluorescent protein; H&E: hematoxylin and eosin; MMP-9: matrix metallopeptidase-9; NIR: near-infrared; PDGFRβ: platelet-derived growth factor receptor β.
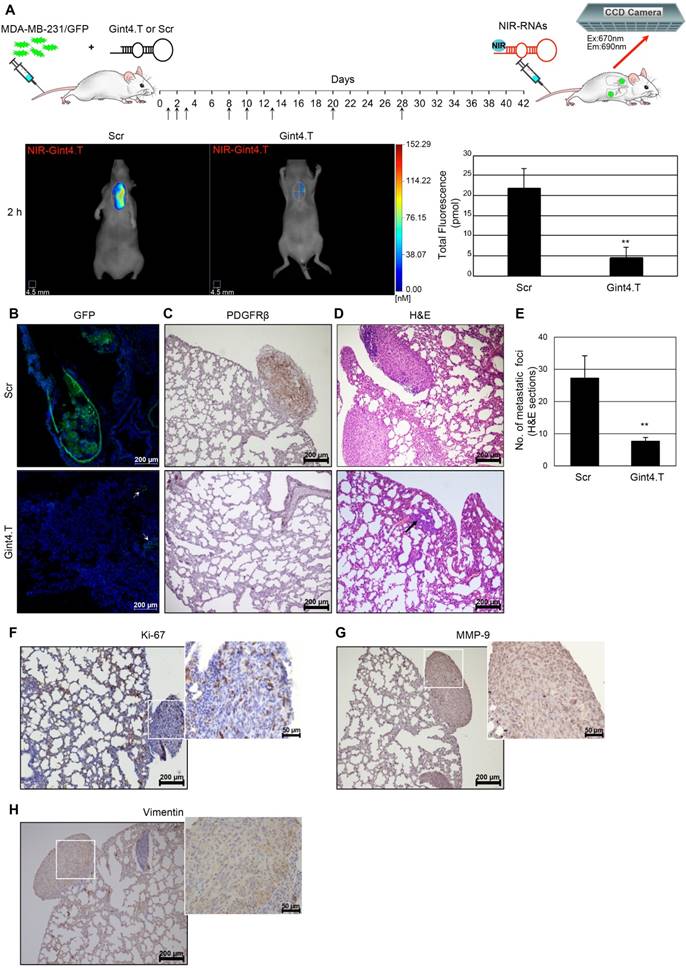
Moreover, H&E staining confirmed the dramatic reduction of the number of metastatic foci upon treatment with PDGFRβ aptamer (Figure 8D-E). Interestingly, large metastases with high proliferative potential (Figure 8F) that were positive for human MMP-9 (Figure 8G) and vimentin (Figure 8H) expression were present in the lungs from the control group and undetectable in those from the Gint4.T group, where just a few cancer cells dispersed in the lung tissue were present (Figure 8B, D, bottom row). Despite the fact that it is well-accepted that tail-vein injection of MDA-MB-231 cells leads to metastases in lungs [46], we also checked that the reduction of lung metastases by Gint4.T did not result in a relocation of metastases to the liver and axillary lymph nodes (Figure S10), two other common metastatic sites of MDA-MB-231 cells when orthotopically injected into the mouse mammary fat pad [49, 50].
Together, these data indicate NIR-Gint4.T as a specific probe for non-invasive detection of lung metastases reduction induced by Gint4.T treatment.
Discussion
TNBC usually affects younger patients and is characterized by tumors that are larger, of higher grade and biologically more aggressive than other breast cancers [51]. While the mortality from breast cancer has recently declined, management of TNBC is still challenging because of a lack of targeted therapy. It has been recognized that tumor-driving pathways significantly differ among TNBC subtypes; thus, deciphering the high heterogeneity of these tumors is imperative to treatment outcomes [52]. Unique to the mesenchymal TNBC subtype are patterns of gene expression associated with EMT and stemness [4, 53], features correlating with metastatic spread and chemoresistance [54]. Consistent with the unique tumor biology of each subtype are differences in tissue tropism of metastasis, with the M and LAR subtypes displaying a significantly higher frequency of lung and bone metastases, respectively, compared to the other subtypes [5]. This suggests that innovative and more personalized imaging approaches are needed for monitoring metastatic disease in newly diagnosed M and LAR patients [5].
In agreement with the role of PDGFRβ in GBM tumors, where the receptor acts as a critical mediator of the stem-like phenotype of these tumors [13], recent studies have suggested that the expression of PDGFRβ in breast cancer may be a unique feature of cancer cells that possess stem cell characteristics and/or that have undergone EMT [19, 55]. Moreover, it has been found that PDGFRβ signaling is involved in mediating the VM of MDA-MB-231 cells, a phenomenon strictly depending on their mesenchymal phenotype. Accordingly, expression of PDGFRβ marks tumor cells engaged in vascular lacunae [21].
A recent study reporting immunohistochemical analyses of a cohort of 550 human breast cancer samples including 30 TNBCs reported expression of PDGFRβ in the stroma and not in tumor cells [56], and suggested that the paracrine crosstalk existing between basal-like mammary cancer cells, expressing the PDGF-CC ligand, and cancer-associated fibroblasts, expressing both PDGFRβ and the cognate PDGFRα [57], could be the most prominent route of PDGFR-dependent signal transduction in breast cancer. Herein, by extending the analysis on a TMA comprising 200 cases of human TNBC, we present novel data showing that PDGFRβ is also expressed in tumor cells belonging to a restricted subgroup of tumors of the mesenchymal subtype expressing either MMP-9 or ALDH proteins that appears as a highly invasive and stem-like phenotype. Standing on the above protein and gene expression data, this study explored the role of PDGFRβ on TNBC lung metastases formation, thus providing the first report on the use of PDGFRβ-targeted aptamer as a selective tool for imaging and inhibition of metastases.
The Gint4.T aptamer is a nuclease-resistant 2′F-Py RNA (33-nucleotide) that we previously selected by a cell-internalization SELEX approach on human GBM cells [25]. It acts as a neutralizing ligand for PDGFRβ in GBM cell lines, primary cultures of GBMs and GBM implanted in mice, discriminating in vivo between PDGFRβ-positive tumors and non-target tumors [25, 31]. Further, given its exquisite targeting efficacy, the aptamer was used to deliver drug-loaded nanoparticles to intracranial GBM by crossing the blood-brain barrier [26]. Recently, according to the tumor stimulatory roles of PDGFRβ expressed by tumor stroma cells [58], we proved the ability of Gint4.T to modulate the interaction between TNBC and its microenvironment. Specifically, in an orthotopic TNBC mouse xenograft model of spontaneous breast metastases, by inhibiting the receptor expressed on BM-MSCs, the aptamer blocked the tropism of BM-MSCs to TNBC, thus counteracting BM-MSCs' role in enhancing multifocal lung metastatic lesions [12].
We now show that the anti-PDGFRβ aptamer strongly reduced the migration and invasion of mesenchymal TNBC cells in culture as well as their growth and acquisition of an invasive phenotype in a 3D culture environment. Importantly, we prove the use of a noninvasive approach, based on NIR fluorescence imaging technology for tracking the aptamer in preclinical mouse models of human TNBC and formation of lung metastases. The use of in vivo NIR fluorescence imaging for sensitive cancer early detection is highly desirable, because biological tissues show very low absorption and autofluorescence in the NIR spectrum window [59]. In comparison with other imaging techniques, NIR fluorescence technology does not require the use of ionizing radiation or radioactive materials and is low-cost, safe and easy to use. Progress in NIR fluorescence for cancer imaging has rapidly increased in recent years thanks to newly developed NIR dyes and optical imaging instruments [60-62]. Although nowadays the main application of NIR probes is to facilitate the identification and resection of malignant lesions during surgery, many studies are attempting to use these molecules to early detect tumor response to treatment as well as early metastasis occurrence upon conjugation to molecular targeted agents [63-66]. In this respect, the highly specific and sensitive PDGFRβ-targeted aptamer represents an ideal candidate for NIR conjugation, thus improving the selectivity of non-targeting NIR probes towards the target sites in living systems. NIR-labeled Gint4.T systemically i.v. administrated to TNBC-bearing mice revealed high target specificity (as assessed by competitive studies with unlabeled aptamer and by injection in mice bearing PDGFRβ-negative BT-474 tumors), rapid tumor uptake (~2% injected dose at 15 min) and durable tumor retention (at least 24 h). Further, it efficiently detected metastatic lungs in a well-established model for lung metastases formation obtained through the tail vein injection of MDA-MB-231 cells into nude mice [47]. Remarkably, Gint4.T PDGFRβ aptamer at a low therapeutic dose (eight treatments at ~0.74 mg/kg) suppressed the formation of lung metastases, and NIR-labeled Gint4.T was able to image the metastatic suppression in mice. Ex vivo fluorescence and histological examination confirmed the in vivo results showing that large metastases, which were evident in control groups, were undetectable in the lung tissue from Gint4.T-treated mice. It is likely that this dramatic anti-metastatic effect of the aptamer may be due both to an impaired cell arrest and invasion and a reduced proliferation of TNBC cells in the metastatic site, taking into account the anti-proliferative effect observed in vitro.
To date, only one antibody (Olaratumab) targeting PDGFRα has entered the clinic for the treatment of certain solid tumors [67], while no anti-PDGFRβ antibody [68-70] has been translated into clinical use.
Importantly, the properties of Gint4.T aptamer qualify this molecule as a very promising agent for imaging and therapy of PDGFRβ-positive TNBC. The aptamer exhibits high tissue penetration due to its small size and does not elicit immune stimulation upon administration to immune-competent mice [25]. Further, the potential clinical impact of Gint4.T is strongly highlighted by the undoubted advantages of aptamers. Indeed, because of their nucleic acid nature, the rational design of advanced strategies to manipulate aptamers for biosensing, bioimaging, and targeted cancer therapy is greatly simplified over antibodies [71, 72]. Such advantages include: 1) easier manufacturing, greater reproducibility, lower costs of aptamers compared to protein antibodies; 2) structural stability; 3) ready chemical modifications for overcoming nuclease susceptibility and rapid excretion through renal filtration [24]. In addition, because of its ability to rapidly internalize into target cancer cells, Gint4.T aptamer appears as a prime candidate tool for delivering existing or novel drugs (or molecular imaging probes) specifically to the diseased site in TNBC and other PDGFRβ-positive human cancers [26]. Recently, aptamers have been proven superior to antibodies in cancer theranostics due to their better tumor penetration and longer retention in tumor sites [73]. Future investigation will explore whether Gint4.T aptamer also displays superior performance over antibody as a therapeutic/imaging agent.
Overall, the work described herein is important since we provide new insights into the role of PDGFRβ in driving TNBC cell invasiveness and suggest a novel combination of NIR fluorescence imaging and RNA aptamer targeting to human PDGFRβ for recognition and inhibition of TNBC metastases in lung. These findings may significantly expand the current repertoire of strategies used to manage patients with mesenchymal aggressive TNBC.
Conclusions
Previous genomic studies have advanced the understanding of biologic heterogeneity of TNBC. Nevertheless, actionable biomarkers for subtype stratification and treatment of TNBC patients still need to be identified. Here, we investigated 200 TNBC samples using protein expression and found that PDGFRβ expression, previously reported only on endothelial and tumor-associated stromal cells, marks tumor cells belonging to a subgroup of mesenchymal TNBC with stem-like and invasive phenotype. Importantly, we prove that aptamer-mediated PDGFRβ inhibition combined to molecular fluorescence imaging using the NIR dye-conjugated version of the aptamer could be a very effective approach for non-invasive imaging and suppression of TNBC lung metastases.
Abbreviations
ALDH1A1: aldehyde dehydrogenase 1‑A1; BLIA: basal-like immune activated; BLIS: basal-like immune-suppressed; BM-MSCs: bone marrow mesenchymal stem cells; DAPI: 4′,6-Diamidino-2-phenylindole; DPBS: Dulbecco's phosphate-buffered saline; EC: extracellular domain; Em: emission; EMT: epithelial-to-mesenchymal transition; ENG: endoglin; ESI: electron spray ionization; Ex: excitation; FBS: fetal bovine serum; 2′F-Py: 2′ fluoro-pyrimidines; FMT: fluorescence molecular tomography; GBM: glioblastoma; GFP: green fluorescent protein; H&E: hematoxylin and eosin; HER2: human epidermal growth factor receptor 2; i.v.: intravenously; Kd: dissociation constant; LAR: luminal-androgen receptor; LNM: metastatic lymph nodes; MES: mesenchymal; MFI: mean fluorescence intensity; ML: mesenchymal-like; MMP-2: matrix metallopeptidase-2; MMP-9: matrix metallopeptidase-9; MS: mass spectrometry; NIR: near-infrared; PAGE: polyacrylamide gel electrophoresis; PDGFRβ: platelet-derived growth factor receptor β; PDGF-BB: platelet derived growth factor-BB; PK: pharmacokinetic; RT-qPCR: reverse transcription quantitative polymerase chain reaction; RT: room temperature; s.c.: subcutaneously; THY1: Thy-1 cell surface antigen; TMA: tissue micro array; TNBC: triple-negative breast cancer; TOF: time-of-flight; 3D: three-dimensional; 2D: two-dimensional; VE: vascular endothelial; UV-Vis: ultraviolet-visible; VM: vasculogenic mimicry; VOI: volume-of-interest.
Acknowledgements
This work was supported by Associazione Italiana per la Ricerca sul Cancro (AIRC, IG18753) to L.C.
B.S.H. was supported by INCIPIT (EUH2020-MSCA). The authors wish to acknowledge Giovanni N. Roviello for help with the mass spectrometry analysis of NIR-RNAs, Silvia Esposito for technical assistance with some in vivo experiments, and Francesco Leonetti and Margherita Passariello who kindly assisted in some experimental procedures.
Contributions
S.C. performed the majority of the experiments, interpreted results and assisted with manuscript preparation. B.S.H. and S.G. performed and/or assisted with the animal experiments. F.C. and M.C. performed TMA analysis and contributed to interpreting the results with the human TNBC samples. M.D.B. evaluated tumor morphology and immunohistochemical staining. M.N. contributed to FACS analysis. G.B. provided the human TNBC samples. M.F. performed in silico analysis and contributed to critical data analysis and writing of the manuscript. A.Z. performed and contributed to interpreting the animal experiments and assisted with manuscript preparation. L.C. conceived and designed the study, analyzed the data and coordinated the research, was responsible for funding and wrote the manuscript.
Supplementary Material
Supplementary figures and tables.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Newman LA, Reis-Filho JS, Morrow M, Carey LA, King TA. The 2014 society of surgical oncology Susan G. Komen for the cure symposium: triple-negative breast cancer. Ann Surg Oncol. 2015;22:874-82
2. Gadi VK, Davidson NE. Practical approach to triple-negative breast cancer. J Oncol Pract. 2017;13:293-300
3. Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA. et al. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res. 2007;13:4429-34
4. Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y. et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121:2750-67
5. Lehmann BD, Jovanović B, Chen X, Estrada MV, Johnson KN, Shyr Y. et al. Refinement of triple-negative breast cancer molecular subtypes: implications for neoadjuvant chemotherapy selection. PLoS One. 2016;11:e0157368
6. Burstein MD, Tsimelzon A, Poage GM, Covington KR, Contreras A, Fuqua SA. et al. Comprehensive genomic analysis identifies novel subtypes and targets of triple-negative breast cancer. Clin Cancer Res. 2015;21:1688-98
7. Betsholtz C. Insight into the physiological functions of PDGF through genetic studies in mice. Cytokine Growth Factor Rev. 2004;15:215-28
8. Fredriksson L, Li H, Eriksson U. The PDGF family: four gene products form five dimeric isoforms. Cytokine Growth Factor Rev. 2004;15:197-204
9. Plate KH, Breier G, Farrell CL, Risau W. Platelet-derived growth factor receptor-beta is induced during tumor development and upregulated during tumor progression in endothelial cells in human gliomas. Lab Invest. 1992;67:529-34
10. Bhowmick NA, Neilson EG, Moses HL. Stromal fibroblasts in cancer initiation and progression. Nature. 2004;432:332-7
11. Catena R, Luis-Ravelo D, Antón I, Zandueta C, Salazar-Colocho P, Larzábal L. et al. PDGFR signaling blockade in marrow stroma impairs lung cancer bone metastasis. Cancer Res. 2011;71:164-74
12. Camorani S, Hill BS, Fontanella R, Greco A, Gramanzini M, Auletta L. et al. Inhibition of bone marrow-derived mesenchymal stem cells homing towards triple-negative breast cancer microenvironment using an anti-PDGFRβ aptamer. Theranostics. 2017;7:3595-607
13. Kim Y, Kim E, Wu Q, Guryanova O, Hitomi M, Lathia JD. et al. Platelet-derived growth factor receptors differentially inform intertumoral and intratumoral heterogeneity. Genes Dev. 2012;26:1247-62
14. Akhavan D, Pourzia AL, Nourian AA, Williams KJ, Nathanson D, Babic I. et al. De-repression of PDGFRβ transcription promotes acquired resistance to EGFR tyrosine kinase inhibitors in glioblastoma patients. Cancer Discov. 2013;3:534-47
15. Fujino S, Miyoshi N, Ohue M, Takahashi Y, Yasui M, Hata T. et al. Platelet-derived growth factor receptor-β gene expression relates to recurrence in colorectal cancer. Oncol Rep. 2018;39:2178-84
16. Zhang H, Sun JD, Yan LJ, Zhao XP. PDGF-D/PDGFRβ promotes tongue squamous carcinoma cell (TSCC) progression via activating p38/AKT/ERK/EMT signal pathway. Biochem Biophys Res Commun. 2016;478:845-51
17. Wang F, Remke M, Bhat K, Wong ET, Zhou S, Ramaswamy V. et al. A microRNA-1280/JAG2 network comprises a novel biological target in high-risk medulloblastoma. Oncotarget. 2015;6:2709-24
18. Cortez E, Gladh H, Braun S, Bocci M, Cordero E, Björkström NK. et al. Functional malignant cell heterogeneity in pancreatic neuroendocrine tumors revealed by targeting of PDGF-DD. Proc Natl Acad Sci U S A. 2016;113:E864-73
19. Meng F, Speyer CL, Zhang B, Zhao Y, Chen W, Gorski DH. et al. PDGFRα and β play critical roles in mediating foxq1-driven breast cancer stemness and chemoresistance. Cancer Res. 2015;75:584-93
20. Kuzmanov A, Hopfer U, Marti P, Meyer-Schaller N, Yilmaz M, Christofori G. LIM-homeobox gene 2 promotes tumor growth and metastasis by inducing autocrine and paracrine PDGF-B signaling. Mol Oncol. 2014;8:401-16
21. D'Ippolito E, Plantamura I, Bongiovanni L, Casalini P, Baroni S, Piovan C. et al. miR-9 and miR-200 regulate PDGFRβ-mediated endothelial differentiation of tumor cells in triple-negative breast cancer. Cancer Res. 2016;76:5562-72
22. Cristofanilli M, Morandi P, Krishnamurthy S, Reuben JM, Lee BN, Francis D. et al. Imatinib mesylate (Gleevec) in advanced breast cancer-expressing c-kit or PDGFR-beta: clinical activity and biological correlations. Ann Oncol. 2008;19:1713-9
23. Curigliano G, Pivot X, Cortés J, Elias A, Cesari R, Khosravan R. et al. Randomized phase II study of sunitinib versus standard of care for patients with previously treated advanced triple-negative breast cancer. Breast. 2013;22:650-6
24. Camorani S, Crescenzi E, Fedele M, Cerchia L. Oligonucleotide aptamers against tyrosine kinase receptors: prospect for anticancer applications. Biochim Biophys Acta. 2018;1869:263-77
25. Camorani S, Esposito CL, Rienzo A, Catuogno S, Iaboni M, Condorelli G. et al. Inhibition of receptor signaling and of glioblastoma-derived tumor growth by a novel PDGFRβ aptamer. Mol Ther. 2014;22:828-41
26. Monaco I, Camorani S, Colecchia D, Locatelli E, Calandro P, Oudin A. et al. Aptamer functionalization of nanosystems for glioblastoma targeting through the blood-brain barrier. J Med Chem. 2017;60:4510-6
27. Collina F, Cerrone M, Peluso V, Laurentiis MD, Caputo R, Cecio RD. et al. Downregulation of androgen receptor is strongly associated with diabetes in triple negative breast cancer patients. Am J Transl Res. 2016;8:3530-9
28. R2: genomics analysis and visualization platform. http://r2.amc.nl
29. Guadagno E, Vitiello M, Francesca P, Calì G, Caponnetto F, Cesselli D. et al. PATZ1 is a new prognostic marker of glioblastoma associated with the stem-like phenotype and enriched in the proneural subtype. Oncotarget. 2017;8:59282-300
30. Camorani S, Crescenzi E, Gramanzini M, Fedele M, Zannetti A, Cerchia L. Aptamer-mediated impairment of EGFR-integrin αvβ3 complex inhibits vasculogenic mimicry and growth of triple-negative breast cancers. Sci Rep. 2017;7:46659
31. Camorani S, Crescenzi E, Colecchia D, Carpentieri A, Amoresano A, Fedele M. et al. Aptamer targeting EGFRvIII mutant hampers its constitutive autophosphorylation and affects migration, invasion and proliferation of glioblastoma cells. Oncotarget. 2015;6:37570-87
32. Zannetti A, Iommelli F, Fonti R, Papaccioli A, Sommella J, Lettieri A. et al. Gefitinib induction of in vivo detectable signals by Bcl-2/Bcl-xL modulation of inositol trisphosphate receptor type 3. Clin Cancer Res. 2008;14:5209-19
33. Zannetti A, Iommelli F, Speranza A, Salvatore M, Del Vecchio S. 3'-deoxy-3'-18F-fluorothymidine PET/CT to guide therapy with epidermal growth factor receptor antagonists and Bcl-xL inhibitors in non-small cell lung cancer. J Nucl Med. 2012;53:443-50
34. Zeng Q, Li W, Lu D, Wu Z, Duan H, Luo Y. et al. CD146, an epithelial-mesenchymal transition inducer, is associated with triple-negative breast cancer. Proc Natl Acad Sci U S A. 2012;109:1127-32
35. Pierobon M, Ramos C, Wong S, Hodge KA, Aldrich J, Byron S. et al. Enrichment of PI3K-AKT-mTOR pathway activation in hepatic metastases from breast cancer. Clin Cancer Res. 2017;23:4919-28
36. Le Rhun E, Bertrand N, Dumont A, Tresch E, Le Deley MC, Mailliez A. et al. Identification of single nucleotide polymorphisms of the PI3K-AKT-mTOR pathway as a risk factor of central nervous system metastasis in metastatic breast cancer. Eur J Cancer. 2017;87:189-98
37. Kenny PA, Lee GY, Myers CA, Neve RM, Semeiks JR, Spellman PT. et al. The morphologies of breast cancer cell lines in three-dimensional assays correlate with their profiles of gene expression. Mol Oncol. 2007;1:84-96
38. Ramos-DeSimone N, Hahn-Dantona E, Sipley J, Nagase H, French DL, Quigley JP. Activation of matrix metalloproteinase-9 (MMP-9) via a converging plasmin/stromelysin-1 cascade enhances tumor cell invasion. J Biol Chem. 1999;274:13066-76
39. Balduyck M, Zerimech F, Gouyer V, Lemaire R, Hemon B, Grard G. et al. Specific expression of matrix metalloproteinases 1, 3, 9 and 13 associated with invasiveness of breast cancer cells in vitro. Clin Exp Metastasis. 2000;18:171-8
40. Barrallo-Gimeno A, Nieto MA. The Snail genes as inducers of cell movement and survival: implications in development and cancer. Development. 2005;132:3151-61
41. Yoshio T, Morita T, Kimura Y, Tsujii M, Hayashi N, Sobue K. Caldesmon suppresses cancer cell invasion by regulating podosome/invadopodium formation. FEBS Lett. 2007;581:3777-82
42. Wagenblast E, Soto M, Gutiérrez-Ángel S, Hartl CA, Gable AL, Maceli AR. et al. A model of breast cancer heterogeneity reveals vascular mimicry as a driver of metastasis. Nature. 2015;520:358-62
43. Plantamura I, Casalini P, Dugnani E, Sasso M, D'Ippolito E, Tortoreto M. et al. PDGFRβ and FGFR2 mediate endothelial cell differentiation capability of triple negative breast carcinoma cells. Mol Oncol. 2014;8:968-81
44. Gong C, Qu S, Lv XB, Liu B, Tan W, Nie Y. et al. BRMS1L suppresses breast cancer metastasis by inducing epigenetic silence of FZD10. Nat Commun. 2014;5:5406
45. Keefe AD, Pai S, Ellington A. Aptamers as therapeutics. Nat Rev Drug Discov. 2010;9:537-50
46. Morita Y, Leslie M, Kameyama H, Volk DE, Tanaka T. Aptamer therapeutics in cancer: current and future. Cancers (Basel). 2018;10:80
47. Zhou Y, Shao G, Liu S. Monitoring breast tumor lung metastasis by U-SPECT-II/CT with an integrin α(v)β(3)-targeted radiotracer( 99m)Tc-3P-RGD(2). Theranostics. 2012;2:577-88
48. Kirsch DG, Dinulescu DM, Miller JB, Grimm J, Santiago PM, Young NP. et al. A spatially and temporally restricted mouse model of soft tissue sarcoma. Nat Med. 2007;13:992-7
49. Iorns E, Drews-Elger K, Ward TM, Dean S, Clarke J, Berry D. et al. A new mouse model for the study of human breast cancer metastasis. PLoS One. 2012;7:e47995
50. Kim IS, Baek SH. Mouse models for breast cancer metastasis. Biochem Biophys Res Commun. 2010;394:443-7
51. Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2010;363:1938-48
52. Mayer IA, Abramson VG, Lehmann BD, Pietenpol JA. New strategies for triple-negative breast cancer-deciphering the heterogeneity. Clin Cancer Res. 2014;20:782-90
53. Masuda H, Baggerly KA, Wang Y, Zhang Y, Gonzalez-Angulo AM, Meric-Bernstam F. et al. Differential response to neoadjuvant chemotherapy among 7 triple-negative breast cancer molecular subtypes. Clin Cancer Res. 2013;19:5533-40
54. Fedele M, Cerchia L, Chiappetta G. The epithelial-to-mesenchymal transition in breast cancer: focus on basal-like carcinomas. Cancers (Basel). 2017;9:134
55. Tam WL, Lu H, Buikhuisen J, Soh BS, Lim E, Reinhardt F. et al. Protein kinase C α is a central signaling node and therapeutic target for breast cancer stem cells. Cancer Cell. 2013;24:347-64
56. Jansson S, Aaltonen K, Bendahl PO, Falck AK, Karlsson M, Pietras K. et al. The PDGF pathway in breast cancer is linked to tumour aggressiveness, triple-negative subtype and early recurrence. Breast Cancer Res Treat. 2018;169:231-41
57. Roswall P, Bocci M, Bartoschek M, Li H, Kristiansen G, Jansson S. et al. Microenvironmental control of breast cancer subtype elicited through paracrine platelet-derived growth factor-CC signaling. Nat Med. 2018;24:463-73
58. Östman A. PDGF receptors in tumor stroma: biological effects and associations with prognosis and response to treatment. Adv Drug Deliv Rev. 2017;121:117-23
59. He X, Wang K, Cheng Z. In vivo near-infrared fluorescence imaging of cancer with nanoparticle-based probes. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2010;2:349-66
60. Luo S, Zhang E, Su Y, Cheng T, Shi C. A review of NIR dyes in cancer targeting and imaging. Biomaterials. 2011;32:7127-38
61. Hong G, Antaris AL, Dai H. Near-infrared fluorophores for biomedical imaging. Nat Biomed Eng. 2017;1:10
62. Comegna D, Zannetti A, Del Gatto A, de Paola I, Russo L, Di Gaetano S. et al. Chemical modification for proteolytic stabilization of the selective αvβ3 integrin RGDechi peptide: in vitro and in vivo activities on malignant melanoma cells. J Med Chem. 2017;60:9874-84
63. Santagata S, Portella L, Napolitano M, Greco A, D'Alterio C, Barone MV. et al. A novel CXCR4-targeted near-infrared (NIR) fluorescent probe (Peptide R-NIR750) specifically detects CXCR4 expressing tumors. Sci Rep. 2017;7:2554
64. Gunther JE, Lim EA, Kim HK, Flexman M, Altoé M, Campbell JA. et al. Dynamic diffuse optical tomography for monitoring neoadjuvant chemotherapy in patients with breast cancer. Radiology. 2018;287:778-86
65. Lim W, Sohn H, Ko Y, Park M, Kim B, Jo D. et al. Real-time in vivo imaging of metastatic bone tumors with a targeted near-infrared fluorophore. Oncotarget. 2017;8:65770-7
66. Zhu Q, DeFusco PA, Ricci A Jr, Cronin EB, Hegde PU, Kane M. et al. Breast cancer: assessing response to neoadjuvant chemotherapy by using US-guided near-infrared tomography. Radiology. 2013;266:433-42
67. Rodems TS, Iida M, Brand TM, Pearson HE, Orbuch RA, Flanigan BG. et al. Adaptive responses to antibody based therapy. Semin Cell Dev Biol. 2016;50:153-63
68. Shen J, Vil MD, Prewett M, Damoci C, Zhang H, Li H. et al. Development of a fully human anti-PDGFRbeta antibody that suppresses growth of human tumor xenografts and enhances antitumor activity of an anti-VEGFR2 antibody. Neoplasia. 2009;11:594-604
69. Sano H, Ueda Y, Takakura N, Takemura G, Doi T, Kataoka H. et al. Blockade of platelet-derived growth factor receptor-beta pathway induces apoptosis of vascular endothelial cells and disrupts glomerular capillary formation in neonatal mice. Am J Pathol. 2002;161:135-43
70. Jayson GC, Parker GJ, Mullamitha S, Valle JW, Saunders M, Broughton L. et al. Blockade of platelet-derived growth factor receptor-beta by CDP860, a humanized, PEGylated di-Fab', leads to fluid accumulation and is associated with increased tumor vascularized volume. J Clin Oncol. 2005;23:973-81
71. Meng HM, Liu H, Kuai H, Peng R, Mo L, Zhang XB. Aptamer-integrated DNA nanostructures for biosensing, bioimaging and cancer therapy. Chem Soc Rev. 2016;45:2583-602
72. Liang H, Zhang XB, Lv Y, Gong L, Wang R, Zhu X, Yang R, Tan W. Functional DNA-containing nanomaterials: cellular applications in biosensing, imaging, and targeted therapy. Acc Chem Res. 2014;47:1891-901
73. Xiang D, Zheng C, Zhou SF, Qiao S, Tran PH, Pu C. et al. Superior performance of aptamer in tumor penetration over antibody: implication of aptamer-based theranostics in solid tumors. Theranostics. 2015;5:1083-97
Author contact
![]() Corresponding author: Laura Cerchia, Istituto per l'Endocrinologia e l'Oncologia Sperimentale "G. Salvatore", CNR, Via S. Pansini 5, 80131 Naples, Italy; Tel.: +390815455751; E-mail: cerchiait; Antonella Zannetti, Istituto di Biostrutture e Bioimmagini, CNR, Via T. De Amicis 95, 80145 Naples, Italy; Tel.: +390812203431; E-mail: antonella.zannettiit.
Corresponding author: Laura Cerchia, Istituto per l'Endocrinologia e l'Oncologia Sperimentale "G. Salvatore", CNR, Via S. Pansini 5, 80131 Naples, Italy; Tel.: +390815455751; E-mail: cerchiait; Antonella Zannetti, Istituto di Biostrutture e Bioimmagini, CNR, Via T. De Amicis 95, 80145 Naples, Italy; Tel.: +390812203431; E-mail: antonella.zannettiit.
 Global reach, higher impact
Global reach, higher impact