13.3
Impact Factor
Theranostics 2018; 8(18):5143-5158. doi:10.7150/thno.27310 This issue Cite
Research Paper
Overexpression of HOXC10 promotes angiogenesis in human glioma via interaction with PRMT5 and upregulation of VEGFA expression
1. Program of cancer research, Affiliated Guangzhou Women and Children's Hospital, Department of Biochemistry, Zhongshan School of Medicine, Sun Yat-sen University, China;
2. Guangdong Province Key Laboratory of Brain Function and Disease, Department of Biochemistry, Zhongshan School of Medicine, Sun Yat-sen University, Guangzhou, China;
3. Department of Neurosurgery, The First Affiliated Hospital of Sun Yat-sen University, Guangzhou, Guangdong, China;
4. Department of Neurosurgery, the First Affiliated Hospital of Jinan University, Guangzhou, Guangdong, China;
5. Neurosurgical Research Institute, the First Affiliated Hospital of Guangdong Pharmaceutics University, Guangzhou, Guangdong, China;
6. Clinical Experimental Center, Department of Pathology (Clinical Biobanks), Jiangmen Central Hospital, Affiliated Jiangmen Hospital of Sun Yat-sen University, Jiangmen, Guangdong, China;
7. Department of Endocrinology and Diabetes Center, The First Affiliated Hospital of Sun Yat-sen University, Guangzhou, Guangdong, China;
8. Department of Pharmacology, Zhongshan School of Medicine, Sun Yat-Sen University, Guangzhou, Guangdong, China.
*These authors contributed equally to this work.
Received 2018-5-17; Accepted 2018-9-19; Published 2018-10-6
Abstract
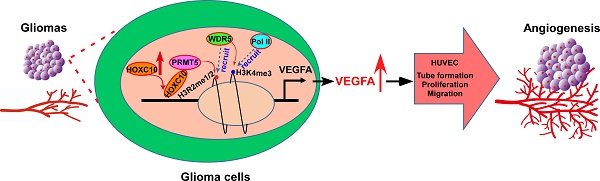
High levels of angiogenesis are associated with poor prognosis in patients with gliomas. However, the molecular mechanisms underlying tumor angiogenesis remain unclear.
Methods: The effect of homeobox C10 (HOXC10) on tube formation, migration, and proliferation of human umbilical vein endothelial cells (HUVECs) and on chicken chorioallantoic membranes (CAMs) was examined. An animal xenograft model was used to examine the effect of HOXC10 on xenograft angiogenesis or the effect of bevacizumab, a monoclonal antibody against vascular endothelial growth factor A (VEGFA), on HOXC10-overexpressing xenografts. A chromatin immunoprecipitation assay was applied to investigate the mechanism in which HOXC10 regulated VEGFA expression.
Results: Overexpressing HOXC10 enhanced the capacity of glioma cells to induce tube formation, migration and proliferation of HUVECs, and neovascularization in CAMs, while silencing HOXC10 had the opposite result. We observed that CD31 staining was significantly increased in tumors formed by HOXC10-overexpressing U251MG cells but reduced in HOXC10-silenced tumors. Mechanistically, HOXC10 could transcriptionally upregulate VEGFA expression by binding to its promoter. Strikingly, treatment with bevacizumab, a monoclonal antibody against VEGFA, significantly inhibited the growth of HOXC10-overexpressing tumors and efficiently impaired angiogenesis. Protein arginine methyltransferase 5 (PRMT5) and WD repeat domain 5 (WDR5), both of which regulate histone post-translational modifications, were required for HOXC10-mediated VEGFA upregulation. Importantly, a significant correlation between HOXC10 levels and VEGFA expression was observed in a cohort of human gliomas.
Conclusions: This study suggests that HOXC10 induces glioma angiogenesis by transcriptionally upregulating VEGFA expression, and may represent a potential target for antiangiogenic therapy in gliomas.
Keywords: HOXC10, glioma, angiogenesis, VEGFA, bevacizumab
Introduction
Gliomas represent the most common primary tumor of the central nervous system [1], accounting for 30% of all brain and central nervous system tumors, and 80% of all malignant brain tumors [2, 3]. Despite advances in surgical techniques and therapeutic strategies over the last two decades, the outcomes of patients harboring malignant gliomas remain very poor, with a cumulative one-year survival rate below 30% [4, 5]. Glioblastoma multiforme (GBM), a grade IV glioma, is the most aggressive type of primary brain tumor [6], with an average survival time following diagnosis of 12 to 15 months [5]. This poor prognosis is largely attributable to aberrant angiogenesis, high invasiveness and therapeutic resistance [1, 7]. Uncontrolled proliferation of glioma cells requires angiogenesis; among different types of cancers, glioma displays the highest degree of vascularization [1, 8, 9]. Increased tumor microvascular density (MVD) has been identified as a hallmark in gliomas with a poor patient prognosis [10, 11]. Therefore, a better understanding of the molecular mechanisms underlying glioma angiogenesis could contribute to the identification of novel therapeutic targets to treat glioma and eventually improve prognosis.
Angiogenesis, which refers to the formation of new blood vessels from pre-existing vasculature, occurs in many physiological and pathological processes, such as development, wound healing, and cancer. Abnormal angiogenesis is necessary for the growth, survival, and metastasis of most solid tumors, and involves the coordinated roles of many bioactive molecules [12]. For instance, an imbalance between pro-angiogenic factors, such as the vascular endothelial growth factor (VEGF) family, and inhibitors of angiogenesis, including endostatin, angiostatin, and other related molecules, results in abnormal angiogenesis [13]. The VEGF family proteins, which include VEGFA, VEGFB, VEGFC, VEGFD, and placental growth factor, are the best-characterized regulators of angiogenesis and metastatic growth in human carcinogenesis [14-16]. However, the precise molecular mechanisms underlying tumor angiogenesis are not completely understood.
There are 39 class I homeobox (HOX) genes in the human genome and they are distributed in four different clusters (A, B, C, and D), located on four chromosomes (2, 7, 12, and 17). HOX genes are evolutionarily conserved genes that make important contributions to cell differentiation and embryonic development [17-19]. Previous studies reported that HOX genes have various influences on tumor cell proliferation, apoptosis, stem cell renewal, differentiation, motility, and angiogenesis [19-22]. HOXC10, a member of HOX gene family, encodes a transcription factor containing a conserved DNA binding homeodomain and is highly expressed in many cancers [23-25]. Feng et al. reported that HOXC10 expression is appreciably increased in human thyroid cancer tissues compared to that in normal human thyroid tissues [24]. Zhai and colleagues reported that elevated HOXC10 is associated with increased invasiveness of cervical cancer-derived cell lines and that knockdown of HOXC10 markedly impaired cervical cancer cell invasiveness [23]. Moreover, HOXC10 expression is dramatically elevated in lung adenocarcinoma compared to normal lung tissues and upregulation of HOXC10 correlated with unfavorable prognosis in patients with lung cancer [25]. Moreover, upregulated HOXC10 expression promotes proliferation, migration, and invasion in gliomas[26]. However, the precise biological role of HOXC10 in glioma angiogenesis remains largely undefined.
In the present study, we reported that HOXC10 was markedly overexpressed in glioma cells and clinical samples compared to primary normal human astrocytes (NHAs) and normal human brain tissues, respectively. Overexpression of HOXC10 significantly promoted, whereas knockdown of HOXC10 impaired, glioma angiogenesis in vitro and in vivo. Furthermore, we demonstrated that HOXC10 upregulated VEGFA expression via directly binding to the VEGFA promoter, eventually promoting angiogenesis in glioma. Therefore, our results establish a vital role for HOXC10 in regulating glioma angiogenesis, suggesting that HOXC10 might serve as a potential target for glioma therapy.
Methods
Cell culture and reagents
Primary NHAs were purchased from Sciencell Research Laboratories (Carlsbad, CA, USA). Glioma cell lines A-172, U87, LN229, and LN-18, were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA) and U251MG cells were obtained from ECACC (Salisbury, UK). All cells were cultured under the conditions recommended by the manufacturer. Cell lines were authenticated using STR method. Bevacizumab (avastin) was purchased from Genentech (San Francisco, CA, USA). OICR-9429 was purchased from Selleck (Houston, TX, USA).
Patient information and tissue specimens
A total of 94 paraffin-embedded and archived glioma samples, including pilocytic astrocytoma, diffuse fibrillary astrocytoma, anaplastic astrocytomas and glioblastoma multiforme, which were histopathologically and clinically diagnosed at the First Affiliated Hospital of Sun Yat-sen University, from January 2008 to December 2014, were examined in this study. Prior informed patient consent and approval from the Institutional Research Ethics Committee were obtained for the use of these clinical materials for research purposes.
Vectors, retroviral infection, and transfection
The human HOXC10 gene and its truncations were amplified from cDNA using PCR and cloned into the pSin-EF2-puro retroviral vector. HOXC10-targeting short hairpin RNA (shRNA) oligonucleotides sequences were cloned into a pSuper-retro-puro vector to generate pSuper-retro-HOXC10-RNAi(s). Transfection of plasmids or siRNAs was conducted using Lipofectamine 3000 reagent (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer's instructions. Stable cell lines expressing HOXC10 or HOXC10 shRNA were selected using 0.5 μg/mL puromycin for 10 days from 48 h after infection.
Western blotting analysis
Western blotting (WB) was performed as previously described [27], using anti-HOXC10, anti-Flag, anti-protein arginine methyltransferase 5 (PRMT5), anti-glutathione-s-transferase (GST), anti-hemagluttinin (HA), anti-VEGFA and anti-WD repeat domain 5 (WDR5) antibodies (Abcam, Cambridge, MA, USA). Membranes were stripped and re-probed with an anti-α-tubulin antibody (Sigma-Aldrich, St Louis, MO, USA) as a protein loading control.
Human umbilical vein endothelial cells (HUVECs) tubule formation assay
Matrigel (200 µL; BD Biosciences, Mountainview, CA, USA) was added into wells of a 24-well plate and polymerized for 30 min at 37 °C. HUVECs (2×104) in 200 µL of conditioned medium were added to each well and incubated at 37 °C in 5% CO2 for 20 h. Images were captured under bright-field illumination using a Zeiss Axio Imager A1 microscope (Carl Zeiss, Jena, Germany) at 100× final magnification. The degree of tube formation in the current study was verified as compared to the formation of cell strings.
Chicken chorioallantoic membrane (CAM) assay
To evaluate the direct effect on angiogenesis, a CAM assay was performed at the 8th day of development of fertilized chicken eggs, as previously described [28]. A 1-cm diameter window was opened in the shell of each egg with an 8-day-old chicken embryo (Yueqin Breeding Co. Ltd, Guangdong, China). The surface of the dermic sheet on the floor of the air sac was removed to expose the CAM. A 0.5 cm diameter filter paper was first placed on top of the CAM, and 100 μL of conditioned medium harvested from transduced glioma cells was added onto the center of the paper. After the window was closed with sterile adhesive tape, the eggs were incubated at 37 °C under 80-90% relative humidity for 48 h. Following fixation with stationary solution (methanol: acetone = 1:1) for 15 min, the CAM was cut and harvested, and gross photos of each CAM were taken using a digital camera (Panasonic, Osaka, Japan). The effect of conditioned media was evaluated by the number of second- and third-order vessels in comparison with that treated with the medium harvested from the vector-control group. Statistical analysis of second- and third-order vessels was carried out using a two-tailed Student's t-test.
Gene set enrichment analysis (GSEA)
Gene set enrichment analysis was performed as previously described [29]. The public microarray data (GSE7696) from the GEO database (https://www.ncbi.nlm.nih.gov/gds) was downloaded and HOXC10 expression was treated as a numeric variable. Then, a continuous-type CLS file of the HOXC10 profile to phenotype labels in GSEA was applied. The metric for ranking genes in GSEA was set as “Pearson”, and the other parameters were set to their default values. GSEA was performed using GSEA 2.0.9 software (http://www.broadinstitute.org/gsea/).
Animal xenografts
Five-week-old female BALB/c nude mice were purchased from Beijing Vital River (Beijing, China). Briefly, 5×105 of the indicated glioma cells stably expressing luciferase were stereotactically injected into the right cerebrum of mice (n = 6/group). Five days after implantation, mice were intraperitoneally treated with IgG or Bevacizumab (10 mg/kg) every three days. Intracranial tumor growth was monitored in vivo using bioluminescence IVIS imaging (Xenogen, Almeda, CA) equipped with Living Image software (Xenogen). Briefly, mice were anesthetized with isofluorane and subsequently received intraperitoneal injection with luciferin (75 μg/g body weight). Images were acquired 5-10 min after luciferin administration. A photographic image of the animal was taken in the chamber under dim illumination, followed by acquisition and overlay of the pseudocolor image representing the spatial distribution of photon counts produced by active luciferase within the animal. A signal averaging time of 1 min was used for luminescence image acquisition. Signal intensity was quantified as the sum of all detected photon counts (photons per second) within a region of interest prescribed over the tumor site using living image software Xenogen. Three brains of each group of mice were harvested on day 21 and subjected to immunohistochemistry (IHC) and hematoxylin and eosin (H&E) staining. IHC was performed using anti-HOXC10 polyclonal antibody (Abcam, ab153904), anti-CD31 polyclonal antibody (Abcam, ab28364), and anti-VEGFA monoclonal antibody (Abcam, ab1316, [VG-1]). Quantification of microvessel density was performed in accordance with a previous study [30]. Briefly, areas of greatest vessel density were then examined under higher magnification (×100) and counted. Any distinct area of positive staining for CD31 was counted as a single vessel. Results were expressed as the mean number of vessels ± SD per high-power field (×100). Experiments associated with animals complied with the Laboratory Animal Care and Use Guide and the Institutional Research Ethics Committee approved these experiments.
3-(4, 5-Dimethyl-2-thiazolyl)-2, 5-diphenyl-2H- tetrazolium bromide (MTT) assay
Cells (2×103) were seeded into 96-well plates and stained at the indicated time point with 100 μL of sterile MTT (Sigma-Aldrich) dye (0.5 mg/mL) for 4 h at 37 °C, followed by removal of the culture medium and the addition of 150 μL of dimethyl sulfoxide (Sigma-Aldrich). The absorbance was measured at 570 nm, with 655 nm used as the reference wavelength.
Transwell migration assay
Cells (2×104) were plated on the top side of a polycarbonate Transwell filter in the upper chamber of a BioCoat Invasion Chamber (BD Biosciences) and incubated at 37 °C for 22 h. The cells remaining on the upper surface were removed with cotton swabs. Cells that had migrated to the lower membrane surface were fixed in 1% paraformaldehyde, stained with crystal violet, and counted under an optical microscope (×200 magnification). Cell counts are expressed as the mean number of cells from 10 random fields per well.
Luciferase reporter assay
Cells were seeded in 48-well plates (3×103 cells/well) in triplicate and allowed to settle for 24 h. Luciferase reporter plasmids (100 ng) with 1 ng of pRL-TK Renilla plasmid (Promega, Madison, WI, USA) were transfected into glioma cells using Lipofectamine 3000 reagent. Luciferase and Renilla signals were determined 24 h after transfection using a Dual Luciferase Reporter Assay Kit (Promega).
Chromatin immunoprecipitation assay
Cells (2×106) plated in a 100 mm culture dish were treated with 1% formaldehyde to cross-link proteins to DNA. The cell lysates were sonicated to shear the DNA into 300-1000 bp lengths. Aliquots containing equal amounts of chromatin supernatants were incubated on a rocking bed at 4 °C overnight with either 1 μg of anti-Flag antibody (Sigma-Aldrich), anti-polymerase II antibodies (Abcam), or 1 μg anti-IgG antibody (Abcam) as a negative control. Following reverse cross-linking of protein-DNA complexes to free the DNA, real-time PCR was carried out. The primers used in this study are listed in the supplementary materials.
Co-immunoprecipitation analysis
The indicated protein in the cell lysates was immunoprecipitated using antibodies including anti-Flag (Sigma-Aldrich), anti-HA (Sigma-Aldrich), and anti-HOXC10 antibody (Novus Biologicals, Littleton, CO, USA) antibodies. The immunocomplexes were then examined using antibodies targeting Flag (Sigma-Aldrich), PRMT5 (Abcam), HOXC10 (Novus Biologicals), and HA (Sigma-Aldrich), and visualized using an ECL detection system.
Far-western blotting analysis
The assay was performed using a standard protocol, according to a previous study [31]. Briefly, a plasmid encoding Flag-tagged HOXC10 was transfected into U251MG cells, and 48 h later, U251MG cells were immunoprecipitated using Flag-tag affinity gel (Sigma-Aldrich). Membranes were incubated with 5 μg/mL of recombinant GST-PRMT5. After TBS-T washes, the polyvinylidene fluoride (PVDF) membrane was incubated with anti-GST antibodies. After development and imaging, the anti-GST antibody was stripped and membranes were incubated with anti-Flag antibodies.
Statistical analysis
Data were evaluated using Fisher's exact test, the log-rank test, the Chi-squared test, and Student's two-tailed t-test. Statistical analyses were performed using the SPSS 21.0 statistical software package (IBM Corp, Armonk, NY, USA). Data are represented as the mean ± SD. P < 0.05 was considered statistically significant.
Results
HOXC10 plays an important role in glioma progression
It has been reported that elevated expression of HOXC10 induced glioma proliferation, migration and invasion[26]. Therefore, we verified whether HOXC10 was upregulated in gliomas via analyzing public glioma datasets from The Cancer Genome Atlas (TCGA) and the Gene Expression Omnibus (GEO; GSE13276 and GSE4290). As expected, HOXC10 was significantly elevated in gliomas, especially in glioblastoma, compared to non-tumor brain tissues and normal brain tissues, respectively (Figure S1A-C). Furthermore, Kaplan-Meier analysis of data from the TCGA and HG-UG133A dataset indicated that GBM or low-grade gliomas (LGG) patients with higher HOXC10 expression presented shorter overall survival than those with lower HOXC10 expression (Figure S1D-F). Moreover, we found that HOXC10 expression was not only correlated with poorer survival of patients with astrocytoma but also correlated with poorer survival of patients with oligodendroglioma analyzed in TCGA dataset, which indicated that HOXC10 is involved in the progression of both astrocytomas and oligodendrogliomas (Figure S1G-H).
To further determine whether HOXC10 expression was correlated with aggressive clinicopathological characteristics of gliomas, we performed western blotting analysis and real-time PCR analysis. The results indicated that the expression levels of HOXC10 in 12 human glioma tissues were higher than those in four normal brain tissues (Figure S2A-B). We also performed western blotting analysis and real-time PCR analysis in NHAs and multiple glioma cell lines, including A-172, U87, LN229, T98G, LN-18, and U251MG. The glioma cell lines showed higher protein levels of HOXC10 than the NHAs (Figure S2C-D). Next, we performed IHC analysis to examine the level of HOXC10 in 94 paraffin-embedded, archived glioma tissues and normal brain tissues (Table S1). As shown in Figure S2E, the average staining intensity of HOXC10 in Grade I to IV tumors was noticeably stronger than that in normal brain tissue. Quantitative analysis of the mean optical density (MOD) indicated that the HOXC10 level increased in parallel with disease grade in glioma, revealing a positive correlation between HOXC10 level and glioma progression. Furthermore, upregulation of HOXC10 correlated significantly with advanced disease grade and poor survival in patients with glioma (Table S2). Based on Kaplan-Meier analysis, higher HOXC10 expression predicted shorter survival of patients with all WHO Grade, WHO Grade I-II, and WHO Grade III-IV gliomas (Figure S2F-H). However, we did not observe HOXC10 level significantly correlated with IDH and 1p/19q status (Table S1 and Table S2). Importantly, univariate and multivariate analysis indicated that HOXC10 might serve as a prognostic factor in gliomas (Table S3). Taken together, these results strongly suggested that HOXC10 might play pivotal roles in glioma progression.
Dysregulation of HOXC10 alters the ability of gliomas to induce angiogenesis
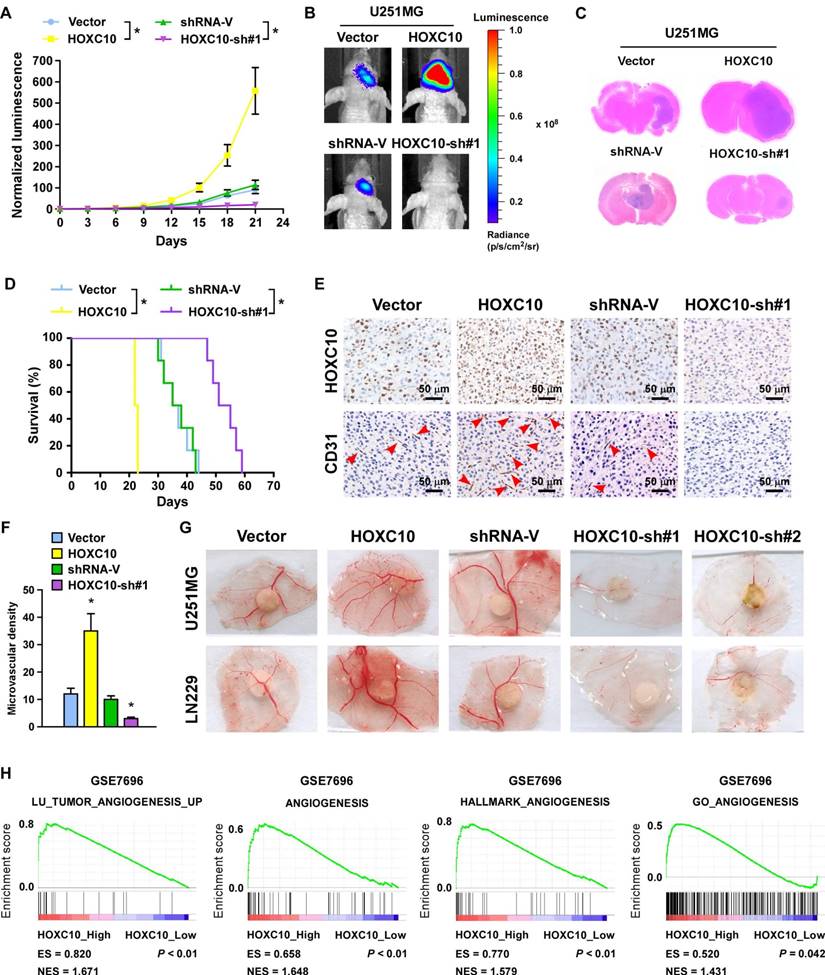
A wealth of literature supports the view that homeobox genes are highly expressed in several types of cancer and play a crucial role in angiogenesis [32-34]. To validate the effect of HOXC10 on angiogenesis of intracranial xenograft, U251MG and LN229 glioma cell lines stably expressing HOXC10 or HOXC10-targeting shRNA were established (Figure S3A). Glioma cells, transduced with the control vector, the HOXC10-overexpression vector, the control shRNA vector, or the HOXC10-sh#1 vector, were stereotactically implanted into mouse brains (n = 6/group). Tumors formed by HOXC10-overexpressing U251MG cells were significantly larger than those formed by the control cells, as indicated by luminescence and H&E staining (Figure 1A-C). Conversely, silencing HOXC10 yielded smaller tumors than the control shRNA vector (Figure 1A-C). Furthermore, overexpression of HOXC10 shortened, while silencing HOXC10 prolonged the survival of tumor-bearing mice compared to the control shRNA group (Figure 1D). Moreover, we found that microvascular density was efficiently enhanced in tumors formed by HOXC10-overexpressing tumors and reduced in HOXC10-silenced tumors, as shown by CD31 staining (Figure 1E-F).
Consistent with the in vivo results, overexpressing HOXC10 significantly promoted, but silencing HOXC10 prominently inhibited, the capability of tube formation, migration ability, and proliferation of HUVECs (Figure S3B-E and Figure S4A-D) and the formation of second- and third-order vessels in the chicken CAM assay (Figure 1G). Gene set enrichment analysis (GSEA) results also revealed that angiogenesis gene signatures (LU_TUMOR_ ANGIOGENESIS_UP, ANGIOGENESIS, HALLMARK_ANGIOGENESIS, GO_ ANGIOGENESIS) were significantly elevated in the higher HOXC10 expression group compared to the lower HOXC10 expression group (Figure 1H). Collectively, our results suggested that HOXC10 overexpression contributes to angiogenesis in gliomas.
Dysregulation of HOXC10 alters the ability of gliomas to induce angiogenesis in vivo. (A) Relative changes in the luminescence of xenograft tumors formed by vector control, HOXC10-transduced, shRNA control, and HOXC10-silenced glioma cells. n = 6/group. Error bars, SD. * P < 0.05. (B) Representative bioluminescence images of the indicated tumor-bearing mice. (C) Representative images of H&E staining of glioma sections from the indicated mice. (D) Kaplan-Meier survival curves of mice (n = 6/group) inoculated with the indicated cells. * P < 0.05. (E) CD31 immunohistochemistry staining (arrow) indicating that overexpressing HOXC10 induced, whereas silencing HOXC10 inhibited, glioma angiogenesis in vivo. (F) Quantification of CD31+ microvessel density. Error bars, SD. * P < 0.05. (G) Representative images of the CAM blood vessels stimulated by CM derived from the vector control, HOXC10-transduced, shRNA control, and HOXC10-silenced glioma cells. The assay was repeated at least three times with similar results. (H) GSEA plots demonstrating a significant correlation between HOXC10 mRNA expression levels in gliomas and gene signatures of angiogenesis from published datasets (GSE7696). ES, enrichment score; NES, normalized enrichment score.
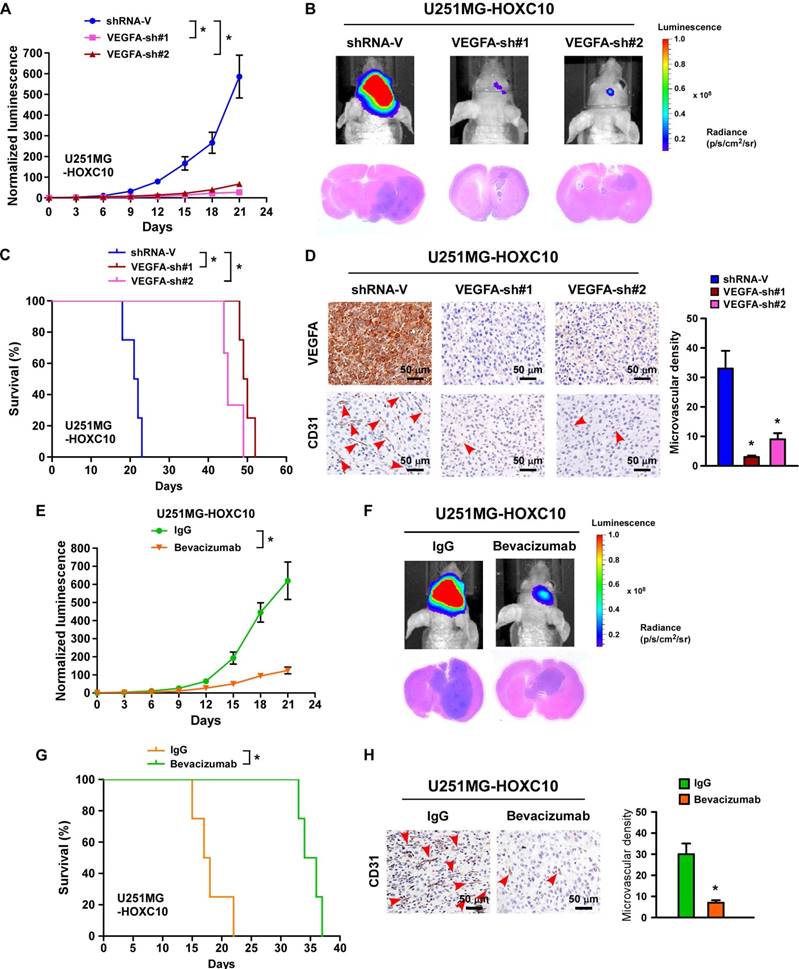
Silencing VEGFA expression or treatment with bevacizumab dramatically inhibits the growth and angiogenesis of tumors overexpressing HOXC10. (A) Relative changes in the luminescence of xenograft tumors formed by control-shRNA-transduced U251MG/HOXC10 cells and VEGFA-shRNA-transduced U251MG/HOXC10 cells. n = 6/group. Error bars, SD, * P < 0.05. (B) Upper panel: representative bioluminescence images of mice bearing tumors formed by control-shRNA-transduced U251MG/HOXC10 cells and VEGFA-shRNA-transduced U251MG/HOXC10 cells. Lower panel: representative images of H&E staining of glioma sections from mice bearing the indicated tumors. (C) Kaplan-Meier survival curves of mice (n = 6/group) inoculated with the indicated cells. * P < 0.05. (D) Left panel: CD31 immunohistochemical staining (arrow) indicating that silencing VEGFA inhibited angiogenesis in tumors overexpressing HOXC10. Right panel: quantification of CD31+ microvessel density. Error bars, SD. * P < 0.05. (E) Relative changes in the luminescence of xenograft tumors formed by HOXC10-overexpressing U251MG cells treated with IgG or bevacizumab. n = 6/group. Error bars, SD. (F) Upper panel: representative images of mice bearing tumors formed by HOXC10- overexpressing U251MG cells treated with IgG or bevacizumab. Lower panel: representative images of H&E staining of glioma sections from mice bearing tumors formed by HOXC10- overexpressing U251MG cells treated with IgG or bevacizumab. (G) Kaplan-Meier survival curves of mice (n = 6/group) bearing tumors formed by HOXC10-overexpressing U251MG cells treated with IgG or bevacizumab. * P < 0.05. (H) Left panel: CD31 immunohistochemical staining (arrow) indicating that bevacizumab inhibited angiogenesis in tumors overexpressing HOXC10. Right panel: quantification of CD31+ microvessel density. Error bars, SD. * P < 0.05.
HOXC10 modulates angiogenesis through regulating VEGFA expression
Previous studies have suggested that the expression of VEGFA is biologically and clinically involved in the angiogenesis of tumor cells [35]. Interestingly, three potential HOXC10 binding sites were predicted in the promoter of VEGFA in the JASPAR database (Figure S5A). As expected, the expression of VEGFA, at both mRNA and protein levels, was consistently upregulated in HOXC10-overexpressing glioma cells, and downregulated in HOXC10-silenced glioma cells compared to control cells (Figure S5B-D). Importantly, silencing VEGFA or treatment with bevacizumab, a monoclonal antibody against VEGFA, could reverse the effects of HOXC10 overexpression-mediated tube formation, migration, and proliferation of HUVECs (Figure S5E-M). Importantly, we found that tumors formed by VEGFA-silenced, HOXC10-overexpressing cells were significantly smaller than those formed by control-shRNA-transduced cells (Figure 2A-B). Mice bearing VEGFA-silenced U251MG/HOXC10 cells had longer survival than those bearing control-shRNA-transduced cells (Figure 2C). The microvascular density was significantly reduced in tumors formed by VEGFA-silenced, HOXC10-overexpressing U251MG cells (Figure 2D). Strikingly, treatment with bevacizumab significantly inhibited the growth of HOXC10-overexpressing tumor and efficiently impaired angiogenesis (Figure 2E-H). These results provide strong evidence that VEGFA is the key effector of HOXC10-induced angiogenesis and bevacizumab might have potential therapeutic value for the treatment of HOXC10-induced glioma.
HOXC10 regulates VEGFA promoter activity in glioma cells
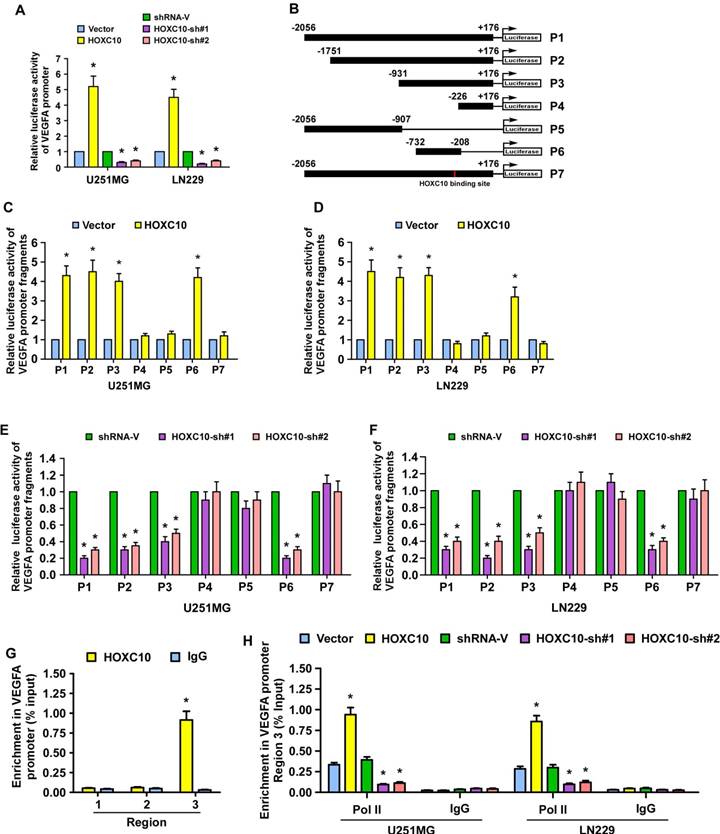
A luciferase reporter assay was performed and showed that overexpression of HOXC10 promoted, while silencing HOXC10 repressed, the luciferase activity of the VEGFA promoter (Figure 3A). To identify the site that HOXC10 binds to in the VEGFA promoter, fragments of the VEGFA promoter, namely P1(nucleotides -2056 to +176), P2 (nucleotides -1751 to +176), P3 (nucleotides -931 to +176), P4 (nucleotides -226 to +176), P5 (nucleotides -2056 to -907) and P6 (nucleotides -732 to -208), were inserted into the luciferase reporter pGL3 vector, respectively (Figure 3B). We observed that overexpression of HOXC10 increased, while silencing of HOXC10 expression decreased, the luciferase activity of VEGFA promoter fragments P1, P2, P3, P6, with the luciferase signals of P4 and P5 being largely unaffected (Figure 3C-F). These results suggest that the HOXC10 binding site may be located in nucleotides -292 to -283 (Figure S5A). Meanwhile, mutation of nucleotides -289 to -286 reversed the effect of overexpression or knockdown of HOXC10 on the luciferase activity of P1 (Figure 3C-F). Moreover, a chromatin immunoprecipitation (CHIP) assay showed that HOXC10 bound to region 3 within the VEGFA promoter region (Figure 3G), which agreed with the luciferase assay results and suggested that HOXC10 upregulates VEGFA expression by binding to the VEGFA promoter. In addition, we found that the recruitment of RNA polymerase II (Pol II) to the VEGFA promoter was enhanced in HOXC10-transduced cells, but impaired in HOXC10-silenced cells (Figure 3H). Taken together, our results provide corroborating evidence that HOXC10 transcriptionally upregulates VEGFA expression.
HOXC10 upregulates VEGFA expression by interacting with PRMT5
To further study the mechanism by which HOXC10 upregulates VEGFA expression, systematic ChIP was performed to probe histone post-translational modifications, including H3R2me1, H3R2me2s, H3R8me2s, H2R3me2s, and H4R3me2s. Interestingly, the enrichment of H3R2me1 and H3R2me2s, both of which are PRMT5-catalyzed histone post-translational modifications, in VEGFA promoter was markedly increased in HOXC10-transduced cells, but strikingly decreased in HOXC10-silenced cells (Figure 4A). Furthermore, we found that recruitment of PRMT5 to the VEGFA promoter was significantly enhanced in HOXC10-overexpressing glioma cells, but markedly suppressed in HOXC10-silenced glioma cells (Figure 4B). Moreover, knockdown of PRMT5 strongly reversed the HOXC10-overexpressing effect on VEGFA enrichment (Figure 4C). These results indicate that PRMT5 might play a role in HOXC10-mediated VEGFA upregulation.
To verify the hypothesis that HOXC10 induced VEGFA expression via forming a complex with PRMT5, co-immunoprecipitation (co-IP) assay was carried out and the results demonstrated that HOXC10 formed a complex with PRMT5 in U251MG and LN229 cells (Figure 4D-E). Far-western blotting further demonstrated that HOXC10 directly interacted with PRMT5 (Figure 4F). To determine the fraction of HOXC10 responsible for their interaction, several HOXC10 truncations were generated (Figure 4G). Co-IP assays showed that PRMT5 interacted with HOXC10-FL (Full Length), HOXC10-F1 and HOXC10-F4, suggesting that a region between aa 77 and aa 197 is required for HOXC10-PRMT5 interaction (Figure 4H). Overexpressing HOXC10-FL and HOXC10-F1 markedly promoted the luciferase activity of VEGFA, while overexpressing HOXC10-F2, HOXC10-F3, HOXC10-F4, all of which lacked the PRMT5 binding site or homeobox, could not activate the VEGFA promoter-dependent luciferase activity (Figure 4I). Consistently, VEGFA expression, at both mRNA and protein levels, was significantly increased in the HOXC10-FL- and HOXC10-F1-overexpressing cells but decreased in the cells transfected with HOXC10-F2, or HOXC10-F3 (Figure S6A-B). Consistent with the inhibitory effect of HOXC10-F4 on the interaction of HOXC10 and PRMT5, we also found that overexpression of HOXC10-F4 peptide significantly abrogated HOXC10-induced VEGFA upregulation and reduced the ability of HOXC10-transduced cells to induce tube formation and proliferation of HUVECs, which provided further evidence that HOXC10 activates VEGFA transcription by interacting with PRMT5 (Figure S6C-F). These results suggested that PRMT5 is required for HOXC10-mediated upregulation of VEGFA.
HOXC10 regulates VEGFA promoter activity in glioma cells. (A) The VEGFA promoter luciferase reporter plasmids and Renilla pRL-TK plasmids were transfected into the empty vector-transduced, HOXC10-transduced, control shRNA-transduced, and the HOXC10-shRNA transduced glioma cells. Forty-eight hours later, the luciferase signal was examined. (B) Schematic illustration of the cloned fragments of the human VEGFA promoter. (C-D) Luciferase activity of VEGFA promoter fragments in control vector cells and HOXC10-overexpressing glioma cells. (E-F) Luciferase activity of VEGFA promoter fragments in shRNA control vector cells and HOXC10-silenced glioma cells. (G) ChIP assays were performed using an anti-HOXC10 antibody to screen three predicted HOXC10-binding sites in the VEGFA promoter. IgG was used as a negative control. (H) ChIP assay showing enrichment of Pol II on region 3 in the VEGFA promoter in the indicated cells. All the assays showed in the figure were independently replicated three times. Error bars, SD. * P < 0.05.
HOXC10 interacts with PRMT5 to promote VEGFA transcription. (A) ChIP-qPCR enrichments of histone post-translational modifications on VEGFA promoter region 3 in the indicated cells, displayed using a heatmap. The color key represents an intensity scale for ratio of HOXC10 versus vector or HOXC10-shRNA versus shRNA control vector, normalized by log2 transformation. (B) ChIP-qPCR enrichments of PRMT5 or lgG on VEGFA promoter region 3 in the indicated cells. (C) ChIP-qPCR enrichments of HOXC10 or lgG on VEGFA promoter region 3 in the control-shRNA-transduced, HOXC10-overexpressing glioma cells or PRMT5-shRNA-transduced, HOXC10-overexpressing glioma cells. (D) Vector or Flag-tagged HOXC10 was transfected into the U251MG or LN229. Co-IP assay showed that HOXC10 interacted with PRMT5. (E) Co-IP assay showed that endogenous HOXC10 interacted with PRMT5 in U251MG and LN229 cells. (F) Far western blotting analysis showed that Flag-tagged HOXC10 in U251MG transfected with Flag-tagged HOXC10 interacted with recombinant GST-PRMT5. (G) Schematic illustration of full length HOXC10 or HOXC10 truncations. (H) Flag-tagged HOXC10 (full length or truncations) and HA-tagged PRMT5 were transfected into U251MG. Co-IP assay revealed that PRMT5 interacted with FL, F1 and F4, but not with F2 and F3. (I) VEGFA promoter luciferase reporter plasmids, Renilla pRL-TK plasmids, vector, or Flag-tagged HOXC10 (full length or truncations) were transfected into U251MG cells. After 48 h, cells were subjected to a luciferase reporter assay. All the assays shown in the figure were independently replicated three times. Error bars, SD. * P < 0.05.
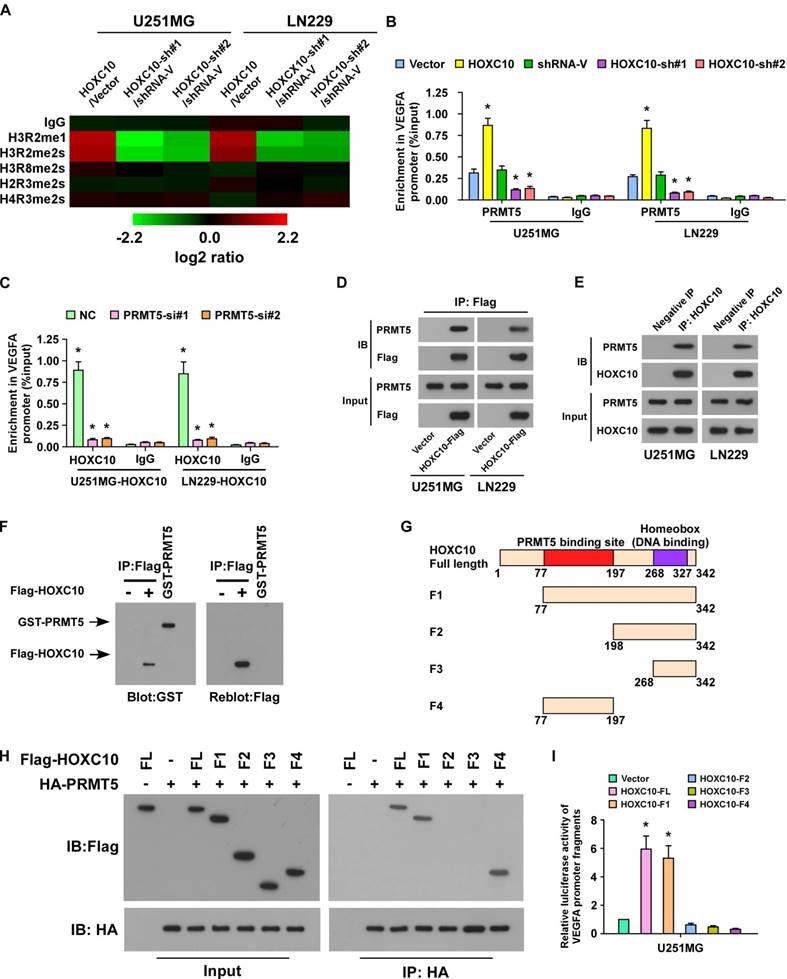
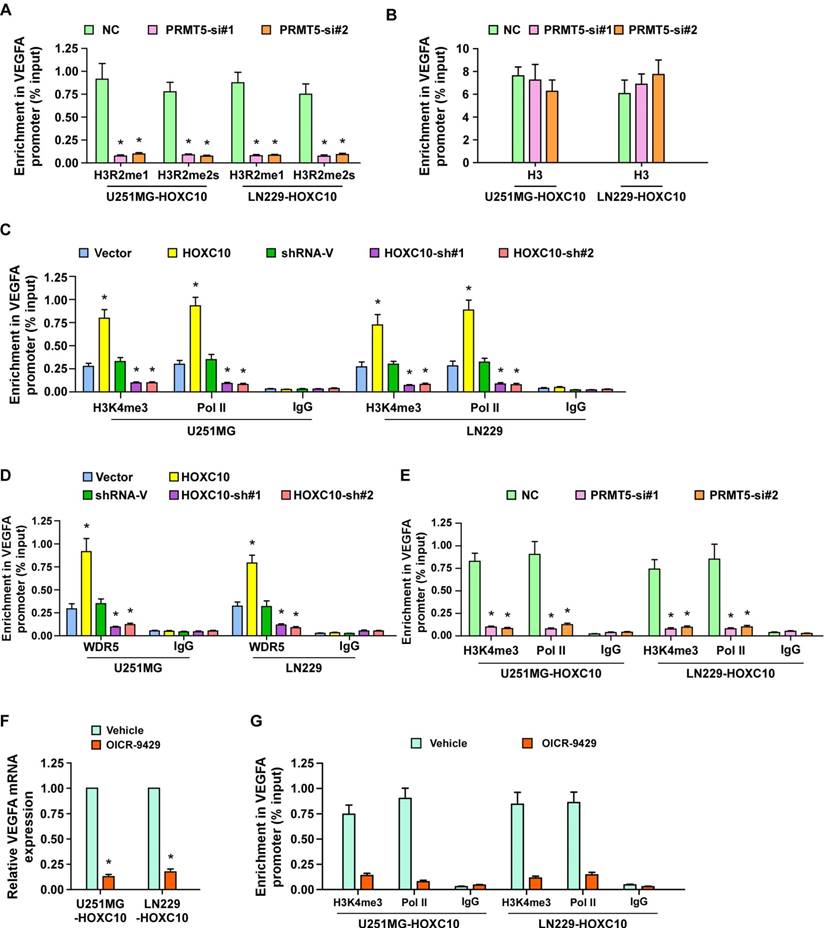
PRMT5 and WDR5 are required for HOXC10-mediated VEGFA transcription
Next, we further investigated the effect of PRMT5 on HOXC10-mediated VEGFA upregulation. As shown in Figure S7A-B and Figure 5A-B, knockdown of PRMT5 significantly reduced VEGFA expression and inhibited the occupancy of H3R2me1 and H3R2me2s on VEGFA in HOXC10-overexpressing glioma cells, with no change in control marker H3. PRMT5-methylated H3R2me1 may lead to WDR5-induced H3K4 methylation and transcriptional activation of target genes [36]. To investigate whether WDR5 and H3K4me3 are involved in HOXC10-mediated VEGFA transcriptional regulation, ChIP-qPCR for H3K4me3, Polymerase II, and WDR5 was conducted. As expected, overexpressing HOXC10 strongly increased, while silencing HOXC10 prominently decreased, the abundance of H3K4me3, Polymerase II, and WDR5 on the VEGFA promoter in U251MG and LN229 cells (Figure 5C-D). Moreover, occupancy of H3K4me3, Polymerase II, and WDR5 on the VEGFA promoter were dramatically attenuated in PRMT5-silenced, HOXC10-transduced glioma cells (Figure 5E and Figure S7C). Furthermore, silencing WDR5 profoundly impaired the stimulatory effect of overexpressing HOXC10 (Figure S7D-F). Importantly, OICR-9429, an inhibitor of WDR5 interaction with the H3 tail, markedly decreased the expression of VEGFA and the occupancy of H3K4me3 and Polymerase II on the VEGFA promoter (Figure 5F-G). These results suggested that HOXC10 interacts with PRMT5, which promotes H3R2me1 and H3R2me2s enrichment on the VEGFA promoter and recruits WDR5 for subsequent methylation of H3K4 at the VEGFA promoter, eventually resulting in VEGFA transcription.
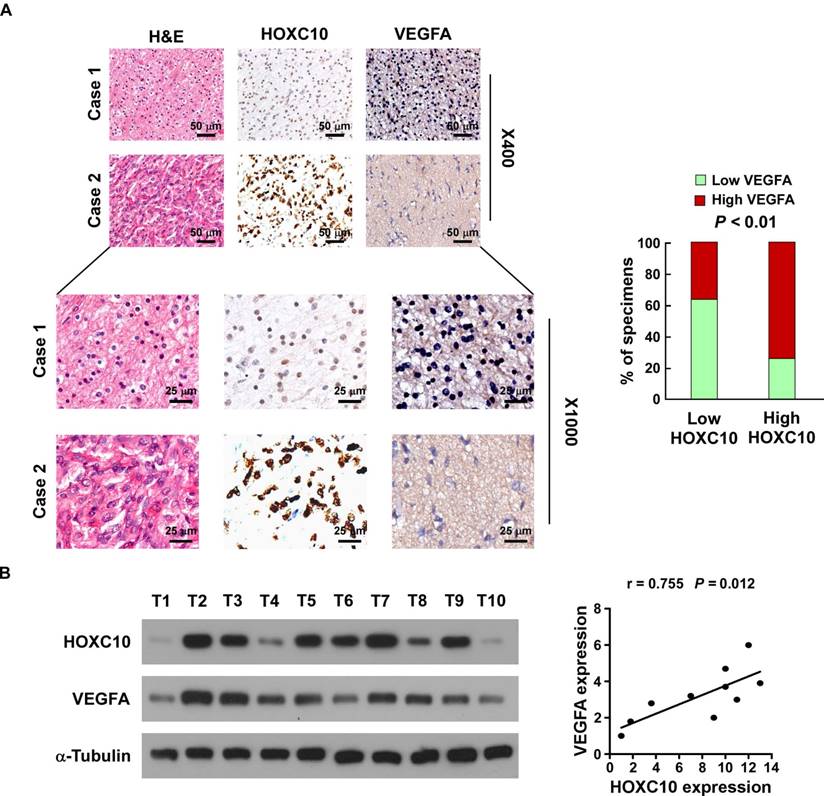
Clinical relevance of HOXC10 and VEGFA expression in human glioma
Finally, we determined whether the HOXC10/VEGFA axis was clinically relevant. As shown in Figure 6A, IHC analysis showed that HOXC10 levels were positively correlated with the expression of VEGFA in human glioma specimens (P < 0.01). Consistent with the IHC results, western blotting analysis demonstrated that levels of HOXC10 also correlated strongly with the levels of VEGFA (r = 0.755, P = 0.012) in 10 freshly collected clinical glioma samples (Figure 6B). Furthermore, statistical analysis revealed that HOXC10 expression was significantly correlated with VEGFA expression in the TCGA-LGG group (n = 516, r = 0.260, P < 0.01). Although HOXC10 expression in the entire TCGA-GBM group was not significantly correlated with VEGFA expression (n = 156, r = 0.146, P = 0.069), we found that HOXC10 expression in the PRMT5 higher-GBM subgroup was significantly correlated with VEGFA levels (n = 78, r = 0.279, P = 0.014), which further supported that PRMT5 contributes to HOXC10-mediated VEGFA upregulation (Figure S8A-C). These results strongly supported the notion that overexpression of HOXC10 transcriptionally upregulates VEGFA, which promotes angiogenesis and ultimately leads to poor survival of patients with glioma.
Discussion
Glioblastoma, the most frequent type of primary brain tumor, is one of the most aggressive and deadly human cancers [37]. Angiogenesis has been reported to be closely involved in glioma growth, invasion, metastasis, and decreased patient survival [38, 39]. Therefore, inhibiting the angiogenic process might be used as a potential therapy to prevent tumor growth and metastasis. However, the molecular mechanisms involved in regulating glioma angiogenesis remain largely unclear. The present study revealed that higher expression of HOXC10 was significantly correlated with a poorer prognosis of patients harboring gliomas. Additionally, our results indicated that upregulated HOXC10 promotes angiogenesis in human gliomas in vitro and in vivo via upregulating VEGFA expression.
Homeobox genes are highly expressed in several types of cancer and play a crucial role in angiogenesis [32-34]. Seki et al. reported that HOXB9-positive breast tumors present a significant increase in vasculature, and Kaplan-Meier analysis revealed that HOXB9-positive breast cancer patients have poor prognosis [32]. Zhu and colleagues reported that HOXB13 expression is correlated positively with tumor VEGF levels and microvessel density in hepatocellular carcinoma (HCC), and univariate and multivariate analysis showed HCC patients with higher HOXB13 expression exhibits poorer overall survival and disease-free survival [33]. Additionally, Storti et al. reported that HOXB7 is overexpressed in more than 40% myeloma cell lines compared to normal plasma cells, and knockdown of HOXB7 expression in multiple myeloma cells represses the production of angiogenic factors and impairs their pro-angiogenic properties [34]. In this study, we demonstrated that higher expression of HOXC10 correlated significantly with advanced grade and poorer prognosis in patients with gliomas. In addition, HOXC10 modulated the VEGFA signaling pathway to mediate an angiogenesis phenotype in human gliomas, further indicating that HOXC10 might represent a promising therapeutic target for anti-angiogenic strategies against gliomas.
PRMT5 and WDR5 are required for HOXC10-mediated VEGFA transcription. (A) ChIP-qPCR enrichments of H3R2me1 and H3R2me2s on VEGFA promoter region 3 in the control-shRNA transduced, HOXC10-overexpressing glioma cells or PRMT5-shRNA transduced, HOXC10-overexpressing glioma cells. (B) Histone H3 served as a control for ChIP experiments. (C) ChIP-qPCR enrichments of H3K4me3, polymerase II or IgG on VEGFA promoter region 3 in the vector control, HOXC10-transduced, control-shRNA transduced, and HOXC10-shRNA transduced glioma cells. (D) ChIP-qPCR enrichments of WDR5 or IgG on VEGFA promoter region 3 in the vector control, HOXC10-transduced, control-shRNA transduced, and HOXC10-shRNA transduced glioma cells. (E) ChIP-qPCR enrichments of H3K4me3, polymerase II or IgG on VEGFA promoter region 3 in the control-shRNA transduced, HOXC10-overexpressing glioma cells or PRMT5-shRNA transduced, HOXC10-overexpressing glioma cells. (F) Real-time PCR analysis showed the VEGFA mRNA expression in the HOXC10-overexpressing glioma cells treated with vehicle or 1 μM OICR-9429 for 2 days. (G) ChIP-qPCR enrichments of H3K4me3, polymerase II or IgG on VEGFA promoter region 3 in the HOXC10-overexpressing glioma cells treated with vehicle or 1 μM OICR-9429 for 2 days. All the assays shown in the figure were independently replicated three times. Error bars, SD. * P < 0.05.
Clinical relevance of HOXC10 and VEGFA expression in gliomas. (A) HOXC10 levels were positively associated with VEGFA expression in 94 primary human glioma specimens. Left panel: two representative cases are shown. Magnification ×400: scale bars, 50 μm (top 2 rows). Magnification ×1,000: scale bars, 25 μm (bottom 2 rows). Right panel: percentage of glioma specimens showing low or high HOXC10 expression relative to the level of VEGFA. (B) Expression analysis (left) and correlation (right) between HOXC10 expression and VEGFA expression in 10 freshly collected human glioma samples. The western blotting assay was replicated independently three times.
Angiogenesis consists of multiple processes, including endothelial cell proliferation, migration, and alignment to form capillary-like structures. It has been reported that anti-angiogenic therapy holds promise for treating malignancies, including gliomas [40]. Although initial responses to anti-angiogenic therapy are often significant, the benefits are transitory and are followed by a restoration of tumor growth and progression[41]. Two large randomized clinical trials of bevacizumab therapy had no significant effect on prolonging survival as part of first line therapy for glioblastomas in humans [42, 43], which suggested that the established glioblastoma cell lines and the employed mouse models might not represent the native characteristics of glioblastoma cells and native environment of glioblastomas in human brain. Indeed, the high passage established glioblastoma cell lines showed substantial alterations of morphology, genome, and functional characteristics compared to the glioblastoma cells present in the glioblastoma mass [44]. It has been reported that the original low-passaged U-251 cells maintained a DNA copy number resembling a typical GBM profile, whereas all long-term subclones lost the typical glioblastoma profile, demonstrated by array comparative genomic hybridization (aCGH). In addition, long-term subclones exhibited variations in phenotypic marker expression and increased growth rate in vitro and a more aggressive growth in vivo [45]. However, glioma primary cultures showed genetic make-ups that were similar to those of patient tumors. For instance, Balbous et al. reported that COL1A1 and IFITM1 genes identified using gliomaspheres derived from glioma patients could be considered for use in stratifying patients with glioblastoma into subgroups for risk of recurrence at diagnosis and prognostic evolution [46]. These results suggested that patient-derived primary glioma cells are more appropriate to be used as models for studies of glioblastoma biology. Meanwhile, the human cell line-derived xenografts showed a level of angiogenesis and circumscribed growth with variable extents of collective tissue invasion, which exhibited limited single cell infiltration in the brain and lacked necrosis and microvascular abnormalities, both of which were characteristics of human glioblastoma [47]. Moreover, glioblastoma PDX models predict poor prognosis of patients with glioblastoma and the model can be used to identify the gene signatures and pathways signatures associated with the clinical aggressiveness of glioblastomas [48]. Therefore, patient-derived glioblastoma cells or patient-derived glioblastoma xenograft models need to be used to further examine the promotory effect of HOXC10 overexpression on angiogenesis and VEGFA upregulation.
The PRMT5 gene encodes an arginine methyltransferase, which catalyzes the transfer of methyl groups to the amino acid arginine in target proteins, including histones, and promotes target gene transcription. A wealth of literature supports the view that PRMT5 plays a vital role in regulating cancer progression via histone methylation [49-51]. PRMT5 promotes the cancer stem cell function of breast cancer by enhancing the enrichment of H3K4me3 on the FOXP1 promoter and subsequently driving FOXP1 transcription and expression [49]. SHARPIN augments invasiveness of lung cancer by interacting with PRMT5, which methylates histones H3 and H4, subsequently upregulating metastasis-related gene expression [50]. Furthermore, PRMT5 is recruited through interaction with Sp1 and activates the androgen receptor by symmetrically dimethylating H4R3, which significantly enhanced the growth of prostate xenograft tumors in mice [51]. Our results showed that HOXC10 interacted with PRMT5, leading to enrichment of H3R2me1, H3R2me2s, and H3K4me3 on the VEGFA promoter, which eventually resulted in increased VEGFA expression and enhances growth of gliomas. Thus, our results provide an additional molecular mechanism in angiogenesis and show that targeting PRMT5 might be an efficient therapeutic approach.
VEGF-A has been demonstrated to show prominent activity with vascular endothelial cells, mainly through its interactions with the VEGFR1 and -R2 receptors. However, bevacizumab, a recombinant humanized monoclonal antibody that blocks angiogenesis by inhibiting secreted VEGF-A, did not improve survival in humans with glioblastoma. Interestingly, it has been recently also reported that the expression levels of different vascular endothelial growth factor A (VEGF) isoforms were associated with angiogenesis and patient prognosis in human cancers [52-54]. Two novel splicing VEGF isoforms L-VEGF144 and L-VEGF138, which were transcripted at the first upstream CUG codon, had nuclear localization ability and were also involved in angiogenic processes in glioblastoma[53, 54]. Herein, we demonstrated that HOXC10 transcriptionally upregulates VEGFA expression by binding to its promoter and silencing HOXC10, which significantly decreased VEGFA expression and angiogenesis, implicating that HOXC10 may regulate expression of various splicing VEGF isoforms, which needs to be further investigated in future studies.
In conclusion, our findings suggest that HOXC10 is overexpressed in malignant glioma tissues and correlates with aggressive clinicopathological characteristics. Overexpression of HOXC10 enhances, while downregulation of HOXC10 impairs, pro-angiogenesis activity in gliomas. Importantly, expression of HOXC10 correlated strongly with the level of VEGFA in clinical glioma samples. Taken together, these findings provide a new strategy to treat malignant glioma, which might lead to improved prognosis in patients with glioma.
Abbreviations
CAM: chicken chorioallantoic membranes; CHIP: chromatin immunoprecipitation; co-IP: co-immunoprecipitation; GBM: glioblastoma multiforme; GEO: Gene Expression Omnibus; GSEA: gene set enrichment analysis; GST: glutathione-s-transferase; HA: hemagluttinin; HCC: hepatocellular carcinoma; HOX: homeobox; HOXC10: homeobox C10; HUVEC: human umbilical vein endothelial cells; H&E: hematoxylin and eosin; IHC: immunohistochemistry; LGG: low-grade gliomas; MOD: mean optical density; MTT: 3-(4, 5-dimethyl-2-thiazolyl)-2, 5-diphenyl-2H- tetrazolium bromide; MVD: microvascular density; NHA: normal human astrocytes; Pol II: RNA polymerase II; PRMT5: protein arginine methyltransferase 5; PVDF: polyvinylidene fluoride; shRNA: short hairpin RNA; TCGA: The Cancer Genome Atlas; VEGF: vascular endothelial growth factor; VEGFA: vascular endothelial growth factor A; WB: Western blotting; WDR5: WD repeat domain 5.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
This work was supported by the Natural Science Foundation of China [No. 81330058, 81830082, 91740119, 91529301, 81621004, 91740118, 81773106 and 81530082]; The Guangzhou Science and Technology Plan Projects [201803010098]; The Natural Science Foundation of Guangdong Province [2018B030311009, 2016A030308002]; The Fundamental Research Funds for the Central Universities [No. 17ykjc02].
Competing Interests
The authors have declared that no competing interest exists.
References
1. Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A. et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683-710
2. Ostrom QT, Gittleman H, Liao P, Rouse C, Chen Y, Dowling J. et al. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2007-2011. Neuro Oncol. 2014;16(Suppl 4):iv1-63
3. Mamelak AN, Jacoby DB. Targeted delivery of antitumoral therapy to glioma and other malignancies with synthetic chlorotoxin (TM-601). Expert Opin Drug Deliv. 2007;4:175-86
4. Reardon DA, Rich JN, Friedman HS, Bigner DD. Recent advances in the treatment of malignant astrocytoma. J Clin Oncol. 2006;24:1253-65
5. Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ. et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987-96
6. Holland EC. Glioblastoma multiforme: the terminator. Proc Natl Acad Sci U S A. 2000;97:6242-4
7. Friedmann-Morvinski D, Narasimamurthy R, Xia Y, Myskiw C, Soda Y, Verma IM. Targeting NF-kappaB in glioblastoma: A therapeutic approach. Sci Adv. 2016;2:e1501292
8. Ribatti D, Vacca A. The role of microenvironment in tumor angiogenesis. Genes Nutr. 2008;3:29-34
9. Cea V, Sala C, Verpelli C. Antiangiogenic therapy for glioma. J Signal Transduct. 2012;2012:483040
10. Leon SP, Folkerth RD, Black PM. Microvessel density is a prognostic indicator for patients with astroglial brain tumors. Cancer. 1996;77:362-72
11. Birlik B, Canda S, Ozer E. Tumour vascularity is of prognostic significance in adult, but not paediatric astrocytomas. Neuropathol Appl Neurobiol. 2006;32:532-8
12. Carmeliet P. Angiogenesis in life, disease and medicine. Nature. 2005;438:932-6
13. Thomas JP, Arzoomanian RZ, Alberti D, Marnocha R, Lee F, Friedl A. et al. Phase I pharmacokinetic and pharmacodynamic study of recombinant human endostatin in patients with advanced solid tumors. J Clin Oncol. 2003;21:223-31
14. Risau W. Mechanisms of angiogenesis. Nature. 1997;386:671-4
15. Carmeliet P, Jain RK. Molecular mechanisms and clinical applications of angiogenesis. Nature. 2011;473:298-307
16. Adams RH, Alitalo K. Molecular regulation of angiogenesis and lymphangiogenesis. Nat Rev Mol Cell Biol. 2007;8:464-78
17. Lawrence HJ, Sauvageau G, Humphries RK, Largman C. The role of HOX homeobox genes in normal and leukemic hematopoiesis. Stem Cells. 1996;14:281-91
18. Alharbi RA, Pettengell R, Pandha HS, Morgan R. The role of HOX genes in normal hematopoiesis and acute leukemia. Leukemia. 2013;27:1000-8
19. Shah N, Sukumar S. The Hox genes and their roles in oncogenesis. Nat Rev Cancer. 2010;10:361-71
20. Abate-Shen C. Deregulated homeobox gene expression in cancer: cause or consequence? Nat Rev Cancer. 2002;2:777-85
21. Samuel S, Naora H. Homeobox gene expression in cancer: insights from developmental regulation and deregulation. Eur J Cancer. 2005;41:2428-37
22. Grier DG, Thompson A, Kwasniewska A, McGonigle GJ, Halliday HL, Lappin TR. The pathophysiology of HOX genes and their role in cancer. J Pathol. 2005;205:154-71
23. Zhai Y, Kuick R, Nan B, Ota I, Weiss SJ, Trimble CL. et al. Gene expression analysis of preinvasive and invasive cervical squamous cell carcinomas identifies HOXC10 as a key mediator of invasion. Cancer Res. 2007;67:10163-72
24. Feng X, Li T, Liu Z, Shi Y, Peng Y. HOXC10 up-regulation contributes to human thyroid cancer and indicates poor survival outcome. Mol Biosyst. 2015;11:2946-54
25. Tang XL, Ding BX, Hua Y, Chen H, Wu T, Chen ZQ. et al. HOXC10 promotes the metastasis of human lung adenocarcinoma and indicates poor survival outcome. Front Physiol. 2017;8:557
26. Li S, Zhang W, Wu C, Gao H, Yu J, Wang X. et al. HOXC10 promotes proliferation and invasion and induces immunosuppressive gene expression in glioma. FEBS J. 2018;285:2278-91
27. Li J, Zhang N, Song LB, Liao WT, Jiang LL, Gong LY. et al. Astrocyte elevated gene-1 is a novel prognostic marker for breast cancer progression and overall patient survival. Clin Cancer Res. 2008;14:3319-26
28. Celerier J, Cruz A, Lamande N, Gasc JM, Corvol P. Angiotensinogen and its cleaved derivatives inhibit angiogenesis. Hypertension. 2002;39:224-8
29. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545-50
30. Kunnumakkara AB, Guha S, Krishnan S, Diagaradjane P, Gelovani J, Aggarwal BB. Curcumin potentiates antitumor activity of gemcitabine in an orthotopic model of pancreatic cancer through suppression of proliferation, angiogenesis, and inhibition of nuclear factor-kappaB-regulated gene products. Cancer Res. 2007;67:3853-61
31. Chen X, Ying Z, Lin X, Lin H, Wu J, Li M. et al. Acylglycerol kinase augments JAK2/STAT3 signaling in esophageal squamous cells. J Clin Invest. 2013;123:2576-89
32. Seki H, Hayashida T, Jinno H, Hirose S, Sakata M, Takahashi M. et al. HOXB9 expression promoting tumor cell proliferation and angiogenesis is associated with clinical outcomes in breast cancer patients. Ann Surg Oncol. 2012;19:1831-40
33. Zhu JY, Sun QK, Wang W, Jia WD. High-level expression of HOXB13 is closely associated with tumor angiogenesis and poor prognosis of hepatocellular carcinoma. Int J Clin Exp Pathol. 2014;7:2925-33
34. Storti P, Donofrio G, Colla S, Airoldi I, Bolzoni M, Agnelli L. et al. HOXB7 expression by myeloma cells regulates their pro-angiogenic properties in multiple myeloma patients. Leukemia. 2011;25:527-37
35. Leung DW, Cachianes G, Kuang WJ, Goeddel DV, Ferrara N. Vascular endothelial growth factor is a secreted angiogenic mitogen. Science. 1989;246:1306-9
36. Chen H, Lorton B, Gupta V, Shechter D. A TGFbeta-PRMT5-MEP50 axis regulates cancer cell invasion through histone H3 and H4 arginine methylation coupled transcriptional activation and repression. Oncogene. 2017;36:373-86
37. Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359:492-507
38. Onishi M, Ichikawa T, Kurozumi K, Date I. Angiogenesis and invasion in glioma. Brain Tumor Pathol. 2011;28:13-24
39. Plate KH, Scholz A, Dumont DJ. Tumor angiogenesis and anti-angiogenic therapy in malignant gliomas revisited. Acta Neuropathol. 2012;124:763-75
40. Castro BA, Flanigan P, Jahangiri A, Hoffman D, Chen W, Kuang R. et al. Macrophage migration inhibitory factor downregulation: a novel mechanism of resistance to anti-angiogenic therapy. Oncogene. 2017;36:3749-59
41. Bergers G, Hanahan D. Modes of resistance to anti-angiogenic therapy. Nat Rev Cancer. 2008;8:592-603
42. Chinot OL, Wick W, Mason W, Henriksson R, Saran F, Nishikawa R. et al. Bevacizumab plus radiotherapy-temozolomide for newly diagnosed glioblastoma. N Engl J Med. 2014;370:709-22
43. Herrlinger U, Schafer N, Steinbach JP, Weyerbrock A, Hau P, Goldbrunner R. et al. Bevacizumab plus irinotecan versus temozolomide in newly diagnosed O6-methylguanine-DNA methyltransferase nonmethylated glioblastoma: the randomized GLARIUS trial. J Clin Oncol. 2016;34:1611-9
44. Hughes P, Marshall D, Reid Y, Parkes H, Gelber C. The costs of using unauthenticated, over-passaged cell lines: how much more data do we need? BioTechniques. 2007;43:575 7-8, 81-2 passim
45. Torsvik A, Stieber D, Enger PO, Golebiewska A, Molven A, Svendsen A. et al. U-251 revisited: genetic drift and phenotypic consequences of long-term cultures of glioblastoma cells. Cancer Med. 2014;3:812-24
46. Balbous A, Cortes U, Guilloteau K, Villalva C, Flamant S, Gaillard A. et al. A mesenchymal glioma stem cell profile is related to clinical outcome. Oncogenesis. 2014;3:e91
47. Huszthy PC, Daphu I, Niclou SP, Stieber D, Nigro JM, Sakariassen PO. et al. In vivo models of primary brain tumors: pitfalls and perspectives. Neuro Oncol. 2012;14:979-93
48. Joo KM, Kim J, Jin J, Kim M, Seol HJ, Muradov J. et al. Patient-specific orthotopic glioblastoma xenograft models recapitulate the histopathology and biology of human glioblastomas in situ. Cell Rep. 2013;3:260-73
49. Chiang K, Zielinska AE, Shaaban AM, Sanchez-Bailon MP, Jarrold J, Clarke TL. et al. PRMT5 is a critical regulator of breast cancer stem cell function via histone methylation and FOXP1 expression. Cell Rep. 2017;21:3498-513
50. Fu T, Lv X, Kong Q, Yuan C. A novel SHARPIN-PRMT5-H3R2me1 axis is essential for lung cancer cell invasion. Oncotarget. 2017;8:54809-20
51. Deng X, Shao G, Zhang HT, Li C, Zhang D, Cheng L. et al. Protein arginine methyltransferase 5 functions as an epigenetic activator of the androgen receptor to promote prostate cancer cell growth. Oncogene. 2017;36:1223-31
52. Rosenbaum-Dekel Y, Fuchs A, Yakirevich E, Azriel A, Mazareb S, Resnick MB. et al. Nuclear localization of long-VEGF is associated with hypoxia and tumor angiogenesis. Biochem Biophys Res Commun. 2005;332:271-8
53. Shen CC, Cheng WY, Chiao MT, Liang YJ, Mao TF, Liu BS. Two novel heparin-binding vascular endothelial growth factor splices, L-VEGF144 and L-VEGF138, are expressed in human glioblastoma cells. Curr Neurovasc Res. 2016;13:207-18
54. Cheng WY, Shen CC, Chiao MT, Liang YJ, Mao TF, Liu BS. et al. High expression of a novel splicing variant of VEGF, L-VEGF144 in glioblastoma multiforme is associated with a poorer prognosis in bevacizumab treatment. J Neurooncol. 2018 [Epub ahead of print]
Author contact
![]() Corresponding author: Jun Li, Ph.D. Program of cancer research, Affiliated Guangzhou Women and Children's Hospital, Department of Biochemistry, Zhongshan School of Medicine, Sun Yat-sen University. Phone: +86(20)87335828; Fax: +86(20)87335828; E-mail: lijun37sysu.edu.cn
Corresponding author: Jun Li, Ph.D. Program of cancer research, Affiliated Guangzhou Women and Children's Hospital, Department of Biochemistry, Zhongshan School of Medicine, Sun Yat-sen University. Phone: +86(20)87335828; Fax: +86(20)87335828; E-mail: lijun37sysu.edu.cn
 Global reach, higher impact
Global reach, higher impact