13.3
Impact Factor
Theranostics 2018; 8(14):4003-4015. doi:10.7150/thno.24106 This issue Cite
Research Paper
circEPSTI1 as a Prognostic Marker and Mediator of Triple-Negative Breast Cancer Progression
Department of Breast Oncology, Sun Yat-Sen University Cancer Center, State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Guangzhou, 510060, China
*These authors contributed equally to this work
Received 2017-11-30; Accepted 2018-5-31; Published 2018-7-1
Abstract
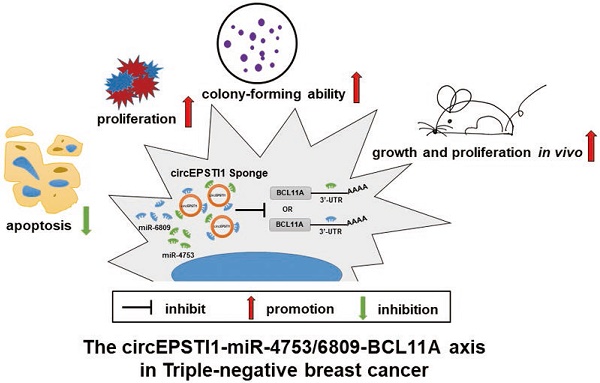
Circular RNAs (circRNAs) represent a class of non-coding RNAs that play a vital role in modulating gene expression and several pathological responses. However, the expression profile and function of circRNAs in triple-negative breast cancer (TNBC) remain unknown. In the current study, we investigated the expression profile of human circRNAs in TNBC tissues and identified circEPSTI1 (hsa_ circRNA_000479) as a significantly upregulated circRNA.
Methods: We performed circular RNA microarray assays to screen circular RNA expression profiles of TNBC and further investigated circEPSTI1. We observed the effect of circEPSTI1 on proliferation, clonal formation and apoptosis in TNBC by knocking downcircEPSTI1 in three TNBC cell lines. Based on the MRE analysis and luciferase reporter assay, we found that circEPSTI1 binds to miRNAs as a miRNA sponge and the co-target genes of miRNAs. We performed xenograft experiments in mice to confirm our findings. We evaluated circEPSTI1 levels in 240 TNBC patients by ISH.
Results: Knockdown of circEPSTI1 inhibits TNBC cell proliferation and induces apoptosis. In vitro and in vivo experiments indicated that circEPSTI1 binds to miR-4753 and miR-6809 as a miRNA sponge to regulate BCL11A expression and affect TNBC proliferation and apoptosis. High levels of circEPSTI1 correlate with reduced survival in TNBC patients.
Conclusions: The circEPSTI1-miR-4753/6809-BCL11A axis affect the proliferation and apoptosis of triple-negative breast cancer through the mechanism of competing endogenous RNAs (ceRNA). In addition, our results identify circEPSTI1 as an independent prognostic marker for survival in patients with TNBC.
Keywords: triple-negative breast cancer, circEPSTI1, circular RNAs, BCL11A, competing endogenous RNAs
Introduction
Triple-negative breast cancer (TNBC) is the most aggressive breast cancer subtype, exhibiting increased recurrence and decreased survival. Approximately 15 to 20 percent of breast cancer cases are categorized as this subtype [1]. Due to absence of druggable molecular targets, TNBC treatment is very limited compared with the treatments for luminal or HER2+ subtypes [2]. Thus, there is considerable interest in obtaining a better understanding of TNBC prognosis, developing novel prognostic markers and identifying new anticancer molecular targets.
Although more than 90% of the human genome is actively transcribed, only 1-2% of the genome sequence encodes proteins, whereas the vast majority of sequences are devoted to the expression of noncoding RNAs (ncRNAs) [3]. During the past two decades, the discovery of non-coding RNA involved in carcinogenesis represents one of the most important scientific discoveries. Numerous studies reflect the broad interest of the scientific community in non-coding RNA. microRNAs (miRNAs) and long non-coding RNAs (lncRNAs) are two well-known subclasses of non-coding RNA [4]. These RNAs modulate the expression of targeted genes and intercellular signaling within the tumor microenvironment. Furthermore, previous studies have analyzed the expression patterns of miRNAs and lncRNAs in TNBC samples, demonstrating that miRNAs and lncRNAs are vital players in TNBC processes [5, 6].
Compared with the familiar non-coding RNA subclass, circular RNA (circRNA) represents a novel research hot spot in the non-coding RNA research field [7]. Circular RNAs are covalently closed, single-stranded transcripts without 5′ caps and 3′ tails that range in length from a few hundred to thousands of nucleotides and are widely express in mammals [8]. Circular RNAs are highly stable compared with linear RNAs and are resistant to RNase R activity [9, 10]. Circular RNAs were initially thought to be artifacts of aberrant RNA splicing and functionless. With the development of bioinformatics and RNA-Seq technology, circRNAs have been shown to play important roles in disease progression, including cancers, as miRNA sponges, RNA-binding protein sponges, gene transcription and expression regulators and protein-coding genes. For example, CiRS‐7, one of the best known circRNAs, functions as a sponge for miR-7 and modulates the expression of several cancer-related genes [11]. The circRNA SRY absorbs miR-138 [12], which is involved in ovarian cancer and colorectal cancer. cir-ITCH suppresses the Wnt/ β-catenin pathway by absorbing miR-7, miR-17 and miR-214 [13]. Circ-Foxo3 is highly expressed in non-cancer cells, and the circ-Foxo3-p21-CDK2 ternary complex inhibits the function of CDK2 and blocks cell cycle progression [14]. Circ-ZNF609 provides an example of a protein-coding circRNA in eukaryotes that is translated into a protein in a splicing-dependent and cap-independent manner [15]. In addition, circRNAs are enriched in exosomes [16], thus laying the foundation for development of circRNAs as a new class of cancer biomarker. Recently, we performed circRNA microarrays to identify circRNAs that are aberrantly expressed in TNBC cell lines and found that circGFRA1 could sponge miR-34a to exert regulatory functions in TNBC [17]. However, no reports describing the role of circRNAs in TNBC based on patient samples are currently available. Potential function and biogenesis processes involving circular RNAs in breast cancer, especially TNBC, remain poorly understood.
In this study, we analyzed the expression profiles of circRNAs in TNBC using microarray data from three TNBC patients. We characterized a significantly upregulated circRNA, circRNA_000479, which is derived from the EPSTI1 (epithelial stromal interaction 1) gene locus, termed circEPSTI1. In functional assays, knockdown of circEPSTI1 inhibits TNBC cell proliferation and induces apoptosis. circEPSTI1 binds to miR-4753 and miR-6809 as a miRNA sponge to regulate BCL11A expression. Both in vitro and vivo experiments indicate that circEPSTI1-miR-4753/6809-BCL11A axis affect TNBC proliferation and apoptosis. circEPSTI1 was further confirmed to be a prognostic marker for survival in TNBC patients. Our research indicates that circRNAs can be used as potential targets in TNBC therapy and prognostic biomarkers.
Methods
Patient samples
We collected tumor tissues and their adjacent normal mammal tissues from TNBC patients who received treatment at Sun Yat-Sen University Cancer Centre (SYSUCC). No patient received neoadjuvant therapy. The resected tissues were immediately cut and stored in RNAlater (Ambion, Austin, Texas). This study was performed in accordance with the ethical standards formulated in the Declaration of Helsinki and approved by the Ethics Committee of Sun Yat-Sen University Cancer Centre Health Authority (RDDB2018000317). All participants provided written informed consent prior to treatment.
Microarray analysis
Three TNBC tumor and matched normal mammalian tissue samples from patients were analyzed using Arraystar Human circRNA Array V2. Total RNA from each sample was quantified using the NanoDrop ND-1000. The sample preparation and microarray hybridization were performed based on the Arraystar's standard protocols. Briefly, total RNAs were digested with Rnase R (Epicentre Technologies, Madison, WI, USA) to remove linear RNAs and enrich circular RNAs. Then, the enriched circular RNAs were amplified and transcribed into fluorescent cRNA utilizing a random priming method (Arraystar Super RNA Labeling Kit; Arraystar). The labeled cRNAs were hybridized onto the Arraystar Human circRNA Array V2 (8x15K, Arraystar). After washing the slides, the arrays were scanned using the Agilent Scanner G2505C. Agilent Feature Extraction software (version 11.0.1.1) was used to analyze acquired array images. Quantile normalization and subsequent data processing was performed using the R software limma package. Differentially expressed circRNAs between two samples were identified through volcano plot filtering and fold change filtering. Fold-changes of ≥ 1.5 and P < 0.05 in microarray data were regarded as significantly differentially expressed. Hierarchical clustering was performed to demonstrate the distinguishable circRNAs expression pattern among samples. We further confirmed the microarray results of four circRNAs in 10 paired TNBC tissues and matched normal tissues by qRT-PCR.
Cell culture and treatment
All cell lines, including normal mammary epithelial cell lines (MCF-10A,184A1), human breast cancer cell lines (HCC38, HCC1806, BT549, MDA-MB-231, MDA-MB-468, MDA-MB-415, MCF-7, T47D, BT474 and Skbr-3) and human embryonic kidney 293T cells (HEK 293T cells), were obtained from the American Type Culture Collection (Manassas, VA, USA). All the cell lines were passaged in our laboratory for less than six months and maintained according to the supplier's instructions, and mycoplasma infection and authenticity were verified by DNA fingerprinting before use.
Quantitative reverse transcription polymerase chain reaction (qRT-PCR)
Total RNA was isolated using TRIzol reagent (Life Technologies, Carlsbad, CA, USA). The nuclear and cytoplasmic fractions were isolated using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Scientific). Complementary DNA was synthesized using the PrimeScript RT reagent kit (Takara Bio Inc., Dalian, China), and RT-PCR were performed using SYBR Premix Ex Taq (Takara Bio Inc.) and All-in-One™ miRNA qRT-PCR Detection Kit (GeneCopoeia, Rockville, MD, USA). Primer information is provided in Table S1.
RNase R digestion
Total RNA (2 μg) was incubated for 20 min at 37 °C with or without 3 U/μg of RNase R. The resulting RNA was purified using an RNeasy MinElute Cleanup Kit (Qiagen).
Actinomycin D assay
MDA-MB-231 cells were exposed to 2 μg /mL actinomycin D (Sigma) to block transcription for 8, 16, and 24 h. The cells were harvested, and the stability of the circEPSTI1 and EPSTI1 mRNA were analysed using qRT-PCR.
Oligonucleotide transfection
Cells were transfected using Lipofectamine 2000 (Invitrogen Life Technologies, Carlsbad, CA, USA). All siRNAs targeting circEPSTI1 were synthesized by GenePharma (Shanghai, China). All miRNA mimics or inhibitors were synthesized by GeneCopoeia (Rockville, MD, USA). The sequences that were used are presented in Table S1.
CCK8 assay
Cell proliferation was assessed using the Cell Counting Kit-8 assay (Dojindo Laboratories, Kumamoto, Japan). The cells (1×103) were seeded into 96-well plates. CCK-8 solution (10 μL) was added to each well on days 1, 2, 3, 4 and 5. After 2 h of incubation at 37 °C, the absorbance at 450 nM was measured using a microtiter plate reader (Bio-Tek EPOCH2, BioTek Instrument, Inc., USA).
5-Ethynyl-2′-deoxyuridine (EdU) assay and apoptosis assay
The IClick™ EdU Andy Fluor™ 488 Flow Cytometry Assay Kit (GeneCopoeia, Rockville, MD, USA) was used to assess the proliferative activity. The 5-ethynyl-2′-deoxyuridine (EdU, 2 μM) was added to MDA-MB-231, BT549 and MDA-MB-468 for 12 h before termination of the cultures. Flow cytometry analysis was performed per the kit's instructions. A total of 100,000 events were recorded for the analysis. Annexin V/propidium iodide staining and flow cytometry were performed using the Andy Fluor 488 Annexin V/PI Kit (GeneCopoeia, Rockville, MD, USA) according to the manufacturer's guidelines.
Colony formation assay
For the colony formation assays, the cells were resuspended and plated in 6-well plates at a density of 1×103 cells/well and incubated at 37 °C for 14 days. Colonies were fixed with absolute methanol and stained with 0.1% crystal violet for 20 min. Cell colonies were then counted and analyzed.
Luciferase reporter assay
HEK 293T cells (5×103) were seeded into 96-well plates and co-transfected with corresponding plasmids and microRNA mimics or inhibitors using the Lipofectamine 2000 transfection reagent. Luciferase activity was measured using the dual-luciferase reporter assay system (Promega, Madison, WI, USA) after 48 h of incubation according to the manufacturer's instructions. Independent experiments were performed in triplicate. Relative luciferase activity was normalized to the Renilla luciferase internal control.
Plasmid constructs
circEPSTI1 was amplified from human genomic DNA and was cloned into the downstream of a CMV promoter-driven firefly luciferase cassette in a pCDNA3.0 vector. Mutations in the miRNA binding sites in the circEPSTI1 sequence were introduced by Fast Site-Directed Mutagenesis Kit (TIANGEN, Beijing, China). The primers are listed in Table S1. All the constructs were confirmed by sequencing.
Western blot analysis
Protein was extracted using RIPA lysis buffer with a proteinase inhibitor and quantified by a BCA kit (Thermo, USA). The proteins in the lysates were separated by 10% SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, USA). The membranes were incubated at room temperature for 1 h with 5% skim milk powder to block nonspecific binding. The membranes were then incubated overnight at 4 °C with antibodies against BCL11A (Abcam, 1:1000), EPSTI1 (Santa cruz,1:500) and β-actin (Cell Signaling, 1:5000). A secondary antibody (1:5000 dilution) and ECL Western blotting detection reagents were used to visualize the target proteins (ECL New England Biolabs, USA).
Mouse xenograft model
Ethical approval was obtained from the Institute Research Ethics Committee of SYSUCC, and all the animal procedures were performed in accordance with institutional guidelines. Approximately 1×107 cells/mL MDA-MB-231, BT549 and MDA-MB-468 cells were subcutaneously injected into the dorsal flanks of 4-week-old female BALB/c nude mice using 1-mL syringes, and mice were treated with intertumoral injection of negative control (40 μL negative control siRNA) and si-crEPSTI1-1 (40 μL cicrEPSTI1 siRNA1) in PBS respectively (five in each group) every four days respectively. Tumor sizes were measured every four days. The mice were euthanized and xenografts were taken out after 28 days. The tumor volumes (mm3) were determined according to the following formula: volume = (shortest diameter)2 × (longest diameter) × 0.5. The tumors from individual mice were determined by immunohistochemistry with antibodies against BCL11A (Abcam, 1:300), Ki-67 (Cell Signaling, 1:300), Caspase-3 (Proteintech, 1:200) and BrdU (Proteintech, 1:200).
In situ hybridization (ISH) and immunohistochemistry (IHC) analysis
The circEPSTI1 detection probe (Exiqon, Vedbaek, Denmark) was used for ISH according to a previously described standard method [18]. The sequence of the circEPSTI1 probe for ISH was Digoxin-5'- AGA GCA TCA GCA ATA CAC AAG TGC ATA CAC -3'-Digoxin. IHC studies were performed using a standard streptavidin-biotin-peroxidase complex method as described previously [19]. The intensities of circEPSTI1 and BCL11A staining were scored using a previously proposed semi-quantitative approach based on staining intensity: 0-1 (no staining), 1-2 (weak staining), 2-3 (medium staining), and 3-4 (strong staining).
Statistical analysis
All statistical analyses were performed using the SPSS22.0 software package (SPSS, Chicago, IL, USA). Means ± standard deviation (SD) were used to present quantitative data. The chi-squared test was used to investigate the significance of the correlation of circEPSTI1 expression with clinicopathological features in TNBC. Survival curves were calculated by the Kaplan-Meier method and compared with the log-rank test. Survival was measured from the day of the surgery. P < 0.05 was considered statistically significant.
Results
Identification of differentially circular RNAs in TNBC
To investigate the potential involvement of circRNAs in TNBC, we performed high-throughput circular RNA microarray assays using three pairs of TNBC patient samples (Table S3). Between TNBC cancerous tissues and non-cancerous matched tissue (NCMT), hierarchical clustering revealed circular RNA expression patterns (Figure 1A shows the top five up-regulated and down-regulated circRNAs). The variation of circular RNA expression was revealed in the volcano (Figure 1B) and scatter plots (Figure 1C). As a result, 173 circular RNAs were up-regulated, whereas 77 circular RNAs were down-regulated based on fold change ≥1.5, P < 0.05 and FDR < 0.05. The distribution of the differentially expressed circular RNAs on the human chromosomes is presented in Figure 1D. In total, 150 exonic, 8 intronic, 12 sense overlapping and 3 antisense circRNAs were noted among increased circRNAs, and 66 exonic, 4 intronic, 6 sense overlapping and 1 antisense circRNAs were noted among decreased circRNAs (Figure 1E and Table S4). Given that circular RNAs are mainly derived from exons, most of the differentially expressed circular RNAs are transcribed from exons. The top ten up-regulated and down-regulated circRNAs are listed in Table S2, and we further confirmed the microarray results of four circRNAs in 10 paired TNBC tissues and matched normal tissues by qRT-PCR (Figure S1).
Identification of differentially circular RNAs in TNBC. (A) Clustered heat map of the differentially expressed circRNAs in three pairs of human TNBC cancerous tissues and adjacent normal tissues. circRNAs with increased (red) or decreased (green) expression are presented. (B) Volcano plots comparing the expression of circRNAs in TNBC cancerous tissues with adjacent normal tissues. The red dots represent the circRNAs up- and down-regulated by 1.5-fold and P-values <0.05. (C) A scatter plot was used to assess the variation in circRNA expression between TNBC cancerous tissues and adjacent normal tissues. The green lines are fold-change lines. (D) The differentially expressed circRNAs distribution rule in chromosomes. (E) Genomic origin of the differentially expressed circRNAs in TNBC. (F) The genomic loci of the EPSTI1 gene and circEPSTI1. The expression of circEPSTI1 was detected by qRT-PCR and validated by Sanger sequencing. (G) Expression levels of circEPSTI1 determined by qRT-PCR in mammary normal cell lines and breast cancer cell lines. Fold changes were normalized based on β-actin, and in each assay the error bars represent standard deviations (SD) from triplicates of one representative experiment. (H) Expression levels of circEPSTI1 in 37 paired TNBC specimens and the corresponding paired normal adjacent tissues. (I) qRT-PCR analysis of circEPSTI1 and EPSTI1 mRNA after treatment with RNase R in MDA-MB-231 cells. (J) qRT-PCR analysis of circEPSTI1 and EPSTI1 mRNA in MDA-MB-231 cells after treatment with Actinomycin D.
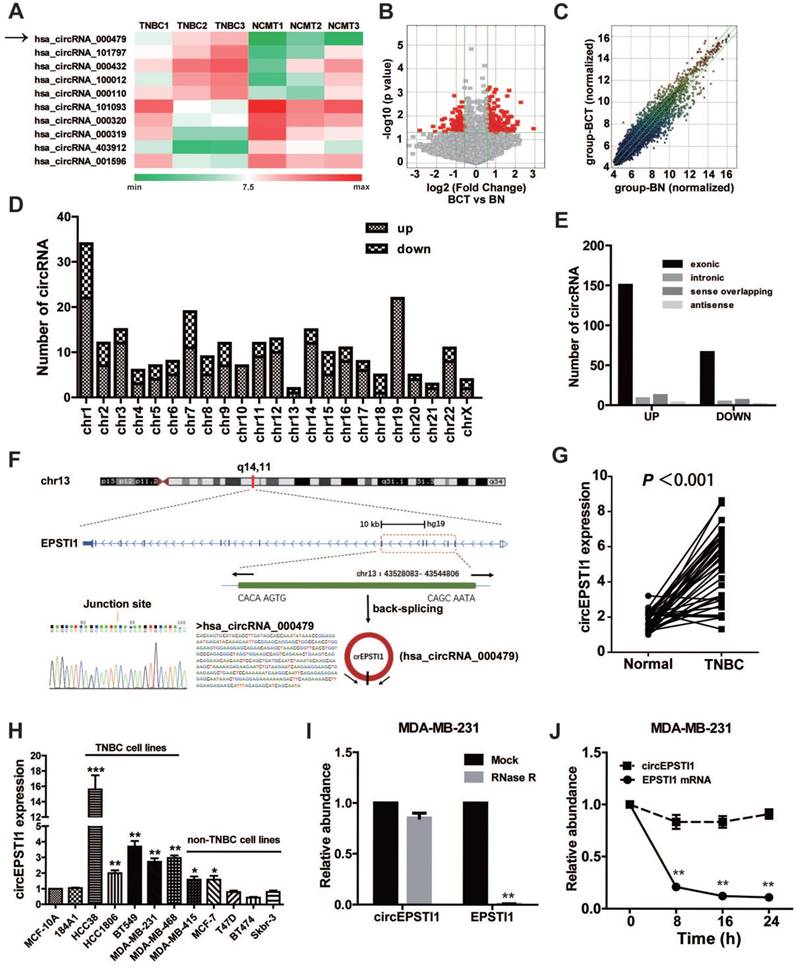
Characterization of circEPSTI1 circular RNA in TNBC
circRNA_000479 (chr13: 43528083- 43544806) was the most up-regulated circRNA, with a greater than sevenfold change. Using the human reference genome (GRCh37/hg19), we further assumed that circRNA_000479 is derived from EPSTI1, which is located on chromosome 13q14. Chromosome 13 carries some important genes associated with breast cancer, including BRCA2 and RB1 genes [20]. We termed circRNA_000479 as “circEPSTI1,” which was amplified by outward-facing primers and confirmed by Sanger sequencing (Figure 1F). We tested circEPSTI1 expression levels in a panel of 12 breast cell lines, including 10 human breast cancer cell lines and 2 normal mammary epithelial cell lines. As shown in Figure 1H, circEPSTI1 levels are up-regulated in 7 human breast cancer cell lines, especially in TNBC cell lines (including HCC38, HCC1806, BT549, MDA-MB-231 and MDA-MB-468). In addition, circEPSTI1 levels were evaluated in tissue samples from 38 pairs of TNBC tissues and their matched adjacent normal tissue. Among those TNBC patients, more than 80% (33/38) of the tumors exhibited increased circEPSTI1 levels (Figure 1G) (P < 0.001). The results indicate that circEPSTI1 was significantly increased in TNBC samples. In addition, circEPSTI1 was preferentially localized within the cytoplasm, as assessed by qRT-PCR analysis of nuclear and cytoplasmic RNAs (Figure S2). Resistance to digestion by RNase R exonuclease further confirmed that this RNA species is circular (Figure 1I). In addition, Actinomycin D assays revealed that the circular RNA isoform circEPSTI1 transcript half-life exceeded 24 h, indicating that this isoform is more stable than the EPSTI1 linear transcript in MDA-MB-231 cells (Figure 1J).
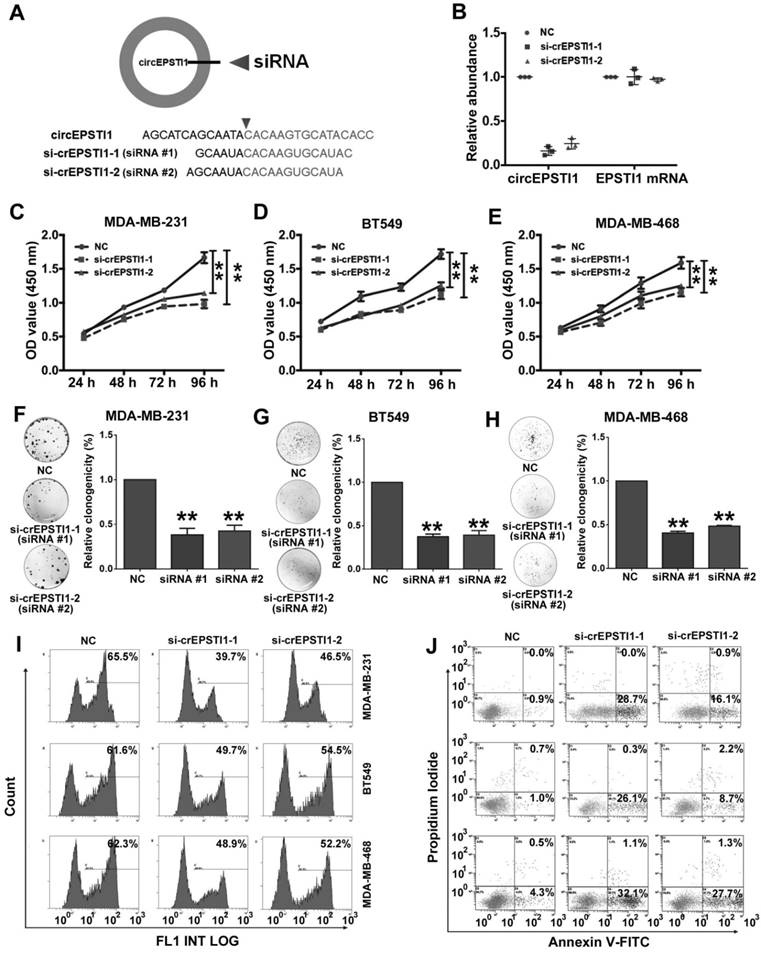
Silencing of circEPSTI1 inhibits TNBC growth and proliferation and induces apoptosis
Next, we used RNA interference to silence the expression of circEPSTI1 in three TNBC cell lines (MDA-MB-231, BT549 and MDA-MB-468) to evaluate its biological functions. We designed two siRNAs to target the back-splice sequence of circEPSTI1 that did not affect the expression of the EPSTI1 linear species (Figure 2A-B) and EPSTI1 protein levels (Figure S3). Subsequent cell proliferation assays revealed that down-regulation of circEPSTI1 significantly suppressed the growth of MDA-MB-231, BT549 and MDA-MB-468 (Figure 2C-E). In colony formation assays, the colony-forming ability of the three TNBC cell lines was significantly reduced after down-regulation of circEPSTI1 expression (Figure 2F-H). Furthermore, we used an EdU assay to evaluate proliferative potential. TNBC cell proliferation was impaired upon the down-regulation of circEPSTI1 expression (Figure 2I and Figure S4A). Next, we evaluated the apoptotic effect of down-regulation of circEPSTI1 in MDA-MB-231, BT549 and MDA-MB-468. Cells were transfected for 48 h. Cell apoptosis was measured by Annexin V-FITC/propidium iodide staining. Our results indicate that circEPSTI1 silencing induces apoptosis in TNBC cell lines (Figure 2J and Figure S4B).
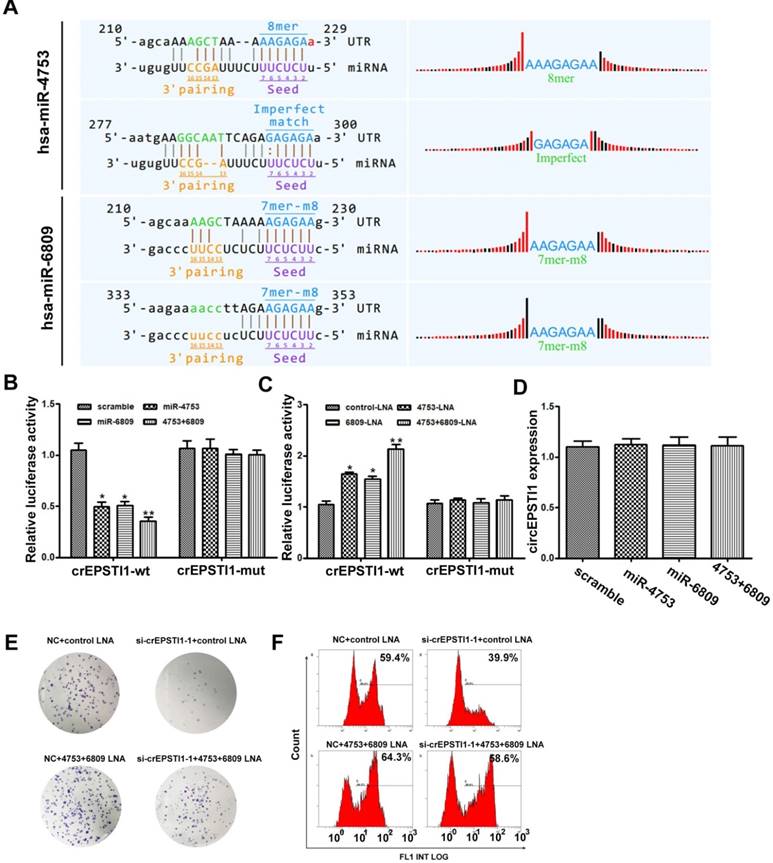
circEPSTI1 serves as a miRNAs sponge for miR-4753 and miR-6809
Subsequently, we explored whether circEPSTI1 binds to miRNAs as a miRNA sponge. According to MREs analysis, the top 5 miRNAs were miR-942, miR-4753, miR-6809, miR-6873 and miR-6739, in accordance with the positions of putative binding sites in the circEPSTI1 sequence. Luciferase reporter assays were applied to determine whether these miRNAs directly target circEPSTI1. Potential binding sites of miR-4753 and miR-6809 were identified within the circEPSTI1 sequence. Each miRNA has two potential binding sites within the circEPSTI1 sequence (Figure 3A). HEK 293T cells were co-transfected with each miRNA mimic and luciferase reporters for luciferase activity assays. Luciferase intensity was reduced by greater than 50% in response to miR-4753 and miR-6809 mimics (Figure 3B). To confirm the direct interaction, the MREs of miR-4753 and miR-6809 were mutated in the luciferase reporter. We found that co-transfection of miRNA mimics and the mutated luciferase reporter had no significant effect on luciferase activity (Figure 3B). We also co-transfected with each miRNA LNA and luciferase reporters for luciferase activity assays to conform our findings (Figure 3C). In addition, qRT-PCR analysis revealed that miR-4753 and miR-6809 did not significantly decrease the level of circEPSTI1, demonstrating that circEPSTI1 is not digested by miR-4753 and miR-6809 (Figure 3D). These data suggest that circEPSTI1 may serve as a sponge for miR-4753 and miR-6809. Moreover, knockdown of circEPSTI1 attenuated the knockdown effects of miR-4753 and miR-6809 in colony formation assays (Figure 3E) and EdU assays (Figure 3F) in MDA-MB-231.
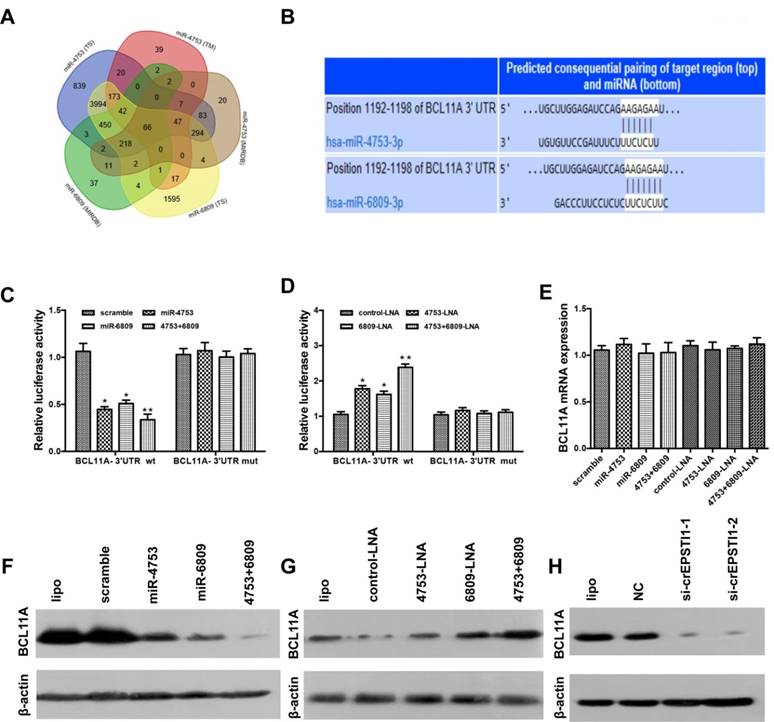
BCL11A is a direct target gene of miR-4753 and miR-6809 and is inhibited via circEPSTI1 knockdown
We used three algorithms (TargetScan [21], MIRDB [22] and TARGETMINER [23]) to identify the putative co-targets genes of miR-4753 and miR-6809 in human breast cancer (Figure 4A). Among these candidate target genes, algorithms predicted BCL11A as a target (Figure 4B). BCL11A is overexpressed in TNBC, and its genomic locus is amplified in up to 38% of basal-like breast cancer tumors [24]. To confirm this finding, we constructed a luciferase reporter vector with the full-length BCL11A 3′-UTR containing target sites for miR-4753 and miR-6809 downstream of the luciferase gene (pGL3-BCL11A wt-3'UTR) and a mutant version of pGL3-BCL11A-3'UTR within the seed region (pGL3-BCL11A mut-3'UTR).
Silencing of circEPSTI1 inhibits TNBC growth and proliferation and induces apoptosis. (A) Schematic representation of the sites of the siRNA specific to the back-splice junction of circEPSTI1. (B) qRT-PCR analysis of circEPSTI1 and EPSTI1 mRNA expression after treatment with two siRNAs. (C-E) CCK-8 assay in MDA-MB-231, BT549 and MDA-MB-468 cells transfected with control or circEPSTI1 siRNAs. (F-H) Colony formation assay of MDA-MB-231, BT549 and MDA-MB-468 cells transfected with control or circEPSTI1 siRNAs. (I) Assessment of DNA synthesis using an EdU assay in MDA-MB-231, BT549 and MDA-MB-468 cells transfected with control or circEPSTI1 siRNAs. (J) Annexin V-FITC/propidium iodide staining and FACS quantification of the number of apoptotic cells in MDA-MB-231, BT549 and MDA-MB-468 cells with indicated treatments. **P < 0.01.
circEPSTI1 serves as a miRNAs sponge for miR-4753 and miR-6809. (A) Schematic of the predicted miR-4753 and miR-6809 sites in the circEPSTI1. (B-C) Luciferase assay of HEK 293T cells co-transfected with a scrambled control, miR-4753 mimics, miR-6809 mimics, miR-4753+ miR-6809 mimics, control-LNA, miR-4753-LNA, or miR-4753+ miR-6809-LNA and a luciferase reporter containing circEPSTI1-wt or mutant constructs with mutated miR-4753 and miR-6809 binding sites (circEPSTI1-mut). *P < 0.05; **P < 0.01. (D) qRT-PCR analysis of circEPSTI1 after transfection with a scrambled control, miR-4753 mimics, miR-6809 mimics or miR-4753+ miR-6809 mimics. (E) Colony formation assay of MDA-MB-231 transfected with NC+ control-LNA, si-crEPSTI1-1+ control-LNA, NC+ miR-4753+ miR-6809-LNA or si-crEPSTI1-1+ miR-4753+ miR-6809-LNA. (F) 5-Ethynyl-2′-deoxyuridine (EdU) assay of MDA-MB-231 cells transfected with NC+ control-LNA, si-crEPSTI1-1+ control-LNA, NC+ miR-4753+ miR-6809-LNA or si-crEPSTI1-1+ miR-4753+ miR-6809-LNA.
Luciferase reporter vector alone or together with miR-4753, miR-6809 or both was transfected into HEK 293T. A significant decrease in luciferase activity was observed when pGL3- BCL11A wt-3'UTR was co-transfected with miR-4753, miR-6809 or both but not with scrambled oligonucleotide (Figure 4C), However, no significant difference in luciferase activity was noted among scrambled oligonucleotide, miR-4753, miR-6809 or both, which were co-transfected with pGL3-BCL11A mut-3'UTR (Figure 4C). In contrast, a significant increase in luciferase activity was observed when pGL3-BCL11A wt-3'UTR was co-transfected with miR-4753 inhibitors, miR-6809 inhibitors or both but not with control-LNA oligonucleotide in HEK 293T cells (Figure 4D). Meanwhile, we tested si-circEPSTI1 effects on BCL11A-3'UTR and found a significant decrease in luciferase activity (Figure S5). qRT-PCR and Western blot analyses were performed to confirm the luciferase assay results. The results revealed that miR-4753 and miR-6809 did not change the mRNA level (Figure 4E), but notable reductions in BCL11A protein levels were noted compared with the negative control (Figure 4F-G). Furthermore, we found that knockdown of circEPASTI1 also reduced the BCL11A protein levels (Figure 4H).
BCL11A is a direct target gene of miR-4753 and miR-6809 and is inhibited by circEPSTI1 knockdown. (A) Venn diagram representing the overlap of co-target genes of miR-4753 and miR-6809 based on three algorithms (TargetScan (TS), MIRDB and TARGETMINER(TM)). The Venn diagram tool is available at http://bioinformatics.psb.ugent.be/webtools/Venn/. (B) Schematic of the predicted miR-4753 and miR-6809 sites in the 3′UTR of BCL11A mRNA. (C-D) Luciferase assay of HEK 293T cells co-transfected with a scrambled control, miR-4753 mimics, miR-6809 mimics, miR-4753+ miR-6809 mimics, control-LNA, miR-4753-LNA, or miR-4753+ miR-6809-LNA and a luciferase reporter containing BCL11A-3'UTR-wt or mutant constructs with mutated miR-4753 and miR-6809 binding sites (BCL11A-3'UTR-mut). (E) qRT-PCR analysis of BCL11A mRNA after transfection with scrambled control, miR-4753 mimics, miR-6809 mimics, miR-4753+ miR-6809 mimics, control-LNA, miR-4753-LNA or miR-4753+ miR-6809-LNA. (F-G) The effect of miR-4753 /miR-6809 mimics or miR-4753 /miR-6809 inhibitors on BCL11A protein expression in MDA-MB-231 cells was determined by Western blot. (H) The effect of knockdown of circEPSTI1 on BCL11A protein expression in MDA-MB-231 cells.
circEPSTI1-miR-4753/6809-BCL11A axis affects TNBC growth and proliferation in vivo
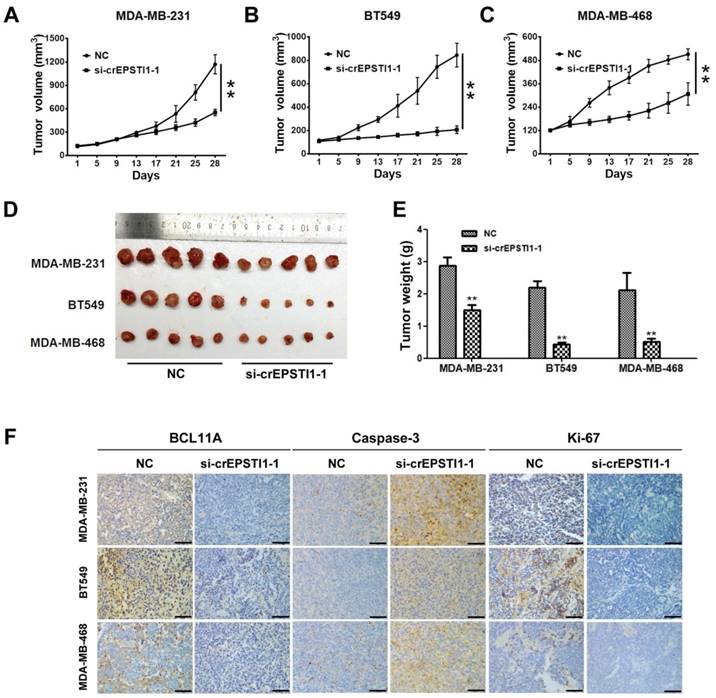
To investigate whether the circEPSTI1-miR-4753/6809-BCL11A axis affects TNBC growth in vivo, MDA-MB-231, BT549 and MDA-MB-468 cells treated with scrambled or si-cEPSTI1-1 siRNAs were subcutaneously injected into nude mice. After continuous intratumoral injection for two weeks, our results revealed that knockdown of circEPSTI1 reduced the tumor volume (Figure 5A-C) and tumor weight (Figure 5D-E) compared with scramble negative control among those three subcutaneous transplantation models. Furthermore, immunohistochemistry was used to detect BCL11A, caspase-3 and Ki67 protein expression in transplanted tumor tissues of each group. The expression of BCL11A and Ki67 were markedly reduced, whereas caspase-3 expression increased in tumor tissues of the si-cEPSTI1-1 groups (Figure 5F).
The circEPSTI1-miR-4753/6809-BCL11A axis affects TNBC growth and proliferation in vivo. (A) MDA-MB-231, (B) BT549 and (C) MDA-MB-468 cells were subcutaneously injected into nude mice and subject to different treatments (i.e., scramble or si-cEPSTI1-1). The growth curves of tumors are plotted. **P < 0.01. (D) Tumor in the mouse xenograft model. (E) The weights of the xenograft tumors. (F) The xenograft tumors were analyzed by immunohistochemistry analysis, and the representative images of BCL11A, caspase-2 and ki-67 expression are presented. Scale bar = 50 μm.
circEPSTI1 is positively correlated with BCL11A expression and serves as a prognosis factor in TNBC
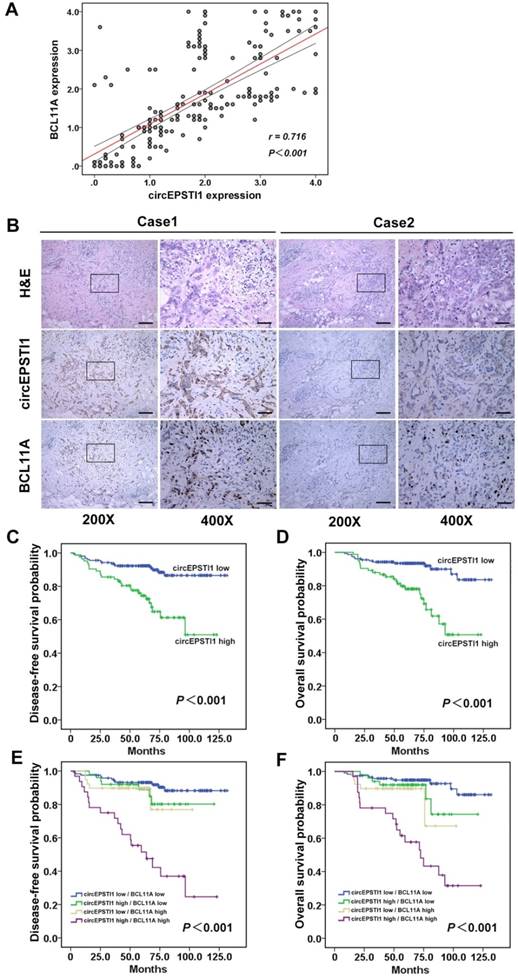
Further, we determined whether circEPSTI1 levels are associated with BCL11A levels in 240 TNBC tissue samples. ISH analysis revealed that 34.6% (83/240) of the tumor samples exhibited high circEPSTI1 expression levels, whereas the IHC analysis revealed that 29.6% (71/240) of the tumor samples exhibited high BCL11A expression. Increased circEPSTI1 expression was positively correlated with BCL11A expression in TNBC tissue (Figure 6A). Figure 6B presents representative H&E and in situ hybridization images of circEPSTI1 and immunohistochemistry images of BCL11A expression in TNBC cases. Then, we analyzed the association of circEPSTI1 expression with the clinicopathological status of TNBC patients. As shown in Table 1, circEPSTI1 expression was positively correlated with the tumor size, infiltrated lymph nodes and TNM stage (P < 0.001, = 0.004 and 0.008, respectively) of TNBC. Moreover, we analyzed the significance of circEPSTI1 and BCL11A in terms of clinical prognosis using patient disease-free survival (DFS) and overall survival (OS) to perform Kaplan- Meier survival analysis. The results revealed that patients with high circEPSTI1 expression exhibited reduced mean months of DFS and OS compared with patients with low circEPSTI1 expression (P < 0.001 for DFS and OS; Figure 6C-D). In addition, patients with high circEPSTI1 and BCL11A expression were significantly associated with reduced DFS and OS (Figure 6E-F).
circEPSTI1 is positively correlated with BCL11A expression and acts as a prognostic factor in TNBC. (A) Analysis of TNBC tissue microarray data (n=240) demonstrating linear regressions and significant Pearson correlations of circEPSTI1 with BCL11A. (B) Representative H&E and in situ hybridization images of circEPSTI1 and immunohistochemistry images of BCL11A expression in two TNBC cases (200× Scale bar = 100 μm; 400× Scale bar =50 μm;). (C-D) High level of circEPSTI1 is correlated with reduced DFS and OS (circEPSTI1 low n=157; circEPSTI1 high n=83). (E-F) The DFS and OS curves for the four possible combinations of circEPSTI1 and BCL11A expression are presented. High levels of circEPSTI1 and BCL11A are correlated with reduced survival (circEPSTI1 low/BCL11A low n=119; circEPSTI1 high/BCL11A low n=50; circEPSTI1 low/BCL11A high n=39; circEPSTI1 high/BCL11A high n=32).
Association between circEPSTI1 and clinicopathological characteristics in triple-negative breast cancer
| Variables | Cases (n=240) | CircEPSTI1 | P value | |
|---|---|---|---|---|
| Low No. (%) | High No. (%) | |||
| Age (years) | 0.285 | |||
| ≤50 | 145 | 91(62.8%) | 54(37.2%) | |
| >50 | 95 | 66(69.5%) | 29(30.5%) | |
| Menopause | 0.375 | |||
| no | 144 | 91(63.2%) | 53(36.8%) | |
| yes | 96 | 66(68.8%) | 30(31.3%) | |
| Tumor size | <0.001* | |||
| ≤2.0 cm | 66 | 59(89.4%) | 7(10.6%) | |
| >2.0 cm | 174 | 98(56.3%) | 76(43.7%) | |
| Lymph node infiltrated | 0.004* | |||
| No | 123 | 91(74.0%) | 32(26.0%) | |
| Yes | 117 | 66(56.4%) | 51(43.6%) | |
| TNM staging | 0.008* | |||
| I- II | 188 | 131(69.7%) | 57(30.3%) | |
| III- IV | 52 | 26(50.0%) | 26(50.0%) | |
| Histological grade | 0.640a | |||
| G1 | 3 | 3(100.0%) | 0(0.0%) | |
| G2 | 141 | 93(66.0%) | 48(34.0%) | |
| G3 | 96 | 61(63.5%) | 35(36.5%) | |
a. Using Fisher's exact test
* P < 0.05, statistically significant.
Discussion
CircRNAs have been increasingly investigated given the development of bioinformatics and high-throughput sequencing technologies. To date, more than ten thousand different circRNAs have been discovered in various organisms. Furthermore, circRNAs have the following characteristics: unique structure, conservation across species, cell type-specific and tissue-specific expression pattern, and stable expression in blood, saliva and exosomes. Thus, circRNAs are a hot topic in cancer research. Although studies on circRNAs are being performed at a rapid pace, many questions remain unanswered. Except for a few well-known circRNAs, such as CiRS‐7 and circSRY, the function of the majority of circRNAs remains unknown. In addition, little is known about the role of circRNAs in breast cancer, especially in triple-negative breast cancer. To our knowledge, this is the first report on the expression profile and regulatory function of circRNAs in TNBC. In this study, 173 up-regulated circRNAs and 77 down-regulated circRNAs were identified in TNBC tumor tissues compared with matched normal mammal tissues samples. Then, we characterized the top up-regulated circRNA, circRNA_000479 (circEPSTI1), which exhibits a miRNA sponge function in TNBC.
CircEPSTI1 is derived from the EPSTI1 gene, which is characterized by extensive epithelial-stromal interactions [25]. Previous studies have demonstrated that the EPSTI1 gene plays potential roles in innate immunity [26], and expression of this gene may be a crucial event in cancer invasion and metastasis [25]. Abnormalities in chromosome 13q14, which harbors EPSTI1 and circEPSTI1, is frequently observed in several types of cancer [27-30]. In our study, we demonstrated that circEPSTI1 was highly expressed in most TNBC cell lines and tissues. Silencing of circEPSTI1 inhibits TNBC cell line proliferation and induces apoptosis as demonstrated by loss-of-function assays. miRNA sponge activity represents the main mechanism of action of circRNAs in cancer. Our further studies revealed that circEPSTI1 exerts its regulatory functions through harboring miR-4753 and miR-6809 to reduce the expression of BCL11A, which is also directly targeted by miR-4753 and miR-6809. Previous studies have indicated that BCL11A is overexpressed in TNBC and the exogenous BCL11A overexpression promotes tumor formation. BCL11A deletion causes a reduction in the number of mammary epithelial stem and progenitor cells [24]. In our study, we demonstrated the presence of circEPSTI1-miR-4753/6809-BCL11A axis in mouse xenograft models of three TNBC cell lines and suggested that circEPSTI1 and BCL11A mRNA are ceRNAs that are associated with miR-4753 and miR-6809.
Emerging evidence indicates that circRNAs may potentially serve as a required novel biomarker and therapeutic target for cancer treatment. Galasso et al. explored the predictive value of circRNAs in breast cancer using a bioinformatics detection tool [31]. Nair et al. found that circRNAs may function as markers of cell proliferation in breast cancer and are associated with breast cancer subtypes [32]. The involvement of circRNAs in breast cancer has been explored but lacks experimental and clinical evidence. We found that high expression of circEPSTI1 was positively correlated with tumor size, lymph node infiltration and TNM stage and associated with poor prognosis as assessed by detecting the expression of circEPSTI1 in 240 cases of triple-breast cancer patients. In addition, circEPSTI1 is positively correlated with BCL11A expression in TNBC. TNBC patients with increased circEPSTI1 and BCL11A levels were significantly associated with reduced DFS and OS. Our findings offer the first evidence that a specific circRNA plays an important role as a prognostic marker in TNBC cases.
Taken together, the evidence indicates that circRNAs may also function in TNBC progression and exert regulatory functions by acting as a miRNA sponge. Through screening of the human circRNA array, we obtained greater than 200 different circular RNAs in TNBC and focused on the most obviously upregulated circRNA. We found that circEPSTI1-miR-4753/6809-BCL11A axis affects the proliferation and apoptosis of triple-negative breast cancer through a mechanism involving ceRNA. In addition, we demonstrated that circEPSTI1 is an independent prognostic marker for TNBC patient survival. Further research and attention on circRNAs will help us to better understand TNBC progression and offer new insight into the identification of biomarkers or potential therapeutic targets of TNBC.
Abbreviations
BCL11A: B cell CLL/lymphoma 11A; ceRNA: competing endogenous RNAs; circRNA: circular RNA; DFS: disease-free survival; EPSTI1: epithelial stromal interaction 1; IHC: immunohistochemistry; ISH: in situ hybridization; lncRNAs: long non-coding RNAs; NCMT: non-cancerous matched tissue; ncRNAs: noncoding RNAs; miRNAs: microRNAs; OS: overall survival. SD: standard deviation; TNBC: triple-negative breast cancer.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
This study was supported by funds from the National Natural Science Foundation of China (81672598 Xiaoming Xie, 81772961, Hailin Tang), the Science and Technology Planning Project of Guangdong and Guangzhou (2016A020214009 and 201607010173, Hailin Tang).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Carey L, Winer E, Viale G, Cameron D, Gianni L. Triple-negative breast cancer: disease entity or title of convenience? Nat Rev Clin Oncol. 2010;7:683-92
2. De Laurentiis M, Cianniello D, Caputo R, Stanzione B, Arpino G, Cinieri S. et al. Treatment of triple negative breast cancer (TNBC): current options and future perspectives. Cancer Treat Rev. 2010;36(Suppl 3):S80-6
3. International Human Genome Sequencing C. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931-45
4. Beermann J, Piccoli MT, Viereck J, Thum T. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev. 2016;96:1297-325
5. Koduru SV, Tiwari AK, Leberfinger A, Hazard SW, Kawasawa YI, Mahajan M. et al. A comprehensive NGS data analysis of differentially regulated miRNAs, piRNAs, lncRNAs and sn/snoRNAs in triple negative breast cancer. Journal of Cancer. 2017;8:578-96
6. Jiang YZ, Liu YR, Xu XE, Jin X, Hu X, Yu KD. et al. Transcriptome analysis of triple-negative breast cancer reveals an integrated mrna-lncrna signature with predictive and prognostic value. Cancer Res. 2016;76:2105-14
7. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A. et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333-8
8. Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J. et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. Rna. 2013;19:141-57
9. Lasda E, Parker R. Circular RNAs: diversity of form and function. Rna. 2014;20:1829-42
10. Chen J, Li Y, Zheng Q, Bao C, He J, Chen B. et al. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett. 2017;388:208-19
11. Weng W, Wei Q, Toden S, Yoshida K, Nagasaka T, Fujiwara T. et al. Circular RNA ciRS-7-A Promising Prognostic Biomarker and a Potential Therapeutic Target in Colorectal Cancer. Clin Cancer Res. 2017;23(14):3918-28
12. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK. et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384-8
13. Li F, Zhang L, Li W, Deng J, Zheng J, An M. et al. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/beta-catenin pathway. Oncotarget. 2015;6:6001-13
14. Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846-58
15. Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O. et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol Cell. 2017;66:22-37 e9
16. Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J. et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981-4
17. He R, Liu P, Xie X, Zhou Y, Liao Q, Xiong W. et al. circGFRA1 and GFRA1 act as ceRNAs in triple negative breast cancer by regulating miR-34a. J Exp Clin Cancer Res. 2017;36:145
18. Tang H, Deng M, Tang Y, Xie X, Guo J, Kong Y. et al. miR-200b and miR-200c as prognostic factors and mediators of gastric cancer cell progression. Clin Cancer Res. 2013;19:5602-12
19. Chen B, Tang H, Liu X, Liu P, Yang L, Xie X. et al. miR-22 as a prognostic factor targets glucose transporter protein type 1 in breast cancer. Cancer Lett. 2015;356:410-7
20. Dunham A, Matthews LH, Burton J, Ashurst JL, Howe KL, Ashcroft KJ. et al. The DNA sequence and analysis of human chromosome 13. Nature. 2004;428:522-8
21. Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015:4 e05005
22. Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015;43:D146-52
23. Bandyopadhyay S, Mitra R. TargetMiner: microRNA target prediction with systematic identification of tissue-specific negative examples. Bioinformatics. 2009;25:2625-31
24. Khaled WT, Choon Lee S, Stingl J, Chen X, Raza Ali H, Rueda OM. et al. BCL11A is a triple-negative breast cancer gene with critical functions in stem and progenitor cells. Nat Commun. 2015;6:5987
25. Nielsen HL, Ronnov-Jessen L, Villadsen R, Petersen OW. Identification of EPSTI1, a novel gene induced by epithelial-stromal interaction in human breast cancer. Genomics. 2002;79:703-10
26. Kosova G, Scott NM, Niederberger C, Prins GS, Ober C. Genome-wide association study identifies candidate genes for male fertility traits in humans. Am J Hum Genet. 2012;90:950-61
27. Heerema NA, Sather HN, Sensel MG, Lee MK, Hutchinson RJ, Nachman JB. et al. Abnormalities of chromosome bands 13q12 to 13q14 in childhood acute lymphoblastic leukemia. J Clin Oncol. 2000;18:3837-44
28. Hayakawa K, Formica AM, Colombo MJ, Shinton SA, Brill-Dashoff J, Morse Iii HC. et al. Loss of a chromosomal region with synteny to human 13q14 occurs in mouse chronic lymphocytic leukemia that originates from early-generated B-1 B cells. Leukemia. 2016;30:1510-9
29. Bandi N, Zbinden S, Gugger M, Arnold M, Kocher V, Hasan L. et al. miR-15a and miR-16 are implicated in cell cycle regulation in a Rb-dependent manner and are frequently deleted or down-regulated in non-small cell lung cancer. Cancer Res. 2009;69:5553-9
30. Neben K, Jauch A, Hielscher T, Hillengass J, Lehners N, Seckinger A. et al. Progression in smoldering myeloma is independently determined by the chromosomal abnormalities del(17p), t(4;14), gain 1q, hyperdiploidy, and tumor load. J Clin Oncol. 2013;31:4325-32
31. Galasso M, Costantino G, Pasquali L, Minotti L, Baldassari F, Corra F. et al. Profiling of the Predicted Circular RNAs in Ductal In Situ and Invasive Breast Cancer: A Pilot Study. Int J Genomics. 2016;2016:4503840
32. Nair AA, Niu N, Tang X, Thompson KJ, Wang L, Kocher JP. et al. Circular RNAs and their associations with breast cancer subtypes. Oncotarget. 2016;7:80967-79
Author contact
![]() Corresponding authors: Xiaoming Xie and Hailin Tang, Department of Breast Oncology, Sun Yat-Sen University Cancer Center, 651 East Dongfeng Road, Guangzhou, 510060, People's Republic of China; Phone: 86-20-87343806; Fax: 86-20-87343805; E-mail: xiexmorg.cn (X. Xie); tanghlorg.cn (H. Tang).
Corresponding authors: Xiaoming Xie and Hailin Tang, Department of Breast Oncology, Sun Yat-Sen University Cancer Center, 651 East Dongfeng Road, Guangzhou, 510060, People's Republic of China; Phone: 86-20-87343806; Fax: 86-20-87343805; E-mail: xiexmorg.cn (X. Xie); tanghlorg.cn (H. Tang).
 Global reach, higher impact
Global reach, higher impact