13.3
Impact Factor
Theranostics 2018; 8(13):3629-3642. doi:10.7150/thno.25955 This issue Cite
Research Paper
Development of a novel peptide aptamer-based immunoassay to detect Zika virus in serum and urine
Zoonosis Research Center, Department of Infection Biology, School of Medicine, Wonkwang University, 460, Iksan-daero, Iksan, 54538, Republic of Korea
‡These authors contributed equally to this manuscript.
Received 2018-3-8; Accepted 2018-5-24; Published 2018-6-7
Abstract
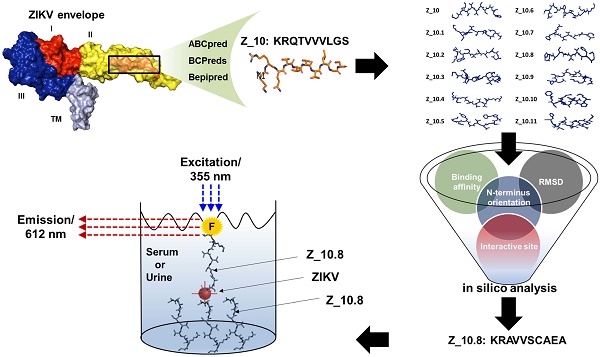
Zika virus (ZIKV) has been identified as a cause of adverse outcomes of pregnancy, including microcephaly and other congenital diseases. Most people infected with ZIKV do not show any symptoms. Development of a method to discriminate dengue virus (DENV) and ZIKV infections has been challenging, and efficient assays for patient management are limited, attributable to high levels of cross-reactivity among co-circulating Flaviviruses. Thus, there is an urgent need for a specific high-throughput diagnostic assay to discriminate ZIKV infections from other Flavivirus infections.
Methods: A novel epitope peptide of the ZIKV envelope protein was predicted using three immune epitope database analysis tools and then further modified. A molecular docking study was conducted using three-dimensional structures of the ZIKV envelope and peptide. Experimentally, interactions between the selected peptides and virus were assessed via a fluorescence-linked sandwich immunosorbent assay (FLISA), and performance of peptide-linked sandwich FLISA was evaluated in virus-spiked human serum and urine.
Results: The Z_10.8 peptide (KRAVVSCAEA) was predicted to be a suitable detector, with a higher binding affinity than other candidates based on four criteria (binding affinity, root mean square deviation, position of amine residue of lysine at the N-terminus, and interactive site) in a docking study. Z_10.8 was significantly more efficient at detecting ZIKV than the other two peptides, as shown in the direct FLISA (P < 0.001). Further, the equilibrium dissociation constant (Kd) for the Z_10.8 peptide was 706.0 ± 177.9 (mean ± SD, nM), with specificity to discriminate ZIKV from DENV. The limit of detection for the sandwich FLISA was calculated as 1×104 tissue culture infective dose (TCID)50/mL. The presence of serum or urine did not interfere with the performance of the Z_10.8-linked sandwich FLISA.
Conclusion: Four criteria are suggested for the development of an in silico modeled peptide aptamer; this computerized peptide aptamer discriminated ZIKV from DENV via immunoassay.
Keywords: Zika virus, peptide aptamer, immunoassay, serum, urine
Introduction
The Zika virus (ZIKV) belongs to the Flavivirus genus of the Flaviviridae family [1] and is similar to the four serotypes of the dengue virus (DENV), with approximately 43% amino acid identity across the viral polyprotein [2]. ZIKV and DENV infectious diseases are currently major global health threats [3], causing similar initial nonspecific symptoms such as systemic febrile illness, but resulting in drastically different potential complications. Infection with DENV can result in life-threatening dengue hemorrhagic fever or dengue shock syndrome [4], whereas ZIKV infection is associated with microcephaly in newborns [5]. Development of a method to discriminate DENV and ZIKV infections has been challenging, and efficient assays for patient management are limited [6].
ZIKV infections are asymptomatic in up to 80% of cases [7]; therefore, blood donors may not be aware of their infection. Viral load during ZIKV infections has previously been reported in plasma and urine samples of asymptomatic blood donors [8, 9]. In August of 2016, the Food and Drug Administration (FDA) issued revised guidance recommending blood centers in all states and U.S. territories to use a blood test to screen individual units of donated whole blood and blood components for ZIKV [10]. Further, the American Red Cross has mandated blood screening for ZIKV via nucleic acid testing (NAT) and serology testing [11]. NAT can reduce the risk of transfusion-transmitted infections [12]; however, a serological assay is needed in NAT-negative samples, attributable to a transient viremia period [6]. Serological assays for the Flaviviridae family in general, and for DENV and ZIKV in particular, are compromised by the high degree of similarities in their proteins, which can lead to cross-reacting antibodies and false-positive test results [13]. Monoclonal antibodies (mAbs) that recognize specific epitopes on ZIKV antigens are required to develop antigen detection assays [14]. Therefore, development of a sensitive and specific diagnostic method may overcome the problems of the currently used blood screening methods.
For efficient ZIKV-specific diagnostic testing, conventional or mAbs requiring hybridoma technology have limitations, including time requirements for animal immunizations, high cost, immunogenicity, and lack of stability [15]. In this context, aptamers derived from nucleic acid ligands have shown numerous advantages [16], and an oligonucleotide has been used to detect the ZIKV NS1 antigen [17]; however, it was only used with a semi pair of aptamer and antibody rather than a pair of aptamers. While oligonucleotide aptamers are diverse and highly sensitive, they are rapidly degraded by nucleases [18]. Modifications can increase the stability of nucleotide aptamers; however, they add to the cost. As an alternative to a DNA aptamer, peptides provide a powerful approach via a peptide-protein interaction (PPI) interface [19]. Charged amino acid residues are often significant contributors to the free energy of binding for PPIs, including antibody-antigen binding and ligand-receptor binding [20].
Previously, we identified an epitope-derived peptide conjugate that was used to detect influenza virus in an immunoassay [21]. In this study, a novel peptide sequence from the ZIKV epitope pool was developed using in silico modeling tools.
Methods
Reagents
Taq polymerase chain reaction (PCR) reagents were acquired from Takara (Kyoto, Japan), and the pET21b (+) plasmid was purchased from Novagen (Birmingham, United Kingdom). Peptides were synthesized by PepTron Inc. (Daejeon, South Korea). P (S/V-COOH) Europium nanoparticle (Eu NP) beads (0.1 µm) were purchased from Bangs Laboratories Inc. (Fishers, IN, USA). N-(3-Dimethylaminopropyl)-N-ethylcarbodiimide hydrochloride (EDC), N-hydroxysulfosuccinimide sodium salt (NHS), and N-hydroxysulfosuccinimide sodium salt (Sulfo-NHS) were acquired from Thermo Scientific (Waltham, MA, USA). Recombinant antigen and mAbs (1A5 and 1G8) were developed and are described in Supplementary Material (Figure S1).
Virus culture
ZIKV and DENV serotype 2 (DENV2) were maintained in C6/36 cells, and chikungunya virus (CHIKV) was grown in Vero cells as previously described [22-24].
Design of peptides
ZIKV-specific peptide candidates were designed against the ZIKV envelope using epitope prediction tools, including ABCpred [25], BCPreds [26], and Bepipred [27]. Three-dimensional (3D) structures of the ZIKV envelope and peptide were generated using an I-tasser server [28]. Binding ability of the epitopes was analyzed using AutoDock Vina in the PyRx virtual screening tool [29, 30].
Peptides with binding energy values lower than -6.5 (kcal/mol) were chosen [31] and modified to increase sensitivity to the ZIKV envelope protein and decrease sensitivity to the DENV envelope protein.
Conjugation of Eu NPs
Conjugation of Eu NPs was conducted as previously described [21]. Antibody and peptides were conjugated with 10 µL Eu NPs based on the carboxylate amine condensation method in 500 µL 2-(N-morpholino)ethanesulfonic acid buffer (MES, 0.05 M, pH 6.1), including 13 µL 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDC, 5 mM) and 100 µL N-hydroxysuccinimide (NHS, 50 mM). After 1 h, the mixture was centrifuged at 27,237 × g for 5 min to remove all non-reacted components. The activated Eu-NPs were then incubated with 45 µL mAbs (1G8, 1 mg/mL) or 60 µL peptide (50 μM). Reactions were performed in 0.1 M sodium phosphate (470 µL, pH 8.0) for 2 h at 25 °C. After washing with storage buffer containing 1% bovine serum albumin (BSA) and 2 mM borax (pH 9.0), the reactions were stored in 200 µL storage buffer at 4 °C.
Direct fluorescent-linked immunosorbent assay (FLISA)
The functionality of candidate peptides was checked by FLISA. Black 96-well plates were coated with ZIKV (105 tissue culture infective dose [TCID]50/mL) or envelope antigen at 4 °C overnight. After washing 3 times with phosphate-buffered saline containing Tween 20 (PBS-T), Eu-NP-conjugated peptides (150 nM peptide) and 1G8 antibodies (150 nM antibodies) were applied to the plates at 37 °C for 1 h. Fluorescence was measured in the presence of PBS using an Infinite F200 microplate reader system (TECAN, Männedorf, Switzerland) (excitation 355 nm/emission 612 nm).
Kinetic dissociation measurements
Black 96-well plates coated with peptide (1 μg/well) were incubated at 4 °C overnight. Blocking buffer containing 5% non-fat milk was applied for 2 h. After washing with PBS-T, plates were treated with ZIKV (104 TCID50/mL) and DENV2 (104 TCID50/mL) and incubated for 1 h. Eu-NP-conjugated peptides (1,500 nM) and 1G8 antibody (1,500 nM) were diluted 2-fold to make serial dilutions from 2-1 to 2-7; diluents were then applied to the plates at 37 °C for 1 h. Fluorescence was measured using an Infinite F200 microplate reader. Equilibrium dissociation constants (Kd) were obtained by fitting fluorescence data directly using a nonlinear analysis [32].
Sandwich FLISA
Peptides (10 μg/mL) and mAbs (10 μg/mL) were coated on 96-black well plates and then incubated overnight at 4 °C. The plates were then washed with PBS-T, blocked with 5% non-fat milk at 37 °C for 2 h, and then washed three times with PBS-T. The different virus titers were incubated for 1 h at 37 °C. Subsequently, the plates were washed 3 times and a dilution of Eu-NP-conjugated peptides (150 nM) or 1G8 (150 nM) was added; the plates were then incubated at 37 °C for 1 h. After stringent washing with PBS-T five times, 100 μL PBS was added to each well. Fluorescence was measured with an Infinite F200 microplate reader.
Real-time reverse transcription PCR (rRT-PCR)
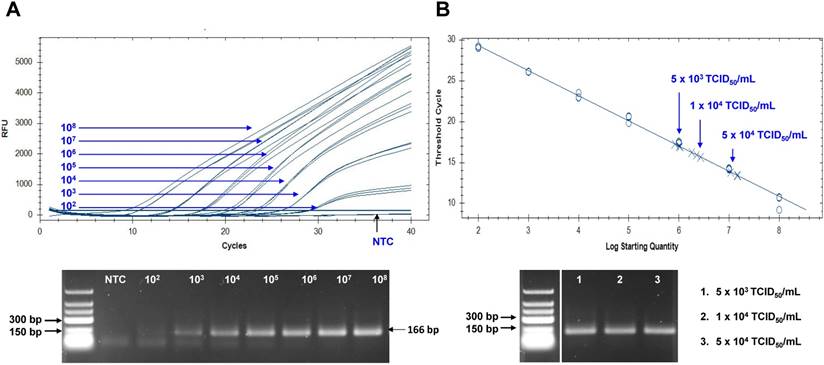
Primers and probe used for ZIKA envelope gene were designed using Primer Express software (Integrated DNA Technologies, Inc. IL, USA). The probe (5'-CCAGCATAGCGGGATGATTGTCAA- 3') was labeled with fluorescein (FAM) at the 5′ terminus and with blackhole quencher 1 (BHQ1) at the 3′ terminus. The forward primer was 5'-TAAGAAGATGACCGGGAAGA-3', and the reverse primer was 5'-TTCCGCTCTTGGTGAATGGAG-3'. rRT-PCR was performed using a Quantitect Probe RT-PCR kit (QIAGEN, Hilden, Germany) to determine cycle threshold (Ct) values in a 20-mL total volume that included 500 nM each of the final concentration of primers and probe. Cycling conditions used for the rRT-PCR were 50 °C for 30 min and 95 °C for 15 min, followed by 40 cycles of 95 °C for 15 s and 56 °C for 60 s. Fluorescence data were collected during the 56 °C annealing/elongation step. The template was generated in plasmid pGEM-T Easy (Promega, Madison, WI, USA), including a 166-base pair (bp) envelope insert for the standard RNA copy number. We used a RiboMax (Promega, Madison, WI, USA) kit for in vitro transcription of envelope RNA to determine the RNA copy number for the FLISA limit of detection (LOD, 1 × 104 TCID50/mL). The standard curve was calculated automatically by plotting the Ct values against each standard of known RNA copies and extrapolating the linear regression line of this curve. The PCR products were analyzed in agarose gels (2%).
Ethical considerations
The study was approved by the Wonkwang University Hospital Institutional Review Board (Approval No. WKIRB-201603-BR-015). Negative specimens were collected at the Wonkwang University Hospital. All patients agreed to participate in the study, and informed consents were processed before taking specimens. All experiments and methods were performed in accordance with relevant guidelines and regulations. Viruses were spiked in a negative human specimen to evaluate assay performance.
Data availability
The datasets generated during the current study are available from the corresponding author on reasonable request.
Statistical analysis
Data (n = 3) are shown as means ± standard deviation (SD) using one-way analysis of variance (ANOVA). All analyses were conducted using GraphPad Prism 5.0 software. All experiments were conducted in three different batches, and each batch was conducted in triplicate.
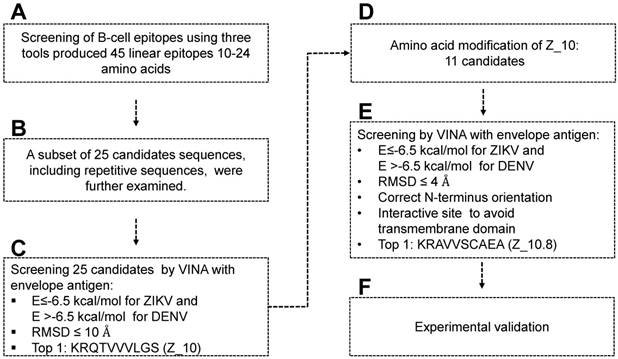
Scheme for designing and screening diagnostic peptides of the Zika virus (ZIKV) envelope protein. (A) Extensive screening of B cell epitope sequences using three tools (BCPreds, ABCpred, and Bepipred) resulted in selection of 45 candidates ranging from 10 to 24 amino acids. (B) Twenty-five sequences found using at least two tools were further examined. (C) Virtual screening using AutoDock Vina identified KRQTVVVLGS (Z_10) as the top sequence. (D) Eleven mutated peptides based on the Z_10 backbone were generated. (E) Binding energy, root-mean-square deviation (RMSD), N-terminus orientation, and interactive site were evaluated via virtual screening using AutoDock Vina. KRAVVSCAEA (Z_10.8) was ranked as the top peptide. (F) Peptides were conjugated with Europium nanoparticles (Eu NPs) to test the reactivity to antigen and virus. E: binding energy; DENV: dengue virus.
Results
Peptide candidates identified using bioinformatics tools
Figure 1 displays a schematic of the peptide design. Peptide aptamers usually consist of short, 5-20 amino acid sequences that are typically embedded as a loop within a stable protein scaffold [33]. In this study, the epitope peptide length was randomly assigned as 12 (median number between 5 and 20 amino acids). Peptides shorter than five or larger than 25 amino acids were removed, as peptide aptamers are rarely outside these boundaries [33] (Figure 1A).
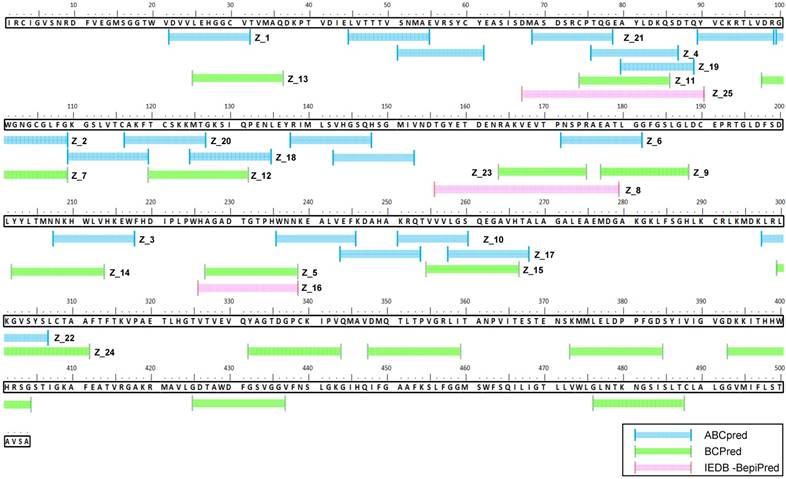
ABCpred, BCPreds server 1.0, and IEDB-BepiPred predicted 21 (threshold: 0.7), 16 (specificity: 75%), and 8 (threshold 0.5) linear epitopes, respectively. Thresholds were defined as described previously [25-27] (Table 1). Common epitopes found by at least 2 different tools were considered potential epitopes; therefore, only 25 sequences were selected for further virtual screening (Figure 1B). Virtual screening using Autodok Vina was conducted to determine binding energies below -6.5 kcal/mol and root-mean-square deviations (RMSD) below 10 Å in the first screening. In this study, KRQTVVVLGS (Z_10) was selected as the template peptide for further modification (Figure 1C); this sequence was conserved in ZIKV species among the Flavivirus genus (Figure S2).
To design a better peptide to discriminate DENV, the template peptide Z_10 (KRQTVVVLGS) was mutated at several amino acids. There were 11 modifications of the Z_10 sequence that predicted different binding energies (Figure 1D). Based on both binding affinity and RMSD, KRAVVSCAEA (Z_10.8) showed lower energy, satisfying the higher binding affinity requirements. Docking status of the amine residue of lysine (K) at the N-terminal was considered as an additional criterion, attributable to the moiety of conjugation with fluorescent material being relatively bulky, with the ability to interfere with interactions of the fluorescent conjugate with proteins. As a last criterion, interactive sites were considered, as it was hard to access the transmembrane domain (TM) (Figure 1E). In this context, the Z_10.8 modes were not predicted to involve the TM. Z_10.8 was experimentally evaluated as an aptamer to determine diagnostic performance via conjugation with Eu NPs (Figure 1F). Figure 2 indicates the position of 25 peptides on the ZIKV envelope protein.
Docking studies for modeling ZIKV envelope protein and peptide complexes
Among 45 linear epitopes containing 10 to 24 amino acids, 25 peptide epitopes were found using at least two tools and were chosen for further docking studies; these were virtually screened using PyRx with the full ZIKV envelope protein (GenBank accession #: YP_009227198.1) and DENV2 envelope protein (GenBank accession #: AHG97600.1). The protein data bank (PDB) file of each protein was modeled using the I-TASSER server, with a 3D structure of each peptide.
Characteristics of B-cell linear epitopes.
| Tools | No | Position | Epitope | Score | Repetition | Name of Sequence |
|---|---|---|---|---|---|---|
| ABCpred | 1 | 258 | LGSQEGAVHT | 0.78 | LGSQEGAVH | Z_17 |
| 2 | 90 | YVCKRTLVDR | 0.77 | |||
| 3 | 22 | DVVLEHGGCV | 0.77 | LEHGGCV | Z_1 | |
| 4 | 143 | VHGSQHSGMI | 0.77 | |||
| 5 | 51 | SNMAEVRSYC | 0.76 | |||
| 6 | 213 | VHKEWFHDIP | 0.76 | |||
| 7 | 125 | MTGKSIQPEN | 0.76 | MTGKSIQ | Z_18 | |
| 8 | 138 | RIMLSVHGSQ | 0.75 | |||
| 9 | 100 | GWGNGCGLFG | 0.75 | GWGNGCGLFG | Z_2 | |
| 10 | 80 | AYLDKQSDTQ | 0.74 | AYLDKQSDTQ | Z_19 | |
| 11 | 208 | NKHWLVHKEW | 0.74 | NKHWLV | Z_3 | |
| 12 | 45 | LVTTTVSNMA | 0.73 | |||
| 13 | 117 | AKFTCSKKMT | 0.73 | TCSKKMT | Z_20 | |
| 14 | 76 | TQGEAYLDKQ | 0.72 | TQGEAYLDKQ | Z_4 | |
| 15 | 244 | EFKDAHAKRQ | 0.72 | |||
| 16 | 236 | WNNKEALVEF | 0.72 | |||
| 17 | 110 | KGSLVTCAKF | 0.72 | |||
| 18 | 69 | ASDSRCPTQG | 0.71 | CPTQG | Z_21 | |
| 19 | 298 | LRLKGVSYSL | 0.71 | RLKGVSYSL | Z_22 | |
| 20 | 251 | KRQTVVVLGS | 0.71 | TVVVLGS | Z_10 | |
| 21 | 172 | NSPRAEATLG | 0.7 | NSPRAEATLG | Z_6 | |
| BCPred | 1 | 98 | DRGWGNGCGLFG | 0.999 | GWGNGCGLFG | Z_7 |
| 2 | 332 | YAGTDGPCKIPV | 0.996 | |||
| 3 | 177 | EATLGGFGSLGL | 0.984 | EATLG | Z_9 | |
| 4 | 227 | AGADTGTPHWNN | 0.974 | AGADTGTPHWNN | Z_5 | |
| 5 | 74 | CPTQGEAYLDKQ | 0.968 | CPTQGEAYLDKQ | Z_11 | |
| 6 | 348 | DMQTLTPVGRLI | 0.948 | |||
| 7 | 164 | RAKVEVTPNSPR | 0.944 | RAKVEVTPNSPR | Z_23 | |
| 8 | 373 | KMMLELDPPFGD | 0.882 | |||
| 9 | 393 | DKKITHHWHRSG | 0.77 | |||
| 10 | 120 | TCSKKMTGKSIQ | 0.758 | TCSKKMTGKSIQ | Z_12 | |
| 11 | 25 | LEHGGCVTVMAQ | 0.758 | LEHGGCV | Z_13 | |
| 12 | 476 | GLNTKNGSISLT | 0.701 | |||
| 13 | 425 | GDTAWDFGSVGG | 0.664 | |||
| 14 | 300 | LKGVSYSLCTAA | 0.549 | LKGVSYSL | Z_24 | |
| 15 | 202 | YYLTMNNKHWLV | 0.443 | NKHWLV | Z_14 | |
| 16 | 255 | VVVLGSQEGAVH | 0.386 | VVVLGS | Z_15 | |
| IEDB-BepiPred | 1 | 36 | QDKPTV | 1.2 | ||
| 2 | 67 | DMASDSRCPTQGEAVLDKQSDTQ | 1.6 | CPTQGEAVLDKQSDTQ | Z_25 | |
| 3 | 125 | MTGKSIQPE | 1.2 | |||
| 4 | 156 | TGYETDENRAKVEVTPNSPRAEAT | 1.8 | RAKVEVTPNSPRAEAT | Z_8 | |
| 5 | 226 | HAGADTGTPHWNN | 1.8 | AGADTGTPHWNN | Z_16 | |
| 6 | 275 | AEMDGA | 1.1 | |||
| 7 | 332 | YAGTDGPCK | 1.5 | |||
| 8 | 364 | VITESTEN | 1.2 |
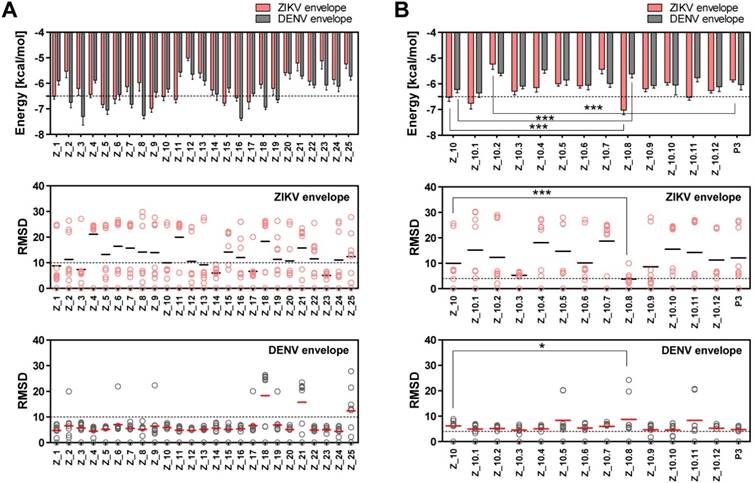
Two epitope sequences (Z_1 and Z_10) out of 25 peptides satisfied the two criteria of a binding energy lower than -6.5 kcal/mol and a RMSD lower than 10 Å. The Z_10 peptide had a lower binding energy with the ZIKV envelope protein (-6.522 ± 0.15, mean ± SD, kcal/mol) than the Z_1 peptide (-6.511 ± 0.11, mean ± SD) (Figure 3A). Binding energy of the Z_10 peptide with the DENV envelope was higher than -6.5 kcal/mol, having a binding energy of -6.222 ± 0.12 (mean ± SD, kcal/mol). RMSDs (mean) of the two peptides were below 10 Å in the ZIKV envelope protein. Using these two criteria (binding energy and RMSD), Z_10 was shown to be more sensitive than Z_1. Therefore, Z_10 was chosen as a template to further modify the sequence.
To increase the binding affinity and decrease the RMSD of Z_10 (KRQVVVLGS) against the ZIKV envelope, 11 peptides were derived from Z_10 by modulating its sequence and then modeled. Table S1 shows docking of 25 peptides, and Table S2 indicates binding energy, RMSD, and location of the N-terminus of 11 modified peptides.
According to a docking study, Z_10.8 (KRAVVSCAEA) showed the strongest binding energy with the ZIKV envelope protein among 11 modified sequences. The binding affinity of Z_10.8 to the ZIKV envelope protein was computed as -7.02 ± 0.15 (mean ± SD) (kcal/mol), representing a significant increase compared to the template sequence (Z_10) (P < 0.001).
In contrast, the binding affinity of Z_10.8 to the DENV envelope was considerably less than that of Z_10 (5.611 ± 0.15, mean ± SD kcal/mol versus -6.222 ± 0.12, mean ± SD kcal/mol, respectively, P < 0.001). The total RMSD average (3.80 Å) for the Z_10.8 peptide against the ZIKV envelope protein was substantially less (below 4 Å) than that of Z_10 (9.97 Å) (P < 0.001). In contrast, the total RMSD average (8.75 Å) for the Z_10.8 peptide against the DENV envelope protein was significantly higher than that of Z_10 (5.81 Å) (P < 0.05). These data indicate that the modulated sequence (Z_10.8) is significantly more sensitive and specific to the ZIKV envelope protein than the template sequence (Z_10) (Figure 3B).
Sequence alignment of epitope candidate peptides on the Zika virus (ZIKV) envelope protein. A total of 45 epitopes predicted using three tools (ABCpred, BCPreds, and immune epitope database and analysis resource [IEDB]-BepiPred) were aligned, and 25 sequences numbered from Z_1 to Z_25 were found via at least two tools.
Docking output of peptides with Zika virus (ZIKV) and dengue virus (DENV) envelope proteins. (A) Binding energy and root-mean-square deviation (RMSD) of 25 candidate epitopes for two viruses, ZIKV and DENV, are plotted. (B) Binding energy and RMSD of candidate peptides derived from Z_10 for two viruses, ZIKV and DENV, are plotted. * P < 0.05; *** P < 0.001.
Docking of peptides on the ZIKA virus (ZIKV) and dengue virus (DENV) envelope proteins. (A) Two native structures of the Z_10 and Z_10.8 peptides were superimposed and then used to compare structural conformations before and after modifications. (B) Two proteins were constructed via 3D modeling. (C) Three peptides (Z_10.2, Z_10.8, and P3) were modeled for active conformations. (D) ZIKV and DENV envelope proteins were superimposed using nine top-scoring docking conformations of three peptides. The binding location of native peptides is shown as a stick in the rectangular image. Envelope protein domains are presented as different colors (i.e., red: domain I; yellow: domain II; blue: domain III; gray: transmembrane domain [TM]).
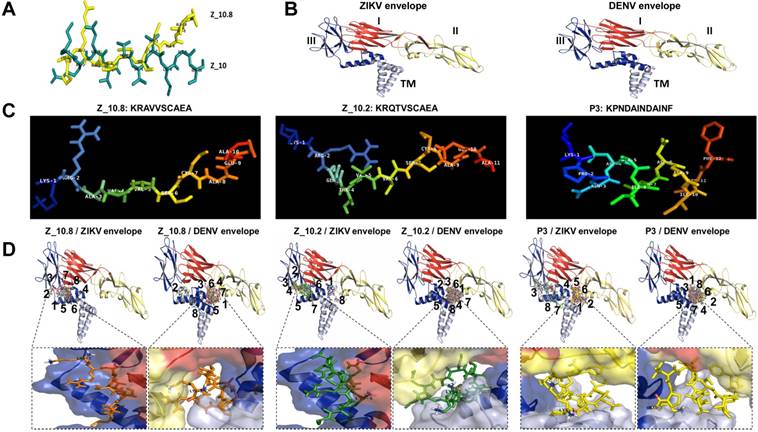
Docking studies of different peptides against envelope proteins
Previously, we showed that peptide aptamers rather than antibodies could be used in diagnostic applications [21]. In the current study, peptide specificity was improved by modifying the binding sites in addition to enhancing the binding affinity. To attain this objective, an active conformation of peptides was modeled in the PyMOL molecular graphics system. Structural modifications of Z_10 to Z_10.8 were confirmed by superimposing two peptides (Figure 4A). While the shape of Z_10 was close to a linear structure, Z_10.8 showed an L shape after modification. The modeling of each protein is shown with domains I, II, and III, as well as the TM (Figure 4B). Native structures of the three peptides used in this study are shown in Figure 4C. Figure 4D indicates nine bound conformations superimposed by peptides on each domain of the envelope proteins. As shown, only two spots were predicted as main interactive sites, one of which was involved in the interaction between the native peptide and the antigen. All Z_10.8 modes were involved in one specific spot between domain I and III of the ZIKV envelope protein, which differed from the site of interaction for the native peptide. Six out of nine modes showed an outward N-terminal amine peptide group for the ZIKA envelope protein, representing the highest frequency of all other peptide insertions (Table S2). In contrast, the main interaction of this ligand against the DENV envelope antigen was found in the pocket surrounded by all four domains, including the TM, implying that insertion of the TM in the lipid layer interferes with the interaction of peptides with the virus particle.
Unlike Z_10.8, two negative peptides showed two interactive sites rather than one against the ZIKV envelope protein. First, like Z_10.8, most Z_10.2 modes were predicted to bind to domain III and domain I; however, the peptide N-terminal amine groups were placed inward of the ZIKV envelope pocket in seven out of nine modes. All Z_10.2 modes were predicted to recognize the pocket of all domains, including the TM of the DENV envelope protein. Second, three P3 modes (3, 7, and 8) recognized the pocket of domain II and the TM of the ZIKA envelope antigen, and, like Z_10.8, other modes (3, 7, and 8) were predicted to bind to the same spots. Like Z_10.2, all modes of P3 recognized the pocket of domain II and TM of the DENV envelope protein (Figure 4D) and showed an inward N-terminal amine of peptide (Table S2). Therefore, P3 was considered a negative peptide against ZIKV based on a low binding affinity, poor RMSD, inward position of the N-terminal amine group, and interactive sites, including the TM.
A representative docking modeling (mode 1) with all modified peptides and the ZIKV envelope is presented in Figure S2.
Assessment of interactions between peptides, protein, and virus. (A) A schematic depiction of the direct fluorescence-linked sandwich immunosorbent assay (FLISA) principle is shown. To validate the interaction of peptides to analytes (virus and antigen) experimentally, peptides were conjugated with Europium nanoparticles (Eu-NPs) and used for detection of analytes coated on 96-well plates. Peptide Eu NP conjugates were applied to react with analytes. Unbound conjugate was removed via stringent washing, and the interaction of peptides to analytes was determined by measuring fluorescence (355 nm/excitation and 612 nm/emission). (B) Serially diluted recombinant Zika virus (ZIKV) envelope proteins (0.16 to 2.5 μg/mL) were tested with 150 nM peptides. (C) Specificity of peptides was determined via direct FLISA after coating with different viruses at 105 tissue culture infective dose (TCID)50/mL. Uninfected: uninfected culture supernatant; RFU: relative fluorescence unit; a.u.: arbitrary unit; DENV: dengue virus; CHIKV: chikungunya virus; * P < 0.05; ** P < 0.01; *** P < 0.001.
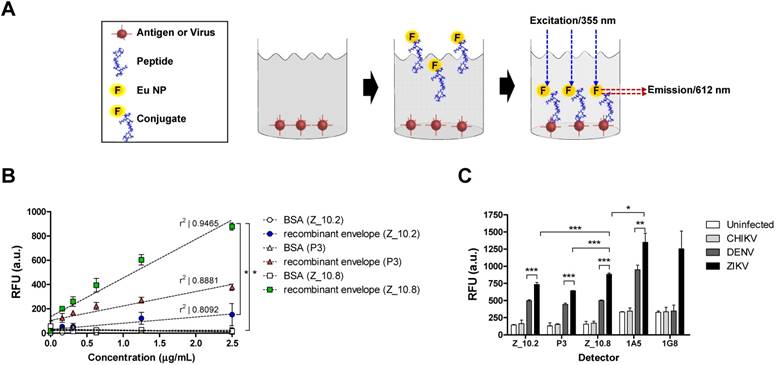
Interaction of peptides with virus and antigen
To evaluate the interaction of candidate peptides with antigen and virus, Eu NPs were conjugated with peptides by forming a stable amide bond, and a FLISA was conducted as previously described [21]. The ZIKV envelope protein (amino acid 1-504) was cloned, and the recombinant antigen was produced in an Escherichia coli expression system; this antigen was used to make mAbs, and the reactivity of antibody to virus was confirmed via enzyme-linked immunoassay (ELISA). Cloning details and development of mAbs are presented in Figure S1.
Figure 5A illustrates the FLISA principles schematically. Briefly, black 96-well plates were coated with the ZIKV envelope protein, and virus and Eu NP-conjugated peptides were applied to measure the bound fluorescence. As shown in Figure 5B, Z_10.8 (KRAVVSCAEA), Z10.2 (KRQTVSCAEA), and P3 (KPNDAINDAINF) differentially detected the recombinant ZIKA envelope protein. Z_10.8 reorganized the recombinant ZIKV envelope protein (P < 0.05) more than the other peptides (in the range of 0.16, 0.32, 0.64, 1.25, and 2.5 μg/mL) and was more highly correlated (r2: 0.9455) than P3 (r2: 0.8881) or Z_10.2 (r2: 0.8092). Therefore, Z_10.8 was more reliable and detected the recombinant ZIKV envelope protein better than the other two peptides. There were no significant differences between the Z_10.2 peptide and the target antigen, whereas the Z_10.8 peptide was significantly different from Z_10.2 (P < 0.05), indicating that the Z_10.8 peptide specifically targeted the ZIKA envelope protein more than the Z_10.2 peptide.
The recombinant antigen had more interactive sites with the three peptides than the virus; therefore, all three peptides could easily but differentially access the interactive sites depending on different dockings. P3 interacted with the recombinant antigen more than Z_10.2, indicating that P3 docking energy was higher than that of Z_10.2 (-5.86 ± 0.07 kcal/mol, mean ± SD versus -5.23 ± 0.2 kcal/mol, mean ± SD, respectively, P < 0.001) (Figure 5B) and may induce stronger interactions with the recombinant antigen.
When the direct FLISA was tested by coating with virus, results indicated that all three peptides (Z_10.2, Z_10.8, and P3) detected ZIKV significantly better than DENV at a titer of 105 TCID50/mL (P < 0.001). However, Z_10.8 was significantly better at detecting ZIKV than the other two peptides (P < 0.001) (Figure 5C). After conjugating two mAbs (1A5 and 1G8) with Eu NPs, a direct FLISA was conducted and compared with a peptide-FLISA. Only the 1A5 mAb had a stronger interaction with ZIKV than Z_10.8 (P < 0.05); however, relative fluorescence units (RFU) associated with 1G8 were not significantly different from those of Z_10.8 against ZIKV, implying that this short peptide may function like an antibody to detect virus.
Performance of the peptide-linked sandwich FLISA
Sandwich assays tend to be more sensitive than direct/indirect ELISAs and are therefore more commonly used [34]. In this study, Z_10.8 was used for both capturing and detection in the sandwich FLISA, which is illustrated schematically in Figure 6A. As is evident from Figure 6B, the Kd (defined as the concentration of ligand that will occupy 50% of the receptors [35]) for the Z_10.8 peptide with ZIKV (104 TCID50/mL) was 706.0 ± 177.9 nM (mean ± SD). Generally, ligand-receptor binding with a Kd of 1 nM or less is considered high affinity binding, whereas ligands with a Kd of 1 µM or more have low affinity binding. Thus, the Z_10.8 peptide displayed high affinity for ZIKV but showed no reactivity with DENV at the same virus titer, even when the concentration was increased to 1,500 nM. Further, Z_10.2 and P3 had no reactivity to ZIKV in the sandwich FLISA, indicating that a sandwich FLISA using Z_10.8 was useful as a ZIKV-specific diagnostic method.
Characteristics of peptide- and fluorescence-linked sandwich immunoassay (FLISA). (A) Experimental validation of peptides was conducted via sandwich FLISA. Peptides were coated on 96-well plates, and after adding the virus, different concentrations of conjugate were applied to evaluate the saturation curve by measuring fluorescence (355 nm/excitation and 612 nm/emission). (B) Serially diluted peptides (1,500 to 10 nM) were tested for the equilibrium dissociation constant (Kd) of peptide using negative antigen (bovine serum albumin, BSA), dengue virus (DENV), and Zika virus (ZIKV) at a 104 tissue culture infective dose (TCID)50/mL. (C) Specificity of the sandwich FLISA was determined by measuring different viruses at 105 TCID50/mL. *** P<0.001.
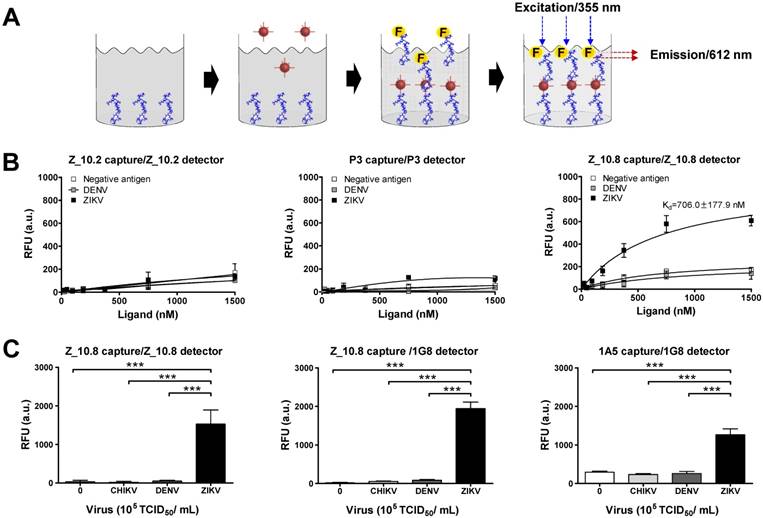
At a higher virus titer (105 TCID50/mL), the fluorescence signals obtained with a pair of Z_10.8 peptides were significantly (P < 0.05) different among ZIKV, DENV, and CHIKV (Figure 6C). The difference in detection between ZIKV and DENV was even more pronounced (P < 0.001) when the Z_10.8 (capture) peptide and 1G8 mAb (for detection) pair was used, implying that there are competitive binding sites by ligands when a peptide is used for both capture and detection. In contrast, each pair of Z_10.2- and P3-linked sandwich FLISAs were unreactive to CHIKV, DENV, and ZIKV at high titers. These results were consistent with those of the docking analysis for parameters of binding affinity, RMSD, location of N-terminal amine, and interactive sites. Taken together, these results suggest that the Z_10.8 peptide-linked sandwich FLISA specifically detects the ZIKV, and performance of peptide-linked sandwich FLISA is comparable to a mAb-linked immunoassay.
Analysis of sandwich FLISA using human specimens (serum and urine)
Proteolytic degradation of peptide-based drugs is often considered a major weakness that limits their systemic therapeutic applications [36]. Hence, we determined the analytical sensitivity of the peptide-linked immunoassay. A quantitative diagnostic performance of the Z_10.8-linked sandwich FLISA was tested in the absence of a specimen. As shown in Figure 7A, performance of the assay significantly increased by at least 5×104 TCID50/mL ZIKV when the virus was diluted in DMEM (P < 0.001). The LOD was determined using the limit of blank (LOB) as previously described [37]. The LOB and LOD formulae were as follows: LOB = mean (blank) + 1.645 (standard deviation (SD) of blank), and LOD = LOB + 1.645 (SD of low concentration sample). The computed values for LOB and LOD are shown in each graph.
Performance of peptide-linked sandwich FLISA for detecting virus specimens. (A) Zika virus (ZIKV) was serially diluted in Dulbecco's modified Eagle's medium (DMEM) from 102 to 106 tissue culture infective dose (TCID)50/mL and applied to the sandwich FLISA. (B) Human serum (1:400) was mixed with the virus at a ratio of 10:1, and human urine stock was mixed with different titers of virus at a ratio of 10:1. After mixing, the samples were tested via sandwich FLISA. ** P < 0.01; *** P < 0.001. Arrows indicate the virus titer corresponding to the limit of detection (LOD) titer. RFU: relative fluorescence units.
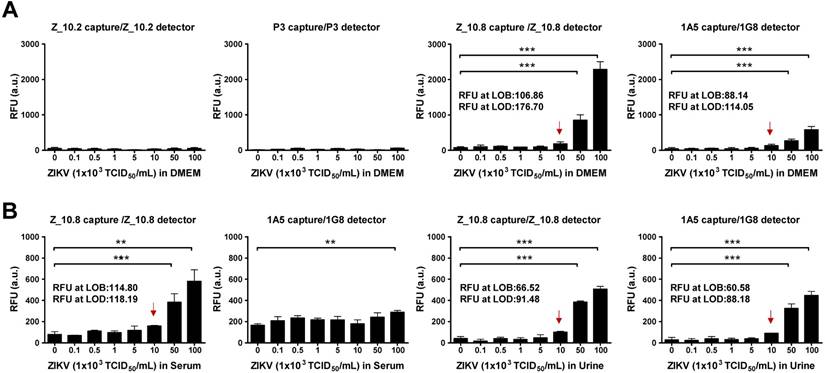
The RFU at the LOB and LOD of the Z_10.8-linked sandwich FLISA were 106.86 and 176.70, respectively. The RFU at the LOD was below the RFU (184 ± 43.15, mean ± SD) at 1×104 TCID50/mL ZIKV in DMEM. Therefore, virus titers corresponding to the LOD were 1×104 TCID50/mL. The 1A5/1G8 pair detected ZIKV at 5×104 TCID50/mL (P < 0.05); however, the overall fluorescence value was lower than that of the Z_10.8-linked sandwich FLISA by about 3-fold. When the same antibodies were paired (i.e., 1A5/1A5 or 1G8/1G8), the FLISA had higher background values than when different antibodies were paired (Figure S3). The RFU values at the LOB and LOD of a pair for the 1A5 capture/1G8 detector sandwich FLISA were 88.14 and 114.05, respectively. The RFU at the LOD was below the RFU (130 ± 32.25, mean ± SD) at 1×104 TCID50/mL for ZIKV in DMEM. Therefore, virus titers corresponding to the LOD were 1×104 TCID50/mL. However, in contrast to the direct FLISA results, Z_10.2- or P3-linked sandwich FLISAs could not detect ZIKV even at a high titer of 1×106 TCID50/mL. Therefore, we confirmed the Z_10.8-linked sandwich FLISA as having an advantage for ZIKV-specific detection. Although the overall fluorescence signal decreased in the presence of serum, the Z_10.8-linked sandwich FLISA detected ZIKV at the same titer (5×104 TCID50/mL) (P < 0.001). RFUs at the LOB and LOD of the Z_10.8-linked sandwich FLISA were 114.80 and 118.19, respectively, while the RFU at the virus titer corresponding to the LOD was below that for 1×104 TCID50/mL ZIKV in serum, indicating that serum did not interfere with the performance of the Z_10.8-linked sandwich FLISA.
In contrast, ZIKV was not detected even at the high titer 1×106 TCID50/mL in human serum when a mAb pair was used (1A5/1G8), indicating that the presence of serum interfered with the ability of the antibodies to recognize the antigen.
In contrast to serum, urine did not interfere with the performance of peptide- or antibody-linked sandwich FLISAs (P < 0.001). RFUs at the LOB and LOD for the Z_10.8-linked sandwich FLISA measured in urine were 66.52 and 91.48, respectively, which were lower than those observed in DMEM, indicating that urine suppressed the fluorescence signal like serum. However, the LOD for both peptide- and antibody-linked sandwich FLISAs was computed as 1×104 TCID50/mL ZIKV in urine. These results clearly indicate that the Z_10.8 peptide has an advantage over antibodies when screening human serum samples for ZIKV.
The Zika viral RNA load in patients has been reported as 5.0×103 to 8.1×106 copies in sera, and 0.7 to 220×106 copies in urine [38, 39]. To assume the practical usefulness of peptide-linked sandwich FLISA for detecting ZIKV in the field, the RNA copy number corresponding to the FLISA LOD was determined (Figure 8). In this study, new primers and probe were developed and applied to determine the RNA copy number. Information regarding primer and probe binding sites is found in Figure S4. To generate a calibration curve, serially-diluted RNA standards were used, and a standard curve was drawn to show each standard RNA copy number vs the Ct (Figure 8A). After preparing three dilutions (5×103, 1×104, and 5×104 TCID50/mL) of ZIKV, samples (75 μL) were used for RNA extraction. Eluted RNA was used for rRT-PCR with the standard using plasmid dilutions. A standard curve of Ct values plotted against the logarithmic dilutions produced a r2 value > 0.998, which was close to the optimized protocol. The 1×104 TCID50/mL of ZIKV corresponded to a Ct value of 15.9 ± 0.21 (mean ± SD) and an RNA copy number of 2.25×106 ± 3.6×104 /mL (Figure 8B). Therefore, peptide-linked sandwich FLISA may be a practical tool for detecting viral load in patient samples.
Assessment of peptide-linked sandwich FLISA performance via real time reverse transcription polymerase chain reaction (rRT-PCR). (A) The linear relationship between the threshold cycle (Ct) and RNA copy number is shown. The bottom panel shows PCR products for different RNA standards. (B) After preparing dilutions of Zika virus (ZIKV), samples were subjected to RNA extraction and used for rRT-PCR. The arrow indicates the point corresponding to the RNA copy number and virus titer. The data (n = 3) are shown as means ± SD. TCID: tissue culture infective dose; RFU: relative fluorescence units; NTC: no template control.
The analytical sensitivity of peptide-linked sandwich FLISA against proteases
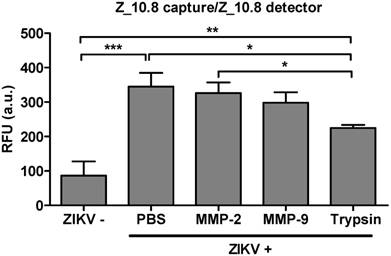
Issues have been raised regarding the analytical sensitivity of peptide therapeutics for systemic use [40]. Therefore, the analytical sensitivity of peptides was tested in the presence of several proteases. A physiological amount of trypsin and matrix metalloproteinases (MMPs) were selected as previously described [41, 42]. ZIKV (5×104 TCID50/mL) was reacted with MMP-2 (251 pg/mL), MMP-9 (318 pg/mL), and trypsin (248 ng/mL) at 37 ℃ for 1 h (Figure 9). The effects of treatment with MMP-2 or MMP-9 were not significantly different from those of PBS-treated virus; however, trypsin significantly affected the assay performance (P < 0.05). Therefore, the Z_10-linked assay may be affected by trypsin but not MMPs.
Effect of a protease inhibitor on performance of peptide- and fluorescence-linked sandwich immunosorbent assay (FLISA). Zika virus (ZIKV, 5×104 tissue culture infective dose [TCID]50/mL) was mixed with matrix metalloproteinase (MMP)2 (251 pg/mL), MMP9 (318 pg/mL), and trypsin (248 ng/mL) and used in the FLISA. * P < 0.05; ** P < 0.01; *** P < 0.001.
Discussion
Currently, there are no licensed vaccines or therapeutics available to combat ZIKV infections; therefore, there has been an urgent call to develop effective and safe vaccines [43]. Further, complications associated with ZIKV infections are poorly understood, making a timely diagnosis important for patient health and to limit its rapid proliferation [44-46]. It is critical to develop ZIKV antibodies that can protect against ZIKV infection and that function as a prophylactic or therapeutic element during pregnancy or in the application of vaccines. However, screening methods for ZIKV antigen have rarely been reported, as it is difficult to develop ZIKV-specific antibodies. Recently, novel strategies have been preferably developed as a diagnostic platform for detecting ZIKV infections using a paper-based platform or plasmonic-gold platform [47, 48].
We sought to develop a detection element against ZIKV that could function as an alternative to antibodies and that provides a new strategy for overcoming the limitations of using antibodies.
As a novel strategy, we used three bioinformatic tools, BCPreds, ABCpred, and Bepipred to select novel template epitope candidate sequences and then modified them to be suitable as diagnostic peptide aptamers. Experimental validation is underdeveloped for peptide aptamers, and our method can help evaluate peptide aptamer performance. Further, the availability of prediction method(s) applying the criteria we established in this study can rapidly aid in simplifying the problem of antibody scarcity. As such, we established a simple experimental method to validate our four docking criteria (binding energy, RMSD, location of N-terminus, and interactive site to avoid the TM).
Interestingly, use of a symmetric pair of peptides to detect ZIKV in a sandwich assay did not interfere with use of the peptide pair for capturing and detection. This is likely attributable to less competition from the short amino acids in the peptide aptamer. The ZIKV surface is arranged with an envelope protein homodimer on the virus surface, and domain III is a good target as a potent neutralizing immune domain [43]. According to the docking study, most Z_10.8 modes were predicted to recognize the stem; however, all modes did not exactly target the same spot, although they appeared to bind the stem (Figure S5). Further, the icosahedral symmetry of ZIKV contains 90 envelope proteins packed tightly as homodimers [49, 50], and short peptide aptamers may have an advantage for recognizing the target site within the binding pocket. Taken together, peptides could cause less interference in an immunoassay than antibodies when used for capture and detection in sandwich assays, giving an advantage to the development of a diagnostic system.
Generally, substances that alter the measurable concentration of an analyte or alter antibody binding can potentially result in immunoassay interference. Interfering substances are natural, polyreactive antibodies or autoantibodies (heterophiles) or human anti-animal antibodies together with other unsuspected binding proteins that lead to interference with the reaction between analyte and reagent antibodies in an immunoassay [51]. We noticed that human serum interfered with the antigen-antibody interaction; however, less interference was observed when using peptides than when using antibodies. This may be attributable to the relatively bulky volume of antibodies with high molecular weights (150 kDa), which hinders access to the same epitope of antigen for other antibodies. In contrast, peptides are about 1 kDa in size; thus, the small peptide size would be beneficial as a diagnostic element, causing less hindrance toward access to the target site.
Serum contains many substances that may interfere with antibody-antigen interactions. To prove that serum interferes with the antibody-antigen interaction, we tested human serum differentially diluted from 1:400 to 1:3200 (Figure S6); background fluorescence responded to serum dilutions. Finally, when we diluted serum 1:3200, the LOD returned to 1×104 TCID50/mL, implicating that peptides may be more useful elements for detecting antigens in the serum than antibodies. In our study, the analytical sensitivity of the peptide-linked immunoassay in the presence of sera and urine was confirmed. In the presence of sera (1:400) for 1 h, the peptide-linked sandwich FLISA was able to detect virus antigen, indicating that peptide aptamers have an advantage for overcoming the limitations of DNA aptamers in clinical applications. In addition, the analytical sensitivity of peptides in the 1 h specific protease treatment was confirmed directly using several proteases. While treatment with MMP-2 or MMP-9 resulted in no significant differences from PBS-treated virus, trypsin significantly affected the assay performance (P < 0.05). However, human serum contains protease and protease inhibitors [52], and trypsin activity may be controlled in serum, giving less influence to peptide activity.
Computational design-derived peptides provide new hope toward completely replacing antibodies in immunoassays, shortening the time to develop a detection element. This approach should be complemented by applying clinically infected sera and urine in the future. Although the presence of ZIKV antigen has not yet been defined in urine, we cannot rule out the possibility of ZIKV antigen in urine, as renal glomeruli serve as an amplification reservoir for ZIKV in the renal compartment [53, 54]. This methodology for developing peptide pair-linked sandwich FLISAs may be useful for diagnosing ZIKV infections in human sera and urine.
FLISAs using fluorescent material are superior to ELISAs; however, there are still disadvantages, as this technology still needs to conjugate peptides to fluorescent material. Peptides can be conjugated to common horseradish peroxidase similar to antibodies [55, 56]; therefore, we believe that peptides could be applied to mainstream uses in a manner similar to that of ELISAs. Thus, peptides can be easily transitioned to an ELISA platform. Further, peptides can be used in immunochromatographic tests (ICT) as a rapid diagnostic system. To succeed in rapid ICT, biotinylated peptides can be further coated with streptavidin.
The limitation of a small number of modified peptide sequences used in this study will be addressed by analyzing more sequences in the future, which is expected to provide greater significance. Further, a current limitation of using DNA aptamers is the lack of high quality options for clinical samples [57]. A DNA aptamer-based ELISA assay has been reported to detect 1 ng/mL in 10% serum; however, it was not a complete antibody-free assay [17].
In summary, we developed a benchmark computational protocol for designing peptide aptamers via docking studies. RMSD, binding energy, location of N-terminus of amine, and considering non-TM domains as interactive sites can be useful parameters when designing peptide aptamers for ZIKV.
Abbreviations
BSA: bovine serum albumin; CHIKV: chikungunya virus; DENV: dengue virus; EDC: N-(3-dimethylaminopropyl)-N-ethylcarbodiimide hydrochloride; Eu NP: europium nanoparticle; FLISA: fluorescence-linked immunosorbent assay; Kd: equilibrium dissociation constant; LOB: limit of blank; LOD: limit of detection; MES: 2-(N-morpholino)ethanesulfonic acid buffer; NAT: nucleic acid testing; NHS: N-hydroxysulfosuccinimide sodium salt; PPI: peptide-peptide interaction; RMSD: root-mean-square deviations; TCID50: 50% tissue culture infective dose; ZIKV: Zika virus.
Acknowledgements
This research was supported by the Priority Research Centers Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education (NRF-2015R1A6A1A03032236).
Author contributions
DTH, DTB, and HP conducted and analyzed all immunoassays under the guidance of SJ. DTH performed 3D modeling, and NM cultured the virus. SJ wrote the paper with input from all other authors.
Supplementary Material
Supplementary methods, figures and tables.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Mukhopadhyay S, Kuhn RJ, Rossmann MG. A structural perspective of the flavivirus life cycle. Nat Rev Microbiol. 2005;3:13
2. Lazear HM, Diamond MS. Zika virus: new clinical syndromes and its emergence in the western hemisphere. J Virol. 2016;90:4864-75
3. Paixao ES, Teixeira MG, Rodrigues LC. Zika, chikungunya and dengue: the causes and threats of new and re-emerging arboviral diseases. BMJ Glob Health. 2018;3:e000530
4. Martina BE, Koraka P, Osterhaus AD. Dengue virus pathogenesis: an integrated view. Clin Microbiol Rev. 2009;22:564-81
5. Rather IA, Lone JB, Bajpai VK, Park YH. Zika Virus Infection during Pregnancy and Congenital Abnormalities. Front Microbiol. 2017;8:581
6. Goncalves A, Peeling RW, Chu MC, Gubler DJ, de Silva AM, Harris E. et al. Innovative and new approaches to laboratory diagnosis of Zika and dengue: a meeting report. J Infect Dis. 2018;217:1060-1068
7. Duffy MR, Chen TH, Hancock WT, Powers AM, Kool JL, Lanciotti RS. et al. Zika virus outbreak on Yap Island, Federated States of Micronesia. N Engl J Med. 2009;360:2536-43
8. Musso D, Richard V, Teissier A, Stone M, Lanteri MC, Latoni G. et al. Detection of Zika virus RNA in semen of asymptomatic blood donors. Clin Microbiol Infect. 2017;23:1001 e1- e3
9. Motta IJ, Spencer BR, Cordeiro da Silva SG, Arruda MB, Dobbin JA, Gonzaga YB. et al. Evidence for transmission of Zika virus by platelet transfusion. N Engl J Med. 2016;375:1101-3
10. CDC: Atlanta, USA. Updated Guidance for US Laboratories Testing for Zika Virus Infection. Revised 24 July 2017. https://www.cdc.gov/zika/laboratories/lab-guidance.html
11. American Red Cross: Infectious Disease Testing. 2018. https://www.redcrossblood.org/biomedical-services/blood-diagnostic-testing/blood-testing.html
12. Chigurupati P, Murthy KS. Automated nucleic acid amplification testing in blood banks: An additional layer of blood safety. Asian J Transfus Sci. 2015;9:9-11
13. Priyamvada L, Quicke KM, Hudson WH, Onlamoon N, Sewatanon J, Edupuganti S. et al. Human antibody responses after dengue virus infection are highly cross-reactive to Zika virus. Proc Natl Acad Sci U S A. 2016;113:7852-7
14. Stettler K, Beltramello M, Espinosa DA, Graham V, Cassotta A, Bianchi S. et al. Specificity, cross-reactivity, and function of antibodies elicited by Zika virus infection. Science. 2016;353:823-6
15. Chames P, Van Regenmortel M, Weiss E, Baty D. Therapeutic antibodies: successes, limitations and hopes for the future. Br J Pharmacol. 2009;157:220-33
16. Kong HY, Byun J. Nucleic Acid aptamers: new methods for selection, stabilization, and application in biomedical science. Biomol Ther (Seoul). 2013;21:423-34
17. Lee KH, Zeng H. Aptamer-Based ELISA Assay for Highly Specific and Sensitive Detection of Zika NS1 Protein. Anal Sci. 2017;89:12743-8
18. Ruscito A, DeRosa MC. Small-Molecule Binding Aptamers: Selection Strategies, Characterization, and Applications. Front Chem. 2016;4:14
19. Yeh JT, Binari R, Gocha T, Dasgupta R, Perrimon N. PAPTi: a peptide aptamer interference toolkit for perturbation of protein-protein interaction networks. Sci Rep. 2013;3:1156
20. Pan K, Long J, Sun H, Tobin GJ, Nara PL, Deem MW. Selective pressure to increase charge in immunodominant epitopes of the H3 hemagglutinin influenza protein. J Mol Evol. 2011;72:90-103
21. Bao DT, Do Thi Hoang Kim HP, Cuc BT, Ngoc NM, Linh NTP, Huu NC. et al. Rapid detection of avian influenza virus by fluorescent diagnostic assay using an epitope-derived peptide. Theranostics. 2017;7:1835
22. Avila-Bonilla RG, Yocupicio-Monroy M, Marchat LA, De Nova-Ocampo MA, Del Angel RM, Salas-Benito JS. Analysis of the miRNA profile in C6/36 cells persistently infected with dengue virus type 2. Virus Res. 2017;232:139-51
23. Kaur P, Lee RC, Chu JJ. Infectious Viral Quantification of chikungunya virus-Virus plaque assay. Methods Mol Biol. 2016;1426:93-103
24. Vicenti I, Boccuto A, Giannini A, Dragoni F, Saladini F, Zazzi M. Comparative analysis of different cell systems for Zika virus (ZIKV) propagation and evaluation of anti-ZIKV compounds in vitro. Virus Res. 2018;244:64-70
25. Saha S, Raghava G. Prediction of continuous B-cell epitopes in an antigen using recurrent neural network. Proteins. 2006;65:40-8
26. EL-Manzalawy Y, Dobbs D, Honavar V. Predicting linear B-cell epitopes using string kernels. J Mol Recognit. 2008;21:243-55
27. Jespersen MC, Peters B, Nielsen M, Marcatili P. BepiPred-2.0: improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res. 2017;45:W24-W29
28. Yang J, Zhang Y. I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Res. 2015;43:W174-W81
29. Dallakyan S: Comparing Virtual Screening with Bioassays Using PyRx. 2010. https://pyrx.sourceforge.io/SI/2010/PyRx2010_rvsnALP_7122010.pdf
30. Latha MS, Saddala MS. Molecular docking based screening of a simulated HIF-1 protein model for potential inhibitors. Bioinformation. 2017;13:388-93
31. Li Y, Liu X, Dong X, Zhang L, Sun Y. Biomimetic design of affinity peptide ligand for capsomere of virus-like particle. Langmuir. 2014;30:8500-8
32. Pollard TD. A guide to simple and informative binding assays. Mol Biol Cell. 2010;21:4061-7
33. Reverdatto S, Burz DS, Shekhtman A. Peptide aptamers: development and applications. Curr Top Med Chem. 2015;15:1082-101
34. Karen L. Cox B, Viswanath Devanarayan, Aidas Kriauciunas, Joseph Manetta, Chahrzad Montrose, and Sitta Sittampalam. Immunoassay Methods. 2014. Bethesda, USA: Eli Lilly & Company and the National Center for Advancing Translational Sciences. 2004
35. Hulme EC, Trevethick MA. Ligand binding assays at equilibrium: validation and interpretation. Br J Clin Pharmacol. 2010;161:1219-37
36. Bottger R, Hoffmann R, Knappe D. Differential stability of therapeutic peptides with different proteolytic cleavage sites in blood, plasma and serum. PloS one. 2017;12:e0178943
37. Armbruster DA, Pry T. Limit of blank, limit of detection and limit of quantitation. Clin Biochem Rev. 2008;29(Suppl 1):S49-52
38. Musso D, Rouault E, Teissier A, Lanteri MC, Zisou K, Broult J. et al. Molecular detection of Zika virus in blood and RNA load determination during the French Polynesian outbreak. J Med Virol. 2017;89:1505-10
39. Gourinat AC, O'Connor O, Calvez E, Goarant C, Dupont-Rouzeyrol M. Detection of Zika virus in urine. Emerg Infect Dis. 2015;21:84-6
40. Jenssen H, Aspmo SI. Serum stability of peptides. Methods Mol. Biol. 2008;494:177-86
41. Thrailkill KM, Moreau CS, Cockrell G, Simpson P, Goel R, North P. et al. Physiological matrix metalloproteinase concentrations in serum during childhood and adolescence, using Luminex Multiplex technology. Clin Chem Lab Med. 2005;43:1392-9
42. Artigas JM, Garcia ME, Faure MR, Gimeno AM. Serum trypsin levels in acute pancreatic and non-pancreatic abdominal conditions. Postgrad Med J. 1981;57:219-22
43. Yang M, Lai H, Sun H, Chen Q. Virus-like particles that display Zika virus envelope protein domain III induce potent neutralizing immune responses in mice. Sci Rep. 2017;7:7679
44. Abbink P, Larocca RA, De La Barrera RA, Bricault CA, Moseley ET, Boyd M. et al. Protective efficacy of multiple vaccine platforms against Zika virus challenge in rhesus monkeys. Science. 2016;353:1129-32
45. Sapparapu G, Fernandez E, Kose N, Bin C, Fox JM, Bombardi RG. et al. Neutralizing human antibodies prevent Zika virus replication and fetal disease in mice. Nature. 2016;540:443-7
46. Wang J, Bardelli M, Espinosa DA, Pedotti M, Ng TS, Bianchi S. et al. A Human Bi-specific Antibody against Zika Virus with High Therapeutic Potential. Cell. 2017;171:229-41 e15
47. Zhang B, Pinsky BA, Ananta JS, Zhao S, Arulkumar S, Wan H. et al. Diagnosis of Zika virus infection on a nanotechnology platform. Nat Med. 2017;23:548-50
48. Pardee K, Green AA, Takahashi MK, Braff D, Lambert G, Lee JW. et al. Rapid, Low-Cost Detection of Zika Virus Using Programmable Biomolecular Components. Cell. 2016;165:1255-66
49. Rey FA, Stiasny K, Vaney MC, Dellarole M, Heinz FX. The bright and the dark side of human antibody responses to flaviviruses: lessons for vaccine design. EMBO Rep. 2018;19:206-24
50. Metz SW, Gallichotte EN, Brackbill A, Premkumar L, Miley MJ, Baric R. et al. In Vitro Assembly and Stabilization of Dengue and Zika Virus Envelope Protein Homo-Dimers. Sci Rep. 2017;7:4524
51. Tate J, Ward G. Interferences in immunoassay. Clin Biochem Rev. 2004;25:105-20
52. Shaternikov VA, Kniazev II. Interaction of trypsin with inhibitors of human blood serum. Vopr Med Khim. 1966;12:496-502
53. Alcendor DJ. Zika Virus Infection of the Human Glomerular Cells: Implications for Viral Reservoirs and Renal Pathogenesis. J Infect Dis. 2017;216:162-71
54. Chen J, Yang YF, Chen J, Zhou X, Dong Z, Chen T. et al. Zika virus infects renal proximal tubular epithelial cells with prolonged persistency and cytopathic effects. Emerg Microbes Infect. 2017;6:e77
55. Laman JD, van den Eertwegh AJ, Deen C, Vermeulen N, Boersma WJ, Claassen E. Synthetic peptide conjugates with horseradish peroxidase and beta-galactosidase for use in epitope-specific immunocytochemistry and ELISA. J Immunol Methods. 1991;145:1-10
56. Hnasko RM. Bioconjugation of Antibodies to Horseradish Peroxidase (HRP). Methods Mol Biol. 2015;1318:43-50
57. Zhou W, Huang PJ, Ding J, Liu J. Aptamer-based biosensors for biomedical diagnostics. Analyst. 2014;139:2627-40
Author contact
![]() Corresponding author: Seon-Ju Yeo, Ph.D., Zoonosis Research Center, Department of Infection Biology, School of Medicine, Wonkwang University, 460, Iksan-daero, Iksan, 54538, Republic of Korea. Tel +82-63-850-6777; Fax +82-63-857-0342; Email: yeosjac.kr
Corresponding author: Seon-Ju Yeo, Ph.D., Zoonosis Research Center, Department of Infection Biology, School of Medicine, Wonkwang University, 460, Iksan-daero, Iksan, 54538, Republic of Korea. Tel +82-63-850-6777; Fax +82-63-857-0342; Email: yeosjac.kr
 Global reach, higher impact
Global reach, higher impact