13.3
Impact Factor
Theranostics 2018; 8(13):3544-3558. doi:10.7150/thno.24607 This issue Cite
Research Paper
A novel HPV16 E7-affitoxin for targeted therapy of HPV16-induced human cervical cancer
1. Institute of Molecular Virology and Immunology, Department of Microbiology and Immunology, School of Basic Medical Sciences, Wenzhou Medical University, Wenzhou 325035, Zhejiang, P. R. China
2. Department of Gynecology, the Second Affiliated Hospital of Wenzhou Medical University, Wenzhou 325035, Zhejiang, China.
* These three authors contributed equally to this research.
Received 2017-12-27; Accepted 2018-4-27; Published 2018-6-7
Abstract
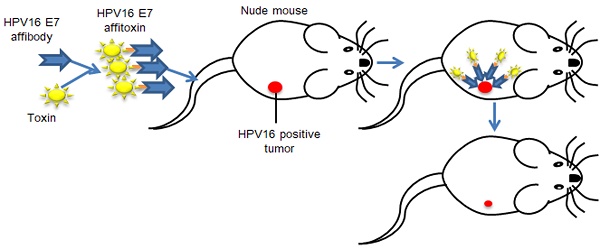
Cervical cancer, the second most common cause of cancer death in women worldwide, is significantly associated with infection of high-risk human papillomaviruses (HPVs), especially the most common genotype, HPV 16. To date, there is no established noninvasive therapy to treat cervical cancer.
Methods: Here, we report a novel affitoxin that targets HPV16 E7 protein, one of the primary target proteins in molecular targeted therapy for HPV-induced cervical cancer. The affitoxin, ZHPV16E7 affitoxin384 was generated by fusing the modified Pseudomonas Exotoxin A (PE38KDEL) to the HPV16 E7-specific affibody. The expressed and purified ZHPV16E7 affitoxin384 was characterized using numerous methods. SPR assay, indirect immunofluorescence assay, and near-infrared (NIR) optical imaging were respectively performed to assess the targeting ability of ZHPV16E7 affitoxin384 to HPV16 E7 protein both in vitro and in vivo. Cell viability assays and SiHa tumor-bearing nude mice were used to evaluate the efficacy of ZHPV16 E7 affitoxin384 in vitro and in vivo, respectively.
Results: Using in vitro methods the SPR assay and indirect immunofluorescence assay showed that ZHPV16E7 affitoxin384 targeted HPV16 E7 with high binding affinity and specificity. Significant reduction of cell viability in HPV16 positive cells was observed in the presence of ZHPV16 E7 affitoxin384. By NIR optical imaging, ZHPV16 E7 affitoxin384 specifically targeted HPV16 positive tumors in vivo. ZHPV16E7 affitoxin384 showed significant in vivo antitumor efficacy in two kinds of tumor-bearing nude mouse models.
Conclusions: ZHPV16E7 affitoxin384 is a potent anti-cervical cancer therapeutic agent that could be effective against HPV16 positive tumors in humans.
Keywords: HPV16, affitoxin, targeted therapy, cervical cancer
Introduction
Cervical cancer is the second most common cause of cancer-related death in women worldwide, especially in developing countries. Approximately 445,000 new cases of cervical cancer are diagnosed each year and 270,000 deaths [1]. Evidence from clinical, epidemiological, and molecular data studies has demonstrated that infections of high-risk types of human papillomaviruses (HPVs) such as HPV16, 18, 31, 33, 35, 45, 52, and 58 are significantly associated with cervical cancer development. Among these high-risk types, HPV16 is the most common worldwide HPV genotype, which is responsible for approximately 50% of cervical cancer cases [2, 3]. Although the commercial HPV vaccines are highly efficient in preventing HPV infection, there is no established noninvasive therapy to treat the HPV-associated lesions such as squamous intraepithelial lesions and cervical cancer. Therefore, the development of effective treatment strategies needs urgent attention.
HPV is a non-enveloped, double-stranded, circular DNA virus. Its genome is composed of about 8000 base pairs, which encodes six to seven early proteins (E1, E2, E4, E5, E6, E7 and E8), and two late structural proteins (L1 and L2). HPV infection can lead to the transformation of basal epithelial cells. In the process of the transformation, up-regulated E6 and E7 oncoproteins principally contribute to malignant transformation and retain the malignant phenotype of cervical cancers [4-6]. Therefore, HPV E6 and E7 proteins may be excellent target proteins in molecular targeted therapy for HPV-induced cervical cancer [7-10].
To date, monoclonal antibodies (mAbs) are the most successful proteins applied in molecular targeted therapy [11-13]. However, mAbs still have some disadvantages (reviewed by Patrick, et al.) [14]. Firstly, it is difficult to produce the active form of mAbs because of their large molecular weight (about 150 kD) and multiple posttranslational modifications such as glycosylation. Secondly, only a small percentage of the administered dose of mAbs can target tumors because most mAbs bind to tumor-specific antigens in the blood. Thirdly, it is difficult for mAbs to reach their target cells in solid tumors because of their poor tissue-penetrating ability. Affibodies, derived from the Z-domain of Staphylococcal protein A (SPA-Z) and based on a 58 amino-acid scaffold, are a new class of affinity proteins with high affinity and specificity [15-19]. Affibodies have the favorable molecular recognition properties of antibodies with improved characteristics, such as small size (~7 kDa) (affibodies are almost 20 times smaller than full-size antibodies and four times smaller than single-chain variable fragment (scFvs)), single domain, high stability, absence of cysteines, fastest folding reaction, high yield bacterial expression and low immunogenicity [20]. Therefore, affibodies and their derivatives are attractive surrogates for antibodies or scFvs in tumor targeted therapy [21-23].
In our previous study, we generated four HPV16 E7-specific affibodies (ZHPV16E7127, ZHPV16E7301, ZHPV16E7384, and ZHPV16E7745) using phage display technology [24]. In order to enhance the cytotoxic efficacy, we connected the modified Pseudomonas Exotoxin A (PE38KDEL) toxin [25] to the N terminal of ZHPV16E7384 by a flexible peptide (Gly4Ser)3 to generate HPV16 E7-specific affibody-PE38KDEL toxin molecule (named as ZHPV16 E7 affitoxin384). In this study, we report the characterization of this novel recombinant protein ZHPV16 E7 affitoxin384 for its binding ability to recombinant and native HPV16 E7 protein, its cytotoxic effect on HPV16 positive cervical cancer cell lines, and the in vivo evaluation of targeted therapy for cervical cancer in tumor-bearing nude mice.
Methods
Animals, cells and vectors
Female athymic nude mice (nu/nu genotype, BALB/c background) and BALB/c mice, 6 to 8 weeks old, were purchased from Shanghai SLAC Laboratory Animal Co. Ltd and kept at the animal facility of Wenzhou Medical University, China. ICR mice, weighing 23-27 g, were purchased from the animal experimental center of Wenzhou Medical University, China. All of the animal procedures were performed according to approved protocols and in accordance with recommendations for the proper use and care of laboratory animals. SiHa (ATCC: HTB-35, HPV16 positive, contains about one to two copies of integrated HPV16 genome), CaSki (ATCC: CRL-1550, HPV16 positive, contains about 600 copies of integrated HPV16 genome), HeLa 229 (ATCC: CCL-2.1, HPV18 positive, used as HPV16 negative control cell line), and melanoma tumor A375 (ATCC: CRL-1619, used as HPV negative control cell line) were obtained from the American Type Culture Collection (ATCC, USA) and cultured as previously described [24]. The pET21a(+) vector and E.coli BL21 (DE3) were purchased from Novagen and ATCC, respectively.
Reagents
The reagents used, including Cell Counting Kit-8 (CCK-8) (Dojindo, Japan), RPMI-1640 (Gibco, USA), fetal bovine serum (FBS) (Gibco, USA), penicillin (Gibco, USA), trypsin-EDTA (Gibco, USA), streptomycin (Sigma Aldrich, Saint Louis, USA), Isopropyl-D-thiogalactopyranoside (IPTG) (Sigma Aldrich, Saint Louis, USA), Ni-NTA agarose (Qiagen Inc., Valencia, CA), and DyLight-755 (Thermo Fisher Scientific, USA), were purchased from commercial sources. The anti-HPV16 E7 rabbit polyclonal antibody and anti-His tag mouse monoclonal antibody (Abcam, Boston, MA, USA), goat anti-rabbit IgG (H+L) conjugated with HRP and goat anti-mouse IgG (H+L) conjugated with HRP, goat anti-rabbit antibody conjugated with FITC and goat anti-mouse antibody conjugated with alexa fluor 647 (MultiSciences Biotech Co., Ltd, China) were purchased from commercial sources. The mouse immune serum anti-PE38KDEL, rabbit immune serum anti-SPA-Z and anti-HPV16 E7 recombinant proteins were prepared in our laboratory [26, 27].
Expression and purification of ZHPV16E7 affitoxin384
A random combinatorial phage display library with 1×108 variants of protein SPA-Z had been prepared by using phage display technology in our laboratory, from which four affibodies specifically binding to HPV16 E7 were selected [24]. The ZHPV16E7384 was selected to construct an affitoxin for further study.
The toxin part used in this study was the mutant fragment of Pseudomonas Exotoxin A with the C-terminal part optimized to KDEL sorting signal and then the toxin was named as PE38KDEL [28, 29]. PE38KDEL was ligated to the C-terminus of ZHPV16E7384 polypeptide by a flexible (Gly4Ser)3-linker domain. In brief, the DNA sequence corresponding to PE38KDEL with (Gly4Ser)3-linker domain modified based on prokaryotic codon usage by the JCat software (Java Codon Adaptation Tool, http://www.jcat.de/) was synthesized by Shanghai Sangon Biotech Co. Ltd and then cloned into vector pET21a (+) between the EcoR I and Xho I sites to generate the recombinant plasmid pET21a (+)/PE38KDEL. The DNA sequence of ZHPV16E7384 was amplified by PCR using the plasmid pet21a (+)/ZHPV16 E7384 as a template [24] and cloned into the pET21a (+)/PE38KDEL vector in frame with the 6×His tag between Nde I and EcoR I sites to generate the recombinant plasmid pET21a (+)/ZHPV16E7 affitoxin384 (The protein encoded by this vector was named as ZHPV16E7 affitoxin384). Meanwhile, SPA-Z molecule (Zwt), was used as a negative control. The plasmid pET21a (+)/Zwt affitoxin (Zwt affitoxin) was constructed in the same method as ZHPV16 E7 affitoxin384. The two recombinant plasmids were confirmed by sequencing.
The expression of ZHPV16 E7 affitoxin384 and Zwt affitoxin in E. coli BL21 (DE3) was induced by 1 mM IPTG (Sigma-Aldrich Co., St. Louis, MO) and verified by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and western blot analysis. Then, the proteins were purified by Ni NTA Sepharose column (Qiagen Inc., Valencia, CA) as described in previous study [24].
Western blot analysis
Western blot analysis was performed to confirm the expression of ZHPV16 E7 affitoxin384 and Zwt affitoxin. Whole cell lysates were run on a 12% SDS-polyacrylamide gel and transferred onto polyvinylidene difluoride membranes (Millipore, USA). Membranes were blocked with 10% skim milk in PBST (1× PBS + 0.1% Tween-20) for 2 h, incubated with indicated primary antibodies, and then incubated with HRP-conjugated secondary antibody. The protein bands were visualized using 0.005% (w/v) 4-chloro-1-naphthol and a 0.015% (v/v) hydrogen peroxidase color development substrate.
Surface plasmon resonance assay
To assess the interaction between the HPV16 E7 protein and ZHPV16 E7 affitoxin384, surface plasmon resonance (SPR) assay was performed with the ProteOn XPR36 protein interaction array system (Bio-Rad Laboratories, USA) as described in a previous study [24]. Zwt affitoxin was used as a negative control. The processed data was fit globally using BIA evaluation software (version 3.0.2; Biacore) for a 1:1 Langmuir model.
Indirect immunofluorescence assay
To assess the in vitro targeting ability of ZHPV16 E7 affitoxin384, indirect immunofluorescence assay was performed as described in a previous study [24]. In brief, SiHa, CaSki, HeLa and A375 cells were seeded in dishes suitable for confocal microscopy observation. After incubation for 24 h, the medium was replaced with fresh challenge media supplemented with 50 μM of ZHPV16 E7 affitoxin384, ZHPV16 E7384 or Zwt affitoxin. Cells were analyzed after treatment for 6 h. The cells were washed with PBS and fixed with 4% paraformaldehyde for 10 min at room temperature. Then, cells were washed with cold 0.01 M PBST, and placed in blocking buffer (PBS containing 5% FBS) at 4 °C overnight, followed by incubation with indicated primary antibodies for 1 h at room temperature. The cells were washed with cold PBST three times and incubated with indicated secondary antibodies for 1 h at room temperature. Cell nuclei were stained with 50 μg/mL propidium iodide (PI) (MultiSciences Biotech Co., Ltd China) at room temperature for 5 min. The cells were analyzed by a confocal fluorescence microscope (TC-1, Nikon, Japan).
To further confirm the specific binding ability of ZHPV16 E7 affitoxin384 to HPV16E7 protein, the co-localization was determined in SiHa cells expressing HPV16 E7 protein by confocal double immunofluorescence assay. The experimental procedure was similar to the description mentioned above.
In vitro efficacy of ZHPV16 E7 affitoxin384
To evaluate the efficacy of ZHPV16 E7 affitoxin384, cell viability assay was performed with CCK-8 kit (Dojindo) as described in a previous study with minor modifications [30]. Briefly, SiHa, CaSki, HeLa and A375 cells were plated onto a 96-well plate at 1×104 cells per well, followed by treatment with ZHPV16 E7 affitoxin384 at different concentrations (0.1, 0.25, 0.5, 1.0 and 2.0 μM). Cells treated with the same concentrations of Zwt affitoxin were used as negative controls. Cell viability was determined after incubation for 0, 1, 3, 6, 12, 24, 36, 48 and 72 h, respectively. According to the manufacturer's instructions, 10 µL of CCK-8 solution was added to each well of the 96-well plate and then incubated for an additional 4 h. Absorbance was measured at 450 nm using a microplate reader (Synergy HT; BioTek, Winooski, VT, USA). The percentage of cell viability was calculated according to the following formula: Cell viability (%) = optical density (OD) of the treatment group/OD of the control group × 100%. The half maximal inhibitory concentration (IC50) values were calculated using GraphPad Prism software (GraphPad Software, Inc.).
Tumor targeting ability of ZHPV16 E7 affitoxin384 in tumor-bearing nude mice
The dynamic distribution and tumor targeting ability of ZHPV16 E7 affitoxin384 were investigated in nude mice using near-infrared (NIR) optical imaging. Briefly, 1×107 of SiHa, CaSki, Hela and A375 cells were respectively injected subcutaneously into the right forelimb of nude mice (n = 5 per group). When the tumor size reached 0.3-0.4 cm in diameter, mice were used for NIR imaging and treatment. ZHPV16 E7 affitoxin384, ZHPV16 E7384 (the positive control) and Zwt affitoxin (the negative control) were labeled with a maleimide derivative of DyLight-755 (Thermo Fisher Scientific, USA, 62278) by attaching the dye to the native lysine residues of affibody molecules according to the manufacturer's protocol. Then, 200 pmol of affibody-DyLight-755 or affitoxin-DyLight-755 dissolved in 100 μL of PBS were injected intravenously. The fluorescence imaging was performed at various time points post-injection using an NIR imaging system (CRi Maestro 2.10, USA). The tumor/muscle ratios (T/M ratio) (T/M ratio = (tumor signal - background signal) / (muscle signal - background signal) × 100%) were analyzed at various time points post injection. The mice were sacrificed at 48 h after the imaging study, and the tumor and dissected organs such as liver, spleen, brain and other organs were analyzed using NIR optical imaging fluorescence analysis.
Evaluation of ZHPV16 E7 affitoxin384 acute toxicity
BALB/c female mice (n = 5 per group) were administered with the indicated doses (5, 10, 20, 50, 100, 200, 300, 400 and 500 nmol/kg) of ZHPV16 E7 affitoxin384 by intravenous injection into the tail vein. Any reported death cases or moribund conditions that occurred within the 2-week post injection period were taken into consideration. All experiments were performed in triplicate. The lethal dose 50% (LD50) value was calculated by GraphPad Prism 5.0 Software.
A total of 96 ICR mice were randomly divided into 4 groups. Mice in group 2, 3, and 4 were respectively administered the indicated doses (20, 100, and 200 nmol/kg) of ZHPV16 E7 affitoxin384 by intravenous injection into the tail vein, while mice from group 1 were administered PBS as a control. Blood samples were collected at day post-injection (DPI) 1, 3, 7, and 14 in EDTA tubes for complete blood count and in plain tubes for liver and kidney function panel test (n=3 mice per time point per test). Then, blood samples in EDTA tubes were analyzed using a Mindray BC-5380 Hematology Analyzer (Shenzhen Mindray Bio-Medical Electronics Co., Ltd, Shenzhen, China) and serums generated from blood samples in plain tubes were analyzed using a Beckman AU680 Chemistry Analyzer (Beckman Coulter, Brea, CA, USA).
Pharmacokinetics of ZHPV16 E7 affitoxin384
Nude mice were divided into 2 groups (n = 3 per group) for intravenous administration of ZHPV16 E7 affitoxin384 (100 nmol/kg) or PBS, respectively. A total of 100 μL of blood was respectively collected at 1, 5, 15, 30, 60 and 120 min post injection from each mouse. At 5 h post injection (hpi), the mice were anesthetized and terminally bled. Blood samples were obtained from the fossa orbitalis and serum levels of ZHPV16 E7 affitoxin384 were measured by recombinant HPV16E7 protein-based ELISA. Briefly, a 96-well plate was coated with purified HPV16E7 protein (100 μg/mL) in carbonate coating buffer (100 μL/well). After an overnight incubation at 4 °C, the plates were rinsed with PBS solution containing 0.05% Tween-20. The coated wells were blocked with 100 μL blocking buffer (PBST containing 5% nonfat dry milk, w/v) at 37 °C for 1 h then washed five times with PBST. Blood samples (at a dilution of 1:100 and 100 μL/well) were added to the wells in triplicate, followed by incubation at 37 °C for 2 h. The rabbit anti-SPA-Z serum polyclonal antibody (at a dilution of 1:2000 and 100 μL/well) and the HRP-conjugated goat anti-rabbit IgG (H1L) (ABR, USA) (at a dilution of 1:5000 and 100 μL/well) were added to the corresponding wells and incubated for 1 h at 37 °C. The wells were washed again, followed by the addition of 100 μL/well of 3, 39, 5, 59-tetramethylbenzidine-H2O2 solution. The reaction was carried out at room temperature for 20 min and halted with 50 μL of 2 M of H2SO4 per well. The absorbance at 450 nm was measured using a Bio-tek ELISA microplate reader. Meanwhile, a second 96-well plate was coated with purified HPV16E7 protein, followed by adding the rabbit serum anti-HPV16E7 or anti-SPA-Z instead of adding the blood sample as positive and negative controls, respectively. To make a standard concentration curve, a 96-well plate was respectively coated with the ZHPV16 E7 affitoxin384 protein at various concentrations (0, 0.002, 0.02, 0.2 and 2 nM) and detected by ELISA as described above. The circulation half-lives and initial concentrations of the proteins were calculated using GraphPad Prism 5.0 Software.
In vivo antitumor efficacy of ZHPV16 E7 affitoxin384
Therapeutic efficacy of ZHPV16 E7 affitoxin384 was studied using SiHa tumor-bearing nude mice by two methods. The first method was that SiHa tumor-bearing mice were prepared in advance, followed by injection of ZHPV16 E7 affitoxin384. Briefly, nude mice were randomly divided into 5 groups (n= 5 per group). The tumors were initiated by subcutaneous injection of 1×107 cells, which were suspended in 0.1 mL of PBS, into the right forelimb of each nude mouse. Tumor dimensions were measured periodically using calipers, and their volumes calculated using the formula: volume = length × width2 × 0.52. When the tumor size reached 0.1-0.2 cm in diameter, mice were treated with 0.2 mL of ZHPV16 E7 affitoxin384 (100 nmol/kg), Zwt affitoxin (100 nmol/kg), ZHPV16 E7384 (100 nmol/kg), PE38KDEL (100 nmol/kg), or PBS, respectively. The indicated agents were injected every three days for six time points via tail vein. The therapeutic efficacies and systemic toxicities of affitoxin proteins were evaluated based on daily measurements of tumor volume and body weight. Tumors from mice in the aforementioned five groups were separated and weighed after all treatments and observations were finished.
The second method involved simultaneous injection of ZHPV16 E7 affitoxin384 and preparation of SiHa tumor-bearing mice. Briefly, nude mice were randomly divided into 5 groups (n= 5 per group). The tumors were initiated by subcutaneous injection of 1×107 cells, which were suspended in 0.1 mL of PBS, into the right forelimb of each nude mouse. Meanwhile, mice were treated with 0.2 mL of ZHPV16 E7 affitoxin384 (100 nmol/kg), Zwt affitoxin (100 nmol/kg), ZHPV16 E7384 (100 nmol/kg), PE38KDEL (100 nmol/kg), and PBS, respectively. The indicated agents were injected every three days for six time points via tail vein. Tumor dimensions were measured as mentioned above. The therapeutic efficacies and systemic toxicities of affitoxin proteins were evaluated based on daily measurements of tumor volume and body weight. Tumors from mice in the above five groups were separated and weighed after all treatments and observations were finished.
Evaluation of ZHPV16E7 affitoxin384 immunogenicity
To evaluate the immunogenicity of ZHPV16E7 affitoxin384 in tumor treatment, we analyzed the levels of ZHPV16E7 affitoxin384-specific antibody by ELISA using the recombinant ZHPV16E7 affitoxin384 as an antigen. Briefly, serums were obtained via tail vein bleedings from all groups of mice in the tumor therapy mentioned above at day 0 and day 35, respectively. Then, 200 pmol of purified recombinant ZHPV16 E7 affitoxin384 was added to each well of a 96-microtiter plate. After an overnight incubation at 4 °C, the plates were rinsed with PBS solution containing 0.05% Tween-20. The coated wells were blocked with 100 μL blocking buffer (PBST containing 5% nonfat dry milk, w/v) at 37 °C for 1 h then plates were washed 5 times with PBST. Serum samples (at a dilution of 1:100 and 100 μL/well) were added to the wells in triplicate, followed by incubation at 37 °C for 2 h. After the plates were washed 5 times with PBST, the HRP-conjugated goat anti-mouse IgG (H+L) (MultiSciences Biotech Co., Ltd, China) (at w dilution of 1:5000 and 100 μL/well) was added to the corresponding wells and incubated for 1 h at 37 °C. The wells were washed again, followed by the addition of 100 μL/well of 3, 39, 5, 59-tetramethylbenzidine-H2O2 solution. The reaction was carried out at room temperature for 20 min and halted with 50 μL of 2 M of H2SO4 per well. The absorbance at 450 nm was measured using a Bio-tek ELISA microplate reader.
Statistical analysis
Data are presented as mean ± SD. Statistical analysis of the significance between groups was conducted using Students t-test, and P < 0.05 was considered to be statistically significant. All calculations were performed with the software SPSS16.0.
Results
Generation of ZHPV16 E7 affitoxin384
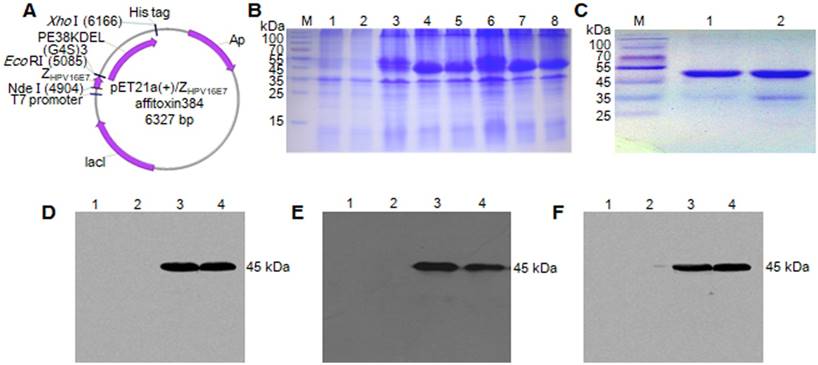
The recombinant plasmids of pET21a (+)/ZHPV16 E7 affitoxin384 and pET21a (+)/Zwt affitoxin were constructed and confirmed by sequencing (Figure 1A). ZHPV16 E7 affitoxin384 and Zwt affitoxin that were expressed in E. coli. BL21 (DE3) were purified and analyzed by SDS-PAGE (Figure 1B-C). The expression of these proteins was confirmed by western blot analysis using anti-His tag mouse monoclonal antibody (Figure 1D), anti-PE38KDEL mouse serum antibody (Figure 1E) and anti-SPA-Z rabbit serum antibody (Figure 1F), respectively. The protein bands at 45 kDa were detected, which were consistent with the expected weights of ZHPV16E7 affitoxin384 and Zwt affitoxin. These results indicated that ZHPV16E7 affitoxin384 or Zwt affitoxin proteins were expressed in the E. coli. BL21 (DE3) strains and purified successfully.
ZHPV16E7 affitoxin384 interacted with HPV16 E7 with high binding affinity
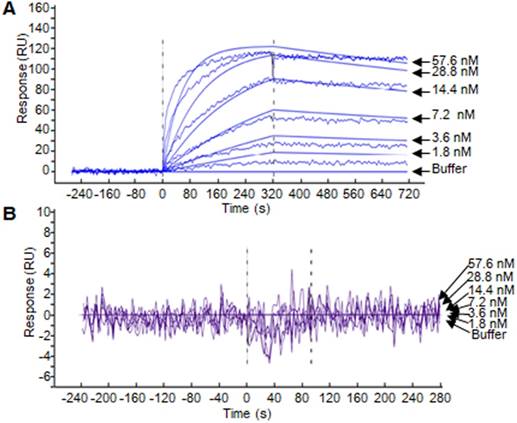
SPR assay was performed to evaluate the binding affinity of the ZHPV16E7 affitoxin384 to HPV16 E7 protein. The data were fit globally using a simple Langmuir binding model (Figure 2 and Table 1). The results showed that the dissociation equilibrium constant (KD) of ZHPV16E7 affitoxin384 was 2.12×10-7 M while the counterpart of Zwt affitoxin was 5.50×10-2 M, which indicated that the binding affinity of ZHPV16 E7 affitoxin384 to HPV16 E7 was significantly higher than that of Zwt affitoxin.
ZHPV16 E7 affitoxin384 interacted with HPV16 E7 with high binding specificity
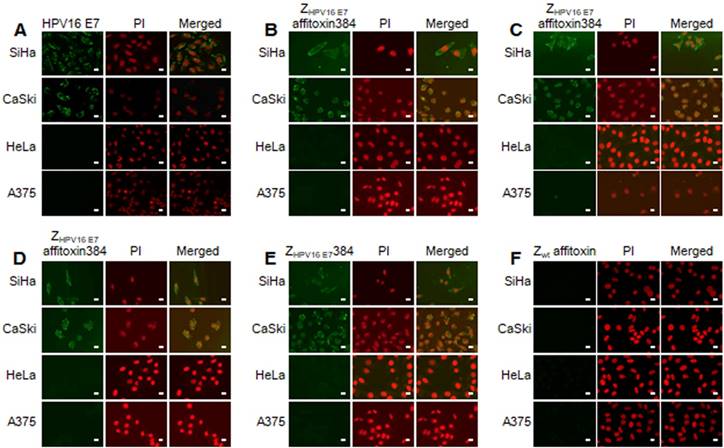
The binding specificity of ZHPV16 E7 affitoxin384 to HPV16 E7 was tested by indirect immunofluorescence assay. SiHa and CaSki cells incubated with ZHPV16 E7 affitoxin384 were compared to HeLa and A375 cells as negative controls. The anti-His tag mouse monoclonal antibody, anti-SPA-Z rabbit immune serum antibody and anti-PE38KDEL mouse immune serum antibody were respectively used as the primary antibodies. Cells incubated with ZHPV16 E7384 and Zwt affitoxin were used as positive and negative controls, respectively. Meanwhile, control cells (no affitoxin incubation), were analyzed using anti-HPV16 E7 rabbit polyclonal antibody as a positive control. As shown in Figure 3A, SiHa and CaSki cells were recognized by anti-HPV16E7 antibody and green fluorescence was observed in these cells whereas HeLa and A375 cells were not recognized by anti-HPV16E7 antibody. When SiHa and CaSki cells were incubated with ZHPV16 E7 affitoxin384, all of the three antibodies (anti-His tag mouse monoclonal antibody, anti-SPA-Z rabbit immune serum antibody and anti-PE38KDEL mouse immune serum antibody) recognized the ZHPV16 E7 affitoxin384. The fluorescence signals were predominantly located around the nucleus and plasma (Figure 3B-D). Similar results were obtained when SiHa and CaSki cells were incubated with ZHPV16 E7384 (Figure 3E). However, there was no visible signal observed when SiHa and CaSki cells were incubated with Zwt affitoxin (Figure 3F). Similar negative results were also obtained when HeLa and A375 cells were incubated with ZHPV16 E7 affitoxin384 (Figure 3B-D, F).
Expression and purification of ZHPV16 E7 affitoxin 384 protein. The pET21a (+)/ZHPV16 E7 affitoxin384 plasmid was transformed into E. coli BL21 (DE3). The protein was expressed and purified by Ni-NTA agarose affinity chromatography. (A) Schematic structure of pET21a (+)/ZHPV16 E7 affitoxin384 plasmid. (B) Comassie blue-stained SDS-PAGE gel of the recombinant proteins. M, protein marker; 1, Empty E.coli BL21 (DE3); 2, E.coli BL21 (DE3) transformed with pET21a empty vector; 3, E.coli BL21 (DE3) transformed with pET21a (+)/ZHPV16 E7 affitoxin384 plasmid; 4-5, E.coli BL21 (DE3) transformed with pET21a (+)/ZHPV16 E7 affitoxin384 plasmid and induced by 1 mM IPTG; 6, E.coli BL21 (DE3) transformed with pET21a (+)/Zwt affitoxin; 7-8, E.coli BL21 (DE3) transformed with pET21a (+)/Zwt affitoxin and induced by 1 mM IPTG. (C) Analysis of the purified ZHPV16 E7 affitoxin384 and Zwt affitoxin recombinant proteins by SDS-PAGE. M, protein marker; 1, ZHPV16 E7 affitoxin384; 2, Zwt affitoxin. (D-F) Confirmation of the expression of ZHPV16 E7 affitoxin384 and Zwt affitoxin recombinant proteins by western blot using the primary antibodies against His tag, PE38KDEL and SPA-Z, respectively. 1, Empty E.coli BL21 (DE3); 2, E.coli BL21 (DE3) transformed with pET21a empty vector; 3, E.coli BL21 (DE3) transformed with pET21a (+)/ZHPV16 E7 affitoxin384 plasmid and induced by 1 mM IPTG; 4, E.coli BL21 (DE3) transformed with pET21a (+)/Zwt affitoxin and induced by 1 mM IPTG.
The kinetic binding constants of ZHPV16 E7 affitoxin384 interacting with HPV16 E7 protein.
| Binding configuration | ka (1/Ms) | kd (1/s) | KD (M) | n |
|---|---|---|---|---|
| ZHPV16 E7 affitoxin384 | 1.84×103 | 3.91×10-4 | 2.12×10-7 | 3 |
| Zwt affitoxin | 3.00×10-5 | 1.65×10-6 | 5.50×10-2 | 3 |
Biosensor binding analysis of the purified ZHPV16 E7 affitoxin384 protein. (A) The binding ability of different concentrations of purified ZHPV16 E7 affitoxin384 protein to HPV16 E7 protein was tested using SPR-based binding assay. (B) The corresponding concentration of Zwt affitoxin protein was used as a negative control. All samples were run in triplicate. The smooth curves represent a global analysis of data using a Langmuir binding model.
Analysis of binding specificity of ZHPV16 E7 affitoxin384 to HPV16 E7 by indirect immunofluorescence assay. SiHa and CaSki cells (HPV16-positive) incubated with ZHPV16 E7 affitoxin384 were used as the tested cells. HeLa and A375 cells (HPV16-negative) with the same incubation were used as control cells. (A) Cells without incubation with affitoxin were analyzed with anti-HPV16 E7 rabbit serum antibody as positive controls. (B-D) Cells incubated with ZHPV16 E7 affitoxin384 were analyzed with three different kinds of primary antibodies against His tag, PE38KDEL and SPA-Z, respectively. (E) Cells incubated with ZHPV16 E7 384 were analyzed with the primary antibody against His tag as positive control. (F) Cells incubated with Zwt affitoxin were analyzed with the primary antibody against SPA-Z as negative control. All corresponding secondary antibodies were labelled with FITC (green). The nuclei of cells were stained with PI (red). Fluorescence signals were observed using a confocal fluorescence microscope. Scale bar = 10 μm.
The subcellular co-localization of ZHPV16 E7 affitoxin384 with HPV16 E7 in SiHa cells. SiHa cells that were respectively incubated with Zwt affitoxin, ZHPV16 E7 384 and ZHPV16 E7 Affitoxin 384 were analyzed by indirect immunofluorescence assay. The anti-HPV16E7 rabbit polyclonal antibody and anti-His tag mouse monoclonal antibody were used as primary antibodies. The goat anti-rabbit antibody conjugated with FITC (green) and goat anti-mouse antibody conjugated with alexa fluor 647 (rose red) were used as secondary antibodies. Cell nucleuses were stained with PI (red). The merged images showed the co-localization of ZHPV16 E7 affitoxin384 with HPV16 E7 (gray white). Scale bar = 10 μm. White boxes represent selected zoomed area.
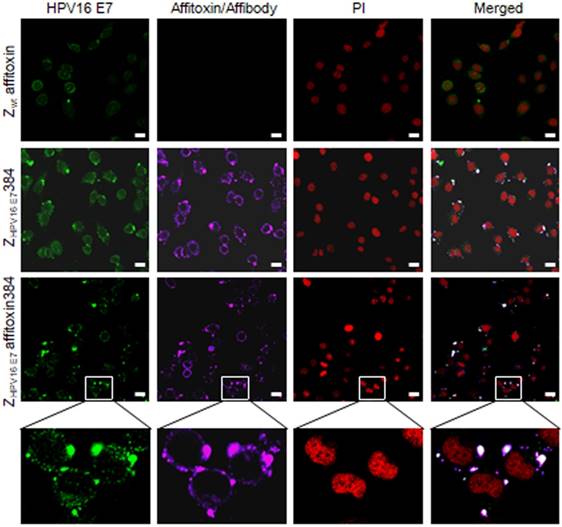
To further confirm the binding specificity of ZHPV16 E7 affitoxin384 to HPV16 E7, a confocal immunofluorescence co-localization assay was performed. As shown in Figure 4, in SiHa cells, the fluorescence signals of HPV16 E7 and ZHPV16 E7 affitoxin384 were co-localized.
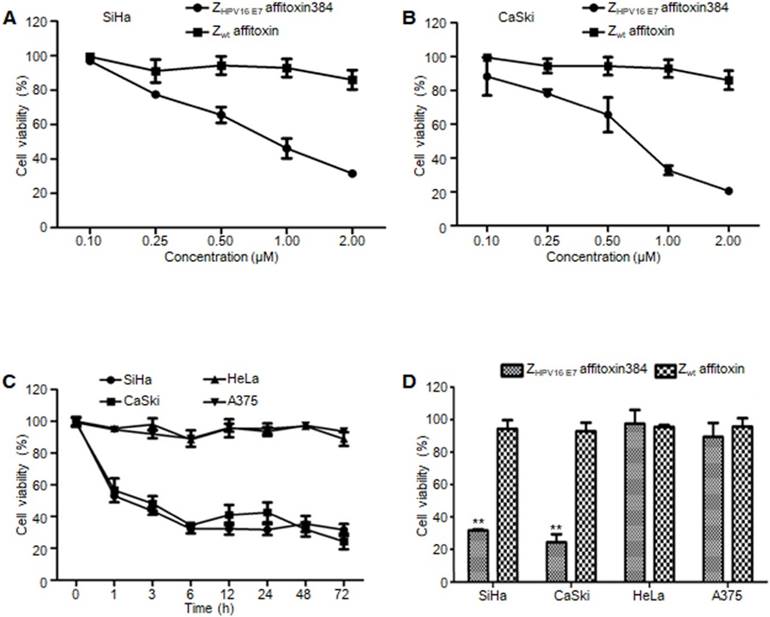
ZHPV16 E7 affitoxin384 significantly reduced the cell viability of HPV16 positive cells
In order to evaluate the efficacy of ZHPV16 E7 affitoxin384 and its potential cytotoxicity, a cell viability assay was performed. Cell viability of SiHa, CaSki, HeLa and A375 cells treated with various concentrations of ZHPV16 E7 affitoxin384 was measured using CCK-8 kit. As shown in Figure 5A-B, the cell viability of SiHa and CaSki cells decreased with the increase of concentration of ZHPV16 E7 affitoxin384. In contrast, the cells treated with various concentrations of Zwt affitoxin remained fully viable. The highest concentration at 2 μM was selected for further investigation. The efficacy of ZHPV16 E7 affitoxin384 was evaluated in a time course (0, 1, 3, 6, 12, 24, 48 and 72 h). As shown in Figure 5C-D, 2 μM of ZHPV16 E7 affitoxin384 significantly reduced the viability of SiHa and CaSki, whereas HeLa and A375 cells treated with the same concentration of ZHPV16 E7 affitoxin384 remained fully viable during the indicated time periods. Zwt affitoxin had no effect on any of the cells tested.
When SiHa and CaSki cells were treated with indicated concentrations of ZHPV16 E7 affitoxin384 for 72 h, IC50 values of ZHPV16 E7 affitoxin384 were 0.28 μM and 0.24 μM, respectively (data not shown).
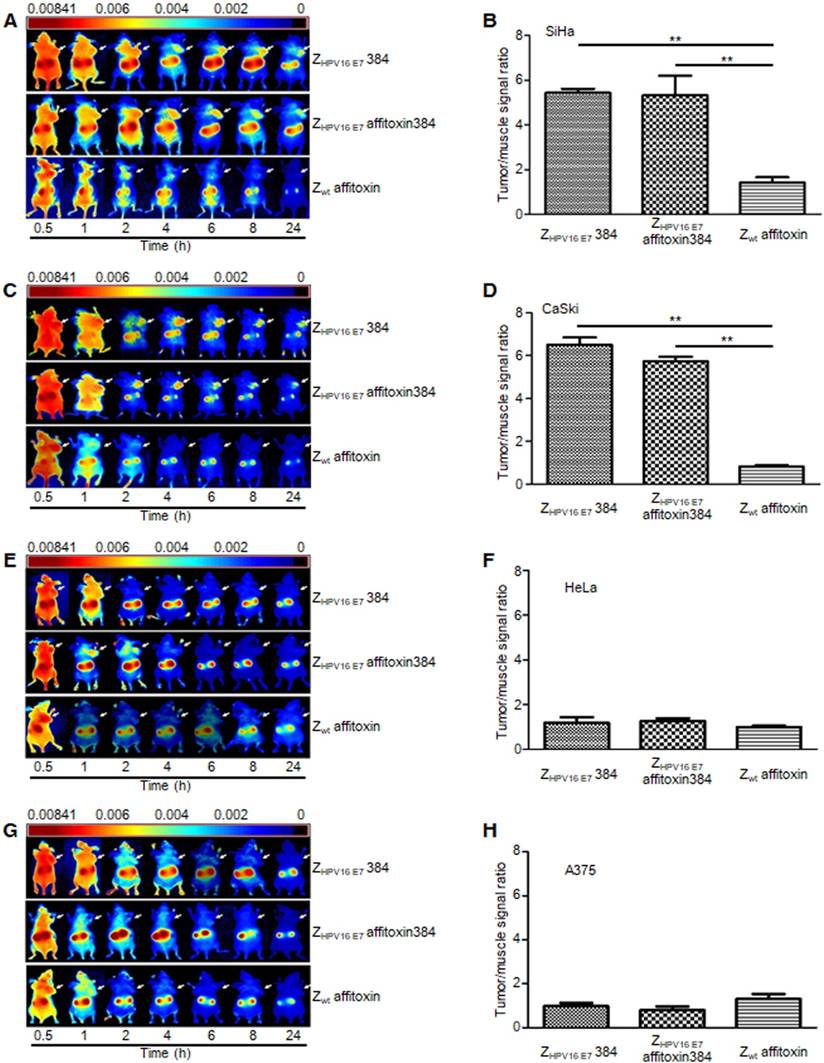
ZHPV16 E7 affitoxin384 specifically targeted HPV16 positive tumors in tumor-bearing nude mice
NIR optical imaging was performed to determine the in vivo distribution and tumor-targeting ability of ZHPV16 E7 affitoxin384. As shown in Figure S1, ZHPV16 E7 affitoxin384 was distributed throughout the body within 30 min after injection, excreted by the kidneys, and mainly cleared from the body within 48 h. At 72 hpi, the fluorescent probes had been thoroughly cleared from the body. In order to evaluate the tumor-targeting ability of ZHPV16 E7 affitoxin384, nude mice bearing tumor xenografts of SiHa, CaSki, HeLa or A375 cells were used. Fluorescence images were obtained at several time points ranging from 0.5 h to 24 h after injection of ZHPV16E7384, ZHPV16 E7 affitoxin384 or Zwt affitoxin. The tumor/muscle ratio at 8 hpi was analyzed. As shown in Figure 6A, C, at 2 hpi, xenografts of SiHa and CaSki cells were identified by the ZHPV16E7384 and ZHPV16 E7 affitoxin384 probes, and the intense signal at tumor sites was maintained up to 24 hpi. Tumors in mice injected with ZHPV16E7384 and ZHPV16 E7 affitoxin384 were observed to have significantly higher fluorescence intensity than those injected with Zwt affitoxin (Figure 6A-D). In contrast, similar tumor-specific fluorescence signals were not detected in nude mice bearing tumor xenografts of HeLa and A375 cells when injected with ZHPV16E7384 and ZHPV16 E7 affitoxin384 (Figure 6E-H).
After imaging for 48 h, the mice were sacrificed and postmortem fluorescence analysis of dissected organs was performed using the NIR imaging system. According to the results, all animals showed similar ZHPV16 E7 affitoxin384 accumulation patterns (data not shown).
The efficacy of ZHPV16 E7 affitoxin384 in vitro. SiHa, CaSki, HeLa and A375 cells were treated with different concentrations of ZHPV16 E7 affitoxin384 for different time periods. Cell viability was measured using CCK-8 assay. Cells treated with Zwt affitoxin were used as control. (A) SiHa cells were treated with indicated concentrations of ZHPV16 E7 affitoxin384 or Zwt affitoxin for 72 h. (B) CaSki cells were treated with indicated concentrations of ZHPV16 E7 affitoxin384 or Zwt affitoxin for 72 h. Compared to the control group, cell viability decreased with the increase of concentration of ZHPV16 E7 affitoxin384. (C) SiHa, CaSki, HeLa and A375 cells were treated with 2 μM of ZHPV16 E7 affitoxin384 for indicated time periods. (D) SiHa, CaSki, HeLa and A375 cells were treated with 2 μM of ZHPV16 E7 affitoxin384 or Zwt affitoxin for 72 h. ZHPV16 E7 affitoxin384 significantly reduced the viability of SiHa and CaSki cells during the indicated time periods, whereas HeLa and A375 cells treated with the same concentration of ZHPV16 E7 affitoxin384 remained fully viable during the indicated time periods. Zwt affitoxin had no effect on any kinds of cells. (E) SiHa cells were treated with indicated concentrations of ZHPV16 E7 affitoxin384 or Zwt affitoxin for 72 h. (F) CaSki cells were treated with indicated concentrations of ZHPV16 E7 affitoxin384 or Zwt affitoxin for 72 h. The IC50 values were calculated using GraphPad Prism software. Data are given as mean ± SD (n=3). **P <0.01 versus the control group. The IC50 values of ZHPV16 E7 affitoxin384 were respectively 3.7×10-5 and 1.3 ×10-4 times lower than that of Zwt affitoxin.
The tumor-targeting ability of ZHPV16 E7 affitoxin384. The xenograft-bearing mice were generated with cell lines of (A) SiHa, (C) CaSki, (E) HeLa, and (G) A375, respectively. NIR imaging was performed at different time points post injection with DyLight-755-labeled ZHPV16 E7 affitoxin384. DyLight-755-labeled ZHPV16 E7 384 and Zwt affitoxin were used as positive and negative controls, respectively. The tumor/muscle ratios were calculated at 8 h post-injection of indicated agents in xenograft-bearing mice generated with cell lines of (B) SiHa, (D) CaSki, (F) HeLa, and (H) A375, respectively. Data are given as mean ± SD (n=5). **P <0.01 versus the control group.
Acute toxity and pharmacokinetics of ZHPV16 E7 affitoxin384
The toxicity of ZHPV16 E7 affitoxin384 was assessed in BALB/c and ICR mice by intravenous injection into the tail vein at the indicated concentrations. As listed in Table S1, a 100% mortality rate was recorded for mice injected with 500 nmol/kg of the protein. Four of 5 mice survived with the treatment of 200 nmol/kg. All death cases were reported within 72 hpi. No mortality was observed in groups treated with 100, 50, 20, 10 or 5 nmol/kg of the protein. The calculated LD50 value was 346.148±83.07 nmol/kg. As listed in Table S2, mice injected with 200 nmol/kg of ZHPV16 E7 affitoxin384 showed significantly higher platelet counts compared with control mice at DPI 7 and 14. In addition, all mice injected with any doses of ZHPV16 E7 affitoxin384 showed significantly larger mean corpuscular volume compared with control mice at DPI 3. No significant difference was shown in other blood test lists between mice injected with ZHPV16 E7 affitoxin384 and control mice. As listed in Table S3, at DPI 1, mice injected with any doses of ZHPV16 E7 affitoxin384 showed a significant increase of concentration of aspartate aminotransferase compared with control mice. Then, at DPI 3, the concentration of aspartate aminotransferase returned to a normal level. Additionally, at DPI 3 and 7, mice injected with any doses of ZHPV16 E7 affitoxin384 showed a significantly lower concentration of urea nitrogen compared with control mice. No significant difference was shown in other liver and kidney panel test lists between mice injected with ZHPV16 E7 affitoxin384 and control mice. Together, these results indicated that injection of ZHPV16 E7 affitoxin384 induced a slight damage to mouse liver and kidney initially.
The standard pharmacokinetic curves were made by recombinant HPV16E7 protein-based ELISA (Figure S2). The concentrations of ZHPV16 E7 affitoxin384 in serum at different time points post injection are shown in Figure S3.
Pharmacokinetics data obtained by protein-based ELISA indicated that the half-life of ZHPV16 E7 affitoxin384 in the bloodstream was 8.95±0.389 min (Figure S3). The initial concentration of ZHPV16 E7 affitoxin384 in the plasma was 2.5±2.47 nM at 1 min post injection, which corresponded to the injected dose.
ZHPV16 E7 affitoxin384 had significant in vivo antitumor efficacy
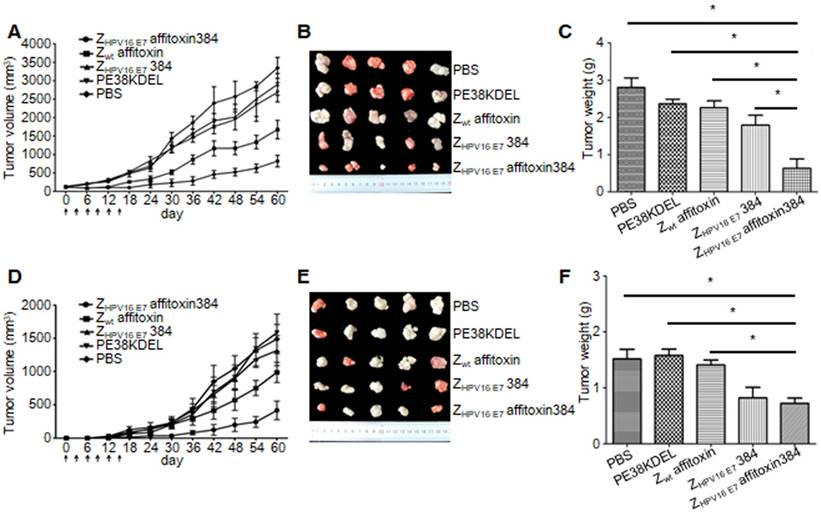
Two methods were used to evaluate the therapeutic efficacy of ZHPV16 E7 affitoxin384 using SiHa tumor-bearing mice. In the first method, SiHa tumor-bearing mice were prepared in advance. When the tumor size reached 0.1-0.2 cm in diameter, ZHPV16 E7 affitoxin384 or other control agents were injected. As shown in Figure 7A, from day 0 to day 15, the tumor sizes from groups injected ZHPV16 E7 affitoxin384 and ZHPV16 E7384 decreased gradually whereas those from the other groups increased gradually. After day 15, tumor sizes from all groups began to rise until day 60. However, the tumors from groups of ZHPV16 E7 affitoxin384 and ZHPV16 E7384 grew much more slowly than those from other groups. As shown in Figure 7B-C, the tumors from the group of ZHPV16 E7 affitoxin384 showed the smallest volume and the lightest weight and there was a significant difference between the tumor weight of ZHPV16 E7 affitoxin384 group and those of other groups.
In the second method, ZHPV16 E7 affitoxin384 and SiHa cells were injected into nude mice at the same time. As shown in Figure 7D, from day 0 to day 15, tumors were observed in mice of all groups except the ZHPV16 E7 affitoxin384 group. Moreover, these tumors grew at a higher speed. At day 30, the tumor sizes from all groups except the ZHPV16 E7 affitoxin384 group were over 200 mm3. As shown in in Figure 7E-F, the tumors from the group of ZHPV16 E7 affitoxin384 also showed the smallest volume and the lightest weight and there was a significant difference between the tumor weight of the ZHPV16 E7 affitoxin384 group and those of the other groups except the ZHPV16 E7384 group.
Immunogenicity of ZHPV16 E7 affitoxin384
As reported in a previous study, although nude mice lack a thymus, some immunologic capabilities still remain [31]. To evaluate the immunogenicity of ZHPV16 E7 affitoxin384 in tumor treatment, the levels of ZHPV16 E7 affitoxin384-specific antibody at day 0 and day 35 were analyzed by ELISA. As shown in Figure S4, compared to day 0, an increase of anti-ZHPV16 E7 affitoxin384 antibody was detected at day 35 after injection of ZHPV16 E7 affitoxin384 every three days for 6 times via tail vein. Meanwhile, an increase of anti-Zwt affitoxin, anti-ZHPV16 E7384 and anti-PE38KDEL antibodies in control groups was also detected.
Discussion
As reported, 99.7% of cervical tumor specimens were detected with HPV infection, about half of which were HPV 16 positive [2, 32, 33]. High-risk HPV E7 was proven to be involved in malignant transformation of cervical cancer and the maintenance of tumor cells was dependent on the constant expression of E7 protein [34-38]. Furthermore, E7 protein could interact with several proteins including pRB, p107, p130, p300, p600, CBP, and pCAF [39, 40], leading to abnormal cell cycle and genetic instability [40, 41].
Although the HPV-associated cancers could be prevented by the developed prophylactic HPV vaccines to a great extent, these prophylactic HPV vaccines have no therapeutic effect on HPV-associated lesions that result from pre-existing HPV infections [42]. In addition, it would take a long time for preventive vaccines to lower the prevalence of cervical cancer because of the limited application of prophylactic HPV vaccines due to medical infrastructure problems and high costs [43]. Therefore, novel and effective treatment strategies for cervical cancer are urgently needed.
In vivo antitumor efficacy of ZHPV16 E7 affitoxin384. Therapeutic efficacy of ZHPV16 E7 affitoxin384 was studied using SiHa tumor-bearing mice by two methods. (A) SiHa tumor-bearing mice were prepared in advance, followed by injection of ZHPV16 E7 affitoxin384, Zwt affitoxin, ZHPV16 E7 384, PE38KDEL, or PBS every three days for 6 times via tail vein. The therapeutic efficacies of affitoxin proteins were evaluated based on daily measurements of tumor volume and body weight. (B) Tumors from mice in (A) were separated. (C) All tumors from (B) were weighed and compared. (D) When SiHa tumor-bearing mice were prepared, cells and ZHPV16 E7 affitoxin384, Zwt affitoxin, ZHPV16 E7 384, PE38KDEL, or PBS were injected at the same time. The therapeutic efficacies of affitoxin proteins were evaluated based on daily measurements of tumor volume and body weight. (E) Tumors from mice in (D) were isolated. (F) All tumors from (E) were weighed and compared. Data are given as mean ± SD (n=5). *P <0.05 versus the control group.
Tumor-specific protein-targeted antibodies have been used in cancer treatment. For instance, the clinical use of a humanized HER2-targeted monoclonal antibody, under the brand name of Herceptin, has significantly improved treatment outcome of breast cancer [11-13]. Therapeutic antibodies have various action mechanisms such as binding to their antigens, thereby interfering with the activity of antigens and blocking interaction with binding partners of antigens, triggering complement-dependent cytotoxicity through their Fc portion, and leading to antibody dependent, cell-mediated cytotoxicity (ADCC) by recruiting and activating macrophages, natural killer cells, dendritic cells, and neutrophils (reviewed by Patrick, et al. [14]). However, there are some limitations during the use of these antibodies such as their inadequate pharmacokinetics, poor penetration in tumors, and elimination by the immune system [14, 44]. Recently, affibody molecules, a novel category of small affinity proteins with 7 kDa of molecular weight, have been developed and successfully applied to detect HER2-positive tumors in vitro and in xenografts [45-55]. Compared with antibodies, affibody molecules have no Fc regions, thereby lacking half-life extension and effector function modules. Thus, their therapeutic action can be executed either by blocking interactions of ligand with receptor directly or by carrying toxic payloads (reviewed by Fredrik, et al. [56]). In subsequent studies, HER2-affitoxin was developed by combining a HER2-specific affibody with modified Pseudomonas aeruginosa Exotoxin A, which was proved to be an attractive agent for treatment of HER2-positive tumors [57, 58]. In a previous study, our group has also produced a series of affibody molecules that can specifically bind with HPV 16 E7 protein both in vitro and in vivo [24]. Given that affibody molecules provide rapid tumor localization, fast clearance from non-specific compartments, small size, and low immunogenicity [20], affibody molecules specifically binding with HPV 16 E7 can be used as an excellent vehicle to deliver other therapeutic agents. The mechanism of ZHPV16 E7 affitoxin384 is different from that of a monoclonal antibody. Thus, ZHPV16 E7 affitoxin384 may serve as a novel and effective treatment strategy for cervical cancer.
As a therapeutic agent for cervical cancer, ZHPV16 E7 affitoxin384 must bind to its target protein, HPV16E7, with high specificity. As shown in SPR assay results, the binding affinity of ZHPV16 E7 affitoxin384 to HPV16 E7 was about 106 times higher than that of Zwt affitoxin (Figure 2). Furthermore, by indirect immunofluorescence assay, not only could ZHPV16 E7 affitoxin384 bind to HPV16E7 positive cell lines, but also HPV16 E7 and ZHPV16 E7 affitoxin384 were co-localized (Figure 3 and Figure 4). The localization of ZHPV16 E7 affitoxin384 was similar to that of ZHPV16 E7384 [24]. In nude mice bearing tumor xenografts, ZHPV16 E7 affitoxin384 could only identify xenografts of HPV16E7 positive cells. The plots of the maximal T/M ratios for all groups of tumor-bearing mice indicated that ZHPV16 E7 affitoxin384 showed good tumor-targeting ability to SiHa and CaSki tumors but not to HeLa and A375 tumors, whereas Zwt affitoxin had no tumor-targeting ability to any kinds of tested tumors (Figure 6). All of these results indicate that ZHPV16 E7 affitoxin384 can bind with HPV16 E7 specifically both in vitro and in vivo.
Another feature of a therapeutic agent is that it must be quickly cleared from non-specific tissues. As shown in Figure S1 and Figure S3, ZHPV16 E7 affitoxin384 was completely cleared from the mouse body at 72 hpi and the circulation half-life of ZHPV16 E7 affitoxin384 in the bloodstream was 8.95±0.389 min, which indicates that ZHPV16 E7 affitoxin384 in non-specific tissues can be cleared at a fast rate, much faster than the clearance of HER2-affitoxin reported in a previous study [58]. Our bio-distribution data showed that ZHPV16 E7 affitoxin384 was cleared through the kidneys, which was consistent with the way of clearance of HER2-Affitoxin [58]. However, there were also some differences between our results and the results from Rafal and colleagues [58]. HER2-Affitoxin partly accumulated in the livers of mice in their study, whereas ZHPV16 E7 affitoxin384 was not observed to accumulate in the liver.
The efficacy and potential cytotoxicity of ZHPV16 E7 affitoxin384 were firstly evaluated in vitro. As shown in Figure 5, treatment with ZHPV16 E7 affitoxin384 significantly reduced the viability of SiHa and CaSki cells, whereas treatment of HeLa and A375 cells with ZHPV16 E7 affitoxin384 and treatment of four kinds of cells with Zwt affitoxin had no effect on cell viability. These results demonstrate that ZHPV16 E7 affitoxin384 specifically affects its target cells and has no detectable cytotoxicity to other unrelated cells.
The efficacy and potential toxicity of ZHPV16 E7 affitoxin384 was also evaluated in vivo. As shown in Table S2 and Table S3, injection of ZHPV16 E7 affitoxin384 caused only minor effects on mouse liver and kidney. As shown in Figure 7, compared to control groups, the tumor growth of mice administered ZHPV16 E7 affitoxin384 was significantly inhibited in both methods we used. However, some difference in the results still existed between the two methods. In the first method, when SiHa tumor-bearing mice were prepared in advance, treatment with ZHPV16 E7 affitoxin384 resulted in the reduction of tumor size early, but increased later. Mice treated with ZHPV16 E7 affitoxin384 did not show significant weight loss and survived more than 2 months without any signs of organ dysfunction. We speculate that ZHPV16 E7 affitoxin384 could kill the outer tumor cells whereas the penetrating ability of ZHPV16 E7 affitoxin384 is not strong enough to kill the deeper tumor cells. As a result, tumors from the group of ZHPV16 E7 affitoxin384 grew significantly slower than those from other groups. In the second method, tumors were observed in mice of all groups except the ZHPV16 E7 affitoxin384 group up to day 15. Tumors grew rapidly in all groups except the ZHPV16 E7 affitoxin384 group and tumor sizes were over 200 mm3 by day 30. These results indicate that injecting a mixture of ZHPV16 E7 affitoxin384 and SiHa cells into nude mice can effectively inhibit tumor growth. The tumor suppression effect of ZHPV16 E7 affitoxin384 in the second method was better than that in the first method. The probable cause may be the complete contact of ZHPV16 E7 affitoxin384 with SiHa cells in the second method.
According to our results of an immunogenicity study, injection of ZHPV16 E7 affitoxin384 multiple times induced humoral immune response (Figure S4), which was consistent with the results of a previous study [58]. The generation of ZHPV16 E7 affitoxin384-specific antibodies may limit the effectiveness of treatment if these antibodies were neutralizing. This may be another explanation for the reduction of tumor size at the early period of treatment and increase later. Therefore, further studies are still needed to develop a less immunogenic version of the ZHPV16 E7 affitoxin384.
In conclusion, our data indicates that ZHPV16 E7 affitoxin384 could target HPV16E7 protein with high affinity and specificity. This affitoxin is proven to be a potent anti-cervical cancer therapeutic agent that might be effective against HPV16E7-expressing solid tumors in humans without apparent cytotoxicity to normal cells. These promising data will have to be further confirmed by clinical trials.
Abbreviations
CCK-8: Cell Counting Kit-8; DPI: day post-injection; FBS: fetal bovine serum; h: hour; hpi: hours post injection; HPV: human papillomavirus; HPVs: human papillomaviruses; IC50: half maximal inhibitory concentration; IPTG: isopropyl-Dthiogalactopyranoside; LD50: lethal dose 50%; M: mol/L; min: minute; NIR: near-infrared; PE38KDEL: modified Pseudomonas Exotoxin A; PI: propidium iodide; SDS-PAGE: sodium dodecyl sulfate-polyacrylamide gel electrophoresis; SPA-Z: Z-domain of staphylococcal protein A; SPR: surface plasmon resonance; T/M ratio: tumor/muscle ratio; ZHPV16 E7 affitoxin384: protein encoded by pET21a (+)/ZHPV16 E7 affitoxin384 vector; Zwt: SPA-Z molecule; Zwt affitoxin: protein encoded by pET21a (+)/Zwt affitoxin vector.
Acknowledgements
We thank Deanna Gotte from NIH/NCI for her helpful comments and revisions. This work was supported by the grant from the National Natural Science Foundation of China (Grant No. 81172463 and Grant No. 81502242), Zhejiang Provincial Natural Science Foundation of China (Grant No. LY15H190009), Wenzhou Science and Technology Bureau of China (Grant No. Y20140659) and the Scientific Research Foundation of Wenzhou Medical University (Grant No. QTJ14040).
Supplementary Material
Supplementary figures and tables.
Competing Interests
The authors have declared that no competing interest exists.
References
1. WHO: Media Centre. Human papillomavirus (HPV) and cervical cancer. Fact sheet. Updated June 2016. http://www.who.int/mediacentre/factsheets/fs380/en/
2. Walboomers JM, Jacobs MV, Manos MM, Bosch FX, Kummer JA, Shah KV. et al. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol. 1999;189:12-9
3. zur Hausen H. Papillomaviruses-to vaccination and beyond. Biochemistry (Mosc). 2008;73:498-503
4. Howley PM, Munger K, Romanczuk H, Scheffner M, Huibregtse JM. Cellular targets of the oncoproteins encoded by the cancer associated human papillomaviruses. Princess Takamatsu Symp. 1991;22:239-48
5. Romanczuk H, Howley PM. Disruption of either the E1 or the E2 regulatory gene of human papillomavirus type 16 increases viral immortalization capacity. Proc Natl Acad Sci USA. 1992;89:3159-63
6. Jabbar SF, Abrams L, Glick A, Lambert PF. Persistence of high-grade cervical dysplasia and cervical cancer requires the continuous expression of the human papillomavirus type 16 E7 oncogene. Cancer Res. 2009;69:4407-14
7. Scholten KB, Schreurs MW, Ruizendaal JJ, Kueter EW, Kramer D, Veenbergen S. et al. Preservation and redirection of HPV16E7-specific T cell receptors for immunotherapy of cervical cancer. Clin Immunol. 2005;114:119-29
8. Welters MJ, Kenter GG, Piersma SJ, Vloon AP, Lowik MJ, Berends-van der Meer DM. et al. Induction of tumor-specific CD4+ and CD8+ T-cell immunity in cervical cancer patients by a human papillomavirus type 16 E6 and E7 long peptides vaccine. Clin Cancer Res. 2008;14:178-87
9. Carter JR, Ding Z, Rose BR. HPV infection and cervical disease: a review. Aust N Z J Obstet Gynaecol. 2011;51:103-8
10. Ratanasiripong NT. A review of human papillomavirus (HPV) infection and HPV vaccine-related attitudes and sexual behaviors among college-aged women in the United States. J Am Coll Health. 2012;60:461-70
11. Pegram MD, Lipton A, Hayes DF, Weber BL, Baselga JM, Tripathy D. et al. Phase II study of receptor-enhanced chemosensitivity using recombinant humanized anti-p185HER2/neu monoclonal antibody plus cisplatin in patients with HER2/neu-overexpressing metastatic breast cancer refractory to chemotherapy treatment. J Clin Oncol. 1998;16:2659-71
12. Cobleigh MA, Vogel CL, Tripathy D, Robert NJ, Scholl S, Fehrenbacher L. et al. Multinational study of the efficacy and safety of humanized anti-HER2 monoclonal antibody in women who have HER2-overexpressing metastatic breast cancer that has progressed after chemotherapy for metastatic disease. J Clin Oncol. 1999;17:2639-48
13. Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A. et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344:783-92
14. Chames P, Van Regenmortel M, Weiss E, Baty D. Therapeutic antibodies: successes, limitations and hopes for the future. Br J Pharmacol. 2009;157:220-33
15. Uhlen M, Guss B, Nilsson B, Gatenbeck S, Philipson L, Lindberg M. Complete sequence of the staphylococcal gene encoding protein A. A gene evolved through multiple duplications. J Biol Chem. 1984;259:1695-702
16. Nilsson B, Moks T, Jansson B, Abrahmsen L, Elmblad A, Holmgren E. et al. A synthetic IgG-binding domain based on staphylococcal protein A. Protein Eng. 1987;1:107-13
17. Nord K, Nilsson J, Nilsson B, Uhlen M, Nygren PA. A combinatorial library of an alpha-helical bacterial receptor domain. Protein Eng. 1995;8:601-8
18. Nord K, Gunneriusson E, Ringdahl J, Stahl S, Uhlen M, Nygren PA. Binding proteins selected from combinatorial libraries of an alpha-helical bacterial receptor domain. Nat Biotechnol. 1997;15:772-7
19. Nygren PA. Alternative binding proteins: affibody binding proteins developed from a small three-helix bundle scaffold. FEBS J. 2008;275:2668-76
20. Lofblom J, Feldwisch J, Tolmachev V, Carlsson J, Stahl S, Frejd FY. Affibody molecules: engineered proteins for therapeutic, diagnostic and biotechnological applications. FEBS Lett. 2010;584:2670-80
21. Eklund M, Axelsson L, Uhlen M, Nygren PA. Anti-idiotypic protein domains selected from protein A-based affibody libraries. Proteins. 2002;48:454-62
22. Tolmachev V, Orlova A, Nilsson FY, Feldwisch J, Wennborg A, Abrahmsen L. Affibody molecules: potential for in vivo imaging of molecular targets for cancer therapy. Expert Opin Biol Ther. 2007;7:555-68
23. Gao J, Chen K, Miao Z, Ren G, Chen X, Gambhir SS. et al. Affibody-based nanoprobes for HER2-expressing cell and tumor imaging. Biomaterials. 2011;32:2141-8
24. Xue X, Wang B, Du W, Zhang C, Song Y, Cai Y. et al. Generation of affibody molecules specific for HPV16 E7 recognition. Oncotarget. 2016;7:73995-4005
25. Kreitman RJ, Pastan I. Importance of the glutamate residue of KDEL in increasing the cytotoxicity of Pseudomonas exotoxin derivatives and for increased binding to the KDEL receptor. Biochem J. 1995;307( Pt 1):29-37
26. Lin X, Hou B, Xiong Y, Ye L, Du W, Chen S. et al. Prokaryotic expression of the Z domain of Staphylococcus protein A and preparation of its poly clonal antibody. Jouranl of Wenzhou Medical University. 2015;45:1-5
27. Cen D, Song Y, Zhang C, Mao S, Ye X, Chen J. et al. Prokaryotic expression of recombinant PE38KDEL protein and preparation of its polyclonal antibody. Jouranl of Wenzhou Medical University. 2017;47:1-6
28. Iglewski BH, Kabat D. NAD-dependent inhibition of protein synthesis by Pseudomonas aeruginosa toxin. Proc Natl Acad Sci U S A. 1975;72:2284-8
29. Siegall CB, Chaudhary VK, FitzGerald DJ, Pastan I. Functional analysis of domains II, Ib, and III of Pseudomonas exotoxin. J Biol Chem. 1989;264:14256-61
30. Liu C, Lin J, Li L, Zhang Y, Chen W, Cao Z. et al. HPV16 early gene E5 specifically reduces miRNA-196a in cervical cancer cells. Sci Rep. 2015;5:7653
31. Wortis HH. Immunological responses of 'nude' mice. Clin Exp Immunol. 1971;8:305-17
32. Durst M, Gissmann L, Ikenberg H, zur Hausen H. A papillomavirus DNA from a cervical carcinoma and its prevalence in cancer biopsy samples from different geographic regions. Proc Natl Acad Sci USA. 1983;80:3812-5
33. Steele C, Shillitoe EJ. Viruses and oral cancer. Crit Rev Oral Biol Med. 1991;2:153-75
34. Chu NR, Wu HB, Wu T, Boux LJ, Siegel MI, Mizzen LA. Immunotherapy of a human papillomavirus (HPV) type 16 E7-expressing tumour by administration of fusion protein comprising Mycobacterium bovis bacille Calmette-Guerin (BCG) hsp65 and HPV16 E7. Clin Exp Immunol. 2000;121:216-25
35. Veldman T, Horikawa I, Barrett JC, Schlegel R. Transcriptional activation of the telomerase hTERT gene by human papillomavirus type 16 E6 oncoprotein. J Virol. 2001;75:4467-72
36. Kaufmann AM, Stern PL, Rankin EM, Sommer H, Nuessler V, Schneider A. et al. Safety and immunogenicity of TA-HPV, a recombinant vaccinia virus expressing modified human papillomavirus (HPV)-16 and HPV-18 E6 and E7 genes, in women with progressive cervical cancer. Clin Cancer Res. 2002;8:3676-85
37. Nishimura A, Nakahara T, Ueno T, Sasaki K, Yoshida S, Kyo S. et al. Requirement of E7 oncoprotein for viability of HeLa cells. Microbes Infect. 2006;8:984-93
38. Liu X, Roberts J, Dakic A, Zhang Y, Schlegel R. HPV E7 contributes to the telomerase activity of immortalized and tumorigenic cells and augments E6-induced hTERT promoter function. Virology. 2008;375:611-23
39. Roman A, Munger K. The papillomavirus E7 proteins. Virology. 2013;445:138-68
40. de Freitas AC, Coimbra EC, Leitao Mda C. Molecular targets of HPV oncoproteins: potential biomarkers for cervical carcinogenesis. Biochim Biophys Acta. 2014;1845:91-103
41. Munger K, Baldwin A, Edwards KM, Hayakawa H, Nguyen CL, Owens M. et al. Mechanisms of human papillomavirus-induced oncogenesis. J Virol. 2004;78:11451-60
42. Lowy DR, Schiller JT. Prophylactic human papillomavirus vaccines. J Clin Invest. 2006;116:1167-73
43. Agosti JM, Goldie SJ. Introducing HPV vaccine in developing countries—key challenges and issues. N Engl J Med. 2007;356:1908-10
44. Bourgeois M, Bailly C, Frindel M, Guerard F, Cherel M, Faivre-Chauvet A. et al. Radioimmunoconjugates for treating cancer: recent advances and current opportunities. Expert Opin Biol Ther. 2017;17:813-9
45. Tolmachev V, Nilsson FY, Widstrom C, Andersson K, Rosik D, Gedda L. et al. 111In-benzyl-DTPA-ZHER2:342, an affibody-based conjugate for in vivo imaging of HER2 expression in malignant tumors. J Nucl Med. 2006;47:846-53
46. Lundberg E, Hoiden-Guthenberg I, Larsson B, Uhlen M, Graslund T. Site-specifically conjugated anti-HER2 Affibody molecules as one-step reagents for target expression analyses on cells and xenograft samples. J Immunol Methods. 2007;319:53-63
47. Orlova A, Tolmachev V, Pehrson R, Lindborg M, Tran T, Sandstrom M. et al. Synthetic affibody molecules: a novel class of affinity ligands for molecular imaging of HER2-expressing malignant tumors. Cancer Res. 2007;67:2178-86
48. Tran T, Engfeldt T, Orlova A, Sandstrom M, Feldwisch J, Abrahmsen L. et al. (99m)Tc-maEEE-Z(HER2:342), an Affibody molecule-based tracer for the detection of HER2 expression in malignant tumors. Bioconjug Chem. 2007;18:1956-64
49. Ekblad T, Tran T, Orlova A, Widstrom C, Feldwisch J, Abrahmsen L. et al. Development and preclinical characterisation of 99mTc-labelled Affibody molecules with reduced renal uptake. Eur J Nucl Med Mol Imaging. 2008;35:2245-55
50. Kramer-Marek G, Kiesewetter DO, Martiniova L, Jagoda E, Lee SB, Capala J. [18F]FBEM-Z(HER2:342)-Affibody molecule-a new molecular tracer for in vivo monitoring of HER2 expression by positron emission tomography. Eur J Nucl Med Mol Imaging. 2008;35:1008-18
51. Lee SB, Hassan M, Fisher R, Chertov O, Chernomordik V, Kramer-Marek G. et al. Affibody molecules for in vivo characterization of HER2-positive tumors by near-infrared imaging. Clin Cancer Res. 2008;14:3840-9
52. Nordberg E, Orlova A, Friedman M, Tolmachev V, Stahl S, Nilsson FY. et al. In vivo and in vitro uptake of 111In, delivered with the affibody molecule (ZEGFR:955)2, in EGFR expressing tumour cells. Oncol Rep. 2008;19:853-7
53. Friedman M, Lindstrom S, Ekerljung L, Andersson-Svahn H, Carlsson J, Brismar H. et al. Engineering and characterization of a bispecific HER2 x EGFR-binding affibody molecule. Biotechnol Appl Biochem. 2009;54:121-31
54. Tolmachev V, Friedman M, Sandstrom M, Eriksson TL, Rosik D, Hodik M. et al. Affibody molecules for epidermal growth factor receptor targeting in vivo: aspects of dimerization and labeling chemistry. J Nucl Med. 2009;50:274-83
55. Nordberg E, Ekerljung L, Sahlberg SH, Carlsson J, Lennartsson J, Glimelius B. Effects of an EGFR-binding affibody molecule on intracellular signaling pathways. Int J Oncol. 2010;36:967-72
56. Frejd FY, Kim KT. Affibody molecules as engineered protein drugs. Exp Mol Med. 2017;49:e306
57. Zielinski R, Lyakhov I, Jacobs A, Chertov O, Kramer-Marek G, Francella N. et al. Affitoxin-a novel recombinant, HER2-specific, anticancer agent for targeted therapy of HER2-positive tumors. J Immunother. 2009;32:817-25
58. Zielinski R, Lyakhov I, Hassan M, Kuban M, Shafer-Weaver K, Gandjbakhche A. et al. HER2-affitoxin: a potent therapeutic agent for the treatment of HER2-overexpressing tumors. Clin Cancer Res. 2011;17:5071-81
Author contact
![]() Corresponding author: Lifang Zhang, Department of Microbiology and Immunology, School of Basic Medical Sciences, Wenzhou Medical University, Wenzhou, 325035, P. R. China, E-mail: wenzhouzlfcom, Tel./Fax: +86-577-86689910
Corresponding author: Lifang Zhang, Department of Microbiology and Immunology, School of Basic Medical Sciences, Wenzhou Medical University, Wenzhou, 325035, P. R. China, E-mail: wenzhouzlfcom, Tel./Fax: +86-577-86689910
 Global reach, higher impact
Global reach, higher impact