13.3
Impact Factor
Theranostics 2018; 8(13):3504-3516. doi:10.7150/thno.24336 This issue Cite
Research Paper
Comparative profiling of analog targets: a case study on resveratrol for mouse melanoma metastasis suppression
1. The State Key Laboratory of Pharmaceutical Biotechnology, School of Life Sciences, Nanjing University, Nanjing, 210023, China
2. Department of Biological Science, National University of Singapore, Singapore, 117543, Singapore
3. School of Pharmaceutical Sciences, Sun Yat-sen University, Guangzhou, 510006, China
4. Artemisinin Research Center, China Academy of Chinese Medical Sciences, Beijing, 100700, China
5. Institute of Chinese Materia Medica, China Academy of Chinese Medical Sciences, Beijing, 100700, China
Received 2017-12-13; Accepted 2018-4-23; Published 2018-6-6
Abstract
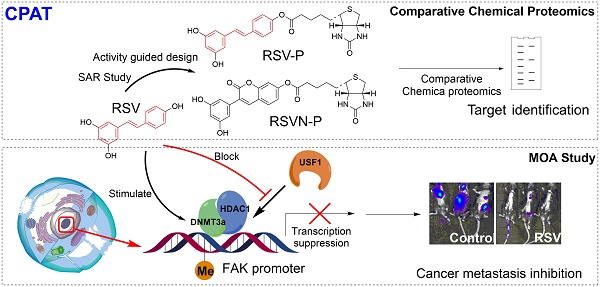
Many plant-specialized metabolites have remedial properties and provide an endless chemical resource for drug discovery. However, most of these metabolites have promiscuous binding targets in mammalian cells and elicit a series of responses that collectively change the physiology of the cells. To explore the potential of these multi-functional and multi-targeted drugs, it is critical to understand the direct relationships between their key chemical features, the corresponding binding targets and the relevant biological effects, which is a prerequisite for future drug modification and optimization.
Methods: We introduced and demonstrated a general workflow, called Comparative Profiling of Analog Targets (CPAT), to connect specific biological effects with defined chemical structures of drugs. Using resveratrol (RSV) as an example, we have synthesized and characterized a series of partial functional analogs of RSV. An analog (named RSVN) that specifically lost the inhibitory effect of RSV in cell migration was identified. The binding targets of RSVN and RSV was profiled and compared.
Results: Comparative profiling of the RSV and RSVN binding targets showed that, unlike RSV, RSVN failed to target specific components involved in DNA methylation (histone deacetylase 1 [HDAC1] and DNA methyltransferase 3 alpha [DNMT3a]), suggesting that RSV suppresses cell migration through epigenetic regulation. Indeed, RSV treatment recruited HDAC1 and DNMT3a to the promoter region of the focal adhesion kinase (FAK), a key factor involved in cell adhesion, enhanced the promoter methylation, and thus attenuated the protein expression. The inhibitory effect of RSV in cell migration was diminished once FAK expression was restored. Thus, the mechanism of RSV in inhibiting cell migration could be largely accounted to epigenetically control of FAK expression.
Conclusion: Our results showed that even though RSV exhibits promiscuous binding, its inhibitory effect on cell migration can be mechanistically understood. First, the presence of 4'-hydroxystilbene within the RSV structure is essential for this activity. Second, it inhibits cell migration through epigenetically based downregulation of FAK expression. Taken together, we propose that CPAT might also be adapted to delineate the specific function of other natural products (NPs) that exhibit binding promiscuity.
Keywords: CPAT, resveratrol, target identification, chemical proteomics, cancer metastasis.
Introduction
For thousands of years, the use of traditional medicines and natural poisons has closely linked natural products (NPs) with medicine [1-4]. Currently, NPs are still a significant source of new drugs, especially in anticancer and antihypertension therapeutic areas [5,6]. As the basis of the discovery of most early medicines, such as aspirin, digitoxin, morphine, quinine and pilocarpine, understanding their clinical, pharmacological and chemical properties as well as the relationship between their chemical structures and the corresponding biological activity is an essential step [7-10], which also contributes to drug modification and optimization [10,11]. However, many NPs exhibit promiscuous binding to multiple targets and possess various drug effects [12-14]. This confounds our understanding of their mechanism of action (MOA).
For example, resveratrol (RSV), a polyphenol that is present in a variety of fruits and vegetables, has been reported to help prevent many diseases such as cancer [15,16], cardiovascular disease [17], and Alzheimer's disease [18]. Moreover, RSV may also have anti-aging effects [19]. Studies have shown that RSV could lead to numerous molecular processes such as apoptosis [20], senescence [21], autophagy [22], and cellular metabolism, proliferation and migration [23] and could also affect the redox status [24] and a wide variety of inflammatory [25] and cell signaling pathways [26]. Recently, it was shown that RSV significantly activates the AMP-activated protein kinase (AMPK) pathway, which partially explains certain functions of RSV [27-29], such as the prevention of diet-induced obesity and enhancement of mitochondrial function, physical stamina, and glucose tolerance in mice. However, the pleiotropic effects of RSV cannot be entirely explained by the activation of the AMPK pathway and the mechanisms behind many of its effects remain obscure. Moreover, the connection between the key chemical structures of RSV and its binding targets and corresponding biological functions remains unknown.
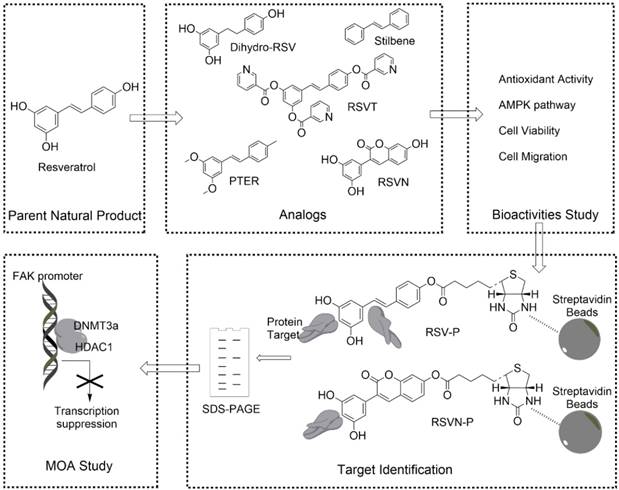
To this end, we have designed a general workflow, named Comparative Profiling of Analog Targets (CPAT). In this scheme (Figure 1), partial functional analogs derived from a parent natural product are synthesized and tested in a variety of functional assays. An analog with the loss of a specific function is selected and its binding targets are profiled together with the fully functional parent compound. Based on their differences in structural elements, binding targets and biological functions, the key chemical features of the parent compound can be connected with a set of defined binding targets and specific biological functions. Here, we have demonstrated this workflow with RSV and found that its inhibition in cell migration, an effect that requiring the presence of 4'-hydroxystilbene within the RSV structure, can be directly attributed to its targeting of components involved in epigenetic regulation. This can serve as a guide for further optimization of RSV or chemical engineering of related compounds. In general, we anticipate that much more structure-function information can be obtained from other natural products by applying the workflow established in this study.
General workflow of CPAT (taking RSV as an example). Analogs that retain part of the parent NP structure are first obtained or synthesized and then their bioactivities are determined. Through a comparative chemical proteomics approach, the targets associated with a specific bioeffect are identified. The MOA related to the bioeffect is also studied with subsequent experiments.
Results and discussion
4'-Hydroxystilbene within the resveratrol structure is essential for its anti-migration activity
To study the structure-activity relationship (SAR) of RSV, we first obtained two RSV analogs, α, β-dihydroresveratrol (dihydro-RSV) and stilbene (Figure 1). In addition, based on our previously published procedures [30,31], we also synthesized another three RSV analogs (Figure 1): RSV trinicotinate (RSVT), pterostilbene (PTER) and 3-(3,5-dihydroxyphenyl)-7-hydroxy-2H-chromen-2-one (RSVN). In stilbene, the hydroxyl groups in the 3, 4' and 5 positions were removed, and the central double bond was replaced with a single bond in dihydro-RSV. In the cases of RSVT and PTER, the hydroxyl groups in the 3, 4' and 5 positions were modified with acylation and alkylation, respectively, while the central double bond was changed into a cyclic structure in RSVN.
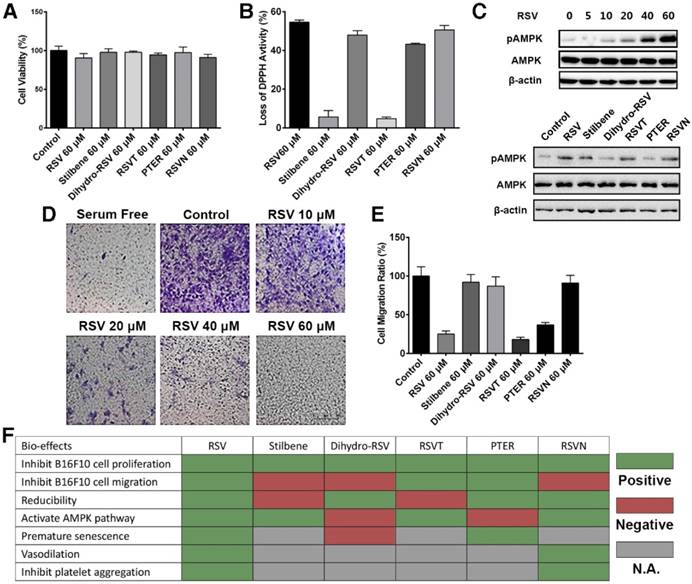
Determination of the bioactivities of RSV and its analogs. (A) B16F10 cells were treated with 60 μM RSV and the RSV analogs for 24 h. Cell viability was measured with cell counting kit (CCK)-8 assays. Data are represented as the mean ± SEM. (B) The reduction activity of 60 μM RSV and analogs was measured with DPPH reduction method. Data are represented as the mean ± SEM. (C) B16F10 cells were treated with different concentrations of RSV and 60 μM RSV analogs for 24 h. The pAMPK expression was detected with western blotting. (D) B16F10 cells were treated with different concentrations of RSV for 16 h. The migration rate of the cells was determined via transwell assays. (E) The migration rates of B16F10 cells treated with 60 μM RSV and the analogs were measured with RTCA assays. Data are represented as the mean ± SEM. (F) Summary of the bioeffects of RSV and the analogs from published reports and assays in the present study.
Here, we tested some bioactivities of RSV and its analogs, including their anti-proliferation, antioxidation, and anti-migration activities, as well as their effects on the AMPK pathway. Melanoma is the most malignant skin cancer, and its occurrence has remarkably increased during the past few decades due to increased UV-ray intensity and artificial skin tanning [32,33]. Conventional treatments, including surgery and chemotherapy, have limited therapeutic uses due to their side effects and low melanoma response rates. Among various melanoma cell lines, B16F10 exhibits high metastatic properties [34]. Therefore, we centralized our experiments on the anti-migratory effects of RSV on this cell line.
We first showed that RSV and its analogs exhibited no cytotoxicity to the B16F10 cells (Figure 2A). Their antioxidant activities were next measured with 2,2-di(4-tert-octylphenyl)-1-picrylhydrazyl (DPPH), which is an oxidizing agent that is dark brown in color [35]. We showed that RSV, dihydro-RSV, PTER and RSVN significantly reduced DPPH to a lighter colored form, while stilbene and RSVT exhibited no obvious effect on DPPH's color (Figure 2B). These results indicate that RSV, dihydro-RSV, PTER and RSVN have strong antioxidant activity while stilbene and RSVT exhibit no reducibility. Subsequently, we conducted western blot analysis to examine the effect of RSV and its analogs on the AMPK pathway. Our results showed that RSV activated the AMPK pathway via phosphorylation of AMPK (pAMPK) in a dose-dependent manner (Figure 2C). Like RSV, stilbene, RSVT and RSVN also significantly increased the phosphorylation of AMPK, while dihydro-RSV and PTER had no obvious effect on AMPK phosphorylation. Next, a transwell assay was performed to examine the effect of different concentrations of RSV on cell migration. As expected, our data showed that RSV can inhibit B16F10 cell migration from the upper chamber to the lower chamber in a dose-dependent manner (Figure 2D). Data acquired with our dual purpose real-time cell analyzer (RTCA) further highlighted that RSV, RSVT and PTER significantly inhibited the migration of B16F10 cells, while dihydro-RSV, stilbene and RSVN exhibited no similar effect (Figure 2E). The biofunctions of RSV and the analogs have also been studied in other published reports as summarized in Figure 2F [36-46]. As with the published data and our functional experimental testing (Figure 2F), the RSVN analog shares similar bioactivities with RSV except for the inhibition of cell migration, which indicates that the presence of 4'-hydroxystilbene within the RSV structure is essential for its anti-migration activity. RSVN is thus a suitable candidate to be used for CPAT in the context of studying the anti-migration activity of RSV.
Synthesis and bioactivity of the RSV and RSVN probes
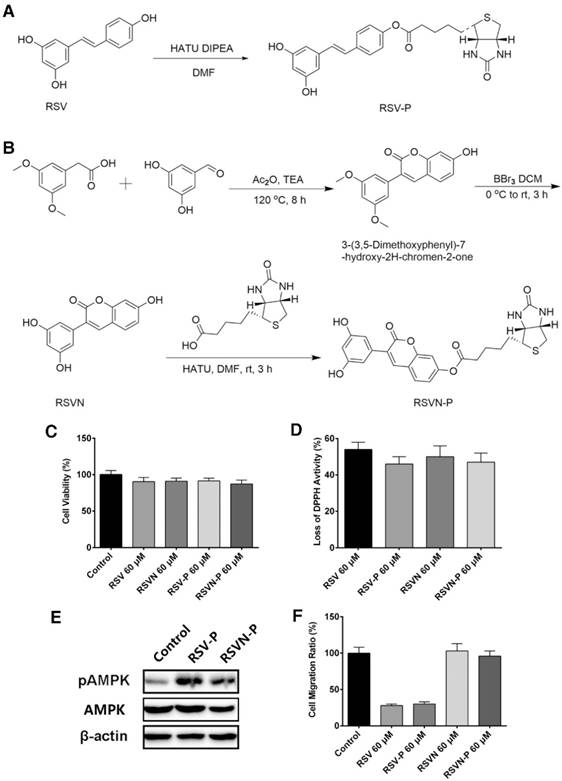
As shown above, RSV and RSVN share many similar functions apart from their effects on cell migration. To identify the functional targets of RSV and RSVN, we synthesized two probes, the RSV probe (RSV-P) and the RSVN probe (RSVN-P; Figure 3A-B). A biotin tag was introduced at the 4' position in both probes, thereby allowing the probes to be enriched by streptavidin beads together with their interactive targets. We also tested their cell migration inhibition activity, antioxidant activity and AMPK activation activity to ensure that the incorporation of the biotin tag did not alter their bioactivities. Both RSV-P and RSVN-P exhibited no cytotoxicity on B16F10 cells (Figure 3C). Furthermore, they exhibited antioxidant activity (Figure 3D) and activated the AMPK pathway (Figure 3E). For their effect on cell migration, the RSV-P showed significant inhibition while RSVN-P did not (Figure 3F), highlighting their close similarity to their respective parent compounds. All the data showed that the incorporation of the biotin tag did not affect the bioactivities of RSV or RSVN.
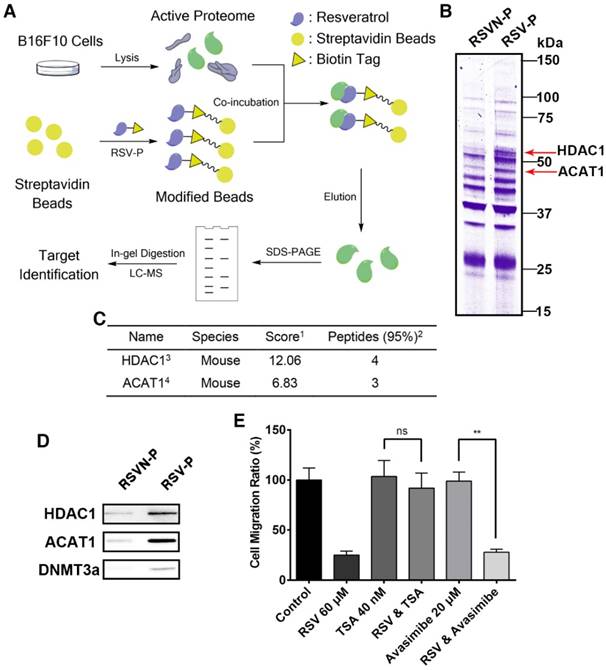
Identifying the anti-migration activity-related targets of RSV by comparative chemical proteomics
We adopted a chemical proteomics approach with the probes to identify the protein targets of RSV and RSVN as illustrated in Figure 4A (RSV as an example) [47]. Briefly, B16F10 cell lysate was incubated with streptavidin beads coated with RSV-P or RSVN-P. After repeatedly washing, the enriched probe-binding proteins were eluted, resolved by SDS-PAGE and stained with Coomassie Brilliant Blue dye. Two major distinct bands at approximately 45 kD and 60 kD in the RSV-P lane were observed but not for the RSVN-P lane (Figure 4B), suggesting that the proteins at 45 kD and 60 kD were possibly related to RSV's anti-migration activity. In addition to the distinct bands, RSV-P and RSVN-P also pulled down many other proteins, which could have included bona fide targets of RSV that accounted for its other functions as well as non-specific binding proteins. The CPAT workflow helped avoid the interference from other non-anti-migration-related protein targets, which might have misled our investigation into RSV's inhibitory effect on cell migration. Excision of the distinct bands followed by in-gel tryptic digestion and subsequent identification of the proteins from the resulting peptide fragments by mass spectrometry revealed the labeled proteins to be histone deacetylase 1 (HDAC1, ~60 kD) and acetyl-coenzyme A acetyltransferase 1 (ACAT1, ~45 kD; Figure 4C). To support this conclusion, the western blotting results showed that RSV-P pulled down greater amounts of HDAC1 and ACAT1 than those by RSVN-P (Figure 4D), which validated that HDAC1 and ACAT1 were the targets of RSV.
As HDAC1 and ACAT1 are the targets of RSV that might be involved in its anti-migration activity, we inferred that application of RSV should either inhibit or increase the targets' activity, leading to inhibition of cell migration. Therefore, when B16F10 cells are treated with the inhibitors of these two targets, RSV should exhibit no effect on cell migration. To confirm whether HDAC1 and/or ACAT1 were confidently related to RSV's anti-migration activity, we used the HDAC1 inhibitor trichostatin A (TSA) and the ACAT1 inhibitor avasimibe. TSA inhibits the activity of deacetylases by binding to the active site of HDACs [48,49], and avasimibe is a sulfamic acid phenyl ester that inhibits ACAT, thereby reducing intracellular cholesterol ester content [50]. Avasimibe did not affect RSV's inhibition of cell migration (Figure 4E), while RSV lost its anti-migration activity in the TSA-treated cells (Figure 4E), indicating that RSV's protein target HDAC1 plays a key role in its anti-migration activity. Hence, we shifted our attention to HDAC1 in our subsequent analysis. In epigenetic regulation, HDAC1 always forms a complex at the promoter region with DNA methyltransferase 3 alpha (DNMT3a, ~100 kD), which is reported to mediate DNA de novo methylation [51]. The complex blocks the binding of transcription factors and leads to gene silencing [52,53]. Therefore, we went on to examine whether DNMT3a could also be co-precipitated by RSV-P using western blotting, and we confirmed that RSV-P can also pull-down DNMT3a (Figure 4D). Due to the possible indirect interaction between RSV and DNMT3a and the low abundance of DNMT3a in the cell, no distinct band at the expected weight of 100 kD in the RSV-P lane was observed (Figure 4B). We also utilized rhodamine-conjugated avidin to label the target-probe complexes in living B16F10 cells. The results showed that the targets of RSV and RSVN were widely distributed in the cytoplasm and nuclei (Figure S1).
Synthesis and bioactivity of the RSV and RSVN probes. (A) Synthesis of the RSV probe (RSV-P). (B) Synthesis of the RSVN probe (RSVN-P). (C) B16F10 cells were treated with 60 μM RSV, RSVN and their probes for 24 h. Cell viability was measured with CCK-8 assays. Data are represented as the mean ± SEM. (D) The reduction activity of 60 μM RSV, RSVN and their probes was measured with the DPPH reduction method. Data are represented as the mean ± SEM. (E) B16F10 cells were treated with 60 μM RSV-P and RSVN-P for 24 h. The pAMPK expression was detected with western blotting. (F) The migration rate of B16F10 cells treated with 60 μM RSV, RSVN and their probes was measured with RTCA assays. Data are represented as the mean ± SEM.
Identification of RSV target proteins with a comparative chemical proteomics approach. (A) Workflow of the comparative chemical proteomics approach. (B) B16F10 lysates were incubated with streptavidin beads modified with RSV-P and RSVN-P for 4 h at room temperature. After washing, the bound proteins were eluted by SDS lysis (5 min, 95 °C) and analyzed by SDS-PAGE followed by Coomassie staining. Two distinct bands were detected. (C) Protein bands of interest (see (B)) were excised, digested, and analyzed by nano-LC-MS/MS. HDAC1 and ACAT1 were identified to be the targets of RSV. (1. Score, unused protein score. For the target identification, a strict total score cut-off of 1.3 was set as the qualification criterion, which corresponded to a protein confidence interval of 95%. 2. Peptides, number of unique peptides identified for a protein. 3. HDAC1, histone deacetylase 1. 4. ACAT1, acetyl-Coenzyme A acetyltransferase 1). (D) The eluted samples were analyzed by western blotting for HDAC1, ACAT1 and DNMT3a. (E) The migration rate of B16F10 cells treated with 60 μM RSV and the inhibitors of ACAT1 and HDAC1 were measured with RTCA assays. Data are represented as the mean ± SEM.
RSV inhibits FAK expression through epigenetic regulation to suppress cancer cell migration
As epigenetic regulation usually relates to gene expression activation or inhibition [50], we inferred that RSV inhibits cell migration through regulating some migration-related protein expression by epigenetic regulation. For decades, focal adhesion kinase (FAK) has been known to have a pivotal role in the regulation of cell adhesion and motility [54,55]. In most experimental systems, increased FAK expression would promote cell motility, while inhibition of FAK signaling would suppress cell migration [56]. Reports have shown that RSV inhibits FAK expression in other cell lines, including VSMC and HO-8910PM cells [57,58]. Here, we applied qPCR and western blot analysis to test the effect of RSV and its analogs on FAK expression in B16F10 cells. RSV, RSVT and PTER inhibited FAK expression in a concentration-dependent manner, while RSVN exhibited minimal inhibition (Figure 5A-B). These observations indicate that FAK expression is involved in RSV's inhibition of cell migration. In addition, transfection with a plasmid carrying the FAK gene into RSV-treated B16F10 cells led to the recovery of FAK expression from the RSV-decreased level to the control level. This led to the complete recovery of cell migration from the RSV-induced inhibition of cell migration, demonstrating that RSV inhibits cell migration through suppressing FAK expression.
The MOA study of RSV inhibiting cell migration. The data are represented as the mean ± SEM. (A) B16F10 cells were treated with different concentrations of RSV and the analogs for 24 h. FAK expression in the cells was measured with qPCR. (B) B16F10 cells were treated with different concentrations of RSV and the analogs for 24 h. FAK expression in the cells was measured with western blotting. (C) B16F10 cells with or without the transfection of the plasmid that overexpresses FAK were treated with 60 μM RSV for 24 h. FAK expression in the cells was measured with western blotting. (D) B16F10 cells with or without the transfection of plasmid that overexpresses FAK were treated with 60 μM RSV for 24 h. Cell migration rate was measured with RTCA assays. (E) B16F10 cells were transfected with the FAK promoter reporter gene plasmid followed by incubation with different concentrations of RSV for 24 h. The FAK promoter activity was measured with dual-luciferase reporter assays. (F) B16F10 cells were treated with RSV, its analogs and the inhibitors of HDAC1 and DNMT3a for 24 h. The binding activity of HDAC1 and DNMT3a with the FAK promoter was measured with ChIP assays. (G) B16F10 cells were treated with RSV, its analogs and the inhibitors of HDAC1 and DNMT3a for 24 h. The methylation level of the FAK promoter in the cells was measured with BSP assays. (H) B16F10 cells were treated with RSV, its analogs and the inhibitors of HDAC1 and DNMT3a for 24 h. The binding activity of USF1 with the FAK promoter was measured with ChIP assays. (I) The methylation conditions of the CpG sites near position -40 of the FAK promoter following treatments with RSV and its analogs. (J) B16F10 cells were treated with RSV and the inhibitors of HDAC1 and DNMT3a for 24 h. The cell migration rates were measured with RTCA assays. The concentrations of DAC and TSA were 200 ng/mL and 40 nM, respectively.
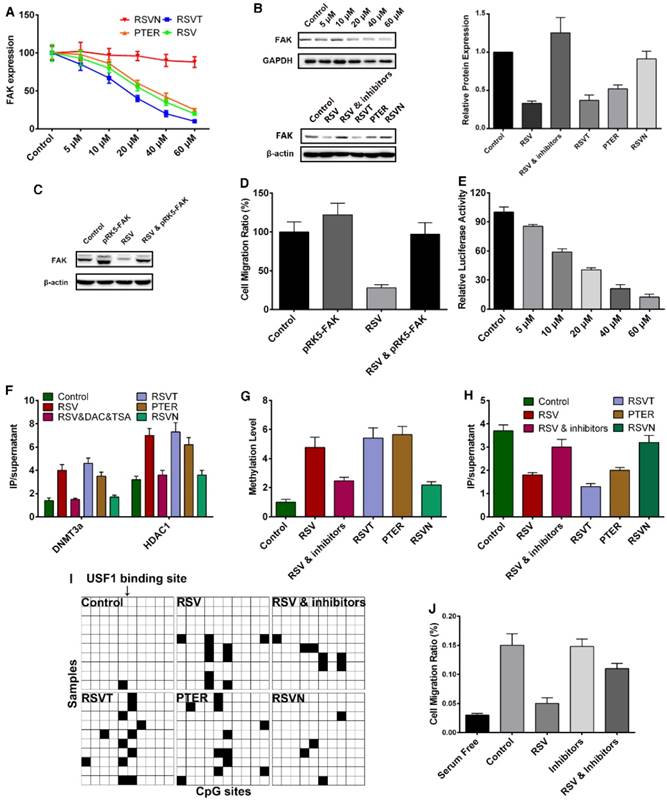
Since epigenetic regulation induces gene silencing at the chromosome level, we utilized a dual-luciferase assay to examine the effect of RSV on FAK promoter activity. RSV significantly inhibited FAK promoter activity in a dose-dependent manner (Figure 5E), revealing that RSV inhibits FAK expression at the chromosome level and might suppress FAK promoter activity through epigenetic regulation. Next, we carefully analyzed the FAK promoter region to validate this hypothesis. To screen out the region of the FAK promoter which was important for FAK transcription, we respectively cloned part of the promoter and created a series of FAK promoter truncations (Figure S2). With the dual-luciferase reporter assay, we tested the transcription activity of the truncated promoters and found that the base pair region from -170 to +44 was the key modulation area (Figure S2). Our conclusion is more specific than that of the previous study, which showed that an ~600 base pair region (-564 to +47) is the key modulation region [59]. As HDAC1 and DNMT3a always form a complex at the promoter region to inhibit expression in epigenetic regulation, we conducted a chromatin immunoprecipitation (ChIP) assay and discovered that RSV, RSVT and PTER significantly enhanced HDAC1 and DNMT3a binding to the FAK promoter, while RSVN had no obvious influence (Figure 5F). Because DNMT3a mediates de novo DNA methylation that contributes to the recruitment of HDAC1, we used bisulfite sequencing PCR (BSP) to examine the methylation status of the FAK promoter in B16F10 cells treated with RSV and the analogs. RSV, RSVT and PTER greatly increased the methylation level of the FAK promoter (Figure 5G).
As epigenetic gene silencing is directly mediated by blocking the binding of a transcription factor to a promoter region, we next sought to screen out the transcription factor that regulates FAK transcription. Via P-Match analysis, we found several transcription binding sites on the promoter, including four upstream stimulatory factor 1 (USF1) binding sites and three NF-κB binding sites (Figure S3). Next, we respectively mutated one or several transcription binding sites and obtained a series of gene reporter plasmids (Figure S3). Through dual-luciferase reporter assays, we discovered that a mutation to the USF1 binding site at position -40 exhibited the lowest FAK transcriptional activity (Figure S4). siRNA of USF1 was then designed to suppress USF1 expression, and we found that the suppression of USF1 expression led to the silencing of the FAK gene (Figure S5). Hence, the binding of USF1 to position -40 of the FAK promoter region is required for FAK transcription. With the ChIP assay, we discovered that RSV, RSVT and PTER did greatly suppress the binding of USF1 to the FAK promoter, while RSVN did not affect USF1 binding (Figure 5H). Notably, the CpG sites close to position -40 within the FAK promoter were highly methylated by RSV, RSVT and PTER (Figure 5I). All the data indicate that RSV enhanced the binding of DNMT3a and HDAC1 to block USF1 binding, thereby decreasing FAK promoter activity and inhibiting FAK expression. To further verify the conclusion, we introduced a DNMT inhibitor decitabine (DAC), a cytidine analog that competes with cytidine during DNA synthesis and disrupts DNA methylation [53]. In supporting the conclusion, the addition of TSA and DAC attenuated the RSV-induced effects (Figure 5B, E-H, J).
Validating the conclusions through in vivo assays
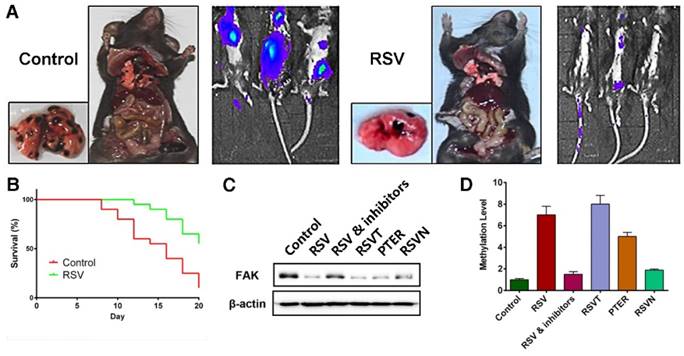
Finally, we validated our conclusions in an animal model. In our B16F10 cell passive transfer model, we observed that RSV significantly suppressed the transfer of the B16F10 tumor cells to the lungs (Figure 6A). Moreover, RSV improved the survival rate of the tumor-bearing mice (Figure 6B). We also injected groups of C57B6 mice with B16F10 tumor cells. Six groups were respectively treated with a series of intraperitoneal injections of 25% PEG400 (the control group), RSV, RSV coupled with DAC and TSA, RSVT, PTER, and RSVN. Growth of the tumors was measured with calipers each day, and the mice were sacrificed when the tumors grew to approximately 500 mm3. Genomes and proteins were extracted from the tumors for subsequent experiments. From our western blot analysis as shown in Figure 6C, we found that RSV, RSVT and PTER remarkably inhibited the expression of FAK, and the inhibitors of DNMT3a and HDAC1 reversed the trend. Moreover, the methylation condition of the FAK promoter (Figure 6D) in the animal model was in line with the in vitro experimental data.
To conclude, we utilized CPAT to elucidate the relationships among chemical structure, protein targets and anti-migration activity of RSV. We first obtained or synthesized five RSV analogs and discovered that RSVN exhibited similar bioactivities to RSV except for its anti-metastasis activity. This meant that the presence of 4'-hydroxystilbene within the RSV structure is essential for RSV's anti-migration activity. A comparative chemical proteomics approach was then applied to identify the specific targets of RSV that are related only to its anti-migration activity. Based on the targets, we discovered a novel mechanistic insight that RSV enhances the binding of the FAK promoter with HDAC1 and DNMT3a, leading to a reduction in the binding of USF1 to the FAK promoter. This loss of USF1 binding to the FAK promoter leads to the inhibition of FAK expression, resulting in a perturbation of cancer cell migration and tumor metastasis. All the data on the RSV analogs confirmed that the relationships among chemical structure, protein targets and anti-migration activity of RSV through CPAT were accurate and reliable. Although RSV is well known for its binding promiscuity with multiple target proteins and biological functions, we were able to rapidly screen out the specific chemical structure and targets of RSV that are solely involved in its anti-migration activity with CPAT. As CPAT is a simplified and precise approach, it might also be adapted to delineate the specific function of other promiscuously binding NPs.
Validating the conclusion with in vivo assays. (A) The effect of RSV on B16F10 cell migration in a passive transfer model. The transfer rate was determined by detecting the distribution of cancer cells and counting the tumor metastasis on the lung. (B) The effect of RSV on the survival rate of tumor-bearing mice. B16F10 cells were hypodermically injected, and mice of RSV group were treated with 50 mg/kg RSV each day. (C) FAK expression in the tumor tissues of mice treated with 25% PEG400 (the control group), RSV, RSV coupled with DAC and TSA, RSVT, PTER, and RSVN. (D) The methylation levels of the FAK promoter in the tumors (see (C)) were measured with BSP assays. Data are represented as the mean ± SEM.
Methods
Cell lines
B16F10 melanoma cells were obtained from the American Type Culture Collection (ATCC, Philadelphia, PA, USA). The cells were maintained in tissue culture using Dulbecco's minimum essential medium (DMEM) (HyClone, Logan, UT, USA) containing 10% fetal calf serum (HyClone, Logan, UT, USA) and 1% penicillin-streptomycin (Invitrogen, Carlsbad, CA, USA). All cells were cultured in a humidified CO2 incubator at 37 °C.
Animals
Seven-week-old C57BL/6 healthy female mice were grown and treated under procedures authorized by Nanjing University Animal Care and Use Committee (20130118).
B16F10 cell passive transfer model
7-week-old female mice were injected with 200 μL 2.5×106/mL B16F10 cells that express luciferase from caudal vein and divided into two groups. Control group were injected with PBS intraperitoneally each day, while RSV group were injected with RSV (50 mg/kg) intraperitoneally each day. After 20 days, the distribution of cancer cells was detected with in vivo imaging system. The two groups of mice were killed, and the tumor metastasis on the lung were counted statistically.
Tumor-bearing mouse model
7-week-old female mice were hypodermically injected with 1×105 B16F10 cells and divided into six groups. Six groups were treated with a series of intraperitoneal injections of 25% PEG400 (control group), RSV, RSV coupled with DAC and TSA, RSVT, PTER, and RSVN respectively. Growth of tumors was measured with calipers each day, and the mice were killed when the tumors of control group grew to about 500 mm3. Genomes and proteins were extracted from the tumors.
RTCA assay
The RTCA system was applied to monitor cell migration by using cell invasion/migration (CIM)-plates. B16F10 cells were cultured in DMEM with 10% fetal bovine serum for 24 h. Medium containing 10% fetal serum or serum-free medium was added into the lower chamber. Then, the upper chamber was mounted, and the CIM-plate was settled for 30 min at room temperature in a sterile condition. Serum-free medium (25 μL) was added to each well of the upper chambers, and the plate was left to equilibrate in the incubator for 1 h at 37 °C and 5% CO2. After equilibrium, the background reading of each well was determined. B16F10 cells were prepared in serum-free medium, 40000 cells and compounds were added into the upper chamber of each well, and serum-free medium was added to a total volume of 180 μL. The first 25 readings were recorded every 5 min followed by scans at 10 min intervals until the end of the experiment (up to 24 h).
DPPH reduction method
RSV and analogs (200 μL of 60 μM) were mixed with 35 μL of a freshly prepared ethanolic solution containing 0.4 mg/mL DPPH in microplate wells. The solutions were incubated at 37 °C for 30 min, and the absorbance was measured at 492 nm with a microplate reader. The percentage inhibition for each compound was calculated by the following formula:
% loss of DPPH activity = (Acontrol - Acompound) / Acontrol × 100,
where Acontrol is the absorbance of the control (0.2 mL of ethanol plus 0.0035 mL of freshly prepared ethanolic solution of DPPH) and Acompound is the absorbance of a sample at 492 nm.
Transwell assay
The cell migration assay was performed using transwell inserts (8.0 mm pore size, Millipore, Billerica, MA, USA). Before the experiment, B16F10 cells were cultured in serum-free medium with RSV at a concentration range from 5 μM to 60 μM for 16 h. Then, the cells were harvested and re-suspended in medium containing the respective concentrations of RSV. The upper chambers were filled with 100 μL of medium containing 1×105 cells and the lower chambers with 0.6 mL of medium containing 20% fetal calf serum. After incubation at 37 °C for 9 h, cells on the upper surface of the membrane were removed. The cells attached to the lower surface (i.e., the migrant cells) were fixed with 10% formalin at room temperature for 30 min and stained for 20 min with 100 mM borate buffer (pH 9.0) containing 1% crystal violet and 2% ethanol. The number of migrant cells on the lower surface was recorded in five fields with a microscope.
Cell viability assay
Cells (5×103) in 50 μL of medium per well were seeded on a 96-well plate and incubated at 37 °C overnight then treated with the compounds for 24 h. Ten microliters of CCK-8 solution per well was added, and the plate was incubated for 4 h at 37 °C. Absorbance at 650 nm was measured.
Western blotting analysis
Total proteins were extracted from cells and separated using 10% SDS-PAGE (80 V for 1 h and 120 V for the subsequent 2 h) and then electrophoretically transferred (300 mA for 1.5 h) to a nitrocellulose membrane (Millipore; HATF00010). The membrane was blocked with 5% milk in PBST for 1 h and incubated with primary antibodies (1:1000) at 4 °C overnight and secondary antibodies (1:2000) for 1 h at room temperature. Target proteins were detected with enhanced chemiluminescence (ECL) detection reagent (Thermo Scientific; 34075). The antibodies used in the study were anti-DNMT3a antibody (Abcam; ab13888), anti-HDAC1 antibody (Abcam; ab7028), anti-ACAT1 antibody (Abcam; ab168342), anti-FAK antibody (BD; 610088), anti-β-actin antibody (CST; 12262), anti-AMPKα antibody (CST; 2532), and anti-phospho-AMPKα antibody (CST; 2535).
Luciferase reporter assays
Cells seeded in 24-well plates (4×104 cells per well) were co-transfected with the reporter plasmid and an internal control plasmid (pRL-TK-Renilla; Promega). After 24 h, the compounds were added into the medium, and the cells were incubated for another 24 h at 37 °C. Then, the cells were harvested and assayed for luciferase activity. The luciferase assays were performed using a dual-luciferase reporter assay system (Promega; E1910). Luciferase activity was measured by a GloMax 96 Microplate Luminometer (Promega). Firefly luciferase activity was normalized to Renilla luciferase activity for each sample.
In vitro labeling of B16F10 cell lysates by RSV-P and RSVN-P
B16F10 cells were seeded in 150-mm dishes until the confluency reached 80-90%. The medium was discarded, and the cells were washed twice with PBS. The cells were lysed with 20 mM Tris (pH 7.5) containing 150 mM NaCl and 1% Triton X-100. Subsequently, the cell lysates were centrifuged at 11200 rcf for 45 min at 4 °C to remove the insoluble fraction. The protein content of the lysates was quantified with BCA protein quantitation kit. 4 mg of protein was treated with streptavidin beads attached by RSV-P or RSVN-P for 4 h at room temperature to enrich the labeling proteins. The beads were washed with PBS with 0.1% SDS, PBS and then double-distilled water several times to remove non-specific binding proteins. Then, the labeled proteins were solubilized with 100 μL of 1× SDS loading buffer, 15 μL of each sample was loaded into the wells of a 10% polyacrylamide gel, and the proteins were separated through SDS gel electrophoresis (80 V for 1 h and 120 V for 2 h). Coomassie Brilliant Blue Staining was applied to visualize the labeled proteins.
Distribution of RSV and RSVN binding proteins in living cells
B16F10 cells were cultured in six-well plates with sterilized coverslips at the base until 40-50% confluence was reached. Then, the cells were washed three times with PBS. RSV-P and RSVN-P (60 μM) in 2 mL of medium with a final dimethyl sulfoxide (DMSO) concentration of 1% was added, and the cells were incubated at 37 °C with 5% CO2 for 4 h. The cells were washed three times with PBS and fixed with 4% paraformaldehyde for 15 min. The cells were washed three times with PBS and incubated with 0.5% Triton X-100 (in PBS) for 20 min at room temperature. The cells were washed three times and incubated with rhodamine-conjugated avidin (Thermo Scientific; A6378) for 2 h at room temperature in the dark. The distribution of protein targets was detected with a confocal microscope.
Mass spectrometry for protein identification
A TripleTOF 5600 system (AB SCIEX, Foster City, CA, USA) was used to obtain a mass spectrum in high-resolution mode with a resolution of more than 30000 (250 ms accumulation time per spectrum and a mass range of 400-1250 m/z), and a tandem mass spectrum was acquired in high-sensitivity mode with a resolution of more than 15000. For each mass spectrum, a maximum of 20 precursors with a charge state between 2 and 4 were chosen for fragmentation. Additionally, the signals were accumulated for a minimum of 100 ms per spectrum, and the dynamic exclusion time was set at 15 s.
Immunofluorescence
Cells seeded on glass coverslips in 24-well plates at a density of 4×104 cells per well were treated with RSV at a concentration range from 5 μM to 60 μM for 24 h. Then, the cells were fixed with 4% paraformaldehyde in PBS for 10 min at room temperature and permeabilized with 0.1% Triton X-100 in PBS for 15 min at room temperature. After being washed with PBS three times, the cells were consecutively stained with primary and secondary antibodies. The primary antibody used was anti-NF-κB mouse monoclonal IgG (Santa Cruz; sc-8008). The secondary antibody used was goat anti-mouse IgG (H + L) Alexa Fluor 488-conjugated (Invitrogen; A-11034). The cells were washed with PBS, the nuclei were stained with DAPI (Invitrogen; D1306) for 30 s, and the cells were mounted on glass slides and observed under a confocal laser scanning microscope (Leica TCS SP8).
Bisulfite sequencing PCR
Genomic DNA was obtained from B16F10 cells or tumor tissues in tumor-bearing mice using a GeneJET Genomic DNA Purification Kit (Thermo Scientific; K0721) and converted with bisulfite using the Zymo EZ DNA Methylation Golden Kit (Zymo; D5005) according to manufacturers' specifications. The following primers were used for FAK promoter PCR amplification: 5'-GTGAAGTTTGGAGAAGATTAGGGT-3' and 5'-CAATACCTCACCTCAA-3'. PCR products were gel-purified and cloned into a pMD19-T Vector (TaKaRa, 6013) according to the manufacture's protocol. For each sample, 10 monoclonal colonies were picked and sequenced with the M13F primer after transformation. The sequencing results were analyzed using a BiQ Analyzer for the methylation level, which was calculated using the following formula: methylation level = (number of methylated CpG sites) / (number of all the detected CpG sites).
ChIP assays
Briefly, cell extracts were prepared from crosslinked cells that were incubated with anti-DNMT3a (Abcam; ab13888), anti-HDAC1 (Abcam; ab7028) and anti-USF1 (Santa Cruz; sc-229) antibodies. The immune complexes were collected via protein A-agarose and washed thoroughly. The DNA present in the cell extracts and in the immunoprecipitants were purified and quantified by real-time qPCR. The sequences of the PCR primers for the FAK promoter were 5'-CGCACAGCTGGGATACACTTTA-3' and 5'-TCACCTCAGCGCAGAGCTCTA-3'.
Quantification and statistical analysis
GraphPad prism 6.0 was used for statistical analysis. The data are summarized as the mean ± SEM. One-way ANOVA was used to determine the significant differences between the groups. The results were considered to be significant for p-values of < 0.05.
Abbreviations
ACAT1: acetyl-coenzyme A acetyltransferase 1; AMPK: AMP-activated protein kinase; BSP: bisulfite sequencing PCR; ChIP: chromatin immunoprecipitation; CPAT: Comparative Profiling of Analog Targets; DAC: decitabine; dihydro-RSV: α, β-dihydroresveratrol; DNMT3a: DNA methyltransferase 3 alpha; DPPH: 2,2-di(4-tert-octylphenyl)-1-picrylhydrazyl; FAK: focal adhesion kinase; HDAC1: histone deacetylase 1; MOA: mechanism of action; NPs: natural products; PTER: pterostilbene; RSV: resveratrol; RSV-P: resveratrol probe; RSVN: 3-(3,5-dihydroxyphenyl)-7-hydroxy-2H-chromen-2-one; RSVN-P: RSVN probe; RSVT: RSV trinicotinate; RTCA: real-time cell analyzer; SAR: structure-activity relationship; TSA: trichostatin A; USF1: upstream stimulatory factor 1
Acknowledgements
This work was funded by NSFC (81630092, 81421091), MOST (2016YFC0902700, 2014CB744501), Shenzhen Peacock Plan (JCYJ20160331152141936, KQTD20140630165057031), Guangdong NSF (2016A030311033, 201505041557046), National Natural Science Foundation of China (81641002, 81473548), National Major Scientific and Technological Special Project for "Significant New Drugs Development" (2017ZX09101002-00X-00X), and the Fundamental Research Funds for the Central public welfare research institutes (ZZ10-024).
Author Contributions
XC carried out most of the experiments and wrote the manuscript with the inputs from other authors; JW and CX conceived C-PAT; WL assisted with dual luciferase assay; JW and BZ assisted with trans-well assay and CCK-8 assay; QH constructed gene reporter plasmids; DC and YKW analyzed immunofluorescence data; JS and YZ designed and synthesized RSTV, RSVN and PTER; YML and QL designed RSV-P and RSVN-P; RT and PS assisted with RTCA assay; ZH and JW supervised the project.
Supplementary Material
Supplementary figures and methods.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Mann J. Murder, magic, and medicine. Revised edition. New York: Oxford University Press. 2000
2. Newman DJ, Cragg GM, Snader KM. The influence of natural products upon drug discovery. Nat Prod Rep. 2000;17:215-34
3. Shen B. A new golden age of natural products drug discovery. Cell. 2015;163:1297-300
4. Newman DJ, Cragg GM. Natural products as sources of new drugs from 1981 to 2014. J Nat Prod. 2016;79:629-61
5. Newman DJ, Cragg GM, Snader KM. Natural products as sources of new drugs over the period 1981-2002. J Nat Prod. 2003;66:1022-37
6. Kim KB, Crews CM. From epoxomicin to carfilzomib: chemistry, biology, and medical outcomes. Nat Prod Rep. 2013;30:600-4
7. Corson TW, Crews CM. Molecular understanding and modern application of traditional medicines: triumphs and trials. Cell. 2007;130:769-74
8. Leuenroth SJ, Crews CM. Studies on calcium dependence reveal multiple modes of action for triptolide. Chem Biol. 2005;12:1259-68
9. Millard M, Odde S, Neamati N. Integrin targeted therapeutics. Theranostics. 2011;1:154
10. Wang J, Zhang J, Shi Y, Xu C, Zhang C, Wong YK. et al. Mechanistic investigation of the specific anticancer property of artemisinin and its combination with aminolevulinic acid for enhanced anticolorectal cancer activity. ACS Cent Sci. 2017;3:743-50
11. Zhang C-J, Wang J, Zhang J, Lee YM, Feng G, Lim TK. et al. Mechanism-guided design and synthesis of a mitochondria-targeting artemisinin analogue with enhanced anticancer activity. Angew Chemie. 2016;128:13974-8
12. Wu X, Wu X, Sun Q, Zhang C, Yang S, Li L. et al. Progress of small molecular inhibitors in the development of anti-influenza virus agents. Theranostics. 2017;7:826
13. Wang J, Zhang CJ, Chia WN, Loh CC, Li Z, Lee YM. et al. Haem-activated promiscuous targeting of artemisinin in Plasmodium falciparum. Nat Commun. 2015;6:10111
14. Chen X, Wang J, Fu Z, Zhu B, Wang J, Guan S. et al. Curcumin activates DNA repair pathway in bone marrow to improve carboplatin-induced myelosuppression. Sci Rep. 2017;7:17724
15. Aida Zulueta, Anna Caretti, Paola Signorelli. et al. Resveratrol: A potential challenger against gastric cancer. World J Gastroenterol. 2015;21:10636-43
16. Lijun Tan BS, Wang W, Gong HB, Ma RDK, Gabrielle Gossner MD, Kueck AS. et al. Resveratrol inhibits ovarian tumor growth in an in vivo mouse model. Cancer. 2016;122:722-729
17. Zordoky BN, Robertson IM, Dyck JR. Preclinical and clinical evidence for the role of resveratrol in the treatment of cardiovascular diseases. Biochim Biophys Acta - Mol Basis Dis. 2015;1852:1155-77
18. Turner RS, Thomas RG, Craft S, Dyck CH Van, Mintzer J, Reynolds BA. et al. A randomized, double-blind, placebo-controlled trial of resveratrol for Alzheimer disease. Neurology. 2015;85:1383-91
19. Baxter RA. Anti-aging properties of resveratrol: review and report of a potent new antioxidant skin care formulation. J Cosmet Dermatol. 2008;7:2-7
20. Gwak H, Kim S, Dhanasekaran DN, Song YS. Resveratrol triggers ER stress-mediated apoptosis by disrupting N-linked glycosylation of proteins in ovarian cancer cells. Cancer Lett. 2016;371:347-53
21. Kilic Eren M, Kilincli A, Eren Ö. Resveratrol induced premature senescence is associated with DNA damage mediated SIRT1 and SIRT2 down-regulation. Hofmann TG, editor. PLoS One. 2015;10:e0124837
22. Kumar B, Iqbal MA, Singh RK, Bamezai RN. Resveratrol inhibits TIGAR to promote ROS induced apoptosis and autophagy. Biochimie. 2015;118:26-35
23. Li L, Qiang S, Li Y, Yang Y, Yang Y, Tao C. et al. Overexpression of SIRT1 induced by resveratrol and inhibitor of miR-204 suppresses activation and proliferation of microglia. J Mol Neurosci. 2015;56:858-67
24. Pignitter M, Schueller K, Burkon A, Knorr V, Esefelder L, Doberer D. et al. Concentration-dependent effects of resveratrol and metabolites on the redox status of human erythrocytes in single-dose studies. J Nutr Biochem. 2016;27:164-70
25. Said RS, El-Demerdash E, Nada AS, Kamal MM. Resveratrol inhibits inflammatory signaling implicated in ionizing radiation-induced premature ovarian failure through antagonistic crosstalk between silencing information regulator 1 (SIRT1) and poly(ADP-ribose) polymerase 1 (PARP-1). Biochem Pharmacol. 2016;103:140-50
26. Britton RG, Kovoor C, Brown K. Direct molecular targets of resveratrol: identifying key interactions to unlock complex mechanisms. Ann N Y Acad Sci. 2015;1348:124-133
27. Tennen RI, Michishita-Kioi E, Chua KF. Finding a target for resveratrol. Cell. 2012;148:387-9
28. Park S-J, Ahmad F, Philp A, Baar K, Williams T, Luo H. et al. Resveratrol ameliorates aging-related metabolic phenotypes by inhibiting cAMP phosphodiesterases. Cell. 2012;148:421-33
29. Lewinska A, Adamczyk-Grochala J, Deregowska A, Wnuk M. Sulforaphane-induced cell cycle arrest and senescence are accompanied by DNA hypomethylation and changes in microRNA profile in breast cancer cells. Theranostics. 2017;7:3461
30. Xiao CF, Zou Y, Du JL, Sun HY, Liu XK. Hydroxyl substitutional effect on selective synthesis of cis, trans stilbenes and 3-arylcoumarins through Perkin condensation. Synth Commun. 2012;43:1243-58
31. Zou Y, Huang Q, Huang TK, Ni QC, Zhang ES, Xu TL. et al. CuI/1,10-phen/PEG promoted decarboxylation of 2,3-diarylacrylic acids: synthesis of stilbenes under neutral and microwave conditions with an in situ generated recyclable catalyst. Org Biomol Chem. 2013;11:6967-74
32. Park H-J, Han E su, Park DK. The ethyl acetate extract of PGP (Phellinus linteus grown on Panax ginseng) suppresses B16F10 melanoma cell proliferation through inducing cellular differentiation and apoptosis. J Ethnopharmacol. 2010;132:115-21
33. He J, Duan S, Yu X, Qian Z, Zhou S, Zhang Z. et al. Folate-modified chitosan nanoparticles containing the IP-10 gene enhance melanoma-specific cytotoxic CD8+ CD28+ T lymphocyte responses. Theranostics. 2016;6:752
34. Porte G, Doll J, Brown AE, Tzena J, Zeidman I. Comparison of the metastatic properties of B16 melanoma clones isolated from cultured cell lines, subcutaneous tumors, and individual lung metastases. Cancer Res. 1982;42:2770-8
35. Mensor LL, Menezes FS, Leitão GG, Reis AS, Santos TC dos, Coube CS. et al. Screening of Brazilian plant extracts for antioxidant activity by the use of DPPH free radical method. Phyther Res. 2001;15:127-30
36. Faragher RGA, Burton DGA, Majecha P, Fong NSY, Davis T, Sheerin A. et al. Resveratrol, but not dihydroresveratrol, induces premature senescence in primary human fibroblasts. Age (Omaha). 2011;33:555-64
37. Fan H, Xiong X, Cao W, Li X, Zou Y, Zhang X. et al. Effect of resveratrol trinicotinate versus resveratrol against tumors in vitro and in vivo. Chinese J New Drugs. 2006;11:11
38. Kang SS, Cuendet M, Endringer DC, Croy VL, Pezzuto JM, Lipton MA. Synthesis and biological evaluation of a library of resveratrol analogues as inhibitors of COX-1, COX-2 and NF-κB. Bioorg Med Chem. 2009;17:1044-54
39. Orsini F, Verotta L, Klimo K, Gerhäuser C. Synthesis of resveratrol derivatives and in vitro screening for potential cancer chemopreventive activities. Arch Pharm (Weinheim). 2016;349:414-27
40. Vilar S, Quezada E, Santana L, Uriarte E, Yánez M, Fraiz N. et al. Design, synthesis, and vasorelaxant and platelet antiaggregatory activities of coumarin-resveratrol hybrids. Bioorganic Med Chem Lett. 2006;16:257-61
41. Fais A, Corda M, Era B, Fadda MB, Matos MJ, Santana L. et al. Tyrosinase inhibitor activity of coumarin-resveratrol hybrids. Molecules. 2009;14:2514-20
42. Rimando AM, Nagmani R, Feller DR, Yokoyama W. Pterostilbene, a new agonist for the peroxisome proliferator-activated receptor α-isoform, lowers plasma lipoproteins and cholesterol in hypercholesterolemic hamsters. J Agric Food Chem. 2005;53:3403-7
43. Joseph JA, Fisher DR, Cheng V, Rimando AM, Shukitt-Hale B. Cellular and behavioral effects of stilbene resveratrol analogues: implications for reducing the deleterious effects of aging. J Agric Food Chem. 2008;56:10544-51
44. Cichocki M, Paluszczak J, Szaefer H, Piechowiak A, Rimando AM, Baer-Dubowska W. Pterostilbene is equally potent as resveratrol in inhibiting 12-O-tetradecanoylphorbol-13-acetate activated NFκB, AP-1, COX-2, and iNOS in mouse epidermis. Mol Nutr Food Res. 2008;52:S62-70
45. Antonella Fais, Corda M, Era B, Fadda MB, Matos MJ, Quezada E. et al. Tyrosinase inhibitor activity of coumarin-resveratrol hybrids. Molecules. 2009;14:2514-20
46. Zhou T-J, Zhang S-L, He C-Y, Zhuang Q-Y, Han P-Y, Jiang S-W. et al. Downregulation of mitochondrial cyclooxygenase-2 inhibits the stemness of nasopharyngeal carcinoma by decreasing the activity of dynamin-related protein 1. Theranostics. 2017;7:1389
47. Chen X, Wong YK, Wang J, Zhang J, Lee Y-M, Shen H-M. et al. Target identification with quantitative activity based protein profiling (ABPP). Proteomics. 2017;17:1600212
48. Maria Olzscha, Heidi Thangue L. HDAC inhibitor-based therapies: Can we interpret the code? Mol Oncol. 2012;6:637-656
49. Ueki N, Wang W, Swenson C, McNaughton C, Sampson NS, Hayman MJ. Synthesis and preclinical evaluation of a highly improved anticancer prodrug activated by histone deacetylases and cathepsin L. Theranostics. 2016;6:808
50. Sahi J, Milad MA, Zheng X, Rose KA, Wang H, Stilgenbauer L. et al. Avasimibe induces CYP3A4 and multiple drug resistance protein 1 gene expression through activation of the pregnane X receptor. J Pharmacol Exp Ther. 2003;306:1027
51. El Bahhaj F, Denis I, Pichavant L, Delatouche R, Collette F, Linot C. et al. Histone deacetylase inhibitors delivery using nanoparticles with intrinsic passive tumor targeting properties for tumor therapy. Theranostics. 2016;6:795
52. Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247-257
53. Hamm CA, Costa FF. Epigenomes as therapeutic targets. Pharmacol Ther. 2015;151:72-86
54. Yoon H, Dehart JP, Murphy JM, Lim STS. Understanding the roles of FAK in cancer. Acta Histochem Cytochem Off J Japan Soc Histochem Cytochem. 2014;63:114-28
55. Noh H, Hong S, Huang S. Role of urokinase receptor in tumor progression and development. Theranostics. 2013;3:487
56. Mitra SK, Hanson DA, Schlaepfer DD. Focal adhesion kinase: in command and control of cell motility. Nat Rev Mol Cell Biol. 2005;6:56-68
57. Qin Y, Li K, He T, Mo L, Liang N. Effects of resveratrol on the expression of FAK and the level of phosphorylated FAK in highly metastatic ovarian carcinoma HO-8910PM cell line. Chinese Pharmacol Bull. 2007;23:729
58. Lin YC, Chen LH, Varadharajan T, Tsai MJ, Chia YC, Yuan TC. et al. Resveratrol inhibits glucose-induced migration of vascular smooth muscle cells mediated by focal adhesion kinase. Mol Nutr Food Res. 2014;58:1389-1401
59. Golubovskaya V, Kaur A, Cance W. Cloning and characterization of the promoter region of human focal adhesion kinase gene: nuclear factor kappa B and p53 binding sites. Biochim Biophys Acta. 2004;1678:111-25
Author contact
![]() Corresponding authors: zchuaedu.cn (Z.H.), wangjigangnus.edu (J.W.)
Corresponding authors: zchuaedu.cn (Z.H.), wangjigangnus.edu (J.W.)
 Global reach, higher impact
Global reach, higher impact