13.3
Impact Factor
Theranostics 2018; 8(9):2496-2507. doi:10.7150/thno.24677 This issue Cite
Review
Encoding activities of non-coding RNAs
1. Changhai Hospital, Second Military Medical University, Shanghai 200433, China.
2. Department of Chemistry and Biochemistry, Stephenson Life Sciences Research Center, University of Oklahoma, 101 Stephenson Parkway, Norman, Oklahoma 73019-5300, USA.
3. School of Materials Science and Engineering, Zhejiang University, Hangzhou, Zhejiang 310027, China.
Received 2018-1-1; Accepted 2018-2-25; Published 2018-3-28
Abstract
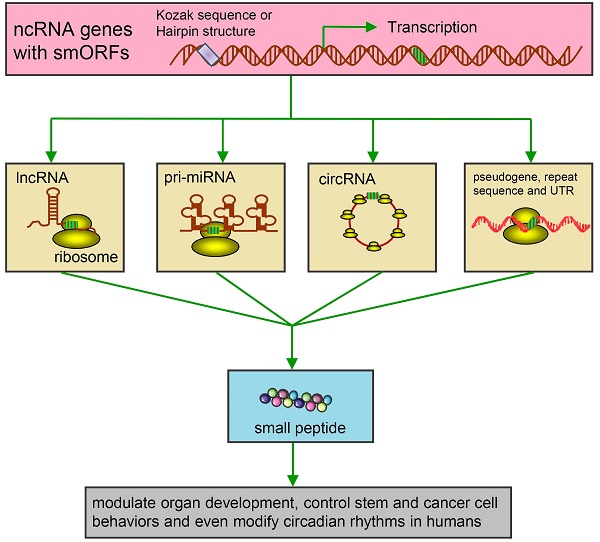
The universal expression of various non-coding RNAs (ncRNAs) is now considered the main feature of organisms' genomes. Many regions in the genome are transcribed but not annotated to encode proteins, yet contain small open reading frames (smORFs). A widely accepted opinion is that a vast majority of ncRNAs are not further translated. However, increasing evidence underlines a series of intriguing translational events from the ncRNAs, which were previously considered to lack coding potential. Recent studies also suggest that products derived from such novel translational events display important regulatory functions in many fundamental biological and pathological processes. Here we give a critical review on the potential coding capacity of ncRNAs, in particular, about what is known and unknown in this emerging area. We also discuss the possible underlying coding mechanisms of these extraordinary ncRNAs and possible roles of peptides or proteins derived from the ncRNAs in disease development and theranostics. Our review offers an extensive resource for studying the biology of ncRNAs and sheds light into the use of ncRNAs and their corresponding peptides or proteins for disease diagnosis and therapy.
Keywords: ncRNAs, translation, coding, peptides, proteins
Introduction
The genome is a cryptic store of genetic information, which encodes the blueprint of life. The product, such as peptide or protein molecules, is a key player in pathological and physiological processes [1]. The central dogma is a widely accepted classical rule in which DNA and protein are correlated by messenger RNA (mRNA) through transcription and translation [2]. However, only ~1.5% of human genome transcripts can be further translated to generate about 300,000 protein molecules, which govern the unlimited activities in human development [3]. Most of the remaining transcripts were once considered useless [4, 5]. A large, but as of yet undetermined, number of non-coding RNAs (ncRNAs) have recently been uncovered. We, along with other groups, have demonstrated that these ncRNAs can regulate normal development and disease occurrence through controlling RNA maturation and protein synthesis [6-12]. Notably, the number of ncRNAs is more than that of protein-coding genes within the genome [13, 14].
More shockingly, it has recently come to light that ncRNAs might have encoding capacity (Figure 1). Many ncRNAs do not overlap with canonical genes and some also contain small open reading frames (smORFs, containing <100 codons), which were previously thought unable to be translated into peptides or proteins [15-18]. The studies of mammalian genomes reveal that hundreds of non-annotated smORFs can be translated into small peptides in mammalian genomes according to Encyclopedia of DNA Elements (ENCODE) [19-21]. Besides 201 cleavage sites of signal peptides and 198 N-termini of proteins, mass spectrometry (MS) analysis showed a total of 808 newly annotated regions within the human genome. These regions are involved in the translation events of 44 novel smORFs, 140 pseudogenes, 106 novel coding regions/exons that originally belonged to the annotated genes, as well as elongation of 110 gene/protein/exons [22]. More importantly, several peptides were detected, which corresponded to 9 transcripts annotated as ncRNAs [23]. Couco et al. further reported that two types of smORFs, which could bind a few ribosomes and go through translation, were mostly found in ncRNAs and 5' untranslated regions (UTRs) [24].
Today the major question and concern lies in whether ncRNAs will be translated into peptides or proteins and whether the translated products will be functional. In particular, whether these peptides or proteins are useful in the diagnosis and treatment of disease, such as cancer, remains to be studied. Here, we summarize the recent progresses within the scope of our comprehension of the coding potential of ncRNAs and discuss the underlying molecular mechanisms. We further provide our perspective about this field in terms of the research tools and methods as well as the role of peptides or proteins derived from ncRNAs in the diagnosis and treatment of disease.
Coding potential of ncRNAs
In terms of length and structure, ncRNAs include two basic types: housekeeping and regulatory ncRNAs [25, 26] (Table 1). Generally, housekeeping ncRNAs are composed of small nuclear RNAs (snRNAs), transfer RNAs (tRNAs) and ribosomal RNAs (rRNAs). The functions of these ncRNAs are relatively clear. The regulatory ncRNAs include long non-coding RNAs (lncRNAs), microRNAs (miRNAs), circular RNAs (circRNAs) and the other transcripts far from known protein-coding regions in the genome [27, 28]. In general, except for mRNAs, transcripts from genomes can be considered as ncRNAs. Such transcripts, from pseudogenes and repeat sequences, were once called “junk genes” due to their non-coding or unproved coding roles. As far as the protein coding was concerned, ncRNAs were generally thought to lack any protein-coding potential. However, lately, a considerable amount of literature has been published on the study of the coding potential of ncRNAs. The epitope tags, such as FLAG, human influenza hemagglutinin (HA), and green fluorescent protein (GFP), have been used to validate the endogenous expression of the predicted smORFs in ncRNAs by detecting the fusion products through western blot (WB) [17, 29]. Anderson et al. inserted FLAG epitope tag into the myoregulin (MLN) locus by CRISPR/Cas9-mediated homologous recombination. Afterwards, the MLN-FLAG fusion peptide was detected by WB directly [30]. For the purpose of testing the in vivo translation and subcellular location of potential peptides or proteins, Magny et al. generated terminal GFP-tagged fusions within predicted smORFs [31]. The start codon of the predicted smORFs could also be mutated with the same approaches to inhibit ncRNA translation [32]. The polyclonal antibodies for predicted smORFs were further raised and then detected by MS [33]. Based upon these approaches, it has been consistently found that ncRNAs can be translated into peptides or proteins (Table 2).
Types of ncRNAs and their basic features.
| Name | Abbreviation | Size | Location | Functions | Examples | Database | References |
|---|---|---|---|---|---|---|---|
| Ribosomal RNAs | rRNAs | 6.9 kb | rDNA | Composition of ribosome | \* | NONCODE database | [14] |
| Transfer RNAs | tRNAs | <0.1 kb | tDNA | Amino acid transport | \* | NONCODE database | [15] |
| Small nucleolar RNAs | snoRNAs | 60 to 300 bp | Intronic | Important function in the maturation of other ncRNAs/Association with development of some cancers | U50, S NORD | Starbase, PITA | [26] |
| Small nuclear RNAs | snRNAs | 100 to 200 bp | \* | mRNAs splicing | U3, U6 | miRanda | [42] |
| MicroRNAs | miRNAs | 19 to 24 bp | Widespread loci | Targeting of mRNAs and many others/ Initiation of various disorders including many cancers | let-7 | miRmap, miRSNP, MirBase | [36] |
| Transcribed ultraconserved regions | T-UCRs | >200 bp | Widespread loci | Regulations of miRNA and mRNA levels/Possible involvement in tumorigenesis | UCR106 | ncRNA.org | [112] |
| PIWI-interacting RNAs | piRNAs | 26 to 31 bp | Clusters, intragenic | Transposon repression or relationship with diseases has not been discovered | MIWI, MILI, MIWI2 | piRNA BANK | [23] |
| Circular RNAs | circRNAs | ~500 bp | Widespread loci | As endogenous competitive RNA/Association with many cancers and diseases | ciRs7, cANRIL | circRNAbase | [25] |
| Long non-coding RNAs | lncRNAs | >200 bp | Widespread loci | Scaffold DNA-chromatin complexes, X-chromosome inactivation/Involved in tumorigenesis and cancer metastasis | H19, XIST, HOTAIR, TSIX | lncRNA database, 3.0, LNCipedia | [19] |
*No reference data.
Long ncRNAs
Transcription of lncRNAs occurs by RNA polymerase II without long and conserved ORFs. Such transcription often occurs in the genomic regions that are far away from the already known protein-coding genes [34, 35]. At the post-transcriptional level, lncRNAs regulate corresponding gene expression. They are also involved in modulating epigenetics and many TF proteins [36]. Notably, some lncRNAs have exogenous features comparable to those of annotated coding genes: they may be capped and polyadenylated. Also, they finally accumulate in the cytoplasm to become functional [19, 37].
It was found that some lncRNAs with smORFs could be translated into peptides [38]. For instance, Jorge Ruiz-Orera et al. found that the translational efficiency of transcripts as both coding RNAs and lncRNAs with smORFs, was significantly increased compared to 3'UTR with smORFs [39]. Furthermore, the sequences of lncRNAs with smORFs presented distinctly higher coding scores than the normal ORFs that were randomly obtained from any kind of ncRNAs. The score was calculated by related software such as phylogenetic analysis of codon substitution frequencies based on sequence alignment (PhyloCSF), coding potential calculator (CPC), sORF finder, coding region identification tool invoking comparative analysis (CRITICA), micro-peptide detection pipeline (micPDP) and so on [20, 40, 41]. More importantly, a number of studies about developmental biology have pointed out that a high fraction of lncRNAs are bound with one or more ribosome [42], indicating that lncRNAs can be translated into peptides or proteins [43]. Bioinformatics analysis indicates that, different from the quintessential protein-coding genes, lncRNA binding with ribosomes tends to be more conserved across different species and overlap with more exon regions. In addition, they are expressed in low abundance and show similar codon usage with annotated genes [44].
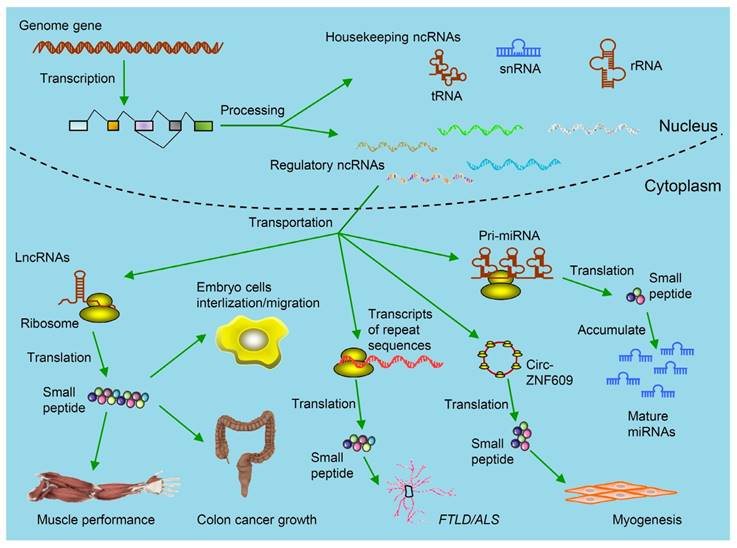
Kinds of ncRNAs-derived small peptides involved in theranostics. Some genome genes are transcribed to be ncRNAs including housekeeping ncRNAs (tRNAs, rRNAs and snRNAs) and regulatory ncRNAs. The functions of these ncRNAs are relatively clear. Regulatory ncRNAs include miRNAs, lncRNAs, circRNAs and transcripts from repeat sequences. As illustrated, by binding with ribosomes in the cytoplasm, these ncRNAs are further translated into small peptides. Circ-ZNF609 is translated and the produced small peptides function in myogenesis. These small peptides are involved in regulating muscle performance, suppressing colon cancer growth, promoting embryo cells internalization/migration, leading to FTLD/ALS and accumulation of mature miRNAs. (FTLD/ALS: frontotemporal lobar degeneration and amyotrophic lateral sclerosis; lncRNAs: long non-coding RNAs; miRNAs: microRNAs; ncRNAs: non-coding RNAs;rRNAs: ribosomal RNAs; snRNAs: small nuclear RNAs; tRNAs: transfer RNAs).
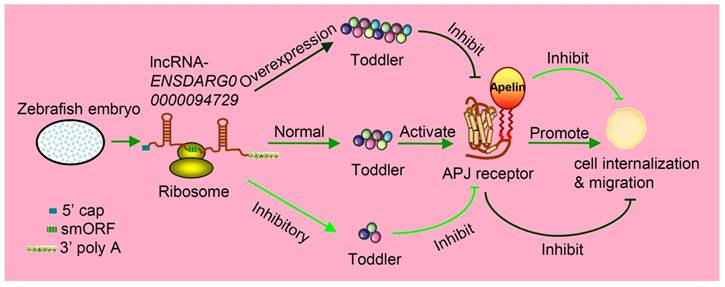
Toddler is an embryonic signal that promotes cell internalization and migration during the embryogenesis of zebrafish. Toddler is annotated as a lncRNA ENSDARG00000094729. It contains a 58-aa smORF and is also capped and polyadenylated, so it can be caught by ribosome and produce peptides named Toddler. The normal expression of Toddler can promote cell internalization and migration during the embryogenesis of zebrafish via the G-protein-coupled APJ/Apelin receptor. However, the overexpression or inhibition of Toddler will inhibit cell internalization and migration.
Peptides or proteins derived from their corresponding ncRNAs.
| ncRNAs | Peptides or Proteins | Length | Reference |
|---|---|---|---|
| ENOD40 | 2 small peptides | 12 and 24 aa | [45] |
| pri | 4 small peptides | 11 to 32 aa | [48] |
| LOC100506013 | Toddler | 58 aa | [52] |
| LINC00948 | MLN | 46 aa | [30] |
| LOC100507537 | DWORF | 34 aa | [54] |
| LINC00961 | SPAR | 90 aa | [32] |
| pri-miR171b | miPEP171b | 9 aa | [58] |
| pri-miR165a | miPEP165a | 18 aa | [58] |
| circ-ZNF609 | ZNF609 peptide | 250 aa | [70] |
| Repeats of GGGGCC | GGGGCC-encoded DPR* | 56 to 290 aa | [84, 85] |
| lncRNA HOXB-AS3 | HOXB-AS3 | 53 aa | [117] |
*DPR: dipeptide-repeat protein.
Increasing studies have measured the protein-coding output from lncRNAs. For example, John et al. discovered that a conserved sequence located at the 5' terminal of the lncRNA was transcribed from gene ENOD40 having smORFs and encoded two small peptides. Both of these small peptides were specially bound to the same sucrose synthase to control sucrose use [45]. Takefumi Kondo et al. reported one lncRNA and named it as pri (polished rice) in Drosophila [46]. In fact, it contains some evolutionarily conserved smORFs that encode four similar peptides, ranging from 11- to 32-aa in length. These small peptides were found to have essential roles in epithelial morphogenesis [47, 48]. In zebrafish, Toddler is annotated as one lncRNA at the very beginning, namely ENSDARG00000094729. The same annotation is found both in mice (Gm10664; also known as Ende [49]), and humans (LOC100506013). Moreover, LOC100506013 is raised in two lncRNA contents [50]. Nevertheless, Pauli et al. found that the ncRNA Toddler is highly conserved among many vertebrates, including homo sapiens. It contains a 58-aa smORF, which can be further translated as a signal peptide [49, 51]. They proved that this small peptide, named the Toddler, was an important signaling molecule expressed during zebrafish embryogenesis. Both the inhibition and overexpression of Toddler reduced the directional movement ability of mesendodermal cells in the period of zebrafish's gastrula stage (Figure 2) [52].
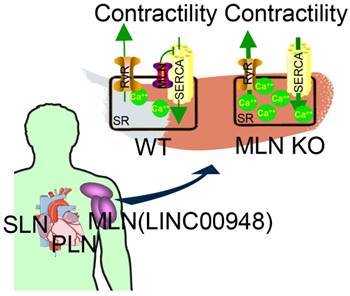
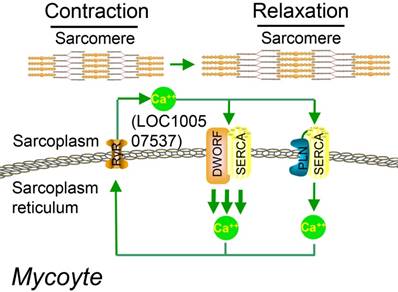
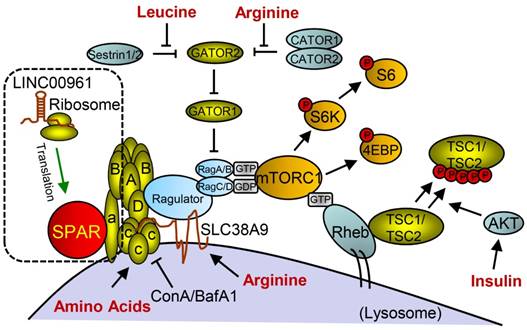
Studies on the whole-genome scale have indicated that lncRNAs can be translated into many functional small peptides for mammlians. For humans, lncRNA LINC00948 is a skeletal muscle-specific RNA. It was demonstrated that this lncRNA encodes a conserved 46-aa small peptide named MLN. MLN bears a function and structure identical to phospholamban (PLN) and sarcolinpin (SLN), which depress sarco endoplasmic reticulum Ca2+-ATPase (SERCA). SERCA is a kind of membrane pump for monitoring the relaxation of muscle by modulating the uptake of calcium ions into the sarcoplasmic reticulum (Figure 3). PLN and SLN are over-expressed in cardiac cells but almost absent in skeletal muscle cells. Unlike them, MLN is stably expressed in skeletal muscle cells [53]. Recently, a putative muscle-specific lncRNA situated in the membrane of SR, which encodes a 34-aa small peptide termed dwarf open reading frame (DWORF), was discovered. Like PLN, SLN and MLN, DWORF could enhance the activity of SERCA by substituting some particular inhibitors of SERCA (Figure 4) [54]. Furthermore, among those previously annotated and intergenic ncRNAs, more than half of them contain predicted longer ORFs (>100 codons), such as HOTAIR and Xist [55]. Another recent study conducted by Matsumoto et al. showed that one putative lncRNA-LINC00961 harboring three smORFs encoded one functional and conserved polypeptide, which is named “small regulatory polypeptide of amino acid response” (SPAR). By localizing to the lysosome/endosome and interacting with the lysosomal v-ATPase, the novel polypeptide SPAR negatively regulated the stimulation of mammalian target of rapamycin complex 1 (mTORC1) (Figure 5) [32]. These data illustrate that many transcripts currently annotated as lncRNAs encode peptides with important biological functions [30]. Moreover, these findings may be useful for the diagnosis and treatment of myopathy.
MLN (from LINC00948) is a skeletal muscle-specific small peptide that regulates muscle performance by modulating intracellular calcium handling. MLN shares structural and functional similarity with PLN and SLN, which inhibit SERCA, the membrane pump that controls muscle relaxation by regulating Ca2+ uptake into SR. (KO: knock out; MLN: myoregulin; PLN: phospholamban; ; SERCA: sarco endoplasmic reticulum Ca2+ -ATPase; SLN: sarcolinpin; SR: sarcoplasmic reticulum; WT: wild type). Reproduced with permission from [30], copyright 2015 Elsevier.
Working model for DWORF (from LOC100507537) function. DWORF localizes to the SR membrane, where it enhances SERCA activity by displacing the SERCA inhibitor PLN. (DWORF: dwarf open reading frame). Adapted with permission from [54], copyright 2016 Springer.
microRNAs (miRNAs)
miRNAs are the most investigated ncRNAs. They are hairpin-derived RNAs transcribed by RNA polymerase II. miRNAs are composed of ~20-24 nucleotides (nt) and regulate post-transcriptional silencing of genes through interacting with the 3' UTRs of mRNA [56]. Generally, miRNA coding regions in the genome can be found in both the intergenic regions and introns [57], so they seemed to be noncoding. However, Lauressegues et al. evaluated the coding potential of pri-miRNAs and demonstrated that pri-miRNAs can produce some peptides that can regulate the accumulation of miRNAs. In this study, some pri-miRNAs in the plant kingdom were found to contain smORFs, which encode regulatory peptides termed miPEP (mi-peptides). Therein, pri-miR171b and pri-miR165a were translated into peptides miPEP171b and miPEP165a, respectively. These miPEPs promote the accumulation of their homologous miRNAs by activating transcriptional activators of their corresponding pri-miRNAs. This resulted in the expression of target genes being down-regulated. Other primary transcripts of some well-studied miRNAs of M. truncatula and A. thaliana were also encoded for functional miPEPs, indicating that miPEPs or the functional peptides may be common [58].
It should be noted that RNA polymerase II transcribes a vast majority of pri-miRNAs, followed by capping, polyadenylatation, and accumulation in the cytoplasm. Their translation may possess the same regulatory patterns of the well-annotated protein-coding genes [59]. Based on these findings, there is no doubt that the traditional concept of miRNAs needs to be redefined in terms of coding potential.
circular RNAs (circRNAs)
circRNAs was first discovered in the 1990s, and had been ignored for a long time until recently [60]. Recently, the abnormal expression of circRNAs was found to be involved in many cell biological behaviors like apoptosis, angiogenesis, epithelial-to-mesenchymal transition, and drug resistance [61]. Fu et al. identified over 500 differentially expressed circRNAs in hepatocellular carcinoma (HCC) tumors with respect to the adjacent tissues. Two circRNAs derived from SMYD4 and FAM53B were found to be related to HCC clinicopathological processing [62]. Generally speaking, circRNAs are very stable. They behave like sponges of miRNAs, sponges or scaffolds of proteins and regulators of splicing activities [63]. Thus, circRNAs may be useful therapeutic targets for cancer and other diseases [64]. Moreover, many circRNAs are highly conserved, tissue-specifically expressed, and can be detected in many kinds of body fluids [65]. Accordingly, circRNAs have potential as non-invasive therapeutic biomarkers for diseases.
As one kind of ncRNAs, circRNAs were also thought unable to be translated. Most of the circRNAs are spliced from exons and are also located in the cytoplasm [66]. Under some certain conditions, many special and endogenous circRNAs with internal ribosome entry sites (IRES) and AUG sites elements have been shown to be translated in some tissues [67]. For example, Yang et al. revealed the widespread N6-methyladenosine (m6A)-driven translation of circRNA, which was further proved by MS, computational prediction and polysome profiling [68]. Meng et al. designed an integrated tool named CircPro to detect circRNAs with protein-coding potential [69]. It was shown that a series of circRNAs was actually translated in a cap-independent manner [67]. The produced small peptides or proteins have specific domains. Legnini et al. profiled the expression of circRNAs to study the differentiation of myoblasts derived from murine and human. They showed a circRNA with smORFs named circ-ZNF609. It can be translated into a small peptide to control myoblast proliferation (Figure 1) [70]. Their studies served as valuable references for the study of circRNA translation. Nevertheless, the study of circRNA translation and the possible function of produced small peptides or proteins is still in its infancy. Furthermore, the cancer relevance of this translation remains to be examined.
Small peptide SPAR derived from LINC00961 involved in working model of mTORC1 activation and signaling with SPAR. With the stimulation of amino acids (aa), Ragulator is released from v-ATPase and then interacts with Rags to facilitate mTORC1 recruitment. Rag proteins are mostly activated by Rheb, but can also be regulated through additional mechanisms involving the aa leucine and arginine. SPAR interacts with v-ATPase to promote and stabilize the interaction between the v-ATPase. (aa: amino acids; AKT: protein kinase B; GDP: guanosine diphosphate; GTP: guanosine triphosphate; SPAR: small regulatory polypeptide of amino acid response). Reproduced with the permission from [32], copyright 2016 Springer.
The transcripts of pseudogenes, repeat sequences and untranslated regions (UTRs)
Compared with their homologous functional genes, pseudogenes are referred to as the same sequences with some defunct genomic loci. Owing to the existence of disruptive mutations (e.g., some frame shifts or premature stop codons), pseudogenes have been thought to be untranslated [71]. The transcripts of pseudogenes are also defined as a sub-class of ncRNAs, and participate in regulating the post-transcriptional process of their homologous genes [72]. However, their connection with RNA polymerase II still needs to be further proved. Studies have shown that pseudogenes can perform crucial roles in the development of some diseases, in particular, cancer. Pandolfi et al. reported that specific transgenic mice were subjected to malignancy analogous to human diffuse large B cell lymphoma due to the overexpression of pseudogenes such as Braf-rs1. Their study indicated the oncogenic potential of pseudogenes [73]. Recently, emerging studies indicated that pseudogenes have a coding potential and can indeed become translated into some reliable peptides or proteins. NANOG is a pluripotent transcript factor and plays a crucial role in self-renewal of embryonic stem cells (ESCs). In cancer cells, however, NANOG mRNA variants include 9 retrotransposed genes, and 8 out of the 9 genes are pseudogenes. The pseudogene NANOG (NANOGP8, gi 47777342) is processed from a retrogene locus and has no structural defects [74]. The expression level of NANOGP8 and NANOG protein is observed to be high in the putative cancer stem cell (CSC) populations, and to have unknown function in the development of tumors [75]. Phosphoglycerate mutase 3 (PGAM3) is an intronless pseudogene and is situated in the Menkes disease gene's first intron. Its transcript has the same length as the mRNA of the homologous gene. PGAM3 also possesses a very short poly-A tail (16bp) at the ending of the 3'UTR. Further, Betran et al. reported that PGAM3 indeed produced a functional protein with the pressure of positive-selection [76]. Moreover, many novel peptides or proteins in the annotated pseudogenes group were searched using the protein basic local alignment search tool (BLAST) [77]. These data indicate that pseudogenes might be not only transcribed but also translated [78].
The repeat sequences occupy about half of the human genome and have often been considered neutral, with no phenotypic consequences [79]. As the main part of non-coding regions in the genome, repeat sequences are also often transcribed as ncRNAs [55]. Recent data indicates that repeat sequences also have the potential to produce proteins for developing frontotemporal lobar degeneration and amyotrophic lateral sclerosis (FTLD/ALS) [80]. In the upstream region of C9orf72, expanding GGGGCC repeat sequences often results in FTLD/ALS [81-83], although the mechanism of pathogenesis remains largely unknown. For these FTLD/ALS patients, researchers also found that the intracellular contents of one misfolded protein are characteristic of C9orf72-associated pathology [84]. Mori et al. found that the majority of these intracellular contents contained not only poly-(Gly-Ala) protein, but also dipeptide-repeat proteins (poly-(Gly-Arg) and poly-(Gly-Pro)) in a small portion. These proteins were assumed to be generated from the expanded GGGGCC repeat sequences by non-ATG-initiated translation. To a large extent, these discoveries directly link the predominant pathology of FTLD/ALS with the coding potential of C9orf72 hexanucleotide expansion [85].
In addition, smORFs also can be found in the 5' UTR of mRNAs and were named as upstream ORFs (uORFs) [86]. Although they have low average conservation, uORFs had been reported in many species [87]. Same as ncRNAs, uORFs in UTR were once also considered to be non-coding. But, a 31-mer peptide translated from the uORF of gene Chop was proved to inhibit the translation of CHOP protein by blocking the peptide exit tunnel of ribosomes [88]. Starck et al. also found notable translation events from uORFs of binding immunoglobulin protein (BiP) mRNA through tracing translation during the integrated stress response (ISR) for T cells. They proposed that the peptides translated from uORFs could serve as primary histocompatibility complex class I ligands to make specified cells identified by the immune system [89].
The underlying coding mechanisms of ncRNAs
Emerging evidence has demonstrated the coding capacity of ncRNAs. However, the underlying mechanisms that result in such coding capacities remain unclear though mRNAs can be translated into peptides or proteins in a relatively known manner. Many ncRNAs such as lncRNAs are also observed to be similar to mRNAs [90]. What's more, most ncRNAs contain at least one smORF [91]. The association of lncRNAs with ribosomes indicates that ncRNAs may be a possible source of novel peptides or proteins [92]. Ingolia et al. reported that, like the coding sequences discussed earlier, some lncRNAs in ESCs can be efficiently associated with one or more ribosome, and, to some extent, their coding capacity was much higher than that of mRNA 3'UTRs [93].
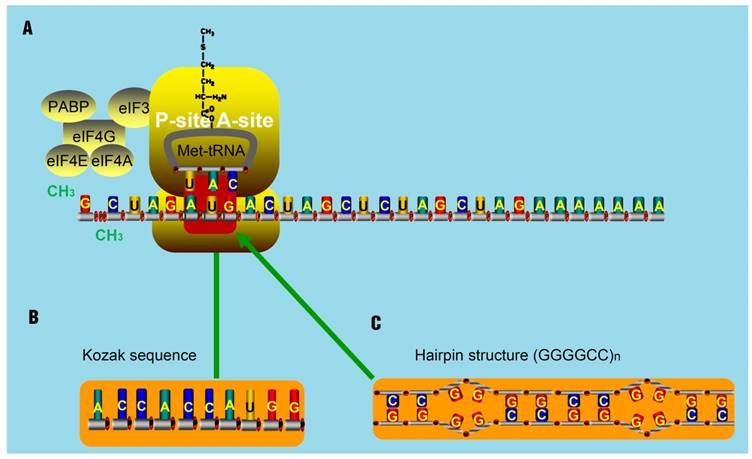
The translation of some smORFs was triggered by mammalian Ste20-like kinase (MST1)-mediated eIF4E phosphorylation. Once phosphorylated, eIF4E could barely interact with the 5' cap of mRNA and then make polyribosomes associated with lncRNAs [94]. Other smORFs used non-ATG start codons [95]. Zu et al. has proved that translational initiation of some ncRNAs, which often occurs at the CAG.CTG expansion sites, is very common [96]. By delaying the binding of 40S ribosomal subunit, it is thought that hairpin structures can initiate translation at some undefined regions [97]. Furthermore, hairpin structures can recruit both the initiation factors and ribosomal subunits to areas known as IRESs. The resultant IRESs-hairpins complexes could act as tRNAiMet and trigger the translation initiation at non-AUG codons. These complexes are stabilized due to the existence of IRES translation-associated factors [98]. But for lncRNAs, they seldom contain highly structured regions like hairpins. They are thought to contain several translatable smORFs ranging from an AUG codon to a stop codon. Occasionally, AUG codons in such smORFs will be replaced as promoters by Kozak sequences that will also initiate the translation [99, 100]. Notably, two remarkable non-AUG-initiated translation mechanisms have been proposed. One mechanism is the translation initiation from an entire Kozak sequence (substitute for the AUG codon) [101-103]. Another is the triggering of non-AUG-associated translation for some repeat sequences, like CAG repeats in ataxin 8 (ATXN8), by hairpin-formation (substitute for the AUG codon) in repeat regions (Figure 6) [104]. For spinocerebellar ataxia type 8 (SCA8) patients, gene ATXN8 encodes a native poly-Q inclusion as a result of the existence of repeat expansion [96, 105, 106].
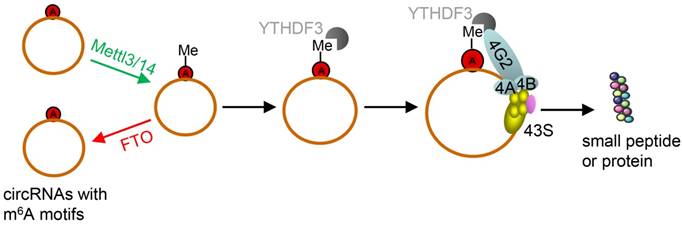
However, due to their unique structure, the translation of circRNAs was promoted by m6A. This is very different from linear ncRNAs. Yang et al. discovered some short sequences in most of circRNAs containing m6A sites [66]. Moreover, studies have reported that m6A in 3'UTRs or 5'UTRs could promote cap-independent translation [107, 108]. Thus, they reported that the translation of circRNAs was triggered by m6A reader YTH domain family protein 3 (YTHDF3). They also found that translation could be initiated by eukaryotic translation initiation factor 4 gamma 2 (eIF4G2) and this process was enhanced by methyltransferase-like 3/14 (METTL3/14) but depressed by demethylase fat mass and obesity-associated protein (Figure 7) [68].
Using a chromatin immunoprecipitation-exonuclease (ChIP-exo) method, Venters et al. identified transcription initiation complexes, as many as 160,000 copies, in the human genome. However, only ~5% of the initiation complex was associated with mRNA genes with the rest belonging to ncRNAs genes. It shows from a side view that ribosomes may occupy any accessible sites in the genome [109]. Hence, translation of “ncRNAs” into peptides or proteins may be fundamentally different from that of mRNAs. Yet, it remains to be studied whether the occupancy of ribosomes on ncRNAs is one of the key features of eukaryotic translation.
The assumed mechanism for translation initiation with special structure. (A) The classical mechanism for translation initiation. The complex composed of eIF4E, eIF4G, eIF4A, binds to the 5' cap of target RNA molecules. The poly A-binding protein (PABP) is associated with eIF4G to circularize the target mRNA molecules. Then, the eIF4F complex recruits the 43S pre-initiation complex (PIC), composed of the 40S ribosomal subunit, 30S ribosomal subunit, and the ternary complex, consisting of initiator methionine-tRNA and GTP. Next, the PIC and the components of the eIF4F complex scan through the 5'UTR in the 5' to 3' direction until encountering an AUG start codon, at which point the translation activity will be triggered by the present AUG codon. (B-C) Studies have shown that some Kozak sequences with AUG codon (B) and hairpin-structures such as (GGGGCC)n (C) can substitute the AUG codon and trigger the translation activity.
Schematic diagram of circRNA translation driven by m6A. The circRNAs here are those with m6A motifs. This m6A driven translation requires initiation factor eIF4G2 and m6A reader YTHDF3, and is enhanced by methyltransferase METTL3/14, inhibited by demethylase FTO. (m6A: N6-methyladenosine; YTHDF3: YTH domain family protein 3; eIF4G2: eukaryotic translation initiation factor 4 gamma 2; METTL3/14: methyltransferase-like 3/14; FTO: fat mass and obesity-associated protein). Adapted with the permission from [68], copyright 2017 Springer.
Unknown worlds for peptides originating from ncRNAs are probably more fantastic than previously assumed. Exploring such worlds may stimulate us to investigate the precise functions of the prevalent translation of ncRNAs. To elucidate the precision functions, we need to not only study the underlying mechanisms by which ncRNAs code for peptides or proteins, but also identify the precise roles of the resultant peptides or proteins, in particular, in disease initiation and development. Knowing these answers will improve our understanding of this biological process and help us develop new therapeutics for disease prevention and therapy.
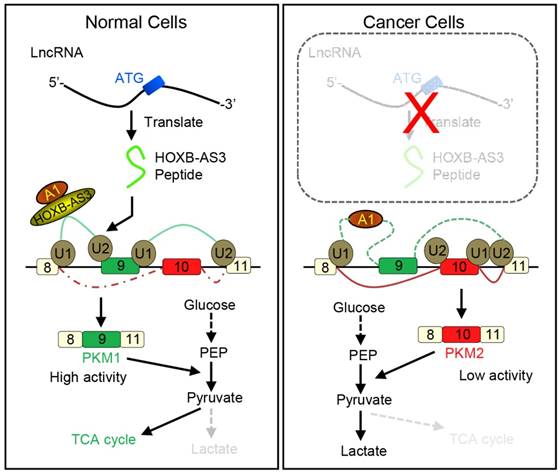
Working model for HOXB-AS3 peptide. Instead of functioning by lncRNA directly, the peptide derived from HOXB-AS3 competitively binds to the arginine residues in RGG motif of hnRNP A1 and antagonizes the hnRNP A1-mediated regulation of pyruvate kinase M (PKM) splicing by blocking the binding of the arginine residues in RGG motif of hnRNP A1 to the sequences flanking PKM exon 9, ensuring the formation of lower PKM2 and suppressing glucose metabolism reprogramming. (hnRNP: heterogeneous nuclear ribonucleoprotein; HOXB-AS3: HOXB cluster antisense RNA 3; PKM: pyruvate kinase M; RGG: Arg-Gly-Gly; TCA cycle: tricarboxylic acid cycle). Adapted with the permission from ref [117], copyright 2016 Elsevier.
Importance of peptides or proteins derived from ncRNAs in theranostics
It has been observed that RNA polymerases tend to trigger transcription at the reputed promoter regions, suggesting that the probability of triggering transcription on reachable DNA sequences is low [110]. However, many random DNA sequences can promote transcription by recruiting TFs. This is consistent with the fact that most of the nucleosome-free DNA could be transcribed in vivo [111]. Like mRNAs, some lncRNAs are capped, spliced and polyadenylated [112]. So, they also have the chance to be caught by ribosomes and translated, though less frequently than most mRNAs [113]. Hence, there is sufficient evidence to believe that ncRNAs may serve as one kind of substrate for the basal translational machinery and could be translated into a random peptide [114].
How novel peptides contribute to normal development and disease will open up new avenues for this field. It is well known that cancer develops by accumulation of driver mutations. Many such mutations occur in non-coding regions of the genome [115]. It is possible that short peptides arising from the so-called non-coding regions may indeed have a higher level of expression in cancers. Moreover, they may also function in the occurrence and development of tumors [116]. An improved understanding of ncRNAs and their corresponding peptides may result in the discovery of new methods to diagnose and treat cancer. For example, Huang et al. reported that a 53-aa peptide encoded by lncRNA HOXB-AS3 is the regulator of pyruvate kinase M (PKM) for alternative splicing and cancer metabolism reprogramming. They found reduced HOXB-AS3 peptide levels were related with a weak prognosis in colon cancer (CRC) patients and the loss of this peptide is one critical oncogenic event. Their findings uncovered a complex regulatory mechanism of cancer metabolism reprogramming orchestrated by a lncRNA-encoded peptide (Figure 8) [117]. Also, ncRNAs and their corresponding peptides may play a significant, but yet to be characterized, role in other diseases.
Small peptides can be used in a variety of ways such as antibacterial agents, cell signaling molecules, and cytoskeletal modulators [118]. For example, dominant-negative peptides are as short as ~100-aa long. Another short peptide of 119-168 aa, termed helix-loop-helix (HLH)-like peptides, could capture basic proteins to form biologically active forms. Specifically, the peptides can modulate organ development, control stem and cancer cell behaviors and even modify circadian rhythms in humans [119]. The study of Huang et al. opened a new prospect of the study of lncRNAs, especially in human development and tumorigenesis. Thus, there are reasons to believe that an increasing number of small peptides, which have been ignored in current clinical medicine and basic biomedical investigations, will be characterized in future studies.
We believe that exploring the pathological and physiological effects of new peptides generated by ncRNAs may unlock a new scientific world. The rules about biogenesis and actions should be more complicated than what we originally believed. We are witnessing a substantial improvement in our understanding of what was once thought to be a fully characterized biological process. With this in mind, researchers should thoroughly study the new coding mechanisms across all genes in the genome and develop new means to purify and identify peptides originating from the ncRNAs. Functional analyses in the future should also focus on the way in which peptides encoded by the ncRNAs with smORFs contribute to the various basic biological processes and mechanisms of some important diseases like cancer. These will very likely lead to new breakthroughs in the development of life science and medicine.
Concluding remarks and future perspectives
The discovery of ncRNAs translation and smORFs in the transcriptome highlights the need for experimental maps of pervasive translation [31, 120]. To date, however, very little, if any, is clear about the biological mechanisms of coding “ncRNAs” as well as the understanding of the biological function of the peptides or proteins derived from them. Future studies need to be performed to enhance our understanding of coding ncRNAs and the role of their corresponding peptides or proteins in disease development and theranostics. The following questions have yet to be answered about translation of putative ncRNAs. First, what is the exact mechanism of smORFs encoding for functional peptides or proteins? Second, what are the functions of these peptides and how do they work? Third, how can we computationally or experimentally identify the translated peptides? And finally, how are new peptides or proteins translated from ncRNAs?
Abbreviations
aa: amino acid; AKT: protein kinase B; ATXN8: ataxin 8; BiP: binding immunoglobulin protein; BLAST: protein basic local alignment search tool; CPC: coding potential calculator; CRC: colon cancer; CRITICA: coding region identification tool invoking comparative analysis; CSC: cancer stem cell; DWORF: dwarf open reading frame; eIF4G2: eukaryotic translation initiation factor 4 gamma 2; ENCODE: Encyclopedia of DNA Elements; ESC: embryonic stem cell; FTLD/ALS: frontotemporal lobar degeneration and amyotrophic lateral sclerosis; FTO: fat mass and obesity-associated; GDP: guanosine diphosphate; GFP: green fluorescent protein; GTP: guanosine triphosphate; HA: human influenza hemagglutinin; HCC: hepatocellular carcinoma; hnRNP: heterogeneous nuclear ribonucleoprotein; HOXB-AS3: HOXB cluster antisense RNA 3; IRESs: internal ribosome entry sites; ISR: integrated stress response; lncRNAs: long non-coding RNAs; m6A: N6-methyladenosine; METTL3/14: methyltransferase-like 3/14; micPDP: micro-peptide detection pipeline; miRNAs: microRNAs; miPEP: mi-peptides; MLN: myoregulin; mRNAs: messenger RNAs; MS: mass spectrometry; ncRNAs: non-coding RNAs; PGAM3: Phosphoglycerate mutase 3; PhyloCSF: phylogenetic analysis of codon substitution frequencies based on sequence alignment; PKM: pyruvate kinase M; PLN: phospholamban; RGG: Arg-Gly-Gly; rRNAs: ribosome RNAs; SCA8: spinocerebellar ataxia type 8; SERCA: sarco endoplasmic reticulum Ca2+ -ATPase; SLN: sarcolinpin; sm ORFs: small open reading frames; snRNAs: small nuclear RNAs; SPAR: small regulatory polypeptide of amino acid response; SR: sarcoplasmic reticulum; TCA cycle: tricarboxylic acid cycle; tRNAs: transfer RNAs; uORFs: upstream ORFs; UTR: untranslated region; YTHDF3: YTH domain family protein 3; WB: western blot.
Acknowledgements
We would like to thank Yijun Hu and Bo Wang for critical reading.
Funding
This work was supported by the China National Funds for Distinguished Young Scientists (81425019), the State Key Program of National Natural Science Foundation of China (81730076), the National Natural Science Foundation (81372763 and 51673168) and the Specially-Appointed Professor Fund of Shanghai (GZ2015009). We would also like to thank the financial support from National Science Foundation (CBET-1512664) and National Institutes of Health (CA200504, CA195607, and EB021339).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Wilkins MR, Sanchez JC, Gooley AA, Appel RD, Humphery-Smith I, Hochstrasser DF. et al. Progress with proteome projects: why all proteins expressed by a genome should be identified and how to do it. Biotechnol Genet Eng Rev. 1996;13:19-50
2. Mattick JS. RNA regulation: a new genetics? Nat Rev Genet. 2004;5:316-23
3. Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A. et al. Landscape of transcription in human cells. Nature. 2012;489:101-8
4. Ohno S. So much "junk" DNA in our genome. Brookhaven Symp Biol. 1972;23:366-70
5. Doolittle WF. Is junk DNA bunk? A critique of ENCODE. Proc Natl Acad Sci U S A. 2013;110:5294-300
6. Maliszewska-Olejniczak K, Gruchota J, Gromadka R, Denby Wilkes C, Arnaiz O, Mathy N. et al. TFIIS-dependent non-coding transcription regulates developmental genome rearrangements. PLoS Genet. 2015;11:e1005383
7. Liu S, Guo W, Shi J, Li N, Yu X, Xue J. et al. MicroRNA-135a contributes to the development of portal vein tumor thrombus by promoting metastasis in hepatocellular carcinoma. J Hepatol. 2012;56:389-96
8. Zhang X, Liu S, Hu T, Liu S, He Y, Sun S. Up-regulated microRNA-143 transcribed by nuclear factor kappa B enhances hepatocarcinoma metastasis by repressing fibronectin expression. Hepatology. 2009;50:490-9
9. Liu S, Li N, Yu X, Xiao X, Cheng K, Hu J. et al. Expression of intercellular adhesion molecule 1 by hepatocellular carcinoma stem cells and circulating tumor cells. Gastroenterology. 2013;144:1031-41.e10
10. Guo W, Liu S, Cheng Y, Lu L, Shi J, Xu G. et al. ICAM-1-Related Noncoding RNA in Cancer Stem Cells Maintains ICAM-1 Expression in Hepatocellular Carcinoma. Clin Cancer Res. 2016;22:2041-50
11. Zhou X, Cao P, Zhu Y, Lu W, Gu N, Mao C. Phage-mediated counting by the naked eye of miRNA molecules at attomolar concentrations in a Petri dish. Nat Mater. 2015;14:1058-64
12. Wang J, Ye H, Zhang D, Cheng K, Hu Y, Yu X. et al. Cancer-derived circulating microRNAs promote tumor angiogenesis by entering dendritic cells to degrade highly complementary microRNAs. Theranostics. 2017;7:1407-21
13. Taft RJ, Pheasant M, Mattick JS. The relationship between non-protein-coding DNA and eukaryotic complexity. Bioessays. 2007;29:288-99
14. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012; 489: 57-74.
15. Eddy SR. Noncoding RNA genes. Curr Opin Genet Dev. 1999;9:695-9
16. Sethi I, Romano RA, Gluck C, Smalley K, Vojtesek B, Buck MJ. et al. A global analysis of the complex landscape of isoforms and regulatory networks of p63 in human cells and tissues. BMC Genomics. 2015;16:584
17. Saghatelian A, Couso JP. Discovery and characterization of smORF-encoded bioactive polypeptides. Nat Chem Biol. 2015;11:909-16
18. Mumtaz MA, Couso JP. Ribosomal profiling adds new coding sequences to the proteome. Biochem Soc Trans. 2015;43:1271-6
19. Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H. et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775-89
20. Pueyo JI, Magny EG, Couso JP. New Peptides Under the s(ORF)ace of the Genome. Trends Biochem Sci. 2016;41:665-78
21. Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH. et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799-816
22. Bullock JM, Schwab J, Thalassinos K, Topf M. The importance of non-accessible crosslinks and solvent accessible surface distance in modelling proteins with restraints from crosslinking mass spectrometry. Mol Cell Proteomics. 2016;15:2491-500
23. Kim MS, Pinto SM, Getnet D, Nirujogi RS, Manda SS, Chaerkady R. et al. A draft map of the human proteome. Nature. 2014;509:575-81
24. Aspden JL, Eyre-Walker YC, Phillips RJ, Amin U, Mumtaz MA, Brocard M. et al. Extensive translation of small open reading frames revealed by Poly-Ribo-Seq. Elife. 2014;3:e03528
25. Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT. et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484-8
26. Goldman A, Capoano CA, Gonzalez-Lopez E, Geisinger A. Identifier (ID) elements are not preferentially located to brain-specific genes: high ID element representation in other tissue-specific- and housekeeping genes of the rat. Gene. 2014;533:72-7
27. St Laurent G, Wahlestedt C, Kapranov P. The landscape of long noncoding RNA classification. Trends Genet. 2015;31:239-51
28. Brown CJ, Hendrich BD, Rupert JL, Lafreniere RG, Xing Y, Lawrence J. et al. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71:527-42
29. Pauli A, Valen E, Schier AF. Identifying (non-)coding RNAs and small peptides: challenges and opportunities. Bioessays. 2015;37:103-12
30. Anderson DM, Anderson KM, Chang CL, Makarewich CA, Nelson BR, McAnally JR. et al. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell. 2015;160:595-606
31. Magny EG, Pueyo JI, Pearl FM, Cespedes MA, Niven JE, Bishop SA. et al. Conserved regulation of cardiac calcium uptake by peptides encoded in small open reading frames. Science. 2013;341:1116-20
32. Matsumoto A, Pasut A, Matsumoto M, Yamashita R, Fung J, Monteleone E. et al. mTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide. Nature. 2016;541:228-232
33. Plaza S, Menschaert G, Payre F. In search of lost small peptides. Annu Rev Cell Dev Biol. 2017;33:391-416
34. Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D. et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223-7
35. Kapusta A, Kronenberg Z, Lynch VJ, Zhuo X, Ramsay L, Bourque G. et al. Transposable elements are major contributors to the origin, diversification, and regulation of vertebrate long noncoding RNAs. PLoS Genet. 2013;9:e1003470
36. Lipovich L, Johnson R, Lin CY. MacroRNA underdogs in a microRNA world: evolutionary, regulatory, and biomedical significance of mammalian long non-protein-coding RNA. Biochim Biophys Acta. 2010;1799:597-615
37. van Heesch S, van Iterson M, Jacobi J, Boymans S, Essers PB, de Bruijn E. et al. Extensive localization of long noncoding RNAs to the cytosol and mono- and polyribosomal complexes. Genome Biol. 2014;15:R6
38. Chew GL, Pauli A, Rinn JL, Regev A, Schier AF, Valen E. Ribosome profiling reveals resemblance between long non-coding RNAs and 5' leaders of coding RNAs. Development. 2013;140:2828-34
39. Ruiz-Orera J, Messeguer X, Subirana JA, Alba MM. Long non-coding RNAs as a source of new peptides. Elife. 2014;3:e03523
40. Kong L, Zhang Y, Ye ZQ, Liu XQ, Zhao SQ, Wei L. et al. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007;35:W345-9
41. Lin MF, Jungreis I, Kellis M. PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics. 2011;27:i275-82
42. Juntawong P, Girke T, Bazin J, Bailey-Serres J. Translational dynamics revealed by genome-wide profiling of ribosome footprints in Arabidopsis. Proc Natl Acad Sci USA. 2014;111:E203-12
43. Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218-23
44. Kutter C, Watt S, Stefflova K, Wilson MD, Goncalves A, Ponting CP. et al. Rapid turnover of long noncoding RNAs and the evolution of gene expression. PLoS Genet. 2012;8:e1002841
45. Rohrig H, Schmidt J, Miklashevichs E, Schell J, John M. Soybean ENOD40 encodes two peptides that bind to sucrose synthase. Proc Natl Acad Sci USA. 2002;99:1915-20
46. Tupy JL, Bailey AM, Dailey G, Evans-Holm M, Siebel CW, Misra S. et al. Identification of putative noncoding polyadenylated transcripts in Drosophila melanogaster. Proc Natl Acad Sci USA. 2005;102:5495-500
47. Kondo T, Hashimoto Y, Kato K, Inagaki S, Hayashi S, Kageyama Y. Small peptide regulators of actin-based cell morphogenesis encoded by a polycistronic mRNA. Nat Cell Biol. 2007;9:660-5
48. Kondo T, Plaza S, Zanet J, Benrabah E, Valenti P, Hashimoto Y. et al. Small peptides switch the transcriptional activity of Shavenbaby during Drosophila embryogenesis. Science. 2010;329:336-9
49. Hassan AS, Hou J, Wei W, Hoodless PA. Expression of two novel transcripts in the mouse definitive endoderm. Gene Expr Patterns. 2010;10:127-34
50. Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147:1537-50
51. Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G. et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295-300
52. Pauli A, Norris ML, Valen E, Chew GL, Gagnon JA, Zimmerman S. et al. Toddler: an embryonic signal that promotes cell movement via apelin receptors. Science. 2014;343:1248636
53. Lagier-Tourenne C, Baughn M, Rigo F, Sun S, Liu P, Li HR. et al. Targeted degradation of sense and antisense C9orf72 RNA foci as therapy for ALS and frontotemporal degeneration. Proc Natl Acad Sci USA. 2013;110:E4530-9
54. Nelson BR, Makarewich CA, Anderson DM, Winders BR, Troupes CD, Wu F. et al. A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science. 2016;351:271-5
55. Hangauer MJ, Vaughn IW, McManus MT. Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS Genet. 2013;9:e1003569
56. Kontomanolis EN, Koukourakis MI. MicroRNA: the potential regulator of endometrial carcinogenesis. Microrna. 2015;4:18-25
57. Gurtan AM, Sharp PA. The role of miRNAs in regulating gene expression networks. J Mol Biol. 2013;425:3582-600
58. Lauressergues D, Couzigou JM, Clemente HS, Martinez Y, Dunand C, Becard G. et al. Primary transcripts of microRNAs encode regulatory peptides. Nature. 2015;520:90-3
59. Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228-34
60. Nigro JM, Cho KR, Fearon ER, Kern SE, Ruppert JM, Oliner JD. et al. Scrambled exons. Cell. 1991;64:607-13
61. Hsiao KY, Lin YC, Gupta SK, Chang N, Yen L, Sun HS. et al. Noncoding effects of circular RNA CCDC66 promote colon cancer growth and metastasis. Cancer Res. 2017;77:2339-50
62. Fu L, Wu S, Yao T, Chen Q, Xie Y, Ying S. et al. Decreased expression of hsa_circ_0003570 in hepatocellular carcinoma and its clinical significance. J Clin Lab Anal. 2018;32:DOI 10.1002/jcla.22239
63. Kristensen LS, Hansen TB, Veno MT, Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37:555-565
64. Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J. et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981-4
65. Kim KM, Abdelmohsen K, Mustapic M, Kapogiannis D, Gorospe M. RNA in extracellular vesicles. Wiley Interdiscip Rev RNA. 2017;8:DOI 10.1002/wrna.1413
66. Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15:409
67. Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L. et al. Translation of circRNAs. Mol Cell. 2017;66:9-21.e7
68. Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y. et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017;27:626-41
69. Meng X, Chen Q, Zhang P, Chen M. CircPro: an integrated tool for the identification of circRNAs with protein-coding potential. Bioinformatics. 2017;33:3314-6
70. Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O. et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell. 2017;66:22-37.e9
71. Pei B, Sisu C, Frankish A, Howald C, Habegger L, Mu XJ. et al. The GENCODE pseudogene resource. Genome Biol. 2012;13:R51
72. Poliseno L. Pseudogenes: newly discovered players in human cancer. Sci Signal. 2012;5:re5
73. Karreth FA, Reschke M, Ruocco A, Ng C, Chapuy B, Leopold V. et al. The BRAF pseudogene functions as a competitive endogenous RNA and induces lymphoma in vivo. Cell. 2015;161:319-32
74. Booth HA, Holland PW. Eleven daughters of NANOG. Genomics. 2004;84:229-38
75. Jeter CR, Badeaux M, Choy G, Chandra D, Patrawala L, Liu C. et al. Functional evidence that the self-renewal gene NANOG regulates human tumor development. Stem Cells. 2009;27:993-1005
76. Betran E, Wang W, Jin L, Long M. Evolution of the phosphoglycerate mutase processed gene in human and chimpanzee revealing the origin of a new primate gene. Mol Biol Evol. 2002;19:654-63
77. Prabakaran S, Hemberg M, Chauhan R, Winter D, Tweedie-Cullen RY, Dittrich C. et al. Quantitative profiling of peptides from RNAs classified as noncoding. Nat Commun. 2014;5:5429
78. Bendz M, Skwark M, Nilsson D, Granholm V, Cristobal S, Kall L. et al. Membrane protein shaving with thermolysin can be used to evaluate topology predictors. Proteomics. 2013;13:1467-80
79. Gemayel R, Vinces MD, Legendre M, Verstrepen KJ. Variable tandem repeats accelerate evolution of coding and regulatory sequences. Annu Rev Genet. 2010;44:445-77
80. Jovicic A, Mertens J, Boeynaems S, Bogaert E, Chai N, Yamada SB. et al. Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nat Neurosci. 2015;18:1226-9
81. DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ. et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245-56
82. Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR. et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257-68
83. van der Zee J, Gijselinck I, Dillen L, Van Langenhove T, Theuns J, Engelborghs S. et al. A pan-European study of the C9orf72 repeat associated with FTLD: geographic prevalence, genomic instability, and intermediate repeats. Hum Mutat. 2013;34:363-73
84. Chew J, Gendron TF, Prudencio M, Sasaguri H, Zhang YJ, Castanedes-Casey M. et al. Neurodegeneration. C9ORF72 repeat expansions in mice cause TDP-43 pathology, neuronal loss, and behavioral deficits. Science. 2015;348:1151-4
85. Mori K, Weng SM, Arzberger T, May S, Rentzsch K, Kremmer E. et al. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013;339:1335-8
86. Andrews SJ, Rothnagel JA. Emerging evidence for functional peptides encoded by short open reading frames. Nat Rev Genet. 2014;15:193-204
87. Johnstone TG, Bazzini AA, Giraldez AJ. Upstream ORFs are prevalent translational repressors in vertebrates. Embo J. 2016;35:706-23
88. Jousse C, Bruhat A, Carraro V, Urano F, Ferrara M, Ron D. et al. Inhibition of CHOP translation by a peptide encoded by an open reading frame localized in the chop 5'UTR. Nucleic Acids Res. 2001;29:4341-51
89. Starck SR, Tsai JC, Chen K, Shodiya M, Wang L, Yahiro K. et al. Translation from the 5' untranslated region shapes the integrated stress response. Science. 2016;351:aad3867
90. Pegueroles C, Gabaldon T. Secondary structure impacts patterns of selection in human lncRNAs. BMC Biol. 2016;14:60
91. Gabellini D. Noncoding RNA interplay with the genome. Methods Mol Biol. 2016;1480:69-72
92. Neme R, Tautz D. Evolution: dynamics of de novo gene emergence. Curr Biol. 2014;24:R238-40
93. Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789-802
94. Min KW, Davila S, Zealy RW, Lloyd LT, Lee IY, Lee R. et al. eIF4E phosphorylation by MST1 reduces translation of a subset of mRNAs, but increases lncRNA translation. Biochim Biophys Acta. 2017;1860:761-72
95. Slavoff SA, Mitchell AJ, Schwaid AG, Cabili MN, Ma J, Levin JZ. et al. Peptidomic discovery of short open reading frame-encoded peptides in human cells. Nat Chem Biol. 2013;9:59-64
96. Zu T, Gibbens B, Doty NS, Gomes-Pereira M, Huguet A, Stone MD. et al. Non-ATG-initiated translation directed by microsatellite expansions. Proc Natl Acad Sci USA. 2011;108:260-5
97. Endoh T, Hnedzko D, Rozners E, Sugimoto N. Nucleobase-modified PNA suppresses translation by forming a triple helix with a hairpin structure in mRNA in vitro and in cells. Angew Chem Int Ed Engl. 2016;55:899-903
98. Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113-27
99. Kall L, Canterbury JD, Weston J, Noble WS, MacCoss MJ. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat Methods. 2007;4:923-5
100. Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283-92
101. Ivanov IP, Firth AE, Michel AM, Atkins JF, Baranov PV. Identification of evolutionarily conserved non-AUG-initiated N-terminal extensions in human coding sequences. Nucleic Acids Res. 2011;39:4220-34
102. Touriol C, Bornes S, Bonnal S, Audigier S, Prats H, Prats AC. et al. Generation of protein isoform diversity by alternative initiation of translation at non-AUG codons. Biol Cell. 2003;95:169-78
103. Peabody DS. Translation initiation at non-AUG triplets in mammalian cells. J Biol Chem. 1989;264:5031-5
104. Green KM, Linsalata AE, Todd PK. RAN translation-What makes it run? Brain Res. 2016;1647:30-42
105. Moseley ML, Zu T, Ikeda Y, Gao W, Mosemiller AK, Daughters RS. et al. Bidirectional expression of CUG and CAG expansion transcripts and intranuclear polyglutamine inclusions in spinocerebellar ataxia type 8. Nat Genet. 2006;38:758-69
106. Pearson CE. Repeat associated non-ATG translation initiation: one DNA, two transcripts, seven reading frames, potentially nine toxic entities!. PLoS Genet. 2011;7:e1002018
107. Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, Qian SB. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature. 2015;526:591-4
108. Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O. et al. 5' UTR m(6)A promotes cap-independent translation. Cell. 2015;163:999-1010
109. Venters BJ, Pugh BF. Genomic organization of human transcription initiation complexes. Nature. 2013;502:53-8
110. Tisseur M, Kwapisz M, Morillon A. Pervasive transcription - Lessons from yeast. Biochimie. 2011;93:1889-96
111. White MA, Myers CA, Corbo JC, Cohen BA. Massively parallel in vivo enhancer assay reveals that highly local features determine the cis-regulatory function of ChIP-seq peaks. Proc Natl Acad Sci USA. 2013;110:11952-7
112. Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339-46
113. Ingolia NT, Brar GA, Stern-Ginossar N, Harris MS, Talhouarne GJ, Jackson SE. et al. Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell Rep. 2014;8:1365-79
114. Chi EY, Krishnan S, Randolph TW, Carpenter JF. Physical stability of proteins in aqueous solution: mechanism and driving forces in nonnative protein aggregation. Pharm Res. 2003;20:1325-36
115. Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546-58
116. Laumont CM, Daouda T, Laverdure JP, Bonneil E, Caron-Lizotte O, Hardy MP. et al. Global proteogenomic analysis of human MHC class I-associated peptides derived from non-canonical reading frames. Nat Commun. 2016;7:10238
117. Huang JZ, Chen M, Chen, Gao XC, Zhu S, Huang H. et al. A peptide encoded by a putative lncRNA HOXB-AS3 suppresses colon cancer growth. Mol Cell. 2017;68:171-84.e6
118. Pueyo JI, Couso JP. The 11-aminoacid long Tarsal-less peptides trigger a cell signal in Drosophila leg development. Dev Biol. 2008;324:192-201
119. Ling F, Kang B, Sun XH. Id proteins: small molecules, mighty regulators. Curr Top Dev Biol. 2014;110:189-216
120. Wethmar K, Barbosa-Silva A, Andrade-Navarro MA, Leutz A. uORFdb—a comprehensive literature database on eukaryotic uORF biology. Nucleic Acids Res. 2014;42:D60-7
Author contact
![]() Corresponding authors: Professor Shanrong Liu, E-mail: liushanrongcom and Professor Chuanbin Mao, Email: cbmaoedu
Corresponding authors: Professor Shanrong Liu, E-mail: liushanrongcom and Professor Chuanbin Mao, Email: cbmaoedu
 Global reach, higher impact
Global reach, higher impact