13.3
Impact Factor
Theranostics 2018; 8(9):2424-2434. doi:10.7150/thno.23852 This issue Cite
Research Paper
High-Discrimination Factor Nanosensor Based on Tetrahedral DNA Nanostructures and Gold Nanoparticles for Detection of MiRNA-21 in Live Cells
1. Key Laboratory of Laboratory Medical Diagnostics, Ministry of Education, Chongqing Medical University, Chongqing 400016, P R China
2. Department of pharmacy, University-Town Hospital of Chongqing Medical University, Chongqing 401331, P R China
3. Clinical Laboratory of Traditional Chinese Medicine Hospital Affiliated to Southwest Medical University, Luzhou 646000, P R China
4. Key Laboratory for Biorheological Science and Technology of Ministry of Education, Bioengineering College of Chongqing University, Chongqing 400030, P R China
Received 2017-11-14; Accepted 2018-1-25; Published 2018-3-27
Abstract
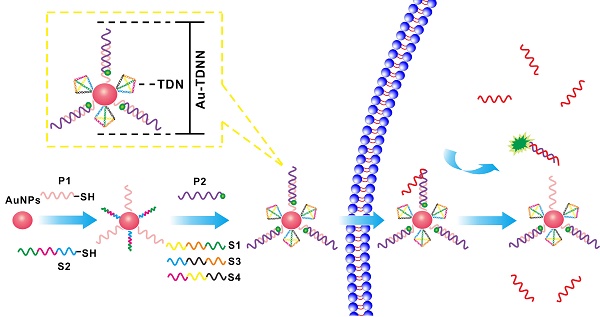
While detection of microRNA with or without signal amplification is highly informative, nanosensors with high specificity for cell-specific RNA detection are rare.
Methods: In this study, a tetrahedral DNA nanostructure (TDN) with a specific function was combined with gold nanoparticles (Au-NP) possessing fluorescence quenching effects and a large surface area to fabricate a fluorescence resonance energy transfer based nanosensor (Au-TDNN). The presence of miR-21 (target) can separate the fluorescent dye-labeled detection probe on Au-TDNNs from Au-NPs, which separates the donor and acceptor, thus inducing an intensive fluorescence signal. High specificity for discerning point mutation targets was achieved by rationally designing the nucleic acid strand displacement reaction to occur spontaneously with ΔG0 ≈ 0 based on thermodynamic parameters; under this condition, slight thermodynamic changes caused by base mismatch exert significant effects on hybridization yield.
Results: Chemically synthesized DNA of three single-base-changed analogues of target, let-7d, and miR-200b were tested. A discrimination factor (DF) of 15.4 was produced by the expected detection probe on Au-NPs for proximal single-base mismatch. As the control group, the DF produced by an ordinary detection probe on Au-NPs only reached 2.4. The feasibility of the proposed strategy was also confirmed using hepatocyte cancer cells (HepG2).
Conclusion: This improved nanosensor opens a new avenue for the specific and easy detection of microRNA in live cells.
Keywords: miR-21, tetrahedral nanostructure, gold nanoparticle, fluorescence resonance energy transfer, strand displacement reaction
Introduction
MicroRNAs (miRNAs) comprise a group of short non-coding RNAs that mediate post-transcriptional gene silencing. Since the first miRNA was reported in 1993 [1], considerable interest has focused on the development of miRNAs as biomarkers for diverse molecular diagnostic applications, including in numerous solid tumors [2, 3], cardiovascular and autoimmune diseases [4], and forensics [5]. miRNAs can be detected in diverse specimen types including plasma, urine, and formalin-fixed tissue blocks, while detection of miRNAs in live cells serves as the foundational tenet in understanding related applications in various aspects of cell biology, disease pathophysiology, drug discovery, medical diagnostics, and therapeutics [7, 8].
Fluorescent quantitative polymerase chain reaction, microarray hybridization, and two-stage isothermal amplification assay [9-12] are techniques commonly used for miRNA detection; however, these methods require killing of cells or extraction of miRNAs from cells before measurement and are unable to capture miRNAs in real time, in their actual locality, and with high precision. Recently, several intracellular nanoprobes for sensing RNA have been reported; these nanoprobes include fluorescence resonance energy transfer (FRET)-based nanoflares and nanobeacons, self-assembled gold nanoparticles (Au-NPs) and upconversion NPs [13-15], nature-inspired nanosnail nucleic acid sensor [8], electrostatic DNA nano-assembly-based hybridization chain reaction, and core-shell nanoprobes [16-18]. However, these probes have only focused on imaging or quantifiable detection of RNA in live cells and paid little attention to the specificity of the nanosensors [19-21]. DNA hybridization loosely follows the Watson-Crick pairing rules [22]. DNAs or RNAs feature the potential to pair with detection probes having one or several mismatches. False-positive signals can also be amplified in various signal amplification methods. Although hybridization near the melting temperature or chemical denaturation [23, 24] are often used to improve specificity of nucleic acid hybridization, this procedure manifests fluctuating feasibility, especially for reactions in live cells.
Recently, rationally designed “toehold exchange” probes have been known as mismatch-sensitive recognition probes [25, 26]. Double-stranded probes were designed to hybridize spontaneously with a single-stranded nucleic acid that proceeds approximately in thermodynamic equilibrium (ΔG0 ≈ 0), at which small thermodynamic changes caused by base mismatch exert significant effect on hybridization yield [27, 28]. Based on this promising property, the thermodynamic-based double-stranded probes can sense nucleic acids with high specificity across diverse conditions (such as temperature or salinity). However, when applied for detection of intracellular miRNA, its usage is still hindered by some limitations, such as the inability to pass through the cell membrane [29] and degradation by intracellular enzymes, which can lead to false positive signals [30, 31].
Nucleic acid-modified Au-NPs have been widely used in diagnosis of diseases [32-35]. In this study, DNA tetrahedral nanostructures (TDNs), mismatch-sensitive recognition probes, and Au-NPs (20 ± 3 nm)-based nanosensors (Au-TDNNs) were constructed for intracellular miRNA detection. TDNs were selected as sensor carriers because of their comparable cellular uptake efficiency with lipid-based transfection reagents [36-38]. Fluorescent dye-labeled double-stranded probes were elaborately designed based on thermodynamics to achieve specific and precise measurement of miR-21 in live cells. Au-NPs were selected as fluorescence acceptors owing to their excellent ability for fluorescence quenching and large surface area for the immobilization of double-stranded probes and TDNs. The ratio of detection probes and TDNs was estimated at 27:1. Chemically synthesized DNA of miR-21 (target) was used to test the in vitro performance of the nanosensors. These well-prepared nanosensors were proved to possess a high-discrimination factor (DF) in detection of single-base mismatch targets. The nanosensors possessed an ability for miR-21 detection in cell extracts of MCF-7 cell lines. In addition, the phosphorothioate-modified nanosensors displayed good stability against enzymatic degradation, and had low false-positive signals. When MCF-7 cell lines were used as in vitro models, the nanosensors displayed an excellent ability to transfect cells. Further, the feasibility of the proposed strategy was also confirmed using hepatocyte cancer cells (HepG2). Thus, we achieved accurate detection of miR-21 in live cells.
Materials and methods
Materials
Chloroauric acid (HAuCl4·3H2O) and tri (2-carboxyethyl) phosphine hydrochloride (TCEP) were obtained from Aladdin (Shanghai, China). Human DNase I and exonuclease III originated from Sangon Biotechnology Co., Ltd (Shanghai, China). All other reagents were of analytical reagent grade and used without further purification. Ultra-pure water (Milli-Q 18.2 MΩ, Millipore System Inc.) was used for all experiments. All oligonucleotide sequences were synthesized and high-performance liquid chromatography-purified by Sangon Biotechnology Co., Ltd (Shanghai, China) and used without further purification.
Preparation of Au-TDNNs
A two-step synthesis method was employed to synthesize Au-TDNNs according to a previously reported method with minor modification [39]. Briefly, 10 µM thiolated ExP1 and 10 µM thiolated S2 were mixed in a NaOH-treated centrifuge tube at a ratio of 24:1, with a total volume of 100 µL. Then, the mixture was activated by 10 µL of 10 mM TCEP at room temperature for 1 h. Afterward, 1 mL of Au-NPs (the synthesis procedure of Au-NPs is supplied in supplementary material) were added and incubated at 4 °C for 12 h. After incubation, 20 µL of 1% sodium dodecyl sulfate was added to stabilize Au-NPs with shaking at room temperature for 1 h. Next, the mixture was aged for another 12 h by slowly adding 100 μL of 0.5 M NaCl solution. The solution was then centrifuged at 16,500 rcf for 20 min. The wine-red Au-NPs@ExP1S2 precipitate was washed thrice by centrifugation with 0.01 M phosphate buffer solution (PBS, pH 7.4; Figure S3). Afterward, 10 µM fluorescent dye-labeled detection probes P2, S1, S3, and S4 were added to the Au-NPs@ExP1S2 with a ratio of 27:1:1:1 and incubated at 37 °C for at least 2 h. To remove excess reagents, the wine-red Au-TDNNs precipitate was washed thrice by centrifugation with PBS and then resuspended in PBS for future use.
Characterization of Au-TDNNs
Au-NPs, TDNs, Au-TDNs, and Au-TDNNs were characterized using transmission electron microscopy (TEM), high-resolution TEM, polyacrylamide gel electrophoresis (PAGE), dynamic light scattering (DLS), and zeta potential analysis. PAGE gel analysis for TDNs was conducted using a 12% nondenaturing polyacrylamide gel in tris-borate-ethylenediaminetetraacetic acid buffer. The gel was run at a constant voltage of 120 V for 1 h. DLS and zeta potential analysis were employed to monitor the hydrodynamic size and potential of step-by-step assembly of the Au-NPs. Briefly, 100 µL (for zeta potential analysis) and 3 mL (for hydrodynamic size analysis) of proper concentrations of newly prepared samples were determined using a zeta potential analyzer (Malvern, Zeta, Worcestershire, UK).
Response of Au-TDNNs to target in vitro
The detection ability of Au-TDNNs for the target was determined using an Agilent fluorescence analyzer. FAM fluorescence signal was determined at excitation and emission wavelengths of 480 nm and 520 nm, respectively. For fluorescence studies, the concentration of Au-TDNNs used measured 2 nM in PBS (pH 7.4). For feasibility analysis of the Au-TDNNs, 50 µL target and miR-200b were added to 150 µL Au-TDNNs solution at a final DNA concentration of 50 nM.
To determine the sensitivity of the nanosensors in vitro, Au-TDNNs in PBS were spiked with different concentrations of target (0, 0.5, 1, 2.5, 10, 25, 40, 50, 80, 100, and 250 nM). After incubation for 30 min at 37 °C, fluorescence signals were monitored.
Enzymatic resistance of Au-TDNNs
Digestion of the two groups of phosphorothioate-protected Au-TDNNs was probed using the Agilent Cary Eclipse to detect generation of nonspecific FAM fluorescence signals. For both human DNase I (Thermo Scientific) and exonuclease III (New England Biolabs) digestion, a common concentration of 3 U/mL was used to digest 2 nM Au-TDNNs samples in PBS. FAM fluorescence signals were monitored and recorded over a period of 3 h at 30 min intervals and at 37 °C.
Cell viability assay
MCF-7 cells were cultured at an initial seeding density of 50,000 cells/cm2 in 96-well plates overnight in Dulbecco's modified Eagle's media (DMEM; Gibco) supplemented with 10% fetal bovine serum (FBS, Thermo Scientific) and 1% penicillin and streptomycin solution (PAA Laboratories Inc.). Thereafter, MCF-7 cells were treated with varying concentrations of Au-TDNNs. After incubation for 24 h, the culture medium was replaced and the cells were treated with Cell Counting Kit-8 for 4 h. Absorption was measured on a microplate reader at 450 nm.
Cellular uptake and cell imaging of Au-TDNNs
The cells were cultured overnight at an initial seeding density of 50,000 cells/cm2 in DMEM (Gibco) supplemented with 10% FBS (Thermo Scientific) and 1% penicillin and streptomycin solution (PAA Laboratories Inc., USA) in confocal dishes, and cells were cultured under standard culture conditions. After incubation with 10 nM of Au-TDNNs, samples were subsequently retrieved and washed thrice with PBS, and fluorescence signals were measured with a Nikon A1*R Laser Scanning Confocal Microscope (Japan) with excitation at 480 nm and emission at 520 nm.
Statistical analysis
A Student's t test was used to analyze differences between groups. Differences were considered statistically significant with values of P < 0.001.
Results and Discussion
Principle of Au-TDNNs
Au-TDNN consists of three main components: (ⅰ) a double-stranded probe, (ⅱ) Au-NPs, and (ⅲ) TDNs (Scheme 1). The double-stranded probe recognition structure is based on a fluorophore-quencher pair. Expected detection probe1 (ExP1) is immobilized on Au-NPs through a Au-S bond, whereas the 3′FAM on expected detection probe2 (ExP2) lies close to Au-NPs by nucleic acid hybridization. The nanosensor was assembled as follows. First, thiolated ExP1 and thiolated S2 were anchored on the Au-NP surface with a Au-S bond. FAM-ExP2 and the remaining three single-stranded nucleic acids (S1, S3, and S4) were then hybridized with the nucleic acid-assembled Au-NPs. The target can hybridize with FAM-labeled ExP2, which separates FAM from Au-NPs to generate an intensive fluorescence signal. In the absence of a target, fluorescence signal was quenched through high FRET efficiency between FAM and Au-NPs.
Working principle of Au-TDNNs for miR-21 detection and the structure of Au-TDNNs.
Rational design of double-stranded detection probes
During design of the detection probes, miR-21 was selected as the target owing to its overexpression levels and biological significance [15, 40]. Exp was designed to hybridize spontaneously with the target at approximately thermodynamic equilibrium (ΔG0 ≈ 0) [28]. As a contrast probe to ExP, the ordinary detection probe (OrP) was designed to maintain the Gibbs free energy change ΔG0 of the OrP2 hybridized with target less than that hybridized with OrP1 (Table S1). Gibbs free energy (∆G0) of the reaction was calculated by ∆G0 = -RTln(Keq), where T refers to the Kelvin temperature, R is the gas constant, and Keq is the equilibrium constant. The rate constant of the toehold exchange reaction was 1×106 M-1s-1 in the presence of a 7 nt toehold [41]. Table S1 summarizes the observed thermodynamic parameters. Figure S1 shows the working principle of ExP and OrP for the single-base mutation target and wild-type target during detection. The oligonucleotide sequences of TDN were newly designed to avoid nonspecific hybridization with detection probes and all the oligonucleotide sequences are provided in Supplementary Material.
Specificity evaluation of the double-stranded detection probe
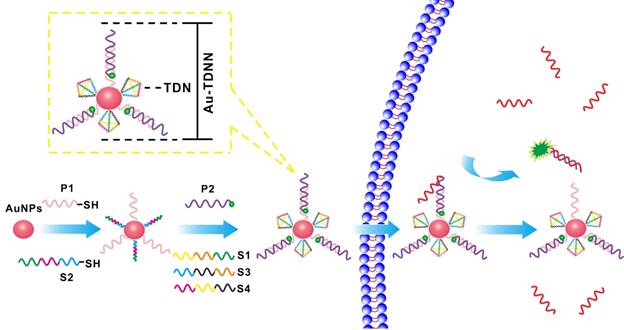
To illustrate the specificity of ExP and OrP, we tested the proximal, middle, and distal mismatch single-base mutation target and wild-type target (Figure S1). Figures 1A and 1B show the kinetic curves of the strand displacement reaction on ExP and OrP, respectively. We observed that the rates of strand displacement reaction of OrP and ExP were nearly the same when detecting target, although their double-chain products feature different complementary base numbers. We attributed this result to the two types of detection probes, which exhibited the same green toehold base number. When background signal was eliminated, ExP became more sensitive to single-base mutation target and wild-type target (Figure 1C). Specificity of the two types of detection probe was evaluated by calculating DF (DF = ∆Ftarget/∆Ftarget analogue), as shown in Figure 1D. ExP exhibited better single-base mutation identification ability than OrP regardless of whether the mutation base existed in the middle, distal, or proximal site of the target. Proximal mismatch DF of ExP reached 13.6, which was higher than that of OrP (P < 0.001).
Structural characterization of Au-TDNNs
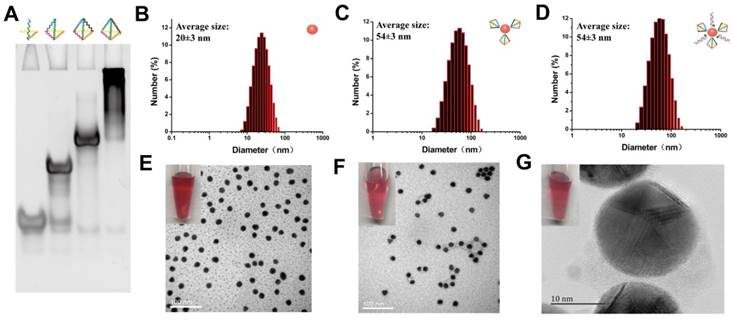
Formation of TDNs was validated by PAGE (Figure 2A). TEM was used to characterize the morphology of Au-NPs (Figure 2E) and Au-TDNNs (Figure 2F). TEM images indicated that Au-NPs were spherical and homogeneously-dispersed. After conjugating TDNs onto the Au-NP surface, a thin and noncrystalline layer could be observed in the high-resolution TEM image (Figure 2G). Immobilization of nucleic acids on the Au-NP surface was validated by UV-vis absorption spectroscopy and zeta potential analysis. UV-vis absorption spectra showed that the plasmon absorption peak of pristine Au-NPs red shifted by approximately 2 nm upon functionalization with nucleic acids (Figure 1E). Zeta potential analysis revealed that the surface zeta potential varied from 0.04 mV for pristine Au-NPs to -40.4 mV for Au-TDNNs (Figure 1F). DLS measurement revealed mean hydrodynamic diameters of 20 nm for Au-NPs (Figure 2B) and 54 nm for Au-TDNNs and Au@TDNs (Figure 2C and 2D, respectively). Au@TDNs structures were assumed spherical. The mean hydrodynamic diameter of TDNs can be calculated by DTDN = (DAu@TDNs - DAu-NPs)/2, where DTDN represents the mean hydrodynamic diameter of TDNs, DAu@TDNs corresponds to the mean hydrodynamic diameter of Au@TDNs, and DAu-NPs denotes the mean hydrodynamic diameter of Au-NPs. Mean hydrodynamic diameter of TDNs was calculated to be 17 nm. By comparing the hydrodynamic diameters of Au@TDNs and Au-TDNNs, we can infer that hydrodynamic diameter of the nanoprobes measured less than that of TDNs. These results indicated the successful assembly of nucleic acids on the Au-NP surface. Stability of Au-TDNNs was also tested in both PBS and culture medium (Figure 1E). The stability experiment used saline-sodium citrate (SSC) hybridization solution. The fabricated Au-TDNNs can maintain their stability in SSC hybridization solution, PBS, and DMEM (10% FBS) after incubation for 12 h in the dark at room temperature. The small red shifts of the plasmon absorption peak of Au-TDNNs in DMEM may be induced by other substances in DMEM (10% FBS).
(A, B) Fluorescence kinetic curves of mutation base in the middle, distal or proximal site of the target and target detection by ExP and OrP, respectively. (C) Relative fluorescence intensity of the ExP and OrP in detecting single-base mutation target and wild-type target. (D) Corresponding discrimination factors (DFs) of ExP and OrP. (E) UV-vis characterization of the stability of Au-TDNNs in SSC, PBS and DMEM (10% FBS). (F) Zeta potential analysis of the step-by-step assembly of the Au-TDNNs. RFI: relative fluorescence intensity. DF = ∆Ftarget/∆Ftarget analogue (∆F = F-F0, F0: background signal.). *P < 0.001.
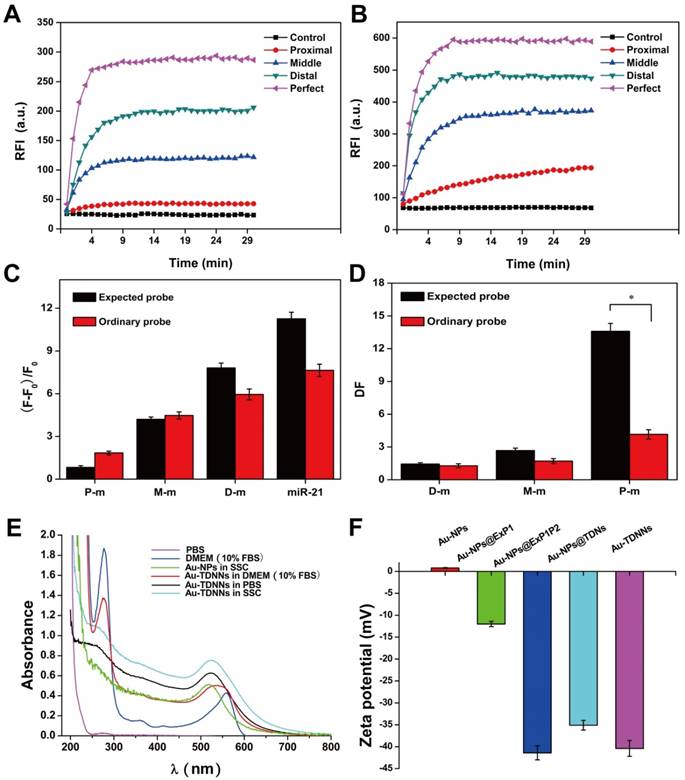
Structural characterization of Au-TDNNs. (A) PAGE analysis shows reduced electrophoretic mobility with the step-by-step assembly of TDNs. (B-D) DLS measurement reveals mean hydrodynamic diameters of 20 ± 3 nm for Au-NPs, and 54 ± 3 nm for Au@TDNs and Au-TDNNs, respectively. (E, F) TEM of Au-NPs and Au-TDNNs. Scale bars = 100 nm. (G) High-resolution TEM of Au@TDNs. Scale bar = 10 nm. Insets in (E-G) are the corresponding photos taken by a cell phone.
Feasibility analysis of the Au-TDNNs
Dabcyl-labeled ExP1 and FAM-labeled ExP2 were used to verify the feasibility of the toehold-mediated strand displacement reaction. As shown in Figure S2, after Dabcyl-labeled ExP1 solution was added to FAM-labeled ExP2 solution, fluorescence signal gradually decreased, indicating that the reaction of step (1) had been accomplished. When target was added to the solution, fluorescence signal gradually recovered, suggesting that the reaction of step (2) had been accomplished. Afterward, Dabcyl-labeled ExP1 was replaced with Au-NPs@ExP1, as shown by the inset in Figure 3A. A growing fluorescence signal could be observed after the target was hybridized with the as-prepared double-stranded detection probe. Extremely weak fluorescence signals were observed in detection of buffers, Au-NPs, probes, target and miR-200b, and two or three combinations of these components. On the other hand, fluorescence signal was significantly high (P < 0.001) in the presence of Au-TDNNs or Au-NPs@ExP1P2 with target (Figure 3A). These findings imply that TDNs caused no significant interference with the assembly of detection probe on Au-NPs. These results also demonstrate that the fabricated Au-TDNNs can be used for target detection.
Analytical performance evaluation of the established nanosensors
The results in Figure 3B reveal the excellent FRET signal changes for different concentrations of target. These changes suggest that fluorescence intensity depended on the concentration of target. To realize resource conservation and environmental protection, the used Au-TDNNs were recycled for detecting target. Au-TDNNs showed good repeatability through a six-repeatability experiment (Figure S4). We also calculated the mean number of FAM-ExP2 on each Au-NP. Approximately 84 FAM-ExP2 were combined on each Au-NP according to the final concentration of Au-NPs and the calibration curve of FAM-ExP2 (Figure S5). Au-NP concentration was calculated by using a previously reported method [42].
The ability of Au-NPs@ExP1P2 to distinguish point mutations of the target and target analogs was evaluated by calculating the DF. DNA strands including 18 deletion, 18 insertion, and 18 mismatch of the target, miR-200b, let-7d, and target were tested by Au-NPs@ExP1P2 and the control group (Au-NPs@OrP1P2). Figure 3C displays the corresponding DFs observed. The calculated DF of Au-NPs@ExP1P2 ranged from 10.1 to 19.6, with a median of 15.1. The median DF produced by Au-NPs@OrP1P2 totaled 6.9. The calculated DFs of Au-NPs@ExP1P2 for the single-base mismatch target was 15.4, whereas that of Au-NPs@OrP1P2 only reached 2.4, suggesting that Au-NPs@ExP1P2 possessed an excellent ability for distinguish point mutations. Although various methods, such as hybridization near the melting temperature, chemical denaturation, and specific recognition by DNAzyme, have been developed for recognizing single-base mutations, none of these methods is applicable for specific intracellular sensing. By contrast, the proposed ExP, owing to its natural characteristics, offers an unprecedented potential for high-specificity intracellular sensing at room temperature without the aid of external conditions. Thus, in the experiment, we assembled ExP on Au-NPs to fabricate Au-TDNNs.
(A) Feasibility of Au-TDNNs was validated by measuring the fluorescence enhancement on hybridizing with target, miR-200b and combination of different components. (B) Relative fluorescence intensity of Au-TDNNs after hybridization with different concentrations of target. Target concentrations were from 1 nM to 100 nM and the fluorescence was collected at 520 nm. (C) The ability of Au-NPs@ExP1P2 and Au-NPs@OrP1P2 to distinguish point mutation target and target analogs were evaluated by DF. (D) MiR-21 detection in MCF-7 cell-derived total RNA samples. *P < 0.001.
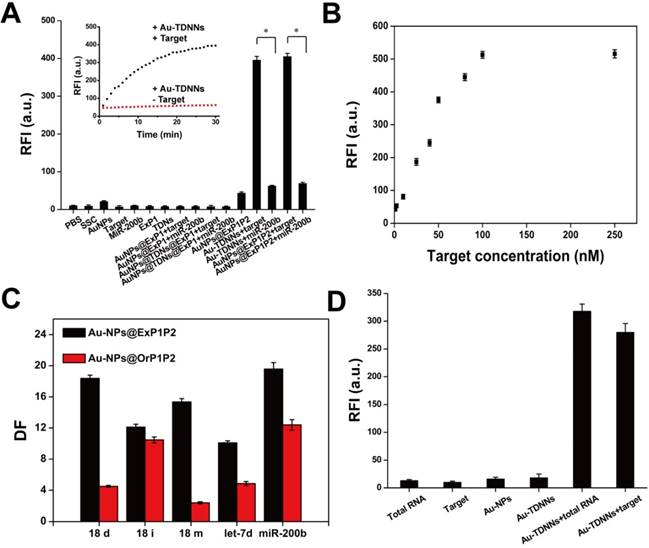
Detection of miR-21 in total RNA extraction solution of MCF-7 cells
Au-TDNNs were used to measure miR-21 in 1.45 ng/mL of total RNA extraction solution of MCF-7 cells (Figure 3D). Negligible fluorescence was detected in the presence of total RNA extraction solution, double-stranded detection probe, and Au-TDNNs alone. By contrast, the Au-TDNNs generated a significantly increased fluorescence signal when incubated with 1 μL of total RNA extraction solution, suggesting that Au-TDNNs can detect miR-21 without any reverse transcription reaction, a condition that bears importance for any real-time intracellular miRNA detection.
Au-TDNNs displayed negligible cytotoxicity and exerted almost no effect on cell viability despite increasing the dose by up to six times (Figure S6). These results provide a solid foundation for real-time miR-21 detection in live cells.
Enzymatic resistance of phosphorothioate-modified Au-TDNNs
To avoid false-positive signals in live cell sensing, probes should be insensitive to both extracellular and intracellular degradation. Two phosphorothioate-modified Au-TDNNs were treated with 3 U/mL of DNase I and Exo III. One was modified with phosphorothioate in the overall detection probe skeleton (overall modified Au-TDNNs), and the other was modified with phosphorothioate in the bare terminal base of the detection probe (terminal modified Au-TDNNs). Figures 4A and 4D show the fluorescence time curves depicting the detection probe degradation induced by DNase I and Exo III. The terminal modified Au-TDNNs were degraded by DNase I, as exemplified by the gradual increase in FAM signal with increasing incubation time (Figure 4A, red curve). Fluorescence signal plateaued at 150 min, corresponding to an approximately eightfold increase. By contrast, DNase I treatment caused no significant fluorescence enhancement in the overall modified Au-TDNNs (Figure 4B, red curve), suggesting that the overall-modified Au-TDNNs were insensitive to DNase I and can prevent the nonspecific signals produced by intracellular nuclease activity.
Enzymatic resistance of phosphorothioate-modified Au-TDNNs. (A, B) Fluorescence time graph depicting terminal-modified Au-TDNN and overall-modified Au-TDNN degradation by DNase I. (C, D) Fluorescence time graph depicting terminal-modified Au-TDNN and overall-modified Au-TDNN degradation by Exo III.
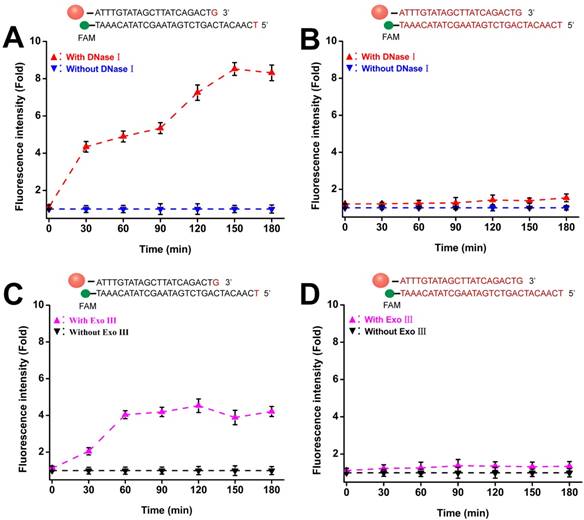
When the two groups were treated with Exo III, the terminal modified Au-TDNNs developed susceptibility to Exo III attack (Figure 4C, pink curve). However, the overall modified Au-TDNNs (Figure 4D, pink curve) were only slightly susceptible to Exo III attack. Thus, we used the overall modified Au-TDNNs to detect miR-21 in live cells.
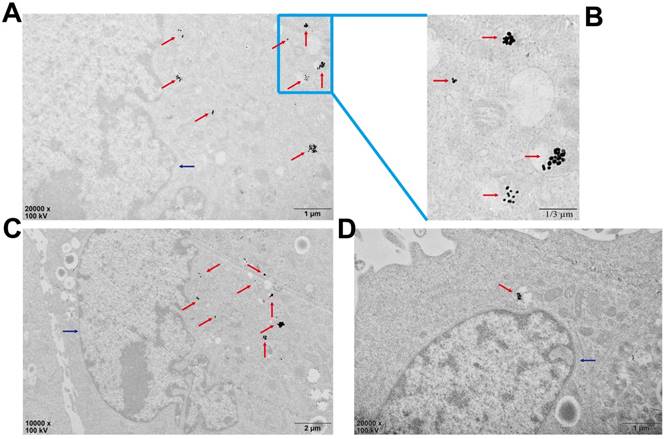
Cellular uptake of Au-TDNNs and live cell miR-21 sensing
Biological TEM was used to verify cellular uptake of Au-TDNNs (Figure 5). As shown in Figure 5A and 5B, Au-TDNNs displayed good dispersibility in MCF-7 cells. Figure 5C and 5D show that Au-TDNNs were mainly dispersed in the cytoplasm.
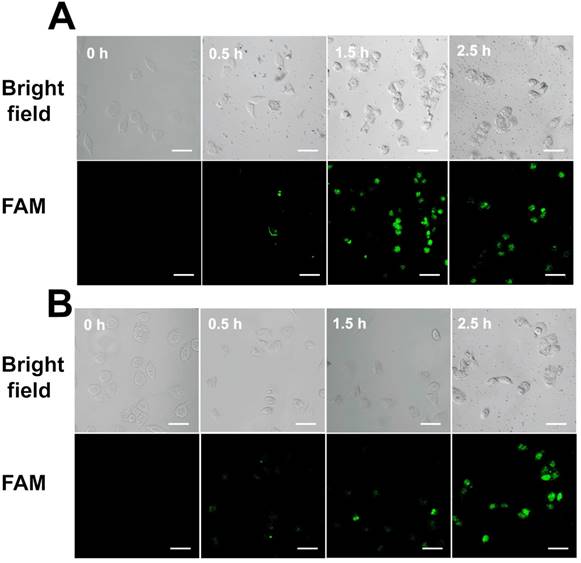
We also monitored the uptake efficiency and dynamics of Au-TDNNs in live cells. TDNs on Au-TDNNs were designed to carry nanosensors into live cells. As the control group, Au-NPs@ExP1P2 was designed without TDNs. Confocal images of the two groups of sensors were captured at different incubation times. In the presence of intracellular miR-21, the green fluorescent signal produced by Au-TDNNs steadily increased, and the strongest fluorescence intensity was obtained at 1.5 h (Figure 6A), beyond which fluorescence intensity decreased. In the control group, the green fluorescent signal similarly increased with increasing incubation time (Figure 6B), and the strongest fluorescence intensity was obtained at 2.5 h. These results suggest that although Au-NPs@ExP1P2 can penetrate MCF-7 cells, attachment of TDNs to the nanosensors significantly enhanced cellular internalization and reduced incubation time.
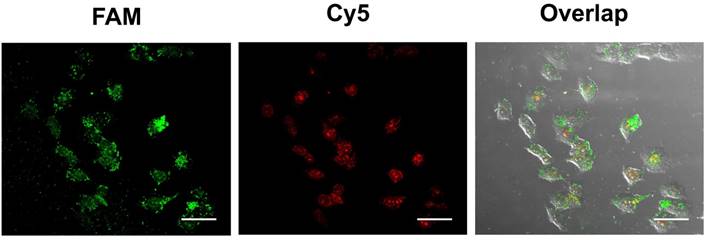
The feasibility of the proposed strategy was further investigated in hepatocytes cancer cells (HepG2). To improve the accuracy of live cell sensing, the 3'FAM-labeled ExP2 was replaced with the 3'Cy5- and 5'FAM-labeled ExP2. Red fluorescence of 3'Cy5 can be quenched by Au-NPs in Au-TDNNs, whereas 5'FAM can release green fluorescence. In the presence of miR-21, ExP2 is separated from Au-NPs, and both FAM and Cy5 can release fluorescence. Green fluorescence can locate Au-TDNNs, whereas red fluorescence can locate miR-21 in live cells. Figure 7 presents the confocal images of improved Au-TDNNs in HepG2 cells. As shown in the figure, strong FRET signal (red fluorescence) was observed for miR-21 in HepG2 cells. The result was consistent with PCR results obtained in previous reports [43,44].
Bio-TEM images of the cell uptake of Au-TDNNs in MCF-7 cells. The blue arrows point to the nucleuses and the red arrows point to Au-TDNNs. (A-C) The uptake of Au-TDNNs in the same perspective, (A, B) Enlarged view of (C). (D) The uptake of Au-TDNNs in the other perspective.
(A) Confocal images of MCF-7 cells with Au-TDNNs at different incubation times. (B) Confocal images of MCF-7 cells with Au-NPs@ExP1P2 at different incubation times. Scale bars = 50 µm.
Confocal images of HepG2 cells with improved Au-TDNNs. The incubation time was 1.5 h. Scale bars = 50 µm
Conclusion
In conclusion, a facile and specific method for miR-21 detection in live cells by Au-TDNNs was developed based on FRET. We proved the chemosynthesis of Au-TDNNs through multistep characterization and calculation. These nanosensors displayed good ability to avoid enzymatic degradation, which can minimize false-positive signals when applied in live cells. This method can rapidly detect miR-21 in MCF-7 cells in 1.5 h. The feasibility of the proposed method was also confirmed in HepG2 cells. Considering these remarkable advantages, we believe that Au-TDNNs will provide new directions for specific detection of DNA or RNA in cell imaging. Combined with PCR, loop-mediated isothermal PCR, or isothermal amplification, the rationally designed detection probes feature the potential for high-specificity detection of DNA or RNA in clinical specimens, such as plasma, urine, and tissue blocks.
Abbreviations
TDN: tetrahedral DNA nanostructure; TDNN: fluorescence resonance energy transfer based nanosensor in this experiment; Au-NP: gold nanoparticles; DF: discrimination factor; miRNAs: microRNAs; FRET: fluorescence resonance energy transfer; TCEP: tri (2-carboxyethyl) phosphine hydrochloride; PBS: phosphate buffer solution; TEM: transmission electron microscopy; PAGE: polyacrylamide gel electrophoresis; DLS: dynamic light scattering; DMEM: Dulbecco's modified Eagle's media; FBS: fetal bovine serum; ExP1: Expected detection probe1; ExP2: expected detection probe2; OrP: ordinary detection probe; SSC: saline-sodium citrate. RFI: relative fluorescence intensity.
Acknowledgements
This work was financially supported by National Natural Science Foundation of China (No. 81672112), Key Project of Health and Family Planning Commission of Sichuan Province (NO. 16ZD036) and Graduate Scientific Research and Innovation Project of Chongqing (No. CYS15119).
Supplementary Material
Supplementary figures and tables.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Lee RC, Feinbaum RL, Ambros V. Feinbaum and V. Ambros. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843-854
2. Cardoso AR, Moreira FTC, Fernandes R, Sales MGF. Novel and simple electrochemical biosensor monitoring attomolar levels of miRNA-155 in breast cancer. Biosens Bioelectron. 2016;80:621-630
3. Zhang X, Cheng R, Shi Z, Jin Y. A PCR-free fluorescence strategy for detecting telomerase activity via double amplification strategy. Biosens Bioelectron. 2016;75:101-107
4. Tili E, Michaille JJ, Costinean S, Croce CM. MicroRNAs, the immune system and rheumatic disease. Nat Clin Pract Rheum Atoll. 2008;4:534-541
5. Courts C, Madea B. Specific micro-RNA signatures for the detection of saliva and blood in forensic body-fluid identification. J Forensic Sci. 2011;56:1464-1470
6. Pritchard CC, Cheng HH, Tewari M. MicroRNA profiling: approaches and considerations. Nat Rev Genet. 2012;13:358-369
7. Hwang DW, Kim HY, Li F, Park JY, Kim D, Park JH, Han HS, Byun JW, Lee YS, Jeong JM, Char K, Lee DS. In vivo visualization of endogenous miR-21 using hyaluronic acid-coated graphene oxide for targeted cancer therapy. Biomaterials. 2017;121:144-154
8. Tay CY, Yuan L, Leong DT. Nature-inspired DNA nanosensor for real-time in situ detection of mRNA in live cells. ACS Nano. 2015;9:5609-5617
9. Git A, Dvinge H, Salmon-Divon M, Osborne M, Kutter C, Hadfield J, Bertone P, Caldas C. Systematic comparison of microarray profiling, real-time PCR, and next-generation sequencing technologies for measuring differential microRNA expression. RNA. 2010;16:991-1006
10. Islam MN, Gopalan V, Haque MH, Masud MK, Hossain MSA, Yamauchi Y, Nguyen NT, Lam AK, Shiddiky MJA. A PCR-free electrochemical method for messenger RNA detection in cancer tissue samples. Biosens Bioelectron. 2017;98:227-233
11. Lee JM, Jung Y. Two-temperature hybridization for microarray detection of label-free microRNAs with attomole detection and superior specificity. Angew Chem Int Ed Engl. 2011;50:12487-12490
12. Yin BC, Liu YQ, Ye BC. One-step, multiplexed fluorescence detection of microRNAs based on duplex-specific nuclease signal amplification. J Am Chem Soc. 2012;134:5064-5067
13. Li N, Chang C, Pan W, Tang B. A multicolor nanoprobe for detection and imaging of tumor-related mRNAs in live cells. Angew Chem Int Ed. 2012;51:7426-7430
14. Song S, Liang Z, Zhang J, Wang L, Li G, Fan C. Gold-nanoparticle-based multicolor nanobeacons for sequence-specific DNA analysis. Angew Chem Int Ed. 2009;48:8670-8674
15. Li S, Xu L, Ma W, Wu X, Sun M, Kuang H, Wang L, Kotov NA, Xu C. Dual-mode ultrasensitive quantification of microRNA in live cells by chiroplasmonic nanopyramids self-assembled from gold and upconversion nanoparticles. J Am Chem Soc. 2016;138:306-312
16. Wu Z, Liu GQ, Yang XL, Jiang JH. Electrostatic nucleic acid nanoassembly enables hybridization chain reaction in live cells for ultrasensitive mRNA imaging. J Am Chem Soc. 2015;137:6829-6836
17. Lin LS, Cong ZX, Cao JB, Ke KM, Peng QL, Gao J, Yang HH, Liu G, Chen X. Multifunctional Fe3O4@polydopamine core-shell nanocomposites for intracellular mRNA detection and imaging-guided photothermal therapy. ACS Nano. 2014;8:3876-3883
18. Ma Q, Nakane Y, Mori Y, Hasegawa M, Yoshioka Y, Watanabe TM, Gonda K, Ohuchi N, Jin T. Multilayered, core/shell nanoprobes based on magnetic ferric oxide particles and quantum dots for multimodality imaging of breast cancer tumors. Biomaterials. 2012;33:8486-8494
19. Asanuma H, Akahane M, Niwa R, Kashida H, Kamiya Y. Highly sensitive and robust linear probe for detection of mRNA in cells. Angew Chem Int Ed. 2015;54:4315-4319
20. Halstead JM, Lionnet T, Wilbertz JH, Wippich F, Ephrussi A, Singer RH, Chao JA. Translation. An RNA biosensor for imaging the first round of translation from single cells to living animals. Science. 2015;347:1367-1671
21. Wu C, Cansiz S, Zhang L, Teng IT, Qiu L, Li J, Liu Y, Zhou C, Hu R, Zhang T, Cui C, Cui L, Tan W. A Nonenzymatic Hairpin DNA Cascade Reaction Provides High Signal Gain of mRNA Imaging Inside Live Cells. J Am Chem Soc. 2015;137:4900-4903
22. Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406-3415
23. DeLong EF, Wickham GS, Pace NR. Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science. 1989;243:1360-1363
24. Saiki RK, Gelfand DH, Stoffel S, Scharf SJ, Higuchi R, Horn GT, Mullis KB, Erlich HA. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487-491
25. Krainer G, Hartmann A, Schlierf M. farFRET: Extending the Range in Single-Molecule FRET Experiments beyond 10 nm. Nano Lett. 2015;15:5826-5829
26. Wang F, Lu CH, Willner I. From cascaded catalytic nucleic acids to enzyme-DNA nanostructures: controlling reactivity, sensing, logic operations, and assembly of complex structures. Chem Rev. 2014;114:2881-2941
27. Chen SX, Zhang DY, Seelig G. Conditionally fluorescent molecular probes for detecting single-base changes in double-stranded DNA. Nat Chem. 2013;5:782-789
28. Zhang DY, Chen SX, Yin P. Optimizing the specificity of nucleic acid hybridization. Nat Chem. 2012;4:208-214
29. Chen YJ, Groves B, Muscat RA, Seelig G. DNA nanotechnology from the test tube to the cell. Nat Nanotechnol. 2015;10:748-760
30. Gilleron J, Querbes W, Zeigerer A, Borodovsky A, Marsico G, Schubert U, Manygoats K, Seifert S, Andree C, Stöter M, Epstein-Barash H, Zhang L, Koteliansky V, Fitzgerald K, Fava E, Bickle M, Kalaidzidis Y, Akinc A, Maier M, Zerial M. Image-based analysis of lipid nanoparticle-mediated siRNA delivery, intracellular trafficking and endosomal escape. Nat Biotechnol. 2013;31:638-646
31. Sahay G, Querbes W, Alabi C, Eltoukhy A, Sarkar S, Zurenko C, Karagiannis E, Love K, Chen D, Zoncu R, Buganim Y, Schroeder A, Langer R, Anderson DG. Efficiency of siRNA delivery by lipid nanoparticles is limited by endocytic recycling. Nat Biotechnol. 2013;31:653-658
32. Pei H, Li F, Wan Y, Wei M, Liu H, Su Y, Chen N, Huang Q, Fan C. Designed diblock oligonucleotide for the synthesis of spatially isolated and highly hybridizable functionalization of DNA-gold nanoparticle nanoconjugates. J Am Chem Soc. 2012;134:11876-11879
33. Jing X, Cao X, Wang L, Lan T, Li Y, Xie G. DNA-AuNPs based signal amplification for highly sensitive detection of DNA methylation, methyltransferase activity and inhibitor screening. Biosens Bioelectron. 2014;58:40-47
34. Chen L, Chao J, Qu X, Zhang H, Zhu D, Su S, Aldalbahi A, Wang L, Pei H. Probing Cellular Molecules with PolyA-Based Engineered Aptamer Nanobeacon. ACS Appl Mater Interfaces. 2017;9:8014-8020
35. Qu X, Zhu D, Yao G, Su S, Chao J, Liu H, Zuo X, Wang L, Shi J, Wang L, Huang W, Pei H, Fan C. An Exonuclease III-Powered, on-Particle Stochastic DNA Walker. Angew Chem Int Ed Engl. 2017;56:1855-1858
36. Liang L, Li J, Li Q, Huang Q, Shi J, Yan H, Fan C. Single-particle tracking and modulation of cell entry pathways of a tetrahedral DNA nanostructure in live cells. Angew Chem Int Ed. 2014;53:7745-7750
37. Schüller VJ, Heidegger S, Sandholzer N, Nickels PC, Suhartha NA, Endres S, Bourquin C, Liedl T. Cellular immunostimulation by CpG-sequence-coated DNA origami structures. ACS Nano. 2011;5:9696-9702
38. Li J, Pei H, Zhu B, Liang L, Wei M, He Y, Chen N, Li D, Huang Q, Fan C. Self-assembled multivalent DNA nanostructures for noninvasive intracellular delivery of immunostimulatory CpG oligonucleotides. ACS Nano. 2011;5:8783-8789
39. Liu J, Lu Y. Preparation of aptamer-linked gold nanoparticle purple aggregates for colorimetric sensing of analytes. Nat Protoc. 2006;1:246-252
40. Patutina OA, Bichenkova EV, MiRNAoshnichenko SK, MiRNAonova NL, Trivoluzzi LT, Burusco KK, Bryce RA, Vlassov VV, Zenkova MA. miRNAes: Novel peptide-oligonucleotide bioconjugates that silence miR-21 in lymphosarcoma cells. Biomaterials. 2017;122:163-178
41. Zhang DY, Winfree E. Control of DNA strand displacement kinetics using toehold exchange. J Am Chem Soc. 2009;131:17303-17314
42. Hurst SJ, Lytton-Jean AK, MiRNAkin CA. Maximizing DNA loading on a range of gold nanoparticle sizes. Anal Chem. 2006;78:8313-8318
43. Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A. et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401-1414
44. Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647-658
Author contact
![]() Corresponding author: G.M.X (guomingxieedu.cn)
Corresponding author: G.M.X (guomingxieedu.cn)
 Global reach, higher impact
Global reach, higher impact