13.3
Impact Factor
Theranostics 2018; 8(5):1301-1311. doi:10.7150/thno.21979 This issue Cite
Research Paper
Non-invasive Prenatal Diagnosis of Chromosomal Aneuploidies and Microdeletion Syndrome Using Fetal Nucleated Red Blood Cells Isolated by Nanostructure Microchips
1. Department of Obstetrics and Gynechology, Zhongnan Hospital of Wuhan University, Wuhan, Hubei, 430071, China
2. Key Laboratory of Artificial Micro- and Nano-Structures of Ministry of Education, School of Physics and Technology, Wuhan University, Wuhan, 430072, Hubei, China
3. Prenatal Diagnostic and Birth Health Clinical Research Center of Hubei Province, Wuhan, Hubei, 430071, China
4. Department of Prenatal Diagnostic Center, Maternal and Child Health Hospital of Hubei Province, Wuhan, Hubei, 430070, China
* equally contributing first author
# equally contributing senior authorship
Received 2017-7-18; Accepted 2017-11-8; Published 2018-2-2
Abstract
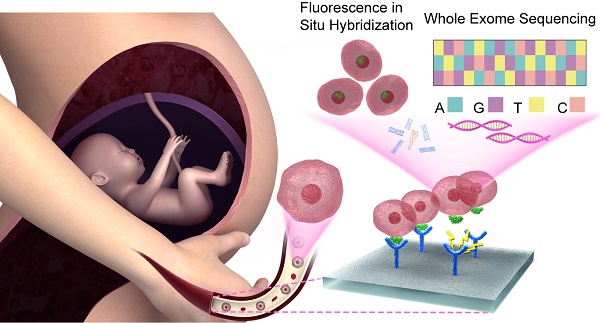
Detection of detached fetal nucleated red blood cells (fNRBCs) in the maternal peripheral blood may serve as a prospective testing method competing with the cell-free DNA, in non-invasive prenatal testing (NIPT).
Methods: Herein, we introduce a facile and effective lab-on-a-chip method of fNRBCs detection using a capture-releasing material that is composed of biotin-doped polypyrrole nanoparticles. To enhance local topographic interactions between the nano-components and fNRBC, a specific antibody, CD147, coated on the nanostructured substrate led to the isolation of fNRBCs from maternal peripheral blood. Subsequently, an electrical system was employed to release the captured cells using 0.8 V for 15 s. The diagnostic application of fNRBCs for fetal chromosomal disorders (Trisomy 13/21/18/X syndrome, microdeletion syndrome) was demonstrated.
Results: Cells captured by nanostructured microchips were identified as fNRBCs. Twelve cases of chromosomal aneuploidies and one case of 18q21 microdeletion syndrome were diagnosed using the fNRBCs released from the microchips.
Conclusion: Our method offers effective and accurate analysis of fNRBCs for comprehensive NIPT to monitor fetal cell development.
Keywords: non-invasive prenatal diagnosis, fetal nucleated red blood cells, nanostructure microchip, chromosomal aneuploidy, microdeletion syndrome
Introduction
Birth defects are a major challenge worldwide, requiring improvements in reproductive healthcare. For example, in 2015, congenital chromosomal abnormalities were one of the leading causes of under-5 mortalities [1]. Currently, the survival rates of trisomy 21, 18 and 13 are still low (1 in 800, 1 in 6000, and 1 in 10 000, respectively) [2], indicating poor quality of life. Therefore, prenatal diagnosis of birth defects is vitally important. However, conventional prenatal diagnostic methods have several limitations and are not adequately reliable or safe. In general, maternal serum biochemical screening (i.e., screening plasma protein A, free beta human chorionic gonadotropin and alpha-fetoprotein in the first or second trimester) and sonographic screening (e.g., measuring nuchal translucency) are reported to have an excessive false positive rate for certain aneuploidies. Other diagnostic techniques such as chorionic villus sampling, cordocentesis, or amniocentesis are all invasive and may cause complications leading to miscarriage, infections or even maternal fatality [3]. Considering that birth defects are largely induced by genetic abnormalities such as fetal chromosomal aneuploidy and genetic aberration, there has been an emphasis on the development of diagnostics using easily accessible maternal peripheral blood that contains abundant fetal genetic materials. Several non-invasive prenatal testing (NIPT) techniques based on blood tests have been established which mainly depend on cell-free fetal DNA (cffDNA) or fetus-derived cells in the maternal peripheral blood.
cffDNA-based NIPT is extensively used in the clinic [4,5] for screening for trisomy 21, 18, and 13 in high-risk gravidas [6,7], presents no risk of pregnancy loss and provides an inexpensive, convenient and effective method compared to other invasive technologies. However, cffDNA-based NIPT suffers from the following limitations [8]: 1) it cannot eliminate chromosomal anomalies like mosaicism, duplication, and deletion; 2) limited data are currently available on the use of NIPT in twins and multiple pregnancies [9]; 3) cell-free DNA cannot be used to distinguish specific abnormalities such as Robertsonian translocation and high-level mosaicism [10]; and, 4) samples from gravidas with a low-level mosaicism or solid tumor as well as a high body mass index (BMI) [11] or early gestational age will result in variations of circulating cffDNA impacting prenatal testing results.
Due to inherent drawbacks of cffDNA in NIPTs, circulating fetus-derived cells in the maternal bloodstream have attracted much attention. Four types of fetal nucleated cells have been reported: trophoblasts, fetal nucleated red blood cells (fNRBCs), hematopoietic progenitor cells, and lymphocytes. Among these, fNRBCs are the preferred choice for NIPTs due to their unique characteristics [12]. First, fNRBCs have intact nuclei containing the total fetal genome for prenatal analysis. And second, fNRBCs have distinct cell markers, such as epsilon hemoglobin transferrin receptor (CD71), thrombospondin receptor (CD36), GPA, and antibody 4B8/4B9 [13-18], enabling isolation of these rare cells from large volumes of maternal blood.
A variety of fNRBC isolation strategies have been developed, such as density gradient centrifugation (DGC) [19], fluorescence-activated cell sorting (FACS) [20], and magnetic-activated cell sorting (MACS) [21]. Recently, fNRBC isolation approaches with better yields and less cell damage have employed microfluidic chips of silicon, glass, and other plastic materials like polymethyl methacrylate (PMMA), polycarbonate (PC) and polydimethylsiloxane (PDMS) [22,23]. Huang et al. [14] and Bhagat et al. [24] developed high throughput microfluidic techniques to isolate fNRBCs from maternal blood based on cell size differences. These strategies enabled enrichment of fNRBCs from maternal peripheral blood with high recovery and purity for clinical applications. Nevertheless, retrieving intact fRNBCs from substrates for subsequent biomedical analysis and directly integrating prenatal diagnostics with the fNRBC isolation platform remains considerable challenges.
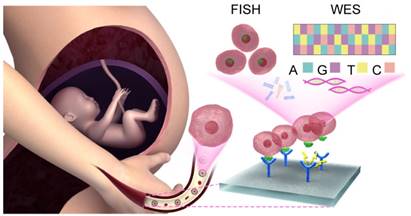
Previously, many methods for detaching fNRBCs have been described [24-26]. In this study, we employed nanostructure microchips to isolate fNRBCs for prenatal diagnosis. Our microchip was made of biocompatible biotin-doped conductive polypyrrole (Ppy) nanoparticles for effective isolation of fNRBCs. These nanoparticles form a topographic three-dimensional (3D) nanostructure to increase the rate of antibody-chip surface bio-conjugation. These nanostructures simulate extracellular matrix (ECM) to enhance the interaction between fNRBCs and microchips [27]. Furthermore, if triggered by electric fields, Ppy can reveal the doped biotin inside and thus help release the antibody-captured fNRBCs from the microchips [28,29]. We used a fNRBC-specific cell membrane antibody, anti-CD147, and validated its specificity by flow cytometry and Western blotting. fNRBCs from maternal peripheral blood at optimum pregnancy age were isolated [30] and validated by three color immunofluorescent staining. Using these cells, prenatal diagnosis based on fluorescence in situ hybridization (FISH) and whole exome sequencing (WES) was performed to diagnose various birth defects (chromosomal aneuploidy and microdeletion syndrome) (Fig. 1). We also investigated the association between fNRBC quantity and fetal abnormality. We believe that our method will benefit non-invasive prenatal diagnostics, especially for screening the complex genetic disorders, which present a challenge for clinical cffDNA-based NIPTs.
Schematic showing NIPT using fNRBCs isolated by our nanostructure microchips. fNRBCs are captured from maternal peripheral blood by the biotin-doped Ppy nanoparticle microchip grafts with anti-CD147 targeting agent, and then retrieved from the substrate through electric stimulation. These isolated fNRBCs are used for chromosome analysis and whole exome sequencing to diagnose birth defects non-invasively.
Materials and Methods
Ethics Statement
This study was approval by the Ethics Committee at Zhongnan Hospital of Wuhan University. Related informed consent was signed before the study, and the study protocols were based on the ethical principles for research that involves human subjects of the Helsinki Declaration for medical research [31].
Materials
Anhydrous ethanol, methanol, acetic acid, and dimethylsulfoxide (DMSO) were purchased from Sinopharm Chemical Reagent Co. Nonyl phenoxypolyethoxylethanol (NP-40), saline sodium citrate (SSC), phosphate buffered saline (PBS), lymphocyte separation medium (LSM), Tween-20, pyrrole, biotin, sodium dodecylbenzene sulfonate (NaDBS), 1-ethyl-3-(3- dimethylaminopropyl) carbodiimide (EDC), N-hydroxysuccinimide (NHS), paraformaldehyde (PFA, 36% in water), Triton X-100, bovine serum albumin (BSA), normal goat serum and 4,6- diamidino-2-phenylindole dihydrochloride (DAPI) were purchased from Sigma-Aldrich. RIPA buffer was purchased from Beyotime Biotechnology Co., Streptavidin and 0.25% Trypsin-ethylenediaminetetraacetic acid (EDTA) (Gibco, 1×) were obtained from Invitrogen. Biotinylated anti-CD147/BSG and human TfR (Transferrin R)/anti-CD71 antibody (goat immunoglobulin G) were acquired from R&D Systems. Phycoerythrin (PE)-labeled anti-fetal hemoglobin-epsilon (anti-ε HbF) antibody, fluorescein isothiocyanate (FITC)-labeled anti-CD71 antibody, fluorescein diacetate (FDA) and propidium iodide (PI) were purchased from BD Biosciences. Prime-Script RT reagent kits were purchased from Takara. FISH X-13, X-21 and FISH Y-13 and Y-21 chromosome probes were obtained from Jinpujia Medical Technology, Inc. Glass slides were cleaned using acetone and then ethanol, and dried using compressed nitrogen.
Cells
Nucleated erythrocyte cell line (CRL_2003, TF-1) was purchased from ATCC Co. and the monocyte cell line was harvested from patients at Union Hospital of Tongji Medical College in Huazhong University of Science and Technology.
Blood samples collection
Blood samples were drawn ahead of any procedure. Approximately 3 mL blood from gravidas who were recruited to this study was collected into anti-coagulant K2-EDTA Vacutainer tubes (Becton-Dickinson) with a proprietary preservative (available from Rare Cyte).
Identification and determination of the specific surface antigen on nucleated red blood cells
Western blots and flow cytometry assay were performed according to standard protocols. Monocytes from normal blood, monocyte from leukemia and nucleated erythrocytes from leukemia were collected to identify their specific surface antigen.
Western blotting was performed according to the routine procedure. Briefly, cells were lysed in RIPA buffer with protease/phosphatase inhibitors. After protein quantification, lysates were resuspended in sample buffer and loaded on the gradient gels (Criterion Tris-HCl Gel, 8-16%). Proteins were transferred onto polyvinyl difluoride (PVDF) membranes. Primary antibody CD147 (Abcam, Ab108317) was used. Blots were incubated with the HRP-conjugated secondary antibody (Boster, BA1054) and developed using Epson V300 scanner. Alpha Innotech software was used to analyze the gray value.
For flow cytometry, cell samples were incubated with combination antibody FITC-labelled anti-CD147 (BD Biosciences, #4192771) at room temperature (RT) for 2 h in the dark. Cells were then centrifuged, resuspended in PBS, and loaded into a FACS Aria flow cytometer. Data were analyzed by FlowJo (Tree Star) software.
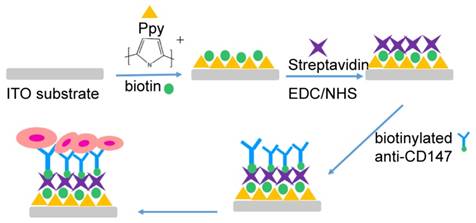
Preparation of biotin-doped Ppy microchips and modification with specific antibody on the microchip
For the preparation of biotin-doped Ppy microchip, first indium tin oxide (ITO) glass substrates were cleaned using acetone and ethanol successively and then dried using compressed nitrogen. Ppy polymerization was carried out in 0.1 M pyrrole and 0.01 M NaDBS aqueous solution containing 1 mM biotin with the application of chronoamperometry (CA) at 0.8 V (versus Ag/AgCl) for 2 min. Subsequently, clean ITO substrates were immersed in the Ppy solution to obtain biotin-doped Ppy microchips. The microchips were immediately washed with ultrapure water 3 times and dried in air for next use.
Surface modification of the microchips for antibody conjugation was carried out according to the protocol reported by Jeon and Nagrath et al [27,32]. In brief, the biotin-doped Ppy-based microchip was modified in an EDC (0.095 g) / NHS (0.061 g) aqueous solution (10 mL) for 45 min, followed by rinsing with ultrapure water 3 times. The chip was further incubated with 50 µL of streptavidin (20 µg/mL in water) for 1 h and then washed with ultrapure water 3 times. The streptavidin-modified microchips could be stored at 4 °C. Before fNRBC isolation, 10 mL biotinylated anti-CD147 (10 µg/ mL in PBS with 1% BSA (w/v)) was added onto the microchip and incubated at RT for 2 h. After washing with PBS, the chip was incubated with 100 µL of 1% BSA (w/v) in 1× PBS to reduce nonspecific binding. The microchip was washed with PBS 3 times and was ready for fNRBC isolation. A general schematic diagram of biotin-doped polypyrrole-based microchip fabrication and the antibody bio-conjugation procedure is depicted in Fig. 2.
Schematic diagram showing the biotin-doped polypyrrole-based microchip fabrication and antibody bio-conjugation procedure.
Blood sample pretreatment
The blood samples were processed according to a density gradient centrifugation (DGC) protocol. First, 1 mL blood sample was 1:1 diluted with PBS. Subsequently, the diluted blood was carefully added onto the surface of LSM in a centrifuge tube to form an obvious liquid-liquid interface. The tube was centrifuged horizontally at 1800 rcf for 30 min to separate the liquid into four layers. The mononuclear cells containing fNRBCs were obtained from two middle layers and suspended in 1mL of PBS for the capture procedure.
fNRBC isolation and release
The mononuclear cell suspensions containing fNRBCs was loaded onto the microchip, which was modified with anti-CD147. After incubation for 60 min, the chip was gently washed five times with PBS to remove nonspecifically adsorbed cells.
To release captured fNRBCs from the Ppy nanostructure substrate, the microchip with the cells was placed into a size-matched container. Subsequently, an electric stimulation system applied the release voltages (0.3 V, 0.5 V and 0.8 V) on the microchip for 15 s, following which the microchip was taken out and washed with PBS. The eluent and the solution in the container were collected in a tube and centrifuged to retrieve the released fNRBCs for subsequent biomedical analysis.
fNRBC capture, immunofluorescent staining, and FISH analysis
Immunofluorescent staining was performed to identify fNRBCs. First, the captured cells were fixed with 1 mL 4 % PFA in PBS at RT for 10 min. The cells were then incubated with 1 mL 0.1 % Triton X-100 in PBS for 10 min to increase cellular permeability. Subsequently, 3 % BSA in PBS was used to treat the cells for 30 min to block non-specific bonding sites. Finally, cells were immunofluorescently stained with the block solution containing 10 mL PE-labelled anti-ε-HbF (10mg/ mL) and 10 mL of FITC-labelled anti-CD71 (10 mg/ mL) and then incubated in the dark at RT for 2 h. Before microscopy, fNRBCs were nucleus-stained with DAPI (0.1 mg/ mL in deionized water) for 10 min and then washed with deionized water.
The FISH analysis [33] was performed according to the kit protocol provided. Briefly, the microchip with captured fNRBCs was immersed in the 2× SSC buffer at RT for 10 min to enhance the permeability of the nuclei. Then the chip was treated with 70 %, 85 % and 100 % ethanol for 2 min dehydration. Subsequently, FISH probes with a volume ratio of 1:4 Chromosome X/Y, 21, 18, or 13 probes or their mixture in the hybridization buffer were added to the chip, which was covered with a coverslip, and sealed with rubber oil. The chip was placed in a 75 ℃ water bath for 5 min and then incubated at 42 ℃ in a dark, wet box for more than 20 h. Subsequently, the coverslip was carefully removed, and the microchip was first soaked in 2 × SSC for 10 min and immersed in NP-40 (0.3 % in 0.4 × SSC) in a 67 ℃ water bath for 90 s, followed by immersion in another NP-40 solution (0.1 % in 2 × SSC) for 30 s at RT. Finally, the chip was dehydrated in 70 % ethanol for 3 min and then air dried. Nuclear staining was done with DAPI (0.1 mg/ mL) for 15 min. After covering with another coverslip, cell imaging and counting were performed on IPP software.
All images were recorded by DP72 charge-coupled devices (CCD, Olympus, Japan), mounted on an IX81 inverted microscope (Olympus, Japan), and analyzed using Image-Pro Plus (IPP) software (Media Cybernetics Inc., USA).
Chromosomal karyotype analysis of amniotic fluid cells
Chromosomal karyotype analysis of amniotic fluid cells, which were compared to fNRBC samples, was done by the Prenatal Diagnostic and Birth Health Clinical Research Center of Hubei Province. All procedures were performed according to the standard protocol. Briefly, amniotic fluid was obtained from women in their second-trimester of pregnancy through amniocentesis. The amniotic fluid was centrifuged at RT for 10 min and cells were recycled and cultured at 37 ℃ for 5-10 days. Adherent viable cells were collected and made into cell slices [34,35]. The cell karyotype analysis was performed by CytoVision analyzer.
Whole exome sequencing analysis of samples
To analyze and assess the diagnostic accuracy of the captured fNRBCs, cell samples from maternal peripheral blood, umbilical cord blood, and maternal peripheral blood after at least 42 days of pregnancy were collected for whole exome sequencing. The sequencing was carried out on genomic DNA using Hiseq-2500 platform (Illumina). The data were obtained after mapping to the reference human genome (Hg 19) and comparing the data during and after pregnancy.
Results and Discussion
Verification of the specificity of anti-CD147
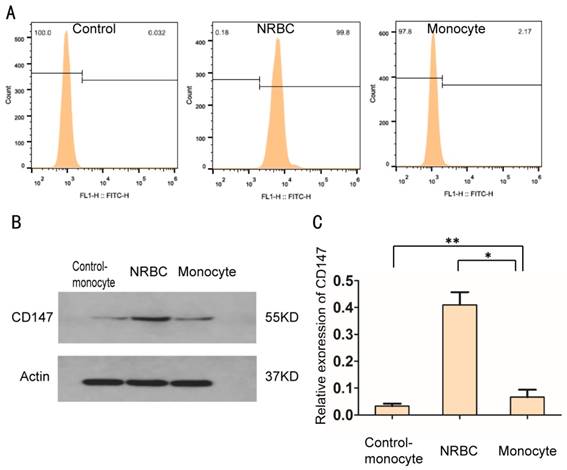
Isolation of fNRBCs from the microchip required a specific antigen on the cell membrane to separate fNRBCs from monocytes. The protein CD147 (also known as basigin, BSG, EMMPRIN and M6, [14]), linked with spermatogenesis, embryo implantation [36], proliferation, invasiveness, migration, and angiogenesis of cancer [37], is expressed on the membrane of nucleated erythrocytes [30, 38]. To investigate the expression of CD147 on the fNRBC membrane, flow cytometry assay and Western blotting were performed. As shown in Fig. 3A, the fluorescence intensity of fNRBC-CD147 was much higher compared with that of monocyte-CD147.
To further validate the high expression of CD147 on the fNRBC membrane, expression of CD147 in the monocyte lineage was compared with that on the primary monocytes collected from nonpregnancy women by Western blotting (Fig. 3B-C). There was no difference in the expression of CD147 in the control group and monocyte cell lineage indicating that CD147 expression was not affected by hematopathy. However, CD147 expression was much higher in NRBCs than in monocytes confirming its utility as an ideal antigen for the separation NRBCs from monocytes.
Characterization and optimization of biotin-doped Ppy-based nanoparticle microchip
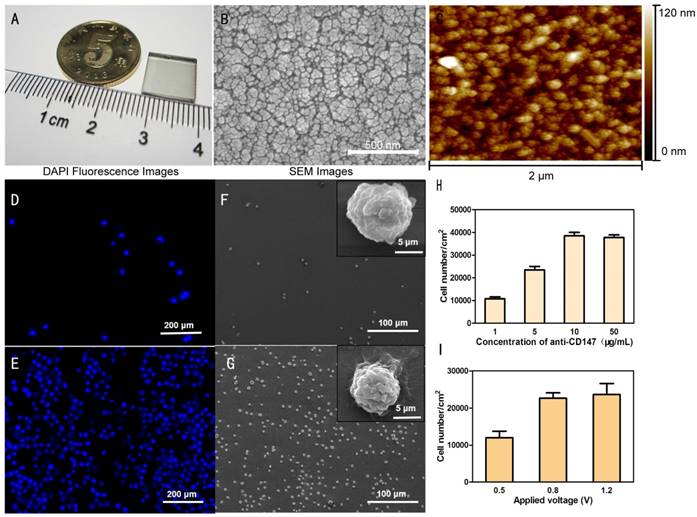
Biotin-doped Ppy-based nanoparticle microchip was fabricated on a conductive indium-tin oxide (ITO) glass and has many excellent characteristics: (1) its good biocompatibility allows biotin-doped Ppy to be widely used in drug delivery systems, (2) its transparency feature facilitates FISH analysis, (3) it possesses a high volume ratio that allows trapping of cell capture agents, (4) it favors release of cells for easy operation with minimal harm to cell integrity.
Fig. 4A displays the mini-size microchip. The captured cell analysis can take advantage of its transparency characteristics. Under scanning electron microscopy (SEM) (Fig. 4B) and atomic force microscopy (AFM) (Fig. 4C), the Ppy-based microchip showed cell-to-chip contact ability due to its raised and hollow surface. When a specific antibody was used, the Ppy-based nanoparticle microchip showed its ability to recognize and capture fNRBCs.
To verify the cell-capture ability of the Ppy-based microchip, the NRBC suspension was introduced onto a 1cm2 biotin-doped Ppy-based microchip, which was modified with biotinylated anti-CD147. As a control, a flat ITO microchip modified with biotinylated anti-CD147 was also examined in parallel. As shown in Fig. 4D-G, the Ppy-based microchip could capture many more cells than the flat ITO microchip. This suggested that the high-volume ratio of Ppy-based microchip was possibly responsible for the enhanced cell-capture numbers. The cells captured on the flat ITO microchip and Ppy-based microchip exhibited vastly different cell numbers and marked morphology differences by SEM observation (Fig. 4F-G). The insets in Fig. 4F-G show the cells captured on the flat ITO appeared more rounded, and no cell filopodia were observed. These results indicated the preferred interaction of NRBCs with the biotin-doped Ppy-based microchip with three-dimensional topography.
Validation of cytomembrane protein CD147 in nucleated red blood cells using flow cytometry assay and western blotting analysis. (A) In flow cytometry analysis, the x-axis represents fluorescence intensity and the y-axis means the number of cells. (B) Representative pictures from protein gel electrophoresis imaging in three kinds of cells. (C) Western blots of protein lysates from control-monocytes, nucleated red blood cells and monocytes; data are representative of 3 independent experiments. No difference between control-monocyte group and monocyte group (**p>0.05) was present; there was a difference between NRBC group and monocyte group/control-monocyte group (*p<0.05).
To optimize the cell-capture agents, the concentration of anti-CD147 used for the surface bio-conjugation on the microchip as well as the electric potential application for electroplating Ppy and biotin on the microchip were investigated. NRBCs were suspended at a density of 2×106 cells/ mL in PBS. Four different concentrations of anti-CD147 were used for cell isolation (Fig. 4H). The results showed that the 10 mg/mL of anti-CD147 concentration had the highest cell capture number. Also, 2 min of electric potential was applied at three different voltages (0.5 V/0.8 V/1.2 V) during electroplating. The results obtained are displayed in Fig. 4I and show that more cells could be captured when the applied voltage was increased from 0.5V to 0.8V with the number of captured cells reaching its maximum at 0.8V or above.
Identification and release of fNRBCs
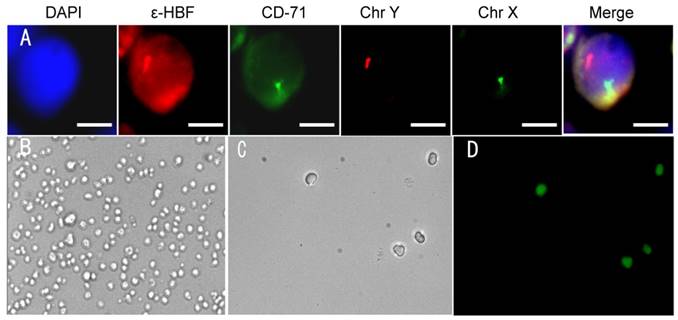
fNRBCs were isolated on microchips using maternal peripheral blood collected from gravidas at optimal gestational age. Tricolor immunofluorescent staining was performed using the biological dye DAPI, and anti-ε-HBF [13] and anti-CD71 [16] antibodies to identify nuclear, cytoplasmic and cytomembrane localization, respectively.
Additionally, chromosome X/Y probes were employed to verify whether the stained cells had maternal origin or stemmed from the fetus.
Fig. 5A presents representative images of isolated fNRBCs from maternal peripheral blood using the nanostructure microchip under fluorescence microscopy. Male fetal nucleated red blood cells were isolated and identified by two FISH signals (chromosome X/Y) and the reliability of anti-CD147 to isolate fNRBCs was validated.
In previous studies, Jeon et al. used electroactive applications to release cancer cells from biotin-doped Ppy films [27], whereas Tao Gao et al. employed an electrochemical system based on redox-control to capture and release cancer cells [39]. Our study used, for the first time, electrical stimulation for releasing fNRBCs from the microchip. Captured nucleated erythrocyte cell line was used in a preliminary study for electric stimulation. NRBCs, isolated from biotin-doped Ppy microchips, were divided into three groups and 0.3 V, 0.5 V and 0.8 V electric stimulation was applied to the chips for 15 s. Subsequently, the viability of the released cells was observed by fluorescence-based live-dead cells assay with green and red fluorescence of FDA and PI, respectively. The microscopic images of NRBCs before and after electric stimulation with different voltages are shown in Fig. S1. Comparison of the cell numbers before and after electric stimulation indicated that higher electric stimulation might enhance cell release without having a noticeable impact on cell viability (Fig. 6A). Therefore, 0.8 V was used for releasing cells from the microchip by electric stimulation. Representative images of NRBCs are shown under the brightfield and fluorescence views before and after 0.8V electric stimulation (Fig. 5B-D).
Microchip characterization. (A) Photograph showing the biotin-doped Ppy nanostructure microchip. Typical SEM image (B) and AFM image (C) showing 3D nanostructures formed by biotin-doped Ppy nanoparticles on the ITO substrate. (D-G) Representative fluorescence images and SEM images of the flat ITO microchip and the biotin-doped Ppy microchip on which NRBCs were captured. The biotin-doped Ppy microchip exhibited considerably better cell capture efficiency than the flat ITO microchip due to its high volume ratio. (H) Comparison of the cell numbers on the nanostructure microchips using different concentrations of anti-CD147 applied for cell capture. (I) Comparison of the cell numbers on the microchips after applying different voltages for electroplating Ppy and biotin.
(A) Identification of fNRBC in maternal peripheral blood using immunofluorescent staining and FISH technique. Representative pictures of fNRBCs from gravida who conceived male infants are shown. The scale bars are 5 μm. (B-C) Representative images of nucleated red blood cells on the microchip before/after electric stimulation by 0.8 V. (D) FDA and PI staining of cells on the microchip after 0.8 V electric stimulation.
(A) Comparison of the average number of NRBCs on the microchip before and after electric stimulation by 0.3 V, 0.5 V and 0.8 V. (B) Using fNRBC to detect releasing efficiency at three different voltages. (C) Comparison of the number of fNRBCs from maternal peripheral blood from abnormal pregnancy (Table S2) and normal pregnancy (Table S1) at 18 weeks of gestation.
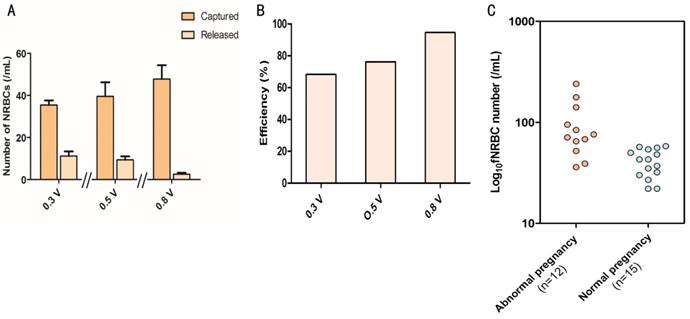
Fifteen maternal peripheral blood samples at the 18th gestational age were collected from gravidas pregnant with a male fetus; the fetus sex was determined by the fNRBCs immunofluorescent staining and FISH analysis. The number of fNRBCs from all samples are presented in Table S2. The samples were divided into three groups of 5 samples/group, and electric stimulation was used for releasing fNRBCs from the microchips. Subsequently, the residual cells on the chips were counted under fluorescence microscopy after electric stimulation. Releasing efficiencies of fNRBCs were 68.3%, 76.2% and 94.6% respectively in the 0.3 V, 0.5 V and 0.8 V groups (Fig. 6B), further indicating that the electric stimulation of 0.8 V for 15 s was sufficient for the maximum fNRBCs release.
Prenatal diagnostics of chromosomal aneuploidies and microdeletion syndrome
To confirm the feasibility of fNRBCs-based NIPT, fetal genetic defects such as chromosomal aneuploidy cases (including autosomal aneuploidy and sexual aneuploidy) and microdeletion syndrome were collected for diagnosis by FISH technique and WES.
Maternal peripheral blood samples were drawn mostly around the 18th week of gestation, which was previously determined to be the optimum time point [30]. fNRBCs were isolated from high risk fetal chromosomal or genetic aberration samples (the data were from cffDNA-based NIPT or B-type ultrasound examination).
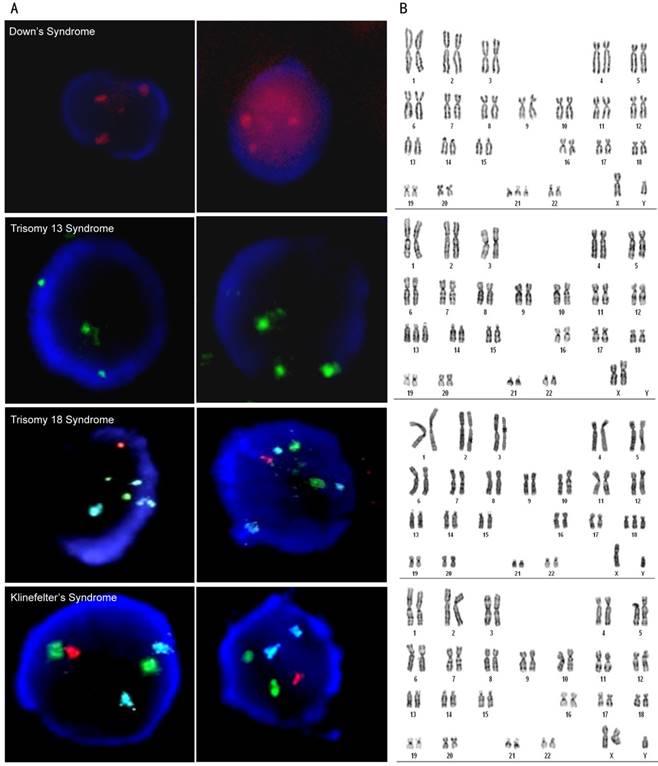
For the diagnosis of fetal chromosomal aneuploidy, three prominent fluorescence staining signals of chromosomes 21/13/18/X were seen in the fNRBCs while using corresponding fluorescent probes (Fig. 7A). Five cases of Down syndrome, three cases of trisomy 13 syndrome, two cases of trisomy 18 syndrome, and two cases of Klinefelter's syndrome were detected. These results were compared with subsequent amniocentesis and G-banding karyotyping analysis (Fig. 7B) and demonstrated that the non-invasive analysis of fNRBCs could diagnose fetal chromosomal aneuploidy malformations in all twelve cases. The numbers of fNRBCs from these samples were counted (Table S3) and compared with fNRBCs isolated from normal pregnancy (Table S2). Fig. 6C shows that the number of fNRBCs from gravidas who conceive a fetus with chromosomal aneuploidy was significantly higher than those with normal chromosomal aneuploidy. These results confirmed that high counts of fNRBCs indicated abnormal gravidity, which was consistent with previously reported studies [40].
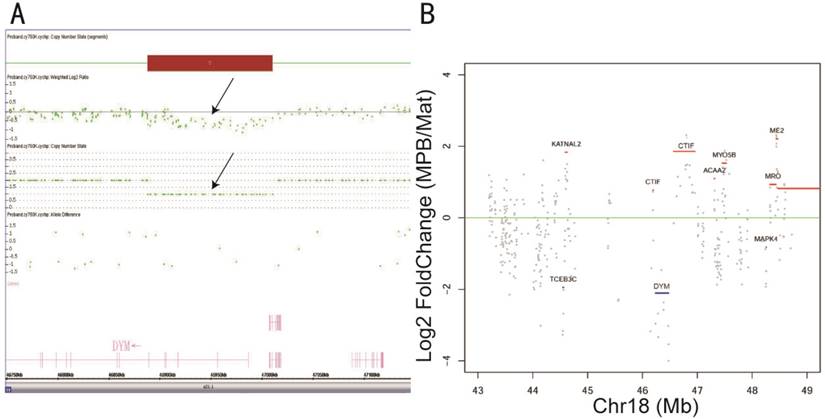
A typical example was from a woman at 21 weeks of gestation whose three-dimensional B-type ultrasound manifested fetus with multiple organ structure abnormities including high airway obstruction syndrome and heart malformation. According to ACOG guidelines [41], amniocentesis and chromosomal microarray (CMA) were performed for prenatal diagnosis. The results showed that the fetus suffered 18q21 microdeletion encompassing a pathogenic gene DYM, which is associated with dysosteogenesis and mental retardation (Fig. 8A). In this case, fNRBCs, representative of fetal genetic material, were isolated from the maternal peripheral blood and released from the nanostructure microchips by electric stimulation. Simultaneously, mouth mucosa exfoliative cells were collected to identify the maternal genetic material and WES of both samples was performed. After the removal of sequencing adapters and trimming of low-quality bases, ~11.93 Gb of cleaned reads data were obtained for each sample (Table S1). More than 99.03% of the sequencing reads were aligned to the human reference genome (hg19), with 49.82% effective reads from target regions obtained after the removal of PCR duplications. The average sequencing depth for each sample was 99.32-fold, with more than 92.06% of target regions having at least a 10-fold coverage. A total of 13683 and 15950 SNVs or InDels were identified, respectively, in the gravida peripheral blood sample and maternal independent sample. The copy ratios (CNVs of gravida peripheral blood sample/maternal independent sample) that manifested genetic amplification or deletion were calculated and 4568 exomes were identified. 101 exomes were present on chr 18, and the copy ratio of DYM gene was 0.262 (1 was the benchmark), which signified microdeletion (Fig. 8B). The WES result conformed with the CMA result. Thus, it was clear that fNRBCs from maternal peripheral blood could be used to diagnose fetal chromosomal aneuploidy and microdeletions without any risk to fetus and mother.
(A) Representative images of diagnosis of chromosome aneuploidy using FISH staining of isolated fNRBCs from maternal peripheral blood. Green fluorescence, red fluorescence and blue fluorescence respectively represent chromosome 13/X, chromosome 21/Y and chromosome 18. The scale bars are 5 μm. (B) Check experiments were performed by amniocentesis and G-banding karyotyping of each sample. Representative pictures of chromosomal karyotype analysis are shown comparatively.
(A) A 122kb chromosome 18q21.1 deletion (Chr 18: 46,888,182-47,010,397) was detected by CytoScanTM HD array. The log 2-based test/reference intensity ratios of DNA clones located on chromosome 18 were below -0.5 (black arrow)—the position of the 18q21.1 deletion within the DYM gene. (B) A picture is demonstrated that the distribution of genetic CNVs surrounding DYM gene in chromosome 18. The log 2-based maternal peripheral blood sample (MPB)/maternal independent sample (Mat) ratio was calculated; >0 means genetic duplication and <0 means deletion. The gene which has a high grey spots coverage part means a high false positive rate.
Conclusion
In this study, we used biocompatible Ppy nanoparticles to fabricate nanostructure microchips and conjugated the device with a highly specific antibody anti-CD147 to isolate fNRBCs from maternal peripheral blood for NIPT of certain birth defects. These biocompatible biotin-doped Ppy nanoparticles formed 3D nanostructures that increase antibody bio-conjugation as well as cell-microchip interactions. Together with highly specific anti-CD147 (validated by flow cytometry and Western blots) as the capture agent, fNRBC in maternal peripheral blood were distinguished from monocytes and effectively captured using our device. Electric stimulation was used to release the captured fNRBCs from the Ppy microchips that could be retrieved for downstream prenatal diagnosis. fNRBCs were validated for their fetal origin using three color immunofluorescent staining and FISH analysis. The fNRBCs could be used to identify fetal abnormalities. 12 cases of chromosome aneuploidy samples were diagnosed using FISH, while WES detected the microdeletion syndrome in fNRBCs. These NIPT results obtained from fNRBCs of maternal peripheral blood were validated by conventional diagnostics. We believe this method has great potential in the development of molecular diagnostics for non-invasive prenatal testing of birth defects, especially for those genetic disorders that are difficult to diagnose by cffDNA-based NIPTs.
Abbreviations
fRNBC: fetal nucleated red blood cell; NIPT: noninvasive prenatal testing; cffDNA: cell-free fetal DNA; BMI: body mass index; CD71: transferrin receptor; CD36: thrombospondin receptor; GPA: glycophorin A; DGC: density gradient centrifugation; FACS: fluorescence-activated cell sorting; MACS: magnetic-activated cell sorting; PMMA: polymethyl methacrylate; PC: polycarbonate; PDMS: polydimethylsiloxane; Ppy: polypyrrole; ECM: extracellular matrix; FISH: fluorescence in situ hybridization; WES: whole exome sequencing; DMSO: dimethyl sulfoxide; NP-40: nonyl phenoxypolyethoxylethanol; SSC: saline sodium citrate; PBS: phosphate buffered saline; LEM: lymphocyte separation medium; NaDBS: sodium dodecylbenzene sulfonate; EDC: 1-ethyl-3-(3- dimethylaminopropyl) carbodiimide; NHS: N-hydroxysuccinimide; PFA: paraformaldehyde; BSA: bovine serum albumin; DAPI: 4,6-diamidino-2-phenylindole dihydrochloride; EDTA: ethylenediaminetetraacetic acid; PE: phycoerythrin; FITC: fluorescein isothiocyanate; FDA: fluorescein diacetate; PI: propidium iodide; PVDF: polyvinyl difluoride; RT: room temperature; ITO: indium tin oxide; CA: chronoamperometry; ε-HbF: εchain of hemoglobin; CD147: a member of a super family in immune globulin; SEM: scanning electron microscopy; AFM: atomic force microscopy; CMA: chromosomal microarray; CNVs: copy number variations.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
We thank Key Laboratory of Artificial Micro- and Nano-Structures of Ministry of Education of Wuhan University and Outpatient Department of Obstetrics and Gynechology of Zhongnan Hospital of Wuhan University for fabrication of nanostructure substrates and samples collection, respectively. This project was funded by National R&D Program for Major Research Instruments (No. 81527801) and Hubei Province Sci-Tech Support Program (No. 2015BCA310).
Competing Interests
The authors have declared that no competing interest exists.
References
1. He C, Liu L, Chu Y. et al. National and subnational all-cause and cause-specific child mortality in China, 1996-2015: a systematic analysis with implications for the Sustainable Development Goals. Lancet Glob Health. 2017;5:e186-e97
2. Driscoll DA, Gross S. Clinical practice: Prenatal screening for aneuploidy. N Engl J Med. 2009;360:2556-62
3. Schueler PA, Yamanishi DT, Pearson J. et al. Inconsistency of fetal trophoblast cells in first trimester maternal peripheral blood prevents non-invasive fetal testing using this cell target. Placenta. 2001;22:702-15
4. Lo YM, Corbetta N, Chamberlain PF. et al. Presence of fetal DNA in maternal plasma and serum. Lancet. 1997;350:485-7
5. Van Schendel RV, Van EI CG, Pajkrt E. et al. Implementing non-invasive prenatal testing for aneuploidy in a national healthcare system: global challenges and national solutions. BMC Health Serv Res. 2017;17:670-9
6. Ashoor G, Syngelaki A, Wagner M, Birdir C, Nicolaides KH. Chromosome-selective sequencing of maternal plasma cell-free DNA for first-trimester detection of trisomy 21 and trisomy 18. Am J Obstet Gynecol. 2012;206:322.e1-5
7. Zhou Q, Pan L, Chen S. et al. Clinical application of noninvasive prenatal testing for the detection of trisomies 21, 18, and 13: a hospital experience. Prenat Diagn. 2014;34:1061-5
8. Liu L, Li K, Fu X, Chung C, Zhang K. A forward look at non-invasive prenatal testing. Trends Mol Med. 2016;22:958-68
9. Du E, Feng C, Cao Y, Yao Y, Lu J, Zhang Y. Massively parallel sequencing (MPS) of cell-free fetal DNA (cffDNA) for trisomies-21,18 and 13 in twin pregnancies. Twin Res Hum Genet. 2017;20:242-9
10. Lench N, Barrett A, Fielding S. et al. The clinical implementation of non-invasive prenatal diagnosis for single-gene disorders: challenges and progress made. Prenat Diagn. 2013;33:555-62
11. Lutgendorf MA, Stoll KA, Knutzen DM, Foglia LM. Non-invasive prenatal testing: limitations and unanswered questions. Genet Med. 2014;16:281-5
12. Kantak C, Chang CP, Wong CC, Mahyuddin A, Choolani M, Rahman A. Lab-on-a-chip technology: impacting non-invasive prenatal diagnostics (NIPD) through miniaturization. Lab Chip. 2014;14:841-54
13. Choolani M, Donoghue KO, Talbert D. et al. Characterization of first trimester fetal erythroblasts for non-invasive prenatal diagnosis. Mol Hum Reprod. 2003;9:227-35
14. Huang R, Barber TA, Schmidt MA. et al. A microfluidics approach for the isolation of nucleated red blood cells (NRBCs) from the peripheral blood of pregnant women. Prenat Diagn. 2008;28:892-9
15. Hromadnikova I, Karamanov S, Houbova B, Hridelova D, Kofer J, Mrstinova M. Non-invasive fetal sex determination on fetal erythroblasts from the maternal circulation using fluorescence in situ hybridization. Fetal Diagn Ther. 2002;17:193-9
16. Al-Mufti R, Hambley H, Farzaneh F. et al. Distribution of fetal and embryonic hemoglobins in fetal erythroblasts enriched from maternal blood. Haematologica. 2001;86:357-62
17. Ziegler BL, Muller R, Valtieri M. et al. Unicellular-unilineage erythropoietic cultures: molecular analysis of regulatory gene expression at sibling cell level. Blood. 1999;93:3355-68
18. Zimmermann S, Hollmann C, Stachelhaus SA. Unique monoclonal antibodies specifically bind surface structures on human fetal erythroid blood cells. Exp Cell Res. 2013;319:2700-7
19. Sitar G, Manenti L, Farina A. et al. Characterization of the biophysical properties of human erythroblasts as a preliminary step to the isolation of fetal erythroblasts from maternal peripheral blood for non-invasive prenatal genetic investigation. Haematologica. 1997;82:5-10
20. Lewis DE, Schober W, Murrell S. et al. Rare event selection of fetal nucleated erythrocytes in maternal blood by flow cytometry. Cytometry. 1996;23:218-27
21. Busch J, Huber P, Pfluger E, Miltenyi S, Holtz J, Radbruch A. Enrichment of fetal cells from maternal blood by high gradient magnetic cell sorting (double MACS) for PCR-based genetic analysis. Prenat Diagn. 1994;14:1129-40
22. Easley CJ, Karlinsey JM, Bienvenue JM. et al. A fully integrated microfluidic genetic analysis system with sample-in-answer-out capability. Proc Natl Acad Sci U S A. 2006;103:19272-7
23. Zhang C, Xing D. Miniaturized PCR chips for nucleic acid amplification and analysis: latest advances and future trends. Nucleic Acids Res. 2007;35:4223-7
24. Bhagat AA, Hou HW, Li LD, Lim CT, Han J. Pinched flow coupled shear-modulated inertial microfluidics for high-throughput rare blood cell separation. Lab Chip. 2011;11:1870-8
25. Lee D, Sukumar P, Mahyuddin A, Choolani M, Xu G. Separation of model mixtures of epsilon-globin positive fetal nucleated red blood cells and anucleate erythrocytes using a microfluidic device. J Chromatogr A. 2010;1217:1862-6
26. Mohamed H, Turner JN, Caggana M. Biochip for separating fetal cells from maternal circulation. J Chromatogr A. 2007;1162:187-92
27. Jeon S, Moon JM, Lee ES, Kim YH, Cho Y. An electroactive biotin-doped polypyrrole substrate that immobilizes and releases EpCAM-positive cancer cells. Angew Chem Int Ed Engl. 2014;53:4597-602
28. Cho Y, Borgens RB. The preparation of polypyrrole surfaces in the presence of mesoporous silica nanoparticles and their biomedical applications. Nanotechnology. 2010;21:205102-11
29. Cho Y, Borgens RB. Biotin-Doped Porous Polypyrrole Films for Electrically Controlled Nanoparticle Release. Langmuir. 2011;27:6316-22
30. He ZB, Guo F, Feng C. et al. Fetal nucleated red blood cell analysis for non-invasive prenatal diagnostics using a nanostructure microchip. J Mater Chem B. 2017 [Epub ahead of print]
31. Salas SP, Russo NM. Analysis of the main ethical conflicts in the 2008 declaration of Helsinki and the proposed changes in the new version. Rev Med Chil. 2014;142:475-80
32. Nagrath S, Sequist LV, Maheswaran S. et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235-9
33. Garimberti E, Tosi S. Fluorescence in situ hybridization (FISH), basic principles and methodology. Methods Mol Biol. 2010;659:3-20
34. Zhou H, Yan Y, Song L. The Clinical Application of Fluorescence in Situ Hybridization in Diagnosing Urothelial Carcinoma. Clin Lab. 2016;62:2001-9
35. Stevens KM, Simons A, Rack K, Hasting RJ. Cytogenetic Nomenclature and Reporting. Methods Mol Biol. 2017;1541:303-9
36. Troeger C, Zhong X, Burgemeister R. et al. Approximately half of the erythroblasts in maternal blood are of fetal origin. Mol Hum Reprod. 1999;5:1162-5
37. Yang S, Qi F, Tang C. et al. CD147 promotes the proliferation, invasiveness, migration and angiogenesis of human lung carcinoma cells. Oncol Lett. 2017;13:898-904
38. Crosnier C, Bustamante LY, Bartholdson SJ. et al. Basigin is a receptor essential for erythrocyte invasion by Plasmodium falciparum. Nature. 2011;480:534-7
39. Gao T, Li L, Wang B, Zhi J, Xiang Y, Li G. Dynamic Electrochemical Control of Cell Capture-and-Release Based on Redox-Controlled Host-Guest Interactions. Anal Chem. 2016;88:9996-10001
40. Hu G, Guan R, Li L. Nucleated red blood cell count in maternal peripheral blood and hypertensive disorders in pregnant women. Am J Med Sci. 2016;351:140-6
41. Practice Bulletin No. 162: Prenatal Diagnostic Testing for Genetic Disorders. Obstet Gynecol. 2016;127:e108-22
Author contact
![]() Corresponding authors: Professor Yuanzhen Zhang, Department of Obstetrics and Gynechology, Zhongnan Hospital of Wuhan University, Wuhan, Hubei, 430071, China. E-mail: zhangyuanzhensina.com; Professor Xingzhong Zhao, Key Laboratory of Artificial Micro- and Nano-Structures of Ministry of Education, School of Physics and Technology, Wuhan University, Wuhan, 430072, Hubei, China. E-mail: xzzhaoedu.cn.
Corresponding authors: Professor Yuanzhen Zhang, Department of Obstetrics and Gynechology, Zhongnan Hospital of Wuhan University, Wuhan, Hubei, 430071, China. E-mail: zhangyuanzhensina.com; Professor Xingzhong Zhao, Key Laboratory of Artificial Micro- and Nano-Structures of Ministry of Education, School of Physics and Technology, Wuhan University, Wuhan, 430072, Hubei, China. E-mail: xzzhaoedu.cn.
 Global reach, higher impact
Global reach, higher impact