13.3
Impact Factor
Theranostics 2018; 8(2):505-517. doi:10.7150/thno.21967 This issue Cite
Research Paper
Magnetic Nanowire Networks for Dual-Isolation and Detection of Tumor-Associated Circulating Biomarkers
1. Biomarker Branch, National Cancer Center, 323 Ilsan-ro, Ilsan-dong-gu, Goyang, Gyeonggi 10408, Korea;
2. Center for Lung Cancer, National Cancer Center, 323 Ilsan-ro, Ilsan-dong-gu, Goyang, Gyeonggi 10408, Korea;
3. Department of Cancer Biomedical Science, Graduate School of Cancer Science and Policy, 323 Ilsan-ro, Ilsan-dong-gu, Goyang, Gyeonggi 10408, Korea;
4. Department of Medical Science, Yonsei University College of Medicine, 50 Yonsei-Ro, Seodaemun-Gu, Seoul 03722, South Korea;
5. Department of Internal Medicine, Seoul National University Hospital, 101 Daehak-ro, Jongno-gu, Seoul 03080, Korea;
6. Genopsy Inc., HongWoo BD 103-B, 373, Kangnamdaero, Seocho-Gu, Seoul 06621, Korea.
* These authors contributed equally to this work.
Received 2017-7-17; Accepted 2017-10-21; Published 2018-1-1
Abstract
Purpose: Recent developments in genomic and molecular methods have revolutionized the range of utilities of tumor-associated circulating biomarkers in both basic and clinical research. Herein, we present a novel approach for ultrasensitive extraction of cfDNA and CTCs, at high yield and purity, via the formation of magnetic nanowire networks.
Materials and Methods: We fabricated and characterized biotinylated cationic polyethylenimine and biotinylated antibody cocktail-conjugated magnetic polypyrrole NWs (PEI/mPpy NW and Ab cocktail/mPpy NW, respectively). We applied these NWs to the extraction of cfDNA and CTC from the blood of 14 patients with lung cancer. We demonstrated reliable detection of EGFR mutations based on digital droplet PCR analysis of cfDNA and CTC DNA from patients with lung cancer.
Results: The NW networks confined with a high density of magnetic nanoparticles exhibited superior saturation magnetization, which enabled rapid and high-yield capture whilst avoiding or minimizing damage and loss. The NW networks enabled the co-isolation of CTCs and cfDNA of high quality and sufficient quantities, thus allowing the amplification of rare and low-prevalence cancer-related mutations.
Conclusion: The simple, versatile, and highly efficient nanowire network tool allows sensitive and robust assessment of clinical samples.
Keywords: Circulating cell-free DNA, circulating tumor cells, lung cancer, blood, plasma, conducting polymer, nanowire.
Introduction
Recently, numerous efforts have been directed towards the development of blood-based diagnostics or “liquid biopsies” to monitor prognostic and predictive responses in cancer [1-3]. Significant advances in molecular genomics have enabled the analysis of tumor-specific circulating biomarkers (i.e., cell-free DNA (cfDNA) and circulating tumor cells (CTCs)), allowing the monitoring of real-time cancer progression and dynamic changes in response to treatment [4-8]. Identifying clinically relevant molecular and genetic alterations through the analysis of blood has great benefits in determining treatment decisions with improved clinical outcomes; therefore, liquid biopsy is of great potential utility in routine clinical applications. In recent years, significant technical advances towards the identification and detection of rare tumor biomarkers have been achieved. Various methods, such as magnetic beads, silicon-column binding, and phenol-chloroform separation, have been used to develop methodologies for the isolation of cfDNA from plasma [9-13]. In addition, a number of techniques, including immuno-magnetic selection, microfluidic devices, and density gradient- or size-based filtration, have been utilized for isolating CTCs from peripheral blood for further molecular characterisation [14-17]. Although CTC and cfDNA extraction techniques have matured, more advanced and reliable methods are still needed for their successful implementation in widespread applications. Currently, despite the potential value of liquid biopsy, tissue biopsy is still considered the gold standard to monitor disease progress and to detect the presence of tumor-associated molecular alterations. However, when the primary tumor tissue is difficult to biopsy, serial repetitive biopsies are not available, or a comprehensive understanding of tumor heterogeneity and evolution is needed, a tissue biopsy may not be practical. Accordingly, to successfully adopt liquid biopsies in clinical practice, a highly efficient platform capable of isolating significantly larger amounts of cfDNA and CTCs from peripheral blood is preferentially required. The lack of reliable and high-yield strategies for extracting rare tumor-specific cfDNA and CTCs has resulted in the inability to acquire useful information about cancer pathogenesis and its treatment, subsequently hampering clinical evaluation and implementation of therapeutic strategies. We anticipate that the availability of high-quality and -quantity cfDNA and CTCs from the peripheral blood of patients with cancer should enable the precise analysis of blood-based molecular and genomic expression profiles. This requires the development of sensitive techniques to efficiently isolate and analyze extremely rare biomarkers without any damage and loss during blood processing. In particular, co-analysis of cfDNA and CTC should enable the acquisition of robust and reliable clinical data and minimize the discrepancies in sensitivity, reproducibility, and concordance between liquid and tissue biopsies, ultimately allowing more in-depth examination of individual patients.
In our previous studies, we developed an electroactive conducting polymer platform (i.e., polypyrrole (Ppy)), where reversible transition in the oxidized and reduced states of the polymeric chains was employed for ultrasensitive isolation of cfDNA and CTCs from blood samples of patients with cancer [18-20]. In more recent studies, we presented a strategy for extracting CTCs using magnetic nanowires (NWs) and demonstrated the excellent performance of these NWs in processing and analyzing CTCs from the blood of patients, even in early-stage non-metastatic breast cancer [21]. By combining two types of epithelial and mesenchymal markers (i.e., EpCAM, epidermal growth factor receptor (EGFR), TROP-2, vimentin, and N-cadherin) on the NW surface, we not only significantly increased the likelihood of capturing CTCs with high phenotypic variation but also minimized the loss of EpCAM-negative CTCs during the process. By extending this principle, we propose a conceptually novel approach for the recovery of tumor-specific cfDNA and CTCs, which involves the use of magnetic NW networks: long and thread-like NWs can move freely in various directions in the blood without being disrupted by steric hindrance, enabling navigation and penetration into even complex blood components (i.e., red blood cells, white blood cells, platelets, etc.). NWs therefore offer the advantage of increased ability to identify, access, and capture rare tumor-specific circulating biomarkers with significantly enhanced contact frequency and duration. In the current study, we developed a highly efficient dual-extraction strategy for tumor-specific cfDNA and CTCs by using NW networks, and we applied it to clinical samples from cancer patients to evaluate overall concordance with tissue analysis data in a proof-of-concept study.
Materials and Methods
Materials
Pyrrole, branched polyethyleneimine (PEI, molecular weight, ~25,000), iron oxide (II, III), poly(sodium-4-styrenesulfonate) (PSS), 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC), N-hydroxysuccinimide (NHS), streptavidin, and Triton X-100 were obtained from Sigma-Aldrich. AAO membrane of 200 nm pore diameter (Anodisc 13) was purchased from Whatman (Maidstone, UK). Biotinylated-anti-EpCAM, biotinylated anti-EGFR, biotinylated-anti-N-cadherin, biotinylated-anti-TROP-2, biotinylated anti-vimentin, anti-EpCAM, and anti-CD44 were obtained from R&D Systems (Minneapolis, MN, USA). CD45 antibody was purchased from Abcam (Cambridge, United Kingdom) and bovine serum albumin (BSA) was obtained from Bovogen (Keilor East VIC, Australia). Alexa 488-goat anti-mouse IgG, Alexa 568-goat anti-rabbit IgG, Alexa 647-goat anti-rabbit IgG, Hoechst 33342, and penicillin/streptomycin (P/S) were purchased from Thermo Scientific (Waltham, MA, USA). Phosphate-buffered saline (PBS), Dulbecco's modified Eagle's medium (DMEM) and Roswell Park Memorial Institute (RPMI)-1640 medium were obtained from WelGene (Daegu, Korea).
Preparation of biotinylated cationic polyethylenimine (PEI/mPpy NW) or biotinylated antibody cocktail (Ab cocktail/mPpy NW)-conjugated magnetic NWs
One side of the AAO template was sputter-coated with an Au layer (~250 nm thick) at 5 × 10-3 mbar and 30 mA for 600 s using a Q150T Modular Coating System (Quorum Technologies, UK). All electrochemical experiments were carried out with a potentiostat/galvanostat (BioLogic SP-50) in which an Au-coated AAO template, platinum wire, and Ag/AgCl (3.0 M NaCl) were employed as the working, counter electrodes, and reference respectively. For preparation of the biotin-doped and magnetic nanoparticle (MNPs)-integrated polypyrrole NWs (biotin-doped mPpy NWs), 30 µL of MNP solution (5 mg/mL; ~10 nm in diameter) was incubated on top of the Au-coated AAO membrane to expedite infiltration into the AAO pores, with aspiration at room temperature (RT). Ppy was then electrochemically polymerized within the pores of the AAO template in a solution of 0.01 M poly(sodium 4-styrenesulfate) and 0.01 M pyrrole containing biotin (1 mg/mL), by applying chronoamperometry (1.5 V vs. Ag/AgCl) for 7 min. The resulting AAO templates were cleaned several times with distilled water, immersed in 2 M NaOH for 3 h, and sonicated in an ultrasonic bath for 10 min at RT to obtain free Ppy NWs doped with MNPs and biotin molecules. Subsequently, the resulting biotin-doped mPpy NWs were immersed into 30 mM N-(3-dimethylaminopropyl)-N-ethylcarbodiimide hydrochloride and 6 mM N-hydroxysuccinimide to activate the carboxylic acid groups. The NWs were then incubated with streptavidin (10 mg/mL) for 45 min and rinsed three times with water. For the preparation of antibody cocktail-conjugated magnetic NWs (Ab cocktail/mPpy NW), streptavidin-labeled and biotin-doped mPpy NWs were incubated for an additional 12 h in biotinylated antibody cocktail (i.e., biotinylated anti-EpCAM, biotinylated anti-EGFR, biotinylated anti-N-cadherin, biotinylated anti-TROP-2, and biotinylated anti-vimentin (10 mg/mL in PBS)) at 4°C. The streptavidin-terminated PpyMNWs were immersed for an additional 1 h in 0.1 mM biotinylated PEI solution for the preparation of cationic polyethylenimine-conjugated NWs (PEI/mPpy NW). After washing with distilled water followed by magnetic separation, the resulting NWs were dispersed in ultrapure water and stored at 4°C until use.
Characterization of the NWs
The morphology of the mPpy NWs was investigated using a field emission scanning electron microscope (JSM 7800F, JEOL) and field emission transmission electron microscope (Tecnai G2 F30ST, FEI). The magnetic characteristics of the PpyMNWs were examined using a SQUID vibrating sample magnetometer (MPMS 3, Quantum Design). The applied magnetic field ranged from 70 to -70 kOe. The transverse relaxation time, T2, was analyzed using a 7 T MRI instrument (Bruker BioSpin MRI GmbH, Billerica, MA, USA; echo time [TE] = 6.5 ms and repetition time [TR] = 1,600 ms).
Evaluation of DNA capture efficiency of PEI/mPpy NW
Initially, various amounts of low- (10-100 bp), mid- (100 bp-2 kb), and high-range (3.5-21 kb) DNA ladders were spiked into Tris- ethylene diaminetetraacetic acid (EDTA) buffer (10 mM) at various concentrations (0.01-1,000 ng/mL). Immediate DNA capture was carried out by adding PEI/mPpy NW (2.5 μg) to the solutions containing the DNA ladder, with shaking, at room temperature for 30 min. Then, the tubes were placed onto a magnetic rack for 15 min to separate unbound or non-specifically bound components and capture DNA-bound magnetic NWs. The isolated DNA was quantified using the Picogreen assay and electrophoresed on a 2% agarose gel; this was followed by staining with ethidium bromide for visualization using a UV transilluminator.
Extraction of cfDNA from patient blood
For extraction of cfDNA, PEI/mPpy NW (150 μg) was added to 300 µL of plasma, followed by micromixing for 30 min at RT. After magnetic separation, the cfDNA-NW complexes were incubated in 500 µL of proteinase K-containing lysis buffer consisting of 1.28 M sucrose, 40 mM Tris-HCl, 20 mM MgCl2, 4% Triton X-100, and 50 mM DTT for 30 min at 60°C. The cfDNA-NW complexes were washed twice with 1× TE buffer, collected by magnetic separation, and incubated in 20 µL of 10 mM Tris-HCl (pH 10) for 30 min to extract cfDNA from MNWs. Finally, the cfDNA was recovered by magnetic separation and preserved at -20°C until use for analysis. We additionally extracted cfDNA from plasma using the QIAamp circulating nucleic acid kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Briefly, 1 mL of plasma sample was mixed with 800 µL of buffer ACL and 100 µL of proteinase K. After incubation at 60°C for 30 min, the mixture was filtered through a silica membrane column by applying vacuum pressure. Following precipitation of the column using buffer AW1, AW2, and EtOH, DNA fragments were eluted into 60 µL of AVE buffer.
Droplet digital PCR (ddPCR) analysis
ddPCR was performed to detect EGFR mutations in cfDNA using a QX200 ddPCR system according to the manufacturer's protocol (BioRad, Hercules, CA, USA). Reactions were carried out in 20 µL volumes using 11 µL of ddPCR supermix no UTP (BioRad), 2 µL of ddPCR probe mix (EGFR L858R or E746-A750 del, BioRad), and 8 µL of template. After generating the droplets, 40 µL of the droplets were transferred into a 96-well PCR plate. PCR was performed using the following protocol: 95°C for 10 min, 40 cycles of 94°C for 30 s and 58°C for 1 min, followed by 98°C for 10 min and holding at 4°C. After PCR, the amplified cfDNA alleles were detected and quantified using QuantaSoft software (BioRad) with the thresholds for detection set manually based on results from negative/positive control using genomic DNA from A549, H1975, or HCC2279 cells. For ddPCR analysis after treatment with DNase or RNase, PEI/mPpy NWs were added into 300 μL of cancer patient plasma to isolate cfDNA and were mixed gently for 30 min at room temperature. The resulting PEI/mPpy NWs were magnetized for 10 min by incubating them on a magnetic separation rack at RT. Subsequently, the samples were precipitated by 200 μL of 1× TE buffer and treated with DNase I (Qiagen, Hilden, Germany) or RNase A (Intron Biotechnology, Seoul, Korea), respectively, prior to ddPCR. In a final reaction volume of 10 μL, the samples were digested for 20 min at 37°C with 3.75 U DNase or 10 μg of RNase, respectively. ddPCR was performed using the following thermal cycling protocol: 95°C for 10 min, 40 cycles of 94°C for 30 s and 55°C for 1 min. Primers and probes were purchased from BioRad Laboratories.
Cell culture and reagents
Colon cancer cells (HCT116 cells), human breast cancer cells (MCF7 cells), and pancreatic cancer cells (MIA PaCa-2 cells) were purchased from the American Type Culture Collection, grown in DMEM or RPMI-1640 medium supplemented with 10% fetal bovine serum and 100 U/mL penicillin/streptomycin, and maintained in a humidified incubator under 5% CO2 at 37°C.
Cell capture and release from PBS and blood samples
We evaluated cell capture by the Ab cocktail/mPpy NW using three different cell lines (HCT116, MCF7, and MIA PaCa-2) that were spiked into 0.1% PBS/BSA or peripheral blood samples from healthy donors. Initially, the Ab cocktail/mPpy NW was added to cell suspensions containing various numbers of cells (10, 20, 50, and 100 cells/mL) at a concentration of 5 µg/mL, followed by shaking for 30 min at RT to promote attachment of the target cells to the NWs. Subsequently, a magnetic field adjusted by a magnet was applied to the sample tubes to efficiently separate the captured cells. After removal of the supernatant, the collected cell complex was washed with 1× PBS, resuspended in 4% PFA solution, and transferred to a glass slide. The isolated cells were incubated at 37°C overnight for immunofluorescence staining.
Collection of plasma from patient blood
For clinical application, whole blood from healthy donors or patients with lung cancer was collected in EDTA Vacutainer tubes (Becton and Dickinson) according to the National Cancer Center Institutional Review Board guidelines. The collected blood was centrifuged at 3,000 × g for 10 min to separate plasma. The plasma was stored in cryostat tubes at -80°C until use.
Isolation of circulating tumor cells from patient blood
Whole blood (1 mL) was diluted 5-fold with 0.1% PBS/BSA. Ab cocktail/mPpy NW (5 µg/mL) were added to the diluted blood followed by shaking for 30 min at room temperature. After magnetic separation for 30 min, the supernatant was removed to isolate CTCs. Subsequently, the isolated CTCs were washed twice with 1× PBS, immersed in 4% PFA, and transferred on to a glass slide. The resulting CTCs were incubated at 37°C overnight for subsequent immunocytochemistry or immunohistochemistry analysis.
Immunofluorescence and immunohistochemistry
After relocating the captured CTCs, the cells were permeabilized with 0.1% Triton X-100 for 7 min. Subsequently, antibodies against EpCAM and Vimentin were incubated on the glass slide for 30 min. Then, Alexa Fluor 488/568-conjugated (Invitrogen; green signal for EpCAM, red signal for Vimentin or CD44) secondary antibodies were added, and the coverslips were incubated for another 15 min. Then antibodies against CD45 were added for 15 min, and Alexa Fluor 647-conjugated (Invitrogen; violet signal for CD45) secondary antibodies were incubated for 7 min. Finally, nuclear DNA (blue signal) was stained with Hoechst 33342 (1 µL/ 10 mL) for 5 min. Labeled cells were examined under a Zeiss Imager. Immunohistochemistry was carried out using the EnVision+ System, Peroxidase (DAKO EnVision+ System, HRP) from DAKO according to the manufacturer's instructions. The blood samples were mounted on glass slides and scanned at 400 × magnification using an Olympus BX51 microscope (Tokyo, Japan) linked with total imaging solution software (IMTcamCCD5 PLUS, IMT i-Solution Inc., ON, Canada).
Results and Discussion
NW network synthesis
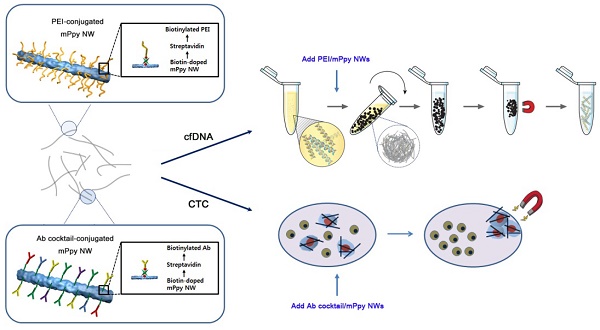
Ppy NWs were synthesized by entrapping large amounts of biotin and magnetic nanoparticles (~10 nm; MNPs) into a polymeric backbone via electrodeposition (i.e., biotin-doped mPpy NWs). The biotin-doped mPpy NWs serve as versatile immobilization matrices for further modification with biotinylated derivatives, such as biotinylated cationic polyethylenimine (PEI/mPpy NW) or biotinylated antibody cocktails (Ab cocktail/mPpy NW), to allow recognition of and interaction with specific targets. Furthermore, the elongated structure of NWs (an average length of 18 μm and diameter of 200 nm) endows large surface-to-volume ratios, which offer great flexibility in incorporating a new assay with the addition of desired target ligands via the biotin-streptavidin interaction. As shown in Figure 1, ultrasensitive isolation of cfDNA and CTCs may be achieved via in situ formation of multiple layers of NW networks. Individual NWs possessing a high density of MNPs exhibited a high level of saturation magnetization (Ms = 84 emu/g) and magnetic resonance imaging contrast (R2 = 45 mMFeS-1) (Figure S1). This finding is attributed to the synergistically enhanced magnetism resulting from confined integration of densely packed MNPs in the NW matrix during electrochemical deposition [22-24].
NW network application to cfDNA extraction from plasma samples
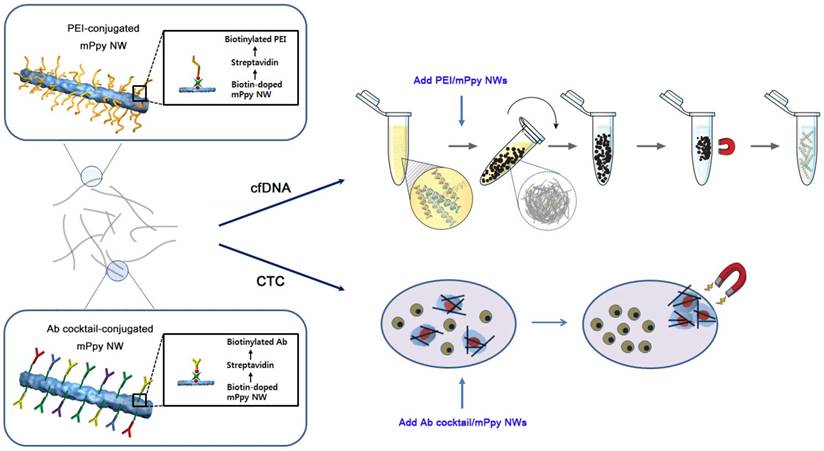
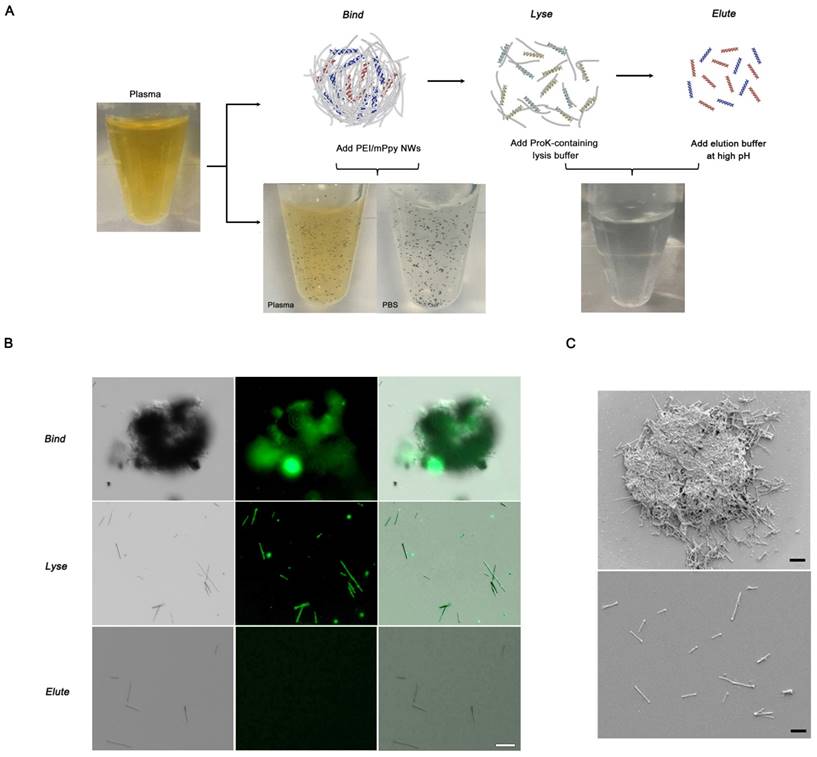
Next, we applied the NW networks to cfDNA extraction from plasma samples from patients with lung cancer through in situ formation of three-dimensional interconnected NW networks. Interestingly, black particle-like structures of various sizes and shapes were immediately observed upon addition of PEI/mPpy NWs to plasma, which could be seen even with the naked eye (Figure 2a). It is likely that positively charged PEI-conjugated NWs induce strong electrostatic binding and condensation of negatively charged DNA. The assembly of DNA-NW conjugates into macro- and micro-aggregates protects the captured and trapped DNA from degradation. We assumed that the spontaneous packaging of these DNA-NW complexes is induced and stabilized by a set of DNA-binding histones and non-histone proteins as a result of an interplay of a number of interactions such as van der Waals forces, hydrogen bonding, and electrostatic interactions. Our results confirm that PEI/mPpy NWs efficiently bind extremely low levels of cfDNA in the plasma, maintaining these as NW networks by tightly organizing them into globular aggregates. The NWs mimic the principle of fishing nets, which are composed of a mesh formed by interconnected thin threads. Unlike conventional DNA extraction procedures that are based on a standard lyse-bind-elute principle, NW networks bind cfDNA and stabilize it into a particulate complex. Interestingly, a round and entangled state was maintained even when the aggregates were transferred into PBS solution.
Successive addition of proteinase K-containing lysis buffer enables the conversion of the agglomerated structures into a homogeneous distribution of individual NWs by eliminating and purifying protein residues around DNA as an efficient lysis step (Figure 2a). The use of elution buffer at high pH results in the release of purified cfDNA from the NWs. As indicated by fluorescence microscopy, the overall sequence of this process was as follows: i) the association of NWs into globular structures in the presence of DNA, ii) the dissociation of the aggregates into individual DNA-NW conjugates in a proteinase K-containing lysis buffer, and iii) the release of DNA molecules from the NWs in Tris-HCl buffer at pH 10.0 (Figure 2b). This modified protocol (i.e., bind-lyse-elute) ensures improved isolation of high-quality, small-fragmented cfDNA while minimizing the possible loss of key components. As DNA-specific dyes (i.e., PicoGreen) selectively bind double-stranded DNA, fluorescent signals indicate the direct attachment of negatively charged cfDNAs via the formation of bead-like structures. As expected, the addition of PEI/mPpy NWs to plasma from patients with lung cancer immediately triggered the aggregation of individual NWs into small masses and globules of different shapes and sizes. However, treatment with proteinase K-containing lysis buffer instantly produced a uniform dispersion of DNA-NW conjugates that exhibited a strong green fluorescence along the entire surface of the NWs. The addition of elution buffer resulted in the immediate cessation of fluorescence emission, indicating complete release and recovery of the cfDNAs attached to the NWs. Similarly, high-resolution SEM images indicated the presence of tightly packed NW networks with relatively smooth exterior surfaces (Figure 2c). Similar to the results described earlier, treatment with lysis buffer resulted in the immediate disassembly of the aggregates into their individual NWs.
Magnetic nanowire (NW) networks for ultrasensitive isolation and analysis of circulating tumor-specific cell-free DNA and circulating tumor cells. Schematic representation depicting co-isolation of circulating cell-free DNA (cfDNA) and circulating tumor cells (CTCs) using NWs synthesized by encapsulating large quantities of biotin and magnetic nanoparticles (~10 nm) into a polypyrrole polymeric backbone during electropolymerisation (i.e., biotin-doped mPpy NWs). The biotin-doped mPpy NWs offer great flexibility in design, allowing them to be functionalized with biotinylated polyethylenimine (PEI/mPpy NW) or biotinylated antibody cocktails (Ab cocktail/mPpy NW) via the biotin-streptavidin interaction.
Assessment of cfDNA capture efficiency of NW networks
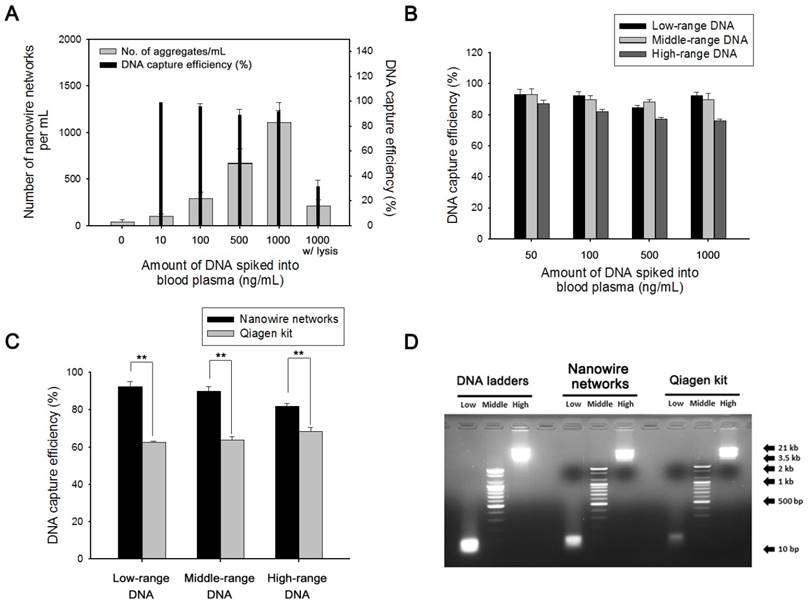
Next, we attempted to elucidate the correlation between the appearance of globular and fibrillar aggregates and DNA capture efficiency as a function of the amounts of DNA ladders (100 bp) that were spiked ex vivo into the plasma of healthy donors. Consistent with the previous results, the addition of the NWs immediately initiated the formation of tightly coiled clumps, where the mean number of aggregates formed was directly proportional to the concentration of DNA, with a recovery rate of over 90% regardless of the amount of the spiked DNA (Figure 3a). As the plasma of a healthy individual contains a small amount of innate DNA, a very small number of aggregates were generated even when no DNA was spiked. Interestingly, pre-treatment with proteinase K-containing lysis buffer led to significant decrease in the formation of aggregates as well as DNA recovery yield, suggesting that proteins wrapped around DNA are involved in the formation of NW networks by helping package the DNA and NWs into coiled structures, rather than interfering with DNA capture.
Additionally, we attempted to explore the relationship between NW length, the formation of NW networks, and DNA capture efficiency by ex vivo spiking DNA ladders (100 bp, 250 ng/mL) in the plasma of three healthy donors. As shown in Figure S2b, the formation of NW networks as well as DNA recovery was strongly correlated with NW length. Indeed, long NWs (~18 µm) possessing a large surface area and surface functional sites permit the attachment of large quantities of DNA fragments and, subsequently, allow DNA-NW conjugates to assemble into a highly tangled network. Meanwhile, 3 µm long NWs did not effectively form globular clusters because of their short length, resulting in relatively low DNA extraction efficiency (Figure S2b, c). Clearly, 18 µm long NWs appeared to be optimal for extracting cfDNA by densely packing DNA-NW conjugates into globular structures for identifying EGFR mutation against a background of wild-type DNA, using droplet digital PCR (ddPCR) (Figure S2d).
We further validated the capability of NW networks in DNA recovery by spiking DNA ladders of three different sizes (i.e., 10-100 bp (low range), 100-2,000 bp (mid-range), and 3.5-21 kb (high range)) into 300 µL of healthy plasma (Figure 3b). As tumor-derived cfDNAs are typically present in the bloodstream, with a size distribution of roughly 100-200 bp, it is necessary to maximize the capture yield of low- and mid-range DNA. [25] Notably, NW networks provided a substantial improvement in DNA adsorption, especially in low- and mid-range DNA ladders, with high recovery rates of >90%, regardless of the amount of DNA spiked into the plasma. The slightly lower binding of high-range DNA fragments is most likely owing to the decrease in binding affinity to cationic NWs as a consequence of natural looped and tangled state of long DNA molecules.
In a comparison of our method with the currently widely used QIAamp circulating nucleic acid kit (Qiagen kit), the performance of the NW networks was found to be far superior to that of the Qiagen kit; this was demonstrated by ex vivo spiking of DNA ladders of various sizes (250 ng/mL) into 1 mL of plasma (Figure 3c and d). With regard to low- and mid-molecular-weight DNA, although some minor loss was observed, NW networks enabled a much higher recovery yield than the Qiagen kit. Furthermore, visualization of the captured DNA by agarose gel electrophoresis demonstrated consistency with the overall results.
cfDNA extraction through in situ formation of three-dimensional interconnected NW networks. (a) The long and thin features of NWs facilitate the development of globular NW networks consisting of cfDNA-NW conjugates. The addition of proteinase K-containing lysis buffer immediately facilitates the dispersion of the NWs coated with cfDNA, promoting the recovery of cfDNA from the NWs in Tris-HCl buffer. The photographs show the presence of NW networks formed in plasma and PBS, and individually dispersed NWs in a proteinase K-containing lysis buffer. (b) Fluorescence images depicting a series of processes: i) NW networks self-assembled in plasma from a patient with lung cancer, ii) uniform distribution of cfDNA-NW conjugates in a proteinase K-containing lysis buffer, and iii) the resulting NWs after efficient elution of cfDNA from the NWs. (c) Scanning electron microscopy image of NW networks spontaneously formed in samples from patients with cancer (top), and uniformly distributed cfDNA-NW conjugates in a proteinase K-containing lysis buffer (bottom) (scale bar, 10 µm).
Validation of NW networks using DNA ladders of different lengths spiked in human plasma. (a) Evaluation of the relationship between the number of NW networks and DNA capture efficiency as a function of the amounts of DNA ladders (100 bp) that are spiked ex vivo into the plasma of healthy donors. (b) DNA capture efficiencies were dependent on the amount of DNA ladders (250 ng/mL) of 3 sizes: 10 bp (low range), 100 bp (mid-range), and 3.5 kb (high range), that were spiked into healthy plasma. (c) Comparison of DNA capture efficiency between NW networks and the Qiagen kit. DNA ladders (250 ng/mL) of 3 sizes were spiked into plasma from a healthy donor. **p < 0.005. (d) Agarose gel electrophoresis of DNA ladders of 3 sizes: 10 bp (low range), 100 bp (mid-range), and 3.5 kb (high range), that were spiked into healthy plasma. NW networks made up of cfDNA-NW conjugates were lysed by treatment with proteinase K-containing lysis buffer and subsequently treated with Tris-HCl to efficiently release cfDNA from the NWs. Eluted cfDNA was collected and analyzed by 2% agarose gel electrophoresis. Control (left lanes), DNA markers; NW networks (middle lanes); Qiagen kit (right lanes). The data shown represent the mean ± SD of data from 5 independent experiments.
Clinical utility of NW networks to isolate cfDNA
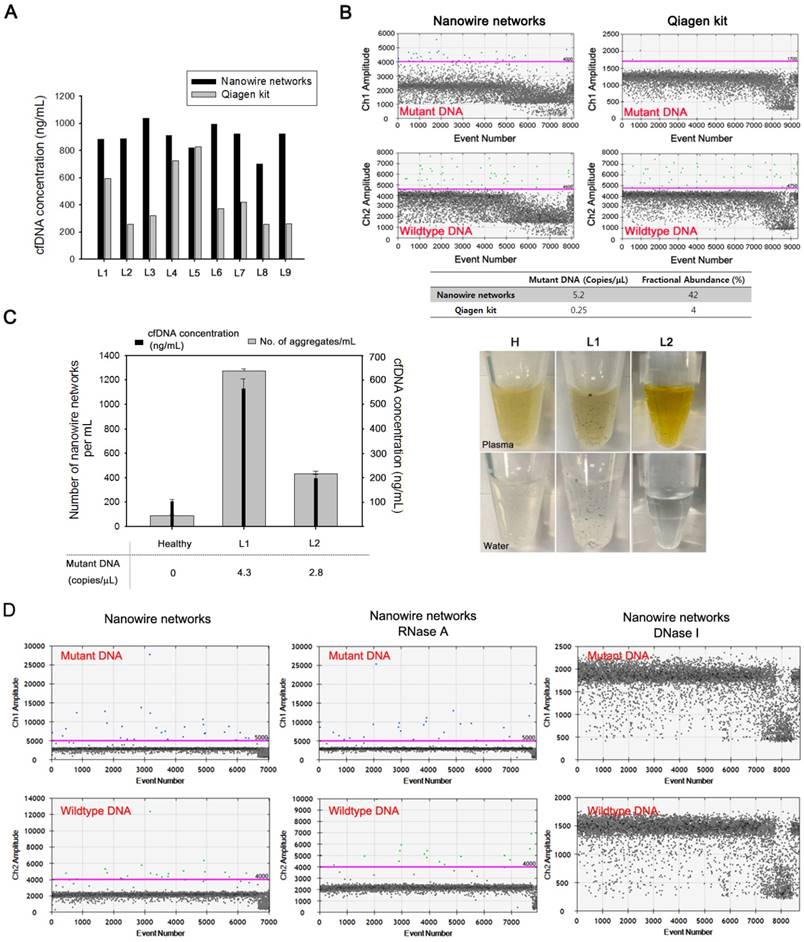
The clinical utility of the NW networks was tested by assessing their ability to isolate cfDNA from the plasma of patients with lung cancer. The DNA yield was directly compared with that obtained using the Qiagen kit (Figure 4a). The NW networks were found to be capable of extracting much higher amounts of cfDNA than the Qiagen kit from 9 lung cancer plasma samples. The availability of a large amount of cfDNA enabled more effective detection of genetic alterations in the EGFR gene of cancer patients by using ddPCR (Figure 4b). Unlike conventional cfDNA isolation methods that guarantee reasonable results only if more than 1 mL of blood plasma and an appropriate amount of cfDNA in the sample are available for analysis, our approach yields reliable results for relatively small clinical samples, even with plasma amounts of as low as 300 µL. The procedure, processing time, cost, and minimum sample volume required for the cfDNA extraction by using NW networks and Qiagen kit are summarized in Table 1.
We subsequently explored the mutation status of cfDNA by directly analyzing cfDNA-NW conjugates by ddPCR to avoid loss of the captured DNA, which generally occurs during the elution process, and to ensure sufficient amplification of target DNA. The same EGFR L858R mutation as in tumor tissue was identified by ddPCR before and after releasing the DNA from the NW in the cfDNA-NW conjugates, as shown in Figure S3. Using this approach, we identified 5.2 copies/µL for EGFR exon 21 (L858R) with a fractional abundance of 42% in cfDNA from patient L6; this is consistent with findings for the corresponding tumor tissue. A relatively low sensitivity for mutations was detected in the cfDNA isolated using the Qiagen kit.
Using the patient's blood, we investigated the relationship between the number of NW networks and cfDNA concentration (Figure 4c). As expected, the concentration of cfDNA recovered directly correlated with the number of black aggregates that could be seen easily with the naked eye in both plasma and water. Therefore, NW networks enable highly efficient isolation of small-fragmented, tumor-specific cfDNA, as well as the identification of specific oncogenic mutations against a background of wild-type DNA, thereby offering a highly clinically relevant and reliable method.
The cfDNA-NW conjugates that are generated as a consequence of a number of synergistic interactions might contain non-specifically bound, negatively charged blood components, including cell-free RNA (cfRNA). To identify whether the presence of cfRNA directly affects mutation detection, we treated cfDNA isolated from the plasma of lung cancer patients by NW networks with enzymes, DNase I or RNase A, which specifically digest either RNA or DNA. The total numbers of mutant and wild-type DNA molecules detected were similar upon the treatment with RNase A, whereas no mutation signal was observed after digestion with DNase I (Figure 4d). These ddPCR results confirmed that the presence of other negatively charged components that are non-specifically adsorbed on the surface of NW networks in the blood do not affect overall sensitivity of EGFR exon 20 (T790M) mutation detection.
Clinical utility of NW networks to isolate rare CTCs
We additionally applied this strategy to the isolation of rare CTCs from peripheral blood of patients with lung cancer. First, we optimized and evaluated the performance of the Ab cocktail/mPpy NW using cell lines (i.e., EpCAM-positive MCF7 cells and EpCAM-negative MIA PaCa-2 cells) that were spiked ex vivo into PBS or whole blood, and we achieved a capture yield of ~90% (Figure S4). Indeed, as demonstrated in a previous study, the use of five different types of antibodies resulted in significantly higher isolation efficiency regardless of the EpCAM expression levels in tumor cells [21]. As with cfDNA extraction, the ability of the NWs to capture CTCs was mainly attributed to their long and thin morphology, which provides the structural and chemical cues required for CTC recovery efficiency: the elongated structure of the NWs not only significantly decreases the inherent steric hindrance, but also increases the duration and surface area of contact with cancer cells as the NWs wrap themselves around the cells. This offers the distinct advantage of very high affinity for cancer cells, resulting in maximum CTC recovery with minimal cell loss (Figure 5a).
CTCs captured by the NW networks were verified by dual-identification immunocytochemistry (ICC) and immunohistochemistry (IHC) (Figure 5b, c). CTCs are identified based on the expression of epithelial origin markers (DAPI+/EpCAM+/CD45- expression), which differentiates these cells from surrounding leukocytes with DAPI+/EpCAM-/CD45+ expression. Interestingly, we found that the majority of CTCs were characterised by the expression of EMT markers (e.g., CD44 or vimentin) whilst retaining their epithelial properties, supporting the occurrence of epithelial-mesenchymal transition in metastatic cells [26, 27]. Cells captured by NW networks were further detected by conventional IHC assessment, in which CTCs were stained with the epithelial marker EpCAM and then counterstained with hematoxylin.
Next, we attempted to verify tumor-specific genomic mutations in CTCs by ddPCR (Figure 5d). In general, genomic DNA (gDNA) from CTCs is often more accessible and reliable for gene expression profiling than cfDNA, as DNA stability and integrity are essential for direct amplification and detection of target mutants. To assess the feasibility of CTCs for the assessment of EGFR mutations, we attempted to identify these mutations (exon 19 deletion), using NW networks, in CTCs extracted from 5 mL of whole blood from patients with advanced lung cancer. The same mutation type was confirmed in both CTCs and the primary tumors, with high abundance of EGFR mutations. mRNA transcripts for the most commonly used epithelial markers, cytokeratin-19 (CK-19) and EGFR, were effectively detected by reverse transcriptase polymerase chain reaction (RT-PCR) and quantitative real-time PCR (qPCR) (Figure 5e).
As a result of tumor heterogeneity across the entire tumor population, the clinical utility of CTCs is limited in addressing the entire tumor population. However, our results confirmed that NW networks are capable of isolating large quantities of CTCs, with high sensitivity and specificity. This enables the collection of pure, intact, and high-quality gDNA, which is crucial for molecular and genomic profiling of primary tumor tissue. Further, we demonstrated ultrasensitive extraction of both cfDNA and CTCs from the plasma of patients with non-small-cell lung cancer through the formation of NW networks. We analyzed CTC gDNA and cfDNA obtained from 14 patients with lung cancer using ddPCR and identified the presence of mutant EGFR alleles, as found in their tumor biopsies in 14 (100%) of the 14 cfDNA and 10 (71%) of the 14 CTC gDNA samples, respectively (Table 2). NW networks offer great advantages over conventional methods in that i) they allow highly sensitive dual extraction of cfDNA and CTCs via flexible and versatile surface modification and ii) they dramatically improve the accuracy and reliability in assessing small quantities of genetic mutants through integrated profiling of cfDNA and CTC gDNA.
NW networks as a highly efficient platform for isolation and analysis of cfDNAs from plasma of patients with lung cancer. (a) Comparison of the concentration of cfDNA eluted from plasma of patients with lung cancer using NW networks and the Qiagen kit. (b) Detection of EGFR exon 21 L858R mutation in cfDNA isolated from plasma of patients with lung cancer, L6, using NW networks and the Qiagen kit. The mutation status of cfDNA was assessed using droplet digital PCR (ddPCR). (c) Assessment of the correlation between the number of NW networks and the concentration of cfDNA isolated, and cfDNA, as measured by ddPCR, in copies of EGFR mutation/µL. The photographs show in situ formation of NW networks in plasma of healthy control (H) and patients with lung cancer (L1, L2). When aggregates formed in plasma are transferred to water, they still do not dissolve. (D) Detection of EGFR exon 20 T790M mutation in cfDNA isolated from plasma of patients with lung cancer using NW networks (left) and evaluation of the mutation status after the introduction of enzymes, RNase (middle) or DNase (Right), in the cfDNA solution. The mutation status of cfDNA was assessed using droplet digital PCR (ddPCR).
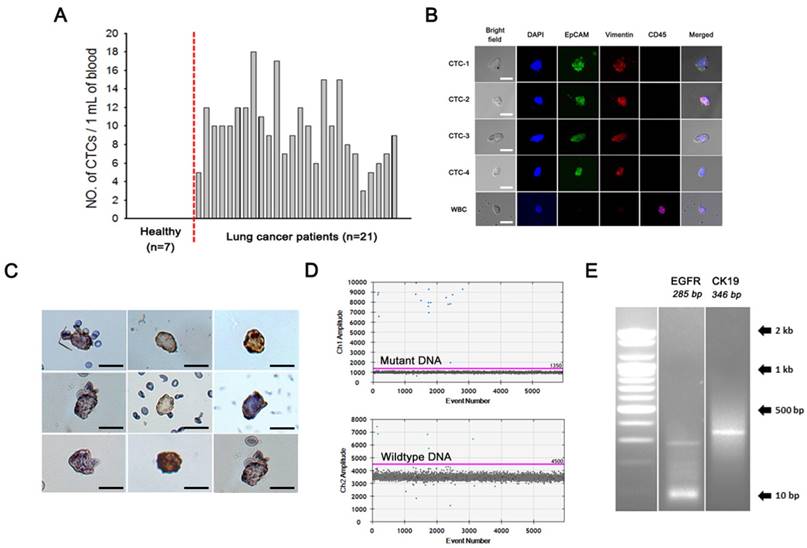
Comparison of processing time, steps, cost, and minimum volume of plasma between the magnetic NW method and a conventional cfDNA sample preparation method.
| Magnetic nanowire | QIAamp Circulating Nucleic Acid kit | |
|---|---|---|
| procedure | 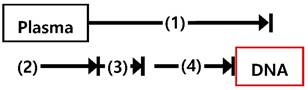 | 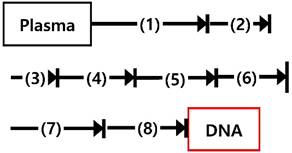 |
| cfDNA capture by nanowire networks, 30 min Place the tube (1) in a magnetic separation rack and wash with lysis buffer, 5 min Place the tube (2) in a magnetic separation rack and wash with TE 1x buffer, 15 min Elute cfDNA from nanowire networks, 5 min | Plasma lysis by buffer ACL for 30 min at 60℃ Add binding buffer and incubation for 5 min on ice Vacuum filtration, 15 min 3 wash steps, 3 min Centrifuge, 3 min Incubate 10 min at 56℃ Immerse into elution buffer and incubate 3 min Centrifuge, 1 min | |
| Processing time | 55 min | 70 min |
| Cost (per sample) | 3 $ | 30 $ |
| Minimum volume of plasma | 300 uL | 1 mL |
Evaluation of CTCs captured by NW networks from the whole blood of patients with lung cancer. (a) Measurement of circulating tumor cells (CTCs) isolated from 1 mL of blood from 7 healthy donors and 21 patients with lung cancer. (b) Immunocytochemistry (ICC) images of CTCs, where CTCs were stained with DAPI (blue), CD45 (violet), EpCAM (green), and vimentin (red) (scale bar, 20 µm). (c) Immunohistochemical (IHC) staining of CTCs using anti-EpCAM with diaminobenzidine (DAB) and hematoxylin staining. Cancer cells show dark brown staining as a result of the DAB-substrate reaction, while nuclei stain blue as a result of hematoxylin counterstaining (scale bar, 20 µm). (d) Representative detection of mutation status (EGFR exon 19 deletion) of genomic DNA in CTCs, as indicated by ddPCR. (e) Assessment of the expression of EGFR and CK19 mRNAs in CTCs isolated from patients with lung cancer.
Proof-of-concept studies of the usability of NW networks to co-isolate and analyze circulating cfDNA and CTCs in the plasma of patients with non-small-cell lung cancer.
| Sample ID | Stage | Primary tumor EGFR mutation | cfDNAs | CTCs | |||||
|---|---|---|---|---|---|---|---|---|---|
| Concentration (ng/mL) | Mutation type | Mutant allele, copies/mL | CTCs / mL | Mutation type | Mutant allele of CTCs, copies/mL | ||||
| NSCLC-1 | IV | L858R | 6687.1 | L858R | 2200 | 10 | L858R | 350 | |
| NSCLC-2 | IV | L858R | 2200.5 | L858R | 900 | 9 | L858R | 800 | |
| NSCLC-3 | III | L858R | 1289.4 | L858R | 550 | 12 | L858R | 500 | |
| NSCLC-4 | IV | Exon 19 Del | 1228.2 | Exon 19 Del | 2900 | 7 | Exon 19 Del | 3400 | |
| NSCLC-5 | IV | L858R | NA | L858R | 1700 | 12 | L858R | 300 | |
| NSCLC-6 | IV | L858R | NA | L858R | 7900 | 10 | L858R | 900 | |
| NSCLC-7 | IV | Exon 19 Del | NA | Exon 19 Del | 280 | 10 | - | 0 | |
| NSCLC-8 | IV | Exon 19 Del | 738.7 | Exon 19 Del | 430 | 12 | - | 0 | |
| NSCLC-9 | III | Exon 19 Del | NA | Exon 19 Del | 650 | 4 | - | 0 | |
| NSCLC-10 | IV | Exon 19 Del | 1486.0 | Exon 19 Del | 2200 | 18 | Exon 19 Del | 490 | |
| NSCLC-11 | III | L858R | 1561.0 | L858R | 520 | 12 | L858R | 2000 | |
| NSCLC-12 | IV | Exon 19 Del | 888.1 | Exon 19 Del | 2900 | 7 | Exon 19 Del | 1200 | |
| NSCLC-13 | IV | L858R | 923.7 | L858R | 1600 | 5 | - | 0 | |
| NSCLC-14 | IV | L858R | 702.6 | L858R | 1200 | 15 | L858R | 600 | |
Conclusion
In the present study, we showed that NW networks represent a straightforward and feasible approach for high recovery of extremely rare circulating biomarkers; the elongated or tube-like morphology of the NWs allows them to actively penetrate complex blood components by significantly promoting accessibility to cfDNA as well as CTCs. Furthermore, NWs confined with a high density of magnetic nanoparticles exhibit superior saturation magnetization, which enables rapid and high-yield capture whilst avoiding or minimizing damage and loss. Accordingly, NW networks enable the co-isolation of CTCs and cfDNA of high quality and sufficient quantities, thus allowing the amplification of rare and low-prevalence cancer-related mutations and greatly improving the concordance rate between data generated from tumor DNA, cfDNA, and CTC gDNA. Rather than relying on the outcome of only one circulating biomarker, the complementary results obtained through co-isolation and -analysis of CTC or cfDNA are likely to yield more accurate and reliable clinical data, which further improves validity and precision in cancer diagnostics and prognostics, and ultimately will help to define the optimal therapy for individual cancer patients.
Abbreviations
CTC: circulating tumor cell; cfDNA: cell-free DNA; NW: nanowire; polyethylenimine: PEI; Ab: antibody; ddPCR: digital droplet PCR; Ppy: polypyrrole; AAO: anionic aluminium oxide; MNP: magnetic nanoparticles; Qiagen kit: QIAamp circulating nucleic acid kit; cfRNA: cell-free RNA; EMT: epithelial-mesenchymal transition; ICC: immunocytochemistry; IHC: immunohistochemistry; RT-PCR: reverse transcriptase polymerase chain reaction; qPCR: quantitative real-time PCR.
Supplementary Material
Supplementary figures.
Acknowledgements
This work was supported by a National Cancer Center grant from the Republic of Korea (1510070-3, 1611170-2) and a grant from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (Grant Number: HI17C0828).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Wan JC, Massie C, Garcia-Corbacho J. et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer. 2017;17:223-238
2. Speicher MR, Pantel K. Tumor signatures in the blood. Nat Biotechnol. 2014;32:441
3. Brock G, Castellanos-Rizaldos E, Hu L, Coticchia C, Skog J. Liquid biopsy for cancer screening, patient stratification and monitoring. Transl Cancer Res. 2015;4:280-290
4. Oellerich M, Schütz E, Beck J. et al. Using circulating cell-free DNA to monitor personalized cancer therapy. Crit Rev Clin Lab Sci. 2017:1-14
5. Maheswaran S, Sequist LV, Nagrath S. et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med. 2008;359:366-377
6. Jamal-Hanjani M, Wilson GA, McGranahan N. et al. Tracking the Evolution of Non-Small-Cell Lung Cancer. N Engl J Med. 2017;376:2109-2121
7. Chi KR. The tumour trail left in blood. Nature. 2016;532:269-271
8. Ignatiadis M, Dawson S-J. Circulating tumor cells and circulating tumor DNA for precision medicine: dream or reality? Ann Oncol. 2014;25:2304-2313
9. Kirsch C, Weickmann S, Schmidt B, Fleischhacker M. An improved method for the isolation of free-circulating plasma DNA and cell-free DNA from other body fluids. Ann N Y Acad Sci. 2008;1137:135-139
10. Beránek M, Vlčková J, Hypiusová V, Živný P, Palička V. Comparison of various methods used for extraction of cell-free genomic DNA from human plasma. Klin Biochem Metab. 2006;14:21-24
11. Sonnenberg A, Marciniak JY, Rassenti L. et al. Rapid electrokinetic isolation of cancer-related circulating cell-free DNA directly from blood. Clin Chem. 2014;60:500-509
12. Xue X, Teare MD, Holen I, Zhu YM, Woll PJ. Optimizing the yield and utility of circulating cell-free DNA from plasma and serum. Clin Chim Acta. 2009;404:100-104
13. Board RE, Williams VS, Knight L. et al. Isolation and extraction of circulating tumor DNA from patients with small cell lung cancer. Ann N Y Acad Sci. 2008;1137:98-107
14. Nagrath S, Sequist LV, Maheswaran S. et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235-1239
15. Yoon HJ, Kim TH, Zhang Z. et al. Sensitive capture of circulating tumour cells by functionalized graphene oxide nanosheets. Nat Nanotechnol. 2013;8:735-741
16. Stott SL, Hsu C-H, Tsukrov DI, Yu M. et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc Natl Acad Sci USA. 2010;107:18392-18397
17. Karabacak NM, Spuhler PS, Fachin F. et al. Microfluidic, marker-free isolation of circulating tumor cells from blood samples. Nat Protoc. 2014;9:694-710
18. Jeon S, Moon JM, Lee ES, Kim YH, Cho Y. An Electroactive Biotin-Doped Polypyrrole Substrate That Immobilizes and Releases EpCAM-Positive Cancer Cells. Angew Chem Int Ed Engl. 2014;126:4685-4690
19. Hong WY, Jeon SH, Lee ES, Cho Y. An integrated multifunctional platform based on biotin-doped conducting polymer nanowires for cell capture, release, and electrochemical sensing. Biomaterials. 2014;35:9573-9580
20. Lee H, Jeon S, Seo J-S, Goh S-H, Han J-Y, Cho Y. A novel strategy for highly efficient isolation and analysis of circulating tumor-specific cell-free DNA from lung cancer patients using a reusable conducting polymer nanostructure. Biomaterials. 2016;101:251-257
21. Hong W, Lee S, Chang HJ, Lee ES, Cho Y. Multifunctional magnetic nanowires: a novel breakthrough for ultrasensitive detection and isolation of rare cancer cells from non-metastatic early breast cancer patients using small volumes of blood. Biomaterials. 2016;106:78-86
22. Park j, Maltzahn G, Zhang L, Schwartz M, Ruoslahti E, Bhatia S, Sailor M. Magnetic iron oxide nanoworms for tumor targeting and imaging. Adv. Mater. 2008;20:1630-1635
23. Huang J, Zhong X, Wang L, Yang L, Mao H. Improving the magnetic resonance imaging contrast and detection methods with engineered magnetic nanoparticles. Theranostics. 2012;2:86-102
24. Chen H, Yeh J, Wang L, Khurshid H, Peng N, Wang A, Mao H. Preparation and control of the formation of single core and clustered nanoparticles for biomedical applications using a versatile amphiphilic diblock copolymer. Nano Res. 2010;3:852-862
25. Jianga P, Chan C, Allen Chan K. et al. Lengthening and shortening of plasma DNA in hepatocellular carcinoma patients. Proc Natl Acad Sci USA. 2015 E1317-E1325
26. Polioudaki H, Agelaki S, Chiotaki R. et al. Variable expression levels of keratin and vimentin reveal differential EMT status of circulating tumor cells and correlation with clinical characteristics and outcome of patients with metastatic breast cancer. BMC Cancer. 2015;15:399
27. Armstrong AJ, Marengo MS, Oltean S. et al. Circulating tumor cells from patients with advanced prostate and breast cancer display both epithelial and mesenchymal markers. Mol Cancer Res. 2011;9:997-1007
Author contact
![]() Corresponding author: Email: ynchore.kr
Corresponding author: Email: ynchore.kr
 Global reach, higher impact
Global reach, higher impact