13.3
Impact Factor
Theranostics 2017; 7(18):4350-4358. doi:10.7150/thno.21727 This issue Cite
Research Paper
Identification and Validation of a Salivary Protein Panel to Detect Heart Failure Early
1. The School of Biomedical Sciences, Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, Queensland, Australia;
2. School of Chemistry and Molecular Biosciences, The University of Queensland, Brisbane, Queensland, Australia;
3. Cardiology Department, Royal Brisbane and Women's Hospital; University of Queensland School of Medicine; School of Nursing, Queensland University of Technology, Brisbane, Australia;
4. Department of Cardiology, Mater Adult Hospital, Brisbane, Queensland, Australia.
Received 2017-7-3; Accepted 2017-8-24; Published 2017-9-26
Abstract
Background: Over 26 million people suffer from heart failure (HF) globally. Current diagnosis of HF relies on clinical evaluation, blood assays and imaging techniques. Our aim is to develop a diagnostic assay to detect HF in at risk individuals within the community using human saliva as a medium, potentially leading to a simple, safe early warning system.
Methods: Saliva samples were collected from healthy controls (n=36) and HF patients (n=75). Salivary proteome profiles were analysed by Sequential Window Acquisition of All Theoretical fragment ion spectra - Mass Spectrometry (SWATH-MS). A total of 738 proteins were quantified and 177 proteins demonstrated significant differences between HF patients and healthy controls. Candidate biomarkers were chosen based on their abundance and difference between the two cohorts. A multi-protein panel was developed using logistic regression analysis. The diagnostic performance of the multi-protein panel was assessed using receiver operative characteristic curves. The candidate proteins were further confirmed, using western blot analysis, and validated technically, using an independent biological cohort.
Results: A group of six proteins were chosen in the discovery phase as potential candidates based on their differences in the abundance between the two cohorts. During the validation phase, two of the proteins were not detected with western blotting and as such were removed. The final panel consists of four proteins with sensitivity of 83.3%, specificity of 62.5% with an area under ROC curve of 0.78 in discriminating healthy controls from NYHA class I/II HF patients, and was validated in a second independent cohort study.
Conclusion: Analysis of salivary proteome using SWATH-MS revealed novel HF-specific protein candidates yielding high diagnostic performance. A multi-centre longitudinal clinical trial will be the next step before clinical implementation of this panel.
Keywords: heart failure, saliva, diagnosis, mass spectrometry, multi-biomarker strategies.
Introduction
Heart Failure (HF) is a clinical syndrome that is associated with cardiac dysfunction or abnormality which impairs the ability of the heart to provide sufficient blood to satisfy the body's needs [1]. HF presents a large social and economic burden due to a high global prevalence, affecting approximately 26 million people worldwide [2]. This becomes a greater problem due to an aging and growing population. The diagnosis of HF is based on clinical criteria, including the history, physical examination and the results of imaging and other investigations [3]. Whilst there are a number of effective treatments [3], given that the symptoms and signs are relatively nonspecific, the diagnosis is often delayed. Hence outcomes remain poor with 17-45% of HF patients dying within 1 year of their first admission to hospital [4]. There is a need for reliable, user-friendly, non-invasive methods to allow detection of HF and ideally cost-effective screening earlier, when treatment may have a more favourable effect on the disease trajectory [5, 6].
In recent years, a number of proteins have been identified and tested as diagnostic or prognostic biomarkers for HF. The plasma/serum concentrations of the N-terminal prohormone of B-type natriuretic peptide (NT-proBNP) and B-type Natriuretic Peptide (BNP) itself are preferred in identifying whether or not dyspnoea is due to HF. However, the use of these natriuretic peptides as a screening assay is not widely accepted, partly due to concerns regarding their specificities, sensitivities, and cost-effectiveness (including reimbursement strategies of medical insurance companies) [7].
Human saliva is gaining attention as an alternative to blood for diagnostics, as it contains a large number of proteins/peptides, which can mirror changes in the circulatory system caused by systemic events. Saliva collection is easy, non-invasive and safe for the collector [8-11]. There is biomolecular transport across salivary acini cells and blood vessel endothelium cells, enabling saliva to be used for detecting systemic events [8]. We have previously detected significantly higher levels of NT-proBNP and galectin-3 in saliva collected from HF patients compared to saliva from healthy controls [12, 13] with acceptable sensitivities and specificities. Unbiased approaches of biomarker discovery have been widely discussed as a means to identify potential candidate biomarkers [14]. It is also important to note that currently there are no means of assessing treatment responses in HF patients in a non-invasive, cost-effective manner [15] and a salivary protein assay could fulfil this unmet clinical need.
Sequential Window Acquisition of All Theoretical fragment ion spectra - Mass Spectrometry (SWATH-MS) is a data independent acquisition (DIA) MS method that allows a complete and permanent recording of all detectable fragment ions from peptide precursors present in a biological sample [16]. SWATH-MS has recently proven successful in fundamental and clinical proteomic research [17-19]. The aims of our study were twofold: to develop a robust SWATH-MS protocol to discover salivary protein candidates in HF and to validate identified proteins using an independent cohort of patients as well as an independent method.
Experimental Procedures
Participants
Research ethics for sample collection have been approved by the human research ethics committee of Mater Adult Hospital (approval number: HREC/13/MHS/142 (1806QA)), The University of Queensland (approval number: 2009000779) and Queensland University of Technology (approval number: 1400000616). Informed consent was obtained from participants before sample collection. Healthy controls, self-reported with no cardiovascular diseases were recruited from university staff, students and the general population.
HF patients were recruited from Mater Adult Hospital and Royal Brisbane and Women's Hospital under the recommendations of cardiologists. HF patients were categorized into New York Heart Association (NYHA) functional class based on clinical symptoms [20] identified by the cardiologist.
Saliva Sample Collection
Participants were asked to refrain from eating and drinking (except for water) for two hours before saliva collection. Resting whole mouth saliva was collected as described previously [21]. In brief, participants were asked to sit in a comfortable position and to rinse their mouths with water. They were asked to tilt their heads down, pool saliva in the mouth for 1 minute and expectorate into a 50 mL Falcon Tube. Saliva samples were aliquoted into 1.5 mL micro centrifuge tubes and stored at -80 °C until analysis.
Saliva Sample Preparation for Mass Spectrometry Analysis
Total protein concentrations of resting whole mouth saliva samples were quantified by PierceTM BCA Protein Assay Kit (Thermo Fisher Scientific Inc, Waltham, MA, USA) as per the manufacturer's protocol.
Saliva samples containing 12.5 µg of total protein were used for the MS analysis as previously described [22]. Proteins in the samples were denatured and reduced by addition of 100 μL of 1% sodium dodecyl sulphate, 100 mM Tris-HCl buffer pH 8.0 and 10 mM Dithiothreitol (DTT), and incubation at 95 °C for 10 min. Cysteines in the samples were then alkylated by addition of 30 mM acrylamide and incubation at room temperature for 30 min. Excess acrylamide was quenched with the addition of DTT to a final concentration of 10 mM. Proteins were then precipitated by addition of 800 µL of 1:1 methanol/acetone, incubation at -20 °C overnight and centrifugation at 18,000 × g for 10 min. The protein pellet was resuspended in 50 µL of 50 mM ammonium acetate and digested with 0.5 µg of proteomics grade trypsin (Sigma-Aldrich, St. Louis, MI, USA) at 37 °C for 16 h. To desalt, clean and concentrate peptides, C18 ZipTips (Merck Millipore, Billerica, MA, USA) were used following manufacturer's instructions.
Salivary Protein Discovery using SWATH-MS
A total of 39 HF patients' (20 NYHA class I/II patients and 19 NYHA class III/IV patients) and 20 healthy controls' saliva samples were analysed in the discovery phase. Liquid chromatography-electrospray ionization-tandem mass spectrometry (LC-MS/MS) was performed using a Prominence nanoLC system (Shimadzu, Kyoto, Japan) coupled to a TripleTof 5600 mass spectrometer with a Nanospray III interface (SCIEX, Framingham, MA, USA) essentially as previously described [23]. In short, approximately 2 µg of peptides were injected and separated on a Vydac EVEREST reversed-phase C18 HPLC column (300 Å pore size, 5 μm particle size, 150 μm i.d. × 150 mm) at a flow rate of 1 μL/min with a gradient of 1-10% buffer B over 2 min followed by 10-60% buffer B over 45 min, with buffer A (1% acetonitrile and 0.1% formic acid) and buffer B (80% acetonitrile with 0.1% formic acid). Gas and voltage setting were adjusted as required. For information-dependent acquisition (IDA), a MS TOF scan from m/z of 350-1800 was performed for 0.5 s followed by IDA of MS/MS with automated Collision Energy selection of the top 20 peptides from m/z of 40-1800 for 0.05 s per spectrum. For SWATH-MS, the LC conditions were the same, and an MS-TOF scan from an m/z of 350-1800 for 0.05 s was performed followed by high-sensitivity IDA with 26 m/z isolation windows with 1 m/z window overlap each for 0.1 s across an m/z range of 400-1250. Collision energy was automatically assigned by the Analyst software (SCIEX) based on m/z window ranges. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [24] partner repository with the dataset identifier PXD007134
Validation of Identity of the Salivary Protein Candidates
Western blotting was performed on six selected proteins. Saliva samples (40 μg total protein) were denatured at 95 °C for 10 min in 1× Laemmli sample Buffer (Bio-Rad, Hercules, CA, USA), loaded into lanes of a 12.5% SDS-PAGE gel and electrophoresed using a Bio-Rad® miniPROTEAN® system with Tris/glycine/SDS buffer system (Bio-Rad). After electrophoresis, the samples were transferred to a PVDF membrane with Mini Trans-Blot wet transfer Module (Bio-Rad). The membranes were blocked with 5% bovine serum albumin in 50 mM Tris-buffered saline containing 0.1% Tween-20 (TBST) for 2 h at RT. Monoclonal antibodies (Anti-Kallikrein-1/KLK1 Antibody, clone 11C7.1, Merck Millipore; Anti-S100A7 Antibody, clone 14F6.1, Merck Millipore; Anti-TCP1 delta antibody [EPR8494(B)], Abcam; Anti-Lipoamide Dehydrogenase antibody [EPR6635], Abcam; Mouse monoclonal A9604D2 Anti-Human IgA2 Fc (HRP), Abcam; Anti-Cathelicidin antibody [mAbcam 58387], Abcam) targeting each candidate protein were used according to manufacturers' recommendations. The membrane was then washed with TBST 3 times for 5 min. Horseradish peroxidase (HRP) -linked goat-anti-mouse IgG secondary antibody or HRP-linked goat-anti-rabbit IgG secondary antibody was used following manufacturer's recommendation expect for A9604D2. The membranes were then washed with TBST 6 times for 5 min. Amersham ECL western Blotting Detection Reagent (GE Healthcare) and a ChemiDoc XRS+ imager was used to visualized the protein bands. IGHA2 was visualized after the first washing process without secondary antibody incubation.
Biological Validation of Identified Salivary Protein Candidates
To determine the biological variability of six selected protein candidates, an independent cohort of samples was included with the same clinical characteristic (n=18 NYHA class I/II, n=18 NYHA class III/IV and n=16 healthy controls). Mass spectrometry with SWATH acquisition was performed on these samples as described above.
Data Analysis
Peptide identification was performed by the Protein Pilot 4.1 software (SCIEX) using the LudwigNR database (downloaded from http://apcf.edu.au as at 27 January 2012; 16 818 973 sequences; 5 891 363 821 residues) with the following setting: Sample Type, identification; cysteine alkylation, acrylamide; instrument, TripleTof 5600; Species, human; ID focus, Biological modification; enzyme, trypsin; Search effort, thorough ID. ProteinPilot was used to perform false discovery rate (FDR) analysis for all searches. Further analysis was performed on peptides identified with greater than 99% confidence and with a local FDR of less than 1%. The abundance of peptides was measured using PeakView Software with standard settings essentially as described [25], summing the integrated areas of up to six fragment ions per peptide. Protein abundance was measured using the sum of the abundances of up to six peptides per protein. The accuracy of peak selection by PeakView was manually confirmed for proteins of interest in each sample.
All statistical analyses were performed using Prism GraphPad (GraphPad Software Inc., La Jolla, CA, USA) and R [26]. Protein significance analysis was performed by applying a linear mixed-effects model using MSstats [27]. This type of analysis is a procedure that combines the quantitative measures for a targeted protein across peptides, charge states, transitions, samples, and conditions; the system detects proteins that change in abundance among conditions more systematically than would be expected by random chance, while controlling the FDR. The differences of the abundance level of proteins between HF patients' and healthy controls' saliva samples were classified and ranked based on the significance and fold-changes of the candidate proteins.
The diagnostic performance of candidate protein was evaluated by receiver operator characteristic (ROC) curve analysis. Multivariate ROC curves [28] were generated to evaluate the diagnostic performance of the multi-proteins panel. A logistic regression predictive model was applied to the candidate markers to calculate a predictive score for each individual sample. The performance of the predictive model was validated with bootstrap validation method by applying the model generated with a random selection of 70% of the data set and applying it to a different set of data generated also by random selection of 80% of the total data set. Western blot analysis was done with Image Lab (Bio-Rad, Version 5.2.1). Band detection was performed with Low Sensitivity setting.
Results
Participants
In total 75 HF patients and 36 healthy individuals were recruited in this study. All study participants were >18 years of age and gave written consent. The clinical characteristics of the participants are listed in Table 1.
Salivary Protein Discovery using SWATH-MS
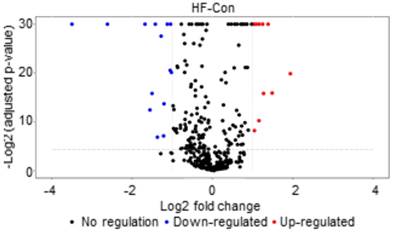
The identified MS/MS spectra were compiled into a spectral library, which contained 738 proteins; 7926 peptides; and 101 234 fragment ions (Supplementary Table 1). In the discovery phase, we identified 430 unique proteins using Information Dependent Acquisition LC-MS/MS (Supplementary Table 2, Total Ion Chromatograph Overlay is provided in Supplementary Figure 1). The protein abundances were then measured with SWATH-MS. When comparing saliva collected from HF patients (NYHA class I/II) with saliva from healthy controls, 177 proteins were found to be significantly (p<0.05) different between the two cohorts. Out of those proteins, 12 proteins were two times more abundant in saliva from HF patients than saliva from controls and 17 were two times more abundant in controls than in HF patients. A volcano plot (Figure 1) was generated by plotting the log-fold changes for the identified proteins against their corresponding adjusted p-value.
The summary of anthropometric parameters for heart fialure patients and healthy controls
| Parameter | NYHA class I/II | NYHA class III/IV | Healthy Controls | P |
|---|---|---|---|---|
| n=39 | n=37 | n=36 | ||
| Age | 63 (29-87) | 70 (38-97) | 56 (41-92) | 0.15 |
| Gender (M:F) | 29:10 | 22:15 | 19:17 | 0.17 |
| Body Mass Index (kg/m2) | 31 | 35 | 24 | 0.30 |
| Previous Acute Coronary Syndrome | 8 (20.5%) | 14 (37.8%) | N/A | 0.10 |
| Hypertension | 12 (30.8%) | 16 (43.2%) | N/A | 0.26 |
| Type 2 Diabetes | 6 (15.4%) | 18 (48.6%) | N/A | 0.0032 |
| Chronic Obstructive Pulmonary Disease | 6 (15.4%) | 6 (16.2%) | N/A | 0.92 |
| ST Elevate Myocardial Infraction | 12 (30.8%) | 10 (27.0%) | N/A | 0.72 |
| Non- ST Elevate Myocardial Infraction | 4 (10.3%) | 4 (10.8%) | N/A | 0.94 |
| Creatinine (mg/dL) | 95 (78-128) | 104 (77-134) | N/A | 0.59 |
The P values refer to the comparison between NYHA class I/II HF patients and NYHA class III/IV patients using Mann-Whitney rank test, unpaired t-test, or Pearson chi-squared test, as appropriate. The statistically significant P values are bolded.
Volcano plot comparing NYHA class I/II HF patients and healthy controls. X-axis: model-based estimate of log-fold change. Y-axis: statistical significance, FDR-adjusted p-values on the negative log2 scale. The dot lines represent the cut off (p value<0.01, fold change >1.5).
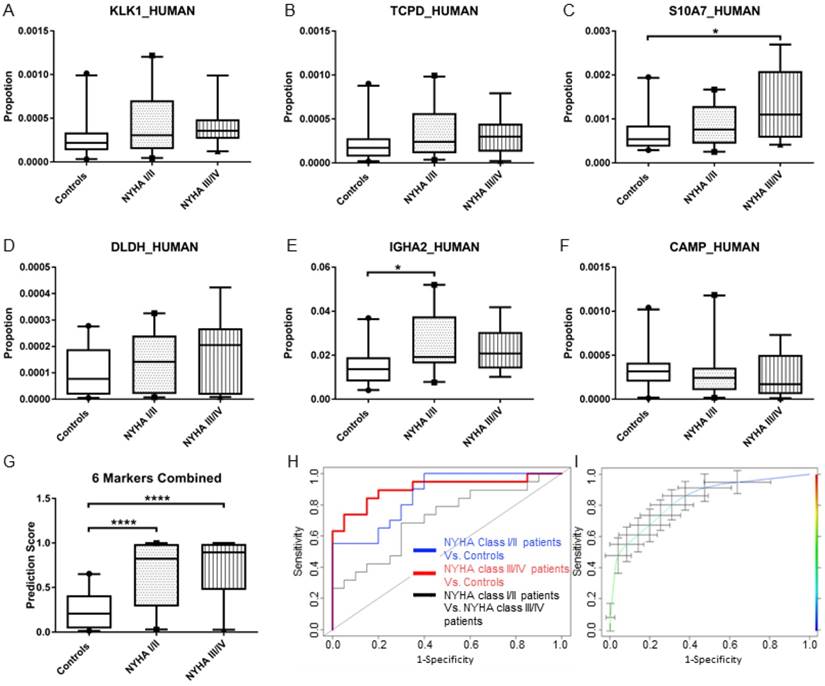
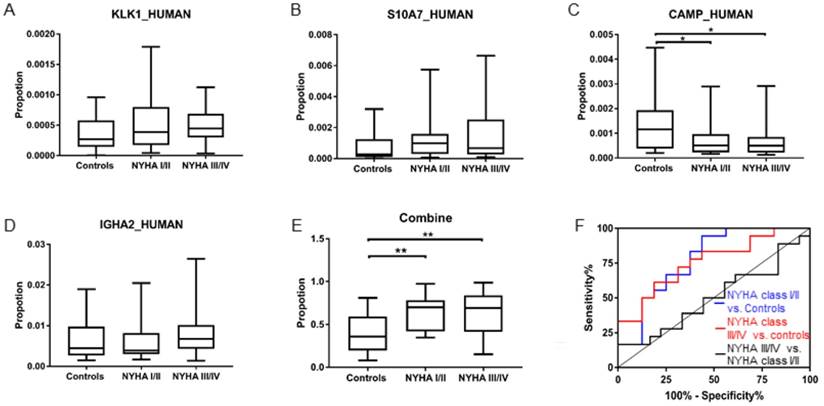
Potential protein candidates Kallikrein-1 (KLK1), T-complex protein 1 subunit delta (TCPD), Protein S100-A7 (S10A7), Dihydrolipoyl dehydrogenase, mitochondrial (DLDH), Immunoglobulin heavy constant alpha 2 (IGHA2) and Cathelicidin antimicrobial peptide (CAMP) were chosen based on their fold change, adjusted P-value calculated by MSstats using the transition-specific data generated by PeakView, as well as their biological relevance. The protein abundance of the candidates in each sample was normalized to the total protein abundance of the individual samples and was plotted as 5-95 percentile whisker plots (Figure 2A-F). An increase in protein abundance (KLK1, TCPD, S10A7, DLDH and IGHA2) was observed in saliva samples from HF patients compared with saliva from healthy controls, and a decrease in CAMP protein abundance. Statistically significant differences (p<0.05) were observed between saliva samples from early class HF patients (NYHA class I/II) compared with saliva samples from healthy controls for IGHA2. In contrast, the abundance of S10A7 was significantly different between saliva from late class HF patients compared with healthy controls.
Multivariate ROC curve analyses were performed to combine these protein candidates and to generate a prediction score for individual samples. Statistically significant differences in the prediction score values (p<0.0001) were observed in protein abundance differences in saliva from NYHA class I/II HF patients compared with saliva from healthy controls as well as saliva from NYHA class III/IV HF patients and healthy controls (Figure 2G). Area Under Curve (AUC) value of the multivariate ROC curve (Figure 2H) for NYHA class I/II HF patients vs. controls and NYHA class III/IV HF patients vs. controls were 0.82 and 0.91 respectively. The diagnostic performance of the multi-marker protein panel was further refined using the bootstrap validation method (Figure 2I). The average AUC value, sensitivity, specificity, positive and negative predictive value at the optimal cut-off point (where sensitivity and specificity are closest) were 0.84, 77.1%, 71.4%, 73.2% and 76.0% respectively.
(A)-(F) Box and whisker (5-95 percentile) plot of the protein abundance of the candidate biomarkers. (G) Box and whisker (5-95 percentile) plot of the prediction score of the multi-biomarker panel generated by logistic regression. (H) Multivariate ROC curve of the multi-biomarker (KLK1, TCPD, S10A7, DLDH, IGHA2 and CAMP) panel for the comparison between the three different cohorts. (I) Bootstrap validation of the multi-biomarker (KLK1, TCPD, S10A7, DLDH, IGHA2 and CAMP) panel. Average sensitivity and specificity with standard error was plotted.
Validation of Identity of the Salivary Protein Candidates
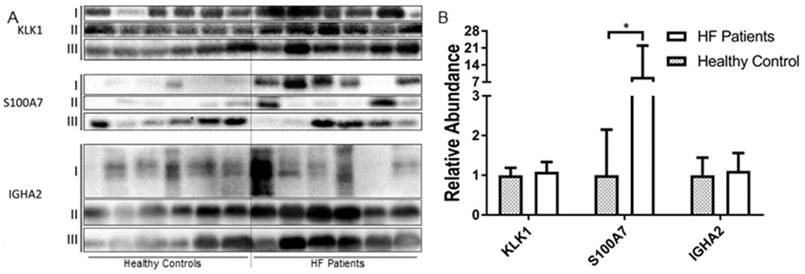
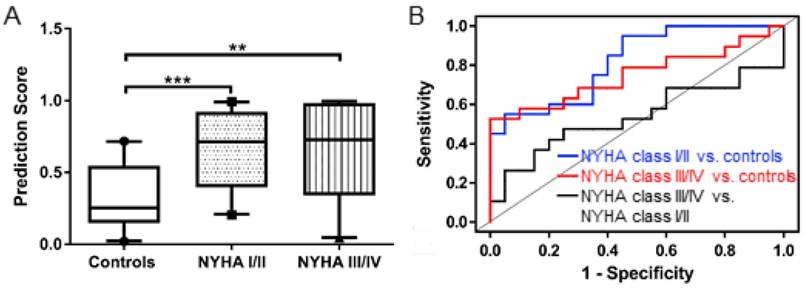
To validate the candidate proteins, western blotting analysis was performed using randomly collected saliva samples from healthy controls (n=18) and and HF patients (n=9 NYHA class I/II patients and n=9 NYHA class III/IV patients). The intensity of each sample was normalized against the average band intensity of the loading control (β-actin) (Figure 3B). As shown in Figure 3A, S10A7 and IGHA2 were detected in individual saliva samples collected from both patients and controls. S10A7 was detected in 13 out 18 HF patients' saliva samples and only 10 out 18 healthy control saliva samples (with weak diffuse bands detected in the remaining samples). Similar to the SWATH-MS data, KLK1, S10A7 and IGHA2 showed higher protein abundance in the saliva samples from HF patients compared to saliva samples from healthy controls. The average band intensity for S10A7 was 9 times higher (p<0.05) in saliva samples from HF patients compared with saliva from healthy controls. IGHA2 had higher abundance in saliva samples from HF patients compared to healthy controls (1.12:1) but no significant difference was observed. In contrast to our findings from the initial screening (Figure 2A), the abundance of KLK1 in saliva from HF patients compared with the saliva from healthy controls gave similar results (1.087:1, Figure 3). CAMP abundance was also different, with higher abundance in saliva from HF patients compared with saliva from controls (1.43:1). TCPD and DLDH were not detected with western blotting (Supplementary Figure 2). After removal of TCPD and DLDH from the multi-marker protein panel the prediction score generated by the remaining four protein candidates (KLK1, S10A7, IGHA2 and CAMP) showed significant differences between NYHA class I/II HF patients vs healthy controls (p<0.001) and NYHA class III/IV HF patients vs healthy controls (p<0.01, Figure 4A). AUC values of the multivariate ROC curve (Figure 4B) for NYHA class I/II HF patients vs. controls and NYHA class III/IV HF patients vs. controls were 0.82 and 0.80 respectively.
(A) Western Blotting of KLK1, S10A7 and IGHA2 in the saliva sample of 18 healthy controls and 18 HF patients in three blots. (B) Average relative band intensity with standard error of KLK1, S10A7 and IGHA2 in healthy controls and HF patients' samples.
(A) Box and whisker (5-95 percentile) plot of the prediction scores of the multi-biomarker (KLK1, S10A7, IGHA2 and CAMP) panel in the validation cohorts. (B) Multivariate ROC curve of the multi-biomarker (KLK1, S10A7, IGHA2 and CAMP) panel in the validation cohorts.
Biological Validation of Identified Salivary Proteins Candidates
We validated the multimarker panel using an independent cohort of HF patients and healthy controls. The normalized protein abundance is plotted with box and 5-95 percentile whisker plots in Figure 5A-D. The prediction model generated by the four-protein panel (KLK1, S10A7, IGHA2 and CAMP) showed significant differences when comparing NYHA class I/II HF patients to healthy controls (p<0.01) and comparing NYHA class III/IV HF patients to healthy controls (p<0.01, Figure 5D). AUC value of the multivariate ROC curve (Figure 5E) for NYHA class I/II HF patients vs. controls and NYHA class III/IV HF patients vs. controls were 0.78 and 0.76 respectively.
Discussion
Currently, diagnosis of HF remains heavily based on clinical evaluation, blood-based assays and imaging techniques, which rely on the presence of trained staff at the time of clinical evaluation and the early diagnosis and monitoring of HF, especially in individuals living in remote areas and in emerging economies. Previous studies have already reported that blood-based HF proteins can be detected in human saliva and can be used as diagnostic markers [12, 13, 29-32]. These findings indicate that even though saliva is not in direct contact with the heart, it may still be an important medium for detecting systemic conditions such as HF. Saliva may overcome these shortcomings by providing an alternate source for diagnosing and monitoring HF realising precision medicine.
To our knowledge, this is the first study to use SWATH-MS and western blotting to discover and validate salivary proteins with potential diagnostic utility in HF. In the present study, we investigated whether saliva contains HF related proteins that can be used as potential diagnostic biomarkers in HF. In the discovery phase, we identified six potential protein candidates in saliva collected from HF patients compared with saliva from healthy controls. Re-examination of these six proteins using an independent cohort and an independent method, we were able to validate four of the six proteins discovered. Previous research has shown that an increase in the diagnostic performance is achieved when multiple proteins are combined into a single panel [33, 34]. After excluding DLDH and CAMP from the panel, we re-established the multi-protein panel using KLK1, S10A7, IGHA2 and CAMP. This four-protein panel gave relatively good diagnostic performance. The AUC was 0.82, similar to that obtained when using six proteins as a panel. By reducing the number of protein candidates from the diagnostic panel, we also reduce the future cost of using this assay in a multiplex-format in a large multi-centre clinical trial. Our findings indicated that SWATH-MS is a useful and rapid method to discern proteins in complex biological matrices such as saliva.
(A)-(D) Box and whisker (5-95 percentile) plot of the protein abundance of the KLK1, S10A7, CAMP and IGHA2 in the validation cohort. (E) Box and whisker (5-95 percentile) plot of the prediction score of the 4-biomarker panel in the validation cohorts. (F) Multivariate ROC curve of the 4-biomarker panel for the comparison between the three different cohorts.
Among the four proteins validated in this study, KLK1 has previously been associated with HF. KLK1 is a kinin forming serine protease synthesized in many organs, such as the salivary glands, kidneys, pancreas, colon, prostate gland and testis. There are 2 isoforms of KLK1 commonly present, with isoform 2 usually expressed in the salivary glands [35]. Emanueli et al. found that the expression of human KLK1 induced angiogenesis and arteriogenesis in CD1 mice model [36]. A lot of research has focused on the relationship between KLK1 and kinins. Kinins are vasodilators and inhibitors of blood coagulation [37]. Studies [38, 39] investigating the kinins as potential therapeutic targets have found that KLK1 provides cardio-protection in both acute cardiac ischemia and post ischemic HF patients.
The levels of S10A7 was significantly higher in saliva from HF patients compared to the saliva collected from controls (P<0.05). S10A7 has not previously been associated with cardiovascular disease (CVD). It was reported that a high level of S10A7 expression was found to be related with increased angiogenesis in breast cancer patients [40]. This pro-angiogenesis property of S100A7 in HF has not been studied. However, its anti-bacterial activity has been reported [41]. Patients with CVD have poor oral health compared to healthy individuals [42]. The presence of S10A7 in HF patients' saliva is therefore consistent with previous studies. However, the underlying mechanisms relating to S10A7 in the pathogenesis of HF is not clear and warrants further investigation.
Western blotting data revealed, no significant differences in the abundance of IGHA2, KLK1, TCPD, DLDH or CAMP in the saliva samples collected from HF patients compared with saliva from healthy controls. The level of CAMP showed a contrasting trend to SWATH-MS (CAMP in HF cohorts has higher abundance than in controls, p<0.01) when compared to the results of SWATH-MS data). TCPD and DLDH were not detected by western blot. The inconsistencies between western blotting data and SWATH-MS analysis for these proteins may be due to the fact that the epitopes detected by antibodies used in western blotting may not overlap with the sequences detected by mass spectrometry. As such, we have eliminated these two proteins from the panel.
The quest for biomarkers is focused on early diagnosis, disease surveillance, and appropriate delivery of targeted therapies. Nevertheless, the translation of biomarkers from discovery to clinical use has been hampered due to the lack of standardised diagnostic biomarker platforms, the long path from discovery to clinical assays, limited sample availability, and diverse sample preparation strategies. This initial study is hypothesis-generating. In order to successfully translate the findings of the present study into clinical tools, the protein panel will need to be tested in a multi-centre clinical setting using samples collected from individuals at risk of developing HF. As Point-of-Care diagnostic strategies evolve, large numbers of proteins can be measured in a simple device with minimal human intervention [43]. The protein panel in the present study could be translated into such a device once its diagnostic performance is established. Although further validation with a large clinical cohort is necessary, the methodology used in this study is powerful and could easily be adapted for biomarker discovery using other biological fluids.
Abbreviations
HF: Heart failure; Brain Natriuretic Peptide: BNP; N-terminal prohormone of Brain Natriuretic Peptide: NT-proBNP; data independent acquisition: DIA; Sequential Window Acquisition of all Theoretical Mass Spectrometry: SWATH-MS; Dithiothreitol: DTT; New York Heart Association: NYHA; Liquid chromatography-electrospray ionization-tandem mass spectrometry: LC-MS/MS; TBST: Tris-buffered saline containing 0.1% Tween-20: TBST; Horseradish peroxidase: HRP; False discovery rate: FDR; Receiver operator characteristic: ROC.
Supplementary Material
Supplementary table 1.
Supplementary table 2.
Supplementary Figure 1.
Acknowledgements
This study was supported by the Queensland University of Technology VC's start-up funds for CP. CP also holds a QUT VC fellowship. BLS is supported by a National Health and Medical Research Council Career Development Fellowship APP1087975. A QUT PhD scholarship was awarded to XZ. We also thank the clinical staff at the Royal Brisbane Women's Hospital and the Mater Hospital for the collection of clinical samples. The biological validation data reported in this paper were obtained at the Central Analytical Research Facility within the Institute for Future Environments (QUT). We would also like the thank Dr. Dimitrios Vagenas (IHBI, QUT) and Dr. Florian Rohart (Institute for Molecular Bioscience, UQ) for statistical consultation.
Competing Interests
XZ, BS and CP are inventors of a PCT patent application Australian provisional patent application number is 2016903387.
References
1. McDonagh TA, Gardner RS, Clark AL. et al. Oxford Textbook of Heart Failure. OUP Oxford. 2011
2. Bui AL, Horwich TB, Fonarow GC. Epidemiology and risk profile of heart failure. Nat Rev Cardiol. 2011;8:30-41
3. Krum H, Jelinek MV, Stewart S. et al. 2011 update to National Heart Foundation of Australia and Cardiac Society of Australia and New Zealand Guidelines for the prevention, detection and management of chronic heart failure in Australia, 2006. Med J Aust. 2011;194:405-9
4. Ponikowski P, Anker SD, AlHabib KF. et al. Heart failure: preventing disease and death worldwide. ESC Heart Failure. 2014;1:4-25
5. McGrady M, Krum H. Screening: The New Frontier in Heart Failure Management. Cardiovasc Ther. 2009;27:1-3
6. Atherton JJ. Stage B heart failure: rationale for screening. Heart Fail Clin. 2012;8:273-83
7. Raymond I, Groenning BA, Hildebrandt PR. et al. The influence of age, sex and other variables on the plasma level of N-terminal pro brain natriuretic peptide in a large sample of the general population. Heart. 2003;89:745-51
8. Pfaffe T, Cooper-White J, Beyerlein P. et al. Diagnostic potential of saliva: current state and future applications. Clin Chem. 2011;57:675-87
9. Ovchinnikov DA, Cooper MA, Pandit P. et al. Tumor-suppressor Gene Promoter Hypermethylation in Saliva of Head and Neck Cancer Patients. Transl Oncol. 2012;5:321-6
10. Zhang X, Wan Y, Cooper-White J. et al. Quantification of D-dimer levels in human saliva. Bioanalysis. 2013;5:2249-56
11. Zhang X, Dimeski G, Punyadeera C. Validation of an immunoassay to measure plasminogen-activator inhibitor-1 concentrations in human saliva. Biochemia medica. 2014;24:258-65
12. Foo J, Wan Y, Kostner K. et al. NT-ProBNP Levels in Saliva and Its Clinical Relevance to Heart Failure. PLoS ONE. 2012;7:e48452
13. Zhang X, Wan Y, Chata R. et al. A pilot study to demonstrate diagnostic potential of galectin-3 levels in saliva. J Clin Pathol. 2016;69:1100-4
14. Mebazaa A, Vanpoucke G, Thomas G. et al. Unbiased plasma proteomics for novel diagnostic biomarkers in cardiovascular disease: identification of quiescin Q6 as a candidate biomarker of acutely decompensated heart failure. Eur Heart J. 2012;33:2317-24
15. Chow SL, Maisel AS, Anand I. et al. Role of Biomarkers for the Prevention, Assessment, and Management of Heart Failure: A Scientific Statement From the American Heart Association. Circulation. 2017;135:e1054-e91
16. Gillet LC, Navarro P, Tate S. et al. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol Cell Proteomics. 2012;11:O111.016717
17. Chang RY, Etheridge N, Nouwens AS. et al. SWATH analysis of the synaptic proteome in Alzheimer's disease. Neurochem Int. 2015;87:1-12
18. Ortea I, Rodriguez-Ariza A, Chicano-Galvez E. et al. Discovery of potential protein biomarkers of lung adenocarcinoma in bronchoalveolar lavage fluid by SWATH MS data-independent acquisition and targeted data extraction. J Proteomics. 2016;138:106-14
19. Hou G, Lou X, Sun Y. et al. Biomarker Discovery and Verification of Esophageal Squamous Cell Carcinoma Using Integration of SWATH/MRM. J Proteome Res. 2015;14:3793-803
20. Dolgin M, Committee NYHAC. Nomenclature and Criteria for Diagnosis of Diseases of the Heart and Great Vessels. Little, Brown. 1994
21. Navazesh M. Methods for collecting saliva. Ann N Y Acad Sci. 1993;694:72-7
22. Xu Y, Bailey UM, Punyadeera C. et al. Identification of salivary N-glycoproteins and measurement of glycosylation site occupancy by boronate glycoprotein enrichment and liquid chromatography/electrospray ionization tandem mass spectrometry. Rapid Commun Mass Spectrom. 2014;28:471-82
23. Xu Y, Bailey UM, Schulz BL. Automated measurement of site-specific N-glycosylation occupancy with SWATH-MS. Proteomics. 2015;15:2177-86
24. Vizcaino JA, Csordas A, del-Toro N. et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44:D447-56
25. Zacchi LF, Schulz BL. SWATH-MS Glycoproteomics Reveals Consequences of Defects in the Glycosylation Machinery. Mol Cell Proteomics. 2016;15:2435-47
26. R Development Core Team. R. Vienna, Austria: R Foundation for Statistical Computing. 2013
27. Clough T, Thaminy S, Ragg S. et al. Statistical protein quantification and significance analysis in label-free LC-MS experiments with complex designs. BMC Bioinformatics. 2012;13(Suppl 16):S6
28. Shultz EK. Multivariate receiver-operating characteristic curve analysis: prostate cancer screening as an example. Clin Chem. 1995;41:1248-55
29. Rahim MAA, Rahim ZHA, Ahmad WAW. et al. Can Saliva Proteins Be Used to Predict the Onset of Acute Myocardial Infarction among High-Risk Patients? Int J Med Sci. 2015;12:329-35
30. Foley JD, Sneed JD, Steinhubl SR. et al. Oral Fluids that Detect Cardiovascular Disease Biomarkers. Oral Surg Oral Med Oral Pathol Oral Radiol. 2012;114:207-14
31. Floriano PN, Christodoulides N, Miller CS. et al. Use of saliva-based nano-biochip tests for acute myocardial infarction at the point of care: a feasibility study. Clin Chem. 2009;55:1530-8
32. Punyadeera C, Dimeski G, Kostner K. et al. One-step homogeneous C-reactive protein assay for saliva. J Immunol Methods. 2011;373:19-25
33. Tonne JM, Campbell JM, Cataliotti A. et al. Secretion of glycosylated pro-B-type natriuretic peptide from normal cardiomyocytes. Clin Chem. 2011;57:864-73
34. Wan Y, Xhang X, Atherton JJ. et al. A multimarker approach to diagnose and stratify heart failure. Int J Cardiol. 2014;181c:369-75
35. Chen LM, Murray SR, Chai KX. et al. Molecular cloning and characterization of a novel kallikrein transcript in colon and its distribution in human tissues. Braz J Med Biol Res. 1994;27:1829-38
36. Emanueli C, Salis MB, Van Linthout S. et al. Akt/Protein Kinase B and Endothelial Nitric Oxide Synthase Mediate Muscular Neovascularization Induced by Tissue Kallikrein Gene Transfer. Circulation. 2004;110:1638-44
37. Leeb-Lundberg LM, Marceau F, Muller-Esterl W. et al. International union of pharmacology. XLV. Classification of the kinin receptor family: from molecular mechanisms to pathophysiological consequences. Pharmacol Rev. 2005;57:27-77
38. Koch M, Wendorf M, Dendorfer A. et al. Cardiac kinin level in experimental diabetes mellitus: role of kininases. Am J Physiol Heart Circ Physiol. 2003;285:H418-23
39. Tang SC, Leung JC, Lai KN. The kallikrein-kinin system. Contrib Nephrol. 2011;170:145-55
40. Krop I, Marz A, Carlsson H. et al. A putative role for psoriasin in breast tumor progression. Cancer Res. 2005;65:11326-34
41. Glaser R, Harder J, Lange H. et al. Antimicrobial psoriasin (S100A7) protects human skin from Escherichia coli infection. Nat Immunol. 2005;6:57-64
42. Kholy KE, Genco RJ, Van Dyke TE. Oral infections and cardiovascular disease. Trends Endocrinol Metab. 2015;26:315-21
43. Sharma S, Zapatero-Rodriguez J, Estrela P. et al. Point-of-Care Diagnostics in Low Resource Settings: Present Status and Future Role of Microfluidics. Biosensors (Basel). 2015;5:577-601
Author contact
![]() Corresponding author: Chamindie Punyadeera, The School of Biomedical Sciences, Institute of Health and Biomedical Innovations, Queensland University of Technology, 60 Musk Avenue, GPO Box 2434, Brisbane QLD 4001, Australia. T: +61 7 3138 0830 | F: +61 7 3138 6030. Email: chamindie.punyadeeraedu.au
Corresponding author: Chamindie Punyadeera, The School of Biomedical Sciences, Institute of Health and Biomedical Innovations, Queensland University of Technology, 60 Musk Avenue, GPO Box 2434, Brisbane QLD 4001, Australia. T: +61 7 3138 0830 | F: +61 7 3138 6030. Email: chamindie.punyadeeraedu.au
 Global reach, higher impact
Global reach, higher impact