13.3
Impact Factor
Theranostics 2017; 7(16):3842-3855. doi:10.7150/thno.19764 This issue Cite
Research Paper
A Circular RNA Binds To and Activates AKT Phosphorylation and Nuclear Localization Reducing Apoptosis and Enhancing Cardiac Repair
1. Sunnybrook Research Institute, Sunnybrook Health Sciences Centre, Toronto, Canada;
2. Department of Laboratory Medicine and Pathobiology, University of Toronto, Toronto, Canada;
3. Department of Cardiovascular Medicine, Second Xiangya Hospital of Central South University, 139 Middle Ren-Min Road, Changsha, Hunan, P.R. China 410011, China;
4. State Key Laboratory of Applied Microbiology Southern China, Guangdong Institute of Microbiology, Guangzhou, 510070, China;
5. The First Hospital, Sun Yat-Sen University, Guangzhou, China;
6. Guangdong Laboratory Animals Monitoring Institute, Guangzhou 510663, Guangdong, China;
7. Toronto General Research Institute, University Health Network, Toronto, Canada;
8. Department of Biology, York University, Toronto, Canada;
9. Institute of Medical Science, University of Toronto, Toronto, Canada.
* These authors contributed equally.
Received 2017-2-21; Accepted 2017-5-1; Published 2017-8-29
Abstract
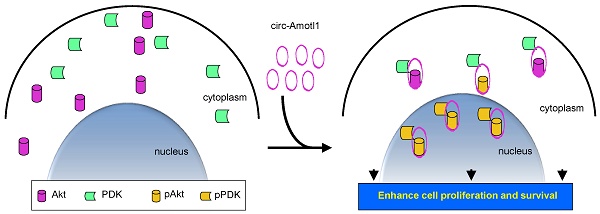
As central nodes in cardiomyocyte signaling, nuclear AKT appears to play a cardio-protective role in cardiovascular disease. Here we describe a circular RNA, circ-Amotl1 that is highly expressed in neonatal human cardiac tissue, and potentiates AKT-enhanced cardiomyocyte survival. We hypothesize that circ-Amotl1 binds to PDK1 and AKT1, leading to AKT1 phosphorylation and nuclear translocation. In primary cardiomyocytes, epithelial cells, and endothelial cells, we found that forced circ-Amotl1 expression increased the nuclear fraction of pAKT. We further detected increased nuclear pAKT in circ-Amotl1-treated hearts. In vivo, circ-Amotl1 expression was also found to be protective against Doxorubicin (Dox)-induced cardiomyopathy. Putative PDK1- and AKT1-binding sites were then identified in silico. Blocking oligonucleotides could reverse the effects of exogenous circ-Amotl1. We conclude that circ-Amotl1 physically binds to both PDK1 and AKT1, facilitating the cardio-protective nuclear translocation of pAKT.
Keywords: circ-Amotl1, AKT, PDK, apoptosis, heart repair.
Introduction
Ischemic heart disease is the leading cause of death according to WHO statistics. Although the myocardium has some ability to regenerate following infarction, this limited remodeling is typically inadequate. Myocardial injury can induce cell death and premature senescence in cardiomyocytes and fibroblasts 1. The dead cells are gradually replaced by a fibrotic scar, which disrupts normal ventricular structure and function. Critically, as hearts age or become senescent, their capacity to remodel and regenerate is reduced, which is further exacerbated following cardiovascular injury and disease 2. Understanding the molecular underpinnings of cardiac remodeling and cardiac senescence will be critical in developing therapeutics for this serious disease.
AKT plays a key role in multiple cellular processes such as cell proliferation, survival, and apoptosis 3-5. It can function as a cardio-protective molecule in cardiovascular disease facilitating tissue remodeling 6. The AKT pathway is highly regulated, starting by activation of PI3K and AKT 7. Normally, AKT resides in the cytoplasm as an inactive form. Upon activation, AKT translocates to plasma membrane where it is phosphorylated by its activating kinases, phosphoinositide dependent kinase (PDK). The activated AKT, becoming pAKT, can translocate to nucleus to activate substrates. It can phosphorylate a wide range of proteins 8. The PI3K-AKT signal pathway is essential for cell survival as activated AKT influences many factors involved in proliferation and apoptosis. Thus, it plays important roles in tissue growth, remodeling and cancer development 3, 9-11.
We recently found that a newly detected class of non-coding RNAs may be crucial for cell cycle progression and cardiac remodeling 12, 13. These RNAs form covalently closed loops, thereby joining the 5' and 3' ends and named circular RNAs or circRNAs 12, 14-20. Although some circRNAs have been shown by our group and others to sponge miRNAs 13, 16, their physiologic roles remain largely unknown 21. We found that circ-Foxo3 was highly expressed in ageing and injured hearts and could induce a senescent phenotype in vivo 12. We hypothesize that some circular RNAs are differentially expressed between neonatal and ageing hearts, since neonatal hearts lost the capacity for regeneration quickly postnatally and circular RNAs may regulate tissue regeneration.
Results
Expression of circ-Amotl1 in neonatal hearts
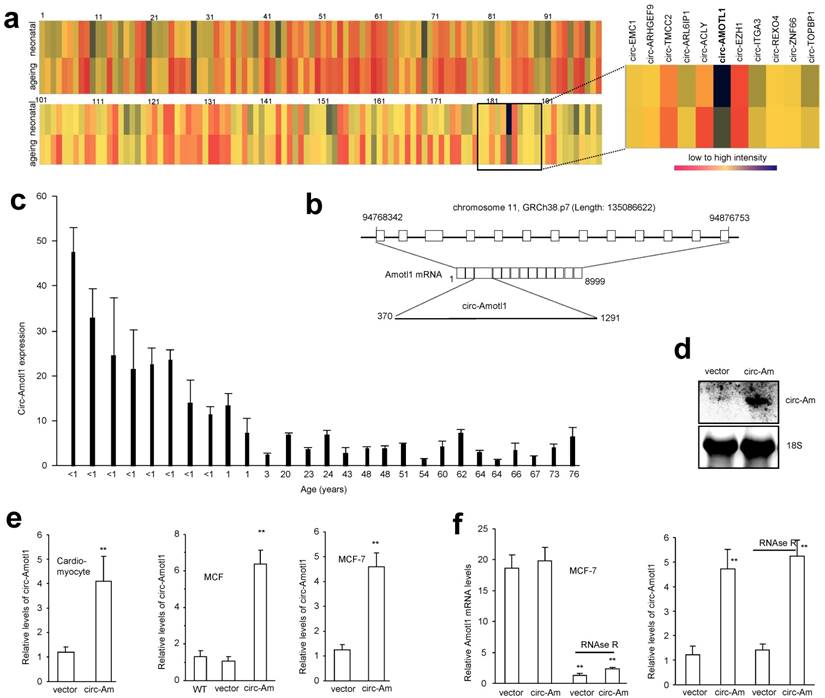
By microarray, we analyzed the expression levels of different circular RNAs in neonatal and mature postnatal human cardiac tissue samples. Although most of the reported circular RNAs could be detected by microarray, only a small portion was differentially expressed with 2-fold cut-off. We searched for those, from which the parental genes are known to function in cardiovascular system. Amongst these differentially expressed circular RNAs, we found that circ-Amotl1 was preferentially expressed in neonate cardiac tissue (Fig 1a, names of circRNAs provided in Fig S1a). Human angiomotin-like 1 gene (Amotl1) codes a peripheral membrane protein that comprises tight junctions, which form the apical junctional structures involved in controlling paracellular permeability and cell polarity 22-24. Amotl1 plays important roles in cardiovascular functioning by regulating endothelial cell migration and capillary formation 23.
Although human Amotl1 is highly conserved to mouse and rat Amotl1, sequence in the junction area of circ-Amotl1 is different from the other species. It appears that circ-Amotl1 is specifically expressed in human, but not in murine nor rat cells. The circ-Amotl1 is derived from Exon 3 of the parental transcript (Fig 1b). We further examined circ-Amotl1 expression using total RNA samples derived from cardiac samples of patients in different age groups. Real-time PCR analysis showed that neonatal cardiac tissues expressed higher levels of circ-Amotl1 relative to those from post-natal patients (Fig 1c).
To explore whether circ-Amotl1 could be developed as an agent for molecular therapy in the cardiovascular system, especially in heart tissues, we generated an expression construct expressing circ-Amotl1 and a mock control. By Northern blotting, we confirmed that cells transfected with the construct expressed higher level of circ-Amotl1 than the vector control (Fig 1d). We also stably transfected a number of cell lines including primary cardiomyocytes, mouse cardiac fibroblasts (MCF), endothelial cell line YPEN, and human MCF-7 cells, allowing us to examine the role of circ-Amotl1 comprehensively. We confirmed expression of circ-Amotl1 in these cells (Fig 1e, Fig S1b). Circularization of the expressed circ-Amotl1 was confirmed by treating the samples with RNase R to digest linear Amotl1 mRNA but not circ-Amotl1 (Fig 1f).
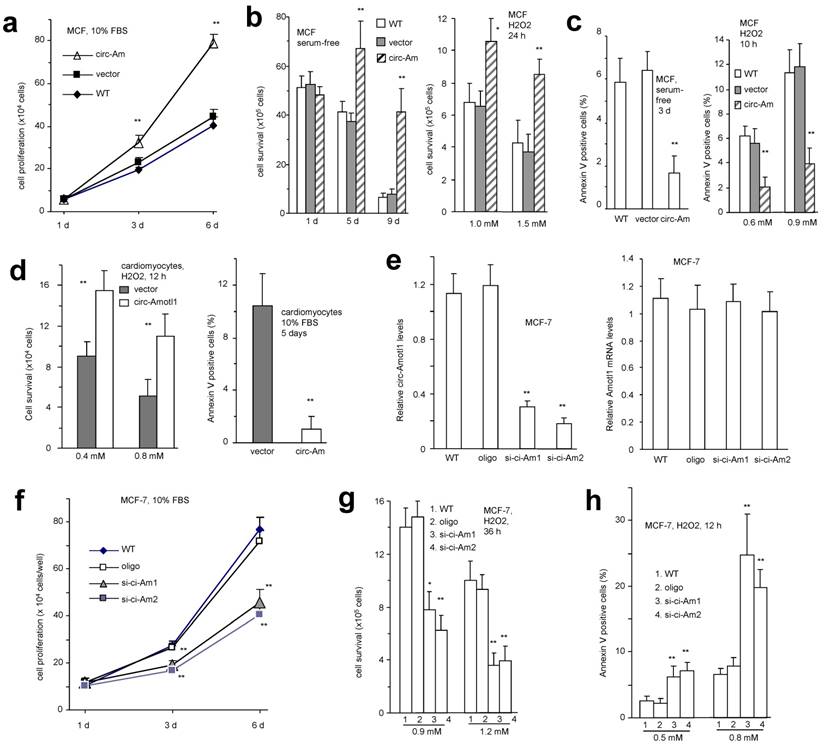
We analyzed the phenotypic effects of circ-Amotl1 expression and found that ectopic circ-Amotl1 increased proliferation (Fig 2a), survival (Fig 2b), and apoptosis (Fig 2c) of MCF cells. It also enhanced primary cardiomyocyte survival and decreased apoptosis (Fig 2d). Similar results were obtained in YPEN cells (Fig S1c-f).
To test whether this interaction occurred in human cells, we designed 2 siRNAs specifically targeting circ-Amotl1. Transfection with these siRNAs silenced endogenous circ-Amotl1 levels but not the linear Amotl1 mRNA (Fig 2e). Silencing circ-Amotl1 decreased cell proliferation (Fig 2f), survival (Fig 2g) and increased cell apoptosis (Fig 2h, Fig S2a). Similar results were obtained in YPEN cells (Fig S2b).
Effect of circ-Amotl1 on Dox-induced cardiomyopathy in mice
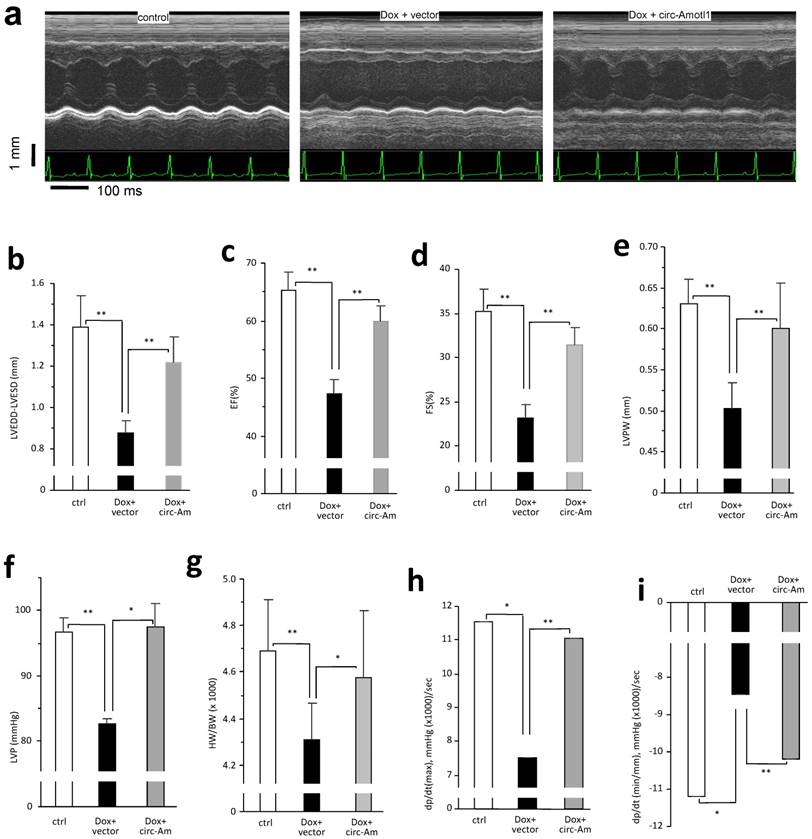
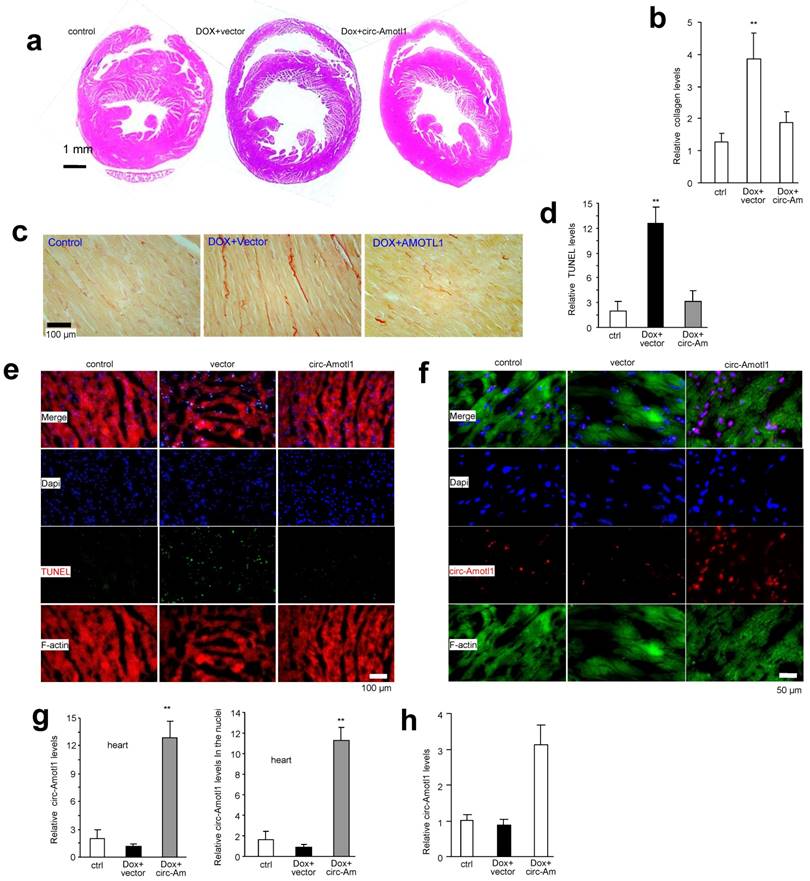
The anthracycline Doxorubicin (Dox) is a chemotherapeutic drug that induces cardiomyopathy following long-term treatment. We sought to determine whether circ-Amotl1 expression would affect a mouse model of Dox-induced cardiomyopathy. After treating the mice for 14 days, we obtained M-mode figures by echocardiography, which confirmed the mouse model of Dox-induced cardiomyopathy (Fig 3a). We detected a significant decrease when the Left Ventricular End Diastolic Diameter (LVEDD) subtracted Left Ventricular End-Systolic Diameters (LVESD) in the Dox-treated mice compared to control (Fig 3b). We further detected a significant decrease in the Left Ventricular Ejection Fraction (EF), Left Ventricular Fractional Shortening (FS), LVPW, LVP, and HW/BW (Fig 3c-g). The Rate of Rise of Left Ventricular Pressure (dp/dt, max and min) were significantly decreased in the Dox-treated mice compared with the control (Fig 3h-i). When a circ-Amotl1 expression plasmid was delivered to the Dox-treated mice, the cumulative effects of Dox were relieved. HE staining revealed a dilated left ventricle in Dox-treated mice (Fig 4a). Ectopic expression of circ-Amotl1 reduced the enlarged left ventricle. Fibrotic remodeling of the LV was then analyzed by Sirius-Red staining and quantified. Dox treatment increased collagen staining, which was abrogated by ectopic circ-Amotl1 (Fig 4b-c). TUNEL staining further showed increased apoptosis in the Dox-treated animals, but this effect was abolished by circ-Amotl1 delivery (Fig 4d-e). This phenotype was correlated with circ-Amotl1 RNA expression, as examined by in situ hybridization (Fig 4f-g) and real-time PCR (Fig 4h) of the cardiac tissue samples. These results demonstrated that circ-Amotl1 expression was protective against Dox-induced cardiomyopathy.
Expression of circ-Amotl1 (a) Comparison of circular RNA expression in human neonatal and ageing hearts by using the Human circular RNA microarray. Names of circular RNAs are provided in Supplementary Fig S1a. (b) Structures of Amotl1 genome and transcript. Circ-Amotl1 is produced by exon 3. (c) Real-time PCR showed that expression of circ-Amotl1 was much higher in human neonatal hearts than the ageing hearts. (d) Northern blot showing transfection with circ-Amotl1 increased levels of circ-Amotl1. (e) Transfection with circ-Amotl1 increased expression of circ-Amotl1 in cardiomyocytes, MCFs and MCF-7 cell line. (f) Total RNA extracted from mock- or circ-Amotl1-transfected MCF-7 cells was incubated with or without RNase R followed by real-time PCR. While RNase R treatment decreased Amotl1 linear RNA levels, it did not affect circ-Amotl1 levels. **, p<0.01. Error bars, SD (n=4).
Function of circ-Amotl1 in cell proliferation, survival and apoptosis (a) Mouse cardiac myocytes (MCF) transfected circ-Amotl1 or a control vector were cultured in 10% FBS/DMEM. Circ-Amotl1 expression promoted cell proliferation (n=4). (b) Ectopic expression of circ-Amotl1 enhanced MCF cell survival with or without H2O2 treatment (n=4). (c) Ectopic expression of circ-Amotl1 decreased MCF cell apoptosis with or without H2O2 treatment (n=4). (d) Primary cardiomyocytes transfected with circ-Amotl1 or the vector were treated with or without H2O2 followed by survival and apoptotic assays. Expression of circ-Amotl1 increased cell survival and decreased apoptosis (n=4). (e) Real-time PCR showed that expression of circ-Amotl1 siRNAs decreased circ-Amotl1, but not Amotl1 mRNA levels in MCF-7 cells (n=6). (f) MCF-7 cells transfected with circ-Amotl1 siRNAs or a control oligo were cultured in 10% FBS/DMEM. Silencing circ-Amotl1 repressed cell proliferation (n=6). In all experiments, asterisks indicate significant differences, where * indicates p<0.05 and ** indicates p<0.01. Error bars means SD. (g) Silencing circ-Amotl1 decreased cell survival (n=6). (h) Silencing circ-Amotl1 increased Annexin V positive (apoptotic) cells (n=4).
Circ-Amotl1 facilitated nuclear translocation of AKT and PDK1
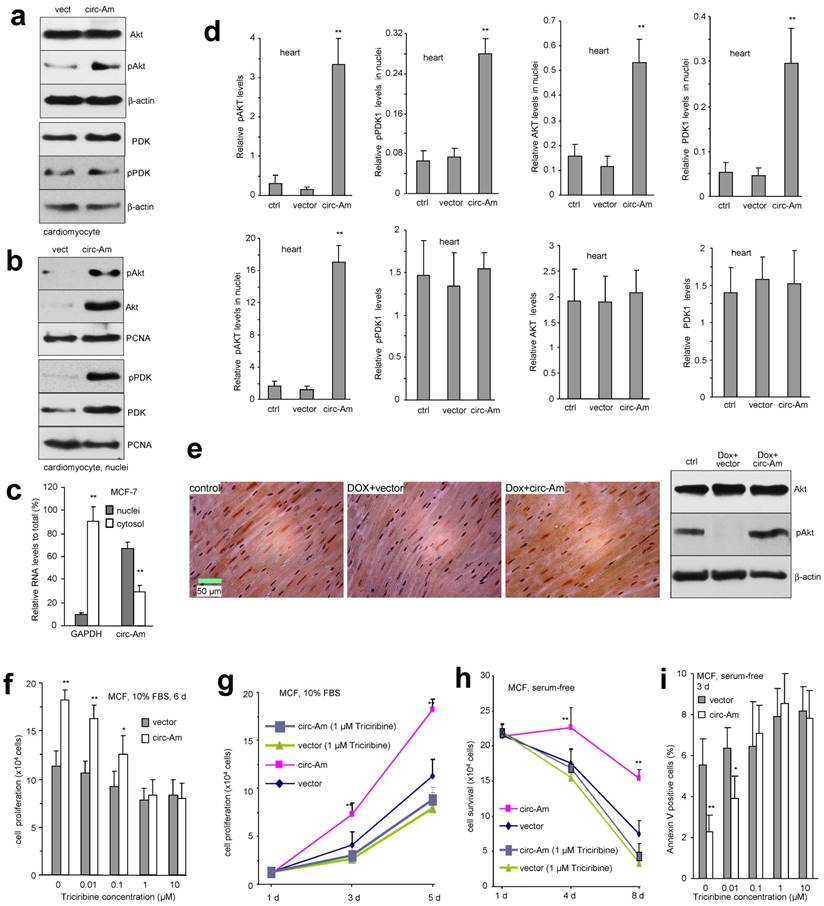
Since AKT is a major signaling pathway in regulating cell proliferation and survival, we examined whether circ-Amotl1 affected AKT expression and found that circ-Amotl1 increased pAKT in primary cardiomyocytes (Fig 5a). Since PDK1 is a kinase that transiently interacts with and activates AKT 25, we examined the levels of PDK1 and pPDK1, but little difference was detected (Fig 5a). Similar results were obtained with YPEN cells (Fig S3a). In the tissue samples of mice treated with circ-Amotl1, we also detected increased pAKT, but not AKT, PDK1 or pPDK1 (Fig S3b). In addition, we detected co-localization of circ-Amotl1 and pAKT with DAPI. It suggested that circ-Amotl1 might have facilitated nuclear translocation of pAKT. Primary cardiomyocytes were transfected with circ-Amotl1 or the vector, followed by nuclear isolation and Western blotting. Increased levels of pAKT, AKT, PDK1, and pPDK1 were detected in the circ-Amotl1-transfected cells (Fig 5b). We also isolated nuclear and cytosolic fractions from MCF-7 cells and analyzed circ-Amotl1 levels. We detected significantly higher circ-Amotl1 levels in the nuclei than in the cytosolic fraction (Fig 5c). We observed similar results of pAKT upregulation (Fig S4a) and nuclear translocation (Fig S4b-c) in the circ-Amotl1-transfected YPEN and MCF-7 cells. We then quantified nuclear translocation of pAKT, AKT, pPDK1 and PDK1 in the heart sections and confirmed nuclear translocation of these proteins in the circ-Amotl1 plasmid group (Fig 5d). Immuno-histochemical staining of the heart sections also revealed increased levels of pAKT in the nuclei (Fig 5e).
Delivery of circ-Amotl1 attenuated Dox-induced cardiomyopathy in mice (a) Representative echocardiography photographs showing heart function in different groups. (b) Dox treatment decreased mouse LVEDD-LVESD. Ectopic expression of circ-Amotl1 reversed Dox-induced effect (Error bars, SD, n=10 mice). (c-i) Dox treatment reduced mouse Ejection fraction (EF, c), Fractional shortening (FS, d), Left ventricular posterior wall (LVPW, e), Left ventricular pressure (LVP, f), Ratio of heart weight and body weight (HW/BW g), Left ventricular contractility dP/dt (max, h), and dP/dt (min, i). Ectopic expression of circ-Amotl1 reversed these effects. (Error bars, SD, n=10 mice).
Delivery of circ-Amotl1 decreased Dox-induced cardiomyocytes death in mice (a) Heart sections were subject to H&E staining. Shown are representative photos. (b) Quantification of Sirius red staining showing that Dox treatment induced mouse myocadium collagen expression, which was inhibited by ectopic expression of circ-Amotl1. (c) Typical photos of Sirius red staining. (d) Image J analyses showed that injection of Dox increased TUNEL staining in heart tissues which was attenuated by delivery of circ-Amotl1 (n=6). (e) Mouse heart tissues were stained with phalloidins (red), DAPI (blue), and TUNEL (green) showing application of Dox increased TUNEL staining which could be attenuated by delivery of circ-Amotl1. (f) Heart tissues were stained with phalloidins (green), DAPI (blue), and circ-Amotl1 (red) showing delivery of circ-Amotl1 increased circ-Amotl1 levels. (g) After in situ hybridization, Image J analyses showed that delivery of circ-Amotl1 enhanced circ-Amotl1 total levels (left) and circ-Amotl1 levels in the nuclei of heart tissues (n=6). (h) Real-time PCR showed that delivery of circ-Amotl1 enhanced circ-Amotl1 levels in heart (n=6).
To confirm the role of AKT in mediating circ-Amotl1 functions, we employed AKT inhibitor Triciribine to block AKT signalling pathway. We found that the effect of circ-Amotl1 on MCF cell proliferation was abolished when the cells were treated with 1 or 10 μM Triciribine for 6 days (Fig 5f). At the concentration of 1 μM, Triciribine was able to abolish the effect of circ-Amotl1 after 3-day incubation (Fig 5g). In serum-free medium, although 0.1 μM Triciribine was able to abolish the effect of circ-Amotl1 on extending cell survival after 8 day treatment (Fig S4d), 1 μM Triciribine was required to abolish circ-Amotl1 function after 4 day treatment (Fig 5h). However, it only took 3 days for 0.1 μM Triciribine to abolish the effect of circ-Amotl1 on repressing cell apoptosis (Fig 5i). When the cells were treated with hydrogen peroxide, 0.1 μM Triciribine was sufficient to abolish the effects of circ-Amotl1 on extending cell survival (Fig S4e) and repressing apoptosis (Fig S4f).
Nuclear translocation of AKT and PDK1 with circ-Amotl1 (a) Western blot showed that transfection with circ-Amotl1 enhanced pAKT levels in primary cardiomyocytes. (b) RNAs extracted from cytoplasm and nuclei of MCF-7 cells were subject to real-time PCR. Significantly higher levels of circ-Amotl1 were detected in the nuclei relative to cytoplasm, while GAPDH was mainly expressed in the cytoplasm (n=6). (c) Western blot showed that circ-Amotl1-transfected mouse cardiomyocytes expressed higher levels of pAKT, AKT, pPDK1 and PDK1 in the nuclei. (d) Image J analyses showed that delivery of circ-Amotl1 increased pAKT levels in heart tissues and promoted pAKT, AKT, pPDK1 and PDK1 nuclear translocation (n=6). (e) Left, Heart sections were immunostained with pAKT. Dox treatment decreased pAKT levels, while delivery of circ-Amotl1 reversed this process. Right, Western blot of the heart tissues. (f-i) Treatment with AKT inhibitor Triciribine abolished the effects of circ-Amotl1 on cell proliferation (f-g), survival (h) and apoptosis (i) (n=4).
Interaction of circ-Amotl1 with PDK1 and AKT1
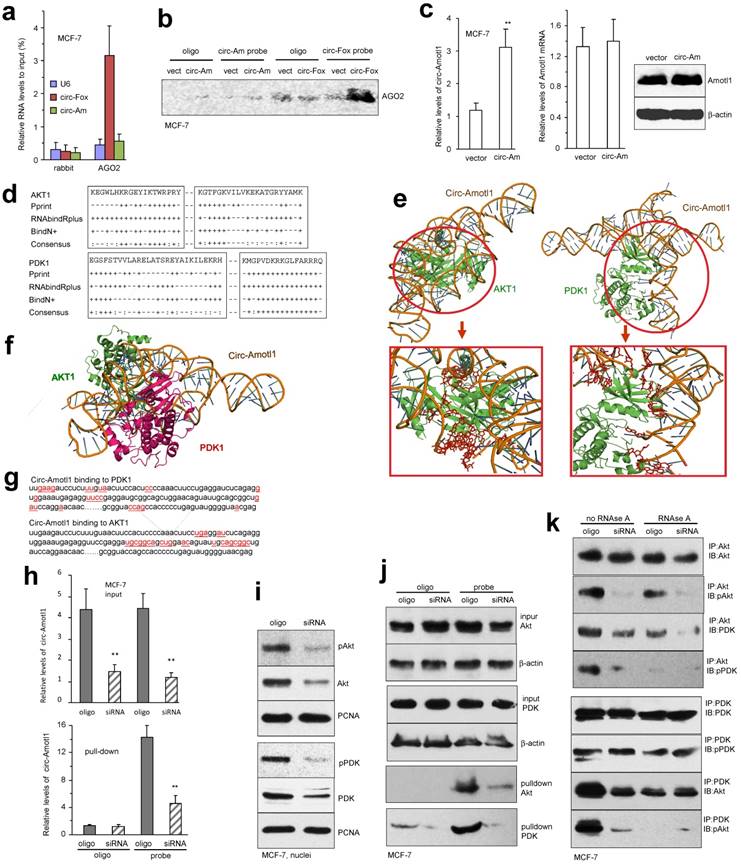
It has been known that circular RNAs can function as sponges for miRNA interaction and we have shown that circ-Foxo3 functions as a sponge to bind a number of miRNAs 26. We tested whether or not circ-Amotl1 possessed this potential. After confirming the binding efficiency of AGO2 antibody (Fig S4g), we showed that AGO2 antibody was able to precipitate circ-Foxo3 but not circ-Amotl1 (Fig 6a). On the other hand, while the probes of both circ-Foxo3 and circ-Amotl1 could pull down the respective circular RNA (Fig S4h), only circ-Foxo3 probe precipitated AGO2 protein (Fig 6b). As a result, transfection with circ-Amotl1 did not affect levels of Amotl1 mRNA and protein, although circ-Amotl1 was up-regulated (Fig 6c). Our results suggest that circ-Amotl1 did not function as a sponge for binding miRNAs.
Using computer algorithm, we predicted probable RNA-binding residues in AKT1 and PDK1 (Fig 6d). Through in silico analyses, the best predicted secondary structure of circ-Amotl1 was derived by analyzing its thermodynamic properties using the formula ΔG=ΔH-TΔS, where ΔG=-52.20 kcal/mol at 37°C, ΔH=-515.00 kcal/mol, and ΔS=-1492.1 cal/(Kmol). The secondary structure defined using dot-bracket notation was then analyzed by the software RNA composer for tertiary structure prediction. NPDock was used to carry out the in-silico molecular docking between circ-Amotl1 and AKT or PDK (Fig 6e). Both PDK1 and AKT1 could dock into one circ-Amotl1 (Fig 6f).
The structure of PDK1 protein used in the docking procedure was derived from Protein Data Bank (PDB) entry 1OKY. The top scoring modeled complex was generated by NPDock which considers the geometric score suggesting good shape complementarity, interface area size, atomic contact energy and potential binding site information along with various favourable interactions. This model showed fit rendering the stability of circ-Amotl1-PDK1 complex. The molecular simulation supports that circ-Amotl1 could perfectly dock PDK1 and predicts a minimal binding region of circ-Amotl1 for PDK1 (Fig 6g). The contact map (Fig S5a), the residue-level resolution contact maps (Fig S5b), the MC score (Fig S5c), the contact distance (Table S1-2), and the Accessible Surface Area (Table S3-6) all supported the conclusion that circ-Amotl1 could dock PDK1 and AKT1.
To test the interaction of endogenous circ-Amotl1 with AKT and PDK1, we used the silencing approach. Silencing endogenous circ-Amotl1 significantly decreased the levels of circ-Amotl1 pulled-down by these proteins (Fig 6h). Silencing endogenous circ-Amotl1 resulted in decreased nuclear translocation of these proteins (Fig 6i) and pulling down decreased levels of AKT and PDK1 (Fig 6j). The interaction was also confirmed in YPEN cells (Fig S6a-f).
We tested whether both AKT and PDK1 could bind to the same circ-Amotl1 forming ternary complexes. Lysates prepared from YPEN cells transfected with circ-Amotl1 or the vector were precipitated with antibody against AKT or PDK1, in the absence or presence of RNase A that cleaved circ-Amotl1, followed by Western blotting probed with antibodies as indicated. Without RNase A treatment, anti-AKT antibody could precipitate PDK1 and pPDK1, while anti-PDK1 antibody could precipitate AKT and pAKT, which was abolished by RNase A treatment (Fig 6k, upper). Silencing circ-Amotl1 or cleaving with RNase A avoided the formation of these ternary complexes (Fig 6k, lower). This result was confirmed in YPEN cells (Fig S6g).
Blocking the interaction of circ-Amotl1 with AKT and PDK1
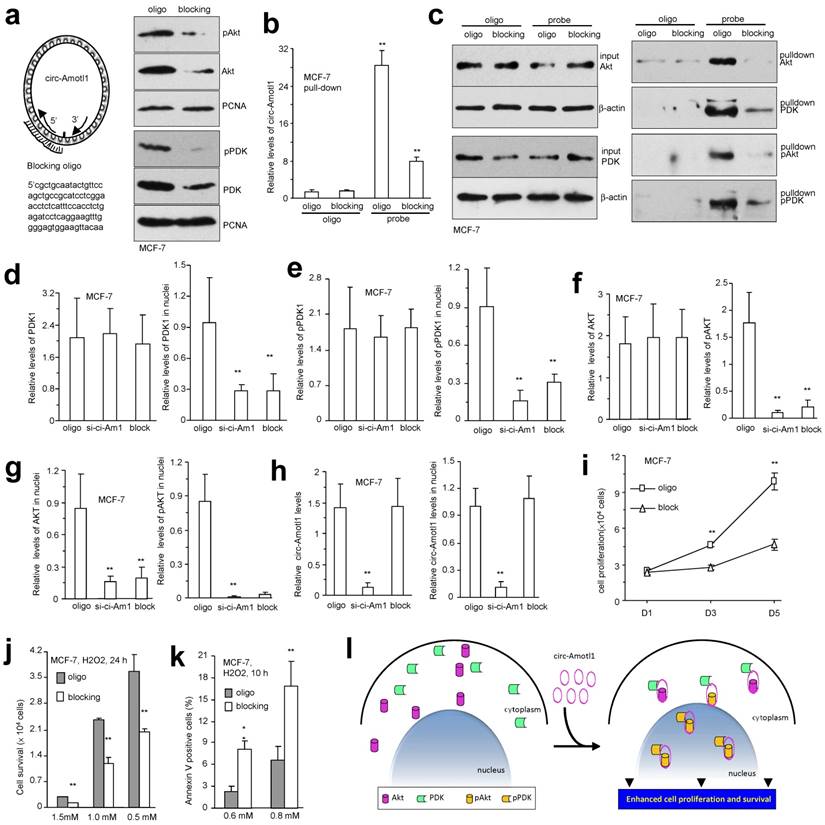
To confirm that the interaction of circ-Amotl1 with AKT and PDK1 was responsible for nuclear translocation of both proteins, we designed antisense oligo complementary to the binding site on circ-Amotl1 for AKT and PDK1 to block the interaction. Transfection with the blocking oligo did not affect expression of circ-Amotl1 (Fig S7a), nor expression of AKT, PDK1 and pPDK1, but decreased AKT activation (Fig S7b) and inhibited nuclear translocation of AKT, pAKT, PDK1, and pPDK1 (Fig 7a). In the pull-down assays, transfection with the blocking oligo decreased precipitation of circ-Amotl1 (Fig 7b) and its interacting proteins (Fig 7c) by the circ-Amotl1 probe.
In MCF-7 cells, transfection with the blocking oligo did not affect PDK expression but decreased PDK nuclear translocation (Fig 7d). Similarly, pPDK levels were not affected but nuclear translocation decreased (Fig 7e). While total levels of AKT were not affected by the blocking oligo, AKT phosphorylation decreased (Fig 7f). As a consequence, nuclear AKT and pAKT decreased (Fig 7g). However, transfection with the blocking oligo did not affect expression and nuclear localization of circ-Amotl1 (Fig 7h). The images were provided in the Supplementary (Fig S7c). We confirmed nuclear translocation of these proteins affected by the blocking oligo in YPEN cells (Fig S8), which was quantified (Fig S9a-b). Functionally, MCF-7 cells transfected with the blocking oligo displayed decreased cell proliferation (Fig 7i) and survival (Fig 7j) but increased apoptosis (Fig 7k). These effects were confirmed in YPEN cells (Fig S9c-e). To confirm the effect of AKT signaling pathway in cell survival, we treated the circ-Amotl1-transfected YPEN cells with AKT inhibitor. AKT activation was inhibited by the inhibitor in a concentration-dependent manner (Fig S9f). Cell survival was inhibited. At high concentration of 2.7 μM, the effect of circ-Amotl1 was completely abolished (Fig S9g).
Interaction of AKT and PDK1 with circ-Amotl1 (a) Ago2 antibody precipitated circ-Foxo3 but not circ-Amotl1 (n=4). (b) Circ-Foxo3 probe could precipitate AGO2, but circ-Amotl1 probe could not. (c) Ectopic expression of circ-Amotl1 (left) had no effect on expression of Amotl1 mRNA (middle) and protein (right). (d) Prediction of probable RNA-binding residues of AKT was carried out by submitting the AKT sequence to Pprint, RNABindRPlus and BindN+ servers. Plus signs indicate the predicted RNA-binding residues. (e) Graphical representation of three-dimensional structures of the docking models of circ-Amotl1 with AKT and PDK1, and zoom-in images of the binding interface done by NPDock. (f) Both AKT and PDK1 could bind to the same circ-Amotl1 molecule. (g) Nucleotides (red) involved in the interaction of circ-Amotl1 with AKT or PDK1. (h) Lysates prepared from MCF-7 cells transfected with circ-Amotl1 siRNA or a control oligo, were subject to pull-down assay. The probe pulled down less circ-Amotl1 in the cells transfected with siRNA. (i) Silencing circ-Amotl1 repressed nuclear translocation of pAKT, AKT, pPDK1 and PDK1. (j) Silencing circ-Amotl1 did not affect AKT and PDK1 expression, but decreased the capacity of the probe pulling down AKT and PDK1. (k) Silencing endogenous circ-Amotl1 decreased the interaction of AKT with PDK1. Treatment with RNase A abolished AKT interacting with PDK1.
Blocking circ-Amotl1 modulated cellular activities. (a) Left, the blocking oligo covered both AKT and PDK1 binding sites. Right, delivery of the blocking oligo repressed nuclear translocation of pAKT, AKT, pPDK1 and PDK1. (b) Lysates prepared from MCF-7 cells transfected with blocking oligo or a control oligo were mixed with the circ-Amotl1 probe or a control oligo. The probe pulled down less circ-Amotl1 in the cells transfected with blocking oligo (n=4). (c) Transfection with the blocking oligo did not affect AKT and PDK1 expression. The probe pulled down decreased levels of pAKT, AKT, pPDK1 and PDK1 in the cells transfected with the blocking oligo. (d) Silencing circ-Amotl1 or transfection with the blocking oligo did not affect PDK expression (left) but decreased PDK nuclear translocation (right) (n=6). (e) Silencing circ-Amotl1 or transfection with the blocking oligo did not affect pPDK levels (left) but decreased pPDK nuclear translocation (right) (n=6). (f) Silencing circ-Amotl1 or transfection with the blocking oligo did not affect AKT levels (left) but decreased AKT phosphorylation (right) (n=6). (g) Silencing circ-Amotl1 or transfection with the blocking oligo decreased nuclear AKT (left) and pAKT (right) levels (n=6). (h) Transfection with the blocking oligo did not affect expression (left) and nuclear localization (right) of circ-Amotl1 (n=6). (i) Transfection with the blocking oligo decreased proliferation of MCF-7 cells. (j) Transfection with the blocking oligo decreased survival of MCF-7 cells. (k) MCF-7 cells transfected with the blocking oligo or a control oligo were cultured in H2O2 for 10 h or 12 h. Transfection with the blocking oligo increased Annexin V positive cells (n=4). (l) Circ-Amotl1 binds AKT and PDK, induces AKT phosphorylation and pAKT nuclear translocation, and enhances cell proliferation and survival.
Taken together, our study demonstrated that circ-Amotl1 is highly expressed in neonatal heart and potentiates cardiomyocyte functions. The circular RNA circ-Amotl1 binds AKT and PDK, induces AKT phosphorylation and pAKT nuclear translocation, enhances cell proliferation and survival, and protects Doxorubicin-induced cardiomyopathy (Fig 7l).
Discussion
We sought to identify circular RNAs that were differentially expressed between neonatal and mature cardiac tissue. Thus we analyzed circular RNA expression in human cardiac tissue samples and found that the circular RNA circ-Amotl1 was more highly expressed in neonatal hearts relative to the mature hearts. Phenotypically, the ability of neonatal cardiomyocytes to proliferate and regenerate is rapidly lost in the postnatal period. It is assumed that some components that are essential for cardiomyocytes proliferation and regeneration in the early stages of heart development become deteriorated in adult hearts. Such components may be strong candidates for potential gene therapy in cardiovascular system. Circ-Amotl1 appeared to be a differentially expressed circular RNA that could underlie this need.
Based on our data, the expression of many circular RNAs may be regulated spatially and temporally. For example, we have recently reported that circ-Foxo3 is highly expressed in an animal model of cardiac senescence and may promote this phenotype 12. Although they use different regenerative mechanisms, we hypothesize that cardiac tissue may express abundance of some circular RNAs in a similar manner, as it has been reported for brain tissue 16, 27-29. Adults fail to regenerate cardiac tissue following injury and this leads to subsequent heart failure and clinical death. Understanding the molecular pathways that allow neonatal cardiomyocytes to proliferate in a controlled manner may allow the development of relevant therapeutic intervention.
We detected increased levels of activated AKT in the circ-Amotl1-transfected cells, particularly in their nuclei. As central nodes in cardiomyocyte signaling, nuclear AKT appears to play a cardio-protective role during cardiovascular disease and injury. Activated AKT mediates downstream responses including cell proliferation, survival, and migration by phosphorylating intracellular proteins 30, 31. This pathway is highly conserved in eukaryotic cells. Normally, AKT is inactive and stays in the cytosol 32. When the cell is stimulated, AKT is translocated to the plasma membrane where it is activated. AKT can be partially activated by phosphorylation of T308 by PDK1, followed by full activation that requires phosphorylation of S473. Although regulation of Ser473 phosphorylation is not fully understood, it may occur through autophosphorylation. Once activated, pAKT translocates to nucleus. In the nucleus, pAKT positively regulates proliferation-associated factors and negatively regulates the expression of pro-apoptotic proteins by direct phosphorylation 33. Although there are many reports of AKT activation and its associated functions, little is known about how activated AKT is translocated to nucleus 34. Our study demonstrates that circ-Amotl1 facilitates AKT activation and nuclear translocation and may play a cardioprotective role in doxorubicin-induced cardiomyopathy.
Our model of doxorubicin-induced cardiomyopathy represents a well-characterized mouse model of myocardial dysfunction following chemotherapeutic treatment. In vivo delivery of circ-Amotl1 shows potential for further exploration as a therapeutic approach for preventing adverse cardiac remodeling. In our animal model, up-take and expression of the plasmids was effective and well-tolerated. We have not detected any side effect of the ectopic circular RNA. This approach could potentially be extended to studying other types of tissues. We have recently found that the injected circular RNA expression plasmids (circ-Foxo3) could be up-taken into tumors and expression of circ-Foxo3 repressed tumor progression 35. Since ectopic expression of circ-Amotl1 induced primary cardiomyocyte proliferation in culture, it would be helpful to find out whether cardiomyocyte proliferation can be induced in vivo and whether this is associated with hypertrophic remodeling.
Methods
Construct generation
The expression plasmid and the control vector were engineered by us previously 13, 26. The circular RNA fragment of circ-Amotl1 was synthesized by two primers (Cir.Amotl1-HindIII and Cir.Amotl1-SalI, Fig S9h) and cloned into the plasmid.
Microarray and animal model of cardiomyopathy
The RNA isolation and microarray analysis of human circular RNAs was performed by KangChen BioTech (Shanghai). Total RNAs were digested with RNase R (Epicentre, Inc.) to remove linear RNAs. The enriched circular RNAs were amplified and transcribed into fluorescent cRNA utilizing a random priming method and hybridized onto the Arraystar Human circRNA Array V2. The sample of neonatal was a mix of tissues from three patients with age under one-year old and the sample of ageing was a mix of tissues from three patients with ages of 61, 67, and 84. Consent for human samples was obtained according to the Declaration of Helsinki.
All animal assays were performed in accordance with guidelines and regulations approved by the Animal Care Committee of Sunnybrook Research Institute. The animal model of Doxorubicin-induced cardiomyopathy was induced in adult mice (8 weeks, BALB/c strain) by intraperitoneal injection of Doxorubicin (Dox, 24 mg/kg in 8 injections over a period of 3 weeks via intraperitoneal administration) as described 12. The tested groups were Dox+circ-Amotl1 plasmid. The control groups were Dox+vector plasmid and buffer vehicle alone using the gender-matched littermate mice. Tissues were harvested for subsequent analyses 2 weeks after treatment. The animals were observed daily. The plasmids (circ-Amotl1/vector) were administered by intraperitoneal injection 1 day before every Dox injection, and continued injection for 2 weeks after Dox treatment (twice a week).
Cardiac function was measured by echocardiography and hemodynamics at 2 weeks post-Dox-treatment to assess percent Ejection Fraction (EF), percent Fractional shortening (FS), left ventricular internal systolic dimension (LVIDS), left ventricular internal diastolic dimension (LVIDD), left ventricular posterior wall (LVPW), Left ventricular pressure (LVP), and Doppler-derived dP/dt(max) and dP/dt(minimum). Heart tissues were harvested for subsequent analysis.
Nanoparticle delivery of plasmids
For generation of circ-Amotl1-PEG conjugate, 500 µg circ-Amotl1 plasmids or vector were dissolved in 1000 µL of RNase -free water. The mPEG-SH (PG1-TH-2k, Nanocs, New York, NY) was mixed with the plasmids at 1:20 molar ratio. Then gold nanoparticles (Cytodiagnostics, Burlington, Ontario, Canada) were mixed with circ-Amotl1-PEG at weight ratio of 1:20 for conjugation. The mixture was gently shaken at 60°C for 30 min and transferred into a syringe. The nanoparticles with circ-Amotl1 plasmids or the vector were administered intraperitoneally in a volume of 200 µl into mice.
Sirius Red staining for collagen visualization
The paraffin sections were de-waxed and hydrated. The slides were stained with Weigert's haematoxylin for 8 min to observe nuclei, and then washed in running tap water. The slides were incubated with 0.1% Sirius Red solution dissolved in aqueous saturated in picric acid for one hour, washed in acidified water (0.5% hydrogen chloride), dehydrated and mounted with DPX Mounting. Collagen and non-collagen components were red- and orange-stained, respectively.
Isolation of mouse cardiomyocytes
The enzymatic dispersion technique was used to isolate mouse cardiomyocytes from mice as described in detail previously 36, 37. Briefly, animals were heparinized, anaesthetized by inhalation of isoflurane and then sacrificed by cervical dislocation. The hearts were removed and washed in PBS solution containing 20 mM BDM. They were then transferred to a drop of hepes-buffered Tyrode solution (130 mM NaCl; 5.4 mM KCl; 1 mM CaCl2; 1 mM MgCl2; 0.33 mM Na2HPO4; 10 mM Hepes; 5.5 mM glucose, pH 7.4) and minced. The tissue fragments were transferred into a new tube and incubated in 25 ml Tyrode solution containing 0.012 g Collagenase D (Roche Diagnostics, 1-088-882), 0.009 g Collagenase B, and 0.001 g Protease XIV at 37°C for 30 min. The digested products were filtered and centrifuged at 600 rpm for 5 min. Cell pellet was re-suspend in DMEM/F12 medium containing 10% FBS and 20 mM BDM and plated onto10-cm cell culture dishes and incubated for 2 h. This pre-plating step removed fibroblasts and endothelial cells, which adhered to the uncoated dish. Non-adherent cardiomyocytes were transferred to cell culture dish coated with 1% gelatin solution.
Cell cultures
Rat endothelial cell line YPEN and mouse cardiac fibroblast (MCF) were cultured in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% fetal calf serum, penicillin (100 U/ ml) and streptomycin (100 μg/ml) and maintained at 37°C in a humidified atmosphere of 5% CO2.
To obtain stable lines expressing circ-Amotl1, the cells were transfected with circ-Amotl1 expression construct and control plasmid, generated by us. Three days after transfection, Geneticin was added to the growth medium at a concentration of 1 mg/ml, and the cells were maintained in this medium until individual colonies were large enough for cloning. Chemically selected stable cell lines were maintained in culture medium containing 0.5 mg/ml Geneticin or stored in liquid nitrogen.
For gene silencing, cells were transfected with siRNAs using Lipofectamine 2000 (Invitrogen). In brief, 4x104 cells in 2 mL of DMEM (10%FBS) were plated in each of five different 35-mm tissue culture dishes and incubated overnight at 37°C and 5% CO2 atmosphere. The cells were cultured in serum-free medium 1 hour before transfection. For each dish, 10 µL siRNA was added into 150 µL of serum-free medium and mixed with 3 µL of Lipofectamine. The mixture was added to cells and incubated for 6 h before replacing the medium with DMEM containing 10% FBS. Cells were harvested for function assay 48 h later. For protein preparation, cell lysates were prepared 72 h after transfection.
Cell proliferation assays
The circ-Amotl1- and vector-transfected cells (2×104 cells) were inoculated on 6-well dishes in 10% FBS/DMEM medium and maintained at 37°C overnight. Cells were harvested daily and cell number was counted. In selected experiments, cell suspensions were cultured with selective AKT inhibitor Triciribine (1 μM).
For single cell proliferation assay, the circ-Amotl1- and vector-transfected YPEN cells were inoculated in Petri dishes in DMEM containing 10% FBS, which allowed the cells to attach but not spread on the plates. Next day, the cell cultures were briefly treated with trypsin/EDTA to obtain single cell suspension. The cell number was determined by continuing dilutions. Fresh medium (DMEM containing 10% FBS) was added to obtain a density of 1 cell per 100 µl followed by immediate distribution into 96-well tissue culture plates at the amount of 100 µl per well. The plates were examined carefully under a light microscope. The wells that contained 1 single cell were marked. Those wells were excluded if more than one or none growing colonies were detected within 2-3 days. The growing colonies were monitored, photographed, and cell number was determined.
Cell survival assay
Cells (2×105 cells) were cultured in 10% FBS/DMEM medium in culture dishes and maintained at 37°C for 12 h. After cell attachment, the cultures were washed with PBS, followed by addition of serum-free DMEM. Cells were harvested after 4 days and cell number was counted to estimate cell survival.
Cell migration assays
Cell migration was tested by scratch and trans-well assays. In the scratch assay, YPEN cells were inoculated onto six-well plates at a density of 3 x105 cells/well for 12 h. The cultures were then scraped linearly with micropipette tips. Cell migration patterns were recorded by light microscopy at 0, 16, and 20 h. Migrated distance was measured and quantified. Trans-well chambers (Coster, Sigma-Aldrich) were placed in 24-well tissue culture dishes and 1×105 cells in 100 µl media were loaded into the upper chamber of the trans-wells. The lower chambers were filled with 600 µl DMEM containing 10% FBS. After 6, 8 and 10 h incubation at 37 °C, non-migrated cells were removed with a cotton swab and invaded cells were fixed by 100% methanol and stained with Coomassie Brilliant blue (Bio-Rad, Hercules, CA) for 10 min. Photos were taken under a light microscope (Zeiss, Carl Zeiss Microscopy).
Western blot
Protein assays on Western blot and immunohistochemistry were performed as described38, 39. Real-time PCR was conducted as described40, 41. Western blotting was performed as described 40.
RNA digestion and pull-down by protein immunoprecipitation
Total RNA was extracted from cell lysate. In brief, 1×107 cells were harvested, washed in ice-cold PBS, lysed in 500 µl co-IP buffer, and isolated using a kit from Qiagen. For RNA digestion assay, 1 µg RNA was incubated with 1 µl RNase R and 1 µl 10 x RNase R buffer in 10 µl total reaction solution (RNase R, RNR07250) at 37 °C for 15 min, followed by real-time PCR analysis of circ-RNA that was resistant to RNase R treatment.
For RNA pull-down assay, the cell lysate was incubated with 5 μg primary antibody at 4°C for 2 h. 50 μl of 50% slurry of protein A-Sepharose was added to each sample, and the mixtures were incubated at 4°C for 4 h followed by brief centrifugation. The pellets were washed 3 times with PBS and resuspended in 0.5 ml Tri Reagent (Sigma-Aldrich) followed by elution. The eluted co-precipitated RNA in the aqueous was subject to real-time PCR to measure the RNA of interest using specific primers.
Protein pull-down by RNA
The pull-down assay was performed using an RNA probe complementary to circ-Amotl1, labelled with biotin using the techniques described 35. Briefly, 1×107 cells were washed in ice-cold phosphate-buffered saline, lysed in 500 µl co-IP buffer, and incubated with 3 µg biotinylated DNA oligo probes at room temperature for 2 h. 50 μl Streptavidin C1 magnetic beads (Invitrogen) were added to each binding reaction and further incubated at room temperature for another one hour. The beads were washed briefly with co-IP buffer for five times. The bound proteins in the pull-down materials were analyzed by Western blotting.
General methods for protein and RNA analyses
Protein assays on Western blot and immunohistochemistry were performed as described38, 39. Real-time PCR was conducted as described 40, 41.
Statistical analysis
All experiments were performed in at least triplicate and numerical data were subject to independent sample t test. The levels of significance were set at *p<0.05 and **p<0.01.
Acknowledgements
This work was supported by Canadian Institutes of Health Research (PJT-149083) to BBY, who is the recipient of a Career Investigator Award (CI7418) from the Heart and Stroke Foundation of Ontario. WWD is supported by a Postdoctoral Fellowship from the Breast Cancer Foundation of Ontario.
Supplementary Material
Supplementary figures and tables.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Zhang FX, Chen ML, Shan QJ, Zou JG, Chen C, Yang B, Xu DJ, Jin Y, Cao KJ. Hypoxia reoxygenation induces premature senescence in neonatal sd rat cardiomyocytes. Acta Pharmacol Sin. 2007;28:44-51
2. Minamino T, Komuro I. Vascular cell senescence: Contribution to atherosclerosis. Circ Res. 2007;100:15-26
3. Gomez-Suarez M, Gutierrez-Martinez IZ, Hernandez-Trejo JA, Hernandez-Ruiz M, Suarez-Perez D, Candelario A, Kamekura R, Medina-Contreras O, Schnoor M, Ortiz-Navarrete V, Villegas-Sepulveda N, Parkos C, Nusrat A, Nava P. 14-3-3 proteins regulate akt thr308 phosphorylation in intestinal epithelial cells. Cell Death Differ. 2016;23:1060-1072
4. Yulyana Y, Ho IA, Sia KC, Newman JP, Toh XY, Endaya BB, Chan JK, Gnecchi M, Huynh H, Chung AY, Lim KH, Leong HS, Iyer NG, Hui KM, Lam PY. Paracrine factors of human fetal mscs inhibit liver cancer growth through reduced activation of igf-1r/pi3k/akt signaling. Mol Ther. 2015;23:746-756
5. Tao SC, Yuan T, Rui BY, Zhu ZZ, Guo SC, Zhang CQ. Exosomes derived from human platelet-rich plasma prevent apoptosis induced by glucocorticoid-associated endoplasmic reticulum stress in rat osteonecrosis of the femoral head via the akt/bad/bcl-2 signal pathway. Theranostics. 2017;7:733-750
6. Kumarswamy R, Lyon AR, Volkmann I, Mills AM, Bretthauer J, Pahuja A, Geers-Knorr C, Kraft T, Hajjar RJ, Macleod KT, Harding SE, Thum T. Serca2a gene therapy restores microrna-1 expression in heart failure via an akt/foxo3a-dependent pathway. Eur Heart J. 2012;33:1067-1075
7. Kuehnert J, Sommer G, Zierk AW, Fedarovich A, Brock A, Fedarovich D, Heise T. Novel rna chaperone domain of rna-binding protein la is regulated by akt phosphorylation. Nucleic Acids Res. 2015;43:581-594
8. Lee JH, Kang BH, Jang H, Kim TW, Choi J, Kwak S, Han J, Cho EJ, Youn HD. Akt phosphorylates h3-threonine 45 to facilitate termination of gene transcription in response to DNA damage. Nucleic Acids Res. 2015;43:4505-4516
9. Yang JL, Chen WY, Chen YP, Kuo CY, Chen SD. Activation of glp-1 receptor enhances neuronal base excision repair via pi3k-akt-induced expression of apurinic/apyrimidinic endonuclease 1. Theranostics. 2016;6:2015-2027
10. Wang C, Jin H, Wang N, Fan S, Wang Y, Zhang Y, Wei L, Tao X, Gu D, Zhao F, Fang J, Yao M, Qin W. Gas6/axl axis contributes to chemoresistance and metastasis in breast cancer through akt/gsk-3beta/beta-catenin signaling. Theranostics. 2016;6:1205-1219
11. Salazar M, Lorente M, Garcia-Taboada E, Perez Gomez E, Davila D, Zuniga-Garcia P, Maria Flores J, Rodriguez A, Hegedus Z, Mosen-Ansorena D, Aransay AM, Hernandez-Tiedra S, Lopez-Valero I, Quintanilla M, Sanchez C, Iovanna JL, Dusetti N, Guzman M, Francis SE, Carracedo A, Kiss-Toth E, Velasco G. Loss of tribbles pseudokinase-3 promotes akt-driven tumorigenesis via foxo inactivation. Cell Death Differ. 2015;22:131-144
12. Du WW, Yang W, Chen Y, Wu ZK, Foster FS, Yang Z, Li X, Yang BB. Foxo3 circular rna promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur Heart J. 2017;38:1402-1412
13. Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular rna retards cell cycle progression via forming ternary complexes with p21 and cdk2. Nucleic Acids Res. 2016;44:2846-2858
14. Zhang XO, Dong R, Zhang Y, Zhang JL, Luo Z, Zhang J, Chen LL, Yang L. Diverse alternative back-splicing and alternative splicing landscape of circular rnas. Genome Res. 2016;26:1277-1287
15. Khan MA, Reckman YJ, Aufiero S, van den Hoogenhof MM, van der Made I, Beqqali A, Koolbergen DR, Rasmussen TB, van der Velden J, Creemers EE, Pinto YM. Rbm20 regulates circular rna production from the titin gene. Circ Res. 2016;119:996-1003
16. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural rna circles function as efficient microrna sponges. Nature. 2013;495:384-388
17. Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL. Circular intronic long noncoding rnas. Mol Cell. 2013;51:792-806
18. Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, Zhu P, Chang Z, Wu Q, Zhao Y, Jia Y, Xu P, Liu H, Shan G. Exon-intron circular rnas regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256-264
19. Holdt LM, Stahringer A, Sass K, Pichler G, Kulak NA, Wilfert W, Kohlmaier A, Herbst A, Northoff BH, Nicolaou A, Gabel G, Beutner F, Scholz M, Thiery J, Musunuru K, Krohn K, Mann M, Teupser D. Circular non-coding rna anril modulates ribosomal rna maturation and atherosclerosis in humans. Nat Commun. 2016;7:12429
20. Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159:134-147
21. Chen L, Shan G. Circular rnas remain peculiarly unclear in biogenesis and function. Sci China Life Sci. 2015;58:616-618
22. Gagne V, Moreau J, Plourde M, Lapointe M, Lord M, Gagnon E, Fernandes MJ. Human angiomotin-like 1 associates with an angiomotin protein complex through its coiled-coil domain and induces the remodeling of the actin cytoskeleton. Cell Motil Cytoskeleton. 2009;66:754-768
23. Zheng Y, Vertuani S, Nystrom S, Audebert S, Meijer I, Tegnebratt T, Borg JP, Uhlen P, Majumdar A, Holmgren L. Angiomotin-like protein 1 controls endothelial polarity and junction stability during sprouting angiogenesis. Circ Res. 2009;105:260-270
24. Couderc C, Boin A, Fuhrmann L, Vincent-Salomon A, Mandati V, Kieffer Y, Mechta-Grigoriou F, Del Maestro L, Chavrier P, Vallerand D, Brito I, Dubois T, De Koning L, Bouvard D, Louvard D, Gautreau A, Lallemand D. Amotl1 promotes breast cancer progression and is antagonized by merlin. Neoplasia. 2016;18:10-24
25. Higuchi M, Onishi K, Kikuchi C, Gotoh Y. Scaffolding function of pak in the pdk1-akt pathway. Nat Cell Biol. 2008;10:1356-1364
26. Yang W, Du WW, Li X, Yee AJ, Yang BB. Foxo3 activity promoted by non-coding effects of circular rna and foxo3 pseudogene in the inhibition of tumor growth and angiogenesis. Oncogene. 2016;35:3919-3931
27. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N. Circular rnas are a large class of animal rnas with regulatory potency. Nature. 2013;495:333-338
28. Jeck WR, Sharpless NE. Detecting and characterizing circular rnas. Nat Biotechnol. 2014;32:453-461
29. Salzman J. Circular rna expression: Its potential regulation and function. Trends Genet. 2016;32:309-316
30. Tang X, Jang SW, Wang X, Liu Z, Bahr SM, Sun SY, Brat D, Gutmann DH, Ye K. Akt phosphorylation regulates the tumour-suppressor merlin through ubiquitination and degradation. Nat Cell Biol. 2007;9:1199-1207
31. Kitamura T, Asai N, Enomoto A, Maeda K, Kato T, Ishida M, Jiang P, Watanabe T, Usukura J, Kondo T, Costantini F, Murohara T, Takahashi M. Regulation of vegf-mediated angiogenesis by the akt/pkb substrate girdin. Nat Cell Biol. 2008;10:329-337
32. Lin HK, Wang G, Chen Z, Teruya-Feldstein J, Liu Y, Chan CH, Yang WL, Erdjument-Bromage H, Nakayama KI, Nimer S, Tempst P, Pandolfi PP. Phosphorylation-dependent regulation of cytosolic localization and oncogenic function of skp2 by akt/pkb. Nat Cell Biol. 2009;11:420-432
33. Manning BD, Cantley LC. Akt/pkb signaling: Navigating downstream. Cell. 2007;129:1261-1274
34. Pekarsky Y, Koval A, Hallas C, Bichi R, Tresini M, Malstrom S, Russo G, Tsichlis P, Croce CM. Tcl1 enhances akt kinase activity and mediates its nuclear translocation. Proc Natl Acad Sci U S A. 2000;97:3028-3033
35. Du WW, Fang L, Yang W, Wu N, Awan FM, Yang Z, Yang BB. Induction of tumor apoptosis through a circular rna enhancing foxo3 activity. Cell Death Differ. 2017;24:357-370
36. Ehler E, Moore-Morris T, Lange S. Isolation and culture of neonatal mouse cardiomyocytes. J Vis Exp. 2013
37. Graham EL, Balla C, Franchino H, Melman Y, del Monte F, Das S. Isolation, culture, and functional characterization of adult mouse cardiomyoctyes. J Vis Exp. 2013:e50289
38. Du WW, Liu F, Shan SW, Ma XC, Gupta S, Jin T, Spaner D, Krylov SN, Zhang Y, Ling W, Yang BB. Inhibition of dexamethasone-induced fatty liver development by reducing mir-17-5p levels. Mol Ther. 2015;23:1222-1233
39. Yang BL, Cao L, Kiani C, Lee V, Zhang Y, Adams ME, Yang BB. Tandem repeats are involved in g1 domain inhibition of versican expression and secretion and the g3 domain enhances glycosaminoglycan modification and product secretion via the complement-binding protein-like motif. J Biol Chem. 2000;275:21255-21261
40. Rutnam ZJ, Du WW, Yang W, Yang X, Yang BB. The pseudogene tusc2p promotes tusc2 function by binding multiple micrornas. Nat Commun. 2014;5:2914
41. Shan SW, Fang L, Shatseva T, Rutnam ZJ, Yang X, Du W, Lu WY, Xuan JW, Deng Z, Yang BB. Mature mir-17-5p and passenger mir-17-3p induce hepatocellular carcinoma by targeting pten, galnt7 and vimentin in different signal pathways. J Cell Sci. 2013;126:1517-1530
Author contact
![]() Corresponding author: BB Yang, S-Wing Research Building, 2075 Bayview Ave, Toronto M4N 3M5 Canada, tel: (416) 480-5874; e-mail: byangutoronto.ca
Corresponding author: BB Yang, S-Wing Research Building, 2075 Bayview Ave, Toronto M4N 3M5 Canada, tel: (416) 480-5874; e-mail: byangutoronto.ca
 Global reach, higher impact
Global reach, higher impact