13.3
Impact Factor
Theranostics 2017; 7(16):3814-3823. doi:10.7150/thno.21098 This issue Cite
Research Paper
Serum Immune Profiling for Early Detection of Cervical Disease
1. Center for Personalized Diagnostics, Biodesign Institute, Arizona State University, Tempe, AZ.
2. Division of High-Consequence Pathogens and Pathology, Centers for Disease Control and Prevention, Atlanta, GA.
Received 2017-5-19; Accepted 2017-7-9; Published 2017-8-23
Abstract
Background: The most recent (2012) worldwide estimates from International Agency for Research on Cancer indicate that approximately 528,000 new cases and 270,000 deaths per year are attributed to cervical cancer worldwide. The disease is preventable with HPV vaccination and with early detection and treatment of pre-invasive cervical intraepithelial neoplasia, CIN. Antibodies (Abs) to HPV proteins are under investigation as potential biomarkers for early detection.
Methods: To detect circulating HPV-specific IgG Abs, we developed programmable protein arrays (NAPPA) that display the proteomes of two low-risk HPV types (HPV6 and 11) and ten oncogenic high-risk HPV types (HPV16, 18, 31, 33, 35, 39, 45, 51, 52 and 58). Arrays were probed with sera from women with CIN 0/I (n=78), CIN II/III (n=84), or invasive cervical cancer (ICC, n=83).
Results: Abs to any early (E) HPV protein were detected less frequently in women with CIN 0/I (23.7%) than women with CIN II/III (39.0%) and ICC (46.1%, p<0.04). Of the E Abs, anti-E7 Abs were the most frequently detected (6.6%, 19.5%, and 30.3%, respectively). The least frequently detected Abs were E1 and E2-Abs in CIN 0/I (1.3%) and E1-Abs in CIN II/III (1.2%) and ICC (7.9%). HPV16-specific Abs correlated with HPV16 DNA detected in the cervix in 0% of CIN 0/I, 21.2% of CIN II/III, and 45.5% of ICC. A significant number (29 - 73%) of E4, E7, L1, and L2 Abs had cross-reactivity between HPV types.
Conclusion: HPV protein arrays provide a valuable high-throughput tool for measuring the breadth, specificity, and heterogeneity of the serologic response to HPV in cervical disease.
Keywords: Antibodies, HPV, cervical cancer, cervical intraepithelial neoplasia, NAPPA, protein microarrays, serology, early detection
Introduction
Human papillomaviruses (HPV) are a family of more than 200 closely related viruses with small (8 kb) circular double-stranded genomes. The virus is restricted to epithelial surfaces where it induces a non-lytic cellular proliferation and minimal immunologic response. The estimated number of new cases of cervical cancer worldwide was 528,000 in 2012, with an annual global mortality rate of 270,000 deaths [1, 2]. Vaccines preventing infection are highly effective for the prevention of type-specific cervical and anogenital cancer precursors (vaginal, vulvar, anal), may reduce relapse after conization [3], but are not a treatment for pre-existing HPV infection. Vaccines are expected to substantially reduce the burden of HPV-associated cancers. In the US, even with low vaccine coverage, vaccination has resulted in a 64% reduction in the prevalence of types 6, 11, 16, and 18 among females aged 14 to 19 years and 34% decrease among those aged 20- 24 years [4].
The epidemiology and natural history of HPV infection has been best characterized in the cervix where precursor lesions are well recognized, using detection of HPV DNA or detection of antibodies (Abs) to HPV as biomarkers of disease pathogenesis [5]. Genital HPV is usually acquired shortly after sexual debut, and prevalence is highest in adolescents and young adults [4, 6, 7]. Cervical cancer is a rare consequence of this common infection, with ~50% of the cases worldwide caused by the HPV16 type. While high risk (HR) HPV infection is considered necessary for cervical carcinogenesis, additional factors are clearly involved. A small fraction of infected women gradually progress to invasive cancer, following a long, histologically well-defined pre-invasive phase (cervical intraepithelial neoplasia; CIN), ranging from low grade (CIN I) to high grade (CIN II and III) [8]. Cervical cancer is preventable because high grade lesions are detectable by clinical, histopathologic, or molecular alterations and can be surgically removed [9]. Current clinical practice in the US relies on regular screening with cytology (Pap test) often combined with HR HPV nucleic acid testing to refer women for colposcopy and biopsy. Recent data have documented that cytology screening is associated with a significantly reduced incidence and risk of death from cervical cancer, with odds ratios ranging from 0.28 to 0.60 [10, 11], despite a reported high false negative rate [12]. Cytologic screening remains subject to sampling errors, problems with cellular preservation, and reader subjectivity. Biomarkers are needed in particular to aid in the selection of patients for colposcopy screening in resource-limited settings in low and middle income countries (LMICs), where nucleic acid and cytology testing are cost-prohibitive. Efforts are underway by the World Health Organization and the Program for Appropriate Technology in Health (PATH) to generate cost-effective HPV DNA testing [13].
Measuring the humoral immune response to HPV antigens (Ags) has been integral to understanding the natural history of infection and efficacy of vaccination [8, 14, 15]. Despite the potential of HPV serology in disease diagnosis and prognosis, its clinical application has been limited by HPV heterogeneity, assay variability, and viral immune evasion. HPV has a limited repertoire of proteins, grouped as early (E1, E2, E4, E5, E6, E7) and late (L1, L2) proteins. The late proteins form the viral protein coat during productive infections. The early proteins interact with host and viral proteins to maintain viral replication. The serologic response to genital HPV infection is primarily directed at conformational epitopes on the viral major capsid protein L1. As the infection is non-lytic, the host Ab response to L1 is weak and may persist for years, as an indication of past infection but not malignancy [16, 17]. Although anti-L1 Abs are an indication of past infection, only 50 - 70% of infected women seroconvert [18, 19].
Abs to both HPV16 E6 and E7 proteins have been detected at low levels in both serum and cervical-vaginal secretions of invasive cervical cancer (ICC) patients [20]. Their levels increase with cervical disease progression but they are not detectable in a subset of patients with cervical cancer [16, 21-23]. They develop later in the course of ICC, and are correlated with disease outcome [21, 24, 25]. Studies of sera collected prior to the diagnosis of cervical cancer have shown that the presence of E6 and E7-specific Abs is associated with an increased relative risk (RR=2.7) for cervical cancer, and can be detected, albeit infrequently, up to 5 years prior to diagnosis [26]. The percentage of women with false negative serology is dependent on the method of Ab detection [16, 27-31].
The diverse array of oncogenic HPV types and the technical limitations of high throughput protein expression and display have been impediments to HPV immune profiling and most research has focused on select Ags from the most common viral types [16, 21, 28]. Nucleic Acid Programmable Protein Arrays (NAPPA) [32, 33] have enabled rapid profiling of the serum Ab response in the settings of infections [34, 35], autoimmune diseases [36, 37] and cancer [38-40]. To measure the serologic responses across multiple HPV types, we adapted NAPPA for the detection of HPV-specific IgG Abs in sera [41]. Full length cDNAs encoding the proteomes of 12 HPV types are expressed as C-terminal GST fusion proteins using mammalian in vitro transcription/translation, and captured onto a glass slide surface [42]. In a pilot study, we demonstrated that HPV protein arrays display immunogenic epitopes that can be detected using HPV-specific monoclonal Abs (MAbs) and with select sera from HPV-specific malignancies [41]. The purpose of this study was to systematically investigate the serologic immune profile to HPV in women with high-grade pre-invasive cervical lesions and ICC, and to identify serologic biomarkers for diagnosis and early detection of cervical cancer.
Materials and methods
Sample selection
We used the Biorepository for Molecular Signatures of Cervical Cancer developed by the Centers for Disease Control and Prevention as a Clinical Epidemiology and Validation Center for NCI's Early Detection Research Network (EDRN). Samples in the biorepository were collected from women attending colposcopy clinics at urban public hospitals in Atlanta, GA, Detroit, MI, or Galveston, TX between 2000 and 2010 and linked to epidemiologic and clinical data, including HPV detected in exfoliated cervical cells, age, race, and tissue confirmation of cervical disease status [43]. For this study, 162 samples from women with cervical intraepithelial neoplasia (CIN) grade 0 (no CIN), I, II, III were selected, of which 78 were CIN 0/I and 84 were CIN II/III. We used 83 archived anonymized plasma samples from women with ICC collected in Atlanta, GA prior to 1997. For convenience, the term serum is used throughout the manuscript. While HPV vaccine history was not collected, HPV vaccination was not introduced before 2006, and it is unlikely that any study participants were vaccinated. Only a subset (n=51) of the ICC samples had information on the HPV DNA status of the tumor, of which 24 (47.1%) were HPV16+. Samples were collected using a standardized sample collection protocol and stored at -80°C until use. Written informed consent was obtained from all subjects under institutional review board approval.
HPV microarray generation and detection of serum antibodies
Production of custom HPV protein arrays and array quality control experiments were performed as previously reported [41] with modifications described here. In brief, arrays displaying codon-optimized proteomes of 2 low risk (HPV6 and 11) and 10 high risk (HPV16, 18, 31, 33, 35, 39, 45, 51, 52, and 58) HPV types were generated. The codon-optimized HPV16 L1 gene previously used [41] was replaced with the non-codon-optimized version, which had higher protein expression. Both codon-optimized and non-optimized HPV16 E6 and E7 were printed on the arrays for direct comparison of Ab reactivity. Since only the non-codon optimized HPV16 E6 and HPV16 E7 [41] showed immunoreactivity, only those results are shown. All non-codon-optimized genes were obtained by nested PCR using gene-specific primers from HPV16 plasmid DNA (American Type Culture Collection, Manassas, VA) as described [44]. All genes were inserted into pDONR221 vector per manufacturer's instructions (Invitrogen, Carlsbad, CA), and were converted to the pANT7_cGST vector (http://dnasu.asu.edu/DNASU/Home.jsp)[32]. Human IgG and the C-terminal portion of the Epstein-Barr virus-derived Ag EBNA-1 were used as positive controls. A set of non-HPV related negative control proteins (n=93) were printed on the arrays and used for array signal intensity normalization and establishment of cut-off values. Arrays were incubated with serum samples diluted 1:50 in 10% E. coli lysate prepared in 5% milk-PBST (0.2% tween) [45] and serum Ab detection was performed as previously described [38, 41, 44, 46].
Protein array image analysis and quantification
After serum binding and IgG detection, arrays were scanned by Tecan PowerScanner (Tecan Group, Männedorf, Switzerland). ArrayPro Analyzer version 6.3 (MediaCybernetics, Bethesda, MD) was used to measure the signal intensity of individual spots on the scanned slides. Normalization of raw intensity values was performed by subtracting the slide background signal and dividing by the background signal subtracted from the median intensity of all spots. The slide background signal was determined by the first quartile of signal intensity of the no-DNA control spots (all material except DNA). In addition, array images were qualitatively inspected to identify and confirm positive responses by adjusting raw images to extreme brightness and contrast using ArrayPro Analyzer and visual analysis of diffused signal (ring) as described previously [34, 40]. Each spot was scored based on the intensity and morphology of the ring on a scale of 0 to 5.
HPV DNA detection by L1 consensus PCR
For all the samples from the biorepository, HPV DNA was detected in extracts of exfoliated cervical cells collected in PreservCyt media as previously described [43]. Briefly, 16 ml of the PreservCyt collection media was extracted using MasterPure Complete DNA and RNA purification kit (Epicentre, Madison, WI). HPV detection and typing was performed using the Roche linear array that detects 37 types. HPV results for the anonymized archived cervical cancer cases (n=83) were based on combined results of colorimetric ISH for HPV16, 18, 31, 33, 35 on formalin-fixed paraffin embedded tissue sections and L1 consensus PCR with MY09/11 primers and type-specific hybridization to 6 HR types (16, 18, 31, 33, 35, 45) on DNA extracts from the same tissues (methods in use at the time of archiving [47]).
Statistical analysis
The correlation of raw signal intensities of protein expression between the arrays randomly selected for quality control was determined with scatter plots and the Pearson correlation coefficient (R) was calculated to assess consistency. Levels of protein expression on the arrays were measured by calculating the mean values of raw signal intensities of duplicate spots from two arrays. For serum Ab reactivity on protein arrays, mean values (of duplicate spots for a given Ag) of normalized signal intensity were compared for different disease groups using Fisher's exact test (Graphpad Prism version 5.0c, San Diego, CA). A p-value of <0.05 was considered significant. Seropositivity for any given Ag was defined as the median of normalized signal intensity values of all negative control proteins (n=93) in all sera (n=234) +3 standard deviations or spots that were positive by visual analysis. A total of 245 serum samples were tested on the arrays, of which 11 (4.5%; n=7 ICC, n=4 CIN) were excluded from the analysis due to high background. High array background was defined as an array with normalized signal intensity values for ≥14 out of 93 negative control spots exceeding the 75th percentile + 1.5*interquartile range of this negative control protein across all arrays.
Results
Characteristics of study samples
Our primary goal was to determine the prevalence and specificity of HPV-specific Ab responses in women with cervical cancer precursors and with ICC. Age, race, and HPV DNA status of patients contributing samples to the study are shown in Table 1. Ab levels were compared in women with CIN 0/I (n=78) and CIN II/III (n=84) who were referred to colposcopy because of abnormalities in cervical cancer screening, and in women with ICC (n=83). Women with CIN 0/I were chosen as the relevant control population to determine the utility of these biomarkers within a high-risk population. As expected, women with CIN 0/I had a lower frequency of cervical high-risk (HR) HPV than women with CIN II/III (57.7% vs. 97.6%, p<0.0001). Infection with 2 or more HPV types was detected in more than 35% of women in both CIN 0/I and CIN II/III (Table 1). Women with CIN II/III were as expected significantly younger than women with ICC (mean 30.0 yrs vs. 52.0 yrs, p<0.0001). There was also a lower frequency (p<0.0015) of HPV16 in CIN 0/I (19.2%) than CIN II/III (63.1%) and ICC (47.1%). The clinics participating in the EDRN study had a high proportion of minority and Hispanic white patients. The racial distribution of the samples occurred by chance.
Production and reproducibility of NAPPA HPV protein arrays
The quality and reproducibility of the array printing were evaluated by picogreen staining of DNA and measuring protein expression with anti-GST Abs (Supplemental Figure 1A). Three arrays were printed on each slide and the correlation coefficients of anti-GST signal intensities were determined for intra-array (R=0.98) and intra-batch replicate arrays (R=0.90) from two subarrays on the same slide or two randomly selected slides within the print batch (Supplemental Figure 1B).
HPV-specific antibody prevalence
There was no significant difference (p=0.46) in the percentages of negative control spots (displaying the non-HPV related proteins; n=93) that exceeded the cut-off value between arrays probed with CIN 0/I (0.71%), CIN II/III (0.81%), and ICC (1.77%) sera.
The prevalence of HPV-specific serum IgG Abs among women with CIN 0/I, CIN II/III, and ICC is summarized in Table 2. At least one of the HPV-specific Abs was detected in serum from women with CIN 0/I (46.1%), CIN II/III (59.8%), and ICC (68.4%). Abs to any early (E) HPV protein were detected more frequently in women with ICC (46.1%) and CIN II/III (39.0%) than women with CIN 0/I (23.7%, p<0.04). Abs to any L1 protein had the highest prevalence (28.9%, 34.1%, and 44.7% in CIN 0/I, CIN II/III, and ICC, respectively). Of the E Abs, anti-E7 Abs were the most frequently detected (CIN 0/I, 6.6%; CIN II/III, 19.5%; and ICC, 30.3%). The least frequently detected Abs were E1 and E2-Abs in CIN 0/I (1.3%) and E1-Abs in CIN II/III (1.2%) and ICC (7.9%). The sensitivity [proportion of cases with at least one HPV Ag-specific Ab detected] was comparable when restricted to cases known to have HPV16 (as opposed to any other oncogenic HPV) DNA detected; 59.6% vs 59.8% for CIN II/III and 81.8% vs 68.4% for ICC, (p-value N.S.). Among all women (irrespective of HPV DNA status), Abs to HPV16 Ags were detected in only 6.6%, 19.5%, and 35.5% in CIN 0/I, CIN II/III, and ICC, demonstrating the importance of multi-antigenic immunoprofiling.
Characteristics of Study Samples.
| Characteristics | Disease Status | ||
|---|---|---|---|
| CIN 0/I N = 78 | CIN II/III N = 84 | ICC N = 83 | |
| N (%) | N (%) | N (%)* | |
| Age in yrs, Mean | 28.7 | 30.0 | 52.0 |
| < 30 | 51 (65.3) | 45 (53.6) | 3/79 (3.8) |
| ≥ 30 | 27 (34.6) | 39 (46.4) | 76/79 (96.2) |
| Race | |||
| Black | 71 (91.0) | 55 (65.5) | 64/79 (81.0) |
| Other | 7 (9.0) | 29 (34.5) | 15/79 (19.0) |
| HPV16 DNA status† | |||
| HPV16+ | 15 (19.2) | 53 (63.1) | 24/51 (47.1) |
| HPV DNA status overall† | |||
| Negative | 23 (29.5) | 2 (2.4) | 12/51 (23.5) |
| 1 HPV type | 29 (37.2) | 46 (54.8) | 39/51 (76.5) |
| 2 HPV types | 10 (12.8) | 19 (22.6) | 0/51 (0) |
| ≥ 3 HPV types | 16 (20.5) | 17 (20.2) | 0/51 (0) |
| Any HR HPV‡ | 45 (57.7) | 82 (97.6) | 39/51 (76.5) |
*N varies for each category because of missing information. The numbers of samples are shown.
†HPV testing methods used for anonymized archived samples differed from those used in biorepository, so results are not directly comparable.
‡The following HPV types were considered as high-risk types for this analysis -HPV16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, 68
Prevalence of positive antibody response (1) to each HPV protein from any HPV type (2).
| No. + (%) | |||||||
|---|---|---|---|---|---|---|---|
| CIN 0/I | CIN II/III | ICC | Total | ||||
| Total | HPV16+(3) | Total | HPV16+(3) | Total | HPV16+(3) | ||
| HPV Antibodies | N=76 | N=14 | N=82 | N=52 | N=76 | N=22(4) | |
| E1 | 1 (1.3) | 0 (0) | 1 (1.2) | 0 (0) | 6 (7.9)* | 1 (4.5) | 8 (3.4) |
| E2 | 1 (1.3) | 0 (0) | 4 (4.9) | 1 (1.9) | 11 (14.5) | 5 (22.7) | 16 (6.8) |
| E4 | 5 (6.6) | 0 (0) | 14 (17.1) | 5 (9.6) | 12 (15.8) | 4 (18.2) | 31 (13.2) |
| E5 | 5 (6.6) | 1 (7.1) | 4 (4.9) | 3 (5.8) | 12 (15.8) | 6 (27.3) | 21 (9.0) |
| E6 | 3 (3.9) | 0 (0) | 8 (9.8) | 5 (9.6) | 12 (15.8)* | 5 (22.7) | 23 (9.8) |
| E7 | 5 (6.6) | 0 (0) | 16 (19.5)* | 10 (19.2) | 23(30.3)* | 8 (36.4)* | 44 (18.8) |
| Any E (2) | 18 (23.7) | 2 (14.3) | 32 (39.0)* | 19 (36.5) | 35 (46.1)* | 15 (68.2)* | 85 (36.3) |
| L1 | 22 (28.9) | 4 (28.6) | 28 (34.1) | 16 (30.8) | 34 (44.7) | 12 (54.5) | 84 (35.9) |
| L2 | 8 (10.5) | 0 (0) | 5 (6.1) | 5 (9.6) | 13 (17.1) | 6 (27.3) | 26 (11.1) |
| Any L (2) | 25 (32.9) | 4 (28.6) | 32 (39.0) | 20 (38.5) | 38 (50.0) | 14 (63.6)* | 95 (40.6) |
| Any E and/or L(2) | 35 (46.1) | 4 (28.6) | 49 (59.8) | 31 (59.6) | 52 (68.4)* | 18 (81.8)* | 136 (58.1) |
| Any HPV16 Ag | 5 (6.6) | 0 (0) | 16 (19.5)* | 11 (21.2) | 27 (35.5)* | 10 (45.5)* | 48 (20.5) |
(1) Cut-off values defined as the median of normalized signal intensity values of all negative control proteins +3 standard deviations in all sera (n=234) or spots that were positive by visual analysis.
(2) Any positive vs. all negative from any of the 12 HPV types tested.
(3) HPV16 DNA detected in cervix. HPV testing methods used for anonymized archived samples differed from those used in biorepository.
(4) HPV DNA status was known for only a subset (n=51) of the ICC samples.
* p<0.05, compared with CIN 0/I
Type-specific antibody response
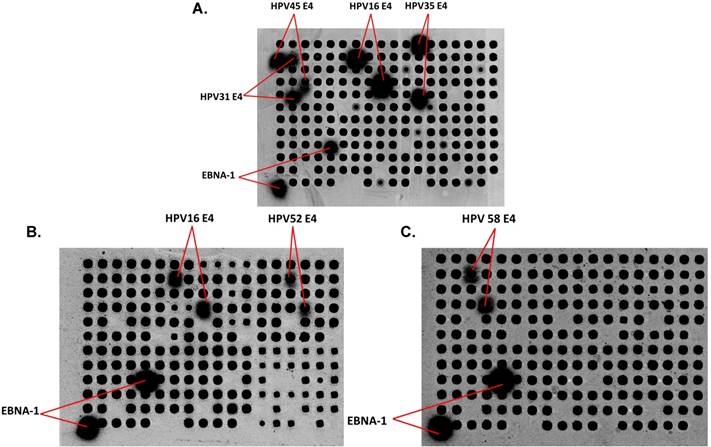
To determine whether patients with a specific HPV infection develop type-specific Abs, there are multiple challenges. First, a significant number of women with CIN have multiple HPV types detected (33.3% of CIN 0/I and 42.8% of CIN II/III, Table 1), and past exposure to other HPV types cannot be excluded. Second, there is likely serologic cross-reactivity across HPV types. Figure 1A shows Abs from an ICC patient reacting with E4 protein from 4 different HPV types (16, 31, 35, and 45). As examples, Figures 1B and 1C show serum from two women with CIN II/III with HPV16 DNA and Abs against HPV16 E4 and HPV52 E4 (B) and HPV 58 E4 (C).
To determine whether there is any correlation between HPV DNA types detected in the patient and type-specific serum Abs, we analyzed the data in two ways. In Table 3A, subjects were stratified based on cervical HPV DNA status. For example, among women with successful Ab testing, cervical HPV16 DNA was detected in 18.4% of those with CIN 0/I; 63.4% of CIN II/III; and 46.8% of ICC. The type-specific Ab detection rate in this group was 0%, 21.2%, and 45.5% for CIN 0/I, CIN II/III and ICC. For the most common HPV DNA detected in cervical samples in CIN II/III, HPV16, 31, 35, and 52, the range of detection of type-specific Abs was 8.3 - 25.0%. In Table 3B, subjects were stratified by type-specific seropositivity to evaluate the proportion with detection of type-specific HPV DNA in the cervix. Women with HPV16 Abs and CIN II/III had the highest type-specific DNA detection, 78.6%, followed by HPV31 (36.4%), 45 (25.0%), and 52 (14.2%).
Prevalence of HPV type-specific IgG Abs in women with known cervical HPV DNA status.
HPV DNA in cervix | No. + (%) | ||||||
|---|---|---|---|---|---|---|---|
| Total Number(1) | CIN 0/I N=76 | CIN II/III N=82 | ICC N=47(2) | ||||
| Total | Ab+(3) | Total | Ab+(3) | Total | Ab+(3) | ||
| HPV6 | 3 | 1 (1.3) | 0 (0) | 2 (2.4) | 0 (0) | 0 (0) | 0 (0) |
| 11 | 2 | 1 (1.3) | 0 (0) | 1 (1.2) | 0 (0) | 0 (0) | 0 (0) |
| 16 | 88 | 14 (18.4) | 0 (0) | 52 (63.4) | 11 (21.2) | 22 (46.8) | 10 (45.5) |
| 18 | 16 | 3 (3.9) | 1 (33.3) | 5 (6.1) | 1 (20.0) | 8 (17.0) | 2 (25.0) |
| 31 | 20 | 2 (2.6) | 0 (0) | 16 (19.5) | 4 (25.0) | 2 (4.3) | 0 (0) |
| 33 | 10 | 2 (2.6) | 0 (0) | 5 (6.1) | 0 (0) | 3 (6.4) | 1 (33.3) |
| 35 | 16 | 5 (6.6) | 2 (40.0) | 10 (12.2) | 2 (20.0) | 1 (2.1) | 1 (100.0) |
| 39 | 5 | 3 (3.9) | 1 (33.3) | 2 (2.4) | 1 (50.0) | 0 (0) | 0 (0) |
| 45 | 10 | 5 (6.6) | 1 (20.0) | 5 (6.1) | 1 (20.0) | 0 (0) | 0 (0) |
| 51 | 7 | 4 (5.3) | 1 (25.0) | 2 (2.4) | 0 (0) | 1 (2.1) | 0 (0) |
| 52 | 21 | 9 (11.8) | 1 (11.1) | 12 (14.6) | 1 (8.3) | 0 (0) | 0 (0) |
| 58 | 14 | 10 (13.2) | 1 (10.0) | 3 (3.7) | 0 (0) | 1 (2.1) | 0 (0) |
Prevalence of type-specific HPV DNA in women with known seropositivity.
HPV -specific Abs | Total Number(4) | No. + (%) | |||||
|---|---|---|---|---|---|---|---|
| CIN 0/I N=76 | CIN II/III N=82 | ICC N=76(2) | |||||
| Total | DNA+(5) | Total | DNA+(5) | Total | DNA+(5) | ||
| HPV6 | 24 | 4 (5.3) | 0 (0) | 10 (12.2) | 0 (0) | 10 (13.2) | 0 (0) |
| 11 | 25 | 5 (6.6) | 0 (0) | 8 (9.8) | 0 (0) | 12 (15.8) | 0 (0) |
| 16 | 46 | 5 (6.6) | 0 (0) | 14 (17.1) | 11 (78.6) | 27 (35.5) | 10 (37.0) |
| 18 | 50 | 11 (14.5) | 1 (9.0) | 15 (18.3) | 1 (6.7) | 24 (31.6) | 2 (8.3) |
| 31 | 37 | 5 (6.6) | 0 (0) | 11 (13.4) | 4 (36.4) | 21 (27.6) | 0 (0) |
| 33 | 58 | 14 (18.4) | 0 (0) | 18 (22.0) | 0 (0) | 26 (34.2) | 1 (3.8) |
| 35 | 78 | 21 (27.6) | 2 (9.5) | 19 (23.2) | 2 (10.5) | 38 (50.0) | 1 (2.6) |
| 39 | 51 | 10 (13.2) | 1 (10.0) | 20 (24.4) | 1 (5.0) | 21 (27.6) | 0 (0) |
| 45 | 26 | 5 (6.6) | 1 (20.0) | 4 (4.9) | 1 (25.0) | 17 (22.4) | 0 (0) |
| 51 | 24 | 5 (6.6) | 1 (20.0) | 6 (7.3) | 0 (0) | 13 (17.1) | 0 (0) |
| 52 | 31 | 5 (6.6) | 1 (20.0) | 7 (8.5) | 1 (14.2) | 19 (25.0) | 0 (0) |
| 58 | 18 | 4 (5.3) | 1 (25.0) | 5 (6.1) | 0 (0) | 9 (11.8) | 0 (0) |
(1) of women with the corresponding HPV DNA type in the cervix.
(2) The ICC samples with unknown tumor DNA status were excluded from the analysis in table 3A.
(3) Positive for any Ab specific to the given HPV type.
(4) of women with serum Abs to any Ag of the corresponding HPV type.
(5) Type-specific HPV DNA positive.
Detection of IgG in human serum using HPV protein arrays. (A) Detection of IgG Abs in serum from a patient with ICC. Immunoreactivity to the positive control EBV EBNA-1 protein, and HPV E4 protein from 4 different HPV types (16, 31, 35, and 45) are detected. (B) and (C) Detection of IgG Abs in sera from two women with CIN II/III. Immunoreactivity to HPV16 E4, HPV52 E4 (B) and HPV58E4 (C) as well as EBNA-1 protein, is shown. Dark spots represent the individual proteins (HPV Ags and non HPV-related controls in random order) displayed on the arrays after adjusting the raw images to extreme brightness and contrast. Positive spots (with diffused signal) are labeled.
Cross-reactivity of serologic responses
We determined the prevalence of Abs against homologous Ags (i.e. all E7 Ags) from more than one HPV type in all sera (n=234) from the three cervical disease groups under investigation (Table 4). Abs against L2 were the most cross-reactive, while anti-E1 Abs were the least cross-reactive. Of sera that had Abs against any L2 Ag, 57.7% were positive for L2 from at least 6 HPV types. For E1, all 8 women who had specific Abs were positive for E1 from only one HPV type. 8.1% of all women had Abs to E7 from at least 2 HPV types. The percentages of sera with cross-reactive Abs to at least one other HPV type were as follows: E2 (6.3%), E4 (29.0%), E5 (14.3%), E6 (17.4%), E7 (43.2%), L1 (39.3%), and L2 (73.1%).
Prevalence of serum Abs to homologous Ags from different HPV types.
| Homologous Abs(1) | E1 | E2 | E4 | E5 | E6 | E7 | L1 | L2 | |
|---|---|---|---|---|---|---|---|---|---|
| No. | 0 | 1 | 7 | 3 | 1 | 10 | 17 | 3 | |
| =2 | % of total(2) | 0.0 | 0.4 | 3.0 | 1.3 | 0.4 | 4.3 | 7.3 | 1.3 |
| % of positive(3) | 0.0 | 6.3 | 22.6 | 14.3 | 4.3 | 22.7 | 20.2 | 11.5 | |
| No. | 0 | 0 | 1 | 0 | 3 | 9 | 9 | 1 | |
| =3 | % of total(2) | 0.0 | 0.0 | 0.4 | 0.0 | 1.3 | 3.8 | 3.8 | 0.4 |
| % of positive(3) | 0.0 | 0.0 | 3.2 | 0.0 | 13.0 | 20.5 | 10.7 | 3.8 | |
| No. | 0 | 0 | 1 | 0 | 0 | 3 | 3 | 0 | |
| =4 | % of total(2) | 0.0 | 0.0 | 0.4 | 0.0 | 0.0 | 1.3 | 1.3 | 0.0 |
| % of positive(3) | 0.0 | 0.0 | 3.2 | 0.0 | 0.0 | 6.8 | 3.6 | 0.0 | |
| No. | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | |
| =5 | % of total(2) | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.9 | 0.0 |
| % of positive(3) | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.4 | 0.0 | |
| No. | 0 | 0 | 0 | 0 | 0 | 1 | 2 | 15 | |
| ≥6 | % of total(2) | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.4 | 0.9 | 6.4 |
| % of positive(3) | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.3 | 2.4 | 57.7 |
(1) Number of homologous Ags from different HPV types to which Abs were detected in sera from all three disease groups (n=234).
(2) Percentage of women who had Abs to a given Ag from one or more HPV types over the total number of women.
(3) Percentage of women who had Abs to a given Ag from multiple HPV types over the number of women who had Abs to this Ag from at least one HPV type.
Discussion
There is a clinical need for circulating biomarkers that identify high-risk HPV infection for early detection and treatment of cervical disease. Here, we have used our custom HPV protein microarrays, displaying the proteomes of two low-risk and ten high-risk HPV types, to characterize the diversity of the immune response in cervical cancer and in pre-invasive cervical disease. We find that 20 - 46% of patients with CIN and ICC have a broad range of Abs to HPV early proteins in their sera and these biomarkers correlate with disease severity.
Up to 80% of patients with HPV-associated oropharyngeal cancer (HPVOPC) have detectable serum HPV16 Abs to E Ags [48-50]. Abs against the oncogenic proteins E6 and E7 are also highly specific to ICC. As a result, most cervical cancer studies have focused on E6, E7, and L1 Abs from HPV16 and 18 [21, 22, 27, 31]. In the only previous study of Abs to entire HPV proteomes using slide-based protein microarrays, high levels of Abs were detected against E7 but not E6 or L1 in ICC sera, possibly due to the difficulty of expressing larger-sized proteins [16]. In that study and others [21, 23, 26], no significant difference in Ab response between CIN II/III and asymptomatic controls was detected.
While L-specific Abs were detected in serum from women in all three groups of cervical disease in our study, Abs against any E protein were, as expected, more prevalent in ICC and CIN II/III than women with CIN 0/I. Anti-E7 Abs were the most frequently detected E-Abs, and our data of E7-specific Abs in ICC is consistent with previous studies. Using ELISA [25, 27, 51] and Luminex bead arrays [22, 52, 53], anti-E7 Abs were detected in 13 - 53% of women with ICC and in ~60% of ICC (and 10% of healthy controls) using protein microarrays [16].
As cervical disease progresses towards malignancy, infectious viral particle production becomes limited to a small area near the surface of the cervical epithelium [54, 55]. E4 plays a role in viral synthesis and possibly viral release [56]. The expression of E4 in CIN II and III is restricted to this subset of cells and is generally lost in ICC [54]. Expression of E4 protein in tissue has been proposed as an early detection marker [55] but specific serum Abs may provide a more convenient detection method. Our data show that E4-specific Abs develop early in disease progression, with 16 - 17% prevalence in both CIN II/III and ICC. Anti-E4 Abs have been reported in sera from women with CIN II/III (34%) and ICC (29%) [57], which is consistent with our findings and the predicted level of E4 expression especially in CIN II/III.
Viral integration into the host genome, with loss of E2 expression, is a frequent hallmark of HPV-associated cancers, leading to derepression of E6 and E7 expression. Since E2 Ag is expressed in CIN II/III [58], we predicted that E2 Abs would also be detected early in CIN II/III. E2-specific Abs were detected in both CIN II/III (4.9%) and at higher (p<0.05) frequency in ICC (14.5%) but were at low prevalence. E2-specific IgG Abs have been reported in 24% of women with CIN I-III (compared to 13% of healthy controls) [59] and in 12% of women with ICC (compared to 2% of healthy controls) [28], consistent with our data. Since the majority (64%) of patients with HPVOPC have Abs against HPV16 E2 [48, 49], our data suggest that these two tumor sites may have significant differences in viral integration and expression of E2 Ag, or exposure of E2 Ag to B cells. In cervical disease, E1 Abs have been reported to have a low prevalence (10% and 0.3% in ICC and healthy controls, respectively) [28]. We also found an overall low prevalence of anti-E1 Abs in cervical disease, as opposed to a 60% prevalence in HPVOPCs [48, 49].
Only a few studies have investigated the immune response against multiple HPV types [16, 21, 28, 31]. Only one study displayed multiple HPV proteomes on protein arrays [16] and the others have used glutathione S-transferase-based multiplex serology to evaluate serum Abs against only E6, E7, and L1 Ags from multiple HPV types [28, 31]. The question of cross-reactivity of Abs against Ags from closely-related viral types has therefore not been adequately addressed. We and others have previously demonstrated that MAbs raised against specific HPV proteins may cross-react with homologous proteins from different HPV types due to sequence similarity [16, 41]. Here, we detected cross-reactive Abs including against the E4 Ag. The E4 ORF is the most divergent between HPV types [60]. The range of amino acid sequence similarity between E4 from the non-HPV16 types detected in the 3 sample sera illustrated in Figure 1 and HPV16 E4 is 42 - 59%. It is therefore not known if Abs to multiple homologous E4 proteins reflect cross-reactivity with conserved epitopes or prior multiple HPV infections. We also observed Abs against L2 from at least 6 HPV types in 57.7% of women who had L2-specific Abs (Table 4), which likely indicates cross-reactivity, given the high (46 - 63%) sequence conservation of the L2 protein, and the interest in developing it as a vaccine [61]. Abs against L1 from three or more HPV types were also detected in 19% of women positive for L1 Abs. This is consistent with previously reported L1 cross-reactivity detected by an HPV16 L1-specific MAb [16]. Overall, these data suggest that these HPV arrays will have limited utility as surrogate markers for HPV typing.
While not directly compared in this study, the signal intensity of Ab binding on the arrays [41] and on RAPID ELISA [48, 49] are consistently weaker in cervical disease than in HPVOPC (unpublished observations). Additionally, despite tissue expression of the oncoproteins E6 and E7 in ICC, we and others [16, 22, 23] have reported specific serum Abs in less than half of the patients, while they are detected in up to 75% of HPVOPC cases [48]. Anti-E6 Abs have been detected in HPVOPC and cervical cancer cases years prior to the establishment of a clinical diagnosis [26, 50], suggesting these may be useful for early detection. The Ab response to the E proteins in high grade pre-invasive cervical lesions, however, has been difficult to detect in previous studies [16, 23, 26]. Even though we detect E-specific Abs in a subset of CIN II/III, the frequencies are low. The presence of both viral DNA and viral oncoproteins in HPVOPC tumors suggest that both cancer sites have similar pathogenetic mechanisms [62]. Therefore, it is likely that the close proximity of lymphoid tissue in the tonsils results in a more potent immune induction in HPVOPC compared with cervical disease.
The lack of infrastructure and resources in LMICs hampers large-scale implementation of Pap test screening [29, 63, 64]. In low-resource environments, visual examination with acetic acid (VIA) is an inexpensive alternative [65]. It results in a 25% reduction in cervical cancer incidence and a 35% reduction in cervical cancer mortality after a single screen [65, 66], with significant downstaging of cervical cancers [66]. However, VIA has low sensitivity in women older than 50 years, poor reproducibility between operators and it requires continuous training and supervision. The absence of HPV nucleic acid in the cervix is a good negative predictor of cervical disease but HPV testing is not recommended for women <30 years old because transient infection reduces specificity [9, 67]. In pooled analyses, HPV testing is more sensitive (90 - 95% for CIN II/III) than cervical cytology alone or VIA but lacks the specificity (89%) for a reliable biomarker [67]. To date, there are no established tissue, blood, or vaginal biomarkers other than HPV nucleic acid and cytology for CIN II/III in high risk patients. Biomarkers such as serology that identify high-risk HPV infection and invasive cervical cancers (ICC) could have an impact on the screening, detection, and treatment of cervical disease.
Abbreviations
Abs, antibodies; Ags, antigens; HPV, human papillomavirus; ICC, invasive cervical cancer; CIN, cervical intraepithelial neoplasia; NAPPA, nucleic acid programmable protein arrays; EBNA-1, epstein-barr virus nuclear antigen-1.
Supplementary Material
Supplementary figure 1.
Acknowledgements
This study was supported by a research grant from the Early Detection Research Network (EDRN) UO1 CA11734 (K.S.A.), the National Cancer Institute 1UG3CA211415 (K.S.A.), EDRN Interagency Agreement Y1-CN-0101-01 (E.R.U) and Y1-CN-5005-01 (E.R.U.), and Arizona State University institutional funds (K.S.A.). The authors thank Jennifer Van Duine, Lisa Miller, Eric Pacheco, and Tirinder Bharaj for technical assistance. We thank Dr. Arvind Varsani for providing the HPV image for the graphical abstract. The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the funding agencies or the official position of the Centers for Disease Control and Prevention.
Competing Interests
Dr. Anderson is a consultant and has stock options with Provista Diagnostics, is a founder of FlexBioTech, and is on institutional patent submissions for HPV serologic testing. Other authors report no conflicts.
References
1. Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893-2917
2. Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M. et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359-386
3. Kang WD, Choi HS, Kim SM. Is vaccination with quadrivalent HPV vaccine after loop electrosurgical excision procedure effective in preventing recurrence in patients with high-grade cervical intraepithelial neoplasia (CIN2-3)? Gynecologic Oncology. 2013;130:264-268
4. Markowitz LE, Liu G, Hariri S, Steinau M, Dunne EF, Unger ER. Prevalence of HPV After Introduction of the Vaccination Program in the United States. Pediatrics. 2016;137:e20151968
5. Crosbie EJ, Einstein MH, Franceschi S, Kitchener HC. Human papillomavirus and cervical cancer. Lancet. 2013;382:889-899
6. Dunne EF, Unger ER, Sternberg M, McQuillan G, Swan DC, Patel SS. et al. Prevalence of HPV infection among females in the United States. JAMA. 2007;297:813-819
7. Markowitz LE, Sternberg M, Dunne EF, McQuillan G, Unger ER. Seroprevalence of human papillomavirus types 6, 11, 16, and 18 in the United States: National Health and Nutrition Examination Survey 2003-2004. J Infect Dis. 2009;200:1059-1067
8. Woodman CB, Collins SI, Young LS. The natural history of cervical HPV infection: unresolved issues. Nat Rev Cancer. 2007;7:11-22
9. Goodman A. HPV testing as a screen for cervical cancer. BMJ. 2015;350:h2372
10. Vicus D, Sutradhar R, Lu Y, Elit L, Kupets R, Paszat L. et al. The association between cervical cancer screening and mortality from cervical cancer: a population based case-control study. Gynecol Oncol. 2014;133:167-171
11. Vicus D, Sutradhar R, Lu Y, Kupets R, Paszat L, Ontario Cancer Screening Research N. Association between cervical screening and prevention of invasive cervical cancer in Ontario: a population-based case-control study. Int J Gynecol Cancer. 2015;25:106-111
12. Soost HJ, Lange HJ, Lehmacher W, Ruffing-Kullmann B. The validation of cervical cytology. Sensitivity, specificity and predictive values. Acta Cytol. 1991;35:8-14
13. Qiao Y-l, Sellors JW, Eder PS, Bao Y-p, Lim JM, Zhao F-h. et al. A new HPV-DNA test for cervical-cancer screening in developing regions: a cross-sectional study of clinical accuracy in rural China. Lancet Oncol. 2008;9:929-936
14. Doorbar J, Quint W, Banks L, Bravo IG, Stoler M, Broker TR. et al. The biology and life-cycle of human papillomaviruses. Vaccine. 2012;30(Suppl 5):F55-70
15. Villa LL, Ault KA, Giuliano AR, Costa RL, Petta CA, Andrade RP. et al. Immunologic responses following administration of a vaccine targeting human papillomavirus Types 6, 11, 16, and 18. Vaccine. 2006;24:5571-5583
16. Luevano M, Bernard HU, Barrera-Saldana HA, Trevino V, Garcia-Carranca A, Villa LL. et al. High-throughput profiling of the humoral immune responses against thirteen human papillomavirus types by proteome microarrays. Virology. 2010;405:31-40
17. Stanley M. HPV - immune response to infection and vaccination. Infect Agent Cancer. 2010;5:19
18. Carter JJ, Koutsky LA, Hughes JP, Lee SK, Kuypers J, Kiviat N. et al. Comparison of human papillomavirus types 16, 18, and 6 capsid antibody responses following incident infection. J Infect Dis. 2000;181:1911-1919
19. Dillner J. The serological response to papillomaviruses. Semin Cancer Biol. 1999;9:423-430
20. Bierl C, Karem K, Poon AC, Swan D, Tortolero-Luna G, Follen M. et al. Correlates of cervical mucosal antibodies to human papillomavirus 16: results from a case control study. Gynecol Oncol. 2005;99:S262-268
21. Gutierrez-Xicotencatl L, Salazar-Pina DA, Pedroza-Saavedra A, Chihu-Amparan L, Rodriguez-Ocampo AN, Maldonado-Gama M. et al. Humoral Immune Response Against Human Papillomavirus as Source of Biomarkers for the Prediction and Detection of Cervical Cancer. Viral Immunol. 2016;29:83-94
22. Reuschenbach M, Waterboer T, Wallin KL, Einenkel J, Dillner J, Hamsikova E. et al. Characterization of humoral immune responses against p16, p53, HPV16 E6 and HPV16 E7 in patients with HPV-associated cancers. Int J Cancer. 2008;123:2626-2631
23. Stanley M. Antibody reactivity to HPV E6 and E7 oncoproteins and early diagnosis of invasive cervical cancer. Am J Obstet Gynecol. 2003;188:3-4
24. Ravaggi A, Romani C, Pasinetti B, Tassi RA, Bignotti E, Bandiera E. et al. Correlation between serological immune response analyzed by a new ELISA for HPV-16/18 E7 oncoprotein and clinical characteristics of cervical cancer patients. Arch Virol. 2006;151:1899-1916
25. Silins I, Avall-Lundqvist E, Tadesse A, Jansen KU, Stendahl U, Lenner P. et al. Evaluation of antibodies to human papillomavirus as prognostic markers in cervical cancer patients. Gynecol Oncol. 2002;85:333-338
26. Lehtinen M, Pawlita M, Zumbach K, Lie K, Hakama M, Jellum E. et al. Evaluation of antibody response to human papillomavirus early proteins in women in whom cervical cancer developed 1 to 20 years later. Am J Obstet Gynecol. 2003;188:49-55
27. Achour M, Zeghal D, Kochbati L, Kahla S, Zouari F, Maalej M. et al. Antibody response for L1, E6, E7 HPV 16, and HPV 18 antigens in Tunisian women with cervical cancer. J Immunoassay Immunochem. 2009;30:82-96
28. Combes JD, Pawlita M, Waterboer T, Hammouda D, Rajkumar T, Vanhems P. et al. Antibodies against high-risk human papillomavirus proteins as markers for invasive cervical cancer. Int J Cancer. 2014;135:2453-2461
29. Kontostathi G, Zoidakis J, Anagnou NP, Pappa KI, Vlahou A, Makridakis M. Proteomics approaches in cervical cancer: focus on the discovery of biomarkers for diagnosis and drug treatment monitoring. Expert Rev Proteomics. 2016;13:731-745
30. Zumbach K, Kisseljov F, Sacharova O, Shaichaev G, Semjonova L, Pavlova L. et al. Antibodies against oncoproteins E6 and E7 of human papillomavirus types 16 and 18 in cervical-carcinoma patients from Russia. Int J Cancer. 2000;85:313-318
31. Waterboer T, Sehr P, Michael KM, Franceschi S, Nieland JD, Joos TO. et al. Multiplex human papillomavirus serology based on in situ-purified glutathione s-transferase fusion proteins. Clin Chem. 2005;51:1845-1853
32. Ramachandran N, Hainsworth E, Bhullar B, Eisenstein S, Rosen B, Lau AY. et al. Self-assembling protein microarrays. Science. 2004;305:86-90
33. Ramachandran N, Raphael JV, Hainsworth E, Demirkan G, Fuentes MG, Rolfs A. et al. Next-generation high-density self-assembling functional protein arrays. Nat Methods. 2008;5:535-538
34. Montor WR, Huang J, Hu Y, Hainsworth E, Lynch S, Kronish JW. et al. Genome-wide study of Pseudomonas aeruginosa outer membrane protein immunogenicity using self-assembling protein microarrays. Infect Immun. 2009;77:4877-4886
35. Prados-Rosales R, Carreno LJ, Batista-Gonzalez A, Baena A, Venkataswamy MM, Xu J. et al. Mycobacterial membrane vesicles administered systemically in mice induce a protective immune response to surface compartments of Mycobacterium tuberculosis. MBio. 2014;5:e01921-01914
36. Bian X, Wasserfall C, Wallstrom G, Wang J, Wang H, Barker K. et al. Tracking the Antibody Immunome in Type 1 Diabetes Using Protein Arrays. J Proteome Res. 2016
37. Miersch S, Bian X, Wallstrom G, Sibani S, Logvinenko T, Wasserfall CH. et al. Serological autoantibody profiling of type 1 diabetes by protein arrays. J Proteomics. 2013;94:486-496
38. Anderson KS, Cramer DW, Sibani S, Wallstrom G, Wong J, Park J. et al. Autoantibody signature for the serologic detection of ovarian cancer. J Proteome Res. 2015;14:578-586
39. Katchman BA, Barderas R, Alam R, Chowell D, Field MS, Esserman LJ. et al. Proteomic mapping of p53 immunogenicity in pancreatic, ovarian, and breast cancers. PROTEOMICS - Clinical Applications. 2016;10:720-731
40. Wang J, Figueroa JD, Wallstrom G, Barker K, Park JG, Demirkan G. et al. Plasma Autoantibodies Associated with Basal-like Breast Cancers. Cancer Epidemiol Biomarkers Prev. 2015;24:1332-1340
41. Ewaisha R, Meshay I, Resnik J, Katchman BA, Anderson KS. Programmable protein arrays for immunoprofiling HPV-associated cancers. PROTEOMICS. 2016;16:1215-1224
42. Wong J, Sibani S, Lokko NN, LaBaer J, Anderson KS. Rapid detection of antibodies in sera using multiplexed self-assembling bead arrays. J Immunol Methods. 2009;350:171-182
43. Rajeevan MS, Swan DC, Nisenbaum R, Lee DR, Vernon SD, Ruffin MT. et al. Epidemiologic and viral factors associated with cervical neoplasia in HPV-16-positive women. Int J Cancer. 2005;115:114-120
44. Anderson KS, Sibani S, Wallstrom G, Qiu J, Mendoza EA, Raphael J. et al. Protein microarray signature of autoantibody biomarkers for the early detection of breast cancer. Journal of proteome research. 2011;10:85-96
45. Wang J, Barker K, Steel J, Park J, Saul J, Festa F. et al. A versatile protein microarray platform enabling antibody profiling against denatured proteins. Proteomics Clin Appl. 2013;7:378-383
46. Qiu J, LaBaer J. Nucleic acid programmable protein array a just-in-time multiplexed protein expression and purification platform. Methods Enzymol. 2011;500:151-163
47. Unger ER, Vernon SD, Lee DR, Miller DL, Reeves WC. Detection of human papillomavirus in archival tissues. Comparison of in situ hybridization and polymerase chain reaction. J Histochem Cytochem. 1998;46:535-540
48. Anderson KS, Dahlstrom KR, Cheng JN, Alam R, Li G, Wei Q. et al. HPV16 antibodies as risk factors for oropharyngeal cancer and their association with tumor HPV and smoking status. Oral Oncol. 2015;51:662-667
49. Anderson KS, Gerber JE, D'Souza G, Pai SI, Cheng JN, Alam R. et al. Biologic predictors of serologic responses to HPV in oropharyngeal cancer: The HOTSPOT study. Oral Oncol. 2015;51:751-758
50. Kreimer AR, Johansson M, Waterboer T, Kaaks R, Chang-Claude J, Drogen D. et al. Evaluation of human papillomavirus antibodies and risk of subsequent head and neck cancer. J Clin Oncol. 2013;31:2708-2715
51. Tjiong MY, Zumbach K, Schegget JT, van der Vange N, Out TA, Pawlita M. et al. Antibodies against human papillomavirus type 16 and 18 E6 and E7 proteins in cervicovaginal washings and serum of patients with cervical neoplasia. Viral Immunol. 2001;14:415-424
52. Castellsague X, Pawlita M, Roura E, Margall N, Waterboer T, Bosch FX. et al. Prospective seroepidemiologic study on the role of Human Papillomavirus and other infections in cervical carcinogenesis: evidence from the EPIC cohort. Int J Cancer. 2014;135:440-452
53. Lang Kuhs KA, Anantharaman D, Waterboer T, Johansson M, Brennan P, Michel A. et al. Human Papillomavirus 16 E6 Antibodies in Individuals without Diagnosed Cancer: A Pooled Analysis. Cancer Epidemiol Biomarkers Prev. 2015;24:683-689
54. Doorbar J. Molecular biology of human papillomavirus infection and cervical cancer. Clin Sci (Lond). 2006;110:525-541
55. Griffin H, Wu Z, Marnane R, Dewar V, Molijn A, Quint W. et al. E4 antibodies facilitate detection and type-assignment of active HPV infection in cervical disease. PLoS One. 2012;7:e49974
56. Doorbar J. The E4 protein; structure, function and patterns of expression. Virology. 2013;445:80-98
57. Pedroza-Saavedra A, Cruz A, Esquivel F, De La Torre F, Berumen J, Gariglio P. et al. High prevalence of serum antibodies to Ras and type 16 E4 proteins of human papillomavirus in patients with precancerous lesions of the uterine cervix. Arch Virol. 2000;145:603-623
58. Xue Y, Bellanger S, Zhang W, Lim D, Low J, Lunny D. et al. HPV16 E2 is an immediate early marker of viral infection, preceding E7 expression in precursor structures of cervical carcinoma. Cancer Res. 2010;70:5316-5325
59. Marais D, Rose RC, Williamson AL. Age distribution of antibodies to human papillomavirus in children, women with cervical intraepithelial neoplasia and blood donors from South Africa. J Med Virol. 1997;51:126-131
60. Bell I, Martin A, Roberts S. The E1^E4 protein of human papillomavirus interacts with the serine-arginine-specific protein kinase SRPK1. J Virol. 2007;81:5437-5448
61. Karanam B, Jagu S, Huh WK, Roden RB. Developing vaccines against minor capsid antigen L2 to prevent papillomavirus infection. Immunol Cell Biol. 2009;87:287-299
62. Gillison ML, D'Souza G, Westra W, Sugar E, Xiao W, Begum S. et al. Distinct risk factor profiles for human papillomavirus type 16-positive and human papillomavirus type 16-negative head and neck cancers. J Natl Cancer Inst. 2008;100:407-420
63. Wentzensen N, von Knebel Doeberitz M. Biomarkers in cervical cancer screening. Dis Markers. 2007;23:315-330
64. Wright TC Jr. HPV DNA testing for cervical cancer screening. FIGO 26th Annual Report on the Results of Treatment in Gynecological Cancer. Int J Gynaecol Obstet. 2006;95(Suppl 1):S239-246
65. Sankaranarayanan R, Esmy PO, Rajkumar R, Muwonge R, Swaminathan R, Shanthakumari S. et al. Effect of visual screening on cervical cancer incidence and mortality in Tamil Nadu, India: a cluster-randomised trial. Lancet. 2007;370:398-406
66. Shastri SS, Mittra I, Mishra GA, Gupta S, Dikshit R, Singh S. et al. Effect of VIA screening by primary health workers: randomized controlled study in Mumbai, India. J Natl Cancer Inst. 2014;106:dju009
67. Arbyn M, Ronco G, Anttila A, Meijer CJ, Poljak M, Ogilvie G. et al. Evidence regarding human papillomavirus testing in secondary prevention of cervical cancer. Vaccine. 2012;30(Suppl 5):F88-99
Author contact
![]() Corresponding author: Karen S. Anderson MD PhD, Associate Professor, Biodesign Institute, Arizona State University, PO Box 876401, Tempe, AZ 85287, Phone: 480-965-6982; Email: Karen.Anderson.1edu
Corresponding author: Karen S. Anderson MD PhD, Associate Professor, Biodesign Institute, Arizona State University, PO Box 876401, Tempe, AZ 85287, Phone: 480-965-6982; Email: Karen.Anderson.1edu
 Global reach, higher impact
Global reach, higher impact