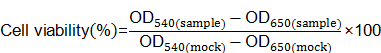13.3
Impact Factor
Theranostics 2017; 7(14):3432-3445. doi:10.7150/thno.20297 This issue Cite
Research Paper
The pH-Triggered Triblock Nanocarrier Enabled Highly Efficient siRNA Delivery for Cancer Therapy
1. Laboratory of Nucleic Acid Technology, Institute of Molecular Medicine, Peking University, Beijing 100871, China;
2. Department of Polymer Science and Technology, Key Laboratory of Systems Bioengineering of the Ministry of Education, School of Chemical Engineering and Technology, Tianjin University, Tianjin 300072, China;
3. Peking-Tsinghua Center for Life Sciences, Academy for Advanced Interdisciplinary Studies, Peking University, Beijing 100871, China;
4. Advanced Research Institute for Multidisciplinary Science, Beijing Institute of Technology, Beijing 100081, China;
5. Collaborative Innovation Center of Chemical Science and Engineering (Tianjin), Tianjin 300072, China.
* These authors contributed equally to this work.
Received 2017-3-28; Accepted 2017-6-15; Published 2017-8-11
Abstract
Small interfering RNA (siRNA) therapies have been hampered by lack of delivery systems in the past decades. Nowadays, a few promising vehicles for siRNA delivery have been developed and it is gradually revealed that enhancing siRNA release from endosomes into cytosol is a very important factor for successful delivery. Here, we designed a novel pH-sensitive nanomicelle, PEG-PTTMA-P(GMA-S-DMA) (PTMS), for siRNA delivery. Owing to rapid hydrolysis in acidic environment, PTMS NPs underwent hydrophobic-to-hydrophilic transition in endosomes that enabled combination of proton sponge effect and raised osmotic pressure in endosomes, resulting in vigorous release of siRNAs from endosomes into cytosol. In vitro results demonstrated that PTMS/siRNA complexes exhibited excellent gene silencing effects in several cell lines. Their gene silencing efficiency could reach ~91%, ~87% and ~90% at the N/P ratio of 50/1 in MDA-MB-231, A549 and Hela cells respectively, which were better than that obtained with Lipofectamine 2000. The highly efficient gene silencing was then proven from enhanced siRNA endosomal release, which is mainly attributed to pH-triggered degradation of polymer and acid-accelerated siRNA release. In vivo experiments indicated that NPs/siRNA formulation rapidly accumulated in tumor sites after i.v. injection. Tumor growth was effectively inhibited and ~45% gene knockdown efficacy was determined at the siRRM2 dose of 1mg/kg. Meanwhile, no significant toxicity was observed during the whole treatment. We also found that PTMS/siRNA formulations could lead to significant gene silencing effects in liver (~63%) and skin (~80%) when injected by i.v. and s.c., respectively. This research work gives a rational strategy to optimize siRNA delivery systems for tumor treatments.
Keywords: siRNA delivery, pH-sensitive, endosomal escape, polymeric nanoparticles, cancer therapy.
Introduction
Since its discovery in 1998 [1], RNA interfering (RNAi) provides huge potentials in treatment of many kinds of diseases, such as cancers [2], glaucoma [3], hyperlipidemia [4] and skin diseases [5]. As the most important effector in RNAi world, small interfering RNA (siRNA) however faces great challenge in realizing such potentials, and the biggest barrier is the lack of effective delivery carriers for siRNA delivery [6, 7]. Recent years, many researchers have attempted to solve the problem and a few delivery nanoparticles (NPs) such as polymer-based [8, 9], lipid-based [10, 11] and conjugation-based vehicles [12] were fabricated and exploited. About twenty nano-based siRNA drugs are under the clinical trials at present [6, 13] (and summarized from https://clinicaltrials.gov), which makes siRNA therapies hopeful in treating human diseases.
Cationic polymers display advantages for siRNA delivery, such as low cost, simple to prepare and high delivery potency [14, 15]. In 2008, the first cationic polymer based siRNA drug (CALAA-01), targeted to rrm2 gene, was used for cancer therapy in clinical trial, and the phase Ia/Ib data was published few years later [16]. Another type of polymer, Dynamic PolyConjugate (DPC), had been deployed for delivery of siRNA tackling HBV infection since 2013 [17] and approved in clinical trial in 2014 (https://clinicaltrials.gov). These developmental milestones provided great affirmation for polymers in siRNA therapies and deeply encouraged their further exploration.
In past several years, a few works revealed that siRNA endosomal escape seemed to be more important than endocytosis for successful delivery. Sahay et al. [18] found that ~70% internalized siRNAs, delivered by LNP (lipid nanoparticles), were exocytosed to extracellular milieu from endosomes/lysosomes, resulting in poor gene silencing effects. However, LNP significantly enhanced silencing efficiency in NPC1-deficient cells, which was believed to increase siRNA retention time in endosomes/lysosomes from knockout of NPC1 gene. Almost the same time, Gilleron et al. [19] demonstrated that only few internalized siRNAs (1~2%) could be released from endosomes into cytosol, and that was very close to ~3.5% revealed by Wittrup et al. in 2015 [20]. Also, several pH-responsive nanoparticles have been successfully developed to enhance cytosolic siRNA delivery for tumor therapy in recent years [21-23]. These works suggested that enhancing siRNA endosomal release would be one of the feasible ways for improving gene silencing potency.
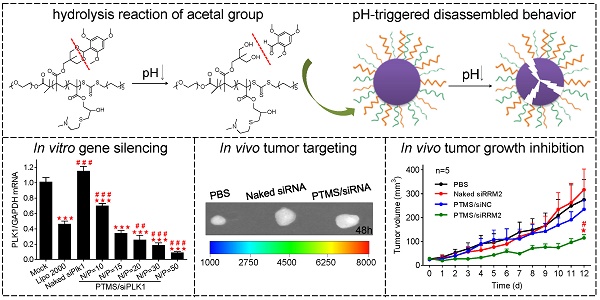
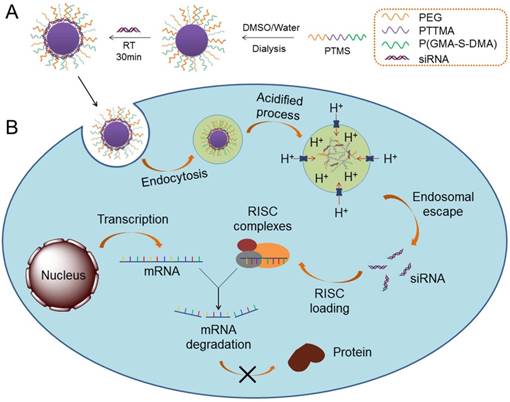
For this reason, we prepared a novel pH-sensitive nanomicelle using poly(ethylene glycol)-co-poly[(2, 4, 6-trimethoxybenzylidene-1, 1, 1-tris(hydroxymethyl)] ethane methacrylate-co-poly(dimethylamino glycidyl methacrylate), PEG-PTTMA-P(GMA-S-DMA), termed PTMS (Fig. 1A). Herein, PTTMA was the hydrophobic core of this amphiphilic nanomicelle. Due to hydrolysis reaction of acetal group, PTTMA polymers underwent hydrophobic-to-hydrophilic transition in acidic buffer, such as pH 5.0 and pH 4.0 [24]. Considering the acidic microenvironment in tumors, few TTMA based delivery systems of chemotherapy drugs had been reported for effective cancer treatment [24, 25]. Therefore, we believed that PTMS NPs would exhibit pH-sensitive property in endosomes/lysosomes (pH ~5.5 [26]), leading to disassembly of NPs there. Combining the collapse of NPs and proton sponge effect triggered by dimethylamino (DMA) groups [15], osmotic pressure of endosomes was dramatically increased. Meanwhile, a large number of hydrolysis products further raised up the pressure of endosomes [27]. Finally, endosomes were swollen, ruptured and siRNAs was heavily released into cytosol, inducing to highly efficient gene knockdown efficiency (Scheme 1). To our knowledge, this is the first research work using TTMA-based polymer for siRNA delivery. In this work, we systematically studied gene knockdown efficiency in vitro and in vivo mediated by PTMS NPs, and explained the delivery mechanism. We hope this work will give a clue for rational design of siRNA delivery systems and make contributions in siRNA therapies.
Schematic illustration of siRNA delivery mediated by PTMS nanomicelles. (A) Formation of PTMS NPs and PTMS/siRNA complexes. (B) Due to pH-sensitive feature, PTMS/siRNA formulation showed highly endosomal escape and effective gene silencing efficiency.
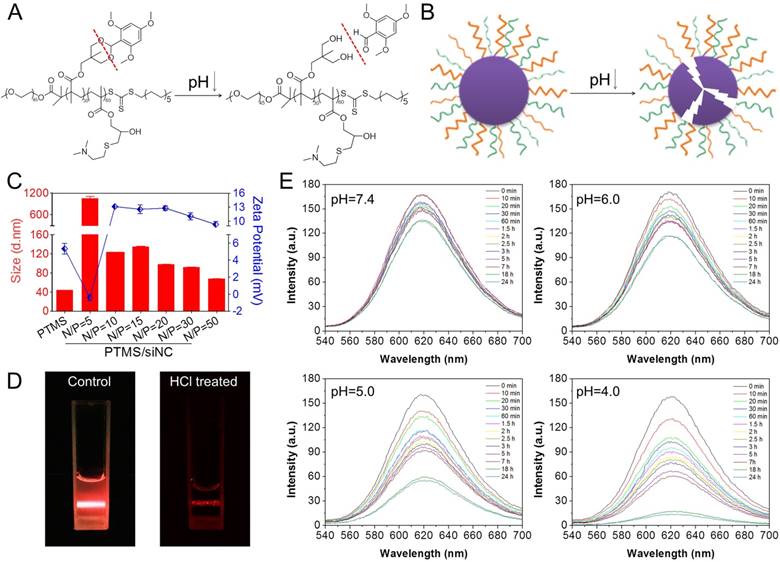
pH-triggered disassembly of PTMS NPs. (A) Hydrolysis reaction of TTMA polymers under acidic buffer. (B) Schematic disintegration of PTMS NPs in acidic buffer due to hydrophobic-to-hydrophilic transition of hydrophobic core (TTMA). (C) Size and Zeta-potential of various PTMS/siNC complexes were detected by Dynamic Light Scattering (DLS) (n=3). Red column and blue dots represented sizes and charges, respectively. (D) Tyndall effect of PTMS NPs was dramatically decreased after treatment with HCl. (E) Nile Red release profile in media with different pH was used to track the disassemble behavior of NPs. Four kinds of solutions, adjusting pH from 7.4 to 4.0, were selected and fluorescence intensity was continuously detected among 24h.
Materials and Methods
Materials
Methoxy poly (ethylene glycol) (mPEG, Mn=2kDa), 1,1,1-tris(hydroxymethyl) ethane, 2,4,6-trimethoxybenzaldehyde, p-toluenesulfonic acid monohydrate, methacryloyl chloride, N,N'-Dicyclohexylcarbodiimide (DCC), 4-dimethylamiopryidine (DMAP), azobisisobutyronitrile (AIBN), glycidyl methacrylate (GMA) and 2-(Dimethylamino) ethanethiol (DMA-SH) were purchased from Sigma-Aldrich. Diethyl ether, tetrahydrofuran (THF), dichloromethane (DCM), triethylamine (TEA) and N,N-Dimethylformamide (DMF) were purchased from Jiangtian company (Tianjin, China). S-1-dodecyl-S′-(α,α′-dimethyl-α″-acetic acid) trithiocarbonate (CTAm) was synthesized according to previous reported [28].
Dulbecco's modified Eagle's medium (DMEM), fetal bovine serum (FBS), lipofectamine 2000 (lipo 2000), trypsin, Opti-MEM, and penicillin-streptomycin were purchased from Invitrogen Corporation (Carlsbad, CA). Agarose was purchased from GEN TECH (Hong Kong, China). Ethidium bromide (EB), 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT), and dimethyl sulfoxide (DMSO) were also purchased from Sigma-Aldrich. Plk1 antibody was obtained from cell Signaling Technology, Inc. (Danvers, MA) and GAPDH antibody was obtained from Zhongshan Goldenbridge Biotechnology Co., Ltd. (Beijing, China). Cy5-labeled siRNA (Cy5-siRNA), Cy3-labeled siRNA (Cy3-siRNA), negative controlled siRNA (siNC), anti-firefly siRNA (siFL), anti-PLK1 siRNA (siPLK1), anti-ApoB siRNA (siApoB), anti-SCD1 siRNA (siSCD1) and anti-RRM2 siRNA (siRRM2) were supplied by Suzhou Ribo Life Science Co., Ltd. (Suzhou, Jiangsu Province, China), Table S1 to check the detailed sequences. All other reagents were purchased from Sigma-Aldrich unless specially marked.
Synthesis of TTMA monomer
2,4,6-Trimethoxybenzylidene-1,1,1-tris(hydroxymethyl) ethane methacrylate (TTMA) was synthesized according to previous works (Fig. S1A) [24, 29]. Briefly, 1,1,1-tris(hydroxymethyl) ethane (12g) and 2,4,6-trimethoxybenzaldehyde (20g) were dissolved in THF (150ml), then p-hydroxybenzenesulfonic acid (1g) as the catalytic reagent was added into the mixture and the reaction mixture was shaken at room temperature for 24h. Hereafter, the dried product (14.9g) and TEA (7.8g) were dissolved in DCM (50ml), methacryloyl chloride (6.84g) was added drop wise to the solution, following to react at room temperature for 24h. The final product TTMA (13.5g) was purified by column chromatography.
Synthesis of PEG-CTAm
Macroinitiator PEG-CTAm was obtained by following steps (Fig. S1B). At the presence of DCC (2.06g) and DMAP (1.22g), dried PEG2000 (10g) and CTAm (3.66g) were dissolved in DCM (50ml) for 48h to completely react. The residue was rinsed three times with diethyl ether and PEG-CTAm was obtained.
Synthesis of PEG-PTTMA (PT)
PEG-PTTMA (PT) was synthesized by Reversible Addition-Fragmentation Chain Transfer (RAFT) polymerization according to published paper (Fig. S1C) [24]. RAFT agent PEG-CTAm (236.5mg), TTMA (1281mg), AIBN (3.28g) and DMF (3ml) were added into a Schlink tube, the solution was degassed three cycles and filled with nitrogen. Then mixtures were placed in a thermostatic water bath at 70ºC for 24h under the protection of nitrogen. At last, product was dialyzed against distilled water and lyophilized.
Synthesis of PEG-PTTMA-PGMA (PTM) and PEG-PTTMA-P(GMA-S-DMA) (PTMS)
PEG-PTTMA-PGMA (PTM) was synthesized as the same as PEG-PTTMA polymer but with PEG-PTTMA (758.8mg) and glycidyl methacrylate (GMA) (427mg) as the reactants. To obtain PEG-PTTMA-P(GMA-S-DMA) (PTMS), PTM (1g) and 2-(dimethylamino) ethanethiol (DMA-SH) (0.5g) were dissolved in DMF (10ml) to react for 24h at 50ºC under the protection of nitrogen (Fig. S1D,E). PTMS polymer was lyophilized after dialysis. To verify the structures, PT, PTM and PTMS polymers were analyzed by 1H-NMR spectra using Varian Unity spectrometer (300 MHz) (Fig. S2).
Agarose gel retardation assay
Gel retardation was tested to evaluate siRNA binding ability of PTMS. As we did in previous work [30], siNC (0.3ug) was incubated with PTMS solutions at room temperature for 20min to achieve various N/P ratios from 1:1 to 8:1. N/P ratio means molar ratio of amino groups (N) of polymer to phosphate groups (N) of siRNA. The NPs/siRNA complexes were adjusted to final volume (16μl) and mixed with 6×loading buffer (4μl, Takara Biotechnology, Dalian, Liaoning Province, China). The mixture was loaded onto 2% agarose gel containing 5μg/ml ethidium bromide (EB). Electrophoresis was carried out at a voltage of 120mV for 20min in 1×TAE running buffer. Finally, the results were recorded at UV light wavelength of 254nm by image master VDS thermal imaging system (Bio-Rad, Hercules, CA).
Nile red release profile in different pH buffer
PTMS nanomicelles were prepared via self-assembly processes. Briefly, dried PTMS polymer (20mg) was dissolved in DMSO (1ml) and dialyzed against distilled water (10ml) for 24h. During this time, dialysate was frequently replaced with fresh water every three hours. After that, PTMS nanomicelles were formed and stored in 4℃. To evaluate pH-sensitive property, PTMS (20mg) and Nile Red (50μg) were dissolved in DMSO (1ml) and formed Nile Red loaded micelles after dialyzed against distilled water. Fluorescence intensities of the PTMS NPs were measured at given time-points by Varian fluorescence spectrophotometer in various buffer, pH from 7.4 to 4.0. The hydrolysis behavior of PTMS NPs was estimated by fluorescent changes.
Size and Zeta-potential of PTMS NPs
The sizes and zeta potential of empty NPs and PTMS/siNC complexes were measured by dynamic light scattering (DLS) (Zetasizer Nano ZS, Malvern, U.K.) at a wavelength of 633nm with a constant angle of 173º. PTMS/siNC complexes (contained 2μg siNC) were prepared as reported in our previous works [31]. Various PTMS/siNC samples were formed with N/P ratios of 10:1 to 50:1 by electrostatic interaction. Before detecting, the sample was adjusted to the final volume (1ml) using distilled water.
Luciferase assay
For all involved in vitro transfection assays in this work, Lipo 2000/siRNA formulation was used as the positive control. Lipo 2000/siRNA formulation was prepared based on standard manufacturer's instructions but fixed weight/volume ratio of siRNA to Lipo 2000 reagent with 1µg/3µl.
MDA-MB-231-Luc (stably expressing luciferase gene) cells were seeded in 24-well plates with 5×104 cells per well. After 24h, the medium was replaced with Opti-MEM (0.5ml) and added PTMS/siFL complexes with different N/P ratios for another 4h, handling the final siRNA transfection concentration of 50nM. Opti-MEM medium was replaced by fresh DMEM (1ml) and further incubated for 20h. Cells were washed three times with 1×PBS (0.5ml) and lyzed with 1×passive lysis buffer (100μl) per well. After violently shaking for 30 min, cell supernatant (10μl) was collected for luminescence measurements with adding substrates (50μl) by Multi-Mode Microplate Reader (Synergy HT, BioTek, USA).
Cell viability
MDA-MB-231-Luc cells were seeded in 96-well plates at 1×104 cells per well the day before transfection. Cells were treated with various complexes for another 24h at the same siRNA concentration as luciferase assay. Ten microliters MTT solution (5mg/ml in 1×PBS) was added into each well to incubate for 4h at 37℃. The solution was removed gently and replaced with DMSO (50μl). After the formazan crystals was dissolved completely, the absorbance of 540nm (OD540) was measured by Multi-Mode Microplate Reader with a reference wavelength of 650nm (OD650). Untreated cells were marked as control sample (mock). Cell viability was calculated according to following equation:
In vitro gene knockdown measurement
Real time PCR (RT-PCR) and western blot were performed to assess gene silencing effect. For RT-PCR test, MDA-MB-231-Luc, A549 and Hela cells were used. Cells were seeded in 6-well plates at 2×105 cells per well. Twenty-four hours later transfected with PTMS/siPLK1, total RNA was extracted using TRIzol Reagent (Invitrogen) according to standard manufacturer's instructions. After finishing reverse transcription with total RNA (1μg), cDNA (50ng) was quantified by RT-PCR system using SYBR Green PCR Master Mix. Herein, we used GAPDH (or β-Actin) gene as the internal control. All primers used in this work were listed in Table S2.
For western blot, MDA-MB-231-Luc cells were seeded in 6-well plates at 2×105 cells per well. After 48h treated with PTMS/siPlk1, total protein was collected and the concentration was measured by BCA protein assay kit. Fifty microgram protein was loaded and separated by SDS-PAGE, following transferred to PVDF membranes. Protein coated membranes were blocked with 5% BSA buffer for 1h then incubated with primary antibody (1:1000) overnight at 4℃. PVDF membrane was probed with horseradish peroxidase (HRP)-labeled secondary antibody (1:5000) for another 1h. At last, the membranes were treated with ECL kit (Pierce, Rockford, IL) and imaged by Bio-Rad Universal Hood Ⅱ (Bio-Rad Laboratory, Bossier City, LA).
Flow cytometry detection
MDA-MB-231-Luc cells were seeded in 6 well-plates at 2×105 cells per well. After 24h, the medium was replaced with Opti-MEM and cells were treated with PTMS/Cy5-siRNA complexes for 4h at the final siRNA concentration of 50nM. Cells were digested by 0.25% trypsin, washed three times by cold 1×PBS (1ml), suspended in 1×PBS (400μl), and detected by flow cytometry (Becton Dickinson, San Jose, CA).
To test endocytic pathway of NPs, pharmacological inhibitors were used as did in our previous work [15]. Briefly, amiloride (100µM), chlorpromazine (30µM) and genistein (1mM) were added into cells for 0.5h before treatment with several formulations. Four hours later, cells were detected by flow cytometry as described above. Here, 4℃ treated sample was the positive control, and normalized results were presented based on non-pretreated cells.
Confocal observation
For studies of the subcellular localization of NPs/siRNA complexes, MDA-MB-231-Luc cells were seeded into 35mm dishes at 2×105 cells each well. Twenty-four hours later, cells were transfected with PTMS/Cy5-siRNA at the final siRNA concentration of 50nM. After 4h and 10h, cells were washed three times with 1×PBS and stained by Lysotracker Green (indicated endosome/lysosome). Then cells were fixed by paraformaldehyde (4%) for 40min and stained by DAPI (indicated nucleus). At last, cells were imaged by Zeiss confocal microscope (LSM700, Carl Zeiss, Germany).
pH-triggered degradation of PTMS
For pH-sensitive measurement of PTMS NPs, 1H NMR was tested first. Briefly, PTMS polymer (20mg) was dissolved in DMSO-d6 (0.6ml) with 5% trifluoroacetic acid (TFA, 30µl) for 2h, following 1H NMR spectra of sample was recorded and analyzed. To determine degradable behavior directly, UV-spectroscopy was measured at the wavelength of 295nm, indicating the relative amount of 2,4,6-trimethoxybenzaldehyde monomer in given solution. PTMS NPs solution (10µg/ml) was prepared as mentioned above and pH was adjusted to 6.0, 5.0 and 4.0, respectively. The solutions were shaken at 37℃, and wavelength of 290-280nm was monitored at each designed time-point within 36h.
Size changes and in vitro siRNA release
The pH of PTMS NPs solution (10µg/ml) was adjusted to 7.4, 6.0, 5.0 and 4.0, respectively, shaking at 37℃. Size changes were recorded and analyzed by DLS in following 32h. TEM images were also observed to verify the size changes treated in pH 5.0 buffer for 24h.
The pH-triggered siRNA release was performed as did as before [31] with slight modification. PTMS/Cy5-siRNA (N/P=15) formulation was prepared as mentioned above, with 40µg of the final amount of Cy5-siRNA per sample. The Cy5-siRNA solution (110µl) was added into PB buffer (1.2ml) with different pH values (n=3), and shaken at the speed of 100rpm at 37℃. At designed time-points, sample was centrifuged for 10min at the speed of 15000g, 200µl supernatant was collected for testing and equal volume of fresh buffer was added. Cy5-siRNA fluorescence was detected with 630nm excitation and 680nm emission. Accumulated release of Cy5-siRNA was finally calculated based on calibration curve.
In vivo tumor accumulation and tumor inhibition activity
All involved animals in this work were maintained in Peking University Laboratory Animal Center (an AAALAC accredited experimental animal facility). All procedures were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee (IACUC) of Peking University.
For tumor targeting evaluation, MDA-MB-231 cells (5×106 cells) were suspended in 1×PBS (100μl) and subcutaneously injected in right axillary fossa of female BALB/c nude mice (16-18g). When the tumor grew to about 400 mm3, the mice were randomly divided into three groups. Mice were i.v. injected with 1×PBS, Free Cy5-siRNA, and PTMS/Cy5-siRNA (N/P=15) at the final siRNA dose of 1mg/kg, respectively. Cy5 signals of mice whole body was captured using animal imaging system among 48h. At the end of experiment, mice were sacrificed by cervical dislocation, main organs were harvested and further imaged.
To evaluate in vivo tumor growth inhibition of PTMS formulation, tumor model of female BALB/c nude mice were prepared as described above. When the tumor grew to about 40mm3, mice were randomly divided into four groups and treated with PBS, naked siRRM2, PTMS/siNC (N/P=15) and PTMS/siRRM2 (N/P=15) by i.v. injection, respectively. Mice were injected every three days for 4 times at the dose of 1mg/kg. Body weight was monitored during entire experiment. Tumor volume was recorded every day until day 12 using caliper and calculated with the following formula: tumor volume (mm3) = 0.5 × length × width2. At the end of experiment, rrm2 knockdown efficiency were tested and four kinds of serum cytokines (IL-2, IL-6, IFN-γ and TNF-α) were measured using Luminex Technology (4A Biotech Co., Ltd., Beijing, China).
In vivo biodistribution and gene knockdown measurement
For biodistribution, male C57BL/6 mice (18-20g) were purchased from the Academy of Military Medical Science of China and divided into three groups. Mice were intravenously (i.v.) injected with PBS, free Cy5-siRNA and PTMS/Cy5-siRNA (N/P=15) at the final siRNA dose of 1mg/kg, respectively. At 6h and 24h after treatment, main organs were harvested and imaged using Kodak in vivo imaging system (Kodak In-vivo Imaging System FX Pro, Carestream Health, USA).
Pharmacokinetics study was performed as did in previous work [31]. Briefly, male C57BL/6 mice were i.v. injected with PBS, naked Cy3-siRNA and PTMS/Cy3-siRNA (N/P=15) at the dose of 1mg/kg. Blood samples were continuously collected at various time points (1, 5, 15, 30, 60 and 120min) post injection. Cy3 fluorescence signal was detected by Multi-Mode Microplate Reader and normalized results were presented based on the starting point (1min).
To assess in vivo gene silencing efficiency, we selected two kinds of injection ways. One was intravenously (i.v.) administration. Male C57BL/6 mice were treated with PTMS/siApoB for 48h. Liver were collected and ApoB mRNA level was detected by RT-PCR. The other one was subcutaneous (s.c.) injection. Mice were administrated with PTMS/siSCD1 for 48h. Skin, at the injected site, was harvested and expression of scd1 gene was tested by RT-PCR. For both experiments, the final injected dose of siRNA was 1mg/kg and the N/P ratio was 15/1.
Statistical analysis
Statistical analysis was taken using GraphPad Prism 5 software. Results were presented as mean ± S.E.M. or mean ± S.D. For statistical comparison, One-Way ANOVA followed by Tukey's test was used and P < 0.05 was considered statistically significant.
Results and Discussions
Formation and characterization of NPs
PTMS co-polymer was synthesized by multistep reactions (Fig. S1). The final structural formula was given out using H1-NMR spectra, PEG45-PTTMA30-P(GMA-S-DMA)60 (Mw=27770), where the number represented degree of polymerization (DP) (Fig. S2) shown by image of agarose gel (Fig. S3), siRNA was loaded easily with increasing N/P ratios and completely captured at the N/P ratio of 5. Therefore, PTMS/siNC formulations with N/P ratios above 5/1 were formed and prepared to use. Size and zeta-potential of PTMS/siNC formulations were analyzed by DLS (Fig. 1C). Interestingly, compared with empty NPs (PTMS) (43.89 ± 0.22nm), the size of PTMS/siNC (N/P=5) was much larger (1052.5 ± 75.66nm). But with N/P ratios further increased, the size gradually decreased, showing about 70-120nm diameters at the higher N/P ratios of 10/1 to 50/1. It was believed that certain surface charge of NPs was helpful to protect NPs from aggregation because of electrostatic repulsion among NPs [31]. From empty NPs to PTMS/siNC (N/P=5), zeta-potential declined from +5.35 ± 0.63mV to -0.14 ± 0.35mV, causing disappearance of electrostatic repulsion and heavy aggregation. When the charge increased to around +10 mV at N/P ratios of 10/1 to 50/1, electrostatic repulsion recovered and made NPs/siRNA stable in solutions.
pH-triggered disassembly of PTMS NPs
As reported, acetal group was sufficiently stable in pH 7.4 but prone to be rapidly hydrolyzed in acidic buffer [24, 25]. We supposed that PTMS NPs were completely destroyed in acidic environment because of hydrophobic-to-hydrophilic transition of hydrophobic core (TTMA) (Fig. 1A, B). To verify this, Tyndall effect was observed by shining a laser beam (Fig. 1D). As expected, Tyndall effect was significantly weakened when treated with HCl, demonstrating PTMS NPs disassembled in the solution. Nile Red release profile was a popular method to track disassemble behavior of NPs [32], the fluorescence signal of Nile Red was strongly influenced by surrounding environment. In hydrophobic environment, fluorescence signal of Nile Red was very strong, but in hydrophilic environment, fluorescence intensity was too weak to be detected. Here, we prepared Nile Red loaded PTMS to test pH-sensitive feature (Fig. 1E). Fluorescence signal was not weakened obviously even after 24h of incubation in pH 7.4 buffer, whereas it gradually decreased in acidic solutions, with pH deceased from 6.0 to 4.0. What's more, almost no signal of Nile Red was detected in pH 4 solution after 20h. These results suggested that PTMS NPs were endowed pH-triggered disassembly behavior in solutions.
In vitro gene knockdown effect
To assess in vitro gene knockdown efficiency, luciferase assay was performed first. MDA-MB-231-Luc (stably expressing luciferase gene) cells were transfected with various PTMS/siFL formulations at the final concentration of 50nM (Fig. 2A). At high N/P ratios (above 20/1), PTMS/siFL complexes exhibited better gene silencing efficiency than Lipo 2000 (30.76 ± 1.74%), even close to 95% at the N/P ratio of 50. We also found that PTMS/siFL (N/P=5) didn't show significant gene silencing activity, that was probably resulted from its heavy aggregation. Importantly, no obvious cytotoxicity was observed for all formulations (Fig. 2B). To further evaluate delivery potency in vitro, Polo-like Kinase 1 (plk1) gene was selected for RT-PCR measurement. As displayed in Fig. 2C, plk1 gene was dramatically inhibited by PTMS/siPLK1 complexes in MDA-MB-231 cells, consistent with luciferase assay. Compared with Lipo 2000 (41.24 ± 18.33%), the remaining activities of PTMS/siPLK1 polyplexes were much lower, that were 25.75 ± 14.53% (N/P=20), 18.64 ± 7.32% (N/P=30) and 9.01 ± 3.32% (N/P=50). Western blot assay also supported results in MDA-MB-231 cells (Fig. 2D). Besides, we expanded plk1 gene knockdown tests in other two cell lines. In both A549 and Hela cells, both RT-PCR (Fig. 2E, F) and western blot (Fig. S4) demonstrated that gene knockdown efficiency mediated by PTMS was much better than Lipo 2000 at high N/P ratios. These results indicated that PTMS nanomicelle was a promising delivery vehicle for siRNA due to its safety and effective gene silencing efficiency in several cell lines.
In vitro cellular uptake
To explore the mechanism why PTMS showed excellent siRNA delivery in vitro, internalization of PTMS/Cy5-siRNA was detected by flow cytometry. Compared with control groups (Lipo 2000 and naked siRNA), the fluorescence peaks of PTMS/Cy5-siRNA complexes were obviously right shifted (Fig. 3A), reflecting PTMS NPs delivered much more siRNAs into cells. It was showed more clearly in quantified data (Fig. 3B), all formulations showed about 100% cellular uptake (blue dots), but mean fluorescence intensity (MFI) of PTMS/Cy5-siRNA were about 4.1 times (N/P=50) to 7.5 times (N/P=10) than Lipo 2000 sample. Interestingly, MFIs of PTMS formulations tended to decline with increasing N/P ratio, which seems to be contrary to gene silencing efficiency results showed in Fig. 2.
To answer this question, endocytic pathway of PTMS/Cy5-siRNA formulation was then evaluated (Fig. 3C). Endocytic inhibitors, including amiloride, chlorpromazine and genistein, were selected to block macropinocytosis, clathrin-mediated endocytosis and caveolae-mediated endocytosis, respectively. Definitely, PTMS/Cy5-siRNA complexes, with various N/P ratios, showed energy-dependent endocytosis as their cellular uptakes were dramatically hindered under cold shock at 4℃. Interestingly, NPs with different N/P ratios exhibited energy dependence: the higher N/P ratio, the less energy-dependency. Also, these polyplexes showed different inhibitors-dependent endocytosis. Showed in Fig. 3C, both chlorpromazine and genistein significantly impeded cellular uptake of PTMS/Cy5-siRNA polyplexes, indicating clathrin and caveolae-mediated endocytosis were two major endocytic pathways. But NPs with different N/P ratios showed different dependency on these two endocytosis pathways. After treatment with chlorpromazine, the inhibition rates were about 89%, 70% and 38% for N/P=10, N/P=20 and N/P=30, respectively, and the inhibition rates were around 18%, 25% and 23% for genistein treated cells. This demonstrated that dependency on different endocytic pathways may contribute partly to the difference of cellular uptake for NPs of various N/P ratios.
Subcellular localization analysis
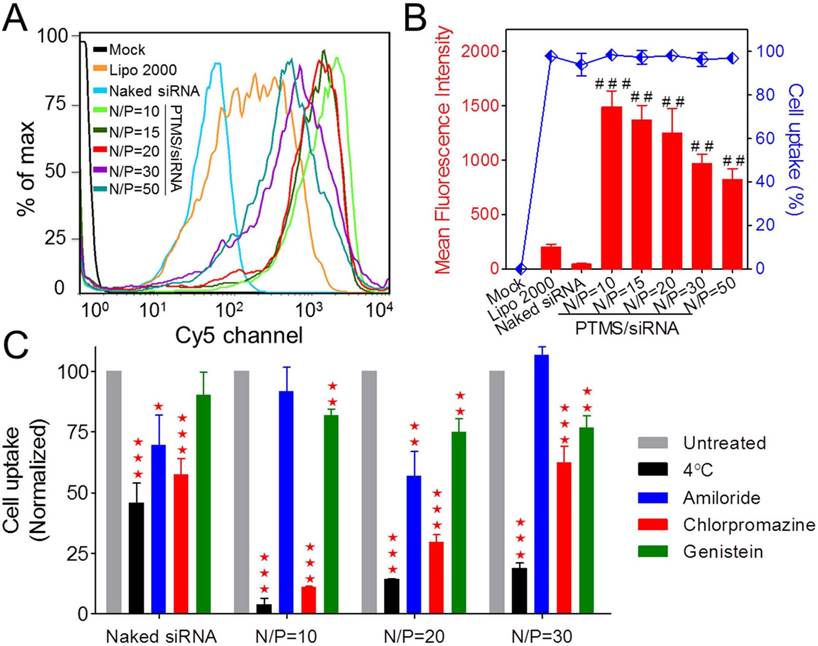
To answer the question why less internalization showed better gene silencing efficacy, subcellular localization of PTMS/Cy5-siRNA was then studied using confocal microscopy (Fig. 4). Cy5 signals (red color) of PTMS/Cy5-siRNA complexes were observed clearly in cytosol at 4h and 10h after treatment. It was observed that signal of lipo 2000 was much weaker than PTMS formulations, consistent with flow cytometry data (Fig. S5). More importantly, we found that Cy5 signals were diffused punctate at 10h rather than aggregated punctate at 4h (indicated by white arrows in Fig. 4A), implying NPs promoted siRNA release from endosomes into cytosol [15, 33]. MFI and co-localization ratios were quantified from randomly selected four images (Fig. 4B). At the same treatment times, PTMS/Cy5-siRNA with higher N/P ratio exhibited lower co-localization ratio. The value of co-localization ratio was significantly declined from 4h to 10h. They were 88.15 ± 0.53% (N/P=10), 76.40 ± 3.85% (N/P=20), 69.02 ± 1.90% (N/P=30) in 4h, and 78.70 ± 3.81% (N/P=10), 64.78 ± 4.44% (N/P=20), 55.13 ± 3.93% (N/P=30) in 10h. As we know, the lower co-localization ratio, the more siRNA endosomal escape, the better gene silencing effect. This might be the reason why PTMS showed better gene knockdown efficiency at higher N/P ratios. Due to rapid hydrolysis of TTMA in acidic environment, PTMS/siRNA formulation disintegrated quickly in endosome, where the pH was about 5.5 [26] and proton sponge effect was induced. At the same time, colloidal osmotic pressure in endosomes was further raised up because of a large number of hydrolyzed products [27]. Finally, endosomes were swollen, ruptured and released siRNA into cytosol. In addition, MFIs of samples were calculated (Fig. 4C). MFI decreased with increasing N/P ratios, exhibiting the similar tendency as observed in flow cytometry. Compared with 4h, the slightly reduction of MFI was detected for each sample in 10h, which may be contributed by exocytosis [18]. Combining consideration of flow cytometry test and confocal images together with gene silencing efficiency results, we supposed that enhancing siRNA endosomal escape was much more important than improving endocytosis for successful delivery.
In vitro cell viability and gene silencing effect of PTMS/siRNA formulations. (A) Luciferase knockdown was tested in MDA-MB-231 cells treated with various polyplexes, and (B) cytotoxicity was measured using MTT assay. PTMS/siPLK1 complexes showed excellent gene knockdown efficiency detected by both (C) RT-PCR and (D) western blot in MDA-MB-231 cells, as well as in (E) A549 and (F) Hela cells tested by RT-PCR. In all tests, cells were treated with different formulations for 24h except for western blot (48h), the final siRNA transfection concentration was 50nM. * represents comparison with mock and # represents comparison with Lipofectamine 2000 (Lipo 2000), respectively. * (#) p<0.05, ** (##) p<0.01 and *** (###) p<0.001 (n=3).
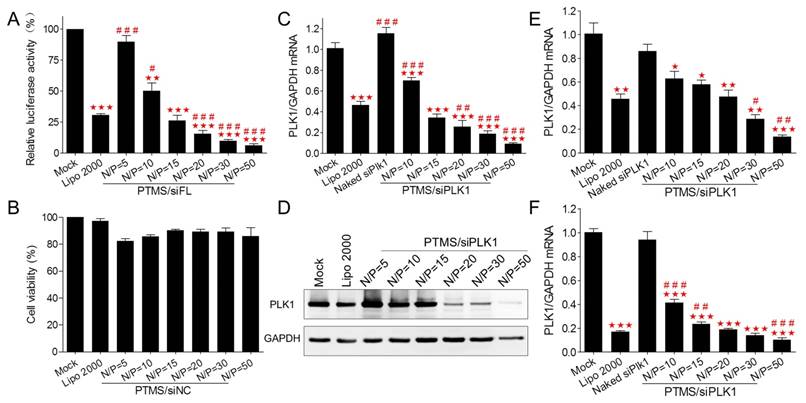
Internalization and cellular uptake pathway of PTMS/Cy5-siRNA were assessed by flow cytometry in MDA-MB-231 cells. Cells were treated with various PTMS/Cy5-siRNA formulations for 4h at the final siRNA concentration of 50nM, following to be analyzed by (A) histogram and (B) quantification. Red columns represent mean fluorescence intensity of Cy5, reflecting the amount of siRNAs in each cell. Blue dots represent cellular uptake, reflecting the percentage of Cy5 positive cells. (C) Cellular uptake pathway was monitored by several endocytic inhibitors and normalized results were showed. # represents comparison with Lipo 2000 and * represents comparison with corresponding non-pretreated sample. * p<0.05, ## (**) p<0.01 and *** p<0.001 (n=4).
pH-triggered degradation of NPs and siRNA release
To further understand high-efficiency siRNA delivery mediated by PTMS, we here analyzed in vitro polymeric degradation and siRNA release in acidic buffer, mimicking endosomal microenvironment. Firstly, hydrolytic study of acetal was performed using 1H NMR spectra according to reported method [34]. As showed in Fig. 5A, the sharp peak at 6.25-6.50 ppm (b') was observed clearly for TFA incubated sample, which was contributed by 2,4,6-trimethoxybenzaldehyde monomer. And the strong signal around at 3.75-4.0 ppm (c') was presented correspondingly, indicating rapid hydrolytic reaction of TTMA under acidic buffer. Similarly, acetal hydrolysis in acidic solutions was determined by UV-spectroscopy with monitoring the wavelength of 295nm that was characteristic absorbance of 2,4,6-trimethoxybenzaldehyde (Fig. 5B). Typically, pH-dependent hydrolytic rate was exhibited, which was agreement with previous reports [24]. The hydrolytic rate of acetal was much faster in pH 4.0 than pH 6.0 and pH 5.0 buffer, further confirming the pH-triggered degradation of PTMS NPs.
In addition, the behavior of PTMS NPs was tracked in pH 5.0 buffer, mimicking endosomal microenvironment. Size changes were tested within 32h (Fig. 5C). Because of hydrophobic-to-hydrophilic transition of PTMS NPs under acidic conditions, significant changes of sizes were observed. Parts of NPs expanded to ~5µm at 8h, and the percentage of larger size increased in 12h and 18h. TEM images also showed dramatically aggregation (Fig. 5D), size changed from ~40nm to ~1µm under pH 5.0 buffer for 24h. Small sizes (~3nm) of NPs were detected after 32h in pH 5.0 buffer showed in Fig. 5C, indicating water-soluble unimers resulted from completely acetal hydrolysis of TTMA [24]. Moreover, size changes exhibited pH-dependency, larger NPs were observed earlier in pH 4.0 buffer than pH 5.0, 4h and 8h, respectively. Few aggregations were detected in pH 6.0 condition and no aggregation was showed in pH 7.4 buffer (Fig. S6), suggesting PTMS NPs was relative stable without acetal hydrolysis in physiological environment. To simulate endosomal escape of siRNA, in vitro siRNA release profile was performed (Fig. 5E). As expected, Cy5-siRNA release was dramatically promoted in acidic conditions compared with neutral buffer, while pH 4.0 showed the fastest. In pH 5.0 condition, about 70% of accumulated release was measured within 10h, which ensured quickly endosomal escape of siRNA after endocytosis. Take these results into consideration, it was believed that high-efficiency silencing effects of PTMS/siRNA formulation were resulted from effective endosomal escape of siRNA driven by fast hydrolysis of acetal.
Subcellular localization study of PTMS/Cy5-siRNA polyplexes. (A) Confocal images of MDA-MB-231 cells transfected by PTMS/Cy5-siRNA formulations for 4h and 10h at the final siRNA concentration of 50nM. From left to right, nucleus was stained by DAPI (blue), siRNA was labeled by Cy5 (red), endosomes/lysosomes were stained by lysotracker green (green) and merged images. Scale bar is 20µm. Statistical (B) co-localization ratios between red and green signal and (C) Cy5 fluorescence intensity of randomly selected four pictures. * represents comparison with N/P=10 sample at the same transfection time and # represents comparison with itself at different treatment time, respectively. ** (##) p<0.01, *** (###) p<0.001 and n.s. means no significance.
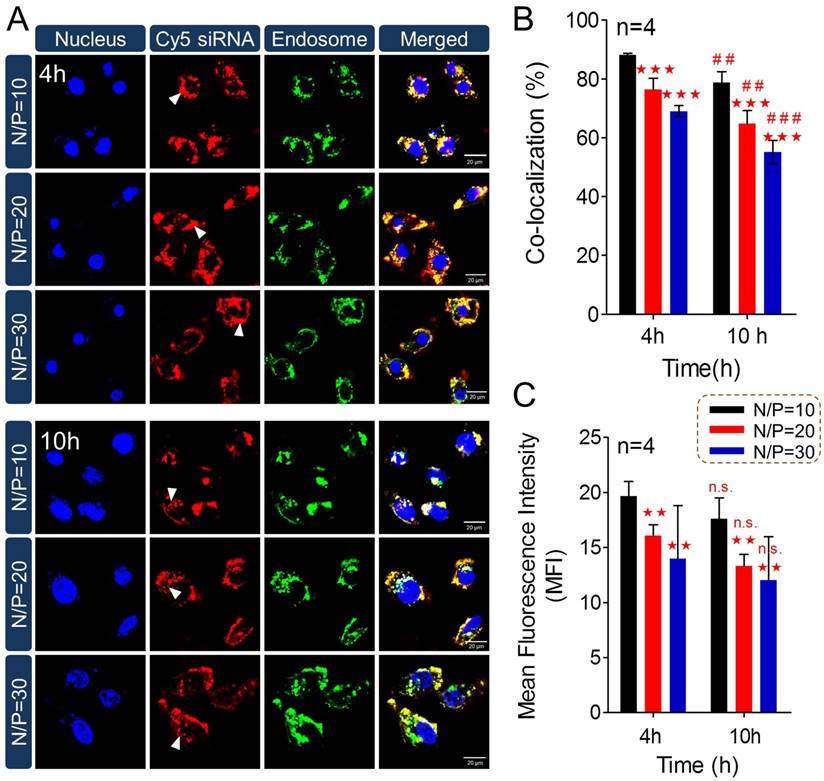
The pH-sensitive properties of PTMS NPs and siRNA release profile in acidic buffer. (A) 1H NMR spectra of PTMS polymer. Before measurement, PTMS was incubated in DMSO-d6 (with 5% TFA) for 2h. (B) Hydrolytic reaction study of PTMS NPs in acidic buffer. Hydrolytic products (2,4,6-trimethoxybenzaldehyde) was monitored by absorbance at the wavelength of 295nm. (C) Size changes of PTMS NPs in pH 5.0 buffer within 32h. Sizes were calculated by volume-weighted. (D) Comparison of TEM images between pH 5.0 treated PTMS NPs and control group. Scale bar are 1µm and 100nm, respectively. (E) In vitro Cy5-siRNA release profile in various pH solutions (n=3). Samples were shaken at 37℃ with the speed of 100rpm, and Cy5-siRNA was monitoring with 630nm excitation and 680nm emission.
In vivo tumor distribution and inhibition
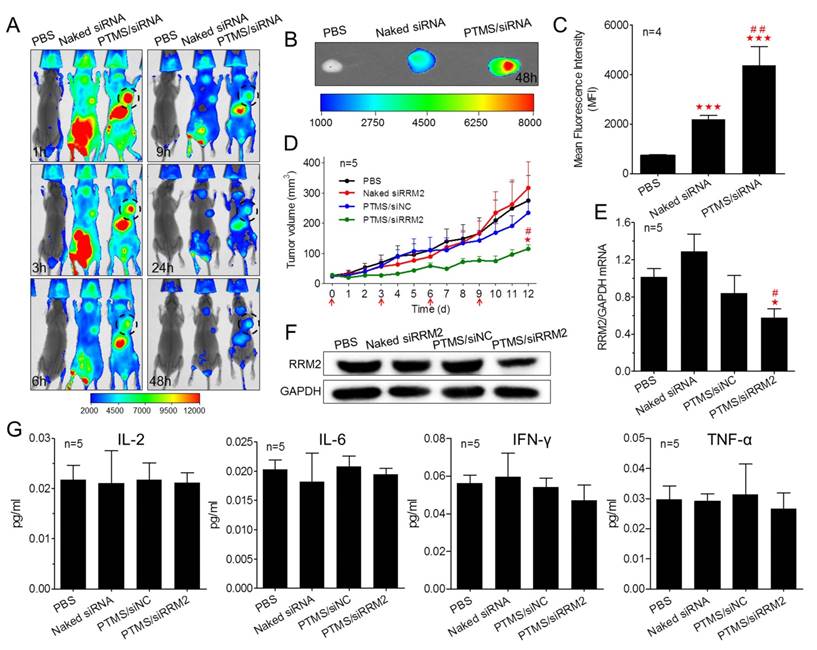
Encouraged by potent gene knockdown efficiency in vitro, In vivo experiments were performed. With the sizes of 70-120nm, PTMS/Cy5-siRNA formulations were endowed with tumor enhanced permeability and retention (EPR) effect [35]. To explore the possibility of cancer therapy in mice level, in vivo tumor accumulation was detected. Cy5 signals of whole body were captured at different time-points during 48h (Fig. 6A). Both naked siRNA and PTMS/Cy5-siRNA complex quickly reached tumor sites (indicated by black circles) within 1h. However, fluorescence intensity of tumor sites fell down with time flies, and it dropped much faster in naked siRNA group. Cy5 signal was too weak to be detected in naked siRNA group after 48h. At the end of experiment, tumors were separated, imaged and analyzed quantification (Fig. 6B, C). Both fluorescent image and quantified data indicated that PTMS NPs delivered much more siRNAs into tumor compared with naked siRNA group. Further in vivo antitumor activity was tested in mice model (Fig. 6D). Significant tumor growth inhibition was observed in PTMS/siRRM2 group, but not shown in other groups. The rrm2 gene expression in tumors was tested later by both mRNA (Fig. 6E) and protein levels (Fig. 6F). Knockdown efficiency was only obtained in PTMS/siRRM2 group (~45%), demonstrating that tumor growth inhibition was resulted from silence of rrm2 gene. Moreover, no significant changes of body weight were observed during entire experiment (Fig. S7) and serum cytokines (Fig. 6G) were not induced significantly, suggesting PTMS/siRRM2 formulation was well-tolerated in mice.
In vivo organ distribution and gene knockdown
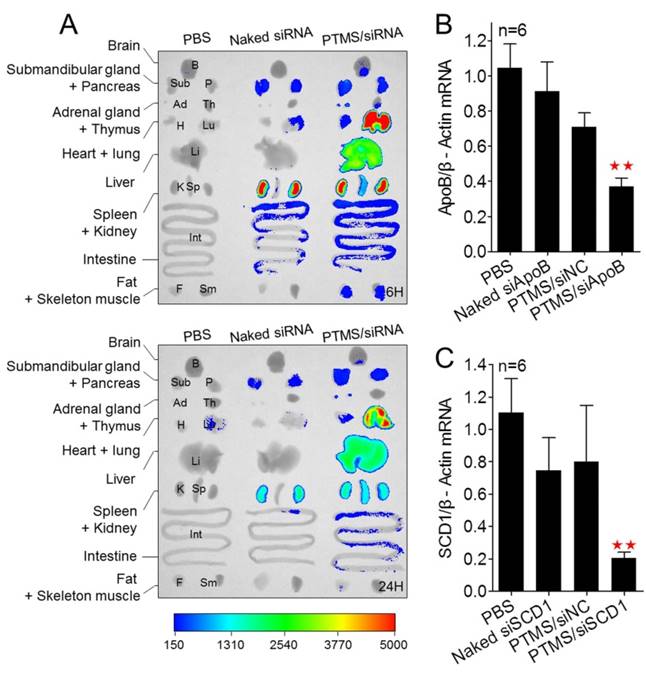
Besides, we tested the biodistribution of PTMS complexes in healthy C57BL/6 mice (Fig. 7A). Cy5 signals of PTMS/Cy5-siRNA were much stronger than naked siRNA group in both monitoring points, suggesting PTMS NPs prolonged siRNA half-life in body. There were at least two factors for this result. One was the PEG motif of PTMS NPs, which was believed to enable NPs to avoid nonspecific absorption of serum proteins and increase blood circulation time [36, 37]. That was indeed observed in pharmacokinetics study (Fig. S8). The half time (T1/2) of naked siRNA was only several minutes (< 5min) but it was around 30min for PTMS formulation. Area under the curve (AUC) of PTMS formulation was also much larger than naked siRNA. The other factor was size distribution of PTMS/Cy5-siRNA. Some works revealed that polyplexes with >10nm size effectively reduced siRNA excretion from kidney [38]. Liver, lung and spleen were observed to show extensive accumulation of polyplexes, indicating PTMS NPs had delivery potential in these organs. We then selected liver for in vivo functional study and apoB (Apolipoprotein B) as the target gene. The remaining activity of apoB gene was 37.1 ± 4.6% for PTMS/siApoB group, much better than naked group (90.98 ± 16.86%) (Fig. 7B). We also tested delivery potency of PTMS formulation by local administration. Recently, siRNA therapies in skin diseases showed huge application prospects and commercial value. It was worth mentioning that therapeutic siRNA, named TD101, has completed phase Ib clinical trial for pachyonychia congenita (PC) treatment [39]. To explore delivery potency of PTMS/siRNA formulation in skin, mice were s.c. injected with PTMS/siSCD1 formulation for 48h. The skin of injected site was then collected and gene silencing efficiency was detected by RT-PCR (Fig. 7C). Stearoyl-CoA desaturase-1 (scd-1) gene expression was dramatically inhibited by PTMS/siSCD1 complexes (~80%). But no significant gene knockdown efficiency was observed in both naked siSCD1 group and PTMS/siNC group. These results were mostly related with high endocytosis and effective siRNA endosomal escape of the formulation.
In vivo evaluation of tumor targeting and tumor growth inhibition. Tumor-bearing BALB/c mice were i.v. injected with PTMS/Cy5-siRNA (N/P=15) complexes at the final siRNA dose of 1mg/kg (n=4), following (A) Cy5 signals of whole body were collected at given time-points. At the end of experiment (48h), tumors were separated, (B) imaged and (C) analyzed quantitatively. For tumor growth inhibition, mice were i.v. injected every three days for 4 times (n=5, red arrows indicate injection times), (D) tumor volumes were measured every day until day 12. At the end of experiment, (E,F) rrm2 mRNA and protein levels of tumor were tested using RT-PCR and western blot. (G) Levels of serum IL-2, IL-6, IFN-γ and TNF-α were detected using Luminex Technology. * represents comparison with PBS group and # represents comparison with naked siRNA group, respectively. ** (##) p<0.01 and *** (###) p<0.001.
In vivo biodistribution and gene silencing detection in C57BL/6 mice. (A) Fluorescence images of major organs. Mice were i.v. injected with PTMS/Cy5-siRNA formulation and sacrificed at given time-points. Major organs were separated and imaged (n=3, showed only one group). (B) Liver apoB gene was measured using RT-PCR after 48h by i.v. injection (n=6). (C) Skin scd1 gene was tested using RT-PCR after 48h by s.c. injection (n=6). In these experiments, N/P=15 was selected and the final siRNA dose was 1mg/kg. * represents comparison with PBS group and ** p<0.01.
Conclusion
In summary, we here reported a novel pH-sensitive nanomicelle, named PTMS. Because of pH-triggered feature of TTMA group, PTMS NPs effectively enhanced siRNA endosomal escape and mediated excellent gene silencing effects in vitro, better than positive control (Lipo 2000). In vivo experiments indicated that PTMS delivered much more siRNA into tumor sites and mediated effective tumor growth inhibition without significant toxicity. Besides, obvious gene inhibitions were detected in liver and skin by i.v. and s.c. injection, respectively. Hence, PTMS/siRNA nanodrugs showed therapeutic prospects in tumors, liver-related diseases and skin-related disorders. In this work, we tried to improve siRNA delivery potency by rational design of carriers, our data revealed that siRNA endosomal release was more important than endocytosis for successful delivery. The pH-sensitive feature is perfectly matched with enhancing siRNA endosomal release, which could be an important principle for optimizing siRNA delivery systems.
Abbreviations
TTMA: 2,4,6-Trimethoxybenzylidene-1,1,1-tris(hydroxymethyl) ethane methacrylate; PTMS: poly(ethylene glycol)-co-poly[(2, 4, 6-trimethoxybenzylidene-1, 1, 1-tris(hydroxymethyl)] ethane methacrylate-co-poly(dimethylamino glycidyl methacrylate); NPs: Nanoparticles; i.v.: Intravenous injection; s.c.: Subcutaneous injection; NPC-1: Niemann-Pick C(NPC)-1; siRNAs: e.g. siApoB, small interfering RNA targeted to ApoB gene; EB: ethidium bromide; DLS: Dynamic Light Scattering; MFI: Mean Fluorescence Intensity.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
This work was supported by grants from the National Basic Research Program of the Chinese Ministry of Science and Technology (No. 2013CB531202), the National Natural Science Foundation of China (No.81473128, 81502586, and 31271073), the National High Technology Research and Development Program of China (2012AA022501) and the National Drug Program of China (No.2012ZX09102301-006).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. nature. 1998;391:806-11
2. Ozpolat B, Sood AK, Lopez-Berestein G. Liposomal siRNA nanocarriers for cancer therapy. Adv Drug Deliv Rev. 2014;66:110-6
3. Martínez T, González MV, Roehl I, Wright N, Pañeda C, Jiménez AI. In vitro and in vivo efficacy of SYL040012, a novel siRNA compound for treatment of glaucoma. Mol Ther. 2014;22:81-91
4. Soh J, Iqbal J, Queiroz J, Fernandez-Hernando C, Hussain MM. MicroRNA-30c reduces hyperlipidemia and atherosclerosis in mice by decreasing lipid synthesis and lipoprotein secretion. Nat Med. 2013;19:892-900
5. Zakrewsky M, Mitragotri S. Therapeutic RNAi robed with ionic liquid moieties as a simple, scalable prodrug platform for treating skin disease. J Control Release. 2016;242:80-8
6. Yin H, Kanasty RL, Eltoukhy AA, Vegas AJ, Dorkin JR, Anderson DG. Non-viral vectors for gene-based therapy. Nat Rev Genet. 2014;15:541-55
7. Wittrup A, Lieberman J. Knocking down disease: a progress report on siRNA therapeutics. Nat Rev Genet. 2015;16:543-52
8. Mokhtarzadeh A, Alibakhshi A, Hashemi M, Hejazi M, Hosseini V, de la Guardia M. et al. Biodegradable nano-polymers as delivery vehicles for therapeutic small non-coding ribonucleic acids. J Control Release. 2017;245:116-26
9. Yu H, Guo C, Feng B, Liu J, Chen X, Wang D. et al. Triple-layered pH-responsive micelleplexes loaded with siRNA and cisplatin prodrug for NF-Kappa B targeted treatment of metastatic breast cancer. Theranostics. 2016;6:14-27
10. Whitehead KA, Dorkin JR, Vegas AJ, Chang PH, Veiseh O, Matthews J. et al. Degradable lipid nanoparticles with predictable in vivo siRNA delivery activity. Nat Commun. 2014;5:4277
11. Lee J, Saw PE, Gujrati V, Lee Y, Kim H, Kang S. et al. Mono-arginine cholesterol-based small lipid nanoparticles as a systemic siRNA delivery platform for effective cancer therapy. Theranostics. 2016;6:192-203
12. Nair JK, Willoughby JL, Chan A, Charisse K, Alam MR, Wang Q. et al. Multivalent N-acetylgalactosamine-conjugated siRNA localizes in hepatocytes and elicits robust RNAi-mediated gene silencing. J Am Chem Soc. 2014;136:16958-61
13. Kanasty R, Dorkin JR, Vegas A, Anderson D. Delivery materials for siRNA therapeutics. Nat Mater. 2013;12:967-77
14. Kozielski KL, Tzeng SY, Hurtado De Mendoza BA, Green JJ. Bioreducible cationic polymer-based nanoparticles for efficient and environmentally triggered cytoplasmic siRNA delivery to primary human brain cancer cells. ACS nano. 2014;8:3232-41
15. Han S, Cheng Q, Wu Y, Zhou J, Long X, Wei T. et al. Effects of hydrophobic core components in amphiphilic PDMAEMA nanoparticles on siRNA delivery. Biomaterials. 2015;48:45-55
16. Zuckerman JE, Gritli I, Tolcher A, Heidel JD, Lim D, Morgan R. et al. Correlating animal and human phase Ia/Ib clinical data with CALAA-01, a targeted, polymer-based nanoparticle containing siRNA. Proc Natl Acad Sci U S A. 2014;111:11449-54
17. Wooddell CI, Rozema DB, Hossbach M, John M, Hamilton HL, Chu Q. et al. Hepatocyte-targeted RNAi therapeutics for the treatment of chronic hepatitis B virus infection. Mol Ther. 2013;21:973-85
18. Sahay G, Querbes W, Alabi C, Eltoukhy A, Sarkar S, Zurenko C. et al. Efficiency of siRNA delivery by lipid nanoparticles is limited by endocytic recycling. Nat Biotechnol. 2013;31:653-8
19. Gilleron J, Querbes W, Zeigerer A, Borodovsky A, Marsico G, Schubert U. et al. Image-based analysis of lipid nanoparticle-mediated siRNA delivery, intracellular trafficking and endosomal escape. Nat Biotechnol. 2013;31:638-46
20. Wittrup A, Ai A, Liu X, Hamar P, Trifonova R, Charisse K. et al. Visualizing lipid-formulated siRNA release from endosomes and target gene knockdown. Nat Biotechnol. 2015;33:870-6
21. Yu H, Zou Y, Wang Y, Huang X, Huang G, Sumer BD. et al. Overcoming endosomal barrier by amphotericin B-loaded dual pH-responsive PDMA-b-PDPA micelleplexes for siRNA delivery. ACS nano. 2011;5:9246-55
22. Xu X, Wu J, Liu Y, Yu M, Zhao L, Zhu X. et al. Ultra-pH-Responsive and Tumor-Penetrating Nanoplatform for Targeted siRNA Delivery with Robust Anti-Cancer Efficacy. Angew Chem Int Ed Engl. 2016;55:7091-4
23. Xu X, Wu J, Liu Y, Saw PE, Tao W, Yu M. et al. Multifunctional Envelope-Type siRNA Delivery Nanoparticle Platform for Prostate Cancer Therapy. ACS nano. 2017;11:2618-27
24. Zhao J, Wang H, Liu J, Deng L, Liu J, Dong A. et al. Comb-like amphiphilic copolymers bearing acetal-functionalized backbones with the ability of acid-triggered hydrophobic-to-hydrophilic transition as effective nanocarriers for intracellular release of curcumin. Biomacromolecules. 2013;14:3973-84
25. Du Y, Chen W, Zheng M, Meng F, Zhong Z. pH-sensitive degradable chimaeric polymersomes for the intracellular release of doxorubicin hydrochloride. Biomaterials. 2012;33:7291-9
26. Wu H, Zhu L, Torchilin VP. pH-sensitive poly (histidine)-PEG/DSPE-PEG co-polymer micelles for cytosolic drug delivery. Biomaterials. 2013;34:1213-22
27. Li J, Chen Y-C, Tseng Y-C, Mozumdar S, Huang L. Biodegradable calcium phosphate nanoparticle with lipid coating for systemic siRNA delivery. J Control Release. 2010;142:416-21
28. Lai JT, Filla D, Shea R. Functional polymers from novel carboxyl-terminated trithiocarbonates as highly efficient RAFT agents. Macromolecules. 2002;35:6754-6
29. Griset AP, Walpole J, Liu R, Gaffey A, Colson YL, Grinstaff MW. Expansile nanoparticles: synthesis, characterization, and in vivo efficacy of an acid-responsive polymeric drug delivery system. J Am Chem Soc. 2009;131:2469-71
30. Cheng Q, Huang Y, Zheng H, Wei T, Zheng S, Huo S. et al. The effect of guanidinylation of PEGylated poly (2-aminoethyl methacrylate) on the systemic delivery of siRNA. Biomaterials. 2013;34:3120-31
31. Cheng Q, Du L, Meng L, Han S, Wei T, Wang X. et al. The promising nanocarrier for doxorubicin and siRNA co-delivery by PDMAEMA-based amphiphilic nanomicelles. ACS Appl Mater Interfaces. 2016;8:4347-56
32. Zhou J, Wu Y, Wang C, Cheng Q, Han S, Wang X. et al. pH-Sensitive Nanomicelles for High-Efficiency siRNA Delivery in Vitro and in Vivo: An Insight into the Design of Polycations with Robust Cytosolic Release. Nano letters. 2016;16:6916-23
33. Morrissey DV, Lockridge JA, Shaw L, Blanchard K, Jensen K, Breen W. et al. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat Biotechnol. 2005;23:1002-7
34. Liu B, Thayumanavan S. Substituent Effects on the pH Sensitivity of Acetals and Ketals and Their Correlation with Encapsulation Stability in Polymeric Nanogels. J Am Chem Soc. 2017;139:2306-17
35. Maeda H, Nakamura H, Fang J. The EPR effect for macromolecular drug delivery to solid tumors: Improvement of tumor uptake, lowering of systemic toxicity, and distinct tumor imaging in vivo. Adv Drug Deliv Rev. 2013;65:71-9
36. Perry JL, Reuter KG, Kai MP, Herlihy KP, Jones SW, Luft JC. et al. PEGylated PRINT nanoparticles: the impact of PEG density on protein binding, macrophage association, biodistribution, and pharmacokinetics. Nano letters. 2012;12:5304-10
37. Wang Q, Jiang J, Chen W, Jiang H, Zhang Z, Sun X. Targeted delivery of low-dose dexamethasone using PCL-PEG micelles for effective treatment of rheumatoid arthritis. J Control Release. 2016;230:64-72
38. Zuckerman JE, Choi CHJ, Han H, Davis ME. Polycation-siRNA nanoparticles can disassemble at the kidney glomerular basement membrane. Proc Natl Acad Sci U S A. 2012;109:3137-42
39. Leachman SA, Hickerson RP, Schwartz ME, Bullough EE, Hutcherson SL, Boucher KM. et al. First-in-human mutation-targeted siRNA phase Ib trial of an inherited skin disorder. Mol Ther. 2010;18:442-6
Author contact
![]() Corresponding authors: Qiang Cheng and Anjie Dong, E-mail: chengqiang115com (Q. Cheng), ajdongedu.cn (A. Dong)
Corresponding authors: Qiang Cheng and Anjie Dong, E-mail: chengqiang115com (Q. Cheng), ajdongedu.cn (A. Dong)
 Global reach, higher impact
Global reach, higher impact