13.3
Impact Factor
Theranostics 2017; 7(9):2537-2554. doi:10.7150/thno.17584 This issue Cite
Research Paper
Regulation on Toll-like Receptor 4 and Cell Barrier Function by Rab26 siRNA-loaded DNA Nanovector in Pulmonary Microvascular Endothelial Cells
1. Institute of Respiratory Diseases, Xinqiao Hospital, Third Military Medical University, Chongqing 400037, China;
2. College of Pharmaceutical Sciences , Southwest University , Chongqing 400715, China;
3. Department of Chemistry, Purdue University, West Lafayette, IN 47907, USA.
* These authors contributed equally.
Received 2016-9-15; Accepted 2017-4-27; Published 2017-6-25
Abstract
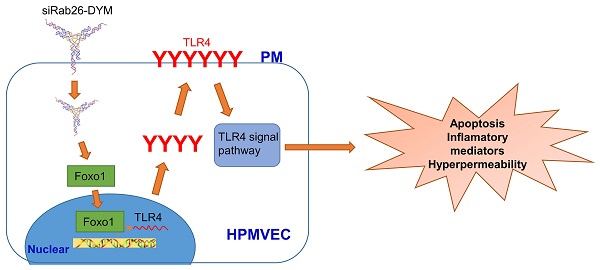
The small GTPase Rab26 is involved in multiple processes, such as vesicle-mediated secretion and autophagy. However, the mechanisms and functions of Rab26 in the human pulmonary microvascular endothelial cells (HPMVECs) are not clear. In this study, we thoroughly investigated the role and novel mechanism of Rab26 in permeability and apoptosis of HPMVECs using a self-assembled Rab26 siRNA loaded DNA Y-motif nanoparticle (siRab26-DYM) and Rab26 adenovirus. We found that siRab26-DYM could be efficiently transfected into HPMVECs in a time- and dose-dependent manner. Importantly, the siRab26-DYM nanovector markedly aggravated the LPS-induced apoptosis and hyper-permeability of HPMVECs by promoting the nuclear translocation of Foxo1, and subsequent activation of Toll-like receptor 4 (TLR4) signal pathway. Overexpression of Rab26 by Rab26 adenoviruses partially inactivated LPS-induced TLR4 signaling pathway, suppressed the cell apoptosis and attenuated the hyperpermeability of HPMVECs. These results suggest that the permeability and apoptosis of HPMVECs can be modulated by manipulating Rab26 derived TLR4 signaling pathway, and that Rab26 can be potential therapeutic target for the treatment of vascular diseases related to endothelial barrier functions.
Keywords: Rab26, DNA nanostructure, TLR4, human pulmonary microvascular endothelial cells, permeability.
Introduction
Human pulmonary microvascular endothelial cells are an important component of the vascular endothelial barrier, a semi-selectively permeable barrier that isolates tissue fluid and blood and regulates the exchange of blood, fluid, electrolytes, and proteins, among other substances, across the vascular wall [1]. Endotoxin-mediated HPMVEC damage induces hyperpermeability and the loss of endothelial barrier function, resulting in acute lung injury (ALI) and acute respiratory distress syndrome (ARDS) [2]. Previous studies have shown that permeability-altering responses are frequently initiated by the regulation of anterograde trafficking, internalization and localization of endothelial surface receptors (such as β2-adrenergic receptor, β2-AR) [3-5], which induces the activation of signaling molecules, including GTPases and their kinases, and consequently affects the expression and degradation of target proteins [6].
Rab26 is a member of the Rab GTPase family, which regulates receptor trafficking in the cytoplasm to control cellular homeostasis, the stress response and survival [7]. Rab26 has been found on secretory granules from parotid acinar cells [8]. Previous studies have demonstrated that Rab26 plays an important role in regulating the transport of α2-ARs from the Golgi to the membrane [9], lysosome trafficking and mitochondrial redistribution [10] and induces the autophagy of neuronal somata [11]. As shown in our previous study, the GTPases Rab1 and Rab5a control the permeability of HPMVECs by regulating the trafficking and localization of the β-AR [3, 4]. However, whether Rab26 regulates the permeability of HPMVECs remains unclear.
TLR4 is localized on the cytoplasm membrane and plays an import role in nonspecific immunity. TLR4 recognizes pathogen-associated molecular patterns (PAMPs) and reacts to damage-associated molecular patterns (DAMPs) to facilitate the release of numerous endogenous antiseptic inflammatory molecules upon injury at the cell surface [12]; thus, the activation of TLR4 and its downstream signaling is one of the earliest responses of an organism to microbial invasion [13]. Previous studies have shown that activation of TLR4 and downstream nuclear factor-κB (NF-κB) signaling promotes lung inflammation, thereby aggravating lung injury. In contrast, TLR4 inhibition in HPMVECs can prevent lung edema and actin cytoskeleton rearrangement [14].
Given that TLR4 can only function appropriately if it is located on the cell surface, the regulation of TLR4 expression on the cytoplasm membrane is a key step in controllingTLR4-mediated inflammation and immune responses [15]. As shown in the study by Wang et al., Rab7b promotes TLR4 translocation into lysosomes from the cytoplasm membrane for degradation and negatively regulates TLR4 signaling in macrophages [16]. Another study illustrated that the up-regulation of Rab10 by Lipopolysaccharide (LPS) facilitated the trafficking of TLR4 from the Golgi to the plasma membrane. This process maintained the surface level of the TLR4 receptor complex and aggravated lung injury. Blocking the function of Rab10 led to a reduction in TLR4 levels at the cell surface and diminished the production of inflammatory cytokines and interferons upon LPS stimulation [17]. Whether the localization of TLR4 controlling by Rab26 in primary HPMVECs is still unknown.
Primary HPMVECs are a valuable tool for studying alterations to the endothelial barrier and the underlying mechanisms. RNA interference technology is typically used to knock down the expression of target genes in primary HPMVECs to determine the effect and potential mechanisms of these genes in cells [18, 19]. Unfortunately, while there are several techniques for transiently transfecting cells, such as viral methods, cationic lipid vectors, and synthetic polymer systems, the efficiency of these methods in knocking down target gene expression in primary cells remains limited [20]. Moreover, these methods damage primary mammalian cells, and the permeability of primary HPMVECs is particularly sensitive to injury [21] due to potential toxicity [22] and immunogenic reactions. Therefore, an ideal gene delivery system for the transfection of primary mammalian cells is needed.
Recently, self-assembled DNA nanostructures have been demonstrated to have great potential for delivering drugs and oligonucleotides into cells, with high stability and transfection efficiency and low toxicity and immunogenicity [23]. Our previous study showed that a delivery vehicle based on programmed self-assembly of mammalian target of rapamycin (mTOR)-siRNA-loaded DNA nanotubes (DNA-NTs) was effectively transfected into primary pulmonary arterial smooth muscle cells (PASMCs), induced obvious autophagy and inhibited cell growth under both normal and hypoxic conditions [24]. Moreover, we designed and constructed a self-assembled RNA-DNA hybrid triangle nanoparticle system that was used to more effectively transfect oligonucleotides into cells using less transfection regent.
Here, we have designed and constructed a Rab26 siRNA loaded DNA nanoparticle to knock down Rab26 expression. This DNA nanoparticle efficiently delivered the siRNA into primary HPMVECs and down-regulated Rab26 expression, thus promoting the localization of TLR4 at the plasma membrane and inducing apoptosis and hypertonicity in HPMVECs. To fully elucidate the Rab26 function in HPMVECs, we further force-expressed Rab26 in vitro with Rab26 adenovirus. Overexpression of Rab26 greatly suppressed the apoptosis of HPMVECs and attenuated the endothelial hyperpermeability.
Materials and Methods
Materials
DNA oligonucleotides were obtained from Sangon Biological Engineering (Shanghai, China); single-stranded siRNA or single-stranded siRNA-Cy3 was purchased from GenePharma (Shanghai, China). The following DNA sequences were designed by the computer program SEQUIN: Y1 sequence, 5'-AGGCACCATCGTAGGTTTAACTTGCCAGGCACCATCGTAGGTTTAACTTGCCAGGCACCATCGTAGGTTTAACTTGCC-3'; Y2 sequence, 5'-AACGACTAGCAACCTGCCTGGCAAGCCTACGATGGACACGGTAA-3'; Y3 sequence, 5'-TACCCATGCCTACTAAATTACCGTGTGGTTGCTAGTCGTT-3'; and Y3' sequence (for the scrambled Rab26 siRNA loaded DNA nanostructure), 5'-GACACGUUCGGAGAAAATTACCGTGTGGTTGCTAGTCGTT-3'. The Rab26 siRNA sequence was 5'-UAGUAGGCAUGGGUAACACTT-3', and the negative control (NC) siRNA sequence was 5'-UUCUCCGAACGUGUCACGUTT-3', the sequence of Foxo1 siRNAs was 5'-GGAGGUAUGAGUCAGUAUATT-3' (sense) and 5'-UAUACUGACUCAUACCUCCTT-3' (antisense). Primary HPMVECs were purchased from CELLBIO (Shanghai, China). LPS from Escherichia coli O111:B4 was obtained from Sigma (USA). The X-tremeGENE siRNA transfection reagent was purchased from Roche (Switzerland). The primary antibodies used in this study were: rabbit anti-Rab26 (Abcam, USA); rabbit anti-TLR4 (Boster, China); rabbit anti-MyD88 (Abcam, USA); rabbit anti-TRAF6 (Abcam, USA); rabbit anti-NF-κB p65 (Abcam, USA); rabbit anti-phosphorylation-Foxo1 (Abcam, USA); rabbit anti-Foxo1 (Abcam, USA); rabbit anti-GAPDH (Abcam, USA); and mouse anti-β-actin (Cell Signaling Technology, USA). Horseradish peroxidase-conjugated AffiniPure goat anti-rabbit IgG and goat anti-mouse IgG secondary antibodies were purchased from Beijing Golden Bridge Biotech (Beijing, China). Anti-human CD284 (TLR4)-PE and isotype control-PE were obtained from eBioscience (USA). FITC-dextran was purchased from Santa Cruz Biotechnology (USA). Endothelial cell medium (ECM), endothelial cell growth supplement (ECGS), penicillin and streptomycin were obtained from ScienCell (USA). Trypsin (0.25%) and fetal bovine serum (FBS) were obtained from Life Technologies (USA). Polyvinylidene difluoride (PVDF) membranes and the enhanced chemiluminescence (ECL) reagents were purchased from Millipore (USA). The reverse transcription polymerase chain reaction (RT-PCR) kit was purchased from Takara Bio, Inc. (Dalian, China). Recombinant Rab26 adenoviruses (Adv. Rab26) and empty adenoviruses (Adv. vector) were obtained from Hanbio Biotechlogy Co., Ltd. (Shanghai, China). All other reagents were commercially available and used as received.
Self-assembly of siRab26-DYM and the negative control (NC)-siRNA-loaded DNA Y-motif (siNC-DYM)
Y1, Y2, Y3 and single-stranded Rab26 siRNA strands were combined according to a molar ratio of 1:3:3:3 in Tris-acetic acid-EDTA-Mg2+ (TAE/Mg2+) buffer (40 mM Tris base, 20 mM acetic acid, 2 mM EDTA, and 12.5 mM Mg(Ac)2, pH 8.0) prepared in nuclease-free water. The mixture was progressively cooled from 95 ℃ to 22 ℃ (95 ℃ for 5 min→65 ℃ for 30 min→50 ℃ for 30 min→37 ℃ for 30 min→22 ℃ for 30 min). Nanoparticles containing Y1, Y2, Y3' and single-stranded negative control siRNA strands were constructed using the same conditions and procedure. The nanoparticles were stored at 4 ℃.
Characterization of the siRab26-DYM nanovector
The siRab26-DYM nanovector was analyzed by native polyacrylamide gel electrophoresis (PAGE). The samples were run on gels containing 6% polyacrylamide (19:1 acrylamide: bisacrylamide) in a Bio-Rad electrophoresis unit at 4°C (120 V, constant voltage) with 1× TAE/Mg2+ running buffer. After electrophoresis, the gels were stained with 0.01% 'Stains-All' (Sigma) for 1 h and scanned.
Moreover, the siRab26-DYM nanovector was analyzed using dynamic light scattering (DLS) measurements. The siRab26-DYM solution was diluted to ~50 nM and measured with a Malvern Zetasizer (Malvern Instruments Ltd, UK) in 1× TAE/Mg2+ buffer. Pure 1× TAE/Mg2+ was used as the blank.
Cell culture
Primary HPMVECs were cultured in ECM containing 10% FBS, 2% ECGS, 100 units/mL penicillin, and streptomycin in a 95% humidified atmosphere of 5% CO2 at 37°C. Cells were trypsinized when they reached 90% confluency, and the cell suspension was then quickly dispersed into three new flasks for further culture.
Experimental and control HPMVECs were grown in plates or flasks to 95% confluence and then treated with 1 µg/mL LPS for different periods of time.
Evaluation of the transfection efficiency of siRab26-DYM by flow cytometry
The transfection efficiency of siRab26-DYM-Cy3 was evaluated by fluorescence activated cell sorting (FACS). Generally, varying concentrations of siRab26-DYM-Cy3 or siRab26-Cy3 were transfected into HPMVECs for 24 h using X-tremeGENE siRNA transfection reagent at a ratio of transfection reagent (µL) to siRNA (µg) of 2:1, and the treated cells were harvested, washed with 1 mL PBS and centrifuged at 1000 rpm for 5 mins. To separate the membrane-bound siRab26-Cy3/siRab26-DYM-Cy3 from the intracellularly internalized molecules, each sample was divided in half. One half was treated with 0.5% trypan blue, an extracellular fluorescence quenching dye, for 5 min and then washed with PBS twice. These samples were then quantified by flow cytometry (Becton Dickinson, San Jose, CA) with an argon-ion laser and 570 nm bandpass filters for emission measurements. The data were analyzed using FlowJo software.
Intracellular uptake study
HPMVECs were seeded on chamber slides at a density of 5×103 cells/mL and pre-cultured for 24 h; the cells were then transfected with either 25 nM siRab26-Cy3 or 25, 50 or 100 nM siRab26-DYM-Cy3. The cells were washed with PBS to remove the excess siRab26-Cy3 or siRab26-DYM-Cy3 after 12 h. The HPMVECs were further cultured in fresh medium for different lengths of time. The nuclei were stained with 4', 6-diamidino-2-phenylindole (DAPI), and the cells were analyzed using confocal laser scanning microscopy (CLSM). To explore the time-dependent cellular uptake of siRab26-DYM, the fixed concentration (50 nM) of siRab26-DYM was transfected into cells for 3 h, 6 h, 12 h, and 24 h. The cells were washed and fixed with a mixture of 4% paraformaldehyde and 4% sucrose in PBS for 20 min, and then were analyzed by CLSM.
RT-PCR analysis of Rab26 mRNA expression
Total RNA was isolated from treated HPMVECs, and cDNAs were synthesized using a PrimeScript RT Reagent Kit according to the instructions. The primers used for the PCR analyses were: Rab26 forward primer, 5'-GCTCTACGATGTCACCAACAAG-3'; Rab26 reverse primer, 5'-TGAAGGGCAGTCCATACTCCT-3'; β-actin forward primer, 5'-GAGCACAGAGCCTCGCCTTT-3'; and β-actin reverse primer, 5'-AGAGGCGTACAGGGATAGCA-3'. The PCR products were separated by electrophoresis on a 2.5% agarose gel containing 0.5% ethidium bromide. The data were analyzed using Image Lab software and normalized to the expression of the β-actin mRNA.
Western blot analysis of Rab26, TLR4, MyD88, TRAF6, NF-κB and Foxo1 protein expression
Western blotts were performed as previously described [9]. Total protein and nuclear protein (using Nuclear and Cytoplasmic Protein Extraction Kit (Beyotime, China)) were extracted from the treated cells, according to manufacturer's instructions [25]. 50 µg of each protein sample was separated by 15% SDS-PAGE and transferred onto PVDF membranes. After blocking with non-fat milk, the membranes were incubated with primary antibodies overnight and then with the appropriate secondary antibody for 1 h. The protein signals were detected by an enhanced chemiluminescence detection system. The densitometric analysis was performed using Quantity One software.
Flow cytometry analysis of cell surface TLR4 expression
To measure the level of TLR4 at the cell surface, treated cells were harvested by trypsinization, incubated with a PE-conjugated anti-human CD284 (TLR4) antibody for 30 mins on ice, and then washed with 1 mL PBS and centrifuged at 1000 rpm for three times. The cell surface expression of TLR4 was determined by measuring the PE fluorescence intensity on a flow cytometer (FACS Calibur; BD Biosciences, San Jose, CA).
Infection of HPMVECs with recombinant Rab26 adenoviruses
Recombinant adenoviruses expressing the human Rab26 cDNA were prepared as described previously [26]. Rab26 adenovirus (Adv. Rab26) infection of HPMVECs was carried out at 37°C and 5% CO2 at MOIs of 100 for 6 h, then cultured for 24 h in fresh medium. The empty adenovirus (Adv. vector) was used as negative control. Twenty-four hours after viral infection, HPMVECs were then treated with indicated dose of LPS for 24 h.
Cell apoptosis analysis by flow cytometry
Apoptosis analysis was carried out as previously study [27]. Briefly, HPMVECs (1x106 cells) were seeded in 25 cm2 flask in ECM for 24 h. After transfection with 50 nM siRab26-DYM/NC-DYM for 12 h and infection with Adv. Rab26/Adv. vector for 24 h, HPMVECs were treated with 1 μg/mL LPS for 24 h. These cells were washed twice with PBS, digested with 0.25% trypsin, harvested for staining Annexin V-FITC and PI in the dark, and then subjected to a flow cytometry measurement.
Endothelial cell permeability assay
HPMVECs were seeded into 0.4 μm Transwell inserts after they were transfected with varying amounts of siRab26-DYM for 24 h. When the HPMVECs became confluent, 1 mg/mL FITC-dextran was added to the upper wells, and the cells were incubated for 0.5 h. Then, 50 µL of the medium from the bottom chamber were transferred to a 96-well plate. Finally, the 96-well plates were evaluated in a fluorescence plate reader at an excitation wavelength of 488 nm and an emission wavelength of 520 nm.
Secreted cytokines test
Supernatants were collected and TNF-α, IL-6 were measured by ELISA Kit (Thermo scientific, Rockford, IL). Detailed experimental procedures refer to the instructions of ELISA Kit.
Statistical analysis
The data are shown as the means ± standard errors (S.E.) of at least three independent experiments. The statistical analysis was performed using a two-tailed Student's t-test for experiments consisting of only two groups. Significant differences were calculated via one-way analysis of variance (ANOVA) for experiments consisting of more than two groups. Statistical significance was defined as a p value less than 0.05.
Results
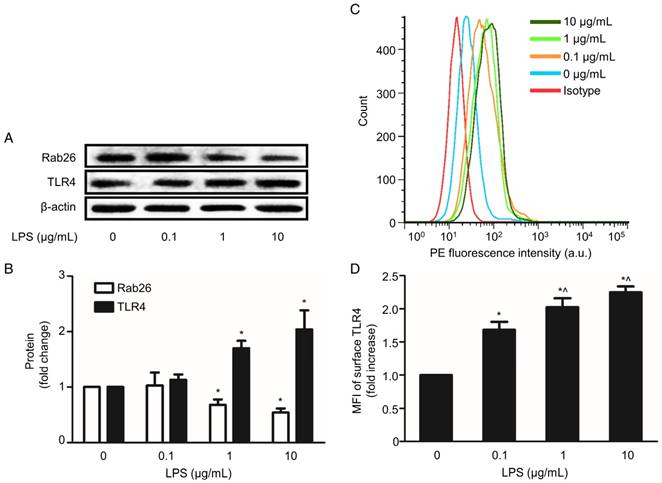
Effect of LPS on Rab26 and TLR4 expression in HPMVECs
LPS-induced endothelial barrier injury, particularly the induction of hypertonicity in HPMVECs, is an initial and important step in the development of ALI. Here, we examined Rab26 and TLR4 expression in LPS-stimulated HPMVECs. As shown in Fig. 1A and B, the Rab26 protein expression was significantly lower in cells treated with 1 and 10 µg/mL LPS than in the control group (p< 0.05). In contrast, the TLR4 protein expression was higher in cells stimulated with 1 and 10 µg/mL LPS than in the control group. Additionally, the surface expression of TLR4 on HPMVECs was elevated after the cells were treated with 0.1, 1 and 10 µg/mL LPS. These data show that LPS can lower down Rab26 protein levels, induce TLR4 overexpression and increase TLR4 localization on the surface of HPMVECs.
Design, construction and characterization of siRab26-DYM
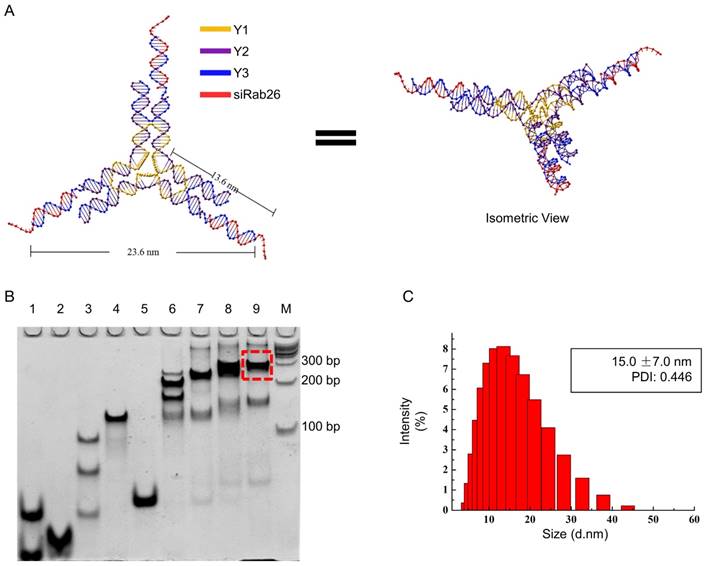
DNA nanomaterials are a convenient delivery carrier, due to their higher transport efficiency, good biocompatibility, and low immunogenicity and nontoxicity. In this study, we constructed a DNA Y-motif nanovector through the self-assembly of three short DNA single strands (Y1, Y2 and Y3) and a Rab26 single-stranded siRNA. As illustrated in Fig. 2A, the DNA single strand Y1 (78 bases) is the core of the DNA Y-motif nanovector. The DNA single strands Y2 (44 bases) and Y3 (40 bases) are the arms of this nanostructure, and at the end of Y3, there are overhangs that can be connected to the Rab26 siRNA (21 bases). All strands were connected by complementary base pairing. As shown in Fig. 2B, all genetic samples appeared as sharp bands with the expected mobility, indicating that the hybrid complexes had been formed, and the yield of the siRab26-DYM nanoparticle was higher (≈80%) when the molar ratio of Y1, Y2, Y3, siRab26 was 1:3:3:3. We found that when the sample that consisted of insufficient Rab26 siRNA (lane 8 in Fig. 2B); the band was broadened and ran slightly faster than target siRab26-DYM band (lane 9). This is likely due to a smaller molecule weight of the partially loaded siRab26 in DYM compared to a complete siRab26-DYM nanoparticle. Similarly, as illustrated in Fig. 2B, we designed siNC-DYM, loaded with a negative control siRNA, as a control and found that it was also produced with a good yield (Fig. S1). DLS measurements further confirmed the formation of siRab26-DYM (Fig. 2C). It had a hydrodynamic diameter of 15.0±7.0 nm, which is in accordance with the theoretical design (Fig. 2A). It should be noted that the three arms of siRab26-DYM are not necessarily in the same plane.
Transfection and cellular uptake of siRab26-DYM in HPMVECs
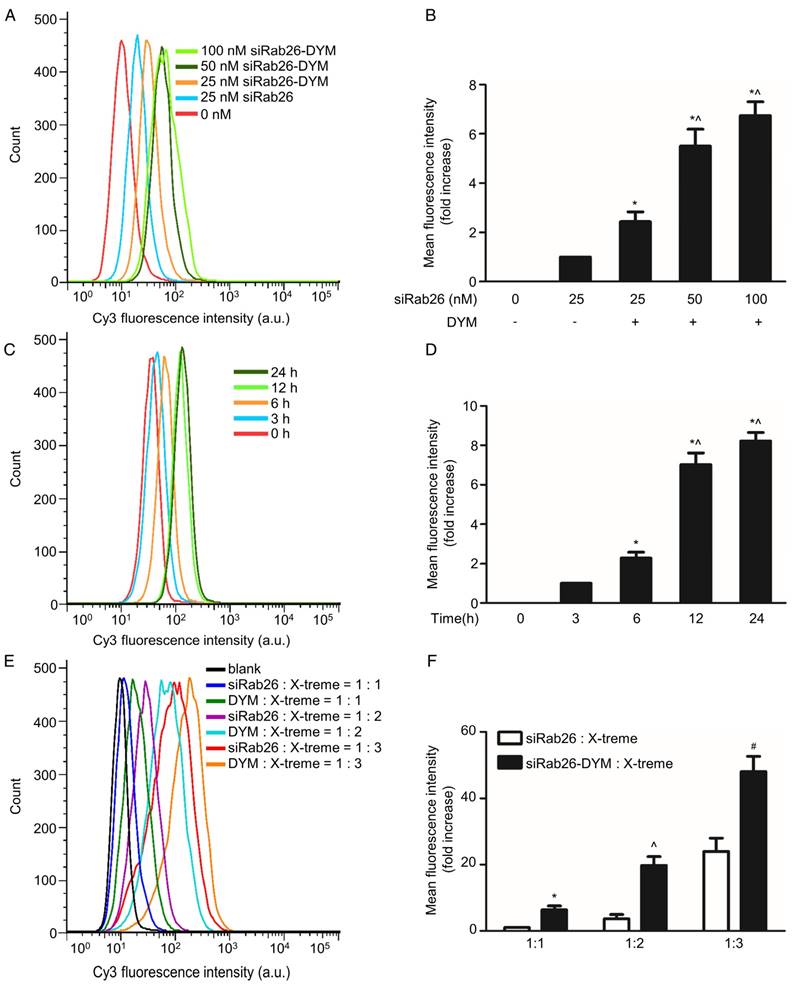
To clarify whether DNA nanoparticles can provide effective delivery of siRab26 into cells, FACS and confocal microscopy were used to analyze the transfection efficiency of siRab26-DYM-Cy3 in HPMVECs. As shown in Fig. 3A and B, after siRab26-DYM was transfected into HPMVECs for 12 h using Roche X-tremeGENE siRNA Transfection Reagent (at a ratio of siRNA (µg) to transfection reagent (µL) of 1:2), the mean fluorescence intensity (MFI) was clearly elevated in the siRab26-DYM-Cy3 and siRab26-Cy3 groups compared to the control group (p< 0.05). The transfection efficiencies of siRab26-DYM-Cy3 at 25, 50, and 100 nM was 22.0%, 59.2%, and 71.5%, respectively, indicating that the efficiency in the siRab26-DYM-Cy3 group was dose dependent. Moreover, the transfection efficiency of 25 nM siRNA alone was approximately only 3.1%, which was significantly lower than the 25 nM siRab26-DYM-Cy3 group (p< 0.05). The time-dependent transfection efficiency of a fixed dose (50 nM) was further examined by varying the incubation time. The transfection efficiency of siRab26-DYM-Cy3 was approximately 5.7%, 21.5%, 62.4% and 70.2% in cells transfected with siRab26-DYM-Cy3 for 0, 3, 6, 12 and 24 h, respectively (Fig. 3C and D). These data demonstrated that the transfection efficiency was enhanced by prolonging the incubation with 50 nM siRab26. Furthermore, we compared the transfection efficiency of siRab26-DYM with bare siRNA (Fig. 3E and F). It was found that the transfection efficiency of siRab26-DYM was much higher than that of siRNA at siRNA/X-treme transfection reagent ratios of 1:1, 1:2 and 1:3 (siRNA (µg) to transfection reagent (µL)).
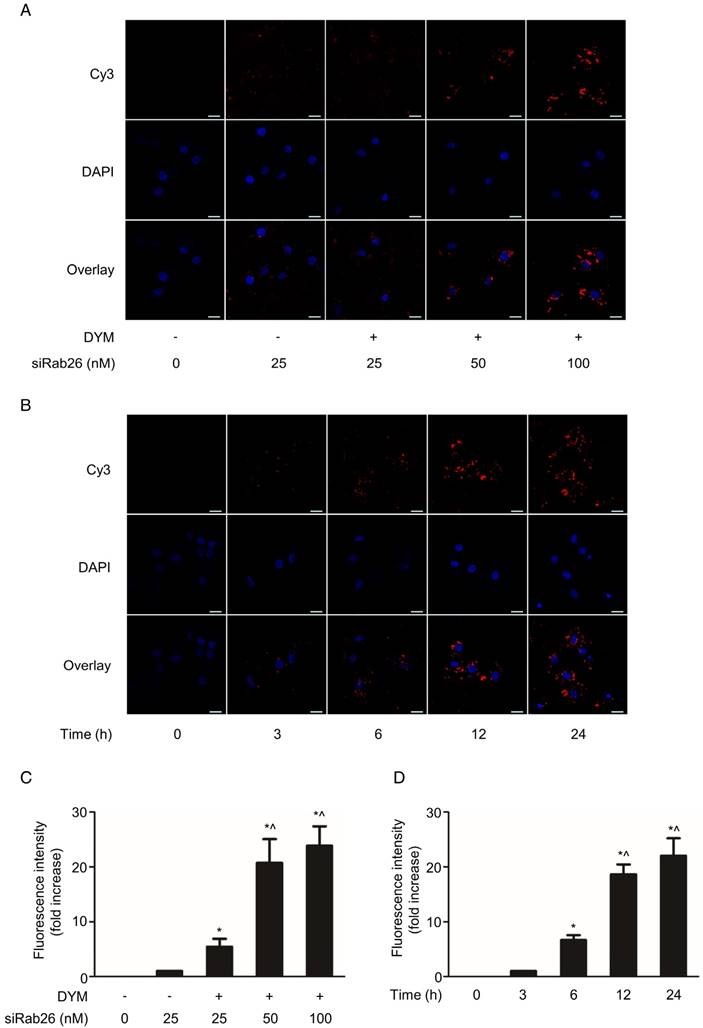
To further investigate the cellular internalization of siRab26-DYM into HPMVECs, we evaluated the cellular uptake of siRab26-DYM-Cy3 using CLSM. As shown in Fig. 4A and C, Cy3 fluorescence intensity was observed in the cells that were transfected with 25, 50 or 100 nM (counting Cy3 tagged Rab26 siRNA) siRab26-DYM-Cy3 for 12 h, and the quantitative analysis showed that the Cy3 fluorescence intensity was elevated with increasing doses of siRab26-DYM-Cy3. Only a small amount of Cy3 fluorescence was observed in cells treated with 25 nM siRab26-Cy3; however, at the same dose, the siRab26-DYM-Cy3 group exhibited a higher Cy3 fluorescence intensity. Quantitative analysis also showed that the Cy3 fluorescence intensity was higher in the siRab26-DYM-Cy3 group than in the siRab26-Cy3 group at a dose of 25 nM (p< 0.05).
The time-dependent cellular uptake and localization were further examined by varying the incubation time. Figure 4B shows CLSM images of HPMVECs incubated with 50 nM Rab26-siRNA-Cy3 for varying times. After 6 h of incubation, Cy3 fluorescence was primarily located in the cytoplasm of HPMVECs. At 12 and 24 h time points, a considerable amount of Cy3 fluorescence was observed in both the cytoplasm and the periphery of the nucleus. Additionally, as shown in Fig. 4D, the Cy3 fluorescence intensity was notably enhanced as the incubation time increased from 3 h, 6 h, 12 h and 24 h compared to that at 0 h (p< 0.05). Overall, these data suggest that siRab26-DYM is an efficient carrier for the delivery of siRab26 into primary cells, and DNA nanostructure carrying Rab26 siRNA is a better way to transfect cells than the siRNA alone.
Inhibition of Rab26 expression by siRab26-DYM
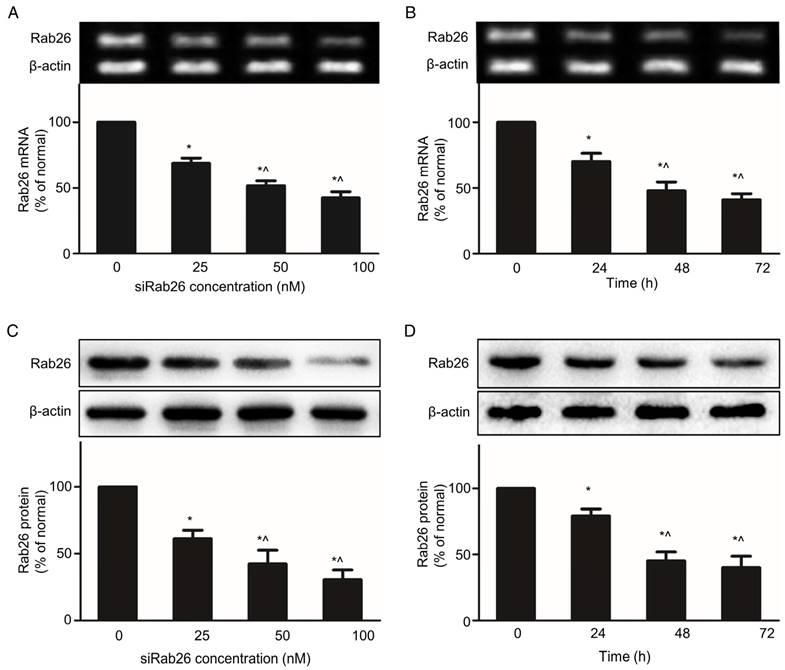
Knocking down target gene is a major function of DNA nanoparticles, which can deliver siRNA into cells. Transfection with siRab26-DYM significantly reduced Rab26 mRNA and protein expression in a dose- and time- dependent manner. As shown in Fig. 5A and C, compared with the 0 nM group, increasing concentrations of siRab26-DYM enhanced the inhibition efficacy. Rab26 mRNA and protein levels decreased to 69.4% and 64.4% of the original levels when cells were incubated with 25 nM siRab26-DYM; and further decreased to 51.2% and 45.7% of the original levels after treatment with 100 nM siRab26-DYM, respectively. Rab26 mRNA and protein levels also decreased to 66.8% and 75.2% of the original levels when cells were transfected with 50 nM siRab26-DYM for 24 h (Fig. 5B and D). At 72 h, Rab26 mRNA and protein levels were further decreased to 41.2% and 44.3%, respectively. Thus, siRab26-DYM efficiently reduced Rab26 mRNA and protein levels in HPMVECs.
Effect of LPS on Rab26 and TLR4 expression in HPMVECs. A. Rab26 and TLR4 proteins expression was detected by western blot analyses after HPMVECs were treated with the indicated doses of LPS for 24 h. The upper, middle and lower panels show representative bands of Rab26, TLR4 and β-actin proteins, respectively. B. The histograms show the quantitative analysis of the Rab26 and TLR4 levels, which were normalized to the β-actin levels. *, p< 0.05 versus the LPS 0 μg/mL group. C. TLR4 surface expression in HPMVECs was analyzed by FACS analysis after the cells were incubated with LPS at the indicated dose for 24 h. D. The histograms show the mean fluorescence intensity (MFI) of surface TLR4 staining. *, p< 0.05 versus the control group. ^, p< 0.05 versus the LPS 0.1 μg/mL group.
Design and characterization of siRab26-DYM. A. Schematic representation of siRab26-DYM. It consists of one central strand Y1 (yellow), three copies of edge strands Y2 (purple) and three copies of peripheral strands Y3 (blue). B. Native PAGE (8%) analysis of the formation of siRab26-DYM nanoparticles. The RNA oligo strands, DNA strands and their molar ratios are: lane “1” - Y1 and Y3, lane “2” - Y2, lane “3” - 1:1 molar ratio of Y1 to Y2, lane “4” - 1:3 molar ratio of Y1 to Y2, lane “5” - 1:1 molar ratio of Y2 to Y3, lane “6” - 1:3:1.5 molar ratio of Y1 to Y2 to Y3, lane “7” - 1:3:3 molar ratio of Y1 to Y2 to Y3, lane “8” - 1:3:3:1.5 molar ratio of Y1 to Y2 to Y3 to siRab26, lane “9” - 1:3:3:3 molar ratio of Y1 to Y2 to Y3 to siRab26, and lane “M”-100 bp DNA ladder. The dashed frame indicates the siRab26-DYM product. C. DLS characterization of siRab26-DYM.
Rab26 regulated the TLR4 signaling pathway in HPMVECs
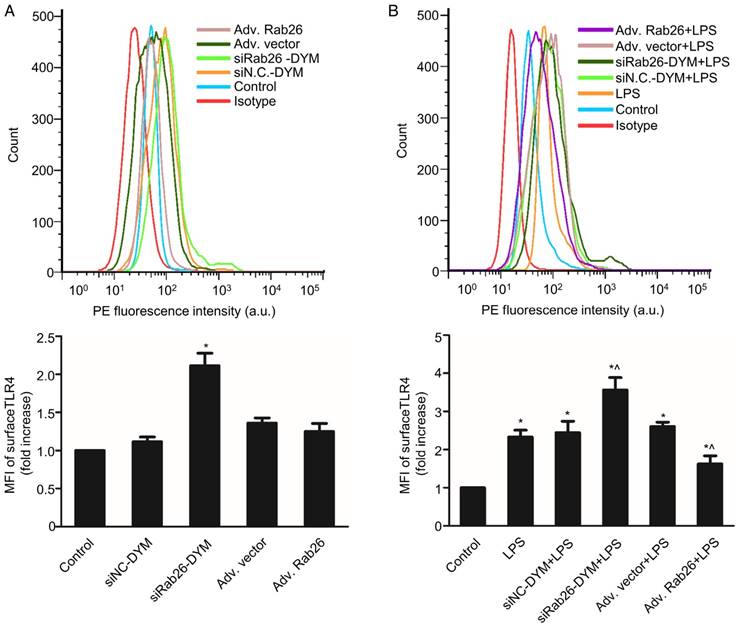
Here, we evaluated the change of TLR4 signaling pathway by regulation of Rab26. As shown in Fig. 6A, the MFI of the siRab26-DYM group was higher than that of the control and siNC-DYM groups (p< 0.05), indicating that the surface level of TLR4 on HPMVECs was increased after siRab26-DYM treatment for 48 h. However, the surface level of TLR4 was not significantly changed after cells were infected with Rab26 adenovirus (Adv. Rab26) compared with empty adenovirus (Adv. vector) and control (p> 0.05). Interestingly, as shown in Fig. 6B, Adv. Rab26 partially reversed the LPS-induced TLR4 accumulation on cell membrane.
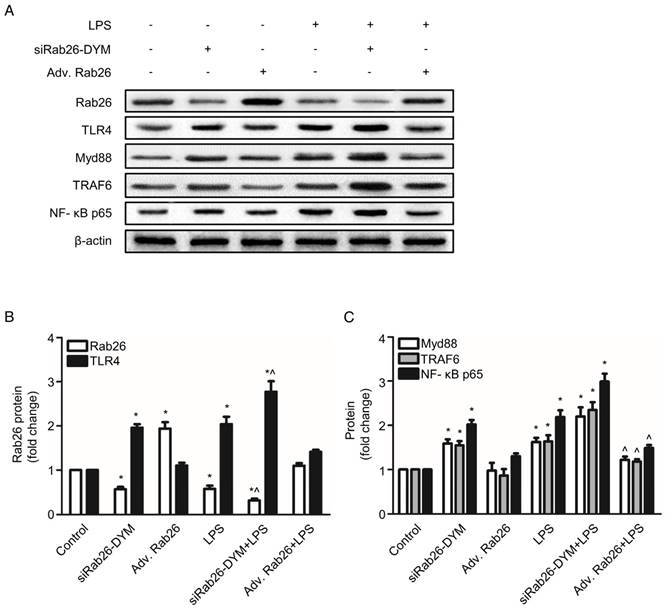
Rab26 silencing not only modulated the surface levels of TLR4 but also activated the TLR4 downstream intermediates MyD88, TRAF6 and NF-κB p65. As illustrated in Fig.7 A, B and Fig. S3, siRab26-DYM significantly decreased the Rab26 protein levels under both normal and LPS stimulated conditions. Compared with LPS treated HPMVECs, the Rab26 level of HPMVECs treated with siRab26-DYM and LPS was much lower (p< 0.05). As illustrated in Fig.7 A and C, the protein expression of the TLR4, MyD88, TRAF6 and NF-κB p65 in siRab26-DYM treated groups were significantly higher than that of control and Adv. Rab26 groups (p< 0.05). The siRab26-DYM transfection elevated the levels of TLR4, MyD88, TRAF6 and NF-κB p65 under LPS stimulation, compared with in the LPS treated alone group (p< 0.05). The expression of TLR4, MyD88, TRAF6 and NF-κB p65 was lower in Adv. Rab26/LPS group than LPS treated group (p< 0.05). These data suggested that Rab26 negatively regulated TLR4 signal pathway in HPMVECs. Additionally, we analyzed the level of inflammatory mediator IL- 6 and TNF-α in medium (Fig. S4) under LPS stimulated conditions. It was found that the level of IL- 6 and TNF-α was dramatically increased upon siRab26-DYM treatment.
Flow cytometry analysis of the transfection efficiency of siRab26-DYM-Cy3 in HPMVECs. A. Typical flow cytometry profiles of HPMVECs after 12 h of incubation with siRab26-DYM-Cy3 or siRab26-Cy3 at the indicated doses. B, Quantitative analysis of the MFI of Cy3 measured in A. The data depict the results of four separate experiments (mean ± S.E., n = 4). *, p< 0.05 versus the 25 nM siRab26 group; ^, p< 0.05 versus the 25 nM siRab26-DYM group. C. Flow cytometry profiles of HPMVECs after various incubation periods with 50 nM siRab26-DYM-Cy3. D. Quantitative analysis of the MFI of Cy3 in C. The data are representative of four separate experiments (mean ± S.E., n = 4). *, p< 0.05 versus the 3 h group; ^, p< 0.05 versus the 6 h group. E. Flow cytometry profiles of HPMVECs after 12 h incubation with siRNA and siRNA/X-treme transfection reagent ratios were varying from 1:1 to 1:3 (siRNA (µg) to transfection reagent (µL)). F. Quantitative analysis of MFI of Cy3 in E. The data are representative of four separate experiments (mean ± S.E., n = 4). *, p< 0.05 versus the siRNA: X-treme transfection reagents =1:1 group ^, p< 0.05 versus the siRNA: X-treme transfection reagents =1:2 group; #, p< 0.05 versus the siRNA: X-treme transfection reagents =1:3 group.
Confocal microscopy analysis of the time- and dose-dependent cellular uptake of siRab26-Cy3 or siRab26-DYM-Cy3 by HPMVECs. A. Fluorescence images showing the cellular uptake of Rab26-siRNA-Cy3 and siRab26-DYM-Cy3 (red) by HPMVECs after 12 h of treatment at the indicated concentrations. The nuclei (blue) were stained with DAPI. Scale bars: 20 μm. B. Fluorescence images illustrating the cellular uptake of siRab26-DYM-Cy3 (red) by HPMVECs after incubation with 50 nM siRab26-DYM-Cy3 (red) for 3, 6, 12 and 24 h. The nuclei (blue) were stained with DAPI. Scale bars represent 20 μm. C. Quantification of the fluorescence intensity of the CLSM images presented in A. The data are presented as the means ± S.E. of three separate experiments. *, p< 0.05 versus the 25 nM siRab26 group. ^, p< 0.05 versus the 25 nM siRab26-DYM group. D. Quantification of the fluorescence intensity of the confocal microscopy images shown in B. The data are presented as the means ± S.E. of three separate experiments. *, p< 0.05 versus the 3 h group. ^, p< 0.05 versus the 6 h group.
RT-PCR and western blot analysis of the Rab26 mRNA and protein levels in HPMVECs. The upper and lower panels illustrate representative blots of Rab26 and β-actin and the quantitative analysis of the Rab26 mRNA and protein levels, respectively. A and C. Rab26 mRNA and protein expression determined by RT-PCR and western blot analyses with varying siRab26-DYM concentrations. The cells were pre-incubated with siRab26-DYM at 0, 25, 50, or 100 nM for 12 h and then incubated with fresh medium for 48 h. The data represent the results of three separate experiments (means ± S.E., n = 4). *, p< 0.05 versus the 0 nM group; ^, p< 0.05 versus the 25nM group. B and D. Rab26 mRNA and protein levels determined by RT-PCR and western blot analyses with different incubation time points. The cells were pre-incubated with siRab26-DYM at 50 nM for 12 h and then incubated for 0, 24, 48 or 72 h in fresh medium. The histograms in the lower panel illustrate the quantitative analysis of the Rab26 levels, which were normalized to the β-actin levels. The data represent the results of three separate experiments (means ± S.E., n = 4). *, p< 0.05 versus the 0 h group; ^, p< 0.05 versus the 24 h group.
Silencing of Rab26 activated TLR4 signal pathway by the nuclear translocation of Foxo1
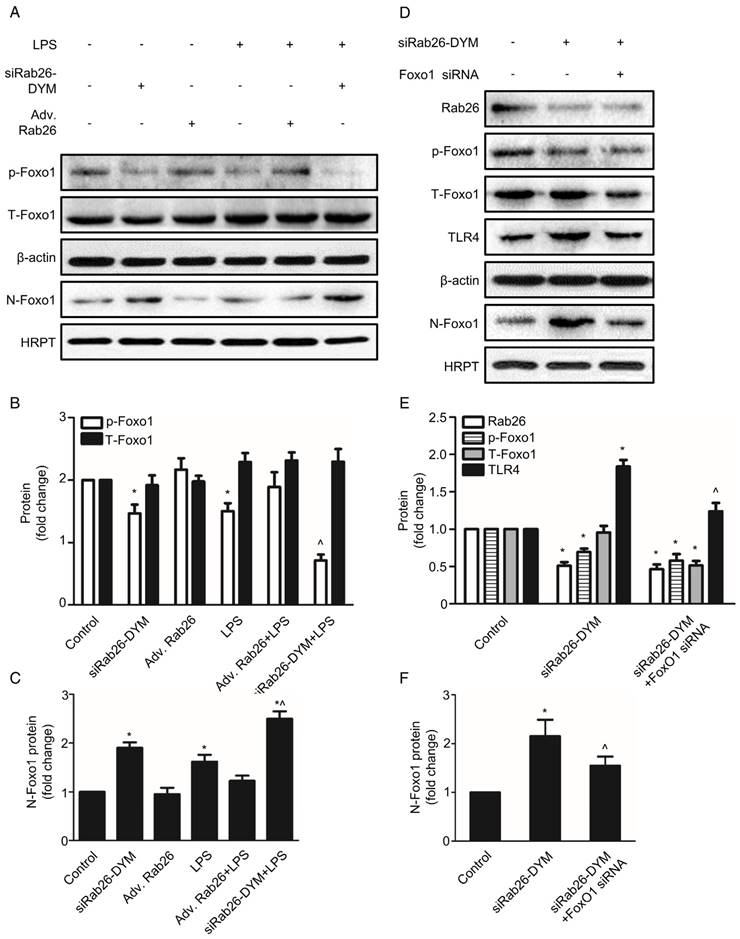
To explore the potential mechanism of down-regulation of Rab26 activated TLR4 signal pathway, we analyzed the expression of Foxo1 at cytoplasm and nuclear, respectively. As illustrated in Fig. 8 A, B and C, the expression of phosphorylation-Foxo1 (p-Foxo1) was lower in siRab26-DYM, LPS, as well as siRab26-DYM/LPS treated groups than control group and Adv. Rab26 groups (p< 0.05). The p-Foxo1 level of Adv. Rab26/LPS group was higher than that of LPS and siRab26-DYM treated groups (p< 0.05). The expression differences of total Foxo1 (T-Foxo1) among these groups (p> 0.05) was negligible. On the contrary, the expression of Foxo1 in cellular nuclear (N-Foxo1) was increased in siRab26-DYM, LPS, as well as siRab26-DYM/LPS treated groups than control and Adv. Rab26 groups (p< 0.05). LPS-induced up-regulation of N-Foxo1 was reversed while HPMVECs was infected with Adv. Rab26. These data demonstrated that Rab26 mediated the nuclear translocation of Foxo1. Next, we further studied whether down-regulation of Rab26 promoted TLR4 expression through Foxo1 nuclear translocation (Fig. 8 D, E and F). The level of p-Foxo1 and total Foxo1 for Foxo1 siRNA and siRab26-DYM treated groups was lower than siRab26-DYM alone group (p< 0.05). Note that all groups were treated with Foxo1 first for 12 h before the transfection of siRab26-DYM. Similarly, the expression of TLR4 and N-Foxo1 was decreased in Foxo1 siRNA and siRab26-DYM groups, compared with siRab26-DYM alone group (p< 0.05). These data revealed that silencing of Foxo1 by Foxo1 siRNA could partially block siRab26-DYM-induced TLR4 expression and TLR4 signal pathway activation. Taken together, silencing of Rab26 by siRab26-DYM activated TL4 signal pathway by the nuclear translocation of Foxo1.
Regulation of the permeability and apoptosis of LPS stimulated HPMVECs by targeting Rab26
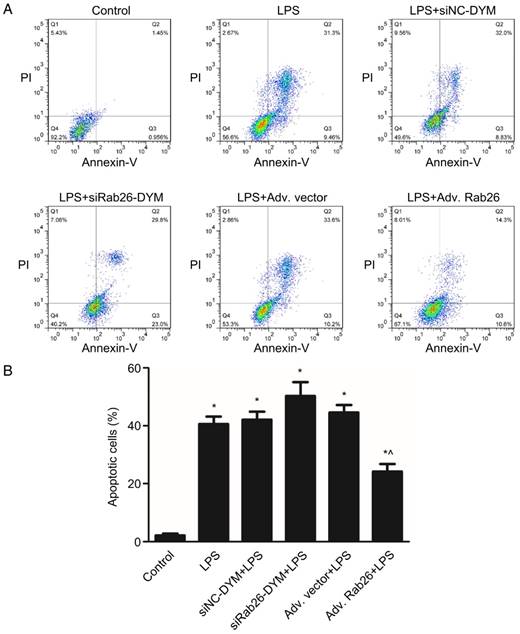
To determine the role of Rab26 in regulating the apoptosis and permeability of HPMVECs under pathological conditions, we first knocked down or overexpressed Rab26, and then analyzed the cellular apoptosis rate and permeability of LPS-treated HPMVECs. As illustrated in Fig. 9, we found that the cell apoptosis also increased upon treatment with LPS, LPS/siRab26-DYM or LPS/Adv. Rab26 (p< 0.05). The apoptosis rate of the LPS/siRab26-DYM treated group was higher than the LPS alone and LPS/Adv. Rab26 group (p< 0.05). Moreover, the apoptosis rate of the LPS /Adv. Rab26 treated HPMVECs was significantly decreased compared with that of the LPS and LPS /siRab26-DYM groups (p< 0.05). A similar trend was observed under optical microscope. (Fig. S5).
Effects of Rab26 on cell surface TLR4 in HPMVECs. A. FACS analysis of the surface level of TLR4. Cells were first cultured in 10% serum ECM for 24 h and then divided into five groups. Two groups were transfected with 50 nM siRab26-DYM or 50 nM siNC-DYM for 12 h; two other groups were infected with an MOI of 100 of Adv. Rab26 or Adv. Vector for 6 h. Then all cells were culture in fresh ECM for 48 h, collected respectively and analyzed using FACS analysis. The histograms show the mean fluorescence intensity of surface TLR4 staining. The data are presented as the means ± S.E. of three separate experiments (n = 3) *, p< 0.05 versus the control group. B. TLR4 expression on the HPMVEC surface when cells were treated with 1 μg/mL LPS for 24 h. Cells were first cultured in 10% serum ECM for 24 h and divided into six groups. Two group were transfected with 50 nM siRab26-DYM or 50 nM siNC-DYM for 12 h and cultured in fresh ECM for 24 h, then treated with 1 μg/mL LPS for 24 h; two other groups were infected with MOI100 Adv. Rab26 or MOI100 Adv. Vector for 6 h and cultured in fresh ECM for 24 h, then treated with 1 μg/mL LPS for 24 h, The last group was cultured in fresh ECM for 36 h and treated with 1 μg/mL LPS for 24 h. All groups were then collected respectively and subjected to FACS analysis. The histograms show the mean fluorescence intensity of surface TLR4 staining. The data are representative of three separate experiments (mean ± S.E., n = 3). *, p< 0.05 versus the control group; ^, p< 0.05 versus the LPS group.
Effect of Rab26 on the TLR4 signaling pathway in HPMVECs. A. HPMVECs were cultured in 10% serum ECM for 24 h and then divided into six groups. The first group was cultured in fresh ECM for 60 h as control. Two groups were transfected with 50 nM siRab26-DYM for 12 h, then cultured in fresh ECM for another 24 h, and treated with or without 1 μg/mL LPS for 24 h. Two groups were infected with MOI100 Adv. Rab26 for 6 h, then cultured in fresh ECM for 24 h, and treated with or without 1 μg/mL LPS for 24 h. The last group was cultured in fresh ECM for 36 h and treated with 1 μg/mL LPS for 24 h. All cells were collected for western blotting detection. These panels show representative bands of the Rab26, TLR4, MyD88, TRAF6, NF-κB p65 and β-actin proteins. B. The histograms show the quantitative analysis of the Rab26 and TLR4 levels, which were normalized to the β-actin levels. *, p< 0.05 versus the control group. ^, p< 0.05 versus the LPS group. C. Quantitative analysis of MyD88, TRAF6 and NF-κB p65 levels, which were normalized to β-actin levels. *, p< 0.05 versus the control group. ^, p< 0.05 versus the LPS group.
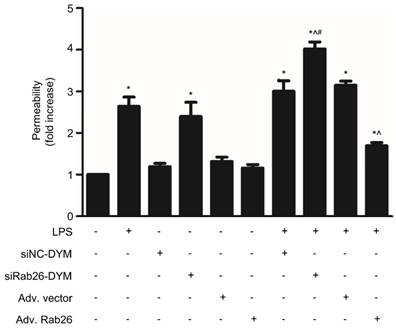
The permeability assay of HPMVECs was showed in Fig. 10. The permeability of both the LPS- and siRab26-DYM-treated groups was increased compared with that of the control and siNC-DYM groups (p< 0.05). Moreover, Rab26 overexpression did not impair the permeability of HPMVECs. The cellular permeability of the LPS, siRab26-DYM/LPS and Adv. Rab26/LPS groups was increased compared with that of the control group (p< 0.05). Importantly, after LPS stimulation, Rab26 silencing with siRab26-DYM increased the hyperpermeability of HPMVECs, and the restoration of Rab26 level with Adv. Rab26 maintained normal cellular permeability. These data suggest that Rab26 play key roles in controlling HPMVEC apoptosis and permeability.
Discussion
The present study was undertaken to construct a self-assembled DNA nanovector with a simple structure, precise programmability, and higher yield, with three copies of Rab26 siRNA loaded onto each nanoparticle. This nanoparticle exhibited useful characteristics, such as a higher transfection efficiency and lower toxicity, for the knockdown of target genes in primary HPMVECs. We also found that siRab26-DYM promoted LPS-induced activation of TLR4 signaling pathway, aggravated cell apoptosis and impaired the endothelial barrier. Rab26 overexpression partially reversed the accumulation of TLR4 at cytoplasm membrane in response to LPS stimulation, resulting in reduced cell apoptosis and partially restored endothelial barrier function. These data suggested that Rab26 played a key role in regulating the endothelial barrier functions of HPMVECs.
Rab26 regulated the nuclear translocation of Foxo1 in HPMVECs. A. HPMVECs were cultured in 10% serum ECM for 24 h and then divided into six groups. One group was cultured in fresh ECM for 60 h as control. Two groups were transfected with 50 nM siRab26-DYM for 12 h, then cultured in fresh ECM for 24 h, and treated with or without 1 μg/mL LPS for 24 h. Two groups were infected with MOI100 Adv. Rab26 for 6 h, then culture in fresh ECM for 24 h, and treated with or without 1 μg/mL LPS for 24 h. The last group was cultured in fresh ECM for 36 h and directly treated with 1 μg/mL LPS for 24 h. All cells were collected for Western blotting detection. Five panels show representative bands of p-Foxo1, T-Foxo1, β-actin, N-Foxo1 and HRPT. B. Quantitative analysis of p-Foxo1, T-Foxo1 levels, which were normalized to β-actin levels. *, p< 0.05 versus the control group. ^, p< 0.05 versus the LPS group. C. Quantitative analysis of N-Foxo1 levels, which were normalized to HRPT levels. *, p< 0.05 versus the control group. ^, p< 0.05 versus the LPS group. D. HPMVECs were cultured in 10% serum ECM for 24 h waiting for cell attachment and then divided into three groups. The first group was cultured in fresh ECM for 60 h as control. The second group was transfected with 50 nM siRab26-DYM for 12 h, then cultured in fresh ECM for 48 h. The last group was treated with Foxo1 siRNA for 12 h first, and then transfected with 50 nM siRab26-DYM, then cultured in fresh ECM for 24 h. All cells were collected as sample for Western blotting detection. Panels show representative bands of Rab26, p-Foxo1, T-Foxo1, TLR4, β-actin, N-Foxo1, HRPT. E. Quantitative analysis of Rab26, p-Foxo1, T-Foxo1, TLR4 levels, which were normalized to β-actin levels. *, p< 0.05 versus the control group. ^, p< 0.05 versus the siRab26-DYM group. F. Quantitative analysis of N-Foxo1 levels, which were normalized to HRPT levels. *, p< 0.05 versus the control group. ^, p< 0.05 versus the siRab26-DYM group.
Rab26 modulated the apoptosis of HPMVECs. HPMVECs were pre-cultured in ECM for 24 h and then divided into six groups. The first group was cultured in fresh ECM for 60 h as control. Two groups were transfected with 50 nM siRab26-DYM or 50 nM siNC-DYM for 12 h, then cultured in fresh ECM for an additional 24 h, and then treated with 1 μg/mL LPS for 24 h. Two groups were infected with MOI100 Adv. Rab26 or MOI 100 Adv. Vector for 6 h, then cultured in fresh ECM for 24 h, and then treated with 1 μg/mL LPS for 24 h. The last group was cultured in fresh ECM for 36 h and treated with 1 μg/mL LPS for 24 h. A. Typical images showing apoptotic HPMVECs induced by LPS, LPS+siNC-DYM, LPS+siRab26-DYM, LPS+Adv. vector or LPS+Adv. Rab26, respectively. The right two quadrants represent apoptotic cells. B. The histograms show the quantitative analysis of apoptotic cells, data are presented as the means ± S.E. of three independent experiments. *, p< 0.05 versus the control group. ^, p< 0.05 versus the LPS group.
Effect of Rab26 on the monolayer permeability of HPMVECs. HPMVECs were divided into 10 groups and grown to confluency in the upper chambers of 0.4-µm Transwell inserts. Four groups were transfected with 50 nM siRab26-DYM or 50 nM siNC-DYM for 12 h, then cultured in fresh ECM for 24 h, and treated with or without 1 μg/mL LPS for 24 h. Four groups were infected with MOI100 Adv. Rab26 or MOI100 Adv. Vector for 6 h, then cultured in fresh ECM for 24 h, and treated with or without 1 μg/mL LPS for 24 h. The last two groups were directly treated with or without 1 μg/mL LPS for 24 h. Next, FITC-dextran was added to the upper chambers at a concentration of 1 mg/mL. After a 0.5 h incubation, 50 µL of medium from the bottom chamber were removed and analyzed in a fluorescence plate reader. The FITC-dextran permeability is expressed as the mean fold increases ± S.E. (n = 3). *, p < 0.05 versus the control group. ^, p < 0.05 versus the LPS group, #, p < 0.05 versus the siRab26-DYM group.
Self-assembled DNA nanoparticles are emerging as a powerful platform for bioimaging [28], delivering therapeutics into cells both in vitro and in vivo [29-33]. A promising function of DNA nanoparticles is the delivery of siRNAs to suppress the expression of target genes in cells, allowing for the study of the cellular response to the loss of target gene functions [34, 35]. A lot of non-viral vectors have been explored as gene delivery tools, such as cationic lipids [36], cationic polymer [37], dendrimers [38], inorganic nanoparticles [39] and even cell penetrating peptides [40]. More recently, it has been consistently observed that self-assembled DNA nanoparticles with a simpler structure are highly effective at delivering siRNAs into mammalian cells and down-regulating the target gene [23]. Moreover, compared with DNA origami-based complex DNA nanomaterials, particles with simpler structures have several advantages, including the ability to be more accurately programmed, higher productivity, a lower mismatch rate and lower cost [23]. Current RNAi strategies using duplex siRNAs suffer from low efficiency and a high cost, particularly for primary cells, and generally at least three different sequences against the target gene need to be designed to ensure knockdown success. We previously found that in the presence of a DNA nanostructure, pulmonary vascular smooth muscle cells needed much less transfection reagent to take up the nanomaterials. Instead of adding two T bases at the three ends of the siRNA or using locked nuclei acid DNA; our simple DNA nanostructure is even more protective of the siRNA in harsh cellular environments. In our study, we developed a simple structure, named siRab26-DYM. This nanovector was constructed with three single strands of DNA and one single stranded siRab26 in a specific molar ratio, and the yield of this structure reached approximately 80%. Transfection and cellular uptake assays showed that this nanovector was more effective at delivering siRab26-Cy3 to primary HPMVECs than transfection of the Rab26 siRNA alone. Furthermore, siRab26-DYM more significantly reduced Rab26 mRNA and protein expression than siRNA alone, largely due to the higher transfection efficiency in primary HPMVECs. To elucidate the advantages of siRab26-DYM over siRNA, we conducted an RNase assay to evaluate the intracellular stability of siRab26-DYM. When subjected to a very high level (extreme) of RNase A, the Rab26 siRNA cargo was degraded in a step-wise manner (Fig. S2). This degradation pattern suggested that the siRab26-DYM releases siRNA in a sustained manner over a much wider time window, thus had a better knockdown effect than the siRNA alone. These results demonstrated that the siRab26-DYM nanoparticle is an efficient method for the delivery of the Rab26 siRNA into primary mammalian cells, which allowed us to further study Rab26 gene function. Notably, siRab26-DYM has one major limitation: the cellular uptake of siRab26-DYM without the assistance of a transfection agent is negligible, and we were therefore unable to completely avoid the potential cytotoxicity of conventional transfection reagents. We attempted to load several different aptamers into the siRab26-DYM structure to allow the nanoparticle to bind to receptors on the surface of HPMVECs and facilitate the internalization of siRab26-DYM. However, CLSM imaging showed that the DNA nanoparticles complexed with aptamers were trapped on the cell membrane and were not taken up by cells (Data not shown). This might be due to the properties of the cell receptor that prevent it from being transported into the cytoplasm. We reasoned that aptamers based on whole cells, instead of certain specific cytoplasm membrane markers, would overcome the internalization issue. We are currently in the process of identifying aptamers based on whole cell selection.
Rab GTPases play important roles in controlling almost every step of vesicle-mediated receptor transport, including the targeting, tethering, and fusion of transport vesicles [41]. Most receptors, such as G-protein-coupled receptors (GPCRs) and TLRs, activate downstream effectors and modulate a variety of cell functions largely [42] by controlling the level of the receptor expression at the cytoplasm membrane at the steady state. This process depends on the ability of Rab GTPases to modulate intercellular trafficking processes, including anterograde receptor trafficking to the cell surface, internalization of the cell membrane receptors into endosomes following agonist stimulation, and receptor transport to lysosomes for degradation [9, 43]. Inactivation of Rab GTPases induces the mistrafficking of GPCRs, which leads to the dysfunction of the receptors that is directly linked to disorder of cell function [3, 4, 44]. Consistent with other studies [3, 4, 44], our study showed that Rab26 silencing with siRab26-DYM in LPS stimulated cells increased the surface levels of TLR4 and subsequently activated TLR4 signal pathway in primary HPMVECs. Up-regulation of Rab26 expression with an adenovirus partially reversed the LPS induced activation of TLR4 signal pathway. However, in normal condition, Rab26 overexpression was neither unable to increase TLR4 levels at the cell membrane, nor to activate TLR4-NF-κB signaling. These data suggest that Rab26 is able to modulate the cellular TLR4 level and distributions, and activates the downstream NF-κB signaling pathway in HPMVECs. Interestingly, unlike previous studies [9] showing that α2-ARs are trapped in the Golgi and fail to be transported to the cell membrane in Rab26 mutants and in cells transfected with a Rab26 siRNA, the siRab26-DYM not only increased the surface levels of TLR4 but also activated TLR4 signaling pathway in primary HPMVECs under both normal and LPS-stimulated conditions in our study. We reasoned that Rab26 indirectly regulated the trafficking of TLR4 from cell organelle to plasma membrane.
Foxo1, also known as FKHR, is one of the Foxo transcription factor family. It contains a conserved 110-amino acid winged helix DNA-binding domain [45]. Previously study showed that nuclear translocation of Foxo1 played a critical role in multiple cellular processes, including metabolism, cell differentiation, apoptosis, proliferation, DNA repair and cellular stress resistance [46, 47]. Moreover, Foxo1 was also critical to the endothelial cell functions at the transcriptional level. Chen et al showed that dehydroepiandrosterone induced the phosphorylation of Foxo1. The phosphorylated Foxo1 was retained in the cytoplasm or degraded through ubiquitination mediation [48], which resulted in the block of nuclear translocation of Foxo1. As a result, the promoter activity and secretion of the vasoconstrictor endothelin-1 (ET-1) were suppressed [49]. Jang and colleagues demonstrated that the phosphorylation status of Foxo1 was consistent with the role of Claudin 5, a critical component of endothelial tight junctions, in acerolein induced endothelial barrier dysfunction in vitro and in vivo [50]. Other studies revealed that stress-induced phosphoinositide 3-kinase (PI3K)/Akt pathway suppresses the nuclear accumulation of Foxo1, thereby promoting cell survival, strengthening intercellular junctions, and suppressing inflammation in endothelial cells [51]. Additionally, because TLR4 is a transcriptional target of Foxo1 [52], Foxo1 may increase the levels of inflammatory mediator, such as interleukin-6 (IL-6), tumor necrosis factor-α (TNF-α) and IL-1β, by upregulation of TLR4 [52]. Knockdown of Foxo1 by a Foxo1 siRNA could abolish LPS-stimulated TLR4 expression and the inflammatory response [53]. It was also reported that Foxo1 regulated TLR4 mediated innate immune in mouse liver ischemia/reperfusion injury [54, 55]. Consistent with documents, we found that the level of p-Foxo1 significantly decreased, and the expression of Foxo1 in nuclear was up-regulated after HPMVECs was transfected with siRab26-DYM under LPS stimulated conditions. Our study further showed that the knock-down of Foxo1 by Foxo1 siRNA partially reversed siRab26-DYM-induced total TLR4 expression. These data suggested that down-regulation of Rab26 promoted the nuclear translocation of Foxo1, which further increased the expression of TLR4.
The activation of the TLR4 signaling pathway by LPS is a key step in inducing an inflammatory injury in cells and impaired the cellular function and induced apoptosis [56]. This process involved in production and release of inflammation mediators and activated apoptosis signaling pathway [57]. In our study, Rab26 silencing with siRab26-DYM activated TLR4 signaling and aggravated cells apoptosis and endothelial hypertonicity under LPS stimulation. Furthermore, the restoration of Rab26 expression in LPS-stimulated cells reduced cell apoptosis, partially improved the permeability of HPMVECs and inhibited the TLR4 signaling pathway. Therefore, we propose that Rab26 might control the survival and permeability of HPMVECs by regulating the TLR4 signal pathway.
Conclusions
In this study, we have knocked down Rab26 expression using Rab26 siRNA-loaded DNA nanoparticles, and overexpressed Rab26 using adenovirus. The siRab26-DYM nanoparticles enhanced normal and LPS-induced TLR4 accumulation on the cell membrane, promoted the nuclear translocation of Foxo1, subsequently activated the TLR4 signaling pathway, and induced the apoptosis and hyperpermeability of HPMVECs. In contrast, Rab26 overexpression reduced the TLR4 levels on cell surface, inactivated the TLR4 signaling pathway, resulting in inhibition of cell apoptosis and attenuated the LPS-stimulated hyperpermeability of HPMVECs. Additionally, siRab26-DYM had a stronger inhibitory effect on Rab26 expression than transfection of the conventional siRNA in HPMVECs. Thus, siRab26-DYM may be a promising delivery system for gene silencing in primary mammalian cells. Our study has also provided evidence that Rab26 play important roles in regulating endothelial barrier permeability and apoptosis through TLR4 and its downstream signaling pathways in HPMVECs. Taken together, Rab26 play a key role in the pathogenesis of ALI and ARDS.
Supplementary Material
Supplementary figures.
Acknowledgements
We thank Zhenghua Wei, Huaping Chen, Jin Li, Zi Wang, Zhou Long and Qing Zhang for technical assistance. This research was supported by NSFC grant 81370168 (GW) and China Joint Research Fund for Overseas Chinese, Hong Kong and Macao Scholars 81429001 (CM and GW).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Qiao RL, Bhattacharya J. Segmental barrier properties of the pulmonary microvascular bed. J Appl Physiol (1985). 1991;71:2152-9
2. Ware LB. Pathophysiology of acute lung injury and the acute respiratory distress syndrome. Semin Respir Crit Care Med. 2006;27:337-49
3. Li Y, Wang G, Lin K, Yin H, Zhou C, Liu T. et al. Rab1 GTPase promotes expression of beta-adrenergic receptors in rat pulmonary microvascular endothelial cells. Int J Biochem Cell Biol. 2010;42:1201-9
4. Yang J, Sun H, Zhang J, Hu M, Wang J, Wu G. et al. Regulation of beta-adrenergic receptor trafficking and lung microvascular endothelial cell permeability by Rab5 GTPase. Int J Biol Sci. 2015;11:868-78
5. Yang J, Yao W, Qian G, Wei Z, Wu G, Wang G. Rab5-mediated VE-cadherin internalization regulates the barrier function of the lung microvascular endothelium. Cell Mol Life Sci. 2015;72:4849-66
6. Sukriti S, Tauseef M, Yazbeck P, Mehta D. Mechanisms regulating endothelial permeability. Pulm Circ. 2014;4:535-51
7. Tian X, Jin RU, Bredemeyer AJ, Oates EJ, Blazewska KM, McKenna CE. et al. RAB26 and RAB3D are direct transcriptional targets of MIST1 that regulate exocrine granule maturation. Mol Cell Biol. 2010;30:1269-84
8. Nashida T, Imai A, Shimomura H. Relation of Rab26 to the amylase release from rat parotid acinar cells. Arch Oral Biol. 2006;51:89-95
9. Li C, Fan Y, Lan TH, Lambert NA, Wu G. Rab26 modulates the cell surface transport of alpha2-adrenergic receptors from the Golgi. J Biol Chem. 2012;287:42784-94
10. Jin RU, Mills JC. RAB26 coordinates lysosome traffic and mitochondrial localization. J Cell Sci. 2014;127:1018-1032
11. Binotti B, Pavlos NJ, Riedel D, Wenzel D, Vorbruggen G, Schalk AM. et al. The GTPase Rab26 links synaptic vesicles to the autophagy pathway. Elife. 2015;4:e05597
12. Schaefer L. Complexity of danger: the diverse nature of damage-associated molecular patterns. J Biol Chem. 2014;289:35237-45
13. Loniewski K, Shi Y, Pestka J, Parameswaran N. Toll-like receptors differentially regulate GPCR kinases and arrestins in primary macrophages. Mol Immunol. 2008;45:2312-22
14. Hoogerwerf JJ, de Vos AF, Bresser P, van der Zee JS, Pater JM, de Boer A. et al. Lung inflammation induced by lipoteichoic acid or lipopolysaccharide in humans. Am J Respir Crit Care Med. 2008;178:34-41
15. Opal SM, Laterre PF, Francois B, LaRosa SP, Angus DC, Mira JP. et al. Effect of eritoran, an antagonist of MD2-TLR4, on mortality in patients with severe sepsis: the ACCESS randomized trial. JAMA. 2013;309:1154-62
16. Wang Y, Chen T, Han C, He D, Liu H, An H. et al. Lysosome-associated small Rab GTPase Rab7b negatively regulates TLR4 signaling in macrophages by promoting lysosomal degradation of TLR4. Blood. 2007;110:962-71
17. Wang D, Lou J, Ouyang C, Chen W, Liu Y, Cao X. et al. Ras-related protein Rab10 facilitates TLR4 signaling by promoting replenishment of TLR4 onto the plasma membrane. Proc Natl Acad Sci U S A. 2010;107:13806-11
18. Patel N, Gonsalves CS, Malik P, Kalra VK. Placenta growth factor augments endothelin-1 and endothelin-B receptor expression via hypoxia-inducible factor-1 alpha. Blood. 2008;112:856-65
19. Bumcrot D, Manoharan M, Koteliansky V, Sah DW. RNAi therapeutics: a potential new class of pharmaceutical drugs. Nat Chem Biol. 2006;2:711-9
20. Mizuno S, Bogaard HJ, Gomez-Arroyo J, Alhussaini A, Kraskauskas D, Cool CD. et al. MicroRNA-199a-5p is associated with hypoxia-inducible factor-1alpha expression in lungs from patients with COPD. Chest. 2012;142:663-72
21. Li J, Zhou J, Zhang D, Song Y, She J, Bai C. Bone marrow-derived mesenchymal stem cells enhance autophagy via PI3K/AKT signalling to reduce the severity of ischaemia/reperfusion-induced lung injury. J Cell Mol Med. 2015;19:2341-51
22. Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov. 2009;8:129-38
23. Wang Y, You Z, Du J, Li H, Chen H, Li J. et al. Self-assembled triangular DNA nanoparticles are an efficient system for gene delivery. J Control Release. 2016;233:126-35
24. You Z, Qian H, Wang C, He B, Yan J, Mao C. et al. Regulation of vascular smooth muscle cell autophagy by DNA nanotube-conjugated mTOR siRNA. Biomaterials. 2015;67:137-50
25. Guo NN, Li BM. Cellular and subcellular distributions of beta1- and beta2-adrenoceptors in the CA1 and CA3 regions of the rat hippocampus. Neuroscience. 2007;146:298-305
26. Mantwill K, Kohler-Vargas N, Bernshausen A, Bieler A, Lage H, Kaszubiak A. et al. Inhibition of the multidrug-resistant phenotype by targeting YB-1 with a conditionally oncolytic adenovirus: implications for combinatorial treatment regimen with chemotherapeutic agents. Cancer Res. 2006;66:7195-202
27. Liu X, Wang G, You Z, Qian P, Chen H, Dou Y. et al. Inhibition of hypoxia-induced proliferation of pulmonary arterial smooth muscle cells by a mTOR siRNA-loaded cyclodextrin nanovector. Biomaterials. 2014;35:4401-16
28. Jungmann R, Avendano MS, Woehrstein JB, Dai M, Shih WM, Yin P. Multiplexed 3D cellular super-resolution imaging with DNA-PAINT and Exchange-PAINT. Nat Methods. 2014;11:313-8
29. Wei JH, Cheang T, Tang B, Xia H, Xing Z, Chen Z. et al. The inhibition of human bladder cancer growth by calcium carbonate/CaIP6 nanocomposite particles delivering AIB1 siRNA. Biomaterials. 2013;34:1246-54
30. Panyam J, Labhasetwar V. Biodegradable nanoparticles for drug and gene delivery to cells and tissue. Adv Drug Deliv Rev. 2003;55:329-47
31. Pedersen RO, Loboa EG, LaBean TH. Sensitization of transforming growth factor-beta signaling by multiple peptides patterned on DNA nanostructures. Biomacromolecules. 2013;14:4157-60
32. Li J, Pei H, Zhu B, Liang L, Wei M, He Y. et al. Self-assembled multivalent DNA nanostructures for noninvasive intracellular delivery of immunostimulatory CpG oligonucleotides. ACS Nano. 2011;5:8783-89
33. Zhang C, Hao L, Calabrese CM, Zhou Y, Choi CH, Xing H. et al. Biodegradable DNA-Brush Block Copolymer Spherical Nucleic Acids Enable Transfection Agent-Free Intracellular Gene Regulation. Small. 2015;11:5360-8
34. Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494-8
35. Oh YK, Park TG. siRNA delivery systems for cancer treatment. Adv Drug Deliv Rev. 2009;61:850-62
36. Felgner PL, Gadek TR, Holm M, Roman R, Chan HW, Wenz M. et al. Lipofection: a highly efficient, lipid-mediated DNA-transfection procedure. Proc Natl Acad Sci U S A. 1987;84:7413-17
37. Boussif O, Lezoualc'h F, Zanta MA, Mergny MD, Scherman D, Demeneix B. et al. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc Natl Acad Sci U S A. 1995;92:7297-301
38. Paleos CM, Tziveleka LA, Sideratou Z, Tsiourvas D. Gene delivery using functional dendritic polymers. Expert Opin Drug Deliv. 2009;6:27-38
39. Wagner DE, Bhaduri SB. Progress and outlook of inorganic nanoparticles for delivery of nucleic acid sequences related to orthopedic pathologies: a review. Tissue Eng Part B Rev. 2012;18:1-14
40. Copolovici DM, Langel K, Eriste E, Langel U. Cell-penetrating peptides: design, synthesis, and applications. ACS Nano. 2014;8:1972-94
41. Hutagalung AH, Novick PJ. Role of Rab GTPases in membrane traffic and cell physiology. Physiol Rev. 2011;91:119-49
42. Pierce KL, Premont RT, Lefkowitz RJ. Seven-transmembrane receptors. Nat Rev Mol Cell Biol. 2002;3:639-50
43. Dong C, Filipeanu CM, Duvernay MT, Wu G. Regulation of G protein-coupled receptor export trafficking. Biochim Biophys Acta. 2007;1768:853-70
44. Yin H, Li Q, Qian G, Wang Y, Li Y, Wu G. et al. Rab1 GTPase regulates phenotypic modulation of pulmonary artery smooth muscle cells by mediating the transport of angiotensin II type 1 receptor under hypoxia. Int J Biochem Cell Biol. 2011;43:401-8
45. Kops GJ, Burgering BM. Forkhead transcription factors: new insights into protein kinase B (c-akt) signaling. J Mol Med. 1999;77:656-65
46. Nakae J, Barr V, Accili D. Differential regulation of gene expression by insulin and IGF-1 receptors correlates with phosphorylation of a single amino acid residue in the forkhead transcription factor FKHR. EMBO J. 2000;19:989-96
47. Barthel A, Schmoll D, Unterman TG. FoxO proteins in insulin action and metabolism. Trends Endocrinol Metab. 2005;16:183-9
48. Kato S, Ding J, Pisck E, Jhala US, Du K. COP1 functions as a FoxO1 ubiquitin E3 ligase to regulate FoxO1-mediated gene expression. J Biol Chem. 2008;283:35464-73
49. Chen H, Lin AS, Li Y, Reiter CE, Ver MR, Quon MJ. Dehydroepiandrosterone stimulates phosphorylation of FoxO1 in vascular endothelial cells via phosphatidylinositol 3-kinase- and protein kinase A-dependent signaling pathways to regulate ET-1 synthesis and secretion. J Biol Chem. 2008;283:29228-38
50. Jang AS, Concel VJ, Bein K, Brant KA, Liu S, Pope-Varsalona H. et al. Endothelial dysfunction and claudin 5 regulation during acrolein-induced lung injury. Am J Respir Cell Mol Biol. 2011;44:483-90
51. Augustin HG, Koh GY, Thurston G, Alitalo K. Control of vascular morphogenesis and homeostasis through the angiopoietin-Tie system. Nat Rev Mol Cell Biol. 2009;10:165-77
52. Fan W, Morinaga H, Kim JJ, Bae E, Spann NJ, Heinz S. et al. FoxO1 regulates Tlr4 inflammatory pathway signalling in macrophages. EMBO J. 2010;29:4223-36
53. Dong H, Zhang X, Dai X, Lu S, Gui B, Jin W. et al. Lithium ameliorates lipopolysaccharide-induced microglial activation via inhibition of toll-like receptor 4 expression by activating the PI3K/Akt/FoxO1 pathway. J Neuroinflammation. 2014;11:140
54. Kamo N, Ke B, Busuttil RW, Kupiec-Weglinski JW. PTEN-mediated Akt/beta-catenin/Foxo1 signaling regulates innate immune responses in mouse liver ischemia/reperfusion injury. Hepatology. 2013;57:289-98
55. Huang J, Yue S, Ke B, Zhu J, Shen XD, Zhai Y. et al. Nuclear factor erythroid 2-related factor 2 regulates toll-like receptor 4 innate responses in mouse liver ischemia-reperfusion injury through Akt-forkhead box protein O1 signaling network. Transplantation. 2014;98:721-28
56. Fiorotto R, Scirpo R, Trauner M, Fabris L, hogue R, Spirli C. et al. Loss of CFTR affects biliary epithelium innate immunity and causes TLR4-NF-kappaB-mediated inflammatory response in mice. Gastroenterology. 2011;141:1498-1508
57. Zhao Y, Zhao Y, Zhang M, Zhao J, Ma X, Huang T. et al. Inhibition of TLR4 signalling-induced inflammation attenuates secondary injury after diffuse axonal injury in rats. Mediators Inflamm. 2016;2016:4706915
Author contact
![]() Corresponding authors: Dr. Guansong Wang, Dr. Hang Qian Tel: 8623-68755644 (o) Fax: 8623-65211653 (o); 8623-88110389 Email: hqianedu.cn, wanggs2003com, wanggsedu.cn
Corresponding authors: Dr. Guansong Wang, Dr. Hang Qian Tel: 8623-68755644 (o) Fax: 8623-65211653 (o); 8623-88110389 Email: hqianedu.cn, wanggs2003com, wanggsedu.cn
 Global reach, higher impact
Global reach, higher impact