13.3
Impact Factor
Theranostics 2017; 7(9):2443-2451. doi:10.7150/thno.18290 This issue Cite
Research Paper
Gut Microbiota-Mediated Personalized Treatment of Hyperlipidemia Using Berberine
1. State Key Laboratory of Bioactive Substance and Function of Natural Medicines, Institute of Materia Medica, Chinese Academy of Medical Sciences/Peking Union Medical College, Beijing 100050, China;
2. The First Hospital of Jilin University, Changchun 130021, China.
* These authors made an equal contribution to this work.
Received 2016-11-10; Accepted 2017-4-24; Published 2017-6-24
Abstract
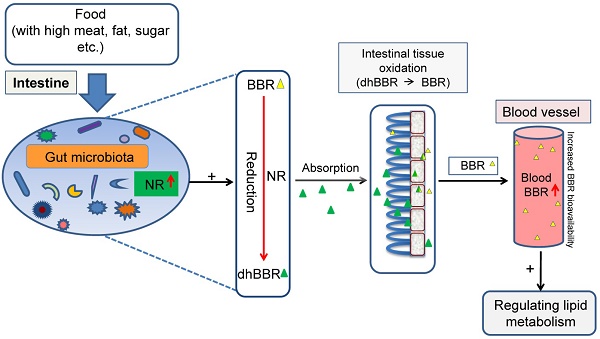
Nitroreductases (NRs) are bacterial enzymes that reduce nitro-containing compounds. We have previously reported that NR of intestinal bacteria is a key factor promoting berberine (BBR) intestinal absorption. We show here that feeding hamsters with high fat diet (HFD) caused an increase in blood lipids and NR activity in the intestine. The elevation of fecal NR by HFD was due to the increase in either the fraction of NR-producing bacteria or their activity in the intestine. When given orally, BBR bioavailability in the HFD-fed hamsters was higher than that in those fed with normal chow (by +72%, *P<0.05). BBR (100 mg/kg/day, orally) decreased blood lipids in the HFD-fed hamsters (**P<0.01) but not in those fed with normal diet. Clinical studies indicated that patients with hyperlipidemia had higher fecal NR activity than that in the healthy individuals (**P<0.01). Similarly, after oral administration, the blood level of BBR in hyperlipidemic patients was higher than that in healthy individuals (*P<0.05). Correlation analysis revealed a positive relationship between blood BBR and fecal NR activity (r=0.703). Thus, the fecal NR activity might serve as a biomarker in the personalized treatment of hyperlipidemia using BBR.
Keywords: Berberine, Hyperlipidemia, Gut microbiota, Nitroreductase, Absorption, Bioavailability.
Introduction
Berberine (BBR) is an alkaloid (Figure 1A, mw 371.8) isolated from plants like Coptis chinensis, and Berberis vulgaris et al. For decades, BBR has been used as an over-the-counter (OTC) drug in China to treat bacteria-caused diarrhea and its safety in humans has been well documented [1-3]. We have previously reported that BBR was effective as a drug for patients with hyperlipidemia and type 2 diabetes [2-3]. The effect of BBR on blood lipids and glucose was associated with several key molecules including low-density lipoprotein receptor (LDLR), adenosine monophosphate-activated protein kinase (AMPK), insulin receptor (InsR), and proprotein convertase subtilisin/kexin type 9 (PCSK9) et al. [2-9]. In the past 10 years, these results have been verified by independent researchers and clinical groups in and outside China and BBR has been approved by the Chinese FDA for clinical studies. However, the unresolved issues for BBR include, for instance, identifying biomarkers that would help in the selection of patients sensitive to BBR treatment. A previous study focusing on genetic variations in the LDLR mRNA 3'UTR region in humans failed to identify signature mutations associated with the treatment outcome of BBR [10], probably because BBR is a multi-target drug [11-12]. Our previous results showed that BBR is mainly metabolized by CYP2D6, CYP1A2, and CYP3A4 in vivo [13]. The low blood concentration of BBR and its pharmacokinetics have been the subject of much research [14, 15]. Also, as described by Deng et al., entero-hepatic circulation might play a role in BBR pharmacokinetics [16]. We have recently demonstrated that an important factor in BBR absorption in the intestine is gut microbiota, which convert BBR into its absorbable form, dihydroberberine (dhBBR) which showed a 5-10 fold higher absorption rate than BBR [17]. The reduction from BBR to dhBBR is mediated through nitroreductase (NR) of gut microbiota [17]. Following absorption in the intestine, dhBBR is quickly oxidized back to BBR and enter blood [17]. Thus, NR in gut microbiota is critical for regulating blood levels of BBR.
NR is a group of enzymes primarily found in bacterial lineages. Members of this family utilize flavin mononucleotide (FMN) as a co-factor to catalyze the reductive reaction of the nitro-group on nitroaromatic and nitroheterocyclic compounds including chemicals with nitro functional groups, for instance the 7-position nitrogen in the BBR's structure [18]. In the present study, we show that high level of NR activity in gut microbiota increased BBR oral bioavailability and might be a potential biomarker to predict the therapeutic efficacy of BBR.
Materials and Methods
Chemicals and reagents
BBR was obtained from the J&K Scientific Ltd (Beijing, China). Tetrahydropalmatine, as the internal standard, was from the National Institute for Food and Drug Control (Beijing, China). Thalifendine (M1), berberrubine (M2), demethyleneberberine (M3), jatrorrhizine (M4) and palmatine (M5), and dihydroberberine (dhBBR) were acquired from the Chengdu Herbpurity Co., Ltd (Chengdu, China). The purity of the standards was above 98% according to the high-performance liquid chromatography (HPLC) assay. Other chemical reagents from the Sino-pharm Chemical Reagent Co, Ltd (Beijing, China) were at chromatographic grade purity.
Assay kits for total cholesterol (TC), low-density lipoprotein cholesterol (LDL-C) and triglycerides (TG) were from the BioSino Bio-technology & Science Inc (Beijing, China). Rat Nitroreductase ELISA kit and Human Nitroreductase ELISA kit were obtained from the Beijing Luyuan Dade Biological Science and Technology Co., Ltd (Beijing, China).
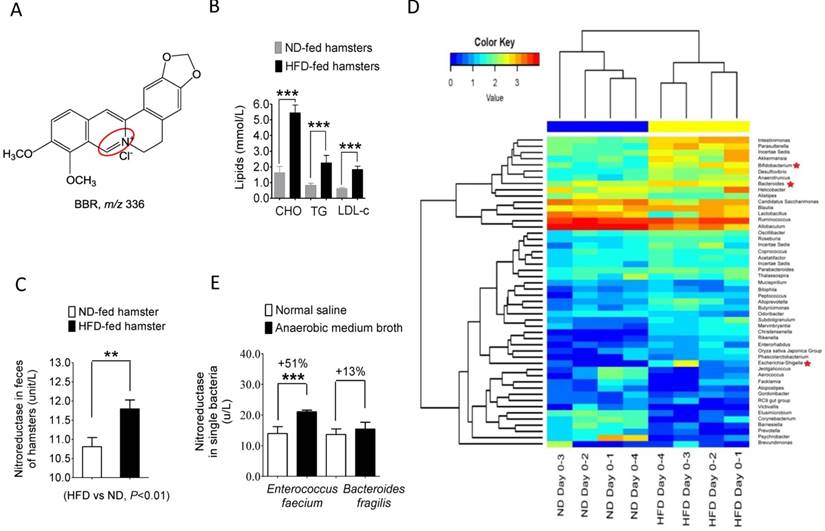
High fat diet (HFD) changed the composition and NR activity of gut microbiota in hamsters. (A) Chemical structure of BBR; (B) the levels of total cholesterol (TC), triglycerides (TG) and low-density-lipoprotein cholesterol (LDL-C) significantly increased in the HFD-fed hamsters (6 weeks), as compared to that in hamsters fed with normal diet (ND) (***P<0.001, n=6); (C) NR activity of intestinal bacteria elevated in HFD-fed hamsters (HFD vs ND, **P<0.01, n=6); (D) Feeding hamsters with HFD for six weeks changed gut microbiota composition in hamsters (n=4 for each group) with an increase in the proportion of NR-producing bacteria; (E) NR activity increased significantly in the in vitro single bacterial cultures (Enterococcus faecium, Bacteroides fragilis) in the presence of the anaerobic medium broth (0.6 ml/1 ml) in culture medium for 24 hrs.
Animals
Male Syrian golden hamsters (8-week old, 110-140 grams, the Vital River Laboratory Animal Technology Co., Ltd., Beijing, China) were housed in the SPF-grade rooms and had free access to food and water.
Hamsters were randomly separated into two groups: the normal controls (26 animals) and hyperlipidemia models (26 animals). The animals fed with normal diet (ND) ad libitum served as controls and those fed with high-fat diet (HFD) for 6 weeks were the hyperlipidemia model hamsters [19].
The animal experiments were approved by the Laboratories' Institutional Animal Care and Use Committee of the Chinese Academy of Medical Sciences (No.003541 and No.00000406). The research was conducted in accordance with the guidelines and ethics of the Chinese Council on Animal Care.
Instruments for detecting BBR and its metabolites
A liquid chromatography instrument coupled to an ion trap time-of-flight mass spectrometer (LC/MSn-IT-TOF) from the Shimadzu Corporation (Kyoto, Japan) was used to identify the chemical structures of BBR and its metabolites. Liquid chromatography with tandem mass spectrometry (LC-MS/MS 8050, Shimadzu Corporation, Kyoto, Japan) and gas chromatography with mass spectrometry (GC-MS QP2010, Shimadzu Cooperation, Kyoto, Japan) were used for the analysis and quantification of BBR and its metabolites in biological samples [14, 17, 20].
Blood lipid test
Blood lipids (TC, TG and LDL-C) were detected in the ND-fed (n=6) and HFD-fed hamsters (n=6) before and after BBR (100 mg/kg/day) oral administration on day 0, 7, and 10 using commercial enzymatic kits [19].
NR activity Determination
For NR activity of gut microbiota in hamsters, 1 gram colon contents were taken and transferred into a flask containing 20 mL anaerobic culture medium. After mixing, the cultures were incubated under anaerobic conditions with a N2 atmosphere at 37 °C for 24 hrs, followed by centrifugation at 5,000 rpm (15 min, 4 °C). The pellet was collected, washed twice in PBS and re-suspended in PBS at a weight/volume ratio of 1:500 (g/μL). Total bacterial protein was extracted via sonication disruptor (Scientiz-ⅡD; Ningbo Scientz Biotechnology Co., Ltd., Ningbo, China). NR activity of the extracted samples was measured with the rat or human ELISA kit. NR activity in human feces was tested using the method above.
For the NR activity in single strain bacteria, the intestinal bacteria Enterococcus faecium ATCC 35667 and Bacteroides fragilis ATCC 25285 (bought from the Guangdong Huankai Microbial Sci. & Tech. Co., Ltd, Guangdong, China) were cultured in an anaerobic condition without or with BBR (10 μg/mL, 24 hrs), respectively [17, 21]. The NR activity was determined using the NR ELISA kit.
Bacterial composition analysis
Gut bacterial composition in the ND- or HFD-fed hamsters was analyzed via 16S rRNA genes analysis. The 16S rRNA genes were amplified using the specific primer for 16S V3-V4: 340F-805R to target the V3-V4 regions of 16S rRNA. PCR products were mixed in equimolar ratios. Subsequently, mixture of PCR products was purified with the GeneJET Gel Extraction Kit (QIAGEN, Germany). Sequencing libraries were generated using the NEXTflex Rapid DNA-seq kit for Illumina (New England Biolabs, USA) following manufacturer's recommendations and index codes were added. The library quality was assessed on the Qubit 2.0 Fluorometer (Thermo Scientific, USA) and Agilent Bioanalyzer 2100 system (Agilent Technologies, USA). Finally, the library was sequenced on the HiSeq2500 (Illumina, USA) platform and 250 bp paired-end reads were generated. Sequences were analyzed using the QIIME software package (Quantitative Insights Into Microbial Ecology). First, the reads were filtered by the QIIME quality filters. Next, we picked representative sequences for each OTU (operational taxonomic units) and used the RDP (ribosomal database project) classifier to annotate taxonomic information for each representative sequence. Sequences with a similarity over 97% were assigned to the same OTUs [22].
Clinical study
Forty individuals were randomly enrolled into the study in the Outpatient Section of the First Hospital of the Jilin University in Changchun in the spring of 2016. The study was approved by the institution ethnic committee of the hospital (clinical study No. 2016-270). Of the 40 individuals, 10 were healthy volunteers (Group 1; 7 males, 3 females; age 46.7 +/- 16.8) with blood lipids and glucose in normal range [blood cholesterol (mmol/L), 4.72 +/- 0.53; triglycerides (mmol/L), 1.09 +/- 0.33; LDL-C (mmol/L), 2.55 +/- 0.35; FBG (mmol/L), 4.90 +/- 0.51)], and 30 were patients (Group 2, 13 males, 17 females; age 58.8 +/- 13.7) with high level of blood lipids [blood cholesterol (mmol/L), 5.59 +/- 0.98; triglyceride (mmol/L) , 2.36 +/- 1.30; LDL-C (mmol/L) , 3.28 +/- 0.73; FBG (mmol/L), 6.56 +/- 2.97]. In the Group 2 individuals, 7 of them had both high blood lipids and glucose. The subjects were not in any drug treatment before enrollment.
Before BBR treatment, blood samples were taken for each individual for grouping purpose (the Group 1 and Group 2); fecal samples were also collected. The subject was orally treated with BBR (500 mg, once). Blood samples were collected 2 hrs after BBR oral administration, and placed at -20゜C. BBR plasma concentration was measured using LC-MS/MS as described [14, 17, 41]. Before BBR treatment (in 24 hrs), 5 grams of fecal samples of the 32 individuals (9 healthy subjects and 23 patients) were collected and immediately prepared for an immediate measurement of NR activity.
Gut microbiota analysis of clinical individuals
Intestinal bacteria composition of healthy volunteers or patients with hyperlipemia / hyperglycemia was analyzed via 16S rRNA genes analysis. The 16S rRNA genes were amplified using the specific primer for 16S V3-V4: 338F-806R to target the V3-V4 regions of 16S rRNA. PCR products were mixed in equimolar ratios. Subsequently, mixture of PCR products was purified with the AxyPrep Gel Extraction Kit (Axygen Biosciences, USA). Sequencing libraries were generated using the NEBNext Ultr DNA Library Prep Kit for Illumina (New England Biolabs, USA) following manufacturer's recommendations and index codes were added. The library quality was assessed on the Qubit 2.0 Fluorometer (Thermo Scientific, USA) and Agilent Bioanalyzer 2100 system (Agilent Technologies, USA). Finally, the library was sequenced on the MiSeq (Illumina, USA) platform and 300 bp paired-end reads were generated. Sequences were analyzed using the QIIME software package (Quantitative Insights Into Microbial Ecology, V1.8.0). First, the reads were filtered by the QIIME quality filters. Next, we picked representative sequences for each OTU (Operational Taxonomic Units) and used the RDP classifier (Ribosomal Database Project) to annotate taxonomic information for each representative sequence via SILVA 119 ribosomal RNA (rRNA) database. Sequences with a similarity over 97% were assigned to the same OTUs.
Pharmacokinetics of BBR in hamsters and BBR concentration in human plasma
The ND-fed or HFD-fed hamsters were orally treated with BBR (100 mg/kg, n=6). Blood samples were collected from the posterior orbital venous plexus at 0, 0.25, 0.5, 1, 2, 4, 6, 8, 12 and 24 hrs. Hamsters were sacrificed at the last time point and brain, heart, kidney, lung and liver samples were collected. BBR was also given to the ND-fed (n=6) or HFD-fed (n=6) hamsters via intravenous injection (iv, 1 mg/kg). Additionally, small intestines were collected at 0.5 hrs after BBR oral administration (n=4). BBR, dhBBR and its metabolites were detected according to previously described methods [14, 17] and pharmacokinetic parameters were calculated by DAS 2.0.
For determining plasma BBR in hamsters treated with BBR (oral, 100 mg/kg/day, for 10 days, n=6), blood samples were collected on day-7 and day-10 0.5 hrs after treating with BBR. Blood BBR was quantified [14, 17, 41]. Plasma (100 μL) was extracted with 1.25 mL ethyl ether after addition of 10 μL internal standard (250 ng/mL of palmatine) and 50 μL 0.5 M sodium hydroxide solution. Following centrifugation and separation, the organic phase (1 mL) was evaporated to dryness in a 40 ºC water bath with a sample concentrator (Nitrogen blowing instrument). The residue was reconstituted with 100 μL mobile phase (a mixture of acetonitrile and 0.5% formic acid with ratio of 1:4). An aliquot of 10 μL was injected into the LC-MS/MS 8050 system for analysis.
Statistical analysis
The statistical analyses were conducted using the two-way ANOVA and Student's t-test with the GraphPad Prism Version 5 (GraphPad Software, CA, USA). The data were expressed as means ± standard deviation. P values less than 0.05 were considered statistically significant.
Results
High-fat diet (HFD) changed gut microbiota composition and NR activity in hamsters
As shown in Figure 1B and C, HFD treatment in hamsters for 10 days significantly increased blood cholesterol, triglycerides, and LDL-C (***P<0.001 for all) while simultaneously elevating NR activity in the HFD-hamster feces as compared to those fed with normal diet (**P<0.01). Composition analysis using microbiomic technique showed an increase in the NR-producing bacteria in the gut microbiota community (Figure 1D) with Bacteroides [23], Escherichia-Shigella [24], and Bifidobacterium [25] et al. showing the highest increase among the top 50 bacterial strains. These results suggested that HFD feeding altered the composition of gut microbiota. Also, incubating NR-producing bacteria Enterococcus faecium [26] and Bacteroides fragilis [23] with broth medium for 12 hrs elevated NR activity in the bacterial culture (Figure 1E). We thus concluded that the HFD or Broth treatment increased either the proportion of the NR-producers in the intestinal bacterial community and/or the NR activity in the bacteria.
Increased level of fecal NR promoted BBR oral bioavailability
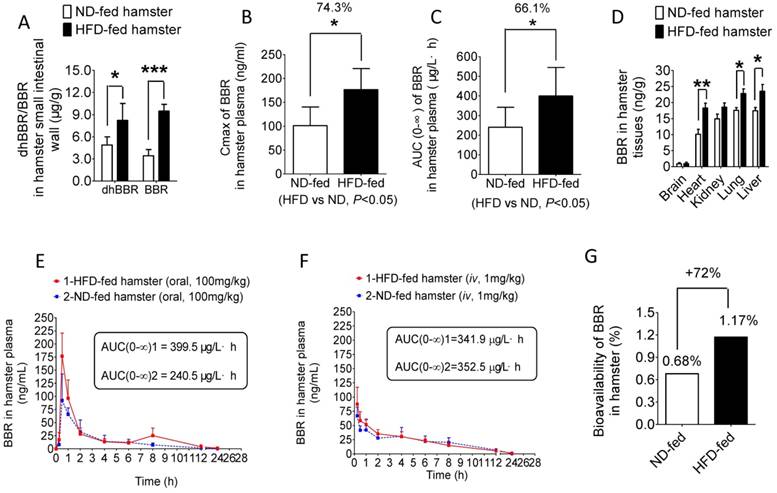
NR is a group of bacterial enzymes that reduces BBR into dhBBR in intestine which has high intestinal absorption [17]. Therefore, the increased activity of NR in the gut microbiota might help BBR to first enter into intestine wall tissue and then blood. As shown in Figure 2A, both dhBBR and BBR were significantly higher in the intestinal tissue of the HFD-fed hamsters as compared to that in the hamsters fed with normal diet (*P<0.05, ***P<0.001, respectively). Consequently, determination of blood concentration of BBR showed that the Cmax (peak concentration) of BBR in the HFD-fed hamsters was about 74.3% higher than that in those fed with normal chow (*P<0.05, Figure 2B). Also, the AUC (Area under the time concentration curve) measurement revealed a pattern similar to that of Cmax (*P<0.05, Figure 2C). Accordingly, BBR content distributed in organs (liver, lung, kidney, heart, brain) of the HFD-hamsters was higher than that in the hamsters fed with normal diet with statistical significance seen in liver, lung and heart (Figure 2D, *P<0.05 or **P<0.01). The distribution of BBR metabolites (M1-M5, Supplementary Figure 1) in organs like intestine and liver revealed a pattern similar to that of BBR (Supplementary Figure 2-3) confirming the results shown in Figure 2D. Although the BBR's AUC value in the HFD-fed hamsters increased by 66% over that in the normal diet controls after BBR oral administration (Figure 2E), difference between the HFD-fed hamsters and normal-diet hamsters in BBR's AUC value was not detectable if BBR was given by iv injection (1 mg/kg, Figure 2F) [27], demonstrating the critical role of gut microbiota in BBR absorption. The BBR iv injection dose was selected based on its bioavailability reported in animals [27]. The oral bioavailability of BBR increased in the HFD-fed hamsters by 72% as compared to that in normal-diet group (Figure 2G). The results supported the role of intestinal NR in BBR absorption [17] and suggested that BBR might be more effective in individuals with high NR activity in intestine.
HFD treatment promotes BBR absorption in intestine and its oral bioavailability. (A) 0.5 hr after oral administration of BBR (100 mg/kg) both dhBBR and BBR were significantly increased in the small intestinal tissue of the HFD-fed hamsters as compared to that of the ND-fed hamsters (*P<0.05 for dhBBR, and ***P<0.001 for BBR, n=4 for both comparison); (B & C) Cmax and AUC of BBR were much higher in the HFD-fed hamsters compared to those in the ND-fed hamsters 24 hrs after oral administration of BBR at 100 mg/kg (*P<0.05 for all; n=5 for the ND-fed group and n=6 for the HFD-fed group); (D) Increased levels of BBR in the main organs of the HFD-fed hamsters compared with that in the ND-fed hamsters 24 hrs after 100mg/kg oral administration of BBR (**P<0.01 and *P<0.05, n=5 for the ND-fed group and n=6 for the HFD-fed group); (E) Concentration curves of BBR in HFD-fed and ND-fed hamsters 24 hrs after oral administration of BBR (100 mg/kg) with a larger BBR AUC value in HFD-fed hamsters (n=5 for the ND-fed group, and n=6 for the HFD-fed group, respectively); (F) 24-hr concentration curves of BBR when given via iv route (1 mg/kg) in the ND-fed or HFD-fed hamsters both groups showing a similar pattern (n=4 for both); (G) BBR oral bioavailability increased in the HFD-fed hamsters by 72% as compared to that in the ND-fed hamsters (iv, 1 mg/kg; oral, 100 mg/kg).
BBR reduced blood lipids in the HFD-fed hamsters but not those fed with normal diet
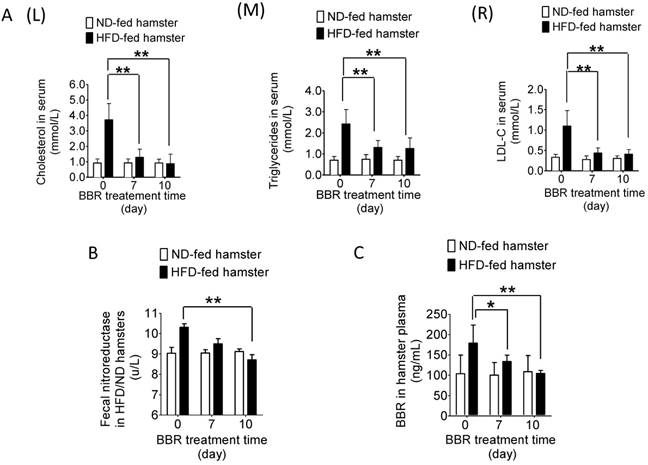
We next extended the test into therapeutic efficacy. As shown in Figure 3A (L, M & R), while BBR treatment did not alter the blood level of lipids in the hamsters fed with normal diet, its administration for 7 days (oral, 100 mg/kg/day) significantly decreased blood cholesterol, triglycerides and LDL-C in the HFD-fed hamsters (**P<0.01). Interestingly, along with the decrease in blood lipids from day 0 to day 10 (Figure 3C, *P<0.05, **P<0.01), the NR activity in intestine also diminished gradually (Figure 3B, **P<0.01). The NR level remained unchanged in the BBR-treated normal-diet hamsters (Figure 3B). It appeared that HFD feeding altered the composition of gut microbiota and caused high blood lipids. Simultaneously, the proportion of the NR-producing bacteria in gut microbiota as well as the NR activity in intestine increased (Figure 1B and C) which, in turn, promoted the conversion of BBR into dhBBR. Thus, there was an increase in oral bioavailability of BBR in the HFD-fed animals resulting in a good therapeutic outcome.
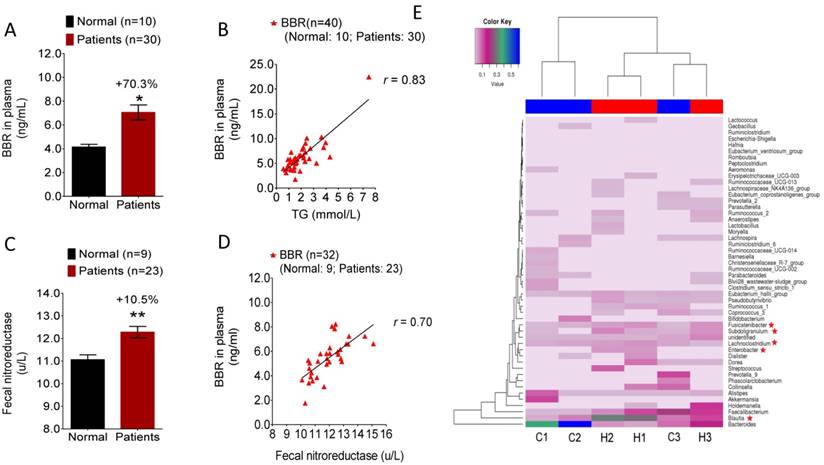
Blood level of BBR positively correlated with fecal NRs activity in clinical subjects
Our results in hamsters suggested that NR activity in gut microbiota might be a biomarker for BBR bioavailability in selecting patients for treatment. To test this hypothesis, 40 individuals were randomly enrolled into the study in the Outpatient Section of the First Hospital of the Jilin University in Changchun in the early spring of 2016 (Clinical approval number 2016-270). Out of the 40 individuals, 10 were healthy volunteers (Group 1), and 30 were patients (Group 2) with high level of blood lipids. As the first step of the trial, blood and fecal samples of the 40 subjects were acquired for blood lipids/glucose measurement and NR activity determination, respectively. BBR was given to the subjects orally followed by examination of blood BBR concentration 2 hrs later. As shown in Figure 4A, individuals in Group 2 had a blood BBR level over 70% higher than that in normal subjects of Group 1 (*P<0.05). Considering the two groups together (n=40), the highest correlation was observed between blood BBR level and blood triglycerides concentration (Figure 4B, r=0.83), suggesting that individuals with higher level of triglycerides might have better oral bioavailability of BBR. Furthermore, the hyperlipidemic patients in Group 2 had a significantly higher fecal NR activity level than that in the healthy individuals of Group 1 (Figure 4C, **P<0.01) which was consistent with the observation in hamsters (Figure 1C). It appears that the elevated blood BBR level in Group 2 patients is the consequence of the increased activity of NR. The correlation analysis between fecal NR and blood BBR was also carried out in individuals from both Group 1 and Group 2. As shown in Figure 4D, although the study size was relatively small (n=32; fecal samples of 8 individuals were not available in the study, with 1 in the Group 1 and 7 in Group 2), the fecal NR activity positively correlated with the blood concentration of BBR (r=0.70). The results demonstrated a significant role of NR in BBR absorption and suggested fecal NR activity as a potential biomarker for determining the therapeutic efficacy of BBR. Also, sex and age appeared to have no effect on the level of blood BBR in this study (Supplementary Figure 4).
To examine the difference in gut flora between hyperlipidemic patients and healthy subjects, composition analysis for gut microbiota was performed by using human fecal samples randomly collected from the hyperlipidemic patients (n=3; labeled H-1, 2, 3) and healthy volunteers (n=3; labeled C-1, 2, 3). The results showed a significant difference in the gut microbiota composition between healthy and hyperlipidemic individuals (Figure 4E). As shown in Figure 4E, Fusicatenibacter, Subdoligranulum, Enterobacter, Blautia and Lachnoclostridium were noticeably increased in the fecal samples of hyperlipidemic patients compared with the samples from normal volunteers. Although the NR information for some of these bacteria is not available, Enterobacter has been reported to be a NR-producer [28]. Indeed, our previous results demonstrated that the Enterobacter cloacae of the gut microbiota efficiently converted BBR to dhBBR [17]. Thus, the composition analysis of gut microbiota confirmed the difference of gut flora between healthy subjects and patients with hyperlipidemia which might contribute to the elevated level of NR in intestine.
Discussion
BBR is known to reduce blood lipids or glucose in patients with hyperlipidemia or type 2 diabetes, respectively [2-4]. Our findings in the present study showed that BBR treatment lowered blood lipids and glucose effectively only in the HFD-fed hamsters with increased level of NR activity in the gut microbiota. On the other hand, BBR's lipid-lowering efficacy was not detectable in the normal diet-fed animals with the normal level of NR in gut microbiota. The results suggested that the intestinal NR activity could be at least part of mechanism for the lipid/glucose lowering activity of BBR. It appeared that HFD treatment resulted either in an increase in the fraction of NR-producing bacteria in gut microbiota and/or elevated the activity of NR in the bacteria. Consequently, the HFD treatment caused an enhanced BBR-to-dhBBR transformation in the intestine and therefore an increased oral bioavailability in vivo. Our clinical study also corroborated the findings in the hamster model and provided strong support for a positive correlation between the fecal NR activity and blood level of BBR, indicating a connection between BBR oral bioavailability and intestinal bacteria. This interesting correlation could be more significant if the size of study population is large. Composition analysis of the gut microbiota showed difference between the NR from healthy and hyperlipidemic individuals. Thus, we consider the fecal NR activity in gut microbiota to be a potential biomarker for the therapeutic efficacy of BBR.
The current concept for bioavailability of oral drugs is based on the evaluation of drug absorption through normal animal intestine [29]. In principle, high oral bioavailability is considered a positive indication for drug therapeutic efficacy as it correlates with drug effectiveness [29]. In the present study, we have shown that the oral bioavailability of BBR in the HFD-fed hamsters was much higher than that in the hamsters fed with normal diet (Figure 2G). This observation indicates that food composition might influence bioavailability of the oral drugs through gut microbiota and sheds new light on the existing knowledge of drug bioavailability when evaluated in appropriate animal models. For treating energy metabolic disorders, we consider the therapeutic selectivity of BBR, which is mediated through NR of gut microbiota, an unique advantage in clinical treatment, because less drug molecules enter the blood of healthy individuals or of those recovered from treatment (Figure 3B & C).
Therapeutic efficacy of drugs is known to be affected by genetic variations in drug targets or drug processing enzymes. A number of examples of genetic variations have been reported for drugs treating a variety of diseases such as hepatitis C infection [30], cancers [31, 32], blood coagulation et al. [33], representing a new frontier in drug therapy and personalized medicine [34, 35]. For example, subtypes of CYP450s differ among races and individuals, resulting in variations in drug degradation rate [36, 37] and thus constituting an important factor in personalized drug therapy. In the present study we showed both in animals and clinical subjects that the individuals with high level of fecal NR activity had more blood BBR as compared to those with regular level of fecal NR, indicating a variation of oral bioavailability of BBR in human. It might cause different therapeutic efficacy in BBR treatment. To the best of our knowledge, this is the first report to show a functional molecule of gut microbiota acts as a biomarker for drug bioavailability and therapeutic efficacy.
BBR reduced blood lipids in the HFD-fed hamsters but not in those fed with normal diet. (A) After treating HFD-fed hamsters with BBR (oral, 100 mg/kg/day, n=6) for 10 days, the levels of total cholesterol (TC) (L), triglycerides (TG) (M) and LDL-C (R) significantly decreased on day 7 and 10 of the treatment course (**P<0.01, vs day 0; n=6) when compared to that in the hamsters fed with normal diet (vs day 0; n=5); (B) The NR activity of gut microbiota in the HFD-fed hamsters decreased on day 10 after BBR treatment (**P<0.01, n=6); the change was not significant in the ND-fed hamsters (n=6); (C) BBR concentration in plasma decreased on day 7 and 10 of the treatment in the HFD-fed hamsters (*P<0.05, **P<0.01, n=6).
Blood level of BBR positively correlated with fecal NR activity in clinical subjects. (A) BBR concentration in the plasma of the hyperlipidemic patients (n=30) was higher by 70.3% as compared to that in the normal subject (n=10) (*P<0.05); (B) Blood BBR level positively correlated with the blood TG level in study subjects (n=40) with an r value of 0.83; (C) NR activity in the feces of the hyperlipidemic patients (n=23) was significantly higher than that in normal subjects (n=9) (**P<0.01); (D) Blood level of BBR positively correlated with the fecal NR activity in the clinical subjects (n=32) with an r value of 0.70; (E) Patients with hyperlipidemia/hyperglycemia showed a change in the gut microbiota composition as compared with that of the normal healthy volunteers (n=3 for each group) showing an increase of the proportion of Fusicatenibacter, Subdoligranulum, Enterobacter, Blautia, and Lachnoclostridium in patients with Enterobacter as an NR-producing bacteria.
Bacterial NR catalyzes the NADPH-dependent reduction of the nitro-group on nitroaromatic and nitroheterocyclic compounds as a mechanism of self-protection [38]. The expression of bacterial NR genes seems to be governed by the MarRA and SoxRS regulatory mechanisms that are associated with bacterial response to antibiotics and chemicals in the environment [39-41]. The NR genes are widely distributed in intestinal anaerobic bacteria [38] causing a steady up-take of BBR in human intestine (Figure 4C) [42]. We show, both in the animal experiment and in the human study, that increasing about 10% of fecal NR activity in gut flora could raise blood BBR concentration by 65-70%, suggesting a highly efficient function of bacterial NR in transforming BBR into dhBBR. In our experiments, we used the NR activity assay to evaluate NR level instead of NR enzyme expression as, despite their high homology, sequence diversity of NR exists in different microorganisms [38]. As fecal NR appears to be a sensitive enzyme for blood level of BBR, it might be a biomarker to predict the therapeutic efficacy of BBR in clinical practice, if verified in large scale clinical studies. Base on the finding in this investigation, we consider gut microbiota to be a new component in personalized medicine.
Abbreviations
NR: Nitroreductase; BBR: berberine; HFD: high fat diet; OTC: over-the-counter; LDLR: low-density lipoprotein receptor; AMPK: adenosine monophosphate-activated protein kinase; InsR: insulin receptor; PCSK9: proprotein convertase subtilisin/kexin type 9; dhBBR: dihydroberberine; Cmax: Peak concentration; AUC: Area under the time concentration curve; M1: Thalifendine; M2: berberrubine; M3: demethyleneberberine; M4: jatrorrhizine; M5: Palmatine; HPLC: high-performance liquid chromatography; TC: total cholesterol; LDL-C: low-density lipoprotein cholesterol; TG: triglycerides; ND: normal diet; LC/MSn-IT-TOF: liquid chromatography with ion trap time-of-flight mass spectrometer; LC-MS/MS: liquid chromatography with tandem mass spectrometry; GC-MS: gas chromatography with mass spectrometry; iv: intravenous injection
Supplementary Material
Supplementary figures.
Acknowledgements
The project was supported by the National Natural Science Foundation of China (Nos. 81321004, 81573493 & 8140130374), CAMS Innovation Fund for Medical Sciences (CIFMS) (Nos. 2016-I2M-3-011 & 2016-I2M-1-011), Beijing Key Laboratory of Non-Clinical Drug Metabolism and PK/PD study (Z141102004414062), National Mega-project for Innovative Drugs (2014ZX09507003-001), and the Fundamental Research Funds (2016GH320002). This study was also supported by the analytical center of the Peking branch of Japanese Shimadzu Corporation.
Author contribution
J-JD and W-Y conceived of and designed the experiments and contributed to the writing and editing of the manuscript. S-JW, Z-ZX, L-XY, Z-XF, F-J, H-CY, L-Y, W-BY, M-SR, and G-F performed the experiments. J-JD and W-Y analyzed the data. J-JD, W-Y, and T-Q contributed to discussions and provided reagents/materials /analysis tools important for completion of the work.
Competing Interests
The authors have declared that no competing interest exists.
References
1. National Pharmacopoeia Committee. Berberine Hydrochloride and Berberine Hydrochloride Tablets. Pharmacopoeia of People's Republic of China/Part II. Beijing: Chemical Industry Press, Beijing. 2015:875-876
2. Kong WJ, Wei J, Abidi P. et al. Berberine is a novel cholesterol-lowering drug working through a unique mechanism distinct from statins. Nat Med. 2004;10:1344-1351
3. Zhang H, Wei J, Xue R. et al. Berberine lowers blood glucose in type 2 diabetes mellitus patients through increasing insulin receptor expression. Metabolism. 2010;59:285-292
4. Derosa G, D'Angelo A, Bonaventura A. et al. Effects of berberine on lipid profile in subjects with low cardiovascular risk. Expert Opin Biol Ther. 2013;13:475-482
5. Lee YS, Kim WS, Kim KH. et al. Berberine, a natural plant product, activates AMP-activated protein kinase with beneficial metabolic effects in diabetic and insulin-resistant states. Diabetes. 2006;55:2256-2264
6. Kong WJ, Zhang H, Song DQ. et al. Berberine reduces insulin resistance through protein kinase C-dependent up-regulation of insulin receptor expression. Metabolism. 2009;58:109-119
7. Li H, Dong B, Park SW. et al. Hepatocyte nuclear factor 1α plays a critical role in PCSK9 gene transcription and regulation by the natural hypocholesterolemic compound berberine. J Biol Chem. 2009;284:28885-28895
8. Chen CH, Zhang YB, Huang C. Berberine inhibits PTP1B activity and mimics insulin action. Biochem Biophys Res Commun. 2010;397:543-547
9. Yin J, Gao Z, Liu D. et al. Berberine improves glucose metabolism through induction of glycolysis. Am J Physiol Endocrinol Meta. 2008;294:E148-E156
10. Chen W, Wang S, Ma Y. et al. Analysis of polymorphisms in the 3' untranslated region of the LDL receptor gene and their effect on plasma cholesterol levels and drug response. Int J Mol Med. 2008;21:345-353
11. Yao J, Kong WJ, Jiang JD. Learning from berberine: Treating chronic diseases through multiple targets. Sci China Life Sci. 2015;58:854-859
12. Zhang Y, Ye JP. Mitochondrial inhibitor as a new class of insulin sensitizer. Acta Pharm Sin B. 2012;2:341-349
13. Li Y, Ren G, Wang YX. et al. Bioactivities of berberine metabolites after transformation through CYP450 isoenzymes. J Transl Med. 2011;15(9):62
14. Tan XS, Ma JY, Feng R. et al. Tissue distribution of berberine and its metabolites after oral administration in rats. PloS One. 2013;8:e77969
15. Liu YT, Hao HP, Xie HG. et al. Extensive intestinal first-pass elimination and predominant hepatic distribution of berberine explain its low plasma levels in rats. Drug Metab Dispos. 2010;38(10):1779-84
16. Deng Y, Liao Q, Li S. et al. Simultaneous determination of berberine, palmatine and jatrorrhizine by liquid chromatography-tandem mass spectrometry in rat plasma and its application in a pharmacokinetic study after oral administration of coptis-evodia herb couple. J Chromatogr B Analyt Technol Biomed Life Sci. 2008;863(2):195-205
17. Feng R, Shou JW, Zhao ZX. et al. Transforming berberine into its intestine-absorbable form by the gut microbiota. Sci Rep. 2015;5:12155
18. Li YH, Yang P, Kong WJ. et al. Berberine analogues as a novel class of the low-density-lipoprotein receptor up-regulators: synthesis, structure-activity relationships, and cholesterol-lowering efficacy. J Med Chem. 2009;52:492-501
19. Li XY, Zhao ZX, Huang M. et al. Effect of berberine on promoting the excretion of cholesterol in high-fat diet-induced hyperlipidemic hamsters. J Transl Med. 2015;27:278
20. Ma JY, Feng R, Tan XS. et al. Excretion of berberine and its metabolites in oral administration in rats. J Pharm Sci. 2013;102:4181-4192
21. He CY, Fu J, Ma JY. et al. Biotransformation and in vitro metabolic profile of bioactive extracts from a traditional miao-nationality herbal medicine, Polygonum capitatum. Molecules. 2014;19:10291-10308
22. Yan Wang, Jia-Wen Shou, Xiao-Yang Li. et al. Berberine-induced bioactive metabolites of the gut microbiota improve energy metabolism. Metabolism. 2017;70:72-84 DOI: http://dx.doi.org/10.1016/j.metabol.2017.02.003
23. Schapiro JM, Gupta R, Stefansson E. et al. Isolation of metronidazole-resistant Bacteroides fragilis carrying the nimA nitroreductase gene from a patient in Washington State. J Clin Microbiol. 2004;42:4127-4129
24. Fu H, Leng W, Wang J. et al. Transcriptional profile induced by furazolidone treatment of Shigella flexneri. Appl Microbiol Biot. 2007;7:657-667
25. Kinouchi T, Manabe Y, Wakisaka K. et al. Biotransformation of 1-Nitropyrene in intestinal anaerobic bacteria. Microbiol and Immunol. 1982;26:993-1005
26. Rafii F, Wynne R, Heinze T M. et al. Mechanism of metronidazole-resistance by isolates of nitroreductase-producing Enterococcus gallinarum, and Enterococcus casseliflavus, from the human intestinal tract. Fems Microbiology Letters. 2003;225(2):195
27. Lee CJ, Wu YT, Hsueh TY. et al. Pharmacokinetics and oral bioavailability of epimedin C after oral administration of epimedin C and Herba Epimedii extract in rats. Biomed Chromatogr. 2014;28:630-636
28. Bryant C, Hubbard L, McElroy WD. Cloning, nucleotide sequence, and expression of the nitroreductase gene from Enterobacter cloacae. Journal of Biological Chemistry. 1991;266(7):4126
29. Lee J, Lee E, Kim D. et al. Studies on absorption, distribution and metabolism of ginseng in humans after oral administration. J. Ethnopharmacol. 2009;122:143-148
30. Eslam M, George J. Genome-wide association studies and hepatitis C: harvesting the benefits of the genomic revolution. Semin Liver Dis. 2015;35:402-420
31. Swanton C, Govindan R. Clinical implications of genomic discoveries in lung cancer. N Engl J Med. 2016;374:1864-1873
32. Wijesinghe P, Bollig-Fischer A. Lung cancer genomics in the era of accelerated targeted drug development. Adv Exp Med Biol. 2016;890:1-23
33. Johnson JA, Cavallari LH. Pharmacogenetics and cardiovascular disease-implications for personalized medicine. Pharmacol Rev. 2016;65:987-1009
34. Chatalic KL, Heskamp S, Konijnenberg M. et al. Towards personalized treatment of prostate cancer: PSMA I&T, a promising prostate-specific membrane antigen-targeted theranostic agent. Theranostics. 2016;6(6):849-61
35. Jo SD, Ku SH, Won YY. et al. Targeted nanotheranostics for future personalized medicine: recent progress in cancer therapy. Theranostics. 2016;6(9):1362-77
36. Dubovsky SL. The usefulness of genotyping cytochrome P450 enzymes in the treatment of depression. Expert Opin Drug Metab Toxicol. 2015;11:369-379
37. Murray M, Petrovic N. Cytochromes P450: decision-making tools for personalized therapeutics. Curr Opin Mol Ther. 2006;8:480-486
38. Roldán MD, Pérez-Reinado E, Castillo F. et al. Reduction of polynitroaromatic compounds: the bacterial nitroreductases. FEMS Microbiol Rev. 2008;32:474-500
39. Liochev SI, Hausladen A, Fridovich I. Nitroreductase A is regulated as a member of the soxRS regulon of Escherichia coli. Proc Natl Acad Sci USA. 1999;96:3537-3539
40. Paterson ES, Boucher SE, Lambert IB. Regulation of the nfsA gene in Escherichia coli by SoxS. J Bacteriol. 2002;184:51-58
41. Barbosa TM, Levy SB. Differential expression of over 60 chromosomal genes in Escherichia coli by constitutive expression of MarA. J Bacteriol. 2000;182:3467-3474
42. Yan HM, Xia MF, Wang Y. et al. Efficacy of berberine in patients with non-alcoholic fatty liver disease. PLoS One. 2015;10:e0134172
Author contact
![]() Corresponding authors: Dr. Yan Wang, Tel.: +86 10 63165238; fax, +86 10 63165238. E-mail address: wangyanac.cn; or Dr. Jian-Dong Jiang, Tel.: +86 10 83160005; fax, +86 10 63017757. E-mail address: jiang.jdongcom
Corresponding authors: Dr. Yan Wang, Tel.: +86 10 63165238; fax, +86 10 63165238. E-mail address: wangyanac.cn; or Dr. Jian-Dong Jiang, Tel.: +86 10 83160005; fax, +86 10 63017757. E-mail address: jiang.jdongcom
 Global reach, higher impact
Global reach, higher impact