13.3
Impact Factor
Theranostics 2017; 7(7):1835-1846. doi:10.7150/thno.18857 This issue Cite
Research Paper
Rapid Detection of Avian Influenza Virus by Fluorescent Diagnostic Assay using an Epitope-Derived Peptide
1. Zoonosis Research Center, Department of Infection Biology, School of Medicine, Wonkwang University, 460, Iksan-daero, Iksan, 54538, Republic of Korea;
2. GenBody Inc, No.206, Biotech Business IC, DanKook University, San-29, Anseo-dong, Dongnam-gu, Cheonan, Republic of Korea;
3. Department of Pediatrics, School of Medicine, Wonkwang University, 460, Iksan-daero, Iksan, 54538, Republic of Korea.
* These authors contributed equally to this manuscript.
Received 2016-12-21; Accepted 2017-2-20; Published 2017-4-10
Abstract
Currently, the point of care testing (POCT) is not fully developed for subtype-specific avian influenza virus detection. In this study, an H5N1 hemaglutinin 1 (HA1) epitope (P0: KPNDAINF) and three modified peptides (P1: KPNTAINF, P2: KPNGAINF, P3: KPNDAINDAINF) were evaluated as POCT elements for rapid detection of avian influenza virus. Based on modeling predictions by Autodock Vina, binding affinity varied depending on alteration of one amino acid in these peptides. The binding energy of P2 indicated its potential for a strong interaction with HA. Fluorescence-linked immunosorbent assay experimentally demonstrated the interaction between these peptides and virus. The four peptides interacted with HA1 of H5N3 with different binding affinities with P2 showing the strongest binding affinity. When P0 and P2 peptides were used in rapid fluorescent immunochromatographic test (FICT) as detection elements, the inter-assay coefficients of variation (CV) indicated that P2-linked FICT was more acceptable than the P0-linked FICT in the presence of human specimens. Antibody pair-linked FICT was influenced by clinical samples more than the P2-linked FICT assay, which showed a 4-fold improvement in the detection limit of H5N3 and maintained H5 subtype-specificity. Compared to the rapid diagnostic test (RDT) which is not specific for influenza subtypes, P2-linked FICT could increase virus detection. In conclusion, results of this study suggest that HA epitope-derived peptides can be used as alternatives to antibodies for a rapid fluorescent diagnostic assay to detect avian influenza virus.
Keywords: Epitope-derived peptide, H5 subtype influenza A virus, Rapid fluorescent immunochromatographic test, Haemagglutinin 1.
Introduction
Avian influenza (AI) A virus with a hemagglutinin (HA) 5 protein and a neuraminidase (NA) 1 protein (subtype H5N1) is a highly pathogenic virus. It causes lower respiratory tract infections associated with a mortality rate of higher than 60% [1]. H5N1 diagnosis should be included in differential diagnosis of acute febrile respiratory illness in areas where H5N1 has been identified in animals [2]. However, rapid point-of-care detection tests (POCT) are not performed for influenza due to sensitivity limitations in detecting H5N1 using the POCT developed by the Centers for Disease Control and Prevention (CDC) [2]. A commercialized H5 subtype-specific rapid diagnostic system is not yet available. HA has been used to categorize influenza A viruses into various subtypes. It shares high amino acid (aa) sequence homologies among all subtypes [3], making the development of subtype-specific antibody based on HA antigen difficult. This is also the reason for the lack of H5 subtype-specific rapid diagnostic system. Therefore, novel approaches are required to develop influenza subtype-specific POCT that can distinguish between H5N1 infection and infections mediated by the influenza virus of other subtypes.
So far, most bioassays used in rapid diagnostic systems are based on specific molecular interactions such as those based on antibody-antigen. Although antibodies have excellent advantages, they also have limitations such as, (i) high labor cost for production; (ii) low stability; (iii) loss of recognition functionality in non-optimal environments; (iv) poor compatibility with various transduction platforms [4]. Given the difficulty in developing novel antibodies, many researchers are seeking alternative agents such as DNA/RNA molecules with major advantages over antibodies for their integration into a rapid diagnostic system [5, 6]. Recently, a major development in influenza A (H5N1) subtype-specific diagnostic system has been achieved by using randomized DNA aptamers [7]. However, it is not very convenient for POCT.
Although the development of DNA/RNA probes has been actively pursued, only a few studies have used peptides as diagnostic agents. Antimicrobial peptides have been used to detect Shiga toxin-producing Escherichia coli bacteria [8]. Also, troponin I-specific peptides have been used to detect heart disease by phage-display and enzyme-linked immunosorbent assay (ELISA) [9].
Normally, epitopes can bind to targets such as immunoglobulin complementarity determining region (CDR) [10] and major histocompatibility complex (MHC) [11]. Currently, scaffolds that function in a manner analogous to immunoglobulin CDR are considered appropriate for creating a peptide aptamer library [12]. Peptide aptamers provide a powerful alternative approach to target the peptide-protein interaction (PPI) interface [13]. PPIs are involved in the precise control of binding properties of proteins. They can regulate many critical protein functions such as their subcellular localization and enzymatic activities [14]. However, peptide epitopes have never been investigated in the context of aptamers or detection agents in rapid diagnostic systems. Developing a rapid diagnostic system using peptides as aptamers will solve the current problem of scarcity of H5 subtype-specific antibodies and facilitate the development of novel and highly sensitive H5 subtype-specific POCT.
Typically, peptide aptamers are defined as affinity binders consisting of short (5-20) aa residues stretching from variable peptide regions constrained within a protein scaffold [12]. In this study, we propose to use a known epitope (KPNDAINF) derived from H5N1 HA [10] as a detection agent. We used the peptide KPNDAINF and its several derivatives to evaluate their usefulness as detection elements in H5 subtype-specific rapid diagnostic systems.
Materials and Methods
Reagents
Peptides were synthesized by PepTron Inc. (Daejeon, South Korea). P (S/V-COOH) Europium nanoparticle (Eu NP) beads (0.2 µm) were purchased from Bangs Laboratories Inc. (Fishers, IN, USA). N-(3-Dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC) and N-hydroxysulfosuccinimide sodium salt (Sulfo-NHS) were purchased from Thermo Scientific (Waltham, MA, USA). All other chemicals were purchased from Sigma-Aldrich (St. Louis, MO, USA). H5 subtype-specific monoclonal antibodies (clones 3F11 and 1C5) were kindly provided by Dr. Chom-Kyu Chong (GenBody Inc., Cheonan, South Korea).
Virus stock and titration
H5N3 virus [influenza A/spot-billed duck/Korea/KNU SYG06/2006(H5N3)] and H7N1 [influenza A/common teal/Korea/KNU YSR12/2012(H7N1)] were cultured by egg inoculation, concentrated, and titrated with HA assay as described previously [15, 16].
Production of peptide conjugates
Peptides were covalently conjugated to Eu NP by a well-established procedure from Bangs Laboratories. Briefly, 10 µL of Eu NP (0.2 µm, 1% w/t) was added to 500 µL of 0.1 M Tris-HCl (pH 7.0) and incubated for 1 h at 25 ºC in the presence of 0.13 mM of EDC and 10 mM Sulfo-NHS. Excess of EDC and Sulfo-NHS was removed by centrifugation at 27,237 × g for 5 min. Activated Eu NP was then mixed with 30 µL of 10 µM peptide in 500 µL of 0.1 M sodium phosphate (pH 8.0) and allowed to react at 30 ºC for 2 h. To indicate proper lateral flow through the strip, mouse IgG (2 pmole) was also co-conjugated to the Eu NP with the peptide. After centrifugation at 27,237 × g for 5 min, the Eu NP-conjugated peptide was collected, washed with 2 mM phosphate-buffered saline (PBS) (pH 8.0), resuspended in 100 µL of storage buffer (1% bovine serum albumin (BSA) in PBS), and stored at 4ºC. To conjugate antibody (3F11) with Eu NP, the same molar ratio of antibody to peptides was used.
Fluorescence-linked immunosorbent assay (FLISA)
FLISA was performed as described previously [16]. Briefly, a black 96-well microtitre plate (Greiner, Germany) was coated with 75 µL/well of various concentrations of H5N3 virus (125, 250, and 500 HAU/mL) and incubated at 37 ºC for 2 h. Subsequently, 75 µL/well of lysis buffer (100 mM Tris-HCl, pH 8.0, 0.1 M EDTA, 0.2% SDS, 0.1% Sodium azide, and 1% Tween 20) was added and incubated at room temperature for 15 min. The plate was washed with 200 μL of PBS, 0.1% Tween 20 (PBS-T, pH 7.4), and then blocked with 5% non-fat dry milk at 37 °C for 2 h. After washing, 200 μL/well of Eu NP-conjugated peptide (36 nM) was added and the plate was incubated at 37 °C to detect antigen. Stringent washing with PBS-T was performed five times to remove unbound peptide followed by the addition of 100 μL PBS/well. Fluorescence (355 nm excitation, 612 nm emission) was then measured with an Infinite F200 microplate reader (TECAN, Männedorf, Switzerland).
Fluorescent immunochromatographic test (FICT)
Test strips consisting of four components (a sample application pad, a conjugate pad, a nitrocellulose membrane, and an absorbent pad) were used. Test line (TL) of the strip was prepared by dispensing 2.5 mg/mL anti-influenza H5 subtype-specific HA1 mouse monoclonal antibody (1C5). Control line (CL) was coated with 0.5 mg/mL of rabbit anti-mouse IgG. The diagnostic strip was tested after drying the membrane at 30 ºC for 2 days. Performance analysis of the peptide aptamer-based diagnostic system was performed using previously developed FICT [15]. After applying 2 µL of Eu NP-conjugated peptide (0.25 pmol) to the conjugate pad, 75 µL of sample and 75 µL of lysis buffer were added to the sample pad and incubated at room temperature for 15 min. During lateral flow on the strip, the influenza virus in the sample reacted with the Eu NP-conjugated peptide. The complexes migrated to the immobilized antibody zone at the TL for 15 min. Results for the ratio of TL/CL were read with a portable strip reader (excitation 355 nm, emission 612 nm) [17]. To evaluate the performance of FICT, the limit of detection (LOD) was compared to that of a commercial influenza virus A/B rapid detection test (RDT) (Asan Pharm.Co., Seoul, South Korea). Tests were performed according to the manufacturer's instructions.
Docking studies for modeling peptide-HA1 complexes
A computational docking approach was used to analyze the structure of complexes between HA1 and each of four different peptides to improve peptide specificity. First, PEP-FOLD3 [18] and I-TASSER [19] were used to model the peptide or HA1 for docking studies. Docking was then carried out by using PEP-FOLD3 [18] and I-TASSER [20] to model the structure of peptide and antigen. PyRx of AutoDock Vina docking software was used to predict binding energies and root-mean-square deviations (RMSD) [21, 22]. PyMOL Molecular Graphics System was used to visualize interactive residues between peptides and antigen [23].
Clinical study
Patient specimens were collected in Wonkwang University Hospital. Influenza-negative nasopharyngeal samples were mixed with the virus for FICT. The study was approved by the Institutional Review Board of Wonkwang University Hospital (Approval No. WKUH201607-HRBR-078).
Statistics
Mean and standard deviation (SD) values were calculated. One-way and two-way analysis of variance (ANOVA) were performed. All analyses were conducted using GraphPad Prism 5.0 software.
Results
Characterization of peptides
Even though previous studies [7, 24] have reported the development of aptamer-based H5 subtype-specific diagnostic systems, their application as rapid diagnostic systems has not been fully explored. Here, we investigated whether peptide epitopes that could recognize HA1, a subtype-specific antigen of influenza A virus, could be used as alternatives to antibodies. For this, we used the low pathogenic H5N3 virus as a representative H5 subtype in this study.
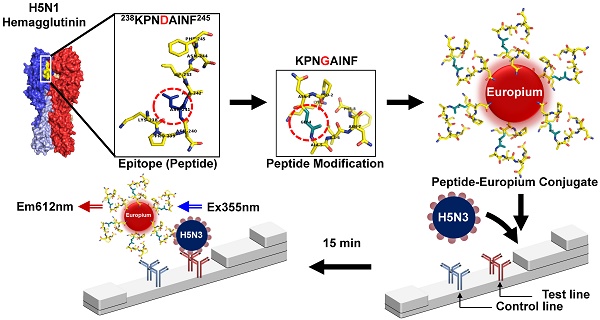
We compared the aa sequence of A/Anhui/1/2005 (H5N1) HA1 (GenBank accession no., ABD28180) [10] to that of H5N3 HA1 (GenBank accession no., AEB89892). As shown in Figure 1, they showed more than 95% sequence similarities among all H5 subtypes. Additionally, the conserved linear epitope KPNDAINF of H5N1 HA1 was found in H5N3 HA1 (aa 249-256) (Figure 1A). An extensive comparison of the epitope aa sequences among several influenza A subtypes was also conducted to determine its diversity (Figure 1B). KPNDAINF was conserved in all H5N1 and H5N3 strains, although other strains than the ones used in this study showed one-aa alteration. By contrast, in other influenza A subtypes (H1N1, H7N1, and H7N9), the epitope showed a low degree of sequence identity (< 50%).
Characterization of the epitope KPNDAINF. The well-known H5N1-specific and conserved epitope sequence (KPNDAINF) is present at aa residues 249-256 of H5N3 HA1(A). Homology of the aa sequence of this epitope was examined by comparing it to other H5 subtypes and representative influenza A subtypes (B). An asterisk (*) indicates the KPNDAINF position in HA1 of each subtype. Red, blue, and green arrows indicate the original HA1 of A/Anhui/1/2005 (H5N1), H5N3, and H7N1 tested in this study. Identical aa sequence is shown in dark cyan. Similarity is indicated in gray. The red-line box indicates the KPNDAINF sequence in subtypes.
Docking studies for modeling HA1 in H5N3-peptide complexes
The goal of this analysis was to determine whether peptide aptamers with relatively conserved sequence could be improved by computational designing to recognize the virus more efficiently and thus be more suitable for developing a rapid diagnostic system based on peptide aptamers instead of antibodies.
As a prerequisite, a peptide aptamer candidate involved in PPIs with HA1 must be defined and a model of the peptide-HA1 complex must be built. Here, we performed a docking study with epitope peptides and HA1 as epitope possessing high binding affinity [25]. It has been reported that pro-region peptides can selectively inhibit the enzymatic activity of cognate antigen [26]. Therefore, H5N3 HA1 aa residues 55-310 (GenBank accession no. AEB89892) were cloned and recombinant HA1 (rHA1) was expressed in E. coli (Figure S1). Information about the cloning primers is listed in Table S1.
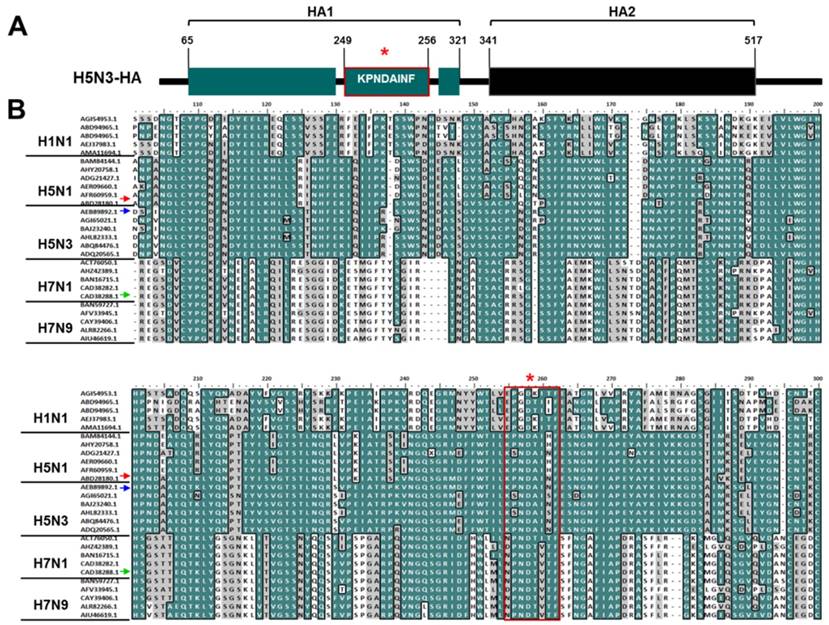
Modeling an active conformation of linear peptides was predicted using PEP-FOLD3 software and visualized in PyMOL Molecular Graphics system (Figure 2A). Final docking models of complexes between peptides (P0, P1, P2, and P3) and rHA1 antigen obtained from a typical VINA run are shown in Figure 2B. Because the binding site was unknown, the docking grid around the rHA1 was chosen. Figure 2B shows nine bound conformations explored by the peptide on rHA1. Two main spots were predicted as interactive sites. One of them was involved in the interaction between the native peptide and rHA1. The site of interaction between the native peptide and rHA1 is shown in Figure 2C. Binding energy and RMSD values as parameters for predicting ligand binding affinity using PyRx and Autodock Vina have been reported elsewhere [22]. The lower and upper RMSD (Å) values for the nine bound configurations were calculated with respect to the native peptide on HA1. The binding energy (VINA docking score) and RMSD values for the nine top scoring peptide conformations are shown in Table 1. Values of binding affinity to rHA1 obtained in this study were -5.6 - -5.0 kcal/mol for the original epitope P0 (KPNDAINF), -5.4 - -5.0 kcal/mol for P1 (KPNTAINF), -6.6 - -6.3 kcal/mol for P2 (KPNGAINF), and -6.3 - -6.1 kcal/mol for P3 (KPNDAINDAINF), with lower energy signifying higher binding affinity. The RMSD cut-off value of 3Å is typically used as the criterion for correct prediction of bound structure [27]. Based on both binding affinity and RMSD, P2 was predicted to have a stronger binding affinity to HA1 than the other peptides tested.
Modeling of the docking interaction between peptides and antigen. 3D modeling was performed for linear peptides and active conformation was generated (A). Each image shows the nine top-scoring docking configurations of each peptide on HA1 of the H5N3 antigen (B). The rectangular image indicates the site of interaction of native peptide on rHA1 of H5N3. Each native peptide (in yellow color) is shown in stick on HA1 (C).
Docking output of peptides tested and HA1 of H5N3.
| Mode | P0 (KPNDAINF) | P1 (KPNTAINF) | P2 (KPNGAINF) | P3 (KPNDAINDAINF) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Affinitya | RMSD l.b.b | RMSD u.b.c | Affinity | RMSD l.b. | RMSD u.b. | Affinity | RMSD l.b. | RMSD u.b. | Affinity | RMSD l.b. | RMSD u.b. | |
| 0 | -5.7 | 0.0 | 0.0 | -5.5 | 0.0 | 0.0 | -7.0 | 0.0 | 0.0 | -6.5 | 0.0 | 0.0 |
| 1 | -5.6 | 9.217 | 15.621 | -5.4 | 6.483 | 9.695 | -6.6 | 23.173 | 27.943 | -6.3 | 24.876 | 32.296 |
| 2 | -5.5 | 2.441 | 7.511 | -5.2 | 22.31 | 26.088 | -6.5 | 25.154 | 30.638 | -6.3 | 3.024 | 12.996 |
| 3 | -5.2 | 2.256 | 6.27 | -5.1 | 22.553 | 26.082 | -6.5 | 6.343 | 12.343 | -6.3 | 23.801 | 29.224 |
| 4 | -5.2 | 3.12 | 8.453 | -5.0 | 27.089 | 30.03 | -6.4 | 2.672 | 4.578 | -6.2 | 25.119 | 31.664 |
| 5 | -5.1 | 3.187 | 7.061 | -5.0 | 22.024 | 26.138 | -6.3 | 3.679 | 11.133 | -6.2 | 2.606 | 10.573 |
| 6 | -5.1 | 3.101 | 8.384 | -5.0 | 22.852 | 26.836 | -6.3 | 5.224 | 9.316 | -6.2 | 25.854 | 32.772 |
| 7 | -5.0 | 18.06 | 22.355 | -5.0 | 22.746 | 26.054 | -6.3 | 22.773 | 27.203 | -6.1 | 21.106 | 28.615 |
| 8 | -5.0 | 2.468 | 7.093 | -5.0 | 6.226 | 9.654 | -6.3 | 4.782 | 7.602 | -6.1 | 24.404 | 28.963 |
a, binding energy (kcal/mol); A typical output from a docking run with Autodock Vina shows the calculated binding affinity in descending order for peptides and HA1
b, root mean square deviation lower bound;
c, root mean square deviation upper bound.
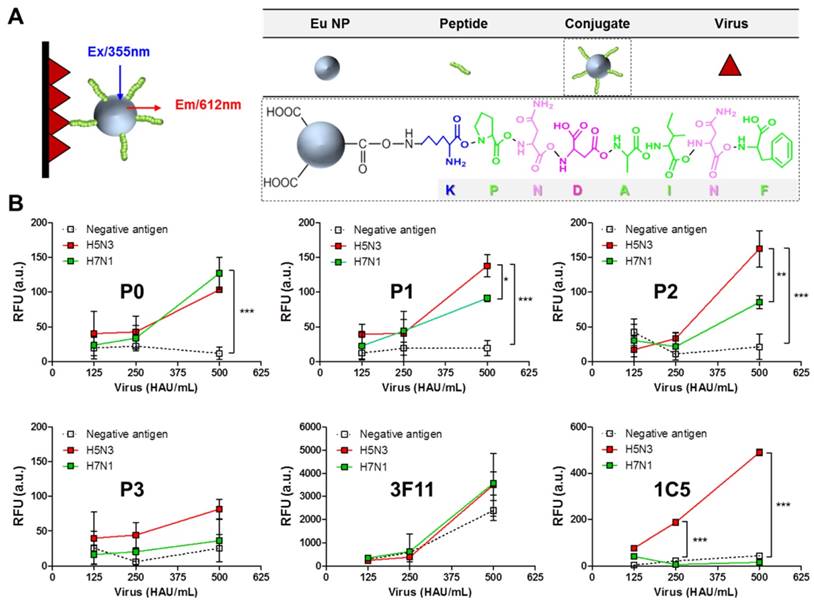
Assessment of the interaction between avian influenza virus and peptides tested in this study. Schematic depiction of the FLISA principle (A). The presence of virus was detected by Eu NP-conjugated peptides or antibody using a fluorescence microplate reader (excitation at 355 nm, emission at 612 nm) (B). Data (n = 3) are shown as mean ± SD (*, P < 0.05; **, P < 0.01; ***, P < 0.001). A pair of H5 subtype-specific antibodies (3F11 and 1C5 clone) was tested in FLISA for comparison with the peptides. P0, KPNDAINF; P1, KPNTAINF; P2, KPNGAINF; P3, KPNDAINDAINF; 3F11, antibody for detection; 1C5, antibody for capturing; RFU, relative fluorescence unit; a.u., arbitrary unit.
Interaction of peptides with the virus and target antigen
Eu NP-conjugated peptides based on the formation of a stable amide bond were generated and used for FLISA as detection elements. To conduct FLISA, black 96-well plates were coated with different amounts of two virus subtypes (H5N3 and H7N1). To evaluate the interaction of peptides with the target antigen, binding of Eu NP-peptide conjugates to avian influenza virus (H5N3 and H7N1) was tested by FLISA. Figure 3A illustrates FLISA schematically. Covalently conjugated peptides to Eu NP were prepared by a well-established procedure based on EDC/NHS chemical reaction, where NHS ester could react with peptide primary amines (N-terminus of K residue) to yield a stable amide bond. Eu-NP-conjugated peptides were used in FLISA to detect the virus.
FLISA results showed that P0 peptide (KPNDAINF) could detect both subtype viruses H5N3 and H7N1 at a titer of at least 500 HAU/mL (Figure 3B). However, there were no significant differences in FLISA results between H5N3 and H7N1, implicating that the epitope was not specific for the detection of influenza A subtypes (Figure 3B, P0). On the other hand, at virus titer of 500 HAU/mL, the fluorescence signals obtained with P1 peptide (KPNTAINF) were significantly (P < 0.05) different between H5N3 and H7N1 (Figure 3B, P1). The difference in detection between the two virus subtypes was even more (P < 0.01) pronounced when P2 peptide (KPNGAINF) was used (Figure 3B, P2), implicating that the one aa difference might be able to induce subtype-specific interactions. P3 peptide (KPNDAINDAINF) showed no subtype-specific binding despite its relatively high binding affinity (Figure 3B, P3). These results were consistent with those of docking analysis regarding both binding affinity and RMSD parameters. Results obtained from peptide-mediated FLISA were compared to those obtained from antibody-mediated FLISA using a pair of H5 subtype-specific antibody comprising an antibody (3F11 clone) for detection and an antibody (1C5 clone) for capturing. When the same molar ratio of antibody to peptides was applied to FLISA, the 3F11 antibody-mediated FLISA showed remarkably strong binding affinity to any virus, even with negative antigen (the protein concentrated from egg allantoic cavity), indicating that specificity was not necessary when the antibody was used as a detection element (Figure 3B, 3F11). In contrast, the H5 subtype-specific antibody (1C5 clone) showed a positive signal only with H5N3 at a titer of 250 HAU/mL (P < 0.001), but not with the H7N1 virus, confirming that it was specific for the H5 subtype (Figure 3B, 1C5). These results also showed that the binding affinity of the studied peptides was at least two-fold lower than that of the 1C5 antibody.
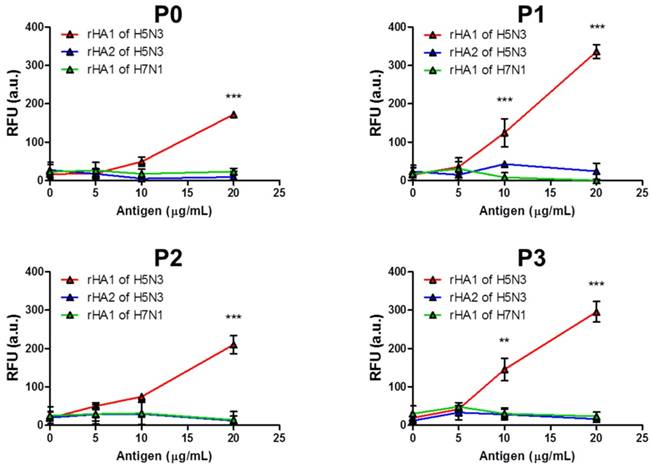
Next, we tested interactions between peptides and recombinant HA (rHA) antigens. The rHA1 from H5N3 (aa 55-310), rHA2 from H5N3 (aa 343-518), and rHA1 from H7N1 (aa 106-295) were expressed in E. coli and purified using Ni-NTA (Figure S1). Interactions between rHA antigens (5, 10, and 20 μg/well) with Eu NP-conjugated peptides were tested by FLISA. Results (Figure 4) showed that all four peptides recognized rHA1 from H5N3 at a concentration of at least 20 μg/mL. Peptides P1 and P3 detected rHA1 of H5N1 at 10 μg/mL. However, peptides P0 and P2 could not detect rHA1 of H5N1 at this concentration. In contrast, none of these peptides could recognize rHA2 from H5N3 or rHA1 from H7N1 at any concentration tested, indicating that the epitope or epitope-derived peptides were specifically targeting HA1 from H5N3.
Performance of peptides on rapid FICT
In peptide-linked FICT, Eu NP-conjugated peptides instead of antibodies were used as the detection elements. To normalize the value of test line (TL) by each control line (CL), minimum mouse IgG (mIgG) was added to peptide conjugates. Optimization of the suitable mIgG was conducted by adding differential molar ratios of mIgG. Finally, 2 pmole of mIgG was chosen for co-conjugation with the peptides. Detailed results of the co-conjugation of mIgG can be found in the Methods section of Supplementary Materials (Figure S2).
Dose-response interactions between peptides and the target antigen. Black 96-well plates were coated with recombinant antigens (5-20 μg/mL) and Eu NP-conjugated peptides were added to detect recombinant antigens. Fluorescence intensity was measured with a microplate reader (excitation at 355 nm, emission at 612 nm). Data (n = 3) are shown as mean ± SD (**, P < 0.01; ***, P < 0.001). P0, KPNDAINF; P1, KPNTAINF; P2, KPNGAINF; P3, KPNDAINDAINF.
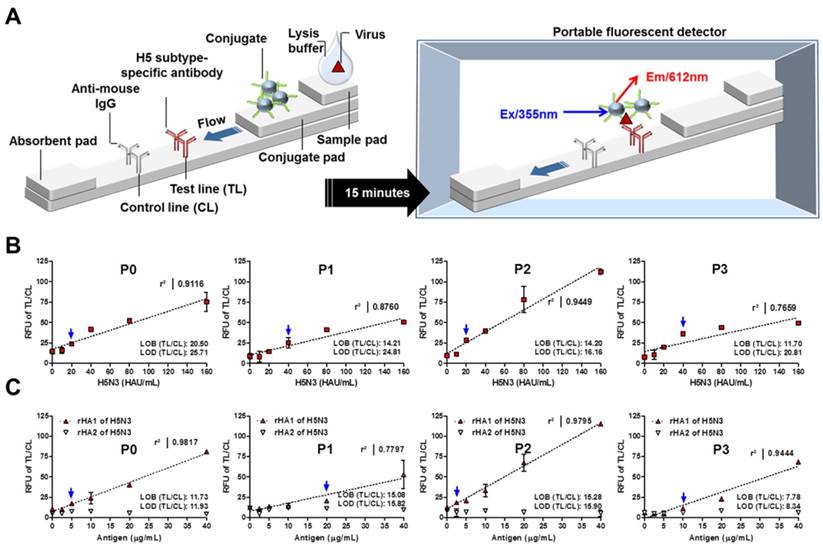
As shown in Figure 5A, the fluorescence signal from TL and CL of the strip was measured with a portable fluorescence detector (excitation at 355 nm and emission at 612 nm).
Using serially diluted H5N3 virus, the FICT LOD was determined using the limit of blank (LOB) as described previously [28]. The computed TL/CL values for LOB and LOD are shown in each graph. The virus titer was analyzed for each LOD value. As seen in Figure 5B, the TL/CL value of LOD was below that of 20 HAU/mL of H5N3 virus for P0 and P2 peptides. Thus, the LOD was determined to be 20 HAU/mL for P0 and P2 peptide-linked FICT. P1 and P3 peptide-linked FICT showed an LOD value of 40 HAU/mL for the virus (Figure 5B). The Kd (equilibrium dissociation constant) value of each peptide was assessed by the experimental application using Eu NP-conjugated peptide against H5N3 virus (saturation binding curve) and the results supported the highest binding affinity of P2 (Figure S3).
For each peptide, quantitative range of FICT was determined using different titers of the virus by linear regression. Results showed that P2 had the highest coefficient of variation (R2 = 0.9449) between 10 and 160 HAU/mL. This indicates that, among the four peptides tested, P2 had the best performance for FICT. P0 showed a fair linearity (R2 = 0. 9116) in this range of virus while P1 and P2 showed moderate (R2 = 0. 8760) and poor linearity (R2 = 0.7659), respectively. Furthermore, serially diluted rHA antigens (2.5-40 μg/mL) were tested in FICT to determine where the peptides potentially recognize HA1 and HA2 in FICT assay. Results are shown in Figure 5C. All peptides could bind to rHA1, with P2 and P0 having high linear regression (R2 = 0.978 and R2 = 0.950, respectively). The TL/CL value of LOD was below 2.5 μg/mL and 5 μg/mL for P2 and P0, respectively. P1 and P3 peptide-linked FICT showed a LOD value of 20 μg/mL of rHA1. Therefore, P0 and P2 were suitable for quantitative detection of the target in FICT. All raw FICT results for virus and antigen are shown in Supplementary Materials (Figure S4 and Figure S5).
Quantitative analysis by rapid FICT. FICT was carried out on diagnostic strips with Eu NP-conjugated peptides. The fluorescence signal was measured with a portable strip reader after 15 min of incubation (A). Serially diluted H5N3 virus (10-160 HAU/mL) (B) or recombinant antigens (2.5-40 μg/mL) (C) were measured by FICT to determine the LOD and linear regression in this range for each peptide conjugate. Data (n = 3) are shown as mean ± SD. Linear regression is shown in dotted line using GraphPad Prism 5.0 software. Blue arrow indicates the virus titer corresponding to the TL/CL value of LOD. P0, KPNDAINF; P1, KPNTAINF; P2, KPNGAINF; P3, KPNDAINDAINF; RFU, Relative fluorescent unit.
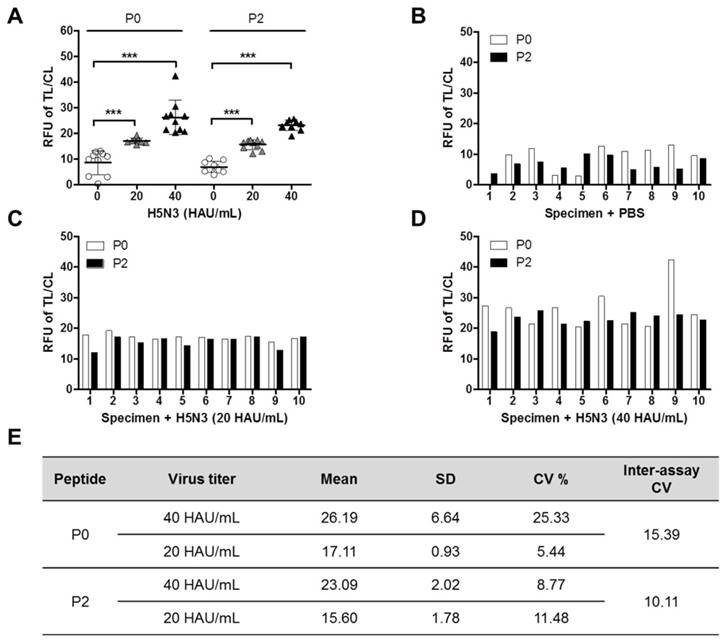
FICT analysis of human nasopharyngeal samples containing H5N3 virus
A variety of proteolytic degrading enzymes are present in human nasopharyngeal specimens [29], implicating that they might affect the stability of peptide. To determine if the proposed diagnostic system could be useful for detecting highly pathogenic HA 5 subtype present in human nasopharyngeal samples [30], the performance of peptide-linked FICT was assessed using H5N3 virus-spiked specimens.
Since P0 and P2 peptide-conjugates showed excellent performance to quantitatively detect the H5N3 virus in FICT than the other two peptides, human nasopharyngeal samples (n = 10) were tested by FICT using these two peptide-conjugates. Negative specimens or spiked with the virus were also used. The virus titer of spiked specimens was adjusted to 20 HAU/mL or 40 HAU/mL, by mixing 10 µL of the appropriate virus stock with 90 µL of nasopharyngeal specimen. The ratios of TL/CL derived from all experiments were plotted in the same graph to allow comparison between negative and virus-spiked specimens (Figure 6A). Comparison of fluorescent signals between negative and virus-spiked specimens showed a significant difference (P < 0.001). However, the TL/CL values measured by P0 peptide-linked FICT had a wider distribution than those measured by P2 peptide-linked FICT (Figure 6B, 6C, and 6D), implicating that P2 peptide might be more suitable for quantitative assay by the peptide-linked FICT than P0 peptide. The analytical error rate was computed by inter-assay coefficients of variation (CV) (Figure 6E). The inter-assay CVs of P0 and P2 were 15. 39 and 10.11, respectively, demonstrating that P2 would be acceptable because the range of inter-assay CVs was below 15% [20]. All raw FICT results are shown in Figure S6.
Efficiency of P0 and P2 as detection elements in FICT for human nasopharyngeal specimens. Ten normal nasopharyngeal samples were chosen and mixed with the virus at a 1:10 dilution ratio. After mixing, 75 μL of the sample was tested by P0- or P2-linked FICT (***, P < 0.001). (A) All data; (B) Nasopharyngeal samples without virus; (C) Nasopharyngeal samples with H5N3 virus (20 HAU/mL); (D) Nasopharyngeal samples with H5N3 virus (40 HAU/mL). (E) Coefficients of variation (CV) of P0- and P2- linked FICT results with virus (20 and 40 HAU/mL) in specimens.
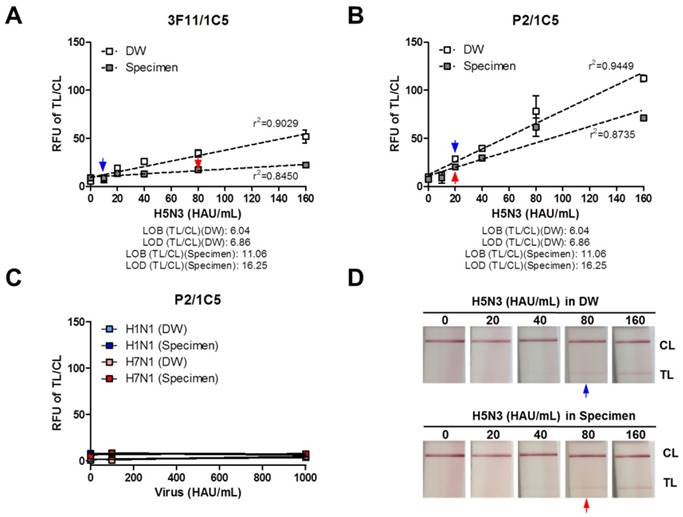
The detection limit of the P2 peptide was evaluated by comparing it to that of an antibody (3F11 clone) using serially diluted H5N3 virus (10-160 HAU/mL) in the human nasopharyngeal specimens. First, the performance of 3F11 antibody-linked FICT was tested with the virus in the absence or presence of clinical samples. As shown in Figure 7A, the fluorescence signal ratio of the TL/CL was linearly increased (R2 = 0.9029) with increasing virus titer when the virus was diluted in distilled water (DW). However, in the presence of a specimen, the signal was decreased by about 2-fold, indicating that the presence of clinical samples interfered with the antigen-antibody reaction, resulting in a moderately linear relationship (R2 = 0.8450). The TL/CL values for LOB and LOD are shown under each graph. The virus titer corresponding to the LOD value was determined. The TL/CL of LOD values of FICT employing 3F11 antibody was below the value corresponding to 10 HAU/mL and 80 HAU/mL for H5N3 virus diluted in DW and clinical specimens, respectively.
P2 peptide-linked FICT was then performed with the same amount of H5N3 in the absence or presence of nasopharyngeal samples. Results are shown in Figure 7B. When the virus was mixed with DW, P2 peptide-linked FICT detected the virus in a linear manner (R2 = 0.9449), but the LOD was 20 HAU/mL, indicating a 2-fold decreased performance compared with antibody-linked regular FICT. In the presence of clinical samples, the fluorescence signals were decreased with increasing virus titre. The linearity between signal and virus titer was also decreased (R2 = 0.8735). However, the LOD was computed to be 20 HAU/mL, indicating that P2 peptide-linked FICT was less influenced by the presence of clinical samples than antibody pair-FICT.
H5 subtype-specificity was confirmed by testing H1N1 and H7N1 virus in DW or clinical specimens at a titer of 100 and 1,000 HAU/mL, showing all negative reactions in P2-linked FICT (Figure 7C). The raw FICT results are presented in Figure S7. The commercial RDT only provided information for influenza virus A/B. Its LOD was 80 HAU/mL for the H5N3 virus in DW or specimen, indicating that P2 aptamer-liked FICT increased the virus detection capacity by at least 4-fold compared to the commercial RDT (Figure 7D). Reliability of this proposed assay was confirmed by applying the P2 conjugate to sandwich FLISA using a 96-well plate assay (Figure S8).
Comparison between regular FICT employing the 3F11 antibody and P2 peptide-FICT. Human nasopharyngeal clinical samples were mixed with the virus at a 1:10 dilution ratio and subjected to FICT test. The ratio of fluorescence (TL/CL) was measured using antibody (3F11) conjugate-linked FICT assay (A) and peptide (P2) conjugate-linked FICT assay (B) in the absence or presence of clinical samples. To confirm the specificity of H5 subtype, other influenza A subtypes (H1N1 and H7N1) were tested at 100 and 1,000 HAU/mL in P2-linked FICT assay (C). Data (n = 3) are shown as mean ± SD. Linear regression is shown by the dotted line using the GraphPad Prism 5 software. A commercialized RDT for influenza A/B was tested with serially diluted H5N3 virus in the absence or presence of nasopharyngeal specimens (D). The blue arrow indicates the LOD in the presence of DW while the red arrow indicates LOD in the presence of specimen.
Discussion
H5-subtype of AI virus has long caused serious health problems both locally and globally. It has the potential to acquire human pandemic dimensions [31, 32]. Since AI viruses could mutate and gain the ability to spread easily between humans, the Centers for Disease Control and Prevention (CDC) considers monitoring of human AI infections and person-to-person transmission of AI to be extremely important for public health [33]. Currently, AI testing depends on molecular and structural studies to determine the subtype of the virus [33]. However, an RDT system that can distinguish between different AI subtypes is not yet commercially available. A faster on-site control/surveillance system for detecting the H5 AI virus subtype is required to prevent the spread of this highly pathogenic avian virus. H5 subtype-specific rapid diagnostic systems are less studied than H7 subtype-specific diagnostic kits due to the difficulties in producing subtype specific-antibodies as there is a relatively high HA1 homology among influenza A subtypes compared to H7 subtype. To overcome this obstacle, a sandwich diagnostic assay based on aptamer-antibody or aptamer-aptamer interaction has been recently developed [7, 34]. However, it utilizes sophisticated equipment for surface plasmon resonance (SPR) analysis, making it incompatible with RDT.
For a successful diagnostic system, development of efficient aptamers is necessary. For novel antibody-based diagnostics and therapeutics, a better understanding of the principle governing antibody affinity and specificity is required. Antibody paratopes are rich in aromatic residues such as Y and W. A binary-code library containing Y and S has been useful in selecting functional antibodies through PPIs [35, 36]. However, another study on serine protease-inhibitor has reported that PPIs are different from the antibody-antigen interface as only 12 interacting residue pairs contribute to over 40% of the interaction energy in antibody-antigen interactions whereas PPIs use a larger set of residing types [37]. Other drawbacks are, PPI characterization is incomplete and the development of specific peptide aptamers is labor-intensive. It has been suggested that in silico prediction of key amino acids in combination with experimental verification by screening library of ligands against the target antigen will help identify better peptide aptamers [38]. Although emphasis has been given to the design of peptide scaffolds as the first step in creating a peptide library, a computational design is still under development and evaluation [12].
In this study, we analyzed the binding potential of a well-conserved epitope peptide, KPNDAINF sequence (P0), to its cognate antigen (HA1) of AI virus despite the lack of information about the function of this epitope as a detection agent. To understand the value of this epitope sequence (P0) as a probe, docking analysis between the epitope and its cognate target antigen (HA1) is essential. The binding affinity of the best scoring model obtained by docking (-5.6 kcal/mol) suggested that the interaction is relatively weak compared with another affinity peptide reported elsewhere [39]. To modify the binding energy, the charged aa D in the middle of the P0 sequence was mutated to T, a polar aa (peptide P1: KPNTAINF). Conversely, when D was mutated to hydrophobic aa G, it contributed to a tight protein (peptide P2: KPNGAINF). Finally, to increase the number of hydrogen bonds between peptide and HA1, P3 (KPNDAINDAINF) was designed to form a helix through the twice repetition of sequence DAIN. Our docking models suggested that peptides P0 and P2 had a similar pocket of HA1, whereas peptides P1 and P3 bound on the opposite site of HA1.
Consistent with a previous report suggesting that polar residues dominate antibody-antigen interactions rather than PPI [37], the energy values calculated from our docking models showed weaker affinity of P1 than P0 but a stronger affinity of P2 than P0. The higher affinity of P2 might be due to a larger surface area of P2 buried in HA1 because of the smaller size of G than D. In contrast, P3 with its large helical structure might not be able to fully access the pocket. This result was confirmed by the saturation binding curve using differential peptide concentration against virus showing that P2 possessed the best Kd value among the peptides tested.
Subsequently, we applied the epitope-derived peptides to a rapid diagnostic system (FICT) using clinical samples. The peptide aptamers showed similar detection limits in clinical specimens suggesting their utility in clinical diagnostics. Notably, P2 peptide-linked FICT had an excellent performance with respect to specific and rapid detection of the H5N3 virus in clinical specimens. Its specificity was 4-fold higher than antibody-linked FICT possessing H5 subtype and an RDT (for influenza A/B) which could not distinguish between different influenza A subtypes.
In summary, we successfully developed an epitope-derived peptide-linked rapid fluorescent diagnostic system for H5 subtype-specific detection of AI virus in this study. Peptide binding energy was used to optimize the peptide sequence. To the best of our knowledge, this is the first report of an epitope-derived peptide to detect a cognate antigen. Furthermore, we demonstrated the great potential of molecular modeling when used in conjunction with experimental results. This approach is expected to contribute to future development of novel peptide aptamers-linked rapid fluorescent diagnostic systems for H5 subtype avian influenza virus.
Supplementary Material
Supplementary figures.
Acknowledgements
This research was supported by Priority Research Centers Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, (NRF-2015R1A6A1A03032236).
Authors' contributions
DTB, DTH, and HP carried out assays and analyzed the results; BT, NM, NT, NC, TT, and NT supported the optimization of the assays and prepared data; CK developed the strip; ST and DY collected specimens and performed assay results analyses; DTH and TD conducted docking analysis for Figure 2. HP and SJ conceived and designed the experiments. SJ wrote the manuscript. All authors read and approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Campitelli L, Mogavero E, De Marco MA, Delogu M, Puzelli S, Frezza F. et al. Interspecies transmission of an H7N3 influenza virus from wild birds to intensively reared domestic poultry in Italy. Virology. 2004;323:24-36
2. WHO: Clinical management of human infection with avian influenza A (H5N1) virus. http://www.who.int/influenza/resources/documents/clinical_management_h5n1_15_08_2007/en/
3. Tharakaraman K, Raman R, Stebbins NW, Viswanathan K, Sasisekharan V, Sasisekharan R. Antigenically intact hemagglutinin in circulating avian and swine influenza viruses and potential for H3N2 pandemic. Sci Rep. 2013;3:1822
4. Tadepalli S, Kuang Z, Jiang Q, Liu KK, Fisher MA, Morrissey JJ. et al. Peptide functionalized gold nanorods for the sensitive detection of a cardiac biomarker using plasmonic paper devices. Sci Rep. 2015;5:16206
5. Bruno JG. Application of DNA Aptamers and Quantum Dots to Lateral Flow Test strips for detection of foodborne pathogens with improved sensitivity versus colloidal gold. Pathogens. 2014;3:341-55
6. Dupont DM, Thuesen CK, Botkjaer KA, Behrens MA, Dam K, Sorensen HP. et al. Protein-binding RNA aptamers affect molecular interactions distantly from their binding sites. PloS one. 2015;10:e0119207
7. Nguyen VT, Seo HB, Kim BC, Kim SK, Song CS, Gu MB. Highly sensitive sandwich-type SPR based detection of whole H5Nx viruses using a pair of aptamers. Biosens Bioelectron. 2016;86:293-300
8. Yonekita T, Ohtsuki R, Hojo E, Morishita N, Matsumoto T, Aizawa T. et al. Development of a novel multiplex lateral flow assay using an antimicrobial peptide for the detection of Shiga toxin-producing Escherichia coli. J Microbiol Methods. 2013;93:251-256
9. Park JP, Cropek DM, Banta S. High affinity peptides for the recognition of the heart disease biomarker troponin I identified using phage display. Biotechnol Bioeng. 2010;105:678-686
10. Xiong F, Xia L, Wang J, Wu B, Wang D, Yuan L. et al. A high-affinity CDR-grafted antibody against influenza A H5N1 viruses recognizes a conserved epitope of H5 hemagglutinin. PloS one. 2014;9:e88777
11. Ferrante A, Gorski J. Cooperativity of hydrophobic anchor interactions: evidence for epitope selection by MHC class II as a folding process. J Immunol. 2007;178:7181-9
12. Reverdatto S, Burz DS, Shekhtman A. Peptide aptamers: development and applications. Curr Top Med Chem. 2015;15:1082-101
13. Yeh JT, Binari R, Gocha T, Dasgupta R, Perrimon N. PAPTi: a peptide aptamer interference toolkit for perturbation of protein-protein interaction networks. Sci Rep. 2013;3:1156
14. Perkins JR, Diboun I, Dessailly BH, Lees JG, Orengo C. Transient protein-protein interactions: structural, functional, and network properties. Structure. 2010;18:1233-43
15. Yeo SJ, Choi K, Cuc BT, Hong NN, Bao DT, Ngoc NM. et al. Smartphone-Based Fluorescent Diagnostic System for Highly Pathogenic H5N1 Viruses. Theranostics. 2016;6:231-42
16. Yeo SJ, Huong DT, Han JH, Kim JY, Lee WJ, Shin HJ. et al. Performance of coumarin-derived dendrimer-based fluorescence-linked immunosorbent assay (FLISA) to detect malaria antigen. Malar J. 2014;13:266
17. Ham JY, Jung J, Hwang BG, Kim WJ, Kim YS, Kim EJ. et al. Highly sensitive and novel point-of-care system, aQcare Chlamydia TRF kit for detecting Chlamydia trachomatis by using europium (Eu) (III) chelated nanoparticles. Ann Lab Med. 2015;35:50-6
18. Lamiable A, Thevenet P, Rey J, Vavrusa M, Derreumaux P, Tuffery P. PEP-FOLD3: faster de novo structure prediction for linear peptides in solution and in complex. Nucleic Acids Res. 2016;44:W449-54
19. Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:725-38
20. Thomsson O, Strom-Holst B, Sjunnesson Y, Bergqvist AS. Validation of an enzyme-linked immunosorbent assay developed for measuring cortisol concentration in human saliva and serum for its applicability to analyze cortisol in pig saliva. Acta Vet Scand. 2014;56:55
21. Dallakyan S, Olson AJ. Small-molecule library screening by docking with PyRx. Methods Mol Biol. 2015;1263:243-50
22. Tiwari G, Mohanty D. An in silico analysis of the binding modes and binding affinities of small molecule modulators of PDZ-peptide interactions. PloS one. 2013;8:e71340
23. Seeliger D, de Groot BL. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J Comput Aided Mol Des. 2010;24:417-22
24. Tseng YT, Wang CH, Chang CP, Lee GB. Integrated microfluidic system for rapid detection of influenza H1N1 virus using a sandwich-based aptamer assay. Biosens Bioelectron. 2016;82:105-11
25. Bernat B, Sun M, Dwyer M, Feldkamp M, Kossiakoff AA. Dissecting the binding energy epitope of a high-affinity variant of human growth hormone: cooperative and additive effects from combining mutations from independently selected phage display mutagenesis libraries. Biochemistry. 2004;43:6076-84
26. Korde R, Bhardwaj A, Singh R, Srivastava A, Chauhan VS, Bhatnagar RK. et al. A prodomain peptide of Plasmodium falciparum cysteine protease (falcipain-2) inhibits malaria parasite development. J Med Chem. 2008;51:3116-23
27. Lorber DM, Udo MK, Shoichet BK. Protein-protein docking with multiple residue conformations and residue substitutions. Protein Sci. 2002;11:1393-408
28. Armbruster DA, Pry T. Limit of blank, limit of detection and limit of quantitation. Clin Biochem Rev. 2008;29(Suppl 1):S49-52
29. El Badry AA, El-Fadle AA, El-Balshy AL. Tissue inhibitor of matrix metalloproteinase-2 in nasopharyngeal carcinoma. MedGenMed. 2007;9:3
30. CDC. Isolation of avian influenza A(H5N1) viruses from humans-Hong Kong, May-December 1997. MMWR Morbidity and mortality weekly report. 1997;46:1204-7 https://www.cdc.gov/mmwr/preview/mmwrhtml/00050459.htm
31. Le MQ, Horby P, Fox A, Nguyen HT, Le Nguyen HK, Hoang PM. et al. Subclinical avian influenza A(H5N1) virus infection in human, Vietnam. Emerg Infect Dis. 2013;19:1674-7
32. Hien ND, Ha NH, Van NT, Ha NT, Lien TT, Thai NQ. et al. Human infection with highly pathogenic avian influenza virus (H5N1) in northern Vietnam, 2004-2005. Emerg Infect Dis. 2009;15:19-23
33. Avian Influenza A Virus Infections in Humans. 2016 CDC. https://www.cdc.gov/flu/avianflu/avian-in-humans.htm
34. Lee SJ, Kwon YS, Lee JE, Choi EJ, Lee CH, Song JY. et al. Detection of VR-2332 strain of porcine reproductive and respiratory syndrome virus type II using an aptamer-based sandwich-type assay. Anal Chem. 2013;85:66-74
35. Fellouse FA, Esaki K, Birtalan S, Raptis D, Cancasci VJ, Koide A. et al. High-throughput generation of synthetic antibodies from highly functional minimalist phage-displayed libraries. J Mol Biol. 2007;373:924-40
36. Moreira IS, Fernandes PA, Ramos MJ. Hot spots-a review of the protein-protein interface determinant amino-acid residues. Proteins. 2007;68:803-12
37. Jackson RM. Comparison of protein-protein interactions in serine protease-inhibitor and antibody-antigen complexes: implications for the protein docking problem. Protein Sci. 1999;8:603-13
38. Sriwastava BK, Basu S, Maulik U, Plewczynski D. PPIcons: identification of protein-protein interaction sites in selected organisms. J Mol Model. 2013;19:4059-70
39. Li Y, Liu X, Dong X, Zhang L, Sun Y. Biomimetic design of affinity peptide ligand for capsomere of virus-like particle. Langmuir. 2014;30:8500-8
Author contact
![]() Corresponding author: Seon-Ju Yeo, Ph.D., Zoonosis Research Center, Department of Infection Biology, School of Medicine, Wonkwang University, 460, Iksan-daero, Iksan, 54538, Republic of Korea. Tel +82-63-850-6777 Fax +82-63-857-0342 Email: yeosjac.kr
Corresponding author: Seon-Ju Yeo, Ph.D., Zoonosis Research Center, Department of Infection Biology, School of Medicine, Wonkwang University, 460, Iksan-daero, Iksan, 54538, Republic of Korea. Tel +82-63-850-6777 Fax +82-63-857-0342 Email: yeosjac.kr
 Global reach, higher impact
Global reach, higher impact